Abstract
Inhibitor of differentiation (Id) proteins are DNA-binding transcription factors involved in cellular proliferation, migration, inflammation, angiogenesis and fibrosis. However, their expression and role in the cornea is unknown. The present study was undertaken to characterize the expression of Id proteins and their interactions with the pro-fibrotic cytokine Transforming Growth Factor β1 (TGFβ1) and anti-fibrotic cytokine, bone morphogenic protein 7 (BMP7) in human cornea. Human donor corneas procured from Eye Bank were used. Id proteins were localized in human corneal sections using immunofluorescence. Primary cultures of human corneal fibroblasts (HCF) were established and treated with either TGFβ1 (5 ng/ml) or BMP7 (10 ng/ml) for 24 h in serum free medium. Expression of Id’s in response to TGFβ1, BMP7 and TGFβ1 + BMP7 was analyzed by quantitative real time PCR (qRT-PCR) and western blot analysis. Id1 and Id2 proteins were ubiquitously expressed in the epithelial cells and stromal keratocytes in human cornea. The Id1 was localized to the basal epithelial cells as seen by immunohistochemistry. HCF expressed all known mammalian Id genes (Id1–Id4). In addition, Id1 and Id2 are selectively expressed in HCF. Treatment of human recombinant TGFβ1 (5 ng/ml) to serum-starved HCF showed a significant increase in Id genes (Id1, Id2 and Id4) at 2 h time point compared to BMP7 treatment, which showed time dependent increase in the expression of Id1–Id3 at 24–48 h. Combined treatment with TGFβ1 + BMP7 to HCF showed a significant increase in Id1 transcript and an increasing trend in Id3 and Id4 expression. The results of this study suggest that Id family of genes (Id1–Id4) are localized in the human cornea and expressed in the corneal fibroblasts. Also, Id’s were differentially regulated with TGFβ1 and/or BMP7 in a time dependent manner and might serve as a therapeutic target in corneal fibrosis.
Keywords: Id proteins, Cornea, Differentiation, Proliferation, Transforming growth factor β1, Bone morphogenic protein 7, Corneal fibrosis
1. Introduction
The corneal wound healing mechanism is a remarkably complex cascade mediated by cytokines, growth factors, and chemokines including transforming growth factor (TGF)-β1, interleukin (IL)-1β, connective tissue growth factor (CTGF), epidermal growth factor (EGF), platelet derived growth factor (PDGF), and so on (Tandon et al., 2010; Stapleton et al., 2008; Gibson et al., 2014; Wang et al., 2013; Kaur et al., 2009). TGFβ1 is arguably the most important cytokine involved in wound repair and many other vital pre- and postnatal physiological processes such as cellular proliferation, migration, differentiation, apoptosis, and extracellular matrix (ECM) production in the corneal tissue (Izumi et al., 2006; Kinoshita et al., 2007; Veerasamy et al., 2013). It is well established that both the corneal epithelium and stroma express TGFβ receptors and their ligands to control variety of cellular functions, the most important being the transdifferentiation of quiescent keratocytes to myofibroblasts during corneal wound healing. Following stromal injury, keratocytes adjacent to the wound migrate to the injury site and differentiate into post-mitotic α-smooth muscle actin (α-SMA) expressing myofibroblasts in the presence of TGFβ1 (Myrna et al., 2009). These activated myofibroblasts have contractile properties that assist in wound closure, and secrete extra cellular matrix proteins that aid in tissue remodeling (Myrna et al., 2009; DelMonte and Kim, 2011). However corneal fibrosis occurs when this process fails to terminate and results in myofibroblast-induced excessive deposition of irregular ECM, scar formation and hence vision impairment (Myrna et al., 2009; Honda et al., 2013).
Bone morphogenetic proteins (BMPs) are members of the TGFβ superfamily involved in the embryonic development and cellular proliferation, differentiation, and apoptosis. Several BMPs and their receptors have been described in human corneal epithelium and stroma (You et al., 1999). Among other BMP family members, BMP7 has been shown to play a pivotal role in eye development during embryogenesis and BMP7-knockout mice exhibits an ophthalmic phenotype. In the cornea, therapeutic effects of BMP7 have been examined via nanoparticle-mediated gene transfer using a rabbit corneal fibrosis model in vivo (Tandon et al., 2013). BMP7 has been reported to oppose TGFβ biological activity via Smad signaling and attenuates TGFβ1 hyperactivity-driven fibrosis (Tandon et al., 2013). Many recent studies suggest that inhibitor of differentiation proteins are the crucial targets of BMP7 to confer anti-fibrotic responses in non-ocular tissues (Miyazono and Miyazawa, 2002).
Till today, four Id genes (Id1–Id4) have been identified in mammals. They have been recognized as the functional inhibitors of the basic helix–loop–helix (bHLH) transcription factors. The crucial biochemical attribute of Id genes to their DNA binding activity is to regulate cell fate, differentiation and proliferation (Norton, 2000). The Id1 and Id3 double knockout mice exhibit small brain, impaired angiogenesis, hemorrhage in forebrain, premature exit of neuroblasts from the cell cycle, and death at 13.5 embryonic day suggesting that Id proteins play an important role in embryonic development and tissue regeneration (Lyden et al., 1999; Miyazono and Miyazawa, 2002). Most importantly, Id proteins have been demonstrated to attenuate fibrosis in a variety of animal models including pulmonary (Izumi et al., 2006), hepatic (Kinoshita et al., 2007), and renal tissues (Veerasamy et al., 2013). In fact, Id gene levels have been shown to be regulated positively by BMP7 and negatively by TGFβ1 but not by TGFβ2 or TGFβ3 (Nagata and Todokoro, 1994; Neuman et al., 1995; Norton, 2000). However, no data exists on Id genes expression in cornea and their role in corneal fibrosis to the best of our knowledge. In this study, we characterized mRNA and protein expression of Id genes in human cornea and their regulation in the presence of TGFβ1 and/or BMP7 stimulation in an in vitro model of corneal fibrosis using cultured primary human corneal fibroblasts.
