Abstract
Inhibition of both the de novo (DNP) and salvage (NSP) pathways of nucleoside synthesis has been demonstrated to impair leukemia cells. We endeavored to determine if this approach would be efficacious in glioblastoma (GBM). To diminish nucleoside biosynthesis, we utilized compound DI-39 that selectively targets NSP, in combination with thymidine (dT) that selectively targets DNP. We employed in vitro and ex vivo models to determine the effects of pre-treatment with dT +DI-39 on brain tumor stem cells (BTSC). Here, we demonstrate that this combinatorial therapy elicits a differential response across a spectrum of human patient derived GBM cultures. As determined by apoptotic markers, most cultures were relatively resistant to treatment, although a subset was highly sensitive. Sensitivity was unrelated to S-phase delay and to DNA damage induced by treatment. Bioinformatics analysis indicated that response across cultures was associated with the transcription factor PAX3 (associated with resistance) and with canonical pathways including the nucleotide excision repair pathway, PTEN (associated with resistance), PI3K/AKT (associated with sensitivity), and ErbB2-ErbB3. Our in vitro assays demonstrated that, in sensitive cultures, clonal sphere formation was reduced upon removal from pre-treatment. In contrast, in a resistant culture, clonal sphere formation was slightly increased upon removal from pre-treatment. Moreover, in an intracranial xenograft model, pre-treatment of a sensitive culture caused significantly smaller and fewer tumors. In a resistant culture, tumors were equivalent irrespective of pre-treatment. These results indicate that, in the subset of sensitive GBM, BTSC are targeted by inhibition of pyrimidine synthesis.
Keywords: pyrimidine, nucleotide, DI-39, glioblastoma, brain tumor stem cell
Introduction
Glioblastoma (GBM) is a devastating disease with no cure, a dire prognosis and a median survival of approximately one year(1). Novel treatments are necessary to reduce the malignancy of these brain tumors. One therapeutic strategy is to target brain tumor stem cells (BTSC) as they are the cells believed to be responsible for tumor formation and recurrence(2–4). Evidence for the existence of BTSC came from several laboratories(5–7) and has been supported by several studies(8, 9). Indeed, further studies have substantiated that BTSC do exist in models of GBM and are responsible for chemoresistance and tumor formation(10–13). Therefore, identifying agents that target BTSC is a promising strategy in the development of chemotherapeutics to treat GBM.
Personalized medicine that tailors therapy to each individual patient based on tumor characteristics is a potential strategy for the treatment of GBM(14, 15). Verhaak et al. has classified GBM tumors into four distinct groups based on gene expression that correlates with specific mutations(16). Whether or not this classification system distinguishes cohorts that respond to targeted therapy is an area of current study. One way to test the hypothesis that expression profiling can assist in tailored therapy is to classify GBM patient derived gliomasphere cultures based on expression profiling and study their response to targeted therapy.
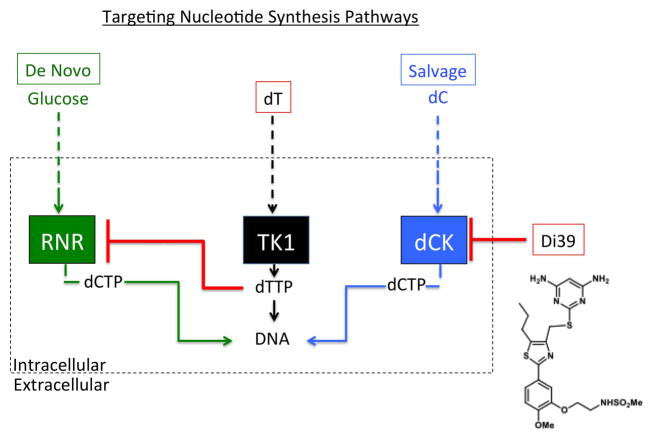
Rapidly dividing cells such as cancer cells require a balanced supply of deoxyribonucleoside triphosphates (dNTPs) in order to replicate and repair DNA(17). Production of nucleotides is carried out by two main pathways, the de novo pathway (DNP), reliant on the enzyme ribonucleotide reductase (RNR), and the salvage pathway (NSP), reliant on the enzyme deoxycytidine (dC) kinase (dCK) (Figure 1). It has been demonstrated that NSP compensates for impaired DNP(18). In fact, this compensation of NSP for impaired DNP may explain why inhibition of the de novo pathway alone, through addition of thymidine (dT), has failed in clinical trials(19–21). The hypothesis that combinatorial targeting of both DNP and NSP may be an efficacious treatment of cancer has been validated in a study on acute lymphoblastic leukemia (ALL) cells(22). This study utilized a novel compound, DI-39, to inhibit dCK and target NSP, as well as thymidine (dT) to inhibit RNR and target DNP (Figure 1). However, this approach has not been tested in solid tumors. In this study, we sought to test whether dual targeting of DNP and NSP would be an effective therapeutic strategy in the treatment of Glioblastoma.
