Abstract
Markov models are ubiquitously used to represent the function of single ion channels. However, solving the inverse problem to construct a Markov model of single channel dynamics from bilayer or patch-clamp recordings remains challenging, particularly for channels involving complex gating processes. Methods for solving the inverse problem are generally based on data from voltage clamp measurements. Here, we describe an alternative approach to this problem based on measurements of voltage traces. The voltage traces define probability density functions of the functional states of an ion channel. These probability density functions can also be computed by solving a deterministic system of partial differential equations. The inversion is based on tuning the rates of the Markov models used in the deterministic system of partial differential equations such that the solution mimics the properties of the probability density function gathered from (pseudo) experimental data as well as possible. The optimization is done by defining a cost function to measure the difference between the deterministic solution and the solution based on experimental data. By evoking the properties of this function, it is possible to infer whether the rates of the Markov model are identifiable by our method. We present applications to Markov model well known from the literature.
1 Introduction
Numerous mechanisms in cells are fueled by the energy contained in concentration gradients that exist across the cell membrane. One such process is the electrical signaling that results from ionic fluxes carried by specialized membrane proteins, collectively termed ion channels. Cell membranes are densely populated by ion channels, which fluctuate between different states in a stochastic manner that depends on the charge difference across the membrane (the membrane potential). These fluctuations between the various states are commonly represented by continuous-time Markov models; see e.g. [1, 2, 3, 4]. Markov models have been successfully used for half a century and offer great flexibility for precisely describing the functional states of a channel. Given the structure and rates of the Markov model, it is straightforward to use simulations to study channel behavior. But the inverse problem, which seeks to infer the structure and rates of the Markov model from single-channel recordings, is much more challenging and remains a field of active research; see e.g. [5, 6, 7, 8, 9].
Single channel recording was first demonstrated in the seminal work by Sakmann and Neher; see [10, 11]. Many of the principles used to deduce the form and rates of Markov models based on single channel data were developed by Colquhoun and Hawkes beginning almost forty years ago; see [12, 13], and are classically summarized in chapter 18 of [14]. More recently, Qin, Auerbach and Sachs [15, 16] developed maximum likelihood (MLE) approaches for defining the most probable hidden Markov model for a given dataset. These algorithms form the core of the open source QuB software package, which is available to the community, and contains tools for automated Markov model construction and parameterization; see Nicolai and Sachs [17]. Furthermore, Markov chain Monte Carlo (MCMC) fitting has been used to solve the inversion problem for models of intracellular calcium channels, which poses the same mathematical problem as the gating of voltage-dependent channels, see [18, 19, 20, 6, 7, 8, 9]. While both the MLE and MCMC approaches have been used to good effect, each has specific shortcomings [7], and a robust method for determining hidden Markov model structure from single-channel data remains a priority for the field.
The problem of identifiability of a model based on recordings of conducting and non-conducting states is of great importance and has been addressed by many authors. Characterizations of identifiable models and non-identifiable models have been provided by e.g. Fredkin and Rice [21] and more recently by Siekmann et al. [7] and Hines et al. [8]. These results, however, address the classical problem based on voltage clamped data and the results are therefore not automatically applicable to the case under consideration in the present paper.
Prior efforts to address the problem of identifying the rates of Markov models have been based on conventional experimental procedures for recording single-channel activity. That is, by fitting the binary changes in transmembrane current associated with channel opening and closure under conditions where transmembrane potential is clamped by the experimenter. The purpose of the present paper is to describe a fundamentally different approach, which involves an analysis procedure for defining a hidden Markov model from measurements of time-varying transmembrane potential in current clamped single ion channels. To the best of our knowledge, this is the first attempt to identify Markov models based on such voltage traces of single channel data. A similar approach has previously been applied for identification of Markov models from voltage recordings made in whole-cells; see e.g. Milescu et al. [22]. Following the tradition in the field, we present the method using simulated data. It turns out that the methodology described here also enable us to address the question of identifiability. We demonstrate use of the method to compute rates of a Markov model and we will show to determine wether the computed rates are unique.
2 Methods
Our aim is to devise an alternative method for inverting Markov models. The inversion will be based on time-traces of the transmembrane potential of single ion-channels. The traces used in our analysis will be pseudo-experimental data generated by a dynamical model formulated in terms of an ordinary differential equation including a stochastic term. The stochastic term is governed by a Markov model with rate-functions expressing the probability of going from one state to another state. The inversion problem is to determine these rate-functions based on observations of recordings of the time-traces of the transmembrane potential.
By running numerous Monte Carlo simulations using the stochastic differential equation, we can compute probability density functions by gathering the simulation results in histograms. The same probability density functions can be computed by solving a deterministic system of partial differential equations, see e.g. Smith [2], Bressloff [3] or [4]. In this report we will use the simulation results based on the stochastic differential equations as pseudo-experimental data in terms of time-dependent voltage traces represented by histograms. The method of inversion is to adjust the parameters of the system of deterministic partial differential equation so that the solution of this system is as close as possible to the histograms representing pseudo-experimental data. The adjustment will be performed in term of minimizing a cost-function.
The method is described for Markov models of the potassium channel, but the method is quite general and can, in principle, be applied to Markov models of any ion channel.
2.1 Markov models
Let us first consider a Markov model on its simplest possible form;
| (1) |
Here C and O denote the closed and open states, respectively. The rates α and β represent the probability of leaving a state in the sense that, for a small time-step Δt, we have
and
respectively. The problem of inversion for this simple model is to find the rates α and β such that the Markov model represents the behavior of the model as good as possible. It is well known that the probabilities o = o(t) and c = c(t) of being in the open (O) or closed state (C) evolve according to the following system of ordinary differential equations (see e.g. Keener and Sneyd [1]),
Since the sum of the probabilities add up to one, we can reduce this system to a single equation that is easily solved. Experimental data on probability of being in the open or closed state can thus be used to find determine the rates α and β.
In principle this approach is straightforward to generalize to much more complex Markov models involving many states. Generally, the system of ordinary differential equations governing the vector v of probabilities of occupying different states is given by
Here the matrix A contains the rates of the Markov model and the problem is to compute these rates. For a matrix with constant coefficients, the solution of this problem can, in principle, always be found on the form
where p0 is the vector of probabilities at time t = 0. In principle, this formula can be used to deduce the rates of the Markov model, but severe difficulties arise due to inherent instabilities in the inversion process. This approach to inversion is based on experimental data with clamped transmembrane potential. Our method is based on data where the transmembrane potential is recorded.