2. Materials and methods
2.1. Human cornea
All experiments in the study adhered to the tenets of the Declaration of Helsinki as a statement of ethical principles for medical research involving human subjects and guidelines of the Institutional Review Board of the University of Missouri. Healthy human corneal rims were procured from the Saving Sight (Kansas City, Missouri) and handled as described previously (Sharma et al., 2009). Corneal rims were either excised for primary human cornea fibroblasts or processed for immunohistochemistry.
2.2. Primary human corneal fibroblasts (HCF)
Human corneal fibroblasts were generated from human donor corneal rims (n = 6). Briefly, corneal tissues were washed with sterile cell culture medium, and the epithelium and endothelium were removed by gentle scraping with a scalpel blade. The corneal stroma was cut into small pieces, placed on culture dish and incubated in humidified 5% CO2 incubator at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum for 2–4 weeks until 100% confluence. The HCF cultures were harvested from corneal buttons, grown in six-well plates in DMEM supplemented with 10% fetal bovine serum to obtain fibroblast cultures. To generate myofibroblasts, HCF were initially seeded in DMEM medium containing 10% serum, switched to serum-free medium containing TGFβ1 (5 ng/ml) after 12–14 h, and grown for 5 days by feeding fresh serum-free medium and TGFβ1 every 24 h. Seventy percent confluent corneal fibroblast and myofibroblast cultures were used for experimentation.
2.3. Immunohistochemistry
Normal human corneas were snap frozen, embedded in optimal cutting temperature (OCT) solution, and cryosectioned at 7 μm thickness using a Microm HM525 cryostat (ThermoFisher Scientific, Waltham, MA). Sections were rinsed in 1 × PBS for 5 min at room temperature, outlined with a pap pen, and blocked using 5% normal donkey serum (Jackson Immuno Research Laboratories, Inc., West Grove, PA). Tissue sections were then subjected to single immunostaining using either anti-Id1 (1:20 dilution, Abcam® Cambridge, MA) or anti-Id2 (1:200 dilution, Abcam® Cambridge, MA). Secondary antibody Alexa Flour® 488 donkey anti-mouse or anti-goat (Invitrogen, Eugene, OR) was used, respectively.
For double immunohistochemistry, HCF were treated with TGFβ1 as described above, and fixed with ice-cold methanol for 15 min. Cells were then washed and blocked in 5% normal donkey Serum (Jackson Immuno Research Laboratories, Inc., West Grove, PA) with 0.1% Tween 20 (Sigma–Aldrich, St Louis, MO) for 1 h at room temperature, followed by Id1 (1:20 dilution, Abcam® Cambridge, MA) or Id2 (1:200 dilution, Abcam® Cambridge, MA) with mouse monoclonal αSMA antibody (1:200 dilution, Dako, Carpinteria, CA) for 90 min. Primary antibodies were removed and cells were rinsed 3 times in 1 × PBS for 10 min s each rinse. Cells were then incubated in secondary antibodies Alexa Flour® 488 donkey anti-mouse (Invitrogen, Eugene, OR) and Alexa Flour® 594 donkey anti-goat (Invitrogen, Eugene, OR) for 1 h at room temperature. The cells were washed three times in PBS, mounted in Vectashield containing 4′-6-diamidino-2-phenylindole (DAPI; Vector Laboratories), and photographed with a Leica DM 4000B fluorescent microscope (Leica, place, state, country) equipped with a digital camera (SpotCam RT KE).
2.4. Human recombinant TGFβ1 and BMP7 treatments
Human corneal fibroblasts were platted in six well culture dishes at density of 7.5 × 104 using medium supplemented with 10% serum, and after 12–14 h switched to serum-free medium. Cultures at 70% confluence were treated either with vehicle, human recombinant TGFβ1 (5 ng/ml) (PeproTech, Inc. Rocky Hills, NJ), or human recombinant BMP7 (10 ng/ml) (Pepro Tech, Inc. Rocky Hills, NJ) or TGFβ1 (5 ng/ml) + BMP7 (10 ng/ml) combination for 2, 12, 24, or 48 h. Thereafter, cultures were washed twice with cold 1× PBS, mRNA were isolated and converted into cDNA for quantitative real-time PCR analysis. Each treatment had 4 replicates in 6-well culture dish. Samples were run in triplicate in every qPCR reaction, and qPCR were repeated thrice.
2.5. Quantitative real-time polymerase chain reaction
Total RNA was isolated using RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. First strand cDNA was synthesized by reverse transcriptase enzyme (Promega, Madison, WI). qRT-PCR was performed using the One Step Plus Real-Time PCR system (Applied Biosystems, Carlsbad, CA). Table 1 lists the gene specific primers used in the reverse transcription PCR and qRT-PCR. 20 μl reaction mixtures containing 1 μl cDNA, 2 μl forward and 2 μl reverse primer, 10 μl iQ™ SYBR® Green Super mix (Bio-Rad Laboratories, Hercules, CA) using the following PCR parameters: 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The fluorescence threshold value (Ct) was calculated to detect differences in signal associated with exponential increase of PCR products in the log linear phase. Relative expression/fold change over the corresponding values for the control was calculated by the 2−ΔΔCt method. Two to three independent experiments were executed, and qRT-PCR reactions were ran in triplicates for each sample, and the average fold changes in mRNA levels were calculated (see Table 2).