Figure 1.
Schematic of targeting the nucleotide synthesis pathways: de novo, and salvage. DI-39 is a compound that inhibits the salvage pathways by blocking the enzyme, dCK. dT is converted to dTTP that inhibits de novo synthesis by blocking the enzyme RNR. Together, dT+DI-39 provide a combinatorial therapy that inhibits nucleotide synthesis from both compensatory pathways.
Materials and Methods
Tumor collection
23 surgical resections were collected under institutional review board-approved protocols and graded by neuropathologists as previously described(23). One widely used(22, 24) tumor sample was obtained from Duke University after it had been resected and placed as a xenograft.
Neurosphere Culturing
Neurospheres were cultured from GBM tumor samples as previously described (6, 23).
In Vitro Removal Assays
We studied the percentage of sphere formation for cultures at clonal density (plating 50 cells/100uL/well of a 96 well plate) after removal from pre-treatment for three days. GBM cells were pre-treated for three days with DMSO, dT(1mM) + dC(2,5uM), DI-39(500nM) +dC, or dT+dC+DI-39. To assess colony formation we chose to use a cell density of 20 cells per well. In preliminary studies using retroviral labeled cells, we found that even in very highly proliferative cultures, the chance of spheres containing contributions from more than one cell were under 10%, with some cultures showing 100% clonal spheres at this cell density. Limiting dilution assays also demonstrated that 20 cells per well was on the linear portion of the curve for the vast majority of cultures tested. We measured sphere diameter using MCID image analysis (http://www.mcid.co.uk/).
Statistics
For comparison of small groups we used a cutoff of P<0.05 to distinguish significant differences. Statistics for comparing cell proliferation, sphere formation, and sphere diameter between groups of treated cells were done in GraphPad Prism software (http://www.graphpad.com/scientific-software/prism/) utilizing the paired Mann-Whitney test. Analysis of tumor formation utilized CHI(2) tests using STATA 8.0 software (StataCorp, http://www.stata.com/). Linear regression of Annexin vs. doubling time for sphere cultues was assessed for significance by GraphPad Prism software.
In Vivo Intracranial Xenotransplantation
Animal experimentation was done with institutional approval following NIH guidelines using non-obese diabetic-severe combined immunodeficiency mice (Jackson Immunoresearch Laboratories, http://www.jacksonimmuno.com). 1×105 of HK308 or HK296 FLUC-GFP GBM cells (constitutively expressing a construct of luciferase (FLUC) and green fluorescence protein (GFP)) were injected intracranially as orthotopic xenotransplants into, female, 2 month old, nod scid, gamma null mice as previously described(23). Each of the two pre-treatment groups (DMSO or dT(1mM) + dC(2.5uM) + DI-39 (500nM)) consisted of 4 mice. After 12–13 weeks, mice underwent optical imaging to assess tumor size. Bioluminescent imaging of the tumors was carried out. Following perfusion with 4% paraformaldehyde, brains were removed, postfixed and sunk in sucrose prior to sectioning on a cryostat.. GFP signal for each tumor section was visualized and photographed in order to validate the bioluminescent imaging.
Optical Imaging
Optical imaging was performed at the Crump Institute for Molecular Imaging at UCLA. Mice were anesthetized by inhalation of isoflurane. Intraperitoneal injection of 100uL of luciferin (30mg/ml) was followed by 10 minutes of live uptake to interact with the luciferase expressing FLUC-GFP cells and produce bioluminescence. The IVIS Lumina 2 imaging system (Caliper life sciences) was utilized for in vivo bioluminescent imaging. A photograph of the mice is overlaid with a color scale of a region of interest representing total flux (photon/second) and quantified with the LivingImage software package (Xenogen).
FACS analysis
For cell cycle analyses, total DNA content was determined using 20 μg/ml propidium iodide containing 5 μg/ml RNase A. Annexin V staining was performed according to the manufacturer’s protocol (Becton Dickinson). All flow cytometry data were acquired on a four-laser LSRII cytometer (Becton Dickinson) and analyzed using FlowJo v7.6.5 (Tree Star). Annexin V flow cytometry data was normalized to the vehicle control for each respective cell line.
Apoptotic response to dual treatment by dT + DI-39
After three days of treatment with dT(1mM) + dC(2.5uM) +DI-39 (500nM), each culture tested was analyzed by FACS analysis for the percentage of cells with cell death as determined by Annexin V staining and propidium iodide. Each value was normalized to the non-treated, dC(2.5uM) only response. Values were plotted using GraphPad Prism software.
