Abstract
Increased osteoclastogenesis is responsible for osteolysis that is a severe consequence of inflammatory diseases associated with bone destruction, such as rheumatoid arthritis (RA) and periodontitis. The mechanisms that limit osteoclastogenesis under inflammatory conditions are largely unknown. We previously identified transcription factor RBP-J as a key negative regulator that restrains TNF-α induced osteoclastogenesis and inflammatory bone resorption. In this study, we tested whether RBP-J suppresses inflammatory osteoclastogenesis by regulating expression of microRNAs (miRNAs) important for this process. Using high throughput sequencing of miRNAs (miRNA-seq), we obtained the first genome-wide profile of miRNA expression induced by TNF-α in mouse bone marrow derived macrophages (BMMs)/osteoclast precursors during inflammatory osteoclastogenesis. We furthermore identified miR-182 as a novel miRNA that promotes inflammatory osteoclastogenesis driven by TNF-α and whose expression is suppressed by RBP-J. Downregulation of miR-182 dramatically suppressed the enhanced osteoclastogenesis program induced by TNF-α in RBP-J-deficient cells. Complementary loss and gain of function approaches showed that miR-182 is a positive regulator of osteoclastogenic transcription factors NFATc1 and Blimp1. Moreover, we identified that direct miR-182 targets Foxo3 and Maml1 play important inhibitory roles in TNF-α mediated osteoclastogenesis. Thus, RBP-J-regulated miR-182 promotes TNF-α induced osteoclastogenesis via inhibition of Foxo3 and Maml1. Suppression of miR-182 by RBP-J serves as an important mechanism that restrains TNF-α induced osteoclastogenesis. Our results provide a novel miRNA mediated mechanism by which RBP-J inhibits osteoclastogenesis and suggest that targeting of the newly described RBP-J-miR-182-Foxo3/Maml1 axis may represent an effective therapeutic approach to suppress inflammatory osteoclastogenesis and bone resorption.
Introduction
Osteoclasts, multinucleated giant cells derived from the monocyte/macrophage lineage, are responsible for bone resorption. As the exclusive bone-degrading cells, osteoclasts play an indispensable role in physiological bone development, remodeling and repair. Osteoclastogenesis is physiologically triggered by RANKL in the presence of M-CSF and ITAM-mediated costimulation. Upon stimulation by these factors, a broad range of signaling cascades is activated, such as NF-κB pathways, protein tyrosine kinases and calcium signaling, and MAPK pathways. These signaling cascades lead to induction of the key transcription factor nuclear factor of activated T cells c1 (NFATc1) that functions in concert with other positive regulators, such as c-Fos and B lymphocyte-induced maturation protein-1 (Blimp1), to drive osteoclast differentiation (1–9). Recent evidence has made it clear that the process of osteoclast differentiation is also delicately controlled by a ‘braking system’, in which negative regulators such as interferon regulatory factor (Irf8), v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B (MafB) and B cell lymphoma (Bcl6) restrain the numbers of osteoclasts that are generated to prevent excessive bone resorption that leads to bone loss (10). Inflammation promotes osteoclastogenesis and thus osteoclasts also function as pathogenic cells leading to excessive bone resorption that is commonly associated with inflammatory bone diseases, such as rheumatoid arthritis (RA), periodontitis and peri-prosthetic osteolysis. The inflammatory cytokine tumor necrosis factor-α (TNF-α) plays a major role, mostly in synergy with RANKL, in promoting pathologic osteoclastogenesis and bone resorption in these inflammatory diseases (2, 9, 11, 12). Compared with RANKL, however, TNF-α alone does not effectively induce osteoclast differentiation. The mechanisms that restrain TNF-α-induced osteoclastogenesis are much less understood than those that promote osteoclasogenesis in response to RANKL (2, 13).
Recently, we have discovered that transcription factor RBP-J functions as a novel osteoclastogenic repressor and plays a critical role in inhibiting TNF-α-induced osteoclast differentiation and bone resorption (13). RBP-J functions as a central transcription factor that receives inputs from several signaling pathways, including the canonical Notch pathway, Wnt-β-catenin, and NF-κB pathways in a context dependent manner to regulate cell differentiation, survival and many other cellular responses and activities (13–19). Distinct from most negative regulators of osteoclast differentiation, a unique feature of RBP-J is that it plays a prominent and selective role in inhibiting TNF-α-induced osteoclastogenesis with minimal effects on RANKL-induced osteoclastogenesis (13). Recent genetic studies have revealed that RBPJ allelic polymorphisms are linked with disease susceptibility of RA (20–22). In parallel, RBP-J expression levels are lower in osteoclast precursors isolated from the synovial fluid of RA patients than healthy donors (19). These studies establish the critical role of RBP-J in restraining TNF-α-mediated inflammatory osteoclastogenesis and support a role of RBP-J in RA disease pathogenesis. Therefore, elucidation of the targets of RBP-J action and mechanisms of its function has the potential to identify novel therapeutic targets for treating excessive osteoclastogenesis and inflammatory bone erosion. The molecular mechanisms by which RBP-J limits TNF-α-induced osteoclast differentiation are not fully understood.
MicroRNAs (miRNAs) are a family of small evolutionarily conserved noncoding single stranded RNAs consisting of ~22 nucleotides that are derived from longer transcribed precursor transcripts. miRNAs repress gene expression by targeting specific mRNAs. They bind specific mRNAs via imperfect complementary binding but with a perfect base pairing between the miRNA “seed region” (nucleotides 2–7 of the miRNA) and the targeted sequences of mRNAs. miRNAs regulate gene expression at the posttranscriptional level by promoting degradation or inhibiting translation of specific target mRNAs, or a combination of both mechanisms. miRNAs account for about 3% of human genome but regulate about 90% of protein coding genes (23–29). The last decade of studies has demonstrated the importance of miRNAs in various biological and pathological settings. As potential therapeutic targets or biomarkers, miRNAs have been gaining much clinical attention, for example, in immunity, cancers, neurological diseases and metabolic disorders, in recent years (23, 28, 30–32). The investigation of the role of miRNAs in bone biology and diseases is just emerging. A few miRNAs have been reported to play important roles in RANKL-induced osteoclast differentiation (33–37). Nonetheless, it remains unclear how TNF-α mobilizes miRNA expression during inflammatory osteoclast differentiation and conversely, how miRNAs regulate TNF-α-induced osteoclastogenesis in an inflammatory setting.
Although miRNAs are involved in almost all major cellular functions, miRNA expression is highly specific to cell and tissue types as well as correlated with various stresses or disease settings (23). The expression patterns and roles of miRNAs involved in TNF-α stimulation are much underexplored so far and remain unknown in osteoclast biology. To address these questions and further explore molecular mechanisms by which RBP-J limits TNF-α-induced osteoclast differentiation, we took advantage of microRNA sequencing (miRNA-seq) technique and performed expression profile analysis of miRNAs in response to TNF-α stimulation during osteoclast differentiation using wild type control and RBP-J deficient bone marrow derived macrophages (BMMs). We have profiled the miRNAs specifically involved in TNF-α-induced osteoclast differentiation. Furthermore, we focused on microRNA-182 (miR-182), which is the most abundant miRNA induced by TNF-α and whose expression is repressed by RBP-J during osteoclast differentiation. The important function of miR-182 in cancer, cell growth and cell fate, and T lymphocyte expansion was just recently appreciated (38–46). In the present study, we identified miR-182 as a novel and critical positive regulator of inflammatory osteoclastogenesis mediated by TNF-α, with minimal effects on RANKL-induced osteoclastogenesis. Moreover, miR-182 is negatively controlled by RBP-J. We also identified two miR-182 targets Maml1 and Foxo3a as negative regulators of TNF-α-induced osteoclast differentiation in the present study. Therefore, our study uncovered a novel regulatory network, where miR-182 functions as an important new node that receives inputs from RBP-J and TNF-α signaling and positively regulates inflammatory osteoclastogenesis. Suppression of miR-182 by RBP-J serves as an important mechanism that restrains TNF-α induced osteoclastogenesis.
Materials and Methods
Mice
RbpjΔM/ΔM (Rbpjflox/floxLysMcre(+)) mice have been described previously (13). Age and gender matched mice with a LysMcre(+) genotype (hereafter referred to as wild type control) are used as controls. 8-week-old male C57/BL6 mice that were obtained from the Jackson Laboratory were used in Maml1 and Foxo3a knockdown experiments. All mouse experiments were approved by the Institutional Animal Care and Use Committee of the Hospital for Special Surgery.
Reagents
Murine TNF-α and sRANKL were purchased from Peprotech. mirVana® miRNA inhibitor mmu-miR-182-5p (Assay ID: MH13088 Cat # 4464084), mirVana™ miRNA Inhibitor, Inhibitor Negative Control #1 (Cat # 4464076), and mirVana™ miRNA mimic, Mimic Negative Control #1 (Cat # 4464058) were purchased from Life Technologies. SignalSilence® Foxo3a siRNA I (Mouse Specific) (Cat# 8620S) was purchased from Cell Signaling and Silencer® Negative Control No. 1 siRNA (AM4611, Life Technologies) was used as a control siRNA. SMARTpool: siGENOME Mouse Maml1 siRNA (Cat# M-059179-02–0005) and corresponding control siRNA were purchased from Dharmacon.
