Abstract
For 200 years, zoologists have relied on phenotypes to learn about the evolution of animals. A glance at the genotype, even through several gene markers, revolutionized our understanding of animal phylogeny. Recent advances in sequencing techniques allow researchers to study speciation mechanisms and the link between genotype and phenotype using complete genomes. We sequenced and assembled a complete genome of the Cloudless Sulphur (Phoebis sennae) from a single wild-caught specimen. This genome was used as reference to compare genomes of six specimens, three from the eastern populations (Oklahoma and north Texas), referred to as a subspecies Phoebis sennae eubule, and three from the southwestern populations (south Texas) known as a subspecies Phoebis sennae marcellina. While the two subspecies differ only subtly in phenotype and mitochondrial DNA, comparison of their complete genomes revealed consistent and significant differences, which are more prominent than those between tiger swallowtails Pterourus canadensis and Pterourus glaucus. The two sulphur taxa differed in histone methylation regulators, chromatin-associated proteins, circadian clock, and early development proteins. Despite being well separated on the whole-genome level, the two taxa show introgression, with gene flow mainly from P. s. marcellina to P. s. eubule. Functional analysis of introgressed genes reveals enrichment in transmembrane transporters. Many transporters are responsible for nutrient uptake, and their introgression may be of selective advantage for caterpillars to feed on more diverse food resources. Phylogenetically, complete genomes place family Pieridae away from Papilionidae, which is consistent with previous analyses based on several gene markers.
Keywords: Phoebis sennae, speciation, introgression, comparative genomics, lepidoptera, phylogeny
Introduction
Butterflies and moths (Lepidoptera) are some of the best-known and best-studied insects. Their colorful wings and complex life cycles attract wide attention from both researchers and the public. Despite this popularity, little is known about the genetic makeup of Lepidoptera, and compete genomes are available for fewer than a dozen species (International Silkworm Genome Consortium 2008; Duan et al. 2010; Zhan et al. 2011; Heliconius Genome 2012; You et al. 2013; Zhan and Reppert 2013; Ahola et al. 2014; Tang et al. 2014; Cong et al. 2015a, 2015b; Nishikawa et al. 2015). However, small genome sizes and extensive knowledge about the morphology and life histories of Lepidoptera offer a promise to further our understanding in genetics, molecular evolution, and speciation through comparative genomics. For instance, genomics studies of Heliconius revealed a new paradigm that gene exchange between species is pivotal in the evolution of adaptation and mimicry (Heliconius Genome 2012). Among butterflies, representative genomes are currently known for only three families: the swallowtails (Papilionidae), the brushfoots (Nymphalidae), and the skippers (Hesperiidae). The brushfoots have been prevalent in genomics studies, with research on Heliconius and the Monarch (Danaus plexippus) leading the field (Nadeau et al. 2014; Zhan et al. 2014). For comparative genomics of butterflies, it is essential to sequence complete genomes of all major phylogenetic groups.
The family Pieridae (Whites and Sulphurs) may be the prototype for the name “butterfly.” A common yellow-toned European species, the Brimstone (Gonepteryx rhamni), was called the “butter-colored fly” by early naturalists (Asher et al. 2001). This family includes some of the very few butterflies known as crop pests, such as the Cabbage Whites (Pieris rapae and Pieris brassicae) and Alfalfa Sulphur (Colias eurytheme). Pierids are particularly well known for using pterins as pigments on their wings (Pfeiler 1968). While most swallowtails diapause as pupae, many pierids overwinter as adults and enter reproductive diapause in the fall. Because of similarities in pupae, pierids were previously hypothesized to be a sister family to the swallowtails (Ehrlich 1958), a view not supported by recent molecular studies (Weller et al. 1996). To help understand genetic bases for morphological traits of Pieridae and to clarify its phylogenetic placement, we sequenced the first complete genome from this family. We chose a large and showy American species, the Cloudless Sulphur (Phoebis sennae), which is similar in size and color to the European Brimstone butterfly.
The Cloudless Sulphur is a large yellow-toned butterfly distributed from the southern regions of the United States through the Neotropics. Its caterpillars feed on Senna plants and close relatives from the Pea family (Fabaceae). Adults are highly vagile but do not survive cold winters. Eastern US populations are known as subspecies Phoebis sennae eubule, and southwestern populations that range throughout Central and most of South America are attributed to subspecies Phoebis sennae marcellina (Brown 1929). Both subspecies are present in Texas. The two subspecies are morphologically distinct, with P. s. eubule being typically less patterned on the underside of the wings and P. s. marcellina females characterized by pronounced dark spots along the margin of hindwings above (fig. 1). In addition, their caterpillars show somewhat different foodplant preferences. Phoebis s. eubule mostly feeds on partridge pea (Chamaecrista fasciculata), whereas P. s. marcellina prefers Senna species. However, their cytochrome c oxidase subunit 1 (COI) mitochondrial DNA sequences show small divergence, no more than 0.6% (Ratnasingham and Hebert 2007). The divergence in nuclear genes, that likely cause the morphological differences, has remained unclear.
Fig. 1.—
Specimens of Phoebis sennae. Males (a–d) and females (e–l) are shown in dorsal (a, b, e, f, i, j) and ventral (c, d, g, h, k, l) aspects. a, c, e, g, i, and k are Phoebis sennae eubule from Texas, Denton Co., Flower Mound, reared from caterpillars, adults eclosed on: a, c September 26, 1996; e, g December 8, 2001; i, k October 24, 1998. b, d, f, h, j, and l are Phoebis sennae marcellina from Texas, Hidalgo Co., 1.5 air mi southeast of Relampago, reared from caterpillars, adults eclosed on: b, d July 8, 2015, and f, g June 10, 2015, and collected on j, l June 14, 2015.
We obtained a complete reference genome of P. s. eubule from a single male collected in southeast Texas. To compare genetic divergence between the North American P. sennae subspecies, we sequenced genomes of two more P. s. eubule specimens (from north Texas and Oklahoma) and of three P. s. marcellina specimens from south Texas. In contrast to mitochondrial DNA, their nuclear genomes revealed unexpectedly large divergence (∼2%), larger than that between the two sister species of Tiger Swallowtails (Pterourus canadensis and Pterourus glaucus). Despite a clear separation on the whole-genome level, there are regions in the genome that show significant (P < 0.0014, false discovery rate [FDR] [20] < 0.05) signs of introgression between the two taxa. This work lays the foundation for Pieridae genomics and provides rich sequence data sets for comparative studies.
Materials and Methods
Library Preparation and Sequencing
We removed and preserved the wings and genitalia of six freshly caught P. sennae specimens (three P. s. eubule: NVG-3314, male, Texas: San Jacinto Co., Sam Houston National Forest, 30.50596, -95.08868, April 12, 2015; NVG-4452, female, Texas: Wise Co., LBJ National Grassland, 33.38401, -97.57381, August 9, 2015; NVG-4541, male, Oklahoma: Atoka Co., McGee Creek Recreation Area, 34.41040, -95.91059, August 22, 2015; and three P. s. marcellina: Hidalgo Co., 1.5 air mi southeast of Relampago, 26.07093, -97.89131: NVG-3356, male, May 23, 2015; NVG-3377, female, May 24, 2015; NVG-3393, male, May 30, 2015), and the rest of the bodies were stored in RNAlater solution. Wings and genitalia of these specimens will be deposited in the McGuire Center for Lepidoptera and Biodiversity, Florida Museum of Natural History, University of Florida, Gainesville, Florida, USA (MGCL).
We used specimen NVG-3314 for the reference genome. We extracted approximately 20 μg of genomic DNA from about 4/5 of specimen NVG-3314 with the ChargeSwitch gDNA mini tissue kit; 250-bp and 500-bp paired-end libraries were prepared using enzymes from NEBNext Modules and following the Illumina TruSeq DNA sample preparation guide. Mate pair libraries (2, 6, and 15 kb) were prepared using a protocol similar to the previously published Cre-Lox-based method (Van Nieuwerburgh et al. 2012). For the 250 bp, 500 bp, 2 kb, 6 kb, and 15 kb libraries, approximately 500 ng, 500 ng, 1.5 μg, 3 μg, and 6 μg of DNA were used, respectively. We quantified the amount of DNA from all the libraries with the KAPA Library Quantification Kit, and mixed 250 bp, 500 bp, 2 kb, 6 kb, and 15 kb libraries at relative molar concentration 40:20:8:4:3. The mixed library was sent to the genomics core facility at UT Southwestern Medical Center to sequence 150 bp at both ends (PE150) using one lane in Illumina HiSeq2500.
The remaining 1/5 of specimen NVG-3314 was used to extract RNA using QIAGEN RNeasy Mini Kit. We further isolated mRNA using NEBNext Poly(A) mRNA Magnetic Isolation Module, and RNA-seq libraries for both specimens were prepared with NEBNext Ultra Directional RNA Library Prep Kit for Illumina following the manufacturer’s protocol. The RNA-seq library was sequenced for 150 bp from both ends using 1/8 of an Illumina lane.
The other five specimens were used to prepare paired-end libraries to map to the reference genome. For each of them, we extracted about 5 μg genomic DNA and used about 500 ng to prepare a 400 bp paired-end library. These paired-end libraries were mixed at equal ratio and sequenced using a similar strategy (PE150), using half of an Illumina lane. The sequencing reads for all the specimens have been deposited in the NCBI SRA database under accession SRP068212.