Microarray
Concentration and quality of RNA samples was examined using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies) and the Agilent 2100 Bioanalyzer (Agilent Technologies). RNA samples were reverse transcribed and labeled according to manufacturer’s instructions and hybridized to Affymetrix high-density oligonucleotide HG-U133A Plus 2.0 Human Arrays. Microarray data analysis was performed as previously described (25). Briefly, array preprocessing was completed in the R computing environment (http://www.r-project.org) using Bioconductor (http://www.bioconductor.org). Raw data was normalized using the robust multi-array (RMA) method (26). To eliminate batch effects, additional normalization was performed using the R package “ComBat” (http://statistics.byu.edu/johnson/ComBat/) (27) with default parameters. Contrast analysis of differential expression was performed using the LIMMA package (28). After linear model fitting, a Bayesian estimate of differential expression was calculated using a modified t-test. The threshold for statistical significance was set at p<0.005 for differential expression analysis and p<0.01 for explorative analyses (gene ontology and pathway analysis). Gene Ontology and Pathway analysis were carried out using the Database for Annotation, Visualization and Integrated Discovery (DAVID) and Ingenuity Pathway Analysis (www.ingenuity.com).
TFacts analysis
Sign sensitive analysis of transcription factor associations with our gene lists of interest was performed at http://www.tfacts.org/TFactS-new/TFactS-v2/index1.html. Transcription factors were considered to be significantly associated with gene lists if E<0.05.
Gene trait correlations
Gene-trait correlations and p-values were obtained using the standard Pearson correlation coefficient r using the cor() function in R. A p<0.001 threshold was used to select the most interesting candidates.
Comet Assay
Cell cultures were treated with either Ctrl-H20, or dT(1mM)+DI-39(500nM) for three days. Comet assays were performed using OxiSelect Comet Assay Kit (Cell Biolabs, INC) according to manufacturer’s protocol. Comet tails were counted and a fraction of nuclei with comet tails was determined and depicted in the results. A minimum of 50 nuclei were counted per condition.
TCGA classification, EGFR amplification, EGFRv3 status, and doubling time (proliferation rate) were determined as previously described (Laks, et al., 2016)(29).
Results
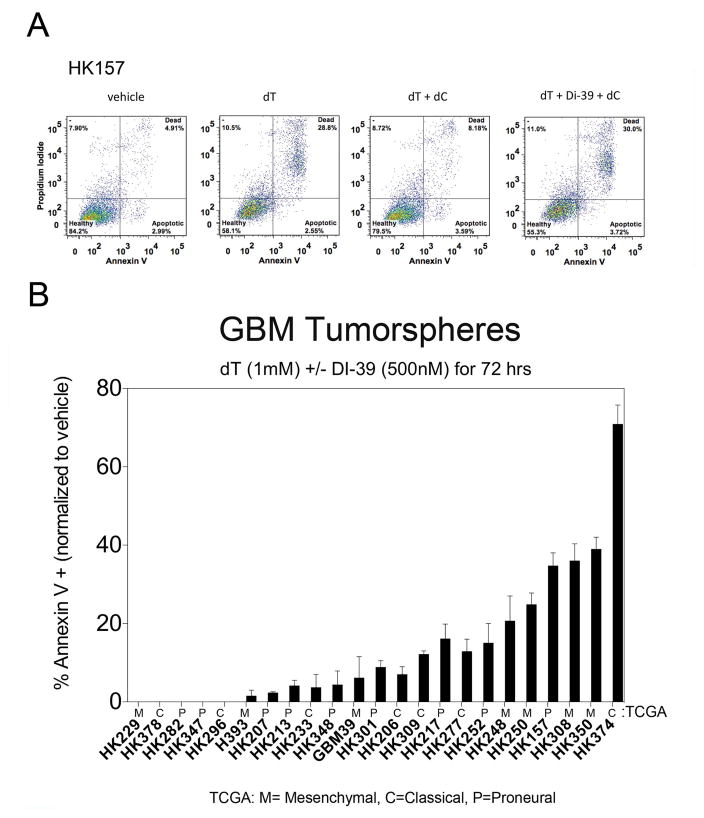
We recently reported that the NSP, through dC metabolism, can compensate for DNP-induced lethality in T-ALL cells(30). To first test whether the NSP can compensate for DNP inhibition in GBM cells, we treated the gliomasphere culture, HK157, with thymidine (dT) in the absence of dC. Upon addition of thymidine (dT), cell death rose to 28.8% (Figure 2A, second panel). However, the addition of dC significantly abrogated apoptosis (8.2%, Figure 2A, third panel). To confirm that the NSP is responsible for mitigating dT-induced apoptosis, we treated HK157 cells with the dCK inhibitor, DI-39, in the presence of dT and dC. The addition of DI-39 restored apoptosis (30.8%), suggesting that, similar to T-ALL, GBM can utilize the salvage pathway to compensate for DNP inhibition. There was no affect of DI-39 monotherapy alone on apoptotic measures (Annexin V) or on cell cycle arrest of either resistant or sensitive cell cultures (data not shown).
Figure 2.