Cell culture
Murine bone marrow cells were harvested from tibiae and femora, and cultured overnight in Petri dishes (BD Biosciences) in α-MEM medium (Invitrogen) with 10% FBS (Invitrogen). Except where stated, CMG14-12 supernatant, which contained the equivalent of 20 ng/ml of recombinant M-CSF, was used as a source of M-CSF as described (13) in experiments. Non-adherent cells were then re-plated into tissue culture dishes and cultured in the same medium for 3 days to obtain bone marrow derived macrophages (BMMs), which are capable of differentiating to osteoclasts, and thus were used as osteoclast precursors. The attached BMMs were scraped, seeded at a density of 4.5 × 104/cm2 and cultured in α-MEM medium with 10% FBS and CMG14-12 supernatant for 1 day. Except where stated, the cells at this time point were used for the basal condition. The cells were then treated without or with TNF-α (40 ng/ml) or RANKL (40 ng/ml) for indicated times in the figure legends. Culture media were exchanged every three days. For osteoclast differentiation assay in 96-well plates, four replicate wells per condition were used. TRAP staining was performed with an acid phosphatase leukocyte diagnostic kit (Sigma-Aldrich) in accordance with the manufacturer’s instructions.
In vitro Inhibition and Overexpression of MicroRNAs and Gene Silencing by siRNAs
MicroRNA antagomirs and mimics were applied to silence and over-express microRNA expression, respectively. Antagomirs were used at concentrations of 40 nM and mimics were used at concentrations of 10 nM. siRNAs were used at concentrations as indicated in the figures. Antagomirs, mimics, siRNAs or their corresponding control oligos were transfected into murine BMMs using TransIT-TKO transfection reagent (Mirus) in accordance with the manufacturer’s instructions.
Cell proliferation and viability assays
BMMs were transfected with miR-182 mimics (10nM) or antagomirs (40nM) or corresponding controls for twenty-four hours. The cells were then re-plated and seeded at a density of 4.5 × 104/cm2 and cultured in α-MEM medium with 10% FBS and CMG14-12 supernatant for the indicated times shown in the figure legends. Four hours after re-seeding, the cells attached to the plate and this time point was regarded as time 0. Cell proliferation and viability were analyzed by CyQUANT Cell Proliferation Assay Kit (Invitrogen) or Cell Proliferation Kit I (MTT assay) (Roche) following the manufacturer’s instructions. Data analyzed by these two methods show similar results. Relative proliferation/viability levels of the cells at each time point in the MTT assay were calculated by the ratio of the absorbance [A550nm-A690nm] at each indicated time point to its corresponding time 0.
Pit formation assay
Pit formation assay was performed as previously described (13). BMMs were transfected with miR-182 mimics (10 nM), antagomirs (40 nM), Foxo3a siRNAs (80 nM), Maml1 siRNAs (80 nM) or corresponding controls for twenty-four hours. The cells were then re-plated on dentin slices (4 mm diam, 0.2 mm thick; 5 × 104 cells/slice) in 96-well culture plates with 200 µl culture medium/well. The cells were cultured for 10 days with TNF-α without or with RANKL priming in the presence of M-CSF with media exchanges every 3 days. The dentin slices were washed with water, and pits formed by mature osteoclasts on the dentin slices were stained with 1% toluidine blue O (Sigma-Aldrich). The total resorptive pit area on each slice was analyzed by Bioquant OsteoII software. The relative pit area (% of slice) was calculated by the ratio of total pit area relative to each dentin slice area.
Reverse transcription and real-time PCR
For quantification of microRNA, total RNA was isolated and the small RNA fraction was enriched with the miRVana miRNA Isolation Kit (Life Technologies) according to the manufacturer’s instructions. For quantitative RT-PCR analysis of miRNA, cDNA was prepared from total RNA with the TaqMan microRNA Reverse Transcription kit (Applied Biosystems). TaqMan MicroRNA assays were used according to the manufacturer's recommendations (Applied Biosystems) for real-time PCR. The TaqMan U6 snRNA assay (Applied Biosystems) was used for normalization of expression values.
For quantification of mRNA, reverse transcription and real-time PCR were performed as previously described (13). The primers for real-time PCR were as follows: Acp5: 5’-ACGGCTACTTGCGGTTTC-3’ and 5’-TCCTTGGGAGGCTGGTC-3’; Ctsk: 5’-AAGATATTGGTGGCTTTGG-3’ and 5’-ATCGCTGCGTCCCTCT-3’; Itgb3: 5’-CCGGGGGACTTAATGAGACCACTT-3’ and 5’-ACGCCCCAAATCCCACCCATACA-3’; Gapdh: 5’-ATCAAGAAGGTGGTGAAGCA-3’ and 5’-AGACAACCTGGTCCTCAGTGT-3’.
Immunoblot analysis
Total cell extracts were obtained using lysis buffer containing 150 mM Tris-HCl (pH 6.8), 6% SDS, 30% glycerol, and 0.03% Bromo Phenol Blue. 10% B-mercaptoethanol was added immediately before harvesting cells. Cell lysates were fractionated on 7.5% SDS-PAGE, transferred to Immobilon-P membranes (Millipore) and incubated with specific antibodies. Western Lightning plus-ECL (PerkinElmer) was used for detection. Densitometry was performed using ImageJ software (National Institutes of Health). NFATc1 antibody was from BD biosciences, Blimp1, GAPDH and p38α antibodies were from Santa Cruz Biotechnology Inc., Foxo3 antibody was from Cell Signaling and Maml1 antibody was from Bethyl Laboratories. p-Akt (serine 473) antibody was obtained from Cell Signaling.
miRNA sequencing (miR-seq)
Total RNA was isolated and the small RNA fraction was enriched with the miRVana miRNA Isolation Kit (Life Technologies) according to the manufacturer’s instructions. miRNA libraries were constructed per the Illumina TruSeq Small RNA Library preparation kit. High-throughput sequencing was performed using the Illumina HiSeq1500. miRNA-seq reads were aligned to the mouse miRNA sequences in the miRBase database (release 21) using miRDeep2. Mature miRNA values were normalized by library size (corresponding to counts per million mapped miRNA reads (CPM)). miRNAs with value smaller than 5 CPM in all conditions were cut off. miRNA-seq data (accession #GSE72966) have been deposited in NCBI’s Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72966).
Statistical analysis
A paired t-test was used to calculate significance between percent difference in number of TRAP-positive cells relative to control in at least three independent experiments. For qPCR experiments, the fold-changes in response to TNF-α were pooled and Student’s t-test was applied using a lognormal distribution. In the case of more than 2 groups of samples, 1-way ANOVA followed by Tukey’s post hoc test was used to calculate significance of differences between any groups of samples. Statistical analysis was carried out using GraphPad Prism. p value < 0.05 was taken as statistically significant; *p value < 0.05 and **p value < 0.01 and unless indicated, all data are presented as the mean ± SD.