Genome and Transcriptome Assembly
We removed sequence reads that did not pass the purity filter and classified the pass-filter reads according to their TruSeq adapter indices to get individual sequencing libraries. Mate pair libraries were processed by the Delox script (Van Nieuwerburgh et al. 2012) to remove the loxP sequences and to separate true mate pair from paired-end reads. All reads were processed by mirabait (Chevreux et al. 1999) to remove contamination from the TruSeq adapters, an in-house script to remove low quality portions (quality scale < 20) at both ends, JELLYFISH (Marcais and Kingsford 2011) to obtain k-mer frequencies in all the libraries (supplementary fig. S1, Supplementary Material online), and QUAKE (Kelley et al. 2010) to correct sequencing errors. The data processing resulted in nine libraries that were supplied to Platanus (Kajitani et al. 2014) for genome assembly: 250 bp and 500 bp paired-end libraries, three paired-end and three mate pair libraries from 2, 6, and 15 kb libraries and a single-end library containing all reads whose pairs were removed in the process (supplementary table S2A, Supplementary Material online).
We mapped these reads to the initial assembly with Bowtie2 (Langmead and Salzberg 2012) and calculated the coverage of each scaffold with the help of SAMtools (Li et al. 2009). Many short scaffolds in the assembly showed coverage that was about half of the expected value (supplementary fig. S2, Supplementary Material online); they likely came from highly heterozygous regions that were not merged to the equivalent segments in the homologous chromosomes. We removed them if they could be fully aligned to another significantly less covered region (coverage > 90% and uncovered region < 500 bp) in a longer scaffold with high sequence identity (>95%). Similar problems occurred in the Heliconius melpomene, Pt. glaucus, and Lerema accius genome projects, and similar strategies were used to improve the assemblies (Heliconius Genome 2012; Cong et al. 2015a, 2015b).
The RNA-seq reads were processed using a similar procedure as the genomic DNA reads to remove contamination from TruSeq adapters and the low quality portion of the reads. Afterward, we applied three methods to assemble the transcriptomes: 1) de novo assembly by Trinity (Haas et al. 2013), 2) reference-based assembly by TopHat (Kim et al. 2013) (v2.0.10) and Cufflinks (Roberts et al. 2011) (v2.2.1), and 3) reference-guided assembly by Trinity. The results from all three methods were then integrated by Program to Assemble Spliced Alignment (PASA) (Haas et al. 2008).
Identification of Repeats and Gene Annotation
Two approaches were used to identify repeats in the genome: The RepeatModeler (Smit and Hubley 2008–2010) pipeline and in-house scripts that extracted regions with coverage four times higher than expected. These repeats were submitted to the CENSOR (Jurka et al. 1996) server to assign them to the repeat classification hierarchy. The species-specific repeat library, repeats we previously identified in other Lepidoptera genomes, and repeats classified in RepBase (Jurka et al. 2005) (V18.12) were used to mask repeats in the genome by RepeatMasker (Smit et al. 1996–2010).
We obtained two sets of transcript-based annotations from two pipelines: TopHat followed by Cufflinks and Trinity followed by PASA. In addition, we obtained five sets of homology-based annotations by aligning protein sets from Drosophila melanogaster (Misra et al. 2002) and four published Lepidoptera genomes (Plutella xylostella, Bombyx mori, H. melpomene, and D. plexippus) to the P. sennae genome with exonerate (Slater and Birney 2005). Proteins from Invertebrate in the entire UniRef90 (Suzek et al. 2007) database were used to generate another set of gene predictions by genblastG (She et al. 2011). We manually curated and selected 1,152 confident gene models by integrating the evidence from transcripts and homologs to train de novo gene predictors: AUGUSTUS (Stanke et al. 2006), SNAP (Korf 2004), and GlimmerHMM (Majoros et al. 2004). These trained predictors, the self-trained Genemark (Besemer and Borodovsky 2005) and a consensus-based pipeline, Maker (Cantarel et al. 2008), were used to generate another five sets of gene models. Transcript-based and homology-based annotations were supplied to AUGUSTUS, SNAP, and Maker to boost their performance. In total, we generated 13 sets of gene predictions and integrated them with EvidenceModeller (Haas et al. 2008) to generate the final gene models.
We predicted the function of P. sennae proteins by transferring annotations and GO terms from the closest BLAST (Altschul et al. 1990) hits (E value < 10−5) in both the Swissprot (UniProt 2014) database and Flybase (St Pierre et al. 2014). Finally, we performed InterproScan (Jones et al. 2014) to identify conserved protein domains and functional motifs, to predict coiled coils, transmembrane helices and signal peptides, to detect homologous 3D structures, to assign proteins to protein families, and to map them to metabolic pathways.
Identification of Orthologous Proteins and Phylogenetic Tree Construction
We identified the orthologous groups from all 11 Lepidoptera genomes using OrthoMCL (Li et al. 2003). In total, 2,106 orthologous groups consisted of single-copy genes from each species, and they were used for phylogenetic analysis. An alignment was built for each universal single-copy orthologous group using both global sequence aligner MAFFT (Katoh and Standley 2013) and local sequence aligner BLASTP. Positions that were consistently aligned by both aligners were extracted from each individual alignment and concatenated to obtain an alignment containing 362,743 positions. The concatenated alignment was used to obtain a phylogenetic tree using RAxML (Stamatakis 2014). Bootstrap was performed to assign the confidence level of each node in the tree. In addition, in order to detect the weakest nodes in the tree, we reduced the amount of data by randomly splitting the concatenated alignment into 100 alignments (about 3,630 positions in each alignment) and applied RAxML to each alignment. We obtained a 50% majority rule consensus tree and assigned confidence levels to each node based on the percent of individual trees supporting this node.
Assembly and Annotation of Mitochondrial Genomes
The mitogenomes of several closely related species, including Catopsilia pomona (Hao et al. 2014), Colias erate (Wu et al. 2015), and Gonepteryx mahaguru (Yang et al. 2014) were used as reference. On the basis of these mitogenomes, we applied mitochondrial baiting and iterative mapping (MITObim) v1.6 (Hahn et al. 2013) software to extract the sequencing reads of the mitogenome in the paired-end libraries for specimen NVG-3314. About 4.3 million reads for the mitogenome were extracted, and they were expected to cover the mitogenome 40,000 times. We used JELLYFISH to obtain the frequencies of 15-mers in these reads and applied QUAKE to correct errors in 15-mers with frequencies lower than 1,000 and excluded reads containing low-frequency 15-mers that cannot be corrected by QUAKE. We used the error-corrected reads to assemble into contigs de novo with Platanus. We manually selected the contig corresponding to the mitogenome (it is the longest one with highest coverage), and manually extended its sequence based on the sequencing reads to obtain a complete circular DNA. In addition, by aligning the protein coding sequences from the mitogenomes of closely related species mentioned above to the P. sennae mitogenome, we annotated the 13 protein coding genes.
Obtaining the Genomes of Six P. sennae Specimens and Phylogenetic Analysis
We mapped the sequencing reads of all six P. sennae specimens to the reference genome using BWA (Li and Durbin 2010) and detected single-nucleotide polymorphisms (SNPs) using the Genome Analysis Toolkit (GATK) (DePristo et al. 2011). We deduced the genomic sequences for each specimen based on the result of GATK. We used two sequences to represent the paternal and maternal DNA in each specimen. For heterozygous positions, each possible nucleotide was randomly assigned to either paternal or maternal DNA. Using the gene annotation of the reference genome, we further deduced the protein-coding sequences of genes in each specimen.
To study the population structure, we selected bi-allelic loci (two nucleotide types in a position of alignment covering all six specimens, coding and noncoding regions). First, we encoded each specimen by a vector consisting of the frequency of a certain nucleotide in each position. For example, if a position is occupied by A and T in all six specimens, then their possible genotypes AA, AT, and TT were represented as 0, 0.5, and 1, respectively. We calculated the covariance between each pair of specimens and obtained a covariance matrix. We performed singular value decomposition on the covariance matrix and visualized the clustering of the six specimens in two-dimensional space defined by the first two singular vectors. Second, we applied fastStructure software (Raj et al. 2014) to analyze the same SNP genotype data. We tested all the possible numbers of model components (from 1 to 6) and selected the population structure with the maximal likelihood.
In order to quantify the divergence between the two P. sennae subspecies, we compared their divergence level in the protein-coding regions to that for a pair of sister species, Pt. glaucus and Pt. canadensis. The transcripts of Pterourus specimens were mapped to the Pt. glaucus reference genome using methods described previously (Cong et al. 2015b). From alignments of Pterourus transcripts to the reference genome, we selected 9,622 nuclear genes for which there are at least 60 aligned positions from at least two Pt. canadensis and two Pt. glaucus specimens. Similarly, we selected 16,137 nuclear genes of P. sennae, requiring the selected genes to have at least 50% coverage for the coding regions in two P. s. marcellina and two P. s. eubule specimens. We extracted the coding regions in the alignments of individual nuclear genes and concatenated them for both Pterourus and Phoebis, respectively. The concatenated alignments were used to build both neighbor-joining trees with PHYLIP (Felsenstein 1989), based on the percentage of different positions between specimens, and maximal-likelihood trees with RAxML (model: GTRGAMMA). Bootstrap resampling was performed to assign confidence levels for nodes in the maximal-likelihood trees.
Identification of Divergence Hotspots and Selection of Nuclear Barcodes
We defined “divergence hotspots” as genes that satisfied the following two criteria: 1) can confidently (bootstrap > 75) separate P. s. eubule and P. s. marcellina specimens into clades in phylogenetic trees by both the DNA sequence and the protein sequence encoded by them, and 2) the divergence within both P. s. eubule and P. s. marcellina specimens is lower than the median divergence level over all the genes. We identified the enriched GO terms associated with these “divergence hotspots” using binomial tests (m = the number of “divergence hotspots” that were associated with this GO term, N = number of “speciation hotspots,” p = the probability for this GO term to be associated with any gene). GO terms with P values lower than 0.01 were considered enriched. We further identified genes that are always more divergent between taxa than within taxa. These genes could be used as nuclear markers to distinguish P. s. eubule and P. s. marcellina.