2.2 Stochastic model of the transmembrane potential
For a single channel, with the initial condition v(0) = v0, the dynamics of the transmembrane potential v̄ can be modeled as follows (see e.g. [3, 2, 4]),
| (2) |
where C is the specific capacitance of the preparation, vK, vS are the Nernst potentials for potassium and for the seal current, respectively, γ is a stochastic variable depending on the state of the channel, and I0 = I0(t) is an applied current; note also that the bar on v̄ is used to indicate that this is a stochastic variable. The dynamics of γ are governed by a Markov model, and in the forward simulations, the rates of the Markov model are assumed to be given. Additionally, the expression for seal current is used to simulate under as realistic clamp conditions as possible, where a proportion of any applied current will be lost to ground through a conductance that exists in parallel to the membrane patch. Intuitively, this conductance represents the current leak between the pipette wall and the membrane, which allows charge to escape directly to ground without traveling through a channel or being directed towards the membrane capacitance. This leak is a major constraint for single channel recordings of all types.
In most computations, we used the following parameters:
In addition the applied current will be specified in each simulation, and it will be in the interval 1000 to 3000 fA. If other parameters are used, it will be stated in the text.
2.3 Voltage clamp and current clamp recordings
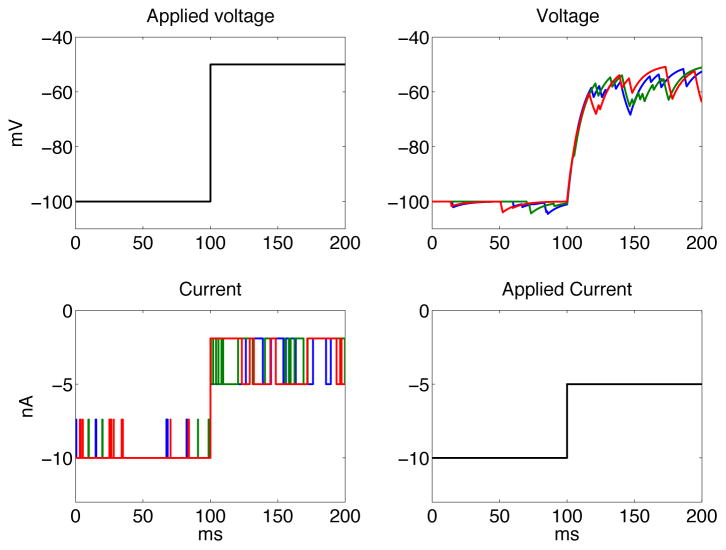
Both voltage and current clamp recordings are based on the same fundamental recording arrangement in which a portion of cell membrane (and included channel or channels) is contained between two electrodes, and as such are elements in a circuit that can be made between the two electrodes. Feedback control can then be used to prescribe either the current passing between the two electrodes (current clamp), or the potential difference between them (voltage clamp). In the case of voltage clamp, the experimenter prescribes a voltage waveform and the feedback amplifier then applies the current required to maintain that control. Because single ion channels open and close in a binary and stochastic fashion the current required to maintain the prescribed voltage reflects those opening and closing events as sharp switch-like changes (Figure 1 Left). In current clamp, the feedback control is modified to prescribe a current waveform and the voltage between the two electrodes (and therefore across the membrane patch) is measured simultaneously. Because the gating of the membrane channels is subject to this voltage, channel opening and closing will occur and result in subsequent changes to measured voltage (as shown at Right in Figure 1). It has become conventional for single channel recordings to be conducted in voltage clamp mode because this provides a direct measurement of single channel current at the prescribed voltage, which is intuitive to experimenters and analysts alike. However, because channel transitions are stochastic and the rates at which individual channels transition between their closed, open, and other functional states (e.g. inactivated) are voltage-dependent, this approach also requires many sweeps at different test voltages to characterize the voltage-dependencies of these transitions. In principle, appropriately designed current clamp recordings should allow these constraints to be partially circumvented because voltage varies continuously with channel opening and closing (thus eliminating discontinuities implicit to choosing discrete test voltages). However, at this time we are not aware of any data that have been collected for single channels in this recording mode. In part it is likely that this is due to technical aspects in making the recordings - specifically the combination of patch capacitance, single channel current, and recording noise are likely to apply significant constraints. Additionally the analytic framework for interpreting such recordings is poorly defined, and this is a significant disincentive for attempting challenging experiments of this type. With this paper we seek to develop that framework. In Figure 1 the two protocols are illustrated.
Figure 1.
Left: Voltage clamped at −100mV and −50mV. Right: Current clamped at −10nA and −5nA. Three realisations were performed for each of the two protocols (color coded).
2.4 Probability density functions for the voltage-gated channel
By running the stochastic model (2) many times, we can summarize the results in probability density functions for the open and closed states. These probability densities can also be computed directly by solving a deterministic system of partial differential equations; see e.g. [3, 2, 4]. The probability of the channel being in the open state or closed state for voltages between v and v + Δv is given, respectively, by
and
where ρo are ρc are the probability density functions of the open and the closed states. The probability density functions are governed by the following deterministic system
| (3) |
where
| (4) |
| (5) |
For a derivation of this system, see e.g. [2, 4]. Numerical methods for solving this system is discussed in detail in [4].
Boundary conditions of this model are set up such that probability do not leak across the boundary. Suppose we solve the system on the interval Ω = [v1, v2], then we require that the flux is zero at both v = v1 and v = v2. Here v1 and v2 define the upper and lower limits of the transmembrane potential governed by the model (2).
2.5 Bounds on the solutions
In order to find the domain Ω = [v1, v2] where we want to solve the system (3) we need to derive bounds on the solution of the model (2). When the channel is open, the model reads
| (6) |
and when the channel is closed (non-conducting) (γ = 0), the model reads
| (7) |
Therefore, the solutions will either converge towards (conducting channel)
| (8) |
or towards (non-conducting channel)
| (9) |
where we have used vS = 0. Furthermore, if the initial condition v0 is in the interval bounded by vo and vn, the solution will always stay in this interval; this holds for any Markov model governing γ. Therefore, by adjusting the value I0 of the applied current and the initial condition v0, we can force the solution of the model (2) to cover the interval of interest. It is useful to note that
which is negative for all values of I0 of interest and therefore v1 = vo and v2 = vn.