Differential response to DI-39 treatment. A. FACS analysis plots of apoptosis (Annexin V) demonstrate an important role for the salvage pathway in GBM. The Y-axis displays propidium iodide that labels DNA and the X-axis display Annexin V that labels apoptotic markers on cells. The first panel (far left) is vehicle (DMSO) treatment of HK157 GBM cells and displays 4.91% basal levels of cell death (upper right quadrant). The next panel is dT (1mM) treated cells and displays 28.8% cell death upon inhibition of de novo nucleotide synthesis (by dT) in the absence of dC which, if present, would activate the salvage pathway. The next panel displays a rescue of the effect (only 8.18% cell death) of dT (inhibition of de novo pathway) by addition of dC (2.5uM) that activates the salvage pathway. The panel on the far right displays the effect of combined inhibition of the de novo and salvage pathways for nucleotide synthesis, with 30% cell death, as the addition of DI-39 (500nM) inhibits the salvage pathway that would otherwise be activated by the addition of dC. B. Differential response across a panel of 23 GBM cultures to combinatorial targeting of de novo and salvage pathways for nucleotide synthesis. The x-axis displays the identification numbers for each patient derived human GBM culture. The y-axis displays the % annexin from FACS analysis, normalized to vehicle treated controls. Each measurement represents the mean +/− SEM from two separate experiments. Each culture was treated for three days with 1mM dT, 2.5 uM dC, and 500nM DI-39. This plot displays cultures that are sensitive to this treatment (on the right side of the plot) and those that are resistant to this treatment (on the left side of the plot).
There is a differential response to combinatorial treatment of dT +DI-39 across a panel of GBM cultures (Figure 2B). The normalized Annexin response (apoptotic index) is plotted with the sensitive cell cultures situated on the right side of the graph, and the resistant cell cultures situated on the left side of the graph (Figure 2B). Apoptotic response (Annexin V) to DI-39 was not related to EGFRv3 in culture (CHI2, P=0.143), EGFR amplification in tumor sample (CHI2, P=0.247), PTEN status in culture (CHI2, P=0.776), or TCGA classification of culture (CHI2, P=0.080). In addition, linear regression indicated that apoptotic response (Annexin V) to DI-39 was not significantly related to the baseline proliferation rate of the cultures in the absence of drug (P=0.0963).
Each cell culture was processed for RNA content, and assessed by microarray analysis to generate gene expression levels prior to treatment with DI-39. We performed gene by gene correlation analysis with the Annexin response (sensitivity) to treatment with dT+DI-39 and generated a list of 1,536 genes that were associated with response (P<0.001). Next we used Qiagen’s Ingenuity Pathway Analysis (IPA, www.qiagen.com/ingenuity) to distinguish canonical pathways associated (P<0.05) with this list of genes. IPA determined 66 canonical pathways associated with response including ErbB2-ErbB3 signaling (undetermined direction of association), PTEN signaling (associated with resistance, Z-score= −1.291), PI3K/AKT signaling (associated with sensitivity, Z-score=0.832), and the nucleotide excision repair pathway (undetermined direction of association, Supplementary Table 1). We analyzed the list of genes associated with DI-39 response in TFacts (www.tfacts.org), in a sign sensitive manner, and the only transcription factor predicted to be associated with this list was PAX3 (E-value=0.01734). PAX3 was predicted to be inhibited in relation to the list of genes, meaning that activation of PAX3 is associated with resistance to treatment by dT+DI-39.
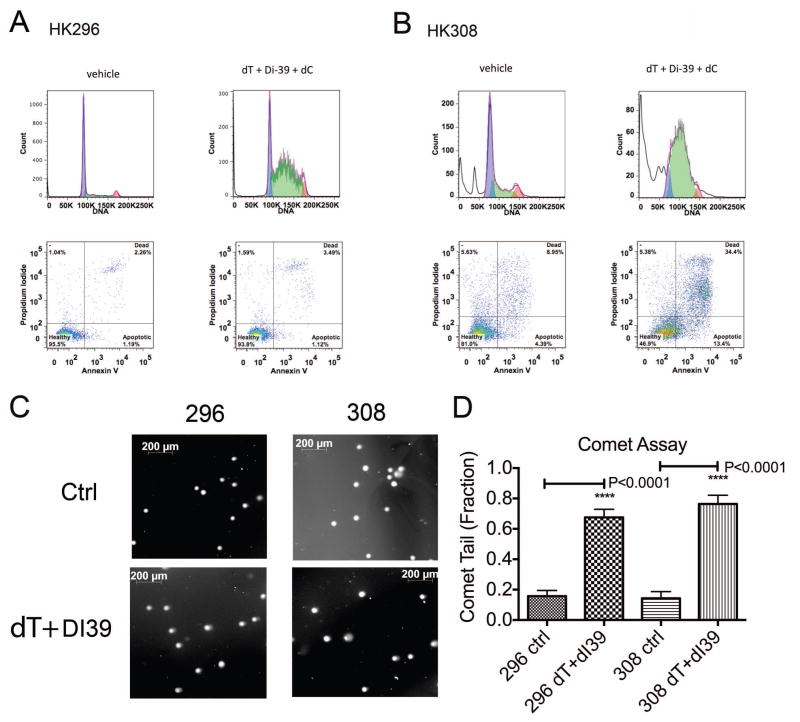
Although dual treatment of DI-39 and dT effectively induced S-phase delay in all cultures treated, only certain sensitive cultures responded with an increase in cell death (Figure 3 A, B). For example, combined targeting of DNP (with dT) and NSP (with DI-39) in HK296 GBM cells promoted S-phase delay (Figure 3A, top right panel) but no lethality (Figure 3A, bottom right panel). In contrast, the sensitive culture, HK308 cells responded to dual treatment with delay in S-phase (Figure 3B, top right panel) and with a dramatic increase in cell death by apoptosis (Figure 3B, bottom right panel). Furthermore, cell death response was not related to the amount of DNA damage induced by treatment (Figure 3 C, D). Both the sensitive culture, HK308, and the resistant culture, HK296, showed equivalent increased levels of DNA damage as demonstrated by comet assay after three days of treatment with DI-39 + dT (P<0.0001 for both increases, Mann-Whitney Test, Figure 3 C,D). For HK296, the resistant culture, there was an increase in comet tails of 51.9% upon treatment as compared to control, and for HK308, the sensitive culture, there was an increase in comet tails of 62.1% upon treatment as compared to control. The fraction of comet tails was not significantly different between HK296 and HK308 treated cultures (P=0.2724, Mann-Whitney test).