Detection of Introgression Between P. s. eubule and P. s. marcellina
To detect introgressed regions in each specimen, we divided the scaffolds in the genome into 20,000 bp windows with 10,000 bp overlaps between neighboring windows. We calculated statistics in each window for each specimen (Vernot and Akey 2014). Briefly, for a specimen from taxon A, the loci with SNPs that are dominant (frequency 100%) in taxon B but rare (show up in no more than in two chromosomes) for taxon A were considered, and the set of these loci is designated as . The summary statistic for the specimen is calculated as , where J is any subset of and maximum is found over all such subsets. is calculated as
where and are loci in that are nearest to each other in the genome, and is the sum of genotype distances over all specimens of taxon A. Genotype distance between loci and for a specimen is defined exactly as in the supplementary material of Vernot and Akey (2014). In the calculation of S(J), only the loci in perfect linkage disequilibrium (genotype distance is 0) are rewarded. Compared to the previously described method, we lowered the genotype distance cutoff (from 5 to 1) for giving infinite penalty, because of the smaller sample size (3 specimens vs. 20 specimens in previous study [Vernot and Akey 2014]).
In order to evaluate the significance of values, we used the ms program (Hudson 2002) to generate genetic variation samples under the null hypothesis, that is, no migration (introgression) between the two taxa. Simulation of these samples was done under constrains of a set of parameters derived from the real data, including a demographic model, population mutation, and recombination rates. The mutation rate for each taxon could vary between different genomic regions. Thus, they are directly estimated from the 20,000 bp windows used to compute using the Watterson estimator: , where is the number of sites with SNPs in the sample, is the number of sampled haplotypes, and .
The demographic parameters, including the effective population sizes and the split time of the two taxa, were deduced based on 400 randomly selected 500 bp segments in the genome using the isolation with migration model (Hey and Nielsen 2007). We required the selected segments to satisfy the following criteria: 1) the sequences for this region were available in all specimens and 2) there was no significant sign of recombination detected by PhiPack (Bruen et al. 2006). We randomly divided these segments into 20 data sets and applied IMa2 (Hey and Nielsen 2007) to each data set. In the simulation, we assumed the mutation rate per base per generation to be 2.9e-9, which is the experimentally determined value for Heliconius and is similar to the rate for Drosophila (Keightley et al. 2015).
The average split time for the 20 data sets, 1.2 million generations ago, was used in the simulation of genetic variation samples. The per generation recombination rate of insects varies broadly (Wilfert et al. 2007), and thus we assumed a large range for this parameter: from 1 to 6 cM/Mb. In a 20,000-bp segment, the recombination rate between the two ends (r) is 0.0002–0.0012. The estimated effective population size () from IMa2 varies from 700,000 to 8,800,000 based on different data sets. Therefore, the recombination rate parameter for the ms program (Hudson 2002), which is defined as should range from 560 to 42,240.
For each 20,000 bp window, we obtained 10,000 simulated samples assuming 1) no introgression, 2) mutation rates as calculated based on the real data, 3) split time determined as above, and 4) a grid of population recombination rate covering all expected values of this parameter. We calculated the for the simulated data in a similar way and thus obtained the distribution of under the null hypothesis. We assigned the P value for introgression in each window as P = n/10,000, where n is the number of simulated samples with higher To control the number of false discoveries resulting from a large number of statistical tests, we applied the FDR test (Storey and Tibshirani 2003) and assigned a Q value (proportion of false discoveries among significant tests) to each window. Genome windows with Q values smaller than 0.05 were considered to represent introgression. Genes mostly (> 50% of the total length of that gene) located in the introgressed regions were extracted, and the functional relevance of these introgressed genes was studied using the GO-term analysis that was applied to the “divergence hotspots” as described above.
Results and Discussion
Genome Quality Assessment and Gene Annotation of the Reference Genome
We assembled a 406-Mb reference genome of P. sennae (Pse) and compared its quality and composition (table 1) with genomes of the following Lepidoptera species (supplementary table S1A, Supplementary Material online): Pl. xylostella (Pxy), B. mori (Bmo), Manduca sexta (Mse), L. accius (Lac), Pt. glaucus (Pgl), Papilio polytes (Ppo), Papilio xuthus (Pxu), Melitaea cinxia (Mci), H. melpomene (Hme), and D. plexippus (Dpl) (International Silkworm Genome Consortium 2008; Duan et al. 2010; Zhan et al. 2011; Heliconius Genome 2012; You et al. 2013; Zhan and Reppert 2013; Ahola et al. 2014; Tang et al. 2014; Cong et al. 2015a, 2015b; Nishikawa et al. 2015). The scaffold N50 of Pse genome assembly is 257 kb. The genome assembly is better than many other Lepidoptera genomes in terms of completeness measured by the presence of Core Eukaryotic Genes Mapping Approach (CEGMA) genes (Parra et al. 2007), cytoplasmic ribosomal proteins, and independently assembled transcripts. The average coverage (87.4%) of CEGMA genes (supplementary table S1B, Supplementary Material online) by single Pse scaffolds is comparable to the coverage by the current Bmo assembly with an N50 of about 4.0 Mb, indicating that the quality of the Pse draft is sufficient for protein annotation and comparative analysis. The genome sequence has been deposited at DDBJ/EMBL/GenBank under the accession LQNK00000000. The version described in this article is version LQNK01000000. In addition, the main results from genome assembly, annotation, and analysis can be downloaded at http://prodata.swmed.edu/LepDB/, last accessed March 11, 2016.
Table 1.
Quality and Composition of Lepidoptera Genomes
| Feature | Pgl | Ppo | Pxu | Dpl | Hme | Mci | Lac | Bmo | Mse | Pxy | Pse |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Size w/o gap (Mb) | 361 | 218 | 238 | 242 | 270 | 361 | 290 | 432 | 400 | 387 | 347 |
| GC content (%) | 35.4 | 34.0 | 33.8 | 31.6 | 32.8 | 32.6 | 34.4 | 37.7 | 35.3 | 38.3 | 39.0 |
| Repeat (%) | 22.2 | NA | NA | 16.3 | 24.9 | 28.0 | 15.5 | 44.1 | 24.9 | 34.0 | 17.2 |
| Exon (%) | 5.11 | 7.79 | 8.59 | 8.41 | 6.19 | 4.34 | 7.24 | 4.07 | 5.34 | 6.47 | 6.20 |
| Intron (%) | 24.8 | 51.6 | 45.5 | 26.6 | 24.1 | 31.2 | 32.3 | 16.1 | 38.3 | 31.3 | 25.5 |
| Genome size (Mb) | 375 | 227 | 244 | 249 | 274 | 390 | 298 | 481 | 419 | 394 | 406 |
| Heterozygosity (%) | 2.3 | NA | NA | 0.55 | NA | NA | 1.5 | NA | NA | ∼2a | 1.2 |
| Scaffold N50 (kb) | 231 | 3,672 | 6,199 | 207 (716) | 194 | 119 | 525 | 27(3,999) | 664 | 734 | 257 |
| CEGMA (%) | 99.6 | 99.3 | 99.6 | 99.6 | 98.2 | 98.9 | 99.6 | 99.6 | 99.8 | 98.7 | 99.3 |
| CEGMA coverage by single scaffold (%) | 86.9 | 85.8 | 88.8 | 87.4 | 86.5 | 79.2 | 86.8 | 86.8 | 86.4 | 84.1 | 87.4 |
| Ribosomal Proteins (%) | 98.9 | 98.9 | 97.8 | 98.9 | 94.6 | 94.6 | 98.9 | 98.9 | 98.9 | 93.5 | 98.9 |
| De novo assembled transcripts (%) | 98 | NA | NA | 96 | NA | 97 | 97∼99 | 98 | NA | 83 | 97 |
| number of proteins (k) | 15.7 | 12.3 | 13.1 | 15.1 | 12.8 | 16.7 | 17.4 | 14.3 | 15.6 | 18.1 | 16.5 |
Note.—NA, data not available.
aEstimated from k-mer frequency histogram.
We assembled the transcriptome of P. sennae from the same specimen. We predicted 16,493 protein-coding genes in the P. sennae genome (supplementary table S2C, Supplementary Material online). Sixty-seven percent of these genes are likely expressed in the adult, as they overlap with the transcripts. We annotated the putative functions for 12,584 protein-coding genes (supplementary table S2D, Supplementary Material online).