Table 1.
Oligonucleotide primers used for RT-PCR.
| Gene | Forward primer sequence 5′–3′ | Reverse primer sequence 5′–3′ | Product size | Accession number |
|---|---|---|---|---|
| Id1 | GTG CGC TGT CTG TCT GAG | CAA CTG AAG GTC CCT GAT GTA G | 228bp | NM_002165.3 |
| Id2 | ATG AAC GAC TGC TAC TCC AAG | GCA AGG ACA GGA TGC TGA TA | 232bp | NM_002166.4 |
| Id3 | CGA CAT GAA CCA CTG CTA CTC | TCG TTG GAG ATG ACA AGT TCC | 213bp | NM_002167.4 |
| Id4 | GTG CGA TAT GAA CGA CTG CTA | AGT GAC GCG AGT TGT GGC | 241bp | NM_001546.3 |
Table 2.
Oligonucleotide primers used for qRT-PCR.
| Gene | Forward primer sequence 5′–3′ | Reverse primer sequence 5′–3′ | Size | Accession number |
|---|---|---|---|---|
| Id1 | GCT GTT ACT CAC GCC TCA A | CAA CTG AAG GTC CCT GAT GTA G | 110bp | NM_002165.3 |
| Id2 | CAA GAA GGT GAG CAA GAT GGA | GGT GAT GCA GGC TGA CAA TA | 104bp | NM_002166.4 |
| Id3 | CGA CAT GAA CCA CTG CTA CTC | GAT GAC GCG CTG TAG GAT TT | 97bp | NM_002167.4 |
| Id4 | GGG AAG AGC AGA AGT TAG AGA AA | ACC TCA GGG TGT TGG TTA TTC | 96bp | NM_001546.3 |
| SMA | TGG GTG ACG AAG CAC AGA GC | CTT CAG GGG CAA CAC GAA GC | 138bp | NM_001613 |
| β-actin | CGG CTA CAG CTT CAC CAC CA | CAG GCA GCT CGT AGC TCT TC | 143bp | x00351 |
Conventional polymerase chain reaction was performed in 50 μl reaction mixtures containing 10 μl buffer (5× green GoTaq® Flexi Buffer, Promega, Madison, WI), 8 μl MgCl2 (Promega, Madison, WI), 1 μl dNTP mix (Promega, Madison, WI), 1 μl forward and 1 μl reverse primers (0.4 μM each), 0.25 μl GoTaq®Flexi DNA (Promega, Madison, WI), 26.75 μl DEPC treated water and 2 μl of cDNA. Cycle details include: 95 °C for 2 min, followed by 40 cycles of 95 °C for 30 s, 50 °C for 30 s, 72 °C for 1 min, 95 °C for 1 min, and a final cycle of 72 °C for 10 min. Primers used in this study are listed in Table 1. β-actin was used as a housekeeping gene. The digital quantification of RT-PCR amplification was performed using NIH Image J software and Image Studio software Version 5.2.
2.6. Western blot analyses
Human corneal fibroblasts were seeded at an initial density of at 7.5 × 104 using medium supplemented with 10% serum, and switched to serum-free medium when reached 50% confluence. Cultures at 70% confluence were treated with either vehicle, recombinant human TGFβ1 (5 ng/ml) (Pepro Tech, Inc. Rocky Hills, NJ) and/or recombinant human BMP7 (10 ng/ml) (Pepro Tech, Inc. Rocky Hills, NJ) for 48 h. Protein lysates were prepared and quantified by Bradford assay as described previously (Sharma et al., 2009; Hogg et al., 2010). Samples were resolved on 4–12% sodium dodecyl sulfate (SDS) polyacrylamide gel, transferred onto polyvinylidene fluoride membrane, incubated with Id1, Id2, β-actin (Santa Cruz Biotechnology, Santa Cruz, CA) and α-SMA (Abcam, Cambridge, MA) antibodies followed by alkaline phosphatase-conjugated anti-mouse secondary antibodies and Nitro-blue tetrazolium chloride and 5-Bromo-4-chloro-3′-indolyphosphate p-toluidine (NBT-BCIP) developing reagents. The digital quantification of western blots was performed using NIH Image J software and Image Studio software Version 5.2.
2.7. Statistical analysis
Data was analyzed for one-way ANOVA using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA) and p < 0.05 was considered to be statistical significant.
3. Results
3.1. Id1 and Id2 are expressed in human cornea and localized in the epithelium and stromal keratocytes
Human corneal sections were subjected to immunohistochemistry with antibodies against Id1 and Id2 (Fig. 1). Id1 and Id2 were detected in keratocytes throughout the stroma in the human cornea as evident from Fig. 1A and B, respectively. Both the proteins were differentially expressed in the corneal epithelial cells where Id1 was expressed in the basal cells of the corneal epithelium (Fig. 1A), while Id2 was distributed throughout the corneal epithelium (Fig. 1B). Fig. 1C and D show expression of Id1 and Id2 in the stroma and corneal endothelium, respectively. Fig. 1E and F show cytoplasmic and nuclear staining of Id1 and Id2 in entire cornea, respectively.
Fig. 1.