Figure 3.
Dual targeting of de novo and salvage pathways for nucleotide synthesis results in S-phase delay and in certain sensitive GBM cultures, apoptosis and cell death. All treatments are three days. A. In a resistant GBM culture, HK296, combinatorial targeting of de novo and salvage pathways results in S-phase delay but not in a substantial increase in cell death. In the top two panels, the x-axis displays propidium iodide that indicates DNA levels in each cell. The y-axis displays the cell count. The first panel on the left hand side displays a normal distribution for the cell cycle with most cells at 2N (85k) and a minority at 4N (170K) with few cells in between (S-phase). However, in the upper panel to the right, upon inhibition of de novo and salvage pathways with the addition of dT and DI-39, the cell cycle is disrupted and there is an S-phase delay with a buildup of cells in between 2N and 4N. Despite this S-phase delay, the FACS analysis plots below display no substantial increase in cell death. B. In contrast, in a sensitive GBM culture, HK308, the S-phase delay is accompanied by a substantial increase in cell death upon combinatorial inhibition by dT and DI-39. C. Representative images of comet assay tails after three day treatment conditions of Ctrl-H20 treated cells, or DI-39 (500nM) +dT (1mM). D. Equivalent increase in DNA damage between sensitive and resistant cell cultures after treatment (both changes were P<0.0001, Mann-Whitney test). Quantification of the fractions of comet tails under each condition (same conditions as in Figure 2D) after three days treatment. Results are Mean +/− standard error of the mean.
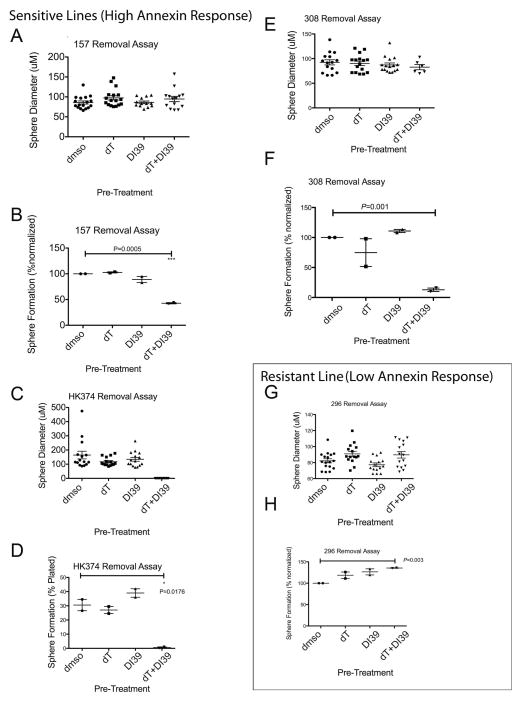
In GBM cultures sensitive to treatment by dT+DI-39, clonal sphere formation was impaired after pre-treatment, whereas in resistant GBM cultures, clonal sphere formation was slightly increased after pre-treatment (Figure 4). We performed removal assays where cells were pre-treated with control dC (2.5uM), dT (1mM) +dC, DI-39 (500nM) +dC, or dT+DI-39+dC. After pre-treatment, cells were seeded at clonal density (20 cells/well of a 96 well plate) and allowed to form sphere colonies in the absence of treatment. Clonal sphere diameter was not changed by pre-treatment in any GBM cell culture tested (Figure 4 A,C,E,G). However, in cultures that were sensitive to dT+DI-39 treatment, sphere formation was significantly reduced after pre-treatment with dT+DI-39 (Figure 4 B, D, F). In the GBM culture tested that was resistant to treatment by dT+DI-39, sphere formation increased after pre-treatment with dT+DI-39 (Figure 4H). These data indicate that dT+DI-39 depletes the cells responsible for sphere formation in the subset of sensitive cultures but slightly enriches for sphere forming cells in at least one culture that was resistant to treatment.
