Summary
Essential gene functions underpin the core reactions required for cell viability, but their contributions and relationships are poorly studied in vivo. Using CRISPR interference, we created knockdowns of every essential gene in Bacillus subtilis and probed their phenotypes. Our high-confidence essential gene network, established using chemical genomics, showed extensive interconnections among distantly related processes and identified modes of action for uncharacterized antibiotics. Importantly, mild knockdown of essential gene functions significantly reduced stationary phase survival without affecting maximal growth rate, suggesting that essential protein levels are set to maximize outgrowth from stationary phase. Finally, high-throughput microscopy indicated that cell morphology is relatively insensitive to mild knockdown but profoundly affected by depletion of gene function, revealing intimate connections between cell growth and shape. Our results provide a framework for systematic investigation of essential gene functions in vivo that is broadly applicable to diverse microorganisms and amenable to comparative analysis.
Introduction
Essential gene functions underpin core cellular processes. Interrogating the relationships among essential gene functions is critical for understanding how bacterial growth is controlled and for facilitating drug development. Yet, few approaches can assess essential gene function in vivo to elucidate their connections. Neither gene-deletion libraries (Baba et al., 2006; Winzeler et al., 1999) nor saturating transposon mutagenesis (Goodman et al., 2009; van Opijnen et al., 2009) can be used to study essential genes, as cells cannot survive without their functions (Christen et al., 2011). Several high-throughput approaches have been used to identify or perturb essential genes in eukaryotes, including destabilizing the 3′ UTR of mRNAs (DaMP alleles; Breslow et al., 2008), CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 gene editing (Blomen et al., 2015; Wang et al., 2015), and CRISPR/dCas9 transcriptional regulation technologies (Gilbert et al., 2014). The only study screening essential genes in bacteria used antisense RNA knockdowns to screen for antibiotic sensitivities (Xu et al., 2010), a method of limited utility due to variable efficacy (Forsyth et al., 2002).
Here we establish a CRISPR interference (CRISPRi) framework for systematic phenotypic analysis of essential genes in bacteria. CRISPRi uses a nuclease-deactivated variant of Streptococcus pyogenes Cas9 (dCas9) paired with a single guide RNA (sgRNA) to sterically hinder transcription at the sgRNA-base pairing genomic locus (Qi et al., 2013) and is a specific and efficient approach for knockdown, with demonstrated applicability in bacteria. We generated a comprehensive essential gene-knockdown library in the Gram-positive model bacterium Bacillus subtilis and used the library to enable drug target discovery, establish a functional network of essential gene processes, characterize how cell morphology and growth rate respond to reductions in essential gene expression, and dissect essentiality in a highly redundant genetic pathway. Our study provides a framework for comprehensive, high-throughput analysis of essential gene functions applicable to diverse bacteria.
Results
CRISPRi is effective, specific, and titratable in B. subtilis
We established a CRISPRi system in B. subtilis, consisting of S. pyogenes dcas9 driven by a xylose-inducible promoter (Pxyl) and sgRNAs expressed from a strong, constitutive promoter (Pveg), both transferred to the chromosome via integrating plasmids (Figure 1A). This system is very efficient, exhibiting 3-fold repression of red fluorescent protein (RFP) without induction and 150-fold repression with full dcas9 induction (Figure 1B, S1A). Although we also report a system with no basal repression based on a weak IPTG-inducible promoter (Figure S1B), we used Pxyl exclusively throughout to consistently sensitize the library with slight knockdown and avoid inconsistencies resulting from slight variations in IPTG induction.