In order to invert the measured data to find the coefficients of the Markov model, it is important to have data covering the values of the transmembrane potential that is of interest in the Markov model. This can be achieved by adjusting the applied current I0, the initial condition v0, or the equilibrium potential vK.
2.6 Inversion based on the output least squares method
Suppose we have pseudo-experimental data generated by repeated runs of the model (2). These data can be combined in order to define the probability density functions for the open state and for the non-conducting states. In the special case of a 2x2 Markov model (see (1)) there is only one non-conducting state (the closed state C), but for more complex Markov models there may be many conducting or non-conducting states.
Real experimental data can only distinguish between conducting (open) states and non-conducting states (closed or inactivated states), and therefore we group the pseudo experimental data into two distributions; ρ̄o = ρ̄o(v, t) and ρ̄n = ρ̄n(v, t) represent the open and non-conductive distributions respectively. We assume that the data (ρ̄o, ρ̄n) are given. The problem is now to adjust the rates r = (α, β) of the system (3) such that the solution of the system satisfies (ρo, ρn) ≈ (ρ̄o, ρ̄n). We do this by minimizing the cost function
| (10) |
Here ρ = ρ(r) = (ρo(r), ρn(r)) and . The norm || · || will be defined below.
The crux of our method is to compute the parameters r by minimizing the cost function J = J(r) given by (10). We will show detailed examples below how this is done and how it works, but first we will characterize what we mean by an optimal solution.
2.7 Characterization of the optimal solution
In general, it is not at all clear that the rates of a Markov model can be computed based on the observations of wether the channel is in the conducting or non- conduction mode. As pointed out in e.g. [7], the situation is often complicated and the problem of deciding wether the rates can be identified is topic for ongoing research. Siekmann et al. [7], use the MCMC method to detect non-identifiable models. We will use a completely different approach but our results coincides with their observations. In order to characterize invertible and non-invertible cases, we need to define invertibility in this context. Roughly speaking, we state that a model is invertible by our method if we can find a unique minimum of the cost function (10).
From classical Calculus (see e.g. Apostol [23]), we may get an indication about the optimality of a given candidate solution r* by invoking the properties of the function J in the vicinity of r*. By Talyor’s theorem, we have
| (11) |
for any vector r and small number ε. Here H is the Hessian matrix of the function J. Clearly, H is symmetric, and assume in addition that it is positive definite. Then, for any vector y, we have
where λ0 > 0 is the smallest eigenvalue of the Hessian matrix H, and | · | is the usual Euclidian norm. Suppose that r* is a stationary point of J, i.e. ∇J(r*) = 0, then, for any non-zero vector r and small non-zero value ε, we have
Since, for all small and non-zero values of ε and any non-zero vector r we have J(r* + εr) > J(r*), the parameters given by r* define a local minimum of the function J. We also note that the smallest eigenvalue is a measure for how well-defined the local minimum is. If the smallest eigenvalue is much larger than zero, any step r away from the minimum will increase the value of the function J considerably. Contrary, when the value of λ0 is very small, the value of J varies little around r* and a well defined minimum is therefore hard to find. In the case of λ0 = 0, we know that there is an associate (non-zero) eigenvector r0 such that H(r*)r0 = 0 and thus
So given the local minimum r*, we can add εr0 for a small value of ε, and the value of J is more or less unchanged; the local minimum r* is not unique. It is also straightforward to see that if the smallest eigenvalue λ0 is negative (then the matrix is not positive definite) at a stationary point r*, that parameter vector is not a local minimum since
Thus, if r* is changed in the direction of the eigenvector r0 associated the negative eigenvalue λ0, the value of the function J is reduced and therefore r* is not a local minimum.
2.8 Definition of identifiability using our method
Based on the consideration above, we are in position to discuss what we mean by identifiability of a Markov model. We will refer to a model as identifiable by our method if the model satisfies three criterions:
Stationary point. If we use the Fminsearch algorithm (a Matlab function based on the Nelder-Mead algorithm) to find an optimal parameter vector r*, this parameter vector must be a stationary point of the cost function J = J(r) in the sense that |∇J(r*)| ≈ 0.
Local minimum. The candidate optimal solution r* must be a local minimum in the sense that all eigenvalues of the Hessian matrix H evaluated at r* are strictly positive.
Global minimum. If the Fminsearch algorithm is run with different initial conditions and result in different local minima r, a candidate optimal solution r* must have a value of the cost function J that is significantly lower than all the other candidates.
This is a pragmatic definition of identifiability which is useful in practical computations. However, the computational procedure outlined here may still fail; it may identify a minimum that is not really the global minimum, and it may indicate that the solution is unique even if it is not. This is the unsatisfying state of matters due to the fact that minimization of complicated non-linear functions is very challenging.
2.9 Defining the norm
In the procedure introduced above, it remains to define the norm used in (10) to measure the difference between the pseudo-experimental data ρ̄ and the probability density functions ρ computed by solving the system (3). The issue here is to compare a numerical solution defined on a uniform mesh representing the solution of a system of partial differential equations of the form (3) with pseudo-experimental data based on repeated solves of stochastic differential equation (2). We do the comparison by introducing an adaptive mesh that reflects the number of samples in the pseudo-experimental data in a given interval, and then we integrate difference between the experimental data and the numerical solutions over the interval.
For m > 0 we define the vector x = [x0, x1, . . . , xm] to represent the boundaries of m bins. The bins are adjusted such that each bin covers approximately the same number of samples in the stochastic simulations. We use about 50 bins to cover the interval [v1, v2], and we impose a lower bound (0.5 mV) on the bin width. Within the bin bounded by xi–1 and xi, the probability density defined by the pseudo-experimental data is given by ρ̄i. We define one set of bins given by xo for the open state, and one set of bins for the non-conducting state given by xn. The norm measuring the difference between the pseudo-experimental data ρ̄ and the probability density functions ρ computed by solving the system (3) can now be defined by
| (12) |
where
and
Here ρ = (ρo(v; r), ρn(v; r)) denotes probability density functions defined by numerical solution of a system of the form (3) for a given rate vector r. The integration of these solutions are performed by integrating a piecewise linear function defined by the numerical solution.