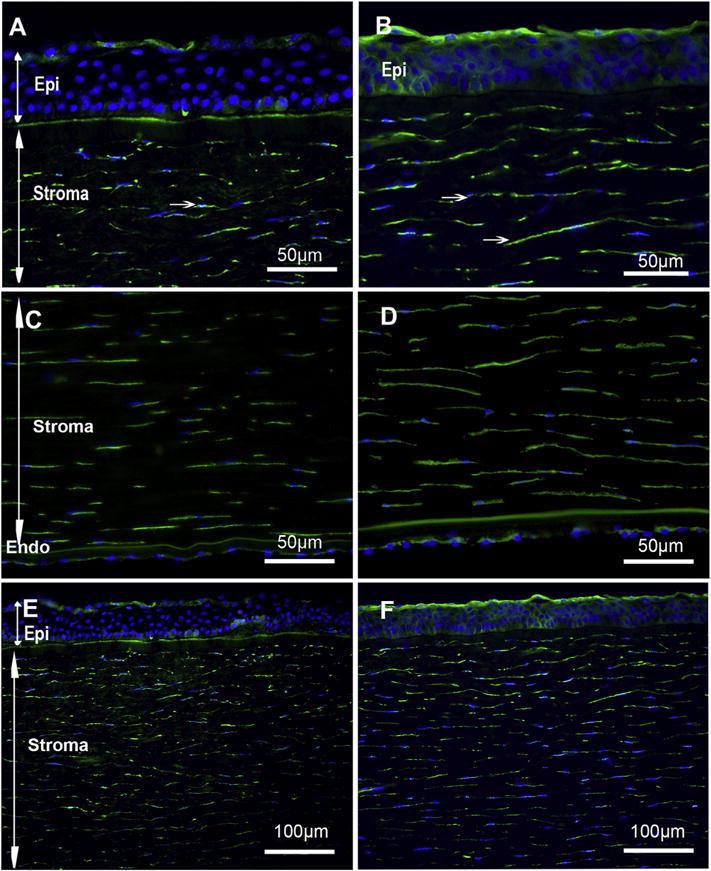
Id1 and Id2 are expressed in human cornea and localized in the epithelium, stromal keratocytes and endothelial layers: Immunofluorescence showing Id1 (A, C, E) and Id2 (B, D, F) expression in normal human corneal cross sections. Id1 and Id2 were detected in epithelial, stromal and endothelial cells of the cornea. Arrows indicate Id1 and Id2 positive keratocytes in the stroma. All panels used DAP1 (blue) for nuclear staining and green + cells show Id1 and Id2.
3.2. Primary human corneal fibroblasts (HCF) expressed all the members of mammalian Id genes
To evaluate the expression of Id genes in corneal fibroblasts, we obtained cDNA from two different batches of cultured primary human corneal fibroblasts (HCF) and analyzed by PCR with primers against the four known Id genes (Id1–Id4). Our results showed that all four Id genes were expressed in corneal fibroblasts (Fig. 2). β-actin was used as an internal control.
Fig. 2.
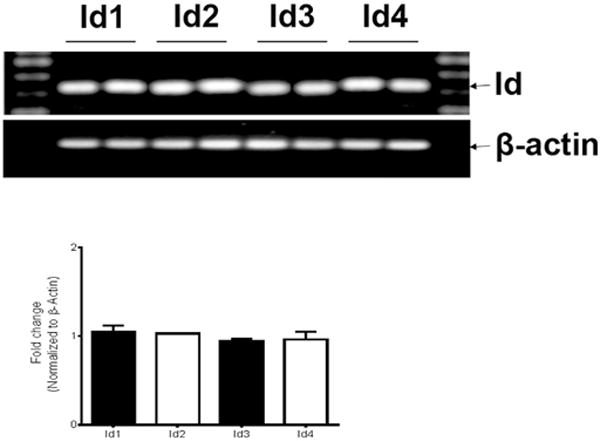
Primary human corneal fibroblasts (HCF) expressed all the members of mammalian Id genes: Representative image showing detection of Id1, Id2, Id3 and Id4 mRNA expression in human corneal fibroblasts with RT-PCR. β-actin was used as internal control. Average of each Id gene expression data by RT-PCR was plotted.
3.3. Id proteins are selectively expressed in corneal fibroblasts and not in myofibroblasts
To explore the role of Id proteins in the differentiation of fibroblast to myofibroblasts, corneal fibroblast cells were treated with TGFβ1 as described, and subjected to double immunostaining by α-SMA and Id1 or Id2 (Fig. 3). Both Id1 and Id2 panels showed expression in the cytoplasm and not co-stained with α-SMA staining. This suggests that differentiated fibroblasts (or myofibroblasts) do not express these Id proteins. Contrary to Id 1–4 mRNA expression in HCF, we were unable to detect Id3 and Id4 in HCF grown with or without TGFβ1 because of the unavailability of suitable antibodies against these proteins.
Fig. 3.
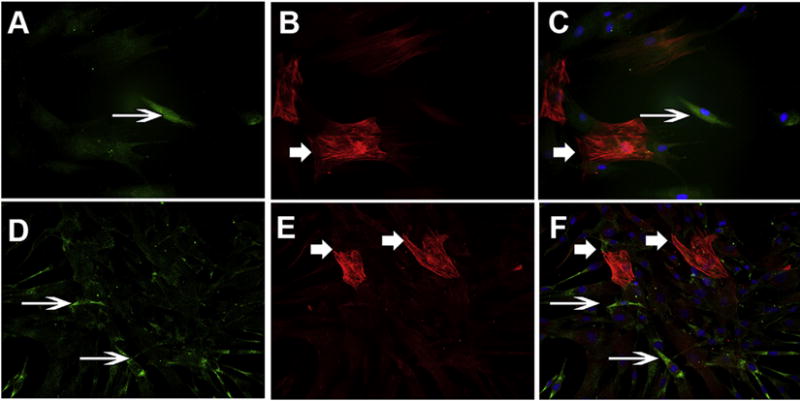
Id proteins are selectively expressed in corneal fibroblasts (thin arrow) and not in myofibroblasts (thick arrow): Human corneal fibroblasts were prepared growing in medium containing serum whereas myofibroblasts were obtained by culturing HCF in the presence of transforming growth factor beta 1 (TGFβ1 5 ng/ml) under serum-free condition. Cells were stained with Id1 (A), α-SMA (B), and Id1 + α-SMA (C) showing distinct staining for each other and no co-localization was observed. Similarly, Id2 (D), α-SMA (E), and Id2 + α-SMA (F) staining showed similar pattern of staining. All panels used DAP1 for nuclear staining (Id1 and Id2 = green, α-SMA = Red, DAPI = Blue).