Phylogeny of Lepidoptera
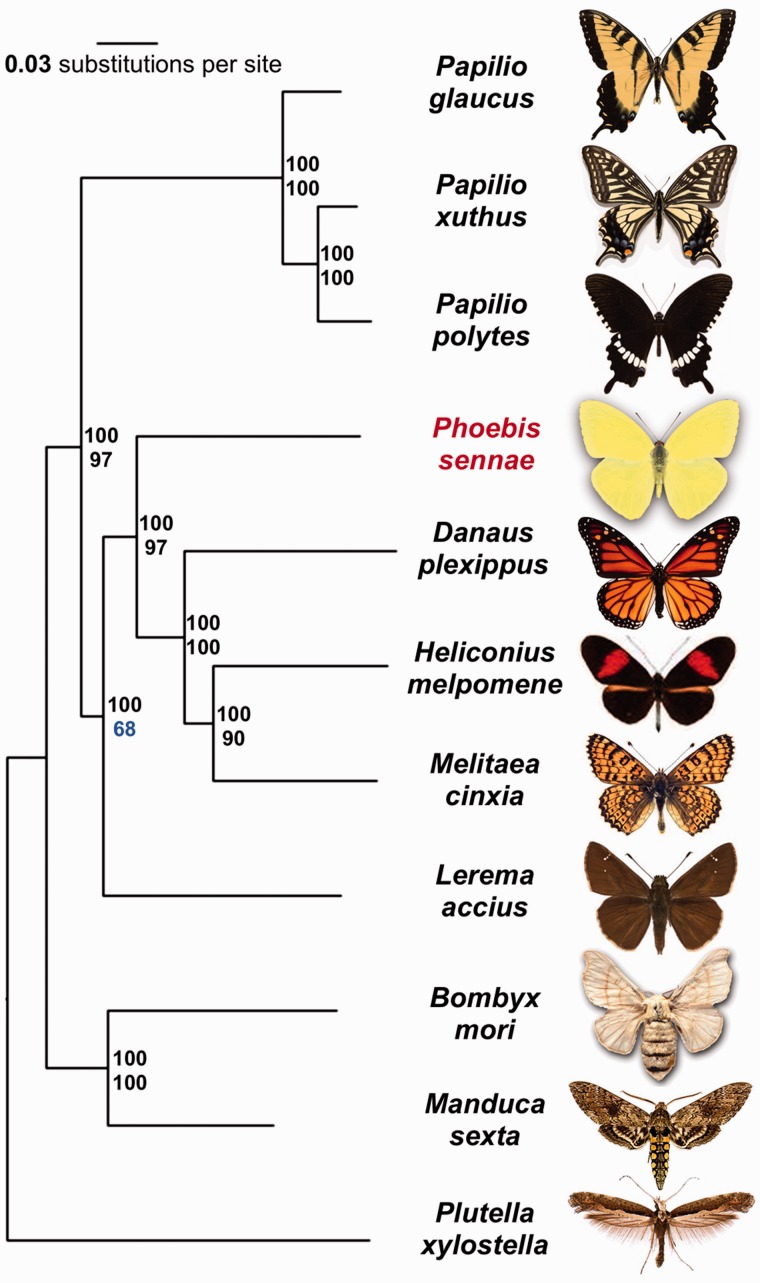
We identified orthologous proteins encoded by 11 Lepidoptera genomes (Pl. xylostella, B. mori, M. sexta, L. accius, Pt. glaucus, Pa. polytes, Pa. xuthus, Me. cinxia, H. melpomene, D. plexippus, and P. sennae) and detected 5,143 universal orthologous groups, of which 2,106 consist of a single-copy gene in each of the species. A phylogenetic tree built on the concatenated alignment of the single-copy orthologous groups using RAxML placed Pheobis as the sister to the Nymphalidae clade (fig. 2). This placement is consistent with the previously published results based on molecular data (Weller et al. 1996; Cong et al. 2015a, 2015b), as expected in the absence of genomes from the families Lycaenidae and Riodinidae.
Fig. 2.—
Phylogenetic tree of the Lepidoptera species with complete genome sequences. Maximal-likelihood tree constructed by RAxML on the concatenated alignment of universal single-copy orthologous proteins. Numbers by the nodes refer to bootstrap support. The numbers above are obtained from random samples of the same size as the complete concatenated alignment, the number below are obtained from random samples of 1% of the data set.
In addition, our analysis placed Papilionidae as a sister to all other butterflies, including skippers (Hesperiidae). Such placement contradicts the traditional view based on morphological studies but is indeed reproduced phylogenetic trees published recently (Kawahara and Breinholt 2014; Cong et al. 2015a, 2015b). All nodes received 100% bootstrap support when the alignment of all the single-copy orthologous groups was used. To find the weakest nodes, we reduced the amount of data by randomly splitting the concatenated alignment into 100 alignments (about 3,670 positions in each alignment). The consensus tree based on these alignments revealed that the node referring to the relative position of skippers and swallowtails has much lower support (68%) compared to all other nodes (above 90%). Thus, the placement of swallowtails and skippers within Lepidoptera tree remains to be investigated further when better taxon sampling by complete genomes is achieved.
Six Genomes of P. sennae
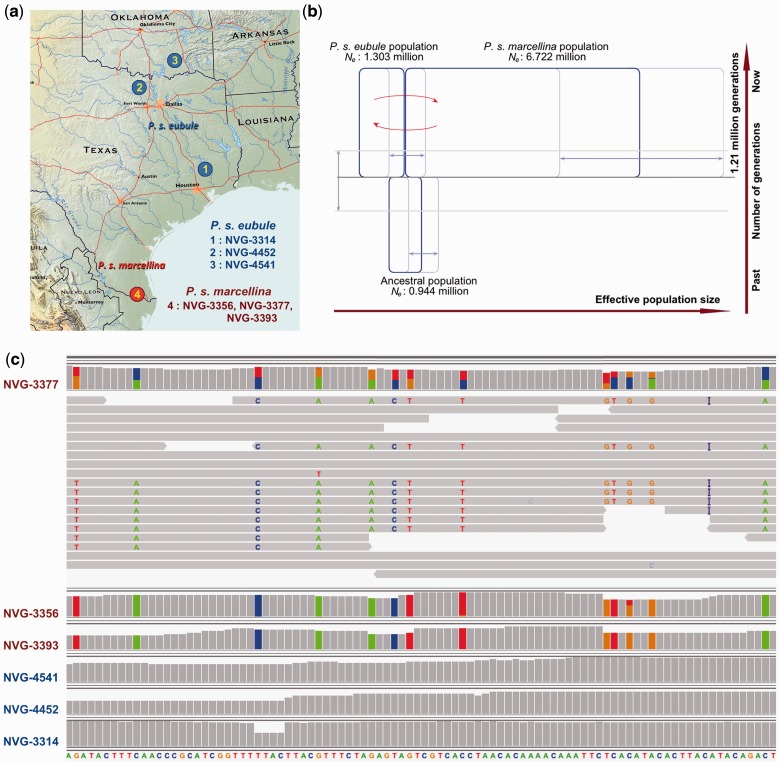
In addition to the reference genome of P. s. eubule from southeast Texas, we sequenced the genomes of five P. sennae specimens and mapped the reads to the reference. Two specimens were P. s. eubule from north-central Texas and southern Oklahoma and three were P. s. marcellina from south Texas (fig. 6a). The coverage by the reads and the completeness of these genomes are summarized in table 2. The sequencing reads for all the specimens are expected to cover the genome 10–12 times, and about 97% of coding regions in the reference genome can be mapped by reads from each specimen. However, fractions of the noncoding region that can be mapped differ significantly (P < 0.001) between specimens. Reads from specimens of the same subspecies as the reference genome can map to 88% of the positions in the reference genome, while reads from the specimens of a different subspecies can map to only 83% of the positions. This indicates a higher divergence in the noncoding region and a substantial difference between the two subspecies in the noncoding region.
Fig. 6.—
Introgression between Phoebis sennae eubule and Phoebis sennae marcellina. (a) Specimens and their locality. The localities where the specimens were collected are marked by dots with numbers inside. The specimens of P. s. eubule are labeled in blue and P. s. marcellina are labeled in red. Map services and data available from US Geological Survey, National Geospatial Program. The USGS home page is http://www.usgs.gov, last accessed March 11, 2016. (b) Evolutionary history for P. s. eubule and P. s. marcellina simulated using the isolation with migration model. The widths of the three blue boxes are correlated with the estimated effective population sizes. The light-blue lines show the confidence intervals (95%). The vertical dimension corresponds to time. The time (1.2 million generations ago) at which the two taxa diverge is marked by the gray solid line and the light-gray lines indicate the 95% confidence interval. The red arrows indicate there is significant (P < 0.001) migration (introgression) between the species. (c) A representative genomic region from the 20,000 bp window with the highest statistic. The figure is a snapshot of the genomic region with mapped reads from all the specimens visualized in Integrative Genomics Viewer (Robinson et al. 2011). The reference genome (NVG-3314) sequence is shown at the bottom. The base composition at each position of each specimen is represented by the bar graph: bases that are the same as in the reference genome are colored in gray and bases that are different from the reference (SNPs) are colored according to the type of the base: A, green; T, red; G, orange; C, blue. For the specimen with significant signs of introgression (NVG-3377), the aligned reads are also shown and each read is represented as a horizontal bar. SNPs in each read are marked using the type of the base in that position. The letter “I” in purple color indicates an insertion compared to the reference. This insertion is present as a homozygous mutation in specimens NVG-3356 and NVG-3393.
Table 2.
Quality of Phoebis Genomes
| Specimen (NVG-) | 3314 | 4452 | 4541 | 3356 | 3377 | 3393 |
|---|---|---|---|---|---|---|
| Coverage | 103 | 11.8 | 10.4 | 12.5 | 12 | 10.8 |
| % Mapped position noncoding region | 99.99 | 87.33 | 87.20 | 82.41 | 82.42 | 81.60 |
| % Mapped position coding region (exon) | 99.98 | 96.78 | 96.47 | 97.41 | 97.33 | 96.92 |
| 100% covered genes | 16,493 | 13,080 | 13,161 | 12,846 | 12,577 | 12,189 |
| 90% covered genes | 16,493 | 14,824 | 14,735 | 14,917 | 14,850 | 14,625 |
| 50% covered genes | 16,493 | 15,987 | 15,897 | 16,146 | 16,158 | 16,067 |
| Heterozygosity | 1.23% | 1.46% | 1.38% | 2.32% | 2.23% | 2.21% |
| Heterozygosity in coding region (exon) | 0.91% | 1.00% | 0.96% | 1.56% | 1.48% | 1.45% |
| Heterozygosity in noncoding region | 1.25% | 1.49% | 1.41% | 2.38% | 2.28% | 2.28% |
We identified SNPs in these genomes compared to the reference genome using GATK (McKenna et al. 2010). There are 1.2% heterozygous positions in the reference genome, and the heterozygosity level (∼1.4%) for the two other P. s. eubule specimens are comparable to the reference genome. The southwestern population shows a higher heterozygosity level (∼2.2%). In all specimens, the overall percentage of heterozygous positions in the exons (0.96 ± 0.02% for each P. s. eubule specimen and 1.50 ± 0.03% for each P. s. marcellina specimen) is lower than that for the noncoding regions (1.38 ± 0.06% for each P. s. eubule specimen and 2.31 ± 0.03% for each P. s. marcellina specimen), which is likely due to the potential deleterious effect of SNPs in the coding regions.