Figure 4.
Combinatorial targeting of de novo and salvage pathways for nucleotide synthesis target brain tumor stem cells in sensitive GBM cultures but not in resistant GBM cultures. Results from removal assays where cells are treated for 3 days and then removed into untreated conditions under clonal conditions (20 cells/well) until spheres form (14–21 days). Each plot represents two separate experiments. Each condition included 2.5uM of dC to activate the salvage pathway. A. Sphere diameter does not change upon treatment in HK157 cells. Sphere diameter (uM) for each measurement is depicted along with the mean and SEM. B. Sphere formation is reduced in HK157 cells upon combinatorial treatment of dT (1mM) + DI-39 (500nM) in the presence of dC (2.5uM) (P=0.0005, T-test). C. Sphere diameter in HK374 cells cannot be measured as no spheres formed. D. Sphere formation is reduced in HK374 cells upon combinatorial treatment of dT (1mM) + DI-39 (500nM) in the presence of dC (2.5uM) (P=0.0176, T-test). E. Sphere diameter does not change in HK308 cells upon treatment. F. Sphere formation is reduced in HK308 cells upon combinatorial treatment of dT (1mM) + DI-39 (500nM) in the presence of dC (2.5uM) (P=0.001, T-test). G. Sphere diameter is not changed upon treatment in HK296 cells. H. Sphere formation is increased upon treatment with dT + DI-39 in HK296 cells (P=0.003, T-test). Results are Mean +/− standard error of the mean.
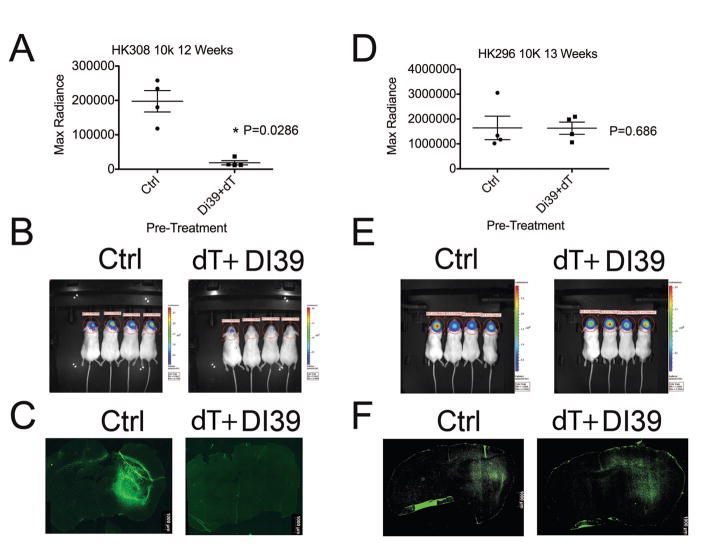
To determine whether in vivo tumor initiation is affected with treatment, we performed intracranial tumor initiating assays after pre-treatment with dT and DI-39. Tumor formation and tumor size were reduced after pre-treatment with dT+DI-39 in a sensitive GBM cell culture but not in a resistant GBM culture (Figure 5). In HK308 cells, a culture that is sensitive to DI-39+dT treatment in vitro, orthotopic xenotransplanted cells that were pre-treated with DI-39+dT formed tumors that were significantly smaller than cells pre-treated with control-dC (P=0.0286, Mann-Whitney Test, Figure 5A). Only one fourth of the mice transplanted with DI-39 pre-treated HK308 cells formed detectable tumors whereas all of the mice transplanted with Control-dC treated HK308 cells formed tumors and this represents a significant reduction in tumor formation due to DI-39 pre-treatment (P=0.028, Chi(2) test, Figure 5B). Figure 5C illustrates the GFP labeled tumor cells at the site of the tumor bulk/injection site. In these representative images of the treatment groups the pre-treatment group formed no discernible tumor. These sections validated the optical imaging in figure 5B. In HK296 culture, a resistant culture in vitro, orthotopic xenotransplanted cells that were pre-treated with DI-39+dT formed tumors that were equivalent in size to cells pre-treated with control-dC (Figure 5D). In addition, all mice formed tumors regardless of pre-treatment (Figure 5E). Sectioning of the tumors validated the measurements generated by optical imaging (Figure 5F). These findings confirm that in vitro treatment with DI-39 plus dT selectively depletes tumor initiating cells in a sensitive culture, but not in resistant culture.
Figure 5.