3.4. Id genes are differentially expressed by TGFβ1 and BMP7 in a time-dependent manner
To assess changes in the Id 1–4 gene expression in response to TGFβ1 and BMP7 in the cornea, HCF cultures at 70% confluence were treated with TGFβ1 (Fig. 4) or BMP7 (Fig. 5) recombinant protein, and harvested at 2, 12, 24, or 48 h. The mRNAs from each time point were isolated, converted to cDNA, and analyzed via qRT-PCR.
Fig. 4.
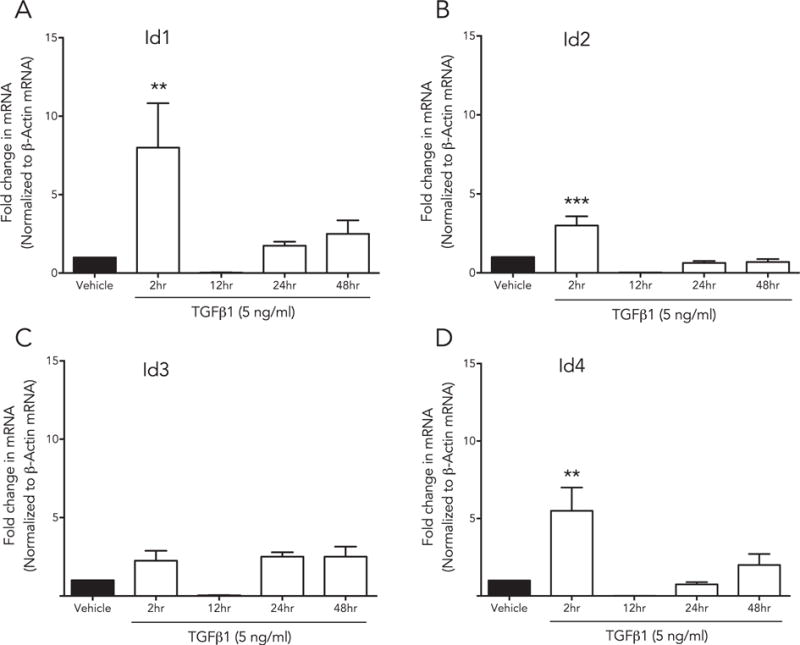
Id genes are differentially expressed by TGFβ in a time-dependent manner: HCF were treated with rhTGFβ1, mRNA was isolated at multiple time points (2 h, 12 h, 24 h and 48 h) and analyzed with qRT-PCR. Each gene [(A) Id1, (B) Id2, (C) Id3, (D) Id4] displayed distinct time-dependent changes in expression (Id1, p < 0.001; Id2, p < 0.01 and Id4, p < 0.001).
Fig. 5.
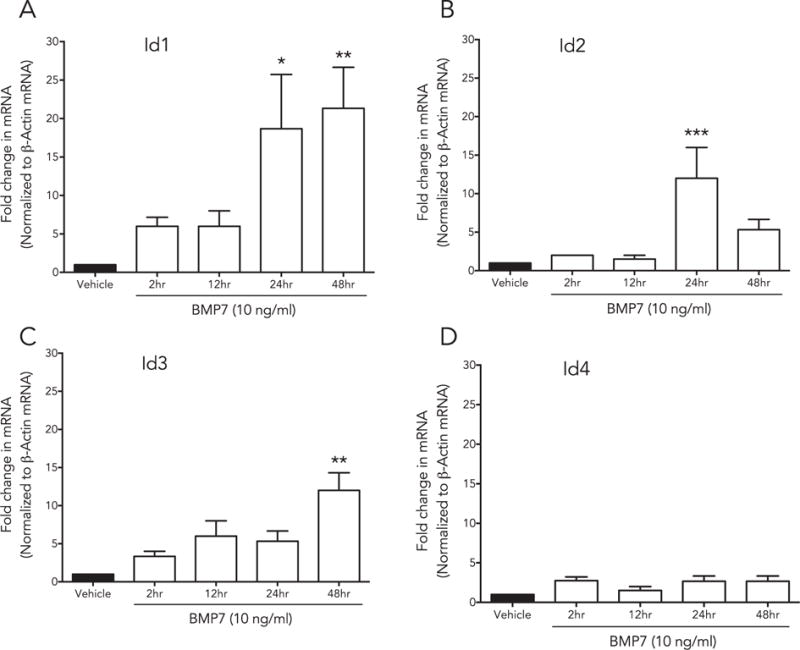
Id genes are differentially expressed by BMP7 in a time-dependent manner: HCF were treated with BMP7, and mRNA was isolated at multiple time points (2 h, 12 h, 24 h and 48 h), and analyzed with qRT-PCR. Each gene [(A) Id1, (B) Id2, (C) Id3, (D) Id4] displayed distinct time dependent changes in expression (24 h, p < 0.005; 48 h, p < 0.001), Id2 (24 h, p < 0.01) and Id3 48 h (p < 0.001).