Results
Profiling of miRNAs regulated by TNF-α and RBP-J during osteoclastogenesis
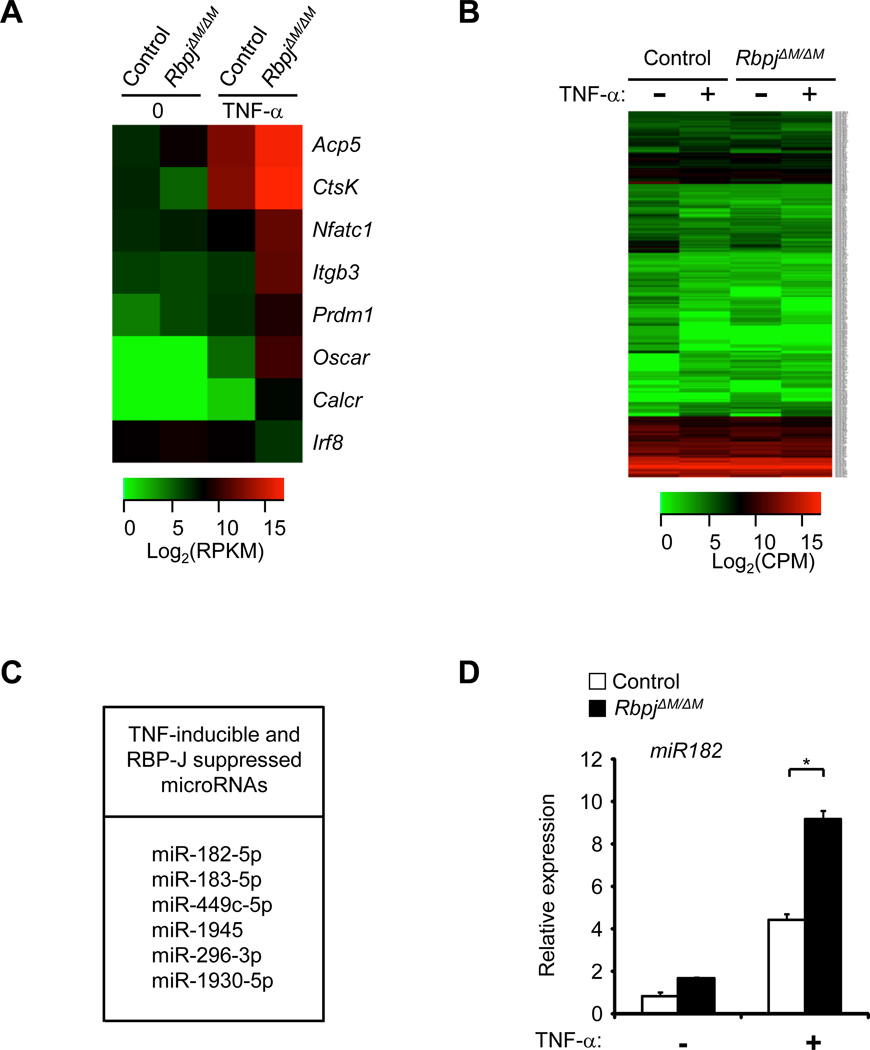
The mechanisms that prevent TNF-α from effectively inducing osteoclast differentiation have remained an enigma. As shown in Figure 1A (column 1 and 3 of a heat map depicting data derived from RNA-seq GSE53218) and literature (13), TNF-α alone has very weak capacity for inducing the expression of osteoclastogenic regulators, such as NFATc1 and Blimp1, and osteoclast marker genes, such as Calcitonin receptor, Oscar, Integrin β3, TRAP and Cathepsin K. In contrast, the induction of all of these positive osteoclastic genes was dramatically enhanced, while expression of negative regulator Irf8 was diminished, in RBP-J deficient cells (Fig. 1A; compare column 3 and 4). This is in alignment with a previous study that RBP-J deficiency enables TNF-α to effectively induce osteoclast differentiation (13). These data together with our previous findings (13) demonstrate that RBP-J is a key inhibitory regulator of inflammatory osteoclastogenesis and bone resorption induced by TNF-α; however, the underlying molecular mechanisms are not fully understood.
Figure 1. RBP-J deficiency enhances TNF-α-induced miR-182 expression in osteoclast precursors.
(A) RNA-seq-based expression heat map of osteoclast marker genes and transcription factors regulated by RBP-J deficiency. BMMs were stimulated without or with TNF-α (40 ng/ml) for 48 hours, and log2 values of RPKMs of a representative of two independent RNA-seq datasets (GSE53218) are shown in the heat map. (B) microRNA-seq-based expression heat map of genome-wide changes of microRNAs in control and RbpjΔM/ΔMBMMs by TNF-α (40ng/mL) after 24 hours of stimulation. Log2 values of counts per million reads (CPMs) of a representative of two independent microRNA-seq experiments are shown in the heat map. (C) Table displaying the 6 microRNAs induced by TNF-α but suppressed by RBP-J based on an overlap of two biological replicates of microRNA-seq experiments. (D) Quantitative real-time PCR analysis of miR-182 in control and RbpjΔM/ΔMmice with and without TNF-α stimulation for 72 hours. Data are representative of and statistical testing was performed on three independent experiments. *P<0.05.
We reasoned that RBP-J regulates the expression of target genes important in osteoclastogenesis, and wondered whether RBP-J could regulate expression of miRNAs that control TNF-α-induced osteoclastogenesis, a possibility that has not been previously considered. We first performed global profiling of miRNA expression using a genome wide approach, high throughput sequencing of microRNAs (microRNA-seq), to determine the miRNAs that are regulated by TNF-α and RBP-J in osteoclast precursors. Bone marrow-derived macrophages (BMMs) obtained from RbpjΔM/ΔM mice (RbpjΔM/ΔM refers to Rbpjflox/floxLysMcre(+), in which RBP-J was specifically deleted in myeloid macrophages) and wild type control mice (13) were used as osteoclast precursors. Surprisingly, TNF-α only significantly changed expression of a small number of miRNAs during osteoclast differentiation (refer to Materials and Methods for cutoff criteria) (Fig. 1B). By overlapping miRNA-seq data from two independent experiments, we found that TNF-α treatment for 24 hours only induced expression of 27 miRNAs and suppressed 12 miRNAs by at least 1.2 fold out of 1915 miRNAs that were expressed in osteoclast precursors (Supplementary Fig. 1). Thus, TNF-α regulated expression of only about 2% of miRNAs in BMMs/osteoclast precursors, indicating a limited and selective regulatory pattern of miRNA expression by long term exposure to TNF-α stimulation in this cell type. Furthermore, we wished to identify miRNAs that are induced by TNF-α but significantly suppressed by the negative regulator RBP-J during osteoclast differentiation. We hypothesized that this group of miRNAs suppressed by RBP-J would have important roles in promoting TNF-α induced osteoclast differentiation. We screened for microRNAs that were induced by at least 1.2 fold by TNF-α in both control and RBP-J deficient cells. From this list, we then searched for microRNAs that were expressed at least 1.2 fold greater in TNF-α stimulated RBP-J deficient cells than in control cells. After overlapping lists derived from two independent miRNA-seq datasets, we came up with a list of six microRNAs that fit our criteria for “TNF-α-induced and RBP-J suppressed” miRNAs (Fig. 1C). In this group, miRNA-182-5p stands out as being expressed the highest in TNF-α stimulated RBP-J deficient cells. We then validated our miRNA-seq data with quantitative real-time PCR (qPCR) (Fig. 1D) and found that TNF-α-induced miR-182 was significantly enhanced by RBP-J deficiency. In addition, in silico analysis of the promoter region of miR-182 revealed three potential RBP-J binding sites within 3kb upstream of its transcription start site (Supplementary Fig. 2A), suggesting that miR-182 is likely a direct target suppressed by RBP-J. The induction pattern of miR-182 expression by TNF-α in the absence of RBP-J is similar to that of the osteoclastogenic genes (Fig. 1A) and correlates with the dramatically enhanced osteoclastogenesis in RBP-J deficient cells (13), which led us to question whether miR-182 is a positive regulator of osteoclast differentiation. To test this hypothesis, we applied both a gain of function approach by overexpressing an miR-182 mimic and a loss of function approach by inhibiting miR-182 function with a specific antagomir.
Inhibition of miR-182 suppresses TNF-α-induced osteoclastogenesis in RBP-J-deficient cells
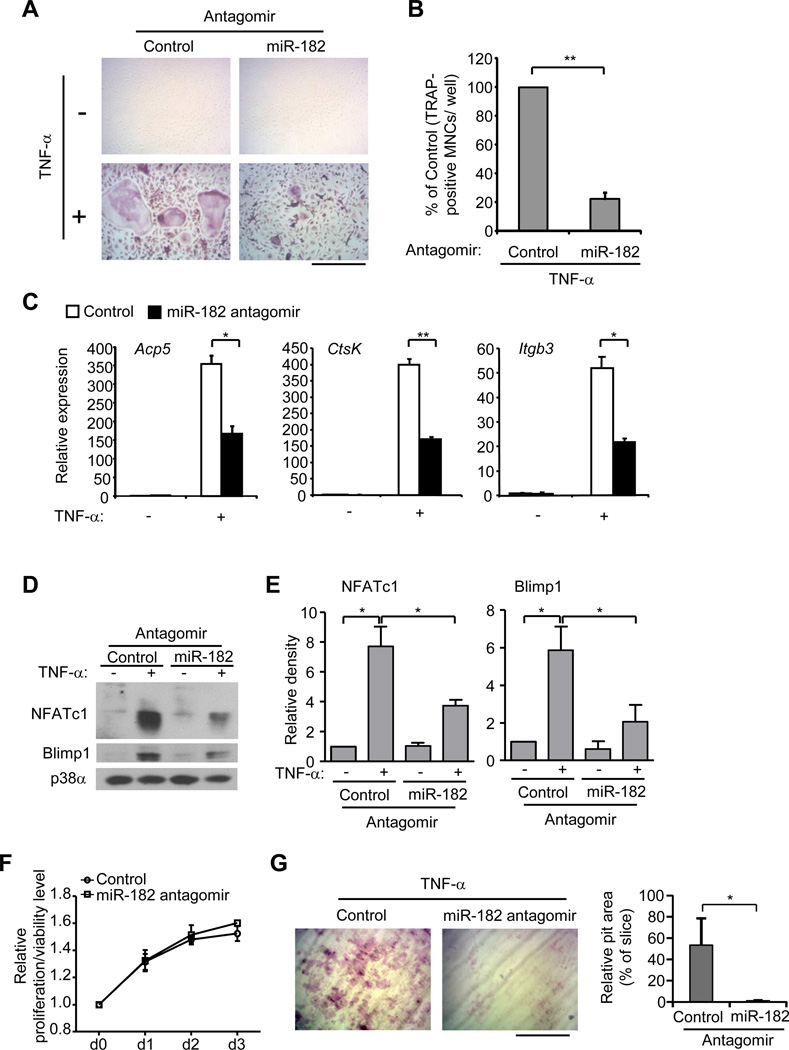
We first examined whether miR-182 contributed to the enhanced osteoclast differentiation in response to TNF-α that is caused by RBP-J deficiency. As miR-182 expression is increased by RBP-J deficiency, we took advantage of an miR-182 antagomir to suppress miR-182 expression. We confirmed the significant downregulation of miR-182 by the antagomir (Supplementary Fig. 2B). As shown in Fig. 2A, RbpjΔM/ΔM BMMs transfected with antagomir control formed large TRAP-positive multinucleated cells (MNCs) in response to TNF-α (Fig. 2A), as expected from our previous work (13). Inhibition of miR-182 by antagomir, however, significantly suppressed the number and size of TRAP-positive MNCs induced by TNF-α in RBP-J deficient cell cultures (Fig. 2A, B). In parallel to the decreased number and size of TRAP-positive MNCs, inhibition of miR-182 substantially suppressed the induction of osteoclast marker genes Acp5 (encoding TRAP), CtsK (encoding Cathepsin K), and Itgb3 (encoding β3) in RBP-J deficient cells relative to those transfected with control antagomir (Fig. 2C), indicating that increased expression of miR-182 significantly contributes to RBP-J deficiency-enhanced osteoclastogenesis. We furthermore investigated whether miR-182 affected the expression of regulators that control osteoclast differentiation. Our previous work showed that RBP-J inhibits induction of positive osteoclastogenic regulators and attenuates down-regulation of negative regulator Irf8. In the present study, we found that inhibition of miR-182 led to a dramatic decrease in the induction of both NFATc1 and Blimp1 in RBP-J-deficient cells (Fig. 2D, E). Down-regulation of Irf8 was not consistently or significantly affected by miR-182 (data not shown), suggesting that miR-182 is mainly involved in the RBP-J regulated NFATc1/Blimp1 axis. Inhibition of miR-182 by the antagomir did not affect cell proliferation/viability (Fig. 2F). Notably, in parallel to the suppression of osteoclast differentiation by miR-182 antagomirs, the resorptive pits generated by the RBP-J deficient osteoclasts on dentin slices in the presence of TNF-α were drastically inhibited by miR-182 antagomirs (Fig. 2G). Collectively, these results indicate that miR-182 plays an important role in enhancing TNF-α induced osteoclastogenesis in the RBP-J deficient cells. Our data also suggest that miR-182 as a downstream factor restrained by RBP-J is a positive regulator of TNF-α induced osteoclast differentiation.