2.10 Computations
The pseudo-experimental data used for inversion data was generated by solving the stochastic equation (2) 100 times for 100 seconds using Δt = 1/1000 ms giving 10 billion samples. In the optimization, the first 10% of each run is discarded in order to obtain steady state behaviour. Two different applied currents are applied; I0 = 1000fA and I0 = 3000fA. For every inversion, 1000 initial guesses were evaluated and the ten best cases (lowest value of J) were used as start vectors for Fminsearch (Matlab). The best of the ten solutions provided by Fminsearch was defined to be the optimal solution. All computations were performed on a standard desktop computer. The PDE problems typically took a few seconds to solve, and the overall optimisation in the order of hours.
The eigenvalues of the Hessian matrix are computed using singular value decomposition (the svd function in Matlab). The Hessian matrix is computed using numerical differentiation of sixth order [24].
3 Results
We will show results of inversion using the method described above for a series of Markov models. The inversion is based on the fact the probability density functions of the open and the non-conducting states can be obtained by pseudo-experimental data and by solving a system of partial differential equations. The parameters defining the Markov model are adjusted in order for the solution of the system of partial differential equations to be as similar to the pseudo-experimental data as possible. We will start by showing that the distribution computed by repeated runs of the stochastic model (2) (Monte Carlo simulations) and the solution of the deterministic system of partial differential equations converge as the mesh parameters are refined, and the number runs of the stochastic model is increased. This result is well known, see e.g. [2, 3, 4].
For ease of reference, we will call the solutions computed by solving the stochastic model (2) Monte Carlo simulations (MC), and the solutions of the associated deterministic system of partial differential equations (PDE) will be called the PDE-solution.
3.1 Comparison of the MC and the PDE solutions
We consider the Markov model
| (13) |
Here the rates (ms−1) are specified on the form
with α1 = −1.0, α2 = 0.02, b1 = −4.5, b2 = −0.05 and I0 = 3000fA. The PDE model associated this Markov model is given by
| (14) |
where ρo, ρc1 and ρc1 denote the probability density function of the states O, C1 and C2, respectively, and the functions αo and αn are given by (4) and (5).
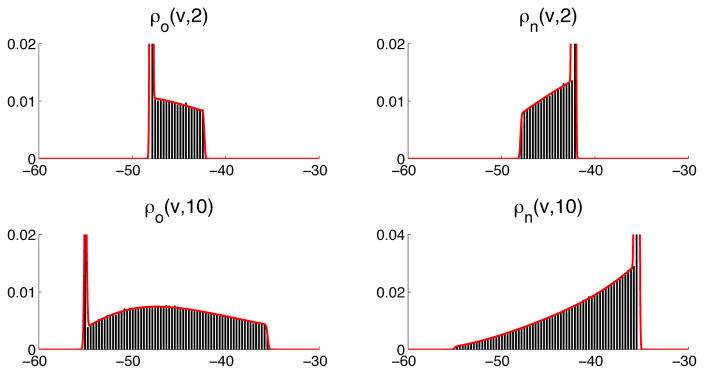
In Figure 2 we compare the results of the solution of the system (14) (red, solid lines) and the solution obtained running MC simulations (histograms) using the stochastic model (2).
Figure 2.
Numerical solution (red lines) of the the PDE system (14) at time t = 2 (upper) and t = 10ms (lower), and histograms computed by MC simulations (units on the horisontal axis in are mV). Probability density function of the open state (left panels) and non-conducting (ρc1 + ρc2 ) probability density function (right panels). The PDE system is solved using Δt = 0.72μs and Δv = 4.7μV, and the MC simulations based on (2) are performed using Δt = 1μs.
3.2 Sensitivity with respect to changes in the rates of the Markov model
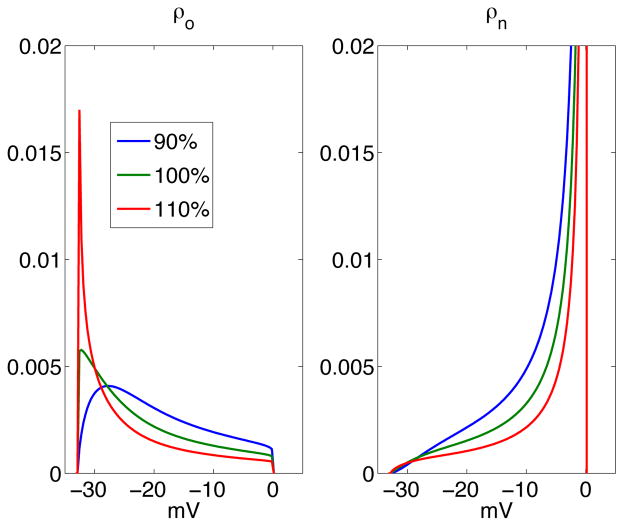
Clearly, in order to be able to identify the rates of the Markov model, the model output must be sensitive with respect to changes in the model. For the Markov model (13) with the rates given above, we illustrate the sensitivity with respect to changes in the rates in Figure 3. The figure shows the steady state probability distribution of the open state (left) and the non-conducting states (right). We observe that by multiplying the rate b1 with either 0.9 or 1.1, the stationary solution changes significantly. This indicates that it is possible to invert this model.
Figure 3.
The stationary probability distributions computed using the PDE model with I0 = 0 using the parameters given above (green), and the solutions obtained when the parameter b1 is multiplied by 1.1 (red) and 0.9 (blue).
3.3 Inversion of three models with no loops
We first apply our inversion procedure to three Markov models with no loop. The form of the models are defined in Table 1 and the results are given in Table 2. The Markov models are based on Scheme 4 in Zhou et al. [25]. The CCCCOI model is identical to their model except that the β rate has been altered to fit the standard Boltzmann form. In the CCCCO model, we have simply removed the inactivated state I, and in the CO model we have kept just one closed and one open state.
Table 3.
The table shows the correct parameters (True value) of the Markov models, the computed (inverted) parameters denoted by r* for the model CCO, the value of the cost function J(r*), the value of the norm |∇J(r*)| and the value of the smallest eigenvalue of the Hessian matrix at r*.
| True value | Found | ||
|---|---|---|---|
| α1 | −2 | −2.0026 | |
| α2 | −1 | −1.0015 | |
| b1 | −4 | −4.0468 | |
| b2 | −3 | −3.0009 | |
| J(r*) | 8.6812 · 10−6 | ||
| |∇J(r*)| | 8.1614 · 10−10 | ||
|
|
6.2461 · 10−4 |
Table 4.