We clustered all six specimens based on their genotype in positions with two possible nucleotides (supplementary fig. S3, Supplementary Material online). The three P. s. eubule specimens formed a tight cluster, indicating high similarity between them. The three P. s. marcellina specimens were more divergent, but they still clustered closer to each other than to the P. s. eubule specimens. In addition, analysis of the same data using fastStructure (Raj et al. 2014) also confirmed this population structure by likelihood calculation: the three P. s. eubule specimens represent one population, while the three P. s. marcellina specimens are from another population (supplementary fig. S4, Supplementary Material online).
Incongruence Between the Divergence in Nuclear and Mitochondrial Genes
COI mitochondrial DNA barcode sequences have been determined for a number of P. sennae specimens across its range (Ratnasingham and Hebert 2007), and they show very little divergence between subspecies. The eastern subspecies in the United States, P. s. eubule, and the southwestern subspecies, P. s. marcellina, differ by only 0.6% (four positions) in their barcode sequences. Barcode differences of 2% and above likely correspond to species-level divergence (Aliabadian et al. 2013). For example, tiger swallowtails Pt. glaucus and Pt. canadensis differ by 2.2% in their barcode sequences. To understand the reasons for apparent morphological and life history differences in the absence of substantial barcode divergence, we compared the nuclear and mitochondrial genomes of all six P. sennae specimens and correlated the results with the complete transcriptome data for Pt. canadensis and Pt. glaucus.
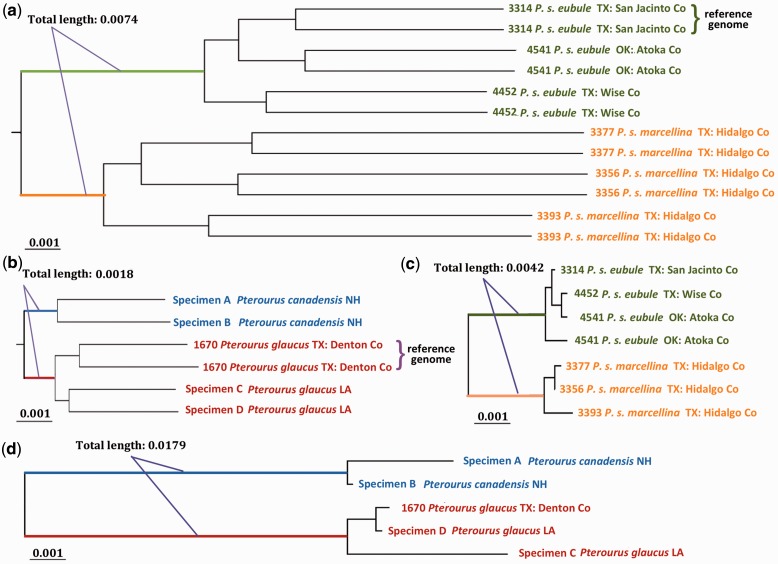
P. s. eubule and P. s. marcellina show low divergence (∼0.5%) not only in the COI barcode but also over all the mitochondrial genes. The mitochondrial genes are very conserved (divergence 0.02 ∼ 0.11%) within each subspecies, and thus the phylogenetic tree based on them clearly separates the two subspecies into clades with branch length between them indicating 0.42% difference (fig. 3c). In contrast, nuclear genes show much higher divergence both within (1.17% for P. s. eubule, 1.78% for P. s. marcellina) and between (1.86%) subspecies. In the phylogenetic tree based on nuclear genes (16,137 genes, 18,877,324 base pairs), the branch length between the two subspecies (branches colored in green and orange in fig. 3a) is 0.7%, twice that of mitochondrial genes.
Fig. 3.—
Incongruence between the speed of evolution for mitochondrial and genomic DNA. Trees were obtained from nuclear (a, b) and mitochondrial (c, d) protein-coding genes of Phoebis (a, c) and Pterourus (b, d). The position of the root in (a) is estimated using Pterourus glaucus as an outgroup in another phylogenetic analysis by RAxML based on the concatenated alignment of single-copy orthologous proteins shared among Ph. sennae and Pt. glaucus. The position of root in (b) is determined using another swallotail genus (Heraclides). Specimen numbers, species names, and localities are labeled in the tree. Specimens with whole-genome sequences are represented by two sequences to reflect the heterozygous positions. Mitochindria of specimen 4541 revealed two distinct types, and therefore, we used two sequences to represent its mitogenome. All trees are shown to scale with the scale bar corrsponding to about 0.1% of sequence divergence. Length of the internal branches that separate the two taxa is measured (approximately) and labeled in the figure.
The higher divergence in nuclear genes compared to mitochondrial genes is unexpected. Mitochondrial DNA usually evolves faster than the nuclear DNA, and thus it is frequently used to resolve relationships of closely related taxa (Brown et al. 1979). Indeed, the divergence level in mitochondrial DNA (∼2.0%) between two Pterourus species is twice that seen in nuclear DNA (∼1.0%). Both nuclear genes (9,622 transcripts, 13,525,930 base pairs) and mitochondrial genes clearly separate the two species in phylogenetic trees, but the internal branch length between the two taxa in the tree based on nuclear DNA (0.18%, fig. 3b) is about 10 times shorter than that for mitochondrial DNA (1.8%, fig. 3d). The clear incongruence between divergence in nuclear and mitochondrial DNA in Phoebis and Pterourus reiterates the need for inclusion of nuclear DNA in phylogenetic studies. Based on the divergence in the nuclear genes (supplementary fig. S5, Supplementary Material online), along with the morphological differences, P. s. eubule and P. s. marcellina may be better treated as two species-level taxa.
We speculate that high nuclear divergence in Phoebis is related to its fast development. While Pt. canadensis breeds only once each year, P. s. marcellina can have up to 15 generations per year. Low divergence in mitochondrial DNA of Phoebis remains a mystery. It might be due to more accurate error-correction machinery during the replication of mitochondrial DNA, keeping the mutation rate very low (Nabholz et al. 2009). Alternatively, a more mundane view is that introgression, population bottlenecks, and mitochondria selective sweeps (Ballard and Whitlock 2004; Bazin et al. 2006; Graham and Wilson 2012; Pons et al. 2014) might result in transfer of mitochondria between taxa or spread of a certain mitochondrial haplotype across all P. sennae populations throughout their vast range.
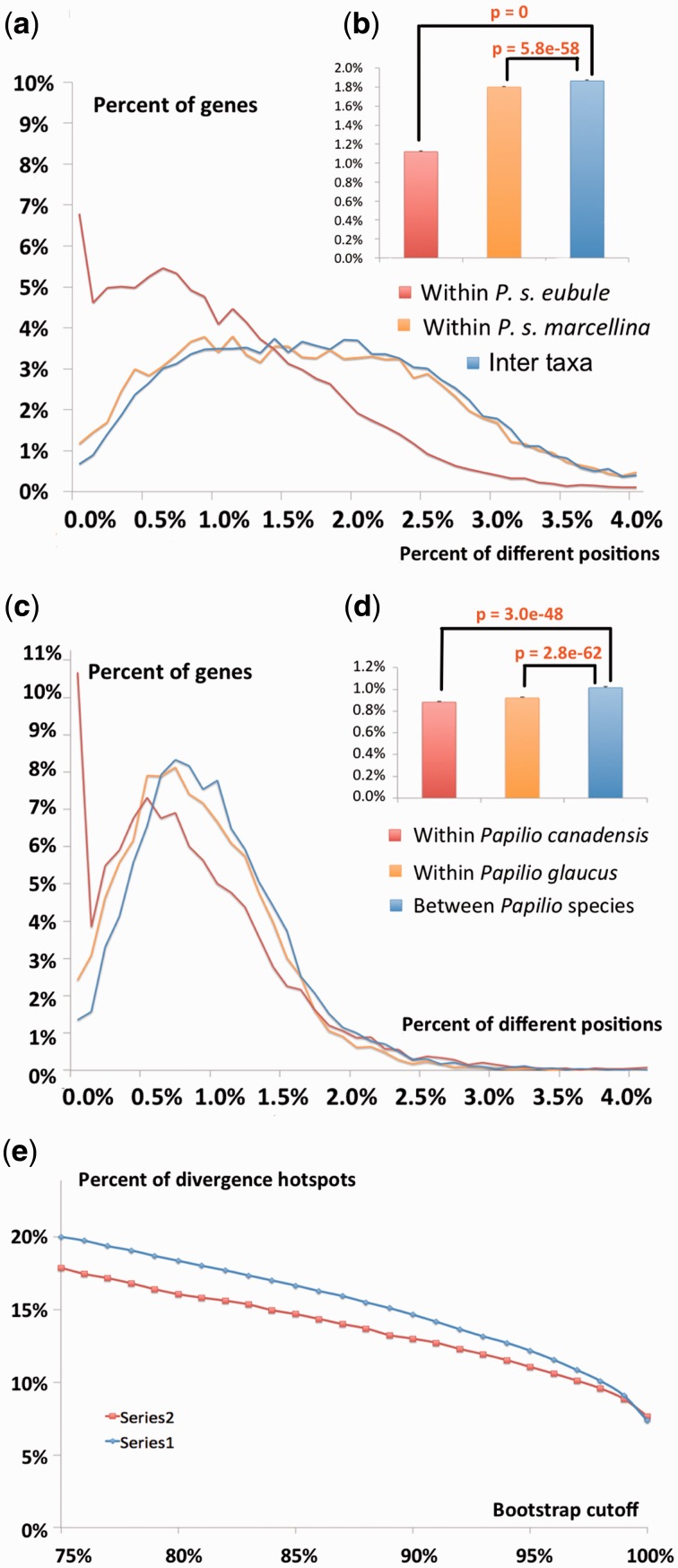
Interestingly, southern taxa of both Phoebis and Pterourus display larger internal divergence than northern taxa (fig. 4). The difference between three specimens of P. s. marcellina (1.80%) collected from the same locality is larger than that between three P. s. eubule specimens (1.12%) collected from different localities that are separated by several hundred miles. The lower sequence variation of P. s. eubule specimens suggests smaller effective population size and possible bottlenecks. Such bottlenecks for northern populations are more likely because Phoebis has low tolerance to subzero temperatures and most individuals do not survive cold winters.