Figure 2. Inhibition of miR-182 significantly suppresses TNF-α-induced osteoclastogenesis in RBP-J-deficient cells.
BMMs derived from RbpjΔM/ΔM mice were transfected with either control or miR-182 antagomir (40 nM) and were stimulated with TNF-α for 3 days. TRAP staining was performed (A) and the number of TRAP-positive MNCs (>= 3 nuclei per cell) per well relative to the control was calculated (B). Data in figure represent average ± SEM of three independent experiments and statistical significance was calculated by paired-t-test. **P<0.01. TRAP-positive cells appear red in the photographs. Bar, 200 µm. (C) Quantitative real-time PCR analysis of mRNA expression of Acp5 (encoding TRAP), Ctsk (encoding cathepsin K) and Itgb3 (encoding β3) in BMMs from RbpjΔM/ΔM mice transfected with control or miR-182 antagomir (40 nM) and stimulated with or without TNF-α for 3 days. Data are representative of and statistical analysis was performed on at least three independent experiments. *P<0.05. **P<0.01. (D) Immunoblot analysis of NFATc1 and Blimp1 expression in whole cell lysates obtained from RbpjΔM/ΔM BMMs transfected with either control or miR182 antagomir (40 nM) in the absence or presence of TNF-α for 3 days. p38α was measured as a loading control. (E) The relative density of the immunoblot bands of NFATc1 and Blimp1 vs. those of loading control p38α from three independent experiments were quantified by densitometry and normalized to the unstimulated control condition. *P<0.05. (F) Cell proliferation/viability at 0, d1, d2 and d3 with control or miR-182 antagomir (40 nM) transfection was examined by MTT assay (refer to Methods for details). (G) Toluidine blue-stained dentin resorption pits (left panel) formed by the osteoclasts derived from RbpjΔM/ΔM BMMs transfected with control or miR-182 antagomir (40 nM) in the presence of TNF-α (40 ng/ml) for 10 days. Bar, 200 µm. Relative pit area (% to slice) is shown in the right panel. Data are representative of and statistical analysis was performed on three independent experiments. *P<0.05.
Increased expression of miR-182 significantly enhances TNF-α-induced osteoclastogenesis
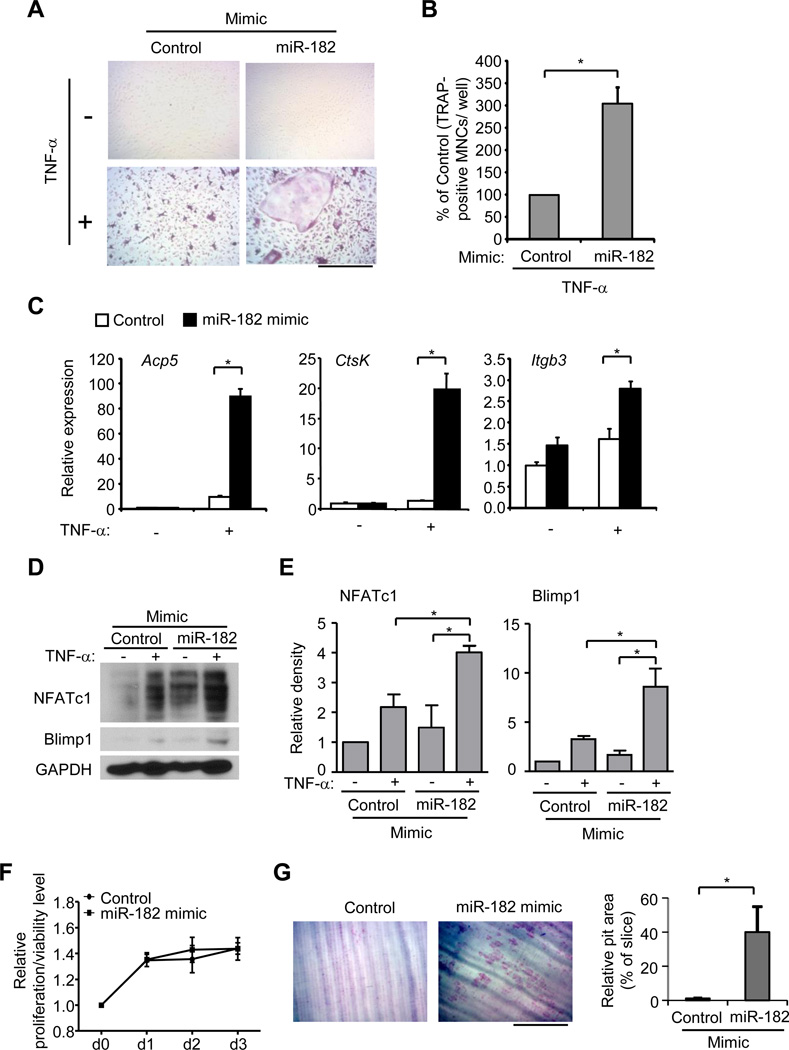
To test our hypothesis that miR-182 functions as a positive regulator in TNF-α mediated inflammatory osteoclastogenesis, we transfected wild type BMMs with an miR-182 mimic to increase miR-182 levels (Supplementary Fig. 2C). Consistent with literature, TNF-α stimulation alone is unable to effectively induce osteoclast differentiation in wild type cells (Fig. 3A, B). Strikingly, forced expression of miR-182 increased the number and size of TRAP-positive MNCs formed upon stimulation by TNF-α (Fig. 3A, B). In parallel, expression of osteoclast marker genes Acp5, CtsK, and Itgb3 was significantly upregulated in the wild type BMMs transfected with miR-182 mimic in response to TNF-α relative to cells transfected with the mimic control (Fig. 3C). Furthermore, we examined whether increasing expression of miR-182 could induce expression of key osteoclastogenic regulators Blimp1 and NFATc1 in wild type control cells upon stimulation with TNF-α alone. Indeed, both Blimp1 and NFATc1 levels were enhanced by the miR-182 mimic (Fig. 3D, E). The cell proliferation and viability were not affected by miR-182 mimic (Fig. 3F). The relative phosphorylation levels of Akt, known to be involved in cell survival pathway, were not significantly changed by miR-182 mimic or antagomir in response to TNF-α (Supplementary Fig. 2D). Importantly, consistent with the enhanced osteoclast differentiation, the increased expression of miR-182 in wild-type BMMs dramatically promoted osteoclastic resorption level on dentin slices (Fig. 3G), indicating that the osteoclasts derived from the miR-182 mimic-transfected precursors are functional and possess bone resorptive capability. These results indicate that increased expression of miR-182 significantly promotes TNF-α-induced osteoclast differentiation in wild type BMMs.
Figure 3. Forced expression of miR-182 significantly enhances TNF-α-induced osteoclastogenesis.