In sensitive GBM cultures HK308, ex vivo treatment with dT + DI-39 depletes cultures of brain tumor stem cells and drastically reduces tumor formation upon intracranial xenotransplantation. In contrast, in resistant culture HK296, there is no difference in tumor formation or tumor size after treatment. A. DI-39 + dT ex vivo pre-treatment results in significantly less tumor burden in the sensitive GBM culture HK308 (P=0.0286, Mann-Whitney test). This plot shows quantification of individual measurements of tumor bioluminescent imaging after 12 weeks with the mean and SEM depicted as bars. B. Images of mice and their bioluminescent signals from transplanted cells expressing luciferase via lentiviral infection. Only one mouse out of four in the treated group has detectable levels of tumor cells while the control group has uniform detection of large tumors. C. Representative sections of GFP expressing tumor cells from Ctrl and dT+DI-39 pre-treated cells display a large tumor in the striatum of the Ctrl and no tumor in the brain of the pre-treated cells. D. Quantification of tumor bioluminescence indicates that DI-39 + dT ex vivo pre-treatment results in no change in tumor burden in the resistant GBM culture HK296 (P=0.686, Mann-Whitney test). This plot shows individual measurements of tumor bioluminescent imaging after 13 weeks with the mean and SEM depicted as bars. E. Images of mice and their bioluminescent signals from transplanted cells expressing luciferase via lentiviral infection. Both the control group and pre-treated group have uniform detection of large tumors. F. Representative sections of GFP expressing tumor cells from Ctrl and dT+DI-39 pre-treated cells display large tumors in the striatum of both the Ctrl and pre-treated cells. Results are Mean +/− standard error of the mean.
Discussion
Effective inhibition of nucleotide synthesis requires combinatorial dT +DI-39 therapy for targeting both the DNP and the compensatory NSP, respectively. In this study, we demonstrate that this therapeutic strategy targets sphere and tumor initiating cells and successfully abrogates tumor formation in a subset of GBM that are sensitive to treatment. This indicates that treatment with dT+DI-39 is effectively targeting BTSC in certain, sensitive GBM. Importantly, the difference between sensitive and resistant GBM cultures is not dependent upon the degree of S-phase delay induced by treatment. Indeed, both sensitive and resistant GBM cell cultures respond to a similar degree with dramatic S-phase delay. Despite the similarity in S-phase delay in response to treatment, a differential response to DI-39 +dT treatment exists across our tested panel of GBM in terms of apoptotic cell death.
Our findings highlight the importance of testing potential therapeutics across a spectrum of GBM. One means that might be used to achieve this goal would be to divide samples according to their classification groups as set out by Verhaak et al.(16) using the TCGA dataset or according to their key oncogenic mutations. However, our findings did not reveal group differences in response according to these schemes, suggesting that other means will need to be used to prospectively identify sensitive or resistant cohorts. A goal of future studies is to distinguish biomarkers that define the subset of GBM which are sensitive to this treatment strategy. Our gene by gene correlation with the percentage Annexin response to treatment indicated that response is related to the nucleotide excision repair pathway. This may provide a clue to the mechanisms of differential response by GBM cultures to this treatment as sensitivity may be dependent upon the cell’s response to DNA damage rather than to the DNA damage itself. In support of this hypothesis, our comet assay data demonstrate an equivalent amount of DNA damage in both sensitive and resistant GBM cultures. Although both sensitive and resistant cultures demonstrate equivalent DNA damage and S-phase delay, the responses to such stress and the ultimate fates of the cells differ considerably.
Our bioinformatics analysis indicated that resistance to treatment was related to activation of the transcription factor, PAX3. PAX3 has been shown to regulate tumorigenicity of GBM and moreover, inhibit apoptosis(31). Perhaps this role of PAX3 in inhibiting apoptosis is associated to response to DNA damage induced by impaired nucleotide synthesis. Further work is necessary to determine if PAX3 serves a functional role in determining response to inhibition of nucleotide synthesis in GBM. A previous study has demonstrated that PAX3 depletion increases cytotoxicity of the DNA damaging agent Cisplatin(32). Our data are consistent with this role for PAX3 in mediating apoptotic cell death in response to impaired DNA.
In our study, we were unable to test these hypotheses that in vivo treatment may reduce malignancy in sensitive GBM but increase malignancy in resistant GBM, as DI-39 has poor blood brain barrier penetration and dT does not sufficiently penetrate in order to achieve therapeutic concentrations in experimental brain tumors (not shown). In order to develop inhibition of nucleotide salvage pathway as a therapeutic approach, we, will need to design replacements for DI-39 and dT that can target brain tumors in vivo. However, the findings presented here, represent a compelling foundation for the role of nucleotide synthesis inhibition in targeting BTSC upon which to build further investigations, with the objective of reducing tumor recurrence and malignancy in human patients.
Supplementary Material
Acknowledgments
Funding: National Institutes of Neurological Disorders and Stroke (NINDS) grant (NS052563), H.I. Kornblum, The Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, H.I. Kornblum and G. Coppola, and a Whitcome Fellowship, D.R. Laks. We acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691), G. Coppola.
The authors thank Dr. Jack Mottahedeh and Andre Gregorian for their aid in culturing brain tumor cells.