Fig. 4.—
Divergences in protein-coding genes within and between taxa. (a, b) Histograms of divergence in protein coding genes (percent of genes for each level of divergence) between the two Phoebis taxa and the two Pterourus taxa, respectively. (c) Percent of protein-coding genes (vertical axis) that can separate the two taxa in phylogenetic analysis with bootstrap support higher than a certain cutoff (horizontal axis).
Molecular Processes Differentiating P. s. eubule and P. s. marcellina
P. s. eubule and P. s. marcellina can be clearly distinguished based on the whole-genome data. The average inter-taxa divergence for protein coding genes is significantly (P = 5.8e-58) higher than the intra-taxa divergence (fig. 4a and b). However, the two taxa are not diverged in most individual genes, and only 20% of genes can confidently (bootstrap ≥ 75%) distinguish them (fig. 4e). The situation is very similar to that of Pt. glaucus and Pt. canadensis (fig. 4c–e).
To further investigate the possible phenotypic consequences caused by genetic divergence between the two Phoebis taxa, we focused on the genes that can clearly distinguish them both by their sequences and by the proteins they encode (i.e., separate the two taxa into clades with bootstrap support no less than 75%). We identified 924 (5.7%) such proteins (supplementary table S3A, Supplementary Material online), but they were significantly enriched (P = 4.6e-24) in nonconserved proteins within each taxon. Out of 710 nonconserved proteins, 314 are enzymes (supplementary table S3B, Supplementary Material online). The functional sites of enzymes are constrained to several catalytically important residues, and therefore the rest of their sequence is likely to be more tolerant to mutations and can undergo faster divergence.
In contrast, the remaining 214 proteins are conserved within each taxon but can clearly distinguish the two taxa (supplementary table S3C, Supplementary Material online). We term these divergence hotspots. Such proteins are candidate loci for Dobzhansky–Muller (DM) hybrid incompatibility (Orr and Turelli 2001) between the two taxa, as the proteins from P. s. eubule may not work well with proteins and genetic materials from P. s. marcellina when functioning together. GO-term analysis (supplementary table S3D, Supplementary Material online) of these divergence hotspots revealed a prevalence of epigenetic mechanisms including histone modification enzymes and chromatin organization (table 3). Variations in epigenetics-related proteins might be an easy source of hybrid incompatibility because these proteins directly interact with the genetic materials, especially the noncoding regions that could evolve rapidly (Sawamura 2012). Epigenetic variation has been shown to be a speciation mechanism in several organisms (Mihola et al. 2009; Durand et al. 2012). Among the genomic regions covered in the mapping results of all six specimens, the noncoding region differs by 3.5% between the two taxa, while the coding region differs by only 1.8%. The actual divergence in the noncoding region should be even larger as the most divergent regions would fail to map to the reference genome (discussed above). Therefore, proteins involved in epigenetic mechanisms from one taxon may not be compatible with the binding sites in the DNA of another taxon, resulting in lower fitness of the hybrids.
Table 3.
Enriched GO Terms for the Divergent Hotspots that are Conserved within Each Subspecies
| GO Term | P | Category | Annotation of the GO Term | Summary |
|---|---|---|---|---|
| GO:0051574 | 7.0E-05 | BP | Positive regulation of histone H3-K9 methylation | Histone methylation and chromatin associated proteins |
| GO:0051570 | 2.8E-04 | BP | Regulation of histone H3-K9 methylation | |
| GO:1900112 | 1.0E-03 | BP | Regulation of histone H3-K9 trimethylation | |
| GO:1900114 | 1.0E-03 | BP | Positive regulation of histone H3-K9 trimethylation | |
| GO:0031062 | 2.0E-03 | BP | Positive regulation of histone methylation | |
| GO:0051571 | 8.7E-03 | BP | Positive regulation of histone H3-K4 methylation | |
| GO:0006325 | 2.6E-03 | BP | Chromatin organization | |
| GO:0042393 | 8.3E-03 | MF | Histone binding | |
| GO:0000791 | 8.6E-03 | CC | Euchromatin | |
| GO:0031519 | 3.0E-03 | CC | PcG protein complex | |
| GO:0044666 | 3.8E-03 | CC | MLL3/4 complex | |
| GO:0035097 | 2.5E-03 | CC | Histone methyltransferase complex | |
| GO:0034708 | 3.5E-03 | CC | Methyltransferase complex | |
| GO:0008607 | 5.4E-03 | MF | phosphorylase kinase regulator activity | |
| GO:2000044 | 5.4E-03 | BP | Negative regulation of cardiac cell fate specification | Early development and cell fate specification |
| GO:2000043 | 8.7E-03 | BP | Regulation of cardiac cell fate specification | |
| GO:0045611 | 6.9E-03 | BP | Negative regulation of hemocyte differentiation | |
| GO:0009997 | 5.4E-03 | BP | Negative regulation of cardioblast cell fate specification | |
| GO:0042686 | 8.7E-03 | BP | Regulation of cardioblast cell fate specification | |
| GO:0061351 | 8.7E-03 | BP | Neural precursor cell proliferation | |
| GO:0045177 | 3.8E-03 | CC | Apical part of cell | |
| GO:0008158 | 8.7E-03 | MF | Hedgehog receptor activity | |
| GO:0090102 | 5.4E-03 | BP | Cochlea development | |
| GO:0042745 | 7.2E-03 | BP | Circadian sleep/wake cycle | Circadian clock |
| GO:0022410 | 6.3E-03 | BP | Circadian sleep/wake cycle process | |
| GO:0050802 | 6.9E-03 | BP | Circadian sleep/wake cycle, sleep | |
| GO:0016469 | 9.5E-03 | CC | Proton-transporting two-sector ATPase complex | Transporter |
| GO:0015399 | 5.9E-03 | MF | Primary active transmembrane transporter activity | |
| GO:0015405 | 5.9E-03 | MF | P-P-bond-hydrolysis-driven transmembrane transporter | |
| GO:0015078 | 6.3E-03 | MF | Hydrogen ion transmembrane transporter activity | |
| GO:0044283 | 5.9E-03 | BP | Small molecule biosynthetic process | Metabolic |
| GO:0016053 | 6.1E-03 | BP | Organic acid biosynthetic process | |
| GO:0046394 | 6.1E-03 | BP | Carboxylic acid biosynthetic process | |
| GO:0008206 | 8.7E-03 | BP | Bile acid metabolic process | |
| GO:0009314 | 6.8E-03 | BP | Response to radiation | Adaptation to sunlight |
| GO:0034644 | 2.0E-03 | BP | Cellular response to UV | |
| GO:0019233 | 6.9E-03 | BP | Sensory perception of pain | Other |
| GO:0005606 | 8.7E-03 | CC | Laminin-1 complex | |
| GO:0043256 | 8.7E-03 | CC | Laminin complex | |
| GO:0009881 | 7.0E-03 | MF | Photoreceptor activity |
BP: biological process; CC: cellular components; MF: molecular function.
Another group of significantly enriched GO terms is related to the circadian sleep/wake cycle (table 3). The divergence hotspots for the two Pterourus species are also enriched in circadian clock related proteins, and in particular, those related to eclosion rhythm (supplementary table S3E, Supplementary Material online). This is consistent with their observed phenotypic divergence in pupal diapause (i.e., the timing of eclosion). The two P. sennae taxa mostly show divergence in proteins that are related to the sleep/wake cycle but not the eclosion rhythm. This might be related to the lack of pupal diapause in P. sennae. However, proteins related to the sleep/wake cycle could have diverged adaptively since the two taxa were partly separated into different latitudes with different levels of sunlight and average temperatures. In addition, proteins associated with early development and cell differentiation are also enriched in the divergence hotspots. Divergence in these proteins may have a profound impact on the morphology and biology of an organism, driving speciation, and adaptation.
Nuclear DNA Markers to Identify P. s. eubule and P. s. marcellina
Eleven out of 13 mitochondrial protein-coding genes can clearly separate P. s. eubule and P. s. marcellina as the maximal intra-taxa divergence is smaller than the minimal inter-taxa divergence. The only two exceptions are the ND4L and ATP8 coding genes, which are identical between the two subspecies. The low divergence in the mitochondrial genes within one taxon could be a result of going through narrower bottlenecks due to their maternal inheritance, and strong selection pressure to function together with the nuclear-encoded proteins and maintain the high efficiency of the mitochondrial electron transport chain.