Wild type BMMs transfected with either control or miR-182 mimic (10 nM) were stimulated with TNF-α for 4 days. TRAP staining was performed (A) and the number of TRAP-positive MNCs (>= 3 nuclei per cell) per well relative to the control was calculated. Data in figure represent average ± SEM of three independent experiments and statistical significance was calculated by paired-t-test *P<0.05 (B). Bar, 200 µm. (C) Quantitative real-time PCR analysis of mRNA expression of Acp5 (encoding TRAP), Ctsk (encoding cathepsin K) and Itgb3 (encoding β3) in BMMs transfected with control or miR-182 mimic and stimulated with or without TNF-α for 3 days. Data are representative of and statistical testing was performed on at least three independent experiments. **P<0.01. (D) Immunoblot analysis of NFATc1 and Blimp1 expression in whole cell lysates obtained from BMMs transfected with control or miR-182 mimic (10 nM) with or without TNF-α for 3 days. GAPDH was measured as a loading control. (E) The relative density of the immunoblot bands of NFATc1 and Blimp1 vs. those of loading control GAPDH from three independent experiments were quantified by densitometry and normalized to the unstimulated control condition. *P<0.05. (F) Cell proliferation/viability at 0, d1, d2 and d3 with control or miR-182 mimic (10 nM) transfection was examined by MTT assay (refer to Methods for details). (G) Toluidine blue-stained dentin resorption pits (left panel) formed by the osteoclasts derived from wild type BMMs transfected with control or miR-182 mimic (10 nM) in the presence of TNF-α (40 ng/ml) for 10 days. Bar, 200 µm. Relative pit area (% to slice) is shown in the right panel. Data are representative of and statistical analysis was performed on three independent experiments. *P<0.05.
Given the positive role of miR-182 in TNF-α mediated inflammatory osteoclastogenesis, we reasoned whether miR-182 played a role in RANKL induced homeostatic osteoclastogenesis as well. miR-182 expression was gradually induced by TNF-α or RANKL (Supplementary Fig. 2E, F). Although RANKL induced miR-182 expression to an even greater level than TNF-α, neither forced expression nor inhibition of miR-182 consistently dramatically affected RANKL-induced in vitro osteoclast differentiation (Supplementary Fig. 2G). Thus, miR-182 appears to play a selective and prominent role in TNF-α-induced osteoclastogenesis. The preference of regulation of TNF-α signaling by miR-182 is similar to that by RBP-J, supporting the idea that RBP-J and miR-182 function in the same axis regulating osteoclast differentiation.
Foxo3 and Maml1 are miR-182 targets that inhibit TNF-α mediated osteoclastogenesis
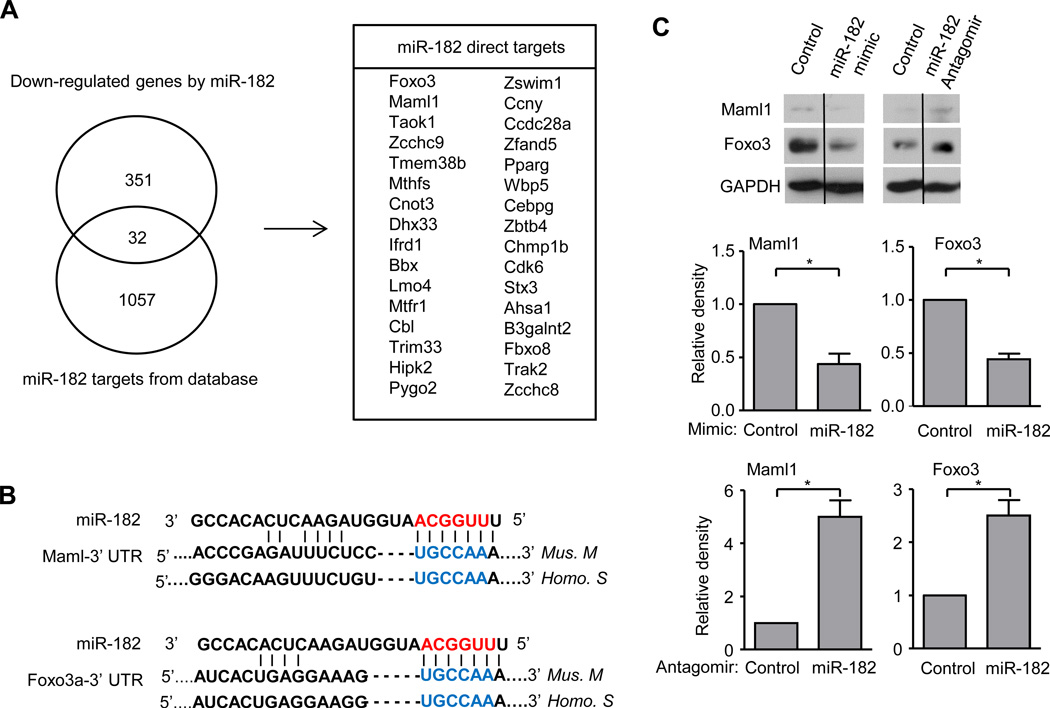
After highlighting the importance of miR-182 in the setting of inflammatory osteoclast differentiation driven by TNF-α, we set out to determine the direct targets of miR-182. We performed two parallel RNA-seq experiments, one with miR-182 gain of function approach using a miR-182 mimic and the other with miR-182 loss of function approach using an antagomir, to first identify genes regulated by miR-182 in osteoclast precursors. In general, miRNAs have modest effects on target mRNA expression, and our first level screen was for genes that were upregulated at least 1.1 fold by the miR-182 antagomir and suppressed at least 1.1 fold by the miR-182 mimic. With this approach we obtained a list of 383 genes, which contained miR-182 potential targets. Next, we took advantage of previous literature that has experimentally identified miR-182 targets by pull down assay of biotin labeled miR-182 (44). We then overlapped these miR-182 targets (44) with the genes regulated by miR-182 in osteoclast precursors obtained from our RNA-seq experiments, and found an overlap of 32 miR-182 direct targets (Fig. 4A). In this group of genes, Forkhead box class O 3a (Foxo3a encoding Foxo3), a member of the Foxo family of transcription factors, has been reported as a direct target of miR-182 in literature (45). Foxo proteins regulate RANKL-induced osteoclastogenesis (47, 48) and FOXO3 activity is associated with outcomes in infectious and inflammatory diseases including RA (49, 50), which suggest its possible relevance in TNF-α-driven osteoclastogenesis. We also selected Mastermind-like 1 (Maml1) as a gene of interest because of its role as a cotranscriptional regulator that is essential for RBP-J signaling (51), which suggests a potential function of Maml1 in osteoclast biology. The miR-182 seed region was identified in the 3’ untranslated region (UTR) of both Maml1 and Foxo3a genes and well conserved between mice and humans (Fig. 4B). To validate the effect of miR-182 on the protein expression of Maml1 and Foxo3, we transfected BMM cells with either miR-182 mimic or antagomir and their corresponding controls. Both Maml1 and Foxo3 expression levels were decreased in cells transfected with miR-182 mimic compared to control. In parallel, the expression of both Maml1 and Foxo3 increased in the cells transfected with miR-182 antagomir relative to its control (Fig. 4C). These results indicate that Maml1 and Foxo3 are miR182 targets in osteoclast precursors.
Figure 4. miR-182 directly targets Maml1 and Foxo3a.
(A) Venn diagram showing the overlap of the down-regulated genes by miR-182 and miR-182 targets obtained from a database (44) in the left panel. The overlapped 32 miR-182 targets in BMMs were shown in the right panel. miR-182 down-regulated genes were identified by overlapping genes that were upregulated by miR-182 antagomir and the genes that were downregulated by miR-182 mimic in BMMs. (B) Seed region (blue color) of miR-182 in the 3’ UTR of Maml1 and Foxo3a. (C) (Top) Immunoblot analysis of Maml1 and Foxo3a expression in whole cell lysates of BMMs transfected with miR-182 mimic or miR-182 antagomir with corresponding controls. GAPDH was measured as a loading control. Lanes separated by a thin black line indicate samples were run on the same gel but were non-contiguous. (Bottom) The relative density of the immunoblot bands of Maml1 and Foxo3a vs. those of loading control GAPDH from three independent experiments were quantified by densitometry and normalized to the control condition. *P<0.05.