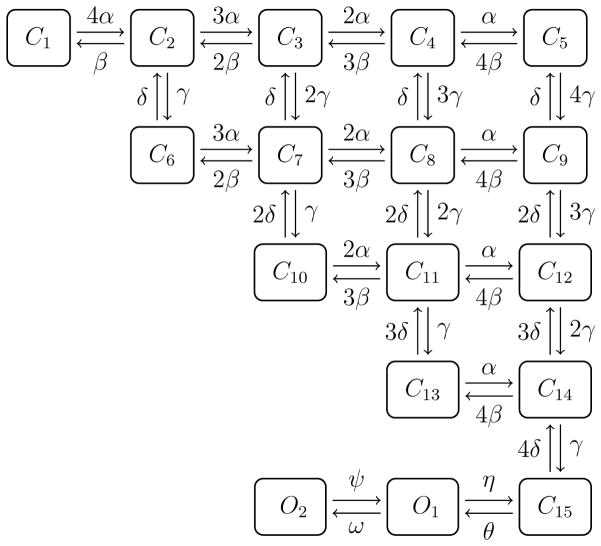
The rates and parameters of the 17 state model.
| α(v) = ea1+a2v | α1 = −7.5617 | α2 = −0.0091 |
| β(v) = eb1+b2v | b1 = −8.3559 | b2 = 0.0916 |
| γ(v) = ec1+c2v | c1 = −4.9268 | c2 = 0.0079 |
| δ(v) = ed1+d2v | d1 = −6.4825 | d2 = −0.0236 |
| θ(v) = ee1+e2v | e1 = −6.9486 | e2 = 0 |
| η(v) = ef1+f2v | f1 = −4.0923 | f2 = −0.0505 |
| φ (v) = eg1+g2v | g1 = −7.9836 | g2 = 0.0467 |
| ω(v) = eh1+h2v | h1 = −8.2472 | h2 = −0.0304 |
We observe that for all the three Markov model (CO, CCCCO and CCCCOI) we are able to compute the rates based on the pseudo-experimental data. The accuracy of the computed parameters are quite good. In the optimum, the value of the cost function is quite small and so is the norm of the gradient of the cost function which means that we are close to a stationary point. Finally, the smallest eigenvalue for all cases are positive. These cases are identifiable according to the definition given above.
In the above problems, all parameters are involved with transitions between conducting and non-conducting states. The rates are also voltage dependent. Next, we try a problem where this is not the case:
with αi = exp(αi) and βi = exp(bi), i = 1, 2. Parameters and results are given in Table 3. We see that the inversion also in this case is successfull, consistent with the relatively large smallest eigenvalue.
Table 5.
Column 1: numbering of eigenvectors. Column 2: Eigenvalues (smallest at the first row and then increasing values). Columns 3, 4 and 5: The value of the cost function J(r* + δri) where the parameter vector r* is given in Table 4 and ri denotes the i–th eigenvector. Recall that J(r*) = 0 so the table illustrates how much the cost function is changed by changing the the parameter vector along and eigenvector of the Hessian matrix.
| i | λi | δ = 10−5 | δ = 10−3 | δ = 10−1 |
|---|---|---|---|---|
| 1 | 7.579e-15 | 3.157e-25 | 5.636e-18 | 5.607e-10 |
| 2 | 1.383e-14 | 9.316e-25 | 5.074e-19 | 5.047e-11 |
| 3 | 7.361e-14 | 3.084e-24 | 2.593e-16 | 2.551e-08 |
| 4 | 1.183e-13 | 2.095e-25 | 1.081e-17 | 9.346e-10 |
| 5 | 1.209e-13 | 3.153e-24 | 1.567e-17 | 1.586e-09 |
| 6 | 1.654e-13 | 4.465e-24 | 7.279e-17 | 7.309e-09 |
| 7 | 1.038e-11 | 5.191e-22 | 6.645e-18 | 1.648e-10 |
| 8 | 1.986e-10 | 9.933e-21 | 1.017e-16 | 2.242e-10 |
| 9 | 7.319e-10 | 3.659e-20 | 3.910e-16 | 1.943e-09 |
| 10 | 7.090e-09 | 3.545e-19 | 3.563e-15 | 2.011e-09 |
| 11 | 1.482e-06 | 7.410e-17 | 7.409e-13 | 3.662e-08 |
| 12 | 3.952e-06 | 1.976e-16 | 1.974e-12 | 1.943e-08 |
| 13 | 3.712e-05 | 1.856e-15 | 1.856e-11 | 1.965e-07 |
| 14 | 1.235e-03 | 6.174e-14 | 6.170e-10 | 5.963e-06 |
| 15 | 2.294e-02 | 1.147e-12 | 1.138e-08 | 6.443e-05 |
| 16 | 3.543e-01 | 1.771e-11 | 1.769e-07 | 1.539e-03 |
3.4 Inversion of a three state model with a loop
It is quite common to use Markov models including a loop of the form illustrated in Figure 4. However, Siekmann et al [7] conclude that this model is not identifiable. They considered a case with constant rates (independent of the trans-membrane potential) and their method is very different from ours. It is therefore interesting to see if the model is identifiable in our framework The rates of the model are parameterized as follows:
Figure 4.
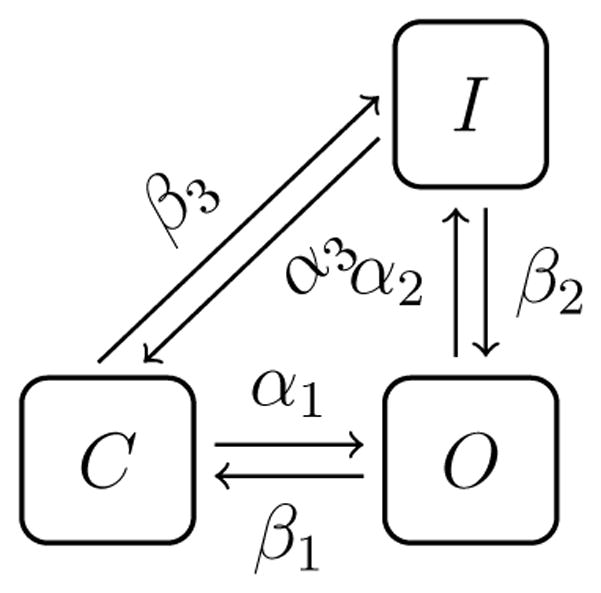
The three state model.