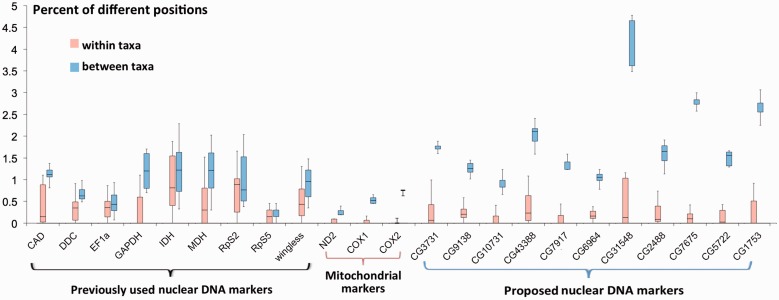
However, the two taxa cannot be clearly identified using the nuclear markers (fig. 5) previously selected for phylogenetic studies of butterflies (Wahlberg and Wheat 2008). This situation is very similar to that of Pt. glaucus and Pt. canadensis (supplementary fig. S6, Supplementary Material online). Out of the 16,137 nuclear genes that are covered by most specimens, only 94 always show higher divergence between P. s. eubule and P. s. marcellina than within either taxon (supplementary table S4, Supplementary Material online). Eleven of the diverging nuclear genes function in biological processes such as histone modification and circadian sleep/wake cycle (discussed in divergent hotspots above). We suggest them as potential nuclear markers (fig. 5) to identify the two taxa, because they may contribute to the reproductive barrier and their exchange between the two taxa may be limited. For example, two of them are related to chromatin remodeling, and they are orthologous to the Drosophila genes Grunge (CG6964) and Nucleoplasmin (CG7917), respectively. Both proteins directly interact with the chromatin and could contribute to a certain level of reproductive isolation as they may not interact well with the genetic material of a different taxon.
Fig. 5.—
Divergence of selected genes (markers) within (red) and between (blue) Phoebis taxa. Nuclear genes commonly used in phylogenetic analysis of Lepidoptera (General nuclear markers) are shown on the left; commonly used mitochondrial markers are shown in the middle; examples of nuclear genes that can unambigously discrimate Phoebis sennae eubule and Phoebis sennae marcellina are shown on the right (specific nuclear markers). These markers are labeled by the flybase IDs of their orthologs in Drosophila, and their IDs in the P. s. eubule gene set and function are as follows. CG3731: pse1226.10, mitochondrial-processing peptidase subunit beta; CG9138: pse132.8, regulator of tracheal tube development; CG10731: pse1425.14, mitochondrial ATP synthase subunit s; CG43388: pse35.12, voltage-gated potassium channel; CG9717: pse730.7, nucleoplasmin; CG6964: pse9575.4, transcriptional repressor; CG31548: pse1095.3, 3-oxoacyl-[acyl-carrier-protein] reductase FabG; CG2488: pse1216.5, cryptochrome-1; CG7675: pse1218.21, retinol dehydrogenase 11; CG5722: pse243.10, Niemann-Pick C1 protein; CG1753: pse42.13, bifunctional L-3-cyanoalanine synthase.
Evolutionary History and Introgression between P. s. eubule and P. s. marcellina
We built isolation-with-migration models (Hey and Nielsen 2007) (supplementary table S5A, Supplementary Material online) for P. s. eubule and P. s. marcellina using genomic regions without any significant sign of recombination. The estimated split time and effective population sizes of the two taxa deduced from these simulations are shown in figure 6b. The two taxa split approximately 1.2 million generations ago. The effective population size of P. s. marcellina is five times larger than that of P. s. eubule, which is consistent with the higher heterozygosity observed in the former. The models detect statistically significant (P < 0.001) migration, that is, introgression, between the two populations in both directions.
In contrast to the rare ancient alleles inherited from the ancestral population or originated by random mutations, recently introgressed alleles tend to show significant linkage disequilibrium (Racimo et al. 2015). Based on this idea, an statistic was proposed to identify genetic regions introgressed from archaic to modern humans (Plagnol and Wall 2006; Vernot and Akey 2014). Using a similar method (see Materials and Methods for details), we found statistical support (P < 0.0014 and FDR < 0.05) for introgression in all specimens: 4–6% of each P. s. eubule genome may have been introgressed from P. s. marcellina, and 1–2% of each P. s. marcellina genome likely represents gene flow from P. s. eubule (supplementary table S5B, Supplementary Material online).
A representative genomic region from the 20,000 bp window with the highest statistic is shown in figure 6c. In this region, the three P. s. eubule specimens (NVG-3314, -4452, and -4541) are identical (every position is a gray bar). Two P. s. marcellina specimens (NVG-3356 and -3393) have a different, but also homozygous, allele containing several SNPs (marked by the colored bars) compared to the reference (NVG-3314). However, the third P. s. marcellina specimen (NVG-3377) is heterozygous (two-toned bars) with alleles from both taxa present. Notably, sequencing reads (gray horizontal bars) mapped to this region show that all the P. s. eubule-type SNPs are linked on the same chromosome (horizontal bars without letters). The linkage and high density (14% base pair difference) of P. s. eubule-type SNPs in the P. s. marcellina specimen suggest recent introgression of this allele, because the P. s. eubule-type SNPs are expected to be randomly distributed between both parental and maternal chromosomes if they were rare SNPs native to the P. s. marcellina population.
We investigated functions of the genes located in the introgressed regions using GO terms. This analysis revealed a significant enrichment in transporters for introgression in both directions: from P. s. eubule to P. s. marcellina (P = 2.0e-4, supplementary table S5C, Supplementary Material online) and from P. s. marcellina to P. s. eubule (P = 7.3e-8, supplementary table S5D, Supplementary Material online). Transporters frequently function by themselves or with closely linked genes (Kihara and Kanehisa 2000; Boll et al. 2003). They may remain fully active after introduction into a different genetic background because they function relatively independently from other proteins and are not likely to cause DM hybrid incompatibility. Moreover, a number of introgressed transporters are responsible for the uptake of nutrients and removal of toxins (supplementary table S5E, Supplementary Material online) and may convey selective advantage by diversifying the gene pool and enabling caterpillars to broaden the food source.
Should P. s. eubule and P. s. marcellina be Treated as Species-Level Taxa?
Comparative analysis of complete genomes of six P. sennae specimens revealed an unexpectedly large divergence between subspecies P. s. eubule and P. s. marcellina in nuclear genes compared to that of mitochondrial genes. This divergence appears more prominent than that between the two swallowtail species Pt. canadensis and Pt. glaucus. The two Phoebis subspecies show significant divergence in epigenetic mechanisms, regulation of the sleep/wake cycle and early development. Multiple proteins participating in each of these processes show clear divergence between the two taxa. It is possible that protein from one taxon may show reduced compatibility with a partner from another taxon, leading to DM hybrid incompatibility.
In addition, inspection of large holdings of P. sennae specimens in the McGuire Center for Lepidoptera and Biodiversity collection shows that both Phoebis subspecies occur in Texas and their ranges partly overlap in central Texas around Austin and San Antonio, where specimens of both subspecies can be found, and P. s. marcellina can stray north into Oklahoma. It is apparent that these butterflies are strong flyers and are known to migrate (Walker 2001). A single individual can fly a hundred miles or more, so there should be ample opportunities for the two taxa to mix. Nevertheless, they remain morphologically identifiable (McGuire Center for Lepidoptera and Biodiversity collection materials) and genetically distinct, which indicates a certain level of reproductive isolation and thus possible genetic incompatibilities. Taken together, the profound genomic divergence, morphological differences, and maintenance of distinctness between eastern and southern populations in Texas, we suggest that it is more meaningful to treat both P. s. eubule and P. s. marcellina as species-level taxa. However, the relationship of each to nominotypical P. sennae sennae from the Caribbean islands remains to be elucidated.
Conclusions
We report six genomes of the Cloudless Sulphur, three of P. s. eubule and three of P. s. marcellina. Being the first sequenced genomes from the family Pieridae, they offer a rich data set for comparative genomics and phylogenetic studies of Lepidoptera. Comparative analyses of Phoebis genomes and Pterourus transcriptomes reveal a remarkable incongruence between relative rates of nuclear and mitochondrial divergence. Phoebis species show low mitochondrial divergence (0.5%) and high nuclear divergence (1.8%). The situation is reversed in Pterourus species. Phoebis s. marcellina and P. s. eubule differ from each other in histone methylation regulators, chromatin associated proteins, circadian clock, and some early developmental proteins. The divergence in these processes, taken together with the unexpectedly high divergence in nuclear genes, suggests a certain level of reproductive isolation between the two taxa, and both P. s. eubule and P. s. marcellina are best treated as species-level taxa.
Supplementary Data
Supplementary tables S1–S5 and figures S1–S6 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).See the Supplemental Information for detailed protocols. Major in-house scripts and intermediate results are available at http://prodata.swmed.edu/LepDB/, last accessed March 11, 2016.
Authors’ Contributions
Q.C. designed the experiments, performed the computational analyses and drafted the manuscript; J.S. carried out the experiments; A.D.W. conceived the project; D.B. and Z.O. designed and supervised experimental studies; N.V.G. directed the project and drafted the manuscript. All authors wrote the manuscript.