Fig. 4 shows that Id genes 1–4 were differentially expressed by the HCF in a time dependent manner in response to TGFβ1 and BMP7 treatments. Shortly after TGFβ1 stimulation (2 h), Id 1–3 gene expression peaked significantly (Id1, p < 0.001; Id2, p < 0.01 and Id4, p < 0.001) followed by a decrease in mRNA expression at tested longer time points (12, 24, and 48 h). There was an increasing trend in Id3 with TGFβ1 treatment but was not significant in any other measured time points. On the other hand as evident from Fig. 5, BMP7 treatment showed a gradual increase in the expression of Id1 (24 h, p < 0.005; 48 h, p < 0.001), Id2 (24 h, p < 0.01) and Id3 48 h (p < 0.001) in a time-dependent manner. Id4 transcripts showed no changes with time after BMP7 treatment.
3.5. TGFβ1 and BMP7 regulates Id gene expression at different time points
Id’s were activated by both TGFβ1 and BMP7 in a time-dependent manner where TGFβ1 acts as early regulator and BMP7 stimulates at later stages suggesting that Id genes responds to growth factors differently based on their function. Next, we studied the effect of co-stimulation of with TGFβ1 and BMP7 treatments for 48 h (Fig. 6). We found a significant increase in the expression of Id1 (p < 0.01). There was an increasing trend in the expression of Id2, Id3 and Id4 but was not significant compared to control. This effect could be due to the late effect of BMP7 observed earlier (Fig. 5). Since TGFβ1 is known activator of α-SMA in corneal fibroblasts and BMP7 counteracts this increase we measured α-SMA transcript and as expected found no increase in its activity (Fig. 6).
Fig. 6.
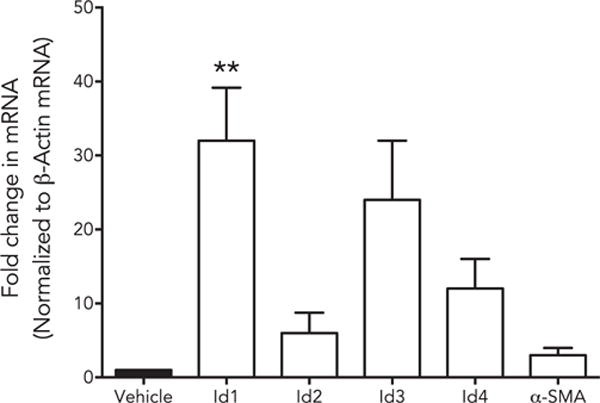
Id genes are differentially expressed by TGFβ and BMP7: Human corneal fibroblasts were treated with TGFβ1 + BMP7, mRNA was isolated, and subjected to qRT-PCR. TGFβ1 + BMP7 showed significant additive effect on Id1 expression (p < 0.01).
3.6. BMP7 stimulates Id protein expression in the presence of TGFβ1
Finally, we examined the Id1 and Id2 protein expression induced by BMP7 in the presence or absence of TGFβ1 stimulation (Fig. 7). Our results showed both Id1 and Id2 were up regulated in response to TGFβ1 and BMP7 treatments. In addition, co-stimulation of HCF significantly increased Id1 and Id2 proteins. On the contrary, TGFβ1 alone increased α-SMA expression and has minimal effects on α-SMA in presence of BMP7 alone or in combination with BMP7 treatments. These results suggest that Id1 and Id2 are dynamically regulated by TGFβ1 and BMP7 in HCF.
Fig. 7.
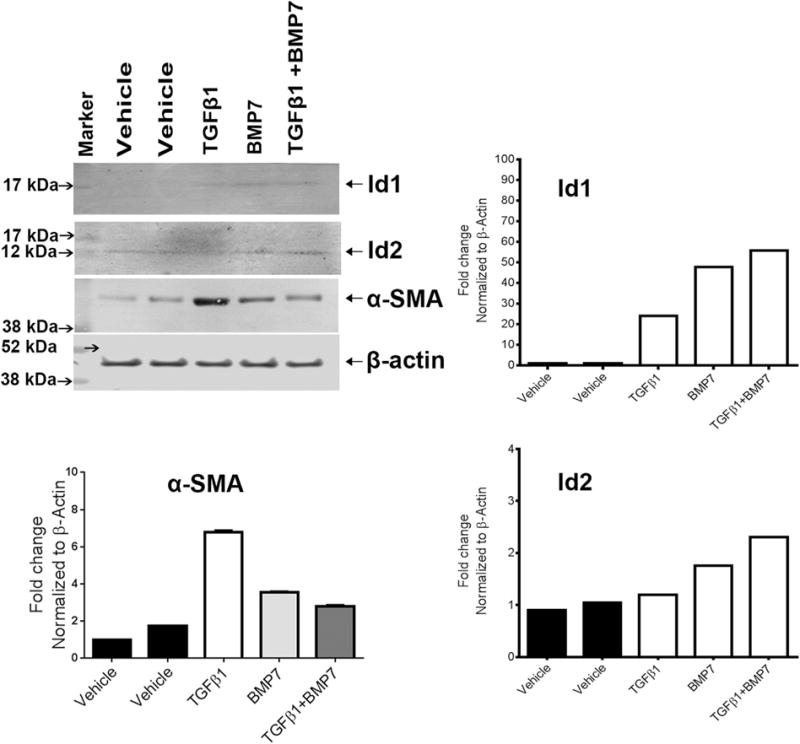
BMP7 stimulates Id protein expression in the presence of TGFβ1: TGFβ1 and BMP7 treatment increases Id1 and Id2 protein expression in HCF. HCF cells treated with TGFβ1, BMP7 and TGFβ1 + BMP7 were analyzed by western blot using anti-Id1, anti-Id2, α-SMA and anti-β-actin antibodies (vehicle denotes no treatment). Quantitation of protein expression data is also presented in this figure.