In order to test if the model is identifiable using our method, we can use data provided by the PDE model an evoke to value of the smallest eigenvalue. We find that which indicate that this case is very hard to invert. Also, running inversion procedure does not result in a converged solution.
We also consider a case where we have removed the voltage dependence of the rates, i.e. we have put αi2 = bi2 = 0 in the model. Using PDE-generated data we find that and thus the model is not invertible. These observations concur with the results of Siekmann et al. [7].
3.5 A special case of the three-state model
For a special case of the three state model shown in Figure 4, we can show mathematically that it is impossible to identify the rates based on observation of (ρ̄o, ρ̄n). To see this, we first note that the associate PDE system is given by
| (15) |
| (16) |
| (17) |
By adding the two latter equations of the system above, we find the system
| (18) |
| (19) |
where ρn = ρc + ρo. If we now consider the special case of α1 = β2 =: μ, we get the system
| (20) |
| (21) |
From this system we notice that we can compute ρo and ρn without any knowledge of the rates α3 and β3 connecting the closed and the inactivated states. These parameters are therefore clearly non-identifiable since they do not in any way affect the probability density functions of the conducting (open) and non-conducting states (closed plus inactivated).
3.6 The size of the capacitance
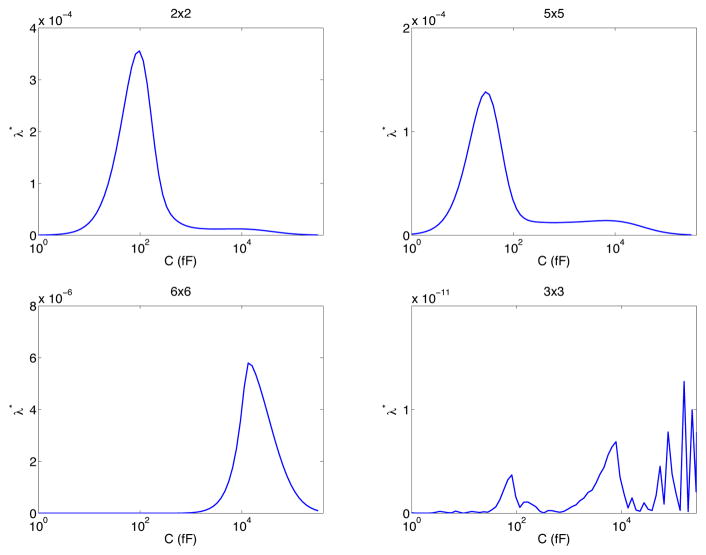
The size of the capacitance influence the identifiability of a Markov model. This is illustrated in Figure 5 for the model CO (upper left) CCCCO (upper right), CCCCOI (lower left) and the three state model including a loop (lower right). The computations is based on data generated by solving the PDE using the correct rates of the Markov model. We observe that for the CO, CCCCO and CCCCOI models, there is a range of values of the capacitance for which the model is invertible. For the three state model with a loop, however, the model is not identifiable for any value of the capacitance.
Figure 5.
Smallest eigenvalues computed using data based on solving the PDE problem for CO (upper left) CCCCO (upper right), CCCCOI (lower left) and, finally (lower right) the three state model including a loop (see Figure 4).
3.7 17 state Markov model of the potassium channel
The method introduced here is easy to work with for Markov models with few states, but in principle it can also be applied to more complex methods. In Figure 6 we show a 17 state model taken from Zhou et al. [25]. The parameters of the model are given in Table 4, and the eigenvalues of the Hessian are given in Table 5. This model has many very small eigenvalues and is therefore clearly not identifiable by our method. In Table 5 we show the change of the cost function when the parameter vector is changed. We change the parameter by a small step along each eigenvector of the Hessian matrix. Each eigenvector is normalized to have Euclidian norm equal to one, and we change each parameter vector by a step of length 10−5, 10−3 and 0.1. We note that for the eigenvectors associated small eigenvalues, the change of the cost function is very small when the parameter vector is changed. This is exactly as expected from the theory discussed above. Finally, large steps in the direction of eigenvectors with larger eigenvalues result in significant changes of the cost function.
Figure 6.
Markov model of the potassium channel taken from Zhou et al. [25]. The rates of the model are given in Table 4.
Table 1.
The table defines the name and the form of three Markov models. The rates are written on the form α(v) = exp(α1 + α2v), β(v) = exp(b1 + b2v). The O–I transition is assumed to be voltage independent; γ = exp(c1), δ = exp(d1). All rates are in units 1/ms.
| CO |
|
|
| CCCCO |
|
|
| CCCCOI |
|
Table 2.
The table shows the correct parameters (True value) of the Markov models, the computed (inverted) parameters denoted by r* for the models CO, CCCCO and CCCCOI, the value of the cost function J(r*), the value of the norm |∇J(r*)| and the value of the smallest eigenvalue of the Hessian matrix at r*.
| True value | CO | CCCCO | CCCCOI | ||
|---|---|---|---|---|---|
| α1 | −1 | −1.0155 | −1.0034 | −0.9996 | |
| α2 | 0.02 | 0.0196 | 0.0199 | 0.0200 | |
| b1 | −4.5 | −4.5077 | −4.5120 | −4.5017 | |
| b2 | −0.05 | −0.0501 | −0.0503 | −0.0501 | |
| c1 | −9 | - | - | −9.2154 | |
| d1 | −9 | - | - | −9.2152 | |
| J(r*) | 1.0488 · 10−5 | 6.5289 · 10−6 | 1.4788 · 10−6 | ||
| |∇J(r*)| | 1.6688 · 10−7 | 3.6612 · 10−8 | 6.5386 · 10−9 | ||
|
|
1.6696 · 10−2 | 1.5840 · 10−2 | 3.0512 · 10−4 |
In Table 6 we consider the reduced case where the O2-state is removed from the model. The smallest eigenvalue does not change much, indicating that the O1-O2 transition is not the most difficult one to identify.
Table 6.