Supplementary Material
Acknowledgments
We acknowledge Texas Parks and Wildlife Department (Natural Resources Program Director David H. Riskind) for the permit no. 08-02Rev that makes research based on material collected in Texas State Parks possible. We thank Lisa N. Kinch, R. Dustin Schaeffer, and Raquel Bromberg for critical suggestions and proofreading of the manuscript. Qian Cong is a Howard Hughes Medical Institute International Student Research fellow. This work was supported in part by the National Institutes of Health (GM094575 to N.V.G.) and the Welch Foundation (I-1505 to N.V.G.).
Literature Cited
- Ahola V, et al. 2014. The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in Lepidoptera. Nat Commun. 5:4737.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliabadian M, et al. 2013. DNA barcoding of Dutch birds. Zookeys 365:25–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Asher J, et al. 2001.The millennium atlas of butterflies in Britain and Ireland. Oxford University Press. Oxford, UK. 433 pp.
- Ballard JW, Whitlock MC. 2004. The incomplete natural history of mitochondria. Mol Ecol. 13(4):729–744. [DOI] [PubMed] [Google Scholar]
- Bazin E, Glemin S, Galtier N. 2006. Population size does not influence mitochondrial genetic diversity in animals. Science 312(5773):570–572. [DOI] [PubMed] [Google Scholar]
- Besemer J, Borodovsky M. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33(Web Server issue):W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll M, et al. 2003. A cluster of proton/amino acid transporter genes in the human and mouse genomes. Genomics 82(1):47–56. [DOI] [PubMed] [Google Scholar]
- Brown FM. 1929. A revision of the genus Phoebis (Lepidoptera). Am Mus Novit. 368:1–22. [Google Scholar]
- Brown WM, George M, Jr, Wilson AC. 1979. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 76(4):1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172(4):2665–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, et al. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18(1):188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. Comput Sci Biol Proc Ger Conf Bioinformatics. 99:45–56. [Google Scholar]
- Cong Q, et al. 2015a. Skipper genome sheds light on unique phenotypic traits and phylogeny. BMC Genomics 16:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, et al. 2015b. Tiger swallowtail genome reveals mechanisms for speciation and caterpillar chemical defense. Cell Rep. 10(6):910–919. [DOI] [PMC free article] [PubMed]
- DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, et al. 2010. SilkDB v2.0: a platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res. 38(Database issue):D453–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, et al. 2012. Rapid establishment of genetic incompatibility through natural epigenetic variation. Curr Biol. 22(4):326–331. [DOI] [PubMed] [Google Scholar]
- Ehrlich PR. 1958. The comparative morphology, phylogeny and higher classification of the butterflies (Lepidoptera: Papilionoidea). Univ Kans Sci Bull. 39:305–370. [Google Scholar]
- Felsenstein J. 1989. PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics. 5:163–166. [Google Scholar]
- Graham RI, Wilson K. 2012. Male-killing Wolbachia and mitochondrial selective sweep in a migratory African insect. BMC Evol Biol. 12:204.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2008. Automated eukaryotic gene structure annotation using evidence modeler and the program to assemble spliced alignments. Genome Biol. 9(1):R7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao JJ, et al. 2014. The complete mitochondrial genomes of the Fenton's wood white, Leptidea morsei, and the lemon emigrant, Catopsilia pomona. J Insect Sci. 14:130.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heliconius Genome, C. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487(7405):94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. 2007. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc Natl Acad Sci U S A. 104(8):2785–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. 2002. Generating samples under a wright-fisher neutral model of genetic variation. Bioinformatics 18(2):337–338. [DOI] [PubMed] [Google Scholar]
- International Silkworm Genome Consortium. 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 38(12):1036–1045. [DOI] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, et al. 1996. CENSOR—a program for identification and elimination of repetitive elements from DNA sequences. Comput Chem. 20(1):119–121. [DOI] [PubMed] [Google Scholar]
- Jurka J, et al. 2005. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110(1–4):462–467. [DOI] [PubMed] [Google Scholar]
- Kajitani R, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24(8):1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara AY, Breinholt JW. 2014. Phylogenomics provides strong evidence for relationships of butterflies and moths. Proc Biol Sci. 281(1788):20140970.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, et al. 2015. Estimation of the spontaneous mutation rate in Heliconius melpomene. Mol Biol Evol. 32(1):239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Schatz MC, Salzberg SL. 2010. Quake: quality-aware detection and correction of sequencing errors. Genome Biol. 11(11):R116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara D, Kanehisa M. 2000. Tandem clusters of membrane proteins in complete genome sequences. Genome Res. 10(6):731–743. [DOI] [PubMed] [Google Scholar]
- Kim D, et al. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14(4):R36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5:59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoros WH, Pertea M, Salzberg SL. 2004. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20(16):2878–2879. [DOI] [PubMed] [Google Scholar]
- Marcais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27(6):764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, et al. 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323(5912):373–375. [DOI] [PubMed] [Google Scholar]
- Misra S, et al. 2002. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 3(12):RESEARCH0083.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabholz B, Glemin S, Galtier N. 2009. The erratic mitochondrial clock: variations of mutation rate, not population size, affect mtDNA diversity across birds and mammals. BMC Evol Biol. 9:54.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau NJ, et al. 2014. Population genomics of parallel hybrid zones in the mimetic butterflies, H. melpomene and H. erato. Genome Res. 24(8):1316–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa H, et al. 2015. A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nature Genet. 47(4):405–409. [DOI] [PubMed] [Google Scholar]
- Orr HA, Turelli M. 2001. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution 55(6):1085–1094. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23(9):1061–1067. [DOI] [PubMed] [Google Scholar]
- Pfeiler EJ. 1968. The effect of pterin pigments on wing coloration of four species of Pieridae (Lepidoptera). J Res Lepidoptera. 7(4):183–189. [Google Scholar]
- Plagnol V, Wall JD. 2006. Possible ancestral structure in human populations. PLoS Genet. 2(7):e105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons JM, et al. 2014. Extensive mitochondrial introgression in North American Great Black-backed Gulls (Larus marinus) from the American Herring Gull (Larus smithsonianus) with little nuclear DNA impact. Heredity 112(3):226–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F, et al. 2015. Evidence for archaic adaptive introgression in humans. Nat Rev Genet. 16(6):359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Stephens M, Pritchard JK. 2014. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197(2):573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnasingham S, Hebert PD. 2007. bold: The Barcode of Life Data System (http://www.barcodinglife.org). Mol Ecol Notes. 7(3):355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, et al. 2011. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27(17):2325–2329. [DOI] [PubMed] [Google Scholar]
- Robinson JT, et al. 2011. Integrative genomics viewer. Nat Biotechnol. 29(1):24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K. 2012. Chromatin evolution and molecular drive in speciation. Int J Evol Biol. 2012:301894.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She R, et al. 2011. genBlastG: using BLAST searches to build homologous gene models. Bioinformatics 27(15):2141–2143. [DOI] [PubMed] [Google Scholar]
- Slater GS, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6:31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R. 2008. –2010. RepeatModeler Open-1.0. Available from: http://www.repeatmasker.org
- Smit AFA, Hubley R, Green P. 1996. –2010. RepeatMasker Open-3.0. Available from: http://www.repeatmasker.org.
- St Pierre SE, et al. 2014. FlyBase 102–advanced approaches to interrogating FlyBase. Nucleic Acids Res. 42(Database issue):D780–D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, et al. 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7:62.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 100(16):9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek BE, et al. 2007. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23(10):1282–1288. [DOI] [PubMed] [Google Scholar]
- Tang W, et al. 2014. DBM-DB: the diamondback moth genome database. Database 2014:bat087.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt, C 2014. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 42(Database issue):D191–D198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nieuwerburgh F, et al. 2012. Illumina mate-paired DNA sequencing-library preparation using Cre-Lox recombination. Nucleic Acids Res. 40(3):e24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B, Akey JM. 2014. Resurrecting surviving Neandertal lineages from modern human genomes. Science 343(6174):1017–1021. [DOI] [PubMed] [Google Scholar]
- Wahlberg N, Wheat CW. 2008. Genomic outposts serve the phylogenomic pioneers: designing novel nuclear markers for genomic DNA extractions of lepidoptera. Syst Biol. 57(2):231–242. [DOI] [PubMed] [Google Scholar]
- Walker TJ. 2001. Butterfly migrations in Florida: seasonal patterns and long-term changes. Environ Entomol. 30(6):1052–1060. [Google Scholar]
- Weller SJ, Pashley DP, Martin JA. 1996. Reassessment of butterfly family relationships using independent genes and morphology. Ann Entomol Soc Am. 89(2):184–192. [Google Scholar]
- Wilfert L, Gadau J, Schmid-Hempel P. 2007. Variation in genomic recombination rates among animal taxa and the case of social insects. Heredity 98(4):189–197. [DOI] [PubMed] [Google Scholar]
- Wu Y, et al. 2015. The complete mitochondrial genome of Colias erate (Lepidoptera: pieridae). Mitochondrial DNA Advance Access published May 26, 2015, doi: 10.3109/19401736.2015.1022743. [DOI] [PubMed] [Google Scholar]
- Yang J, et al. 2014. The complete mitochondrial genome of Gonepteryx mahaguru (Lepidoptera: Pieridae). Mitochondrial DNA 27(2):877–8. [DOI] [PubMed] [Google Scholar]
- You M, et al. 2013. A heterozygous moth genome provides insights into herbivory and detoxification. Nature Genet. 45(2):220–225. [DOI] [PubMed] [Google Scholar]
- Zhan S, Reppert SM. 2013. MonarchBase: the monarch butterfly genome database. Nucleic Acids Res. 41(Database issue):D758–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, et al. 2011. The monarch butterfly genome yields insights into long-distance migration. Cell 147(5):1171–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, et al. 2014. The genetics of monarch butterfly migration and warning colouration. Nature 514(7522):317–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.