4. Discussion
Inhibitor of differentiation genes are helix-loop-helix proteins that bind to nuclear transcription factors and subsequently control gene expression by creating a nuclear environment permissive to cell growth and proliferation (Norton et al., 1998). Previous studies indicated that Id’s are actively involved in the BMP7 signaling pathway and have been implicated for their anti-fibrotic properties in multiple organ systems (Izumi et al., 2006). The role of Id genes in corneal homeostasis and pathology has not yet explored. In this study, we successfully demonstrated the mRNA and protein expression of Id genes in human cornea. Id1 and Id2 proteins are ubiquitously present in the corneal epithelial, stromal kereatocytes and endothelial cell layers. However, the staining pattern differed within the epithelial layer with Id2 being distributed throughout the epithelia whereas Id1 was localized in the basal and squamous cells. While Id expression studies in adult tissues are limited, it has been reported that Id expression levels are distinct, indicating that each Id was expressed at varying levels in a cell-type dependent manner (Hogg et al., 2010; Du and Yip, 2011). Furthermore, it is possible that each Id protein preferentially binds to distinct targets, thus having a specific function, which needs to be seen in the corneal cells.
During corneal wound healing, the activation of fibroblasts by TGFβ1 is accompanied by their transformation into α-smooth muscle actin (α-SMA) expressing contractile myofibroblasts (Serini and Gabbiani, 1999) that are thought to be responsible for extracellular matrix deposition and wound contraction. One of the remarkable findings in this study is the expression of Id1 and Id2 proteins in fibroblasts and not in myofibroblasts. It has been known that overexpression of Id proteins delay the onset of differentiation in muscle cells (Jen et al., 1992). Id genes are mostly expressed in undifferentiated, self-renewing populations, and are downregulated as cells differentiate and exits the cell cycle (Norton et al., 1998). This supports our findings in human cornea where Id1 and Id2 expression was limited to keratocytes and self-renewing epithelial cells.
BMP7 protein has been reported to have anti-fibrotic properties in many organ fibrosis systems including cornea by antagonizing TGFβ1 signaling (Tandon et al., 2013; Weiskirchen and Meurer, 2013). We tested the hypothesis that BMP7 and TGFβ1 signaling pathways regulate transdifferentiation of corneal fibroblast to myofibroblast via Id proteins. We found that HCF expressed all four known mammalian Id genes, Id1–Id4. TGFβ1 treatment to HCF showed an acute response in 2 h with the increase in Id1, Id2 and Id4 genes with progressive decrease within 48 h. Previous studies have shown that quiescent cells expressed low-to-undetectable levels of Id genes but following mitogenic stimulation to fibroblasts, Id expression was rapidly induced (within 2 h) as part of a cascade of ‘delayed’ early response genes. On the contrary, HCF treated with human recombinant BMP7 showed a time-dependent increase in the expression of Id1, Id2 and Id3 transcripts with highest levels observed in 24–48 h. Previous reports indicated that in vitro, BMP7 has been shown to inhibit (Izumi et al., 2006) and reverse (Zeisberg et al., 2003) TGFβ1-mediated myofibroblast activation. In addition, HCF treated with a combination of TGFβ1 + BMP7 treatment showed significant increase in Id1 accompanied by low levels of α-SMA expression suggesting anti-fibrotic effects of BMP7 in the presence of TGFβ1. These intriguing findings led to postulate that Id proteins act as a molecular switch, and determine the cell fate by regulating the timing from cell proliferation to differentiation during active wound healing process in the cornea. Our future studies will test this hypothesis.
In summary, this is the first report of Id proteins (Id1–Id4) in the human corneal tissues. Id1 and Id2 proteins are ubiquitously expressed by the corneal epithelial cells and stromal keratocytes in human cornea except Id1 which was localized in the basal epithelial cells. We also identified selective expression of Id1 and Id2 in undifferentiated corneal fibroblasts. In addition, Id1–Id4 genes were differentially expressed in a time-dependent manner in the presence of TGFβ1 or BMP7 treatments. We found that Id proteins, Id1 and Id2 are activated in the HCF with TGFβ1 treatment in the presence of BMP7 indicating their role in wound healing mechanism. Future studies involving elucidating the role of Id proteins in fibroblast activation or their de-differentiation into myofibroblasts may contribute to the development of therapeutic options for the modulation of corneal fibrosis and vision impairment.
Acknowledgments
This work was supported from 1I01BX000357–01 Veteran Health Affairs Merit grant (RRM), RO1EY017294 National Eye Institute grant (RRM) and Ruth M Kraeuchi Missouri Endowment of Ophthalmology, University of Missouri Columbia fund (RRM). Thanks are due to Prashant R. Sinha and Misha R. Brown for their technical help.