Column 1: numbering of eigenvectors. Column 2: Eigenvalues (smallest at the first row and then increasing values). Columns 3, 4 and 5: The value of the cost function J(r* + δri) where the parameter vector r* is given in Table 4 and ri denotes the i–th eigenvector. Recall that J(r*) = 0 so the table illustrates how much the cost function is changed by changing the the parameter vector along and eigenvector of the Hessian matrix.
| i | λi) | d = 10−5 | d = 10−3 | d = 10−1 |
|---|---|---|---|---|
| 1 | 1.877e-14 | 8.757e-25 | 1.573e-19 | 1.241e-11 |
| 2 | 6.364e-14 | 2.715e-24 | 2.548e-16 | 2.527e-08 |
| 3 | 1.019e-13 | 2.573e-25 | 1.708e-17 | 1.729e-09 |
| 4 | 1.043e-11 | 5.272e-22 | 2.472e-16 | 2.387e-08 |
| 5 | 2.583e-11 | 1.289e-21 | 1.553e-17 | 2.756e-10 |
| 6 | 4.020e-10 | 2.010e-20 | 2.136e-16 | 1.131e-09 |
| 7 | 1.813e-09 | 9.063e-20 | 9.112e-16 | 3.810e-10 |
| 8 | 1.714e-08 | 8.569e-19 | 8.581e-15 | 1.643e-09 |
| 9 | 2.757e-06 | 1.378e-16 | 1.380e-12 | 7.737e-08 |
| 10 | 6.225e-05 | 3.113e-15 | 3.115e-11 | 3.511e-07 |
| 11 | 2.058e-03 | 1.029e-13 | 1.028e-09 | 9.226e-06 |
| 12 | 3.346e-01 | 1.673e-11 | 1.671e-07 | 1.489e-03 |
4 Discussion
The main purpose of this report is to propose a new method for computing rates of Markov models of voltage-gated ion channels. Other methods are based on experimental data obtained by clamping the transmembrane potential and recording whether the channel is in the conducting (open) or non-conducting (closed or inactivated) mode. Our method is based on repeated recordings of traces of the transmembrane potential of the single channel. The method proposed here also enables analysis of identifiability of the model and it may provide a way of quantifying the uncertainty of the resulting rates of the Markov model.
4.1 Inversion
As pointed out by Siekmann et al. [7], it is straightforward to devise procedures that will provide the rates of Markov models based on experimental data. The difficulty involved is to obtain some measure of the reliability of the computed rates. Clearly, if two sets of completely different rates can provide the same results from a model, the model is not suited to give the insight we are ultimately looking for.
4.2 Identifiability using our method
Our method is based on comparing probability density functions from experimental data with probability density functions generated by solving a system of partial differential equations. The experimental data in this study are generated by Monte Carlo (MC) simulations where a given Markov model governs the stochastic state of the ion channel. The challenge is then to tune the parameters of a system of partial differential equations (PDE) such that the solution of the PDE system resembles the solution generated by MC as closely as possible. The model is identifiable if different set of rates for the Markov model gives different probability density distributions.
We find that the simple CO, CCCCO and CCCCOI models are identifiable, and that the looped model, see Figure 4, is not identifiable using our method. For the latter case, one eigenvalue of the Hessian matrix is very small and thus we may change the parameter vector r defining the Markov model a step in the direction of the associated eigenvector and observe little or no change in the cost function.
For the three state Markov model with a loop (Figure 4), we give an example where the rates between the closed and inactivated states simply disappear from the model in the sense that we can compute the probability density distribution of the conducting (open) and non-conducting (closed plus inactivated) states without knowledge of these rates. The model is therefore clearly not identifiable in this particular case.
For the complex Markov model given in Figure 6, we find that eight eigenvalues are very small. We can change the parameter vector in either of the associated eigenvectors and observe very little change of the cost function.
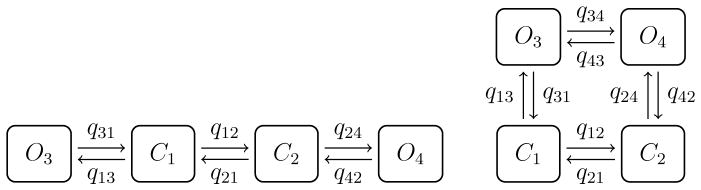
Following Siekmann et al [7] we have considered the Markov models shown in Figure 7. Here data are created using the model without a loop (left panel) and the data are used to invert the model with a loop (right panel). The MCMC method used by Siekmann et al then, correctly, yields results consistent with the model without a loop. Our method similarly provided rates where the transitions between the two open states are virtually zero.
Figure 7.
Two four states models taken from Siekmann et al. [7]; without a loop (left) and with a loop (rihgt).
4.3 The cost function
All the observations regarding identifiability reported in the present paper are based on the particular cost function (10). This cost function can be changed in many different ways; we may vary the applied current I0 in numerous ways, we can add the time course of each trace of pseudo-experimental data, and we can alter the transmembrane concentration gradient and thus change the associated equilibrium (Nernst) potential of the potassium current. None of these changes have significantly altered the observations reported here, and we have therefore chosen to rely on the simplest version of the cost function.
4.4 What is zero?
As demonstrated above, a model is not identifiable if the smallest eigenvalue of the Hessian matrix is zero. In numerical computations, however, you will never get exactly zero – rather a very small number. But by considering the Taylor series (11), it is evident that a very small eigenvalue implies that the cost function changes very little if the parameter vector is changed in the direction of the associated eigenvector. Therefore, we refer to a model as not identifiable when the smallest eigenvalue is sufficiently small. It may be possible to follow this line of reasoning one step further and use the size of the smallest eigenvalue to quantify more precisely the uncertainty of the rates of the Markov model using the method introduced here.
4.5 Errors in computed parameters
The errors of the computed parameters can be caused of variations in the data since the data are generated by a stochastic process, and it can be caused by inaccuracies inherent in the method of inversion. If we do the inversion based on data created by the deterministic system of partial differential equations, there is no randomness involved but still there are errors in the computed parameters. This indicates that there are inherent inaccuracies in the method of inversion (which is very common for methods of solving inverse problems). One source of error is numerical method used to solve the system of partial differential equations. This part of the error is typically reduced by mesh refinement, but extremely fine meshes are unattractive from a computational point of view. On the other hand; when stochastic data are used, the error typically are reduced with larger samples, and also repeated inversion using the same data but different initial values, yields the same results. This confirms that also stochastic variations are source of variations in computed parameters.