Footnotes
Conflict of Interest: The University of California has patented intellectual property for small molecule dCK inhibitors invented by C.G.R. and D.A.N. This intellectual property has been licensed by Trethera Corporation, in which C.G.R., D.A.N., and the University of California hold equity.
References
- 1.Daumas-Duport C, Scheithauer B, O’Fallon J, Kelly P. Grading of astrocytomas. A simple and reproducible method. Cancer. 1988;62:2152–65. doi: 10.1002/1097-0142(19881115)62:10<2152::aid-cncr2820621015>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 2.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–61. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 3.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 4.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 5.Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39:193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 6.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–83. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–8. [PubMed] [Google Scholar]
- 8.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 9.Singh SK, Clarke ID, Hide T, Dirks PB. Cancer stem cells in nervous system tumors. Oncogene. 2004;23:7267–73. doi: 10.1038/sj.onc.1207946. [DOI] [PubMed] [Google Scholar]
- 10.Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z, Khan MA, Weiler M, Blaes J, Jestaedt L, Geibert M, et al. Targeting Self-Renewal in High-Grade Brain Tumors Leads to Loss of Brain Tumor Stem Cells and Prolonged Survival. Cell stem cell. 2014 doi: 10.1016/j.stem.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Zong H, Parada LF, Baker SJ. Cell of Origin for Malignant Gliomas and Its Implication in Therapeutic Development. Cold Spring Harb Perspect Biol. 2015 doi: 10.1101/cshperspect.a020610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ene CI, Holland EC. Personalized medicine for gliomas. Surg Neurol Int. 2015;6:S89–95. doi: 10.4103/2152-7806.151351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Di C, Mattox AK, Wu L, Adamson DC. The future role of personalized medicine in the treatment of glioblastoma multiforme. Pharmacogenomics and personalized medicine. 2010;3:111–27. doi: 10.2147/PGPM.S6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–74. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 18.Austin WR, Armijo AL, Campbell DO, Singh AS, Hsieh T, Nathanson D, et al. Nucleoside salvage pathway kinases regulate hematopoiesis by linking nucleotide metabolism with replication stress. The Journal of experimental medicine. 2012;209:2215–28. doi: 10.1084/jem.20121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiuten DF, Wiernik PH, Zaharko DS, Edwards L. Clinical phase I–II and pharmacokinetic study of high-dose thymidine given by continuous intravenous infusion. Cancer Res. 1980;40:818–22. [PubMed] [Google Scholar]
- 20.Kufe DW, Beardsley P, Karp D, Parker L, Rosowsky A, Canellos G, et al. High-dose thymidine infusions in patients with leukemia and lymphoma. Blood. 1980;55:580–9. [PubMed] [Google Scholar]
- 21.Kufe DW, Wick MM, Moschella S, Major P. Effect of high-dose thymidine infusions in patients with mycosis fungoides. Cancer. 1981;48:1513–6. doi: 10.1002/1097-0142(19811001)48:7<1513::aid-cncr2820480705>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 22.Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–6. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laks DR, Masterman-Smith M, Visnyei K, Angenieux B, Orozco NM, Foran I, et al. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells. 2009;27:980–7. doi: 10.1002/stem.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarkaria JN, Yang L, Grogan PT, Kitange GJ, Carlson BL, Schroeder MA, et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Molecular cancer therapeutics. 2007;6:1167–74. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- 25.Coppola G. Designing, performing, and interpreting a microarray-based gene expression study. Methods Mol Biol. 2011;793:417–39. doi: 10.1007/978-1-61779-328-8_28. [DOI] [PubMed] [Google Scholar]
- 26.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 28.Smyth G. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. Limma: linear models for microarray data. [Google Scholar]
- 29.Laks Dan R, TJC, Shih Michelle YS, Mottahedeh Jack, Gao Fuying, Sperry Jantzen, Garrett Matthew C, Yong William H, Cloughesy Timothy F, Liau Linda M, Lai Albert, Coppola Giovanni, Kornblum Harley I. Large-scale Assessment of the Gliomasphere Model System. Neuro-oncology. 2016 doi: 10.1093/neuonc/now045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nathanson DA, Armijo AL, Tom M, Li Z, Dimitrova E, Austin WR, et al. Co-targeting of convergent nucleotide biosynthetic pathways for leukemia eradication. The Journal of experimental medicine. 2014;211:473–86. doi: 10.1084/jem.20131738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia L, Huang Q, Nie D, Shi J, Gong M, Wu B, et al. PAX3 is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Brain Res. 2013;1521:68–78. doi: 10.1016/j.brainres.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 32.He SJ, Stevens G, Braithwaite AW, Eccles MR. Transfection of melanoma cells with antisense PAX3 oligonucleotides additively complements cisplatin-induced cytotoxicity. Molecular cancer therapeutics. 2005;4:996–1003. doi: 10.1158/1535-7163.MCT-04-0252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.