References
- DelMonte DW, Kim T. Anatomy and physiology of the cornea. J Cataract Refract Surgery. 2011;37:588–598. doi: 10.1016/j.jcrs.2010.12.037. [DOI] [PubMed] [Google Scholar]
- Du Y, Yip HK. The expression and roles of inhibitor of DNA binding helix-loop-helix proteins in the developing and adult mouse retina. Neuroscience. 2011;175:367–379. doi: 10.1016/j.neuroscience.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Gibson DJ, Pi L, Sriram S, Mao C, Petersen BE, Scott EW, Leask A, Schultz GS. Conditional knockout of CTGF affects corneal wound healing. Invest Ophthalmol Vis Sci. 2014;55:2062–2070. doi: 10.1167/iovs.13-12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg K, Etherington SL, Young JM, McNeilly AS, Duncan WC. Inhibitor of differentiation (Id) genes are expressed in the steroidogenic cells of the ovine ovary and are differentially regulated by members of the transforming growth factor-beta family. Endocrinology. 2010;151:1247–1256. doi: 10.1210/en.2009-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda E, Park AM, Yoshida K, Tabuchi M, Munakata H. Myofibroblasts: biochemical and proteomic approaches to fibrosis. Tohoku J Exp Med. 2013;230:67–73. doi: 10.1620/tjem.230.67. [DOI] [PubMed] [Google Scholar]
- Izumi N, Mizuguchi S, Inagaki Y, Saika S, Kawada N, Nakajima Y, Inoue K, Suehiro S, Friedman SL, Ikeda K. BMP7 opposes TGF-beta1-mediated collagen induction in mouse pulmonary myofibroblasts through Id2. Am J Physiol Lung Cell Mol Physiol. 2006;290:L120–L126. doi: 10.1152/ajplung.00171.2005. [DOI] [PubMed] [Google Scholar]
- Jen Y, Weintraub H, Benezra R. Overexpression of Id protein inhibits the muscle differentiation program: in vivo association of Id with E2A proteins. Genes Dev. 1992;6:1466–1479. doi: 10.1101/gad.6.8.1466. [DOI] [PubMed] [Google Scholar]
- Kaur H, Chaurasia SS, Agrawal V, Wilson SE. Expression of PDGF receptor-alpha in corneal myofibroblasts in situ. Exp Eye Res. 2009;89:432–434. doi: 10.1016/j.exer.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Iimuro Y, Otogawa K, Saika S, Inagaki Y, Nakajima Y, Kawada N, Fujimoto J, Friedman SL, Ikeda K. Adenovirus-mediated expression of BMP-7 suppresses the development of liver fibrosis in rats. Gut. 2007;56:706–714. doi: 10.1136/gut.2006.092460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Miyazawa K. Id: a target of BMP signaling. Sci STKE. 2002;151:pe40. doi: 10.1126/stke.2002.151.pe40. [DOI] [PubMed] [Google Scholar]
- Myrna KE, Pot SA, Murphy CJ. Meet the corneal myofibroblast: the role of myofibroblast transformation in corneal wound healing and pathology. Vet Ophthalmol. 2009;12(Suppl. 1):25–27. doi: 10.1111/j.1463-5224.2009.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y, Todokoro K. Activation of helix-loop-helix proteins Id1, Id2 and Id3 during neural differentiation. Biochem Biophys Res Commun. 1994;199:1355–1362. doi: 10.1006/bbrc.1994.1380. [DOI] [PubMed] [Google Scholar]
- Neuman K, Nornes HO, Neuman T. Helix-loop-helix transcription factors regulate Id2 gene promoter activity. FEBS Lett. 1995;374:279–283. doi: 10.1016/0014-5793(95)01128-2. [DOI] [PubMed] [Google Scholar]
- Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000;113:3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- Norton JD, Deed RW, Craggs G, Sablitzky F. Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol. 1998;8:58–65. [PubMed] [Google Scholar]
- Serini G, Gabbiani G. Mechanisms of myofibroblast activity and phenotypic modulation. Exp Cell Res. 1999;250:273–283. doi: 10.1006/excr.1999.4543. [DOI] [PubMed] [Google Scholar]
- Sharma A, Mehan MM, Sinha S, Cowden JW, Mohan RR. Trichostatin-A inhibits corneal haze in vitro and in vivo. Invest Ophthalmol Vis Sci. 2009;50:2695–2701. doi: 10.1167/iovs.08-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton WM, Chaurasia SS, Medeiros FW, Mohan RR, Sinha S, Wilson SE. Topical Interleukin-1 receptor antagonist inhibits inflammatory cell infiltration into the cornea. Exp Eye Res. 2008;86:753–757. doi: 10.1016/j.exer.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon A, Sharma A, Rodier JT, Klibanov AM, Rieger FG, Mohan RR. BMP7 gene transfer via gold nanoparticles into stroma inhibits corneal fibrosis in vivo. PLoS One. 2013;8:e66434. doi: 10.1371/journal.pone.0066434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon A, Tovey JC, Sharma A, Gupta R, Mohan RR. Role of transforming growth factor Beta in corneal function, biology and pathology. Curr Mol Med. 2010;10:565–578. doi: 10.2174/1566524011009060565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerasamy M, Phanish M, Dockrell ME. Smad mediated regulation of inhibitor of DNA binding 2 and its role in phenotypic maintenance of human renal proximal tubule epithelial cells. PLoS One. 2013;8:e51842. doi: 10.1371/journal.pone.0051842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wu X, Shi T, Lu L. Epidermal growth factor (EGF)-induced corneal epithelial wound healing through nuclear factor κB subtype-regulated CCCTC binding factor (CTCF) activation. J Biol Chem. 2013;288:24363–24371. doi: 10.1074/jbc.M113.458141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskirchen R, Meurer SK. BMP7 counteracting TGF-beta1 activities in organ fibrosis. Front Biosci. 2013;18:1407–1434. doi: 10.2741/4189. [DOI] [PubMed] [Google Scholar]
- You L, Kruse FE, Pohl J, Völcker HE. Bone morphogenetic proteins and growth and differentiation factors in the human cornea. Invest Ophthalmol Vis Sci. 1999;40:296–311. [PubMed] [Google Scholar]
- Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, Strutz F, Kalluri R. BMP7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]