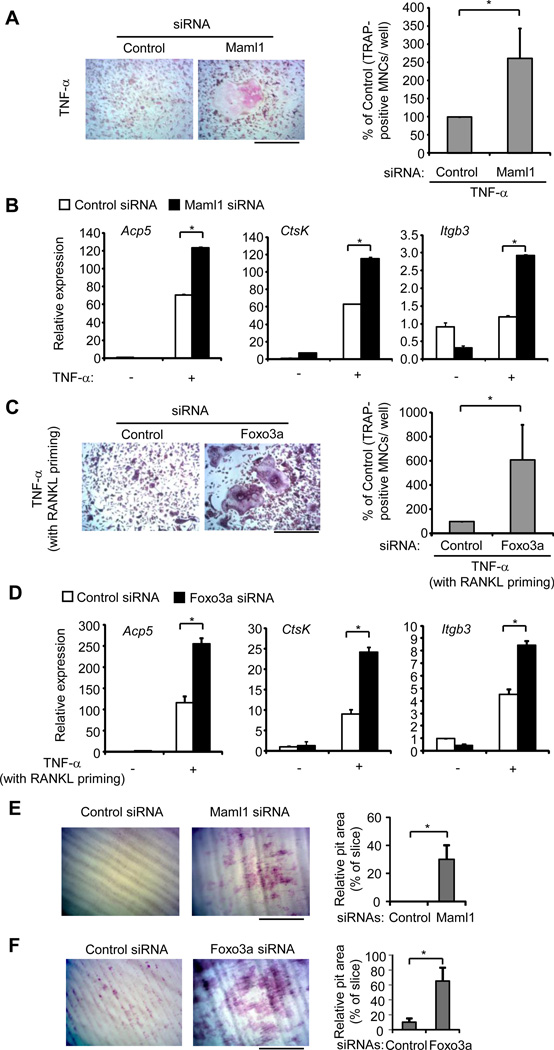
We then asked if Maml1 and Foxo3 are functionally important in TNF-α-mediated inflammatory osteoclastogenesis. To address this question, we knocked down Maml1 or Foxo3a in BMMs (Supplementary Fig. 3). Upon TNF-α stimulation, BMMs transfected with Maml1 siRNA formed a greater number of large TRAP-positive MNCs relative to the cells transfected with the control siRNA (Fig. 5A). Moreover, knockdown of Maml1 markedly enhanced the expression levels of osteoclast marker genes Acp5, CtsK, and Itgb3 during TNF-α induced osteoclastogenesis (Fig. 5B). Knockdown of Foxo3a was insufficient for TNF-α alone to induce osteoclastogenesis; however, with a brief RANKL priming, Foxo3a knockdown significantly promoted TNF-α induced osteoclast differentiation (Fig. 5C) and the expression of osteoclast marker genes Acp5, CtsK, and Itgb3 (Fig. 5D). Furthermore, knockdown of Maml1 or Foxo3a markedly enhanced resorptive pit formation induced by TNF-α (Fig. 5E, F). These results identified miR-182 targets Maml1 and Foxo3 as two novel negative regulators involved in TNF-α-mediated osteoclast differentiation. Our findings highlight that miR-182 functions as a key positive regulator in TNF-α-mediated inflammatory osteoclastogenesis through its inhibition of multiple targets that suppress osteoclastogenesis in inflammatory setting.
Figure 5. miR-182 targets Foxo3a and Maml1 inhibit TNF-α-mediated inflammatory osteoclastogenesis.
(A) BMMs transfected with either control or Maml1 siRNA (80 nM) were stimulated with TNF-α for 6 days. TRAP staining was performed (Left) and the number of TRAP-positive MNCs (>= 3 nuclei per cell) per well relative to the control was calculated. Data in figure represent average ± SEM of four independent experiments and statistical significance was calculated by paired-t-test *P<0.05 (Right). Bar, 200 µm. (B) Quantitative real-time PCR analysis of mRNA expression of Acp5 (encoding TRAP), Ctsk (encoding cathepsin K) and Itgb3 (encoding β3) in BMMs transfected with control or Maml1 siRNA (80nM) and stimulated with or without TNF-α for 3 days. Data are representative of and statistical testing was performed on three independent experiments. *P<0.05. (C) BMMs transfected with either control or Foxo3a siRNA (80 nM) were primed with RANKL (40 ng/mL) overnight followed by TNF-α (40 ng/mL) stimulation for 4 days. TRAP staining was performed (Left) and the number of TRAP-positive MNCs (>= 3 nuclei per cell) per well relative to the control was calculated. Data in figure represent average ± SEM of three independent experiments and statistical significance was calculated by paired-t-test *P<0.05 (Right). Bar, 200 µm. (D) Quantitative real-time PCR analysis of mRNA expression of Acp5 (encoding TRAP), Ctsk (encoding cathepsin K) and Itgb3 (encoding β3) in BMMs transfected with control or Foxo3a siRNA (80nM) and stimulated by RANKL priming and TNF-α for 3 days. Data are representative of and statistical testing was performed on three independent experiments. *P<0.05. (E, F) Toluidine blue-stained dentin resorption pits (left panel) formed by the osteoclasts derived from wild type BMMs transfected with control or Maml1 siRNA (80 nM, E) or Foxo3a siRNA (80 nM, F) in the presence of TNF-α (40 ng/ml) for 10 days without (E) or with RANKL (40 ng/ml) priming for overnight (F). Bar, 200 µm. Relative pit area (% to slice) is shown in the right panel. Data are representative of and statistical analysis was performed on three independent experiments. *P<0.05.
Discussion
miRNAs have emerged as important regulators of various biological processes and disease settings (23, 28, 30–32). Targeting miRNAs as a new treatment approach has shown therapeutic potential in several diseases. For example, a phase II clinical trial using anti-miRNA approach showed promising therapeutic efficacy in the treatment of HCV infection (52), highlighting the promising potential of targeting miRNAs in diseases. Although altered miRNA expression in synovial tissue and synovial fibroblasts in rheumatoid arthritis has been reported (53), the role of miRNAs involved in inflammatory diseases associated with bone destruction, including inflammatory osteoclastogenesis and osteolysis, is still underexplored. In the present study, we applied next generation deep sequencing to profile miRNAs (miRNA-seq) in an inflammatory setting of osteoclastogenesis driven by pro-inflammatory cytokine TNF-α. This miRNA-seq data provided the first genome wide dataset of miRNA profiling in TNF-α induced osteoclastogenesis. Our study showed that TNF-α regulated miRNAs in osteoclast precursors are quite different from the existing few datasets (54–56). The variation could be explained by different cell and tissue types as well as the doses of and exposure times to TNF-α. This underscores the context dependency of miRNA expression (23) and the importance of identifying miRNAs in different scenarios. We also used miRNA-seq to screen the miRNAs regulated by the key osteoclastogenic inhibitor RBP-J in order to fill a gap between RBP-J signaling and its regulation of miRNAs, which was unknown in the context of osteoclastogenesis. By overlapping these datasets, we identified miR-182 as a novel key osteoclastogenic miRNA that is negatively regulated by RBP-J and preferentially promotes TNF-α induced osteoclastogenesis using both loss and gain of miR-182 function approaches. Our data identified an important miRNA player miR-182 with novel osteoclastogenic function that adds a new link between TNF-α signaling and RBP-J signaling. Moreover, we demonstrated that two key targets of miR-182, Maml1 and Foxo3, are new negative regulators of inflammatory osteoclastogenesis. Overall, this newly described RBP-J-miR-182-Maml1/Foxo3 axis identified an miRNA-controlled regulatory pathway in inflammatory osteoclastogenesis, provided important mechanisms by which RBP-J inhibits inflammatory osteoclastogenesis and uncovered a crosstalk connected by an miRNA between TNF-α signaling and RBP-J signaling (Supplementary Fig. 4).
Investigation of osteoclast differentiation is focused on understanding the signaling and function of the key osteoclastogenic cytokine RANKL, which is nonredundant under physiological conditions. Predominant lines of investigation are based upon the hypothesis that RANKL elicits novel cellular signaling events and responses that are not induced by similar cytokines such as TNF-α. Our previous work and this study instead introduce the alternative and not mutually exclusive concept that cytokines such as TNF-α are subject to ‘brakes’ or feedback inhibitory mechanisms that limit their osteoclastogenic potential. A key driver of these negative regulatory mechanisms is the transcription factor RBP-J, and the current study shows that RBP-J ‘applies the brakes’ at least in part by regulating transcription of the gene encoding miR-182, which in turn regulates expression of Foxo3 and Maml1. These findings add a new regulatory circuit to negative regulation of osteoclastogenesis that is orchestrated by RBP-J. Interestingly, the miR-182-Foxo3/Maml axis is more effective in restraining the osteoclastogenic functions of the inflammatory cytokine TNF-α relative to RANKL. This finding suggests that targeting the miR-182-Foxo3/Maml1 axis may be preferentially effective in suppressing osteoclastogenesis at local inflammatory sites while exerting minimal effects on physiological bone remodeling (similar to what we observed with RBP-J, (13)). Our attempts to test this hypothesis in vivo have been limited to date by technical issues related to delivery of antagomirs to osteoclast precursors and difficulty in obtaining knock out mice because they harbor infections. Future work will be needed to test the role of the miR-182-Foxo3/Maml1 axis in pathological inflammatory osteoclastogenesis in vivo.