4.6 Weakness of our approach
Although single channel data has in principle have been available for forty years ([10, 11]), the majority of patch clamp data is still obtained from whole cell recordings. Our method is suitable for inverting data based on single ion channel but is not directly applicable to data from whole cell recording. For a single channel it is relevant to consider the probability density as a function of the trans-membrane potential. For an identifiable Markov model, such distributions can clearly be used to identify the rates of the Markov model. The whole cell situations is completely different since thousands of single channels contribute the transmembrane potential which is therefore a deterministic entity even if the individual channels are stochastic.
References
- 1.Keener J, Sneyd J. Mathematical Physiology. Springer; 2009. [Google Scholar]
- 2.Smith Gregory D. Modeling the stochastic gating of ion channels. In: Fall Christopher P, Marland Eric S, Wagner John M, Tyson John J., editors. Computational Cell Biology, volume 20 of Interdisciplinary Applied Mathematics. chapter 11. Springer; 2002. pp. 285–319. [Google Scholar]
- 3.Bressloff Paul C. Stochastic processes in cell biology. Vol. 41. Springer; 2014. [Google Scholar]
- 4.Tveito Aslak, Lines Glenn Terje. Computing characterizations of drugs for ion channels and receptors using Markov models. Vol. 111. Springer; Heidelberg, Germany: 2016. volume LNCSE. [Google Scholar]
- 5.Shelley Christopher, Magleby Karl L. Linking Exponential Components to Kinetic States in Markov Models for Single-Channel Gating. The Journal of General Physiology. 2008 Jul;132(2):295–312. doi: 10.1085/jgp.200810008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siekmann Ivo, Wagner Larry E, II, Yule David, Fox Colin, Bryant David, Crampin Edmund J, Sneyd James. MCMC Estimation of Markov Models for Ion Channels. Biophysical Journal. 2011 Apr;100(8):1919–1929. doi: 10.1016/j.bpj.2011.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siekmann Ivo, Sneyd James, Crampin Edmund J. MCMC Can Detect Nonidentifiable Models. Biophysical Journal. 2012 Dec;103(11):2275–2286. doi: 10.1016/j.bpj.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hines Keegan E, Middendorf Thomas R, Aldrich Richard W. Determination of parameter identifiability in nonlinear biophysical models: A bayesian approach. The Journal of general physiology. 2014;143(3):401–416. doi: 10.1085/jgp.201311116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Csanády László. Statistical evaluation of ion-channel gating models based on distributions of log-likelihood ratios. Biophysical journal. 2006;90(10):3523–3545. doi: 10.1529/biophysj.105.075135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neher Erwin, Sakmann Bert. A century of Nature: twenty-one discoveries that changed science and the world. 2010. Single-channel currents recorded from membrane of denervated frog muscle fibres; p. 224. [DOI] [PubMed] [Google Scholar]
- 11.Sakmann Bert, Neher Erwin. Patch clamp techniques for studying ionic channels in excitable membranes. Annual review of physiology. 1984;46(1):455–472. doi: 10.1146/annurev.ph.46.030184.002323. [DOI] [PubMed] [Google Scholar]
- 12.Colquhoun D, Hawkes AG. Relaxation and fluctuations of membrane currents that flow through drug-operated channels. Proceedings of the Royal Society of London. Series B, Biological Sciences. 1977;199(1135):231–262. doi: 10.1098/rspb.1977.0137. [DOI] [PubMed] [Google Scholar]
- 13.Colquhoun D, Hawkes AG. On the stochastic properties of bursts of single ion channel openings and of clusters of bursts. Philosophical Transactions of the Royal Society London B. 1982;300:1–59. doi: 10.1098/rstb.1982.0156. [DOI] [PubMed] [Google Scholar]
- 14.Sakmann Bert, Neher Erwin., editors. Single-Channel Recording. 2. Springer; 1995. [Google Scholar]
- 15.Qin Feng, Auerbach Anthony, Sachs Frederick. Estimating single-channel kinetic parameters from idealized patch-clamp data containing missed events. Biophysical Journal. 1996 Jan;70:264–280. doi: 10.1016/S0006-3495(96)79568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin Feng, Auerbach Anthony, Sachs Frederick. A direct optimization approach to hidden markov modeling for single channel kinetics. Biophysical Journal. 2000 Jan;79:1915–1927. doi: 10.1016/S0006-3495(00)76441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicolai Christopher, Sachs Frederick. Solving ion channel kinetics with the QuB software. Biophysical Reviews and Letters. 2013;8(3n4):191–211. [Google Scholar]
- 18.Ball FG, Davies SS. Statistical inference for a two-state markov model of a single ion channel, incorporating time interval omission. Journal of the Royal Statistical Society. Series B (Methodological) 1995:269–287. [Google Scholar]
- 19.Rosales Rafael A. Mcmc for hidden markov models incorporating aggregation of states and filtering. Bulletin of mathematical biology. 2004;66(5):1173–1199. doi: 10.1016/j.bulm.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Gin Elan, Falcke Martin, Wagner Larry E, Yule David I, Sneyd James. Markov chain Monte Carlo fitting of single-channel data from inositol trisphosphate receptors. Journal of Theoretical Biology. 2009 Apr;257(3):460–474. doi: 10.1016/j.jtbi.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 21.Fredkin Donald R, Rice John A. On aggregated markov processes. Journal of Applied Probability. 1986:208–214. [Google Scholar]
- 22.Milescu Lorin S, Yamanishi Tadashi, Ptak Krzysztof, Mogri Murtaza Z, Smith Jeffrey C. Real-time kinetic modeling of voltage-gated ion channels using dynamic clamp. Biophysical journal. 2008;95(1):66–87. doi: 10.1529/biophysj.107.118190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apostol Tom M. Calculus. II. Wiley; 1969. [Google Scholar]
- 24.Fornberg Bengt. Generation of finite difference formulas on arbitrarily spaced grids. Mathematics of Computation. 1988;51(184):699–706. [Google Scholar]
- 25.Zhou Qinlian, Bett Glenna CL, Rasmusson Randall L. Markov Models of Use-Dependence and Reverse Use-Dependence during the Mouse Cardiac Action Potential. PLoS ONE. 2012 Aug;7(8):e42295. doi: 10.1371/journal.pone.0042295. [DOI] [PMC free article] [PubMed] [Google Scholar]