Recognition of the biological importance of miR-182 is just recently emerging. miR-182 has been shown to play key roles in cell differentiation, apoptosis, tumor development and immune responses (38–46). miR-182 is differentially expressed in various cell types in given conditions and tissues/organs, including skeletal system (Supplementary Fig. 2H, ref. 38–46, 57). A recent publication noted that miR-182 negatively regulates osteoblast differentiation (57), which brought attention to the role of miR-182 in bone biology. Foxo1 and Foxo3 are well-defined miR-182 targets in several biological settings (42, 45). We found that Foxo3 is also a key miR-182 target in TNF-α-mediated osteoclast differentiation. Despite different functions for different Foxo family members, Foxo1, 3, and 4 proteins have been reported to regulate RANKL induced osteoclast differentiation (47, 48). Our study is the first one investigating the role of Foxo3 in TNF-α mediated osteoclastogenesis. Interestingly, a brief RANKL priming period is needed to reveal the role of Foxo3 in TNF-α-induced osteoclast differentiation, suggesting that other negative regulator(s) presumably plays an important role in limiting the early phase of osteoclastogenesis induced by TNF-α. The other miR-182 target Maml1 is likely an inhibitory candidate at early phase as deletion of Maml1 was able to turn on osteoclast differentiation induced by TNF-α without a need of RANKL priming. Therefore, the two miR-182 targets Maml1 and Foxo3 likely work coordinately to restrain inflammatory osteoclastogenesis.
In the present study, we identified not only two key targets of miR-182 but also RBP-J as an important upstream inhibitory regulator of miR-182 in response to TNF-α. RBP-J has several well conserved potential binding sites in the upstream regulatory region of the gene encoding miR-182 precursor transcript, and RBP-J deficiency significantly increases miR-182 expression, indicating that miR-182 is a downstream target of RBP-J. Furthermore, our results show that miR-182 plays a key role in RBP-J mediated osteoclast differentiation by contributing to the regulation of NFATc1 and Blimp1. In addition to these positive osteoclastogenic regulators, we have found previously that RBP-J also suppresses negative regulator Irf8 (13). In this study, miR-182 seems to regulate osteoclast differentiation independently of Irf8 (Zhao B, unpublished data). Therefore, our data identified a key RBP-J regulated osteoclastogenic miRNA and also provided an important mechanism for RBP-J mediated inhibition of osteoclastogenesis. Interestingly, transcriptional coactivator Maml1 is also involved in Notch/RBP-J signaling mainly through interaction with RBP-J to mediate its transcriptional activity (51). Down-regulation of Maml1 by miR-182 may attenuate its cotranscriptional activity for RBP-J, which may contribute to RBP-J mediated suppression of osteoclastogenesis. Thus, miR-182 seems to be involved in a reciprocal regulatory circuit with RBP-J signaling in TNF-α-induced osteoclastogenesis and forms a new regulatory network, in which miR-182 is centered to receive an activating signal from TNF-α stimulation and an inhibitory signal from RBP-J as well as to suppress two downstream targets Foxo3 and Maml1 (Supplementary Fig. 4). Suppression of miR-182 by RBP-J serves as an important mechanism that restrains TNF-α induced osteoclastogenesis. Taken together of the genetic evidence that RBPJ and FOXO3 are closely associated with human inflammatory diseases (20–22, 49, 50), such as RA, our findings establishing a key role of miR-182 in TNF-α induced osteoclastogenesis and RBP-J mediated inhibition of osteoclastogenesis support the biological importance of the newly identified RBP-J-miR-182-Maml1/Foxo3 axis and suggest its therapeutic implications for inflammatory osteoclastogenesis and bone lysis.
Supplementary Material
Acknowledgments
We thank Drs. Baosen Jia, Kazuki Inoue and Shiaoching Gong for valuable discussion and technical support.
Abbreviations
- BMMs
bone marrow derived macrophages
- RANK
receptor activator for NF-κB
- RANKL
receptor activator for NF-κB ligand
- siRNA
small interfering RNA
- Blimp1
B lymphocyte-induced maturation protein-1
- Irf8
interferon regulatory factor
- MNCs
multinucleated cells
- TRAP
tartrate resistant acid phosphatase
- RA
rheumatoid arthritis
- miRNAs
microRNAs
Footnotes
This work was supported by NIH grants DE019420, AI044398, AR050401 (to L.I.) and AR062047 and AR068970 (to B.Z.).
Disclosures: The authors have no financial conflict of interest.
References
- 1.Asagiri M, Takayanagi H. The molecular understanding of osteoclast differentiation. Bone. 2007;40:251–264. doi: 10.1016/j.bone.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 2.Boyce BF, Xiu Y, Li J, Xing L, Yao Z. NF-kappaB-Mediated Regulation of Osteoclastogenesis. Endocrinology and metabolism. 2015;30:35–44. doi: 10.3803/EnM.2015.30.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce BF, Yao Z, Zhang Q, Guo R, Lu Y, Schwarz EM, Xing L. New roles for osteoclasts in bone. Ann N Y Acad Sci. 2007;1116:245–254. doi: 10.1196/annals.1402.084. [DOI] [PubMed] [Google Scholar]
- 4.Humphrey MB, Lanier LL, Nakamura MC. Role of ITAM-containing adapter proteins and their receptors in the immune system and bone. Immunol Rev. 2005;208:50–65. doi: 10.1111/j.0105-2896.2005.00325.x. [DOI] [PubMed] [Google Scholar]
- 5.Lorenzo J, Horowitz M, Choi Y. Osteoimmunology: interactions of the bone and immune system. Endocrine reviews. 2008;29:403–440. doi: 10.1210/er.2007-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakashima T, Takayanagi H. New regulation mechanisms of osteoclast differentiation. Ann N Y Acad Sci. 2011;1240:E13–E18. doi: 10.1111/j.1749-6632.2011.06373.x. [DOI] [PubMed] [Google Scholar]
- 7.Negishi-Koga T, Takayanagi H. Ca2+-NFATc1 signaling is an essential axis of osteoclast differentiation. Immunol Rev. 2009;231:241–256. doi: 10.1111/j.1600-065X.2009.00821.x. [DOI] [PubMed] [Google Scholar]
- 8.Xu F, Teitelbaum SL. Osteoclasts: New Insights. Bone research. 2013;1:11–26. doi: 10.4248/BR201301003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novack DV, Teitelbaum SL. The osteoclast: friend or foe? Annu Rev Pathol. 2008;3:457–484. doi: 10.1146/annurev.pathmechdis.3.121806.151431. [DOI] [PubMed] [Google Scholar]
- 10.Zhao B, Ivashkiv LB. Negative regulation of osteoclastogenesis and bone resorption by cytokines and transcriptional repressors. Arthritis Res Ther. 2011;13:234. doi: 10.1186/ar3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schett G, Gravallese E. Bone erosion in rheumatoid arthritis: mechanisms, diagnosis and treatment. Nat Rev Rheumatol. 2012;8:656–664. doi: 10.1038/nrrheum.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teitelbaum SL. Osteoclasts; culprits in inflammatory osteolysis. Arthritis Res Ther. 2006;8:201. doi: 10.1186/ar1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao B, Grimes SN, Li S, Hu X, Ivashkiv LB. TNF-induced osteoclastogenesis and inflammatory bone resorption are inhibited by transcription factor RBP-J. J Exp Med. 2012;209:319–334. doi: 10.1084/jem.20111566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu T, Kagawa T, Inoue T, Nonaka A, Takada S, Aburatani H, Taga T. Stabilized beta-catenin functions through TCF/LEF proteins and the Notch/RBP-Jkappa complex to promote proliferation and suppress differentiation of neural precursor cells. Mol Cell Biol. 2008;28:7427–7441. doi: 10.1128/MCB.01962-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izumiya Y, Izumiya C, Hsia D, Ellison TJ, Luciw PA, Kung HJ. NF-kappaB serves as a cellular sensor of Kaposi's sarcoma-associated herpesvirus latency and negatively regulates K-Rta by antagonizing the RBP-Jkappa coactivator. J Virol. 2009;83:4435–4446. doi: 10.1128/JVI.01999-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang Y, Smith S, Hu X. Role of Notch signaling in regulating innate immunity and inflmmation in health and disease. Protein Cell. 2016;7:159–174. doi: 10.1007/s13238-016-0250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engin F, Lee B. NOTCHing the bone: insights into multi-functionality. Bone. 2010;46:274–280. doi: 10.1016/j.bone.2009.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Miller CH, Giannopoulou E, Hu X, Ivashkiv LB, Zhao B. RBP-J imposes a requirement for ITAM-mediated costimulation of osteoclastogenesis. The Journal of clinical investigation. 2014;124:5057–5073. doi: 10.1172/JCI71882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FA, Zhernakova A, Hinks A, Guiducci C, Chen R, Alfredsson L, Amos CI, Ardlie KG, Barton A, Bowes J, Brouwer E, Burtt NP, Catanese JJ, Coblyn J, Coenen MJ, Costenbader KH, Criswell LA, Crusius JB, Cui J, de Bakker PI, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TW, Kastner DL, Ke X, Lee AT, Liu X, Martin P, Morgan AW, Padyukov L, Posthumus MD, Radstake TR, Reid DM, Seielstad M, Seldin MF, Shadick NA, Steer S, Tak PP, Thomson W, van der Helm-van Mil AH, van der Horst-Bruinsma IE, van der Schoot CE, van Riel PL, Weinblatt ME, Wilson AG, Wolbink GJ, Wordsworth BP, Wijmenga C, Karlson EW, Toes RE, de Vries N, Begovich AB, Worthington J, Siminovitch KA, Gregersen PK, Klareskog L, Plenge RM. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eyre S, Bowes J, Diogo D, Lee A, Barton A, Martin P, Zhernakova A, Stahl E, Viatte S, McAllister K, Amos CI, Padyukov L, Toes RE, Huizinga TW, Wijmenga C, Trynka G, Franke L, Westra HJ, Alfredsson L, Hu X, Sandor C, de Bakker PI, Davila S, Khor CC, Heng KK, Andrews R, Edkins S, Hunt SE, Langford C, Symmons D, Concannon P, Onengut-Gumuscu S, Rich SS, Deloukas P, Gonzalez-Gay MA, Rodriguez-Rodriguez L, Arlsetig L, Martin J, Rantapaa-Dahlqvist S, Plenge RM, Raychaudhuri S, Klareskog L, Gregersen PK, Worthington J. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44:1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orozco G, Viatte S, Bowes J, Martin P, Wilson AG, Morgan AW, Steer S, Wordsworth P, Hocking LJ, Barton A, Worthington J, Eyre S. Novel RA susceptibility locus at 22q12 identified in an extended UK genome wide association study. Arthritis Rheum. 2013 doi: 10.1002/art.38196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olive V, Minella AC, He L. Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Science signaling. 2015;8:re2. doi: 10.1126/scisignal.2005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews. Genetics. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 26.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nature structural & molecular biology. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 27.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh RP, Massachi I, Manickavel S, Singh S, Rao NP, Hasan S, Mc Curdy DK, Sharma S, Wong D, Hahn BH, Rehimi H. The role of miRNA in inflammation and autoimmunity. Autoimmunity reviews. 2013;12:1160–1165. doi: 10.1016/j.autrev.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nature reviews. Genetics. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 30.Srinivasan S, Selvan ST, Archunan G, Gulyas B, Padmanabhan P. MicroRNAs -the next generation therapeutic targets in human diseases. Theranostics. 2013;3:930–942. doi: 10.7150/thno.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends in molecular medicine. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Sonkoly E, Pivarcsi A. Advances in microRNAs: implications for immunity and inflammatory diseases. Journal of cellular and molecular medicine. 2009;13:24–38. doi: 10.1111/j.1582-4934.2008.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim KM, Lim SK. Role of miRNAs in bone and their potential as therapeutic targets. Current opinion in pharmacology. 2014;16:133–141. doi: 10.1016/j.coph.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T, Dawson BC, Munivez E, Tao J, Lee BH. miRNA-34c regulates Notch signaling during bone development. Human molecular genetics. 2012;21:2991–3000. doi: 10.1093/hmg/dds129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei J, Shi Y, Zheng L, Zhou B, Inose H, Wang J, Guo XE, Grosschedl R, Karsenty G. miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. The Journal of cell biology. 2012;197:509–521. doi: 10.1083/jcb.201201057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G, Sood AK, Mendell JT, Wan Y. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014;512:431–435. doi: 10.1038/nature13375. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Nakasa T, Shibuya H, Nagata Y, Niimoto T, Ochi M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. 2011;63:1582–1590. doi: 10.1002/art.30321. [DOI] [PubMed] [Google Scholar]
- 38.Sachdeva M, Mito JK, Lee CL, Zhang M, Li Z, Dodd RD, Cason D, Luo L, Ma Y, Van Mater D, Gladdy R, Lev DC, Cardona DM, Kirsch DG. MicroRNA-182 drives metastasis of primary sarcomas by targeting multiple genes. The Journal of clinical investigation. 2014;124:4305–4319. doi: 10.1172/JCI77116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song L, Liu L, Wu Z, Li Y, Ying Z, Lin C, Wu J, Hu B, Cheng SY, Li M, Li J. TGF-beta induces miR-182 to sustain NF-kappaB activation in glioma subsets. The Journal of clinical investigation. 2012;122:3563–3578. doi: 10.1172/JCI62339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li XL, Hara T, Choi Y, Subramanian M, Francis P, Bilke S, Walker RL, Pineda M, Zhu Y, Yang Y, Luo J, Wakefield LM, Brabletz T, Park BH, Sharma S, Chowdhury D, Meltzer PS, Lal A. A p21-ZEB1 complex inhibits epithelial-mesenchymal transition through the microRNA 183-96-182 cluster. Mol Cell Biol. 2014;34:533–550. doi: 10.1128/MCB.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu B, Qian T, Wang Y, Zhou S, Ding G, Ding F, Gu X. miR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury. Nucleic acids research. 2012;40:10356–10365. doi: 10.1093/nar/gks750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, Panse I, Riedel R, Flossdorf M, Dong J, Fuhrmann F, Heinz GA, Fang Z, Li N, Bissels U, Hatam F, Jahn A, Hammoud B, Matz M, Schulze FM, Baumgrass R, Bosio A, Mollenkopf HJ, Grun J, Thiel A, Chen W, Hofer T, Loddenkemper C, Lohning M, Chang HD, Rajewsky N, Radbruch A, Mashreghi MF. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nature immunology. 2010;11:1057–1062. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 43.Kouri FM, Hurley LA, Daniel WL, Day ES, Hua Y, Hao L, Peng CY, Merkel TJ, Queisser MA, Ritner C, Zhang H, James CD, Sznajder JI, Chin L, Giljohann DA, Kessler JA, Peter ME, Mirkin CA, Stegh AH. miR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes & development. 2015;29:732–745. doi: 10.1101/gad.257394.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishnan K, Steptoe AL, Martin HC, Wani S, Nones K, Waddell N, Mariasegaram M, Simpson PT, Lakhani SR, Gabrielli B, Vlassov A, Cloonan N, Grimmond SM. MicroRNA-182-5p targets a network of genes involved in DNA repair. Rna. 2013;19:230–242. doi: 10.1261/rna.034926.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei R, Tang J, Zhuang X, Deng R, Li G, Yu J, Liang Y, Xiao J, Wang HY, Yang Q, Hu G. Suppression of MIM by microRNA-182 activates RhoA and promotes breast cancer metastasis. Oncogene. 2014;33:1287–1296. doi: 10.1038/onc.2013.65. [DOI] [PubMed] [Google Scholar]
- 47.Bartell SM, Kim HN, Ambrogini E, Han L, Iyer S, Serra Ucer S, Rabinovitch P, Jilka RL, Weinstein RS, Zhao H, O'Brien CA, Manolagas SC, Almeida M. FoxO proteins restrain osteoclastogenesis and bone resorption by attenuating H2O2 accumulation. Nature communications. 2014;5:3773. doi: 10.1038/ncomms4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Dong G, Jeon HH, Elazizi M, La LB, Hameedaldeen A, Xiao E, Tian C, Alsadun S, Choi Y, Graves DT. FOXO1 mediates RANKL-induced osteoclast formation and activity. Journal of immunology. 2015;194:2878–2887. doi: 10.4049/jimmunol.1402211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JC, Espeli M, Anderson CA, Linterman MA, Pocock JM, Williams NJ, Roberts R, Viatte S, Fu B, Peshu N, Hien TT, Phu NH, Wesley E, Edwards C, Ahmad T, Mansfield JC, Gearry R, Dunstan S, Williams TN, Barton A, Vinuesa CG, Consortium UIG, Parkes M, Lyons PA, Smith KG. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell. 2013;155:57–69. doi: 10.1016/j.cell.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gregersen PK, Manjarrez-Orduno N. FOXO in the hole: leveraging GWAS for outcome and function. Cell. 2013;155:11–12. doi: 10.1016/j.cell.2013.08.050. [DOI] [PubMed] [Google Scholar]
- 51.Kovall RA. More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene. 2008;27:5099–5109. doi: 10.1038/onc.2008.223. [DOI] [PubMed] [Google Scholar]
- 52.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV infection by targeting microRNA. The New England journal of medicine. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 53.Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, Detmar M, Gay S, Kyburz D. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 54.Meyer SU, Thirion C, Polesskaya A, Bauersachs S, Kaiser S, Krause S, Pfaffl MW. TNF-alpha and IGF1 modify the microRNA signature in skeletal muscle cell differentiation. Cell communication and signaling : CCS. 2015;13:4. doi: 10.1186/s12964-015-0083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruan W, Xu JM, Li SB, Yuan LQ, Dai RP. Effects of down-regulation of microRNA-23a on TNF-alpha-induced endothelial cell apoptosis through caspase-dependent pathways. Cardiovascular research. 2012;93:623–632. doi: 10.1093/cvr/cvr290. [DOI] [PubMed] [Google Scholar]
- 56.Dong J, Cui X, Jiang Z, Sun J. MicroRNA-23a modulates tumor necrosis factor-alpha-induced osteoblasts apoptosis by directly targeting Fas. Journal of cellular biochemistry. 2013;114:2738–2745. doi: 10.1002/jcb.24622. [DOI] [PubMed] [Google Scholar]
- 57.Kim KM, Park SJ, Jung SH, Kim EJ, Jogeswar G, Ajita J, Rhee Y, Kim CH, Lim SK. miR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2012;27:1669–1679. doi: 10.1002/jbmr.1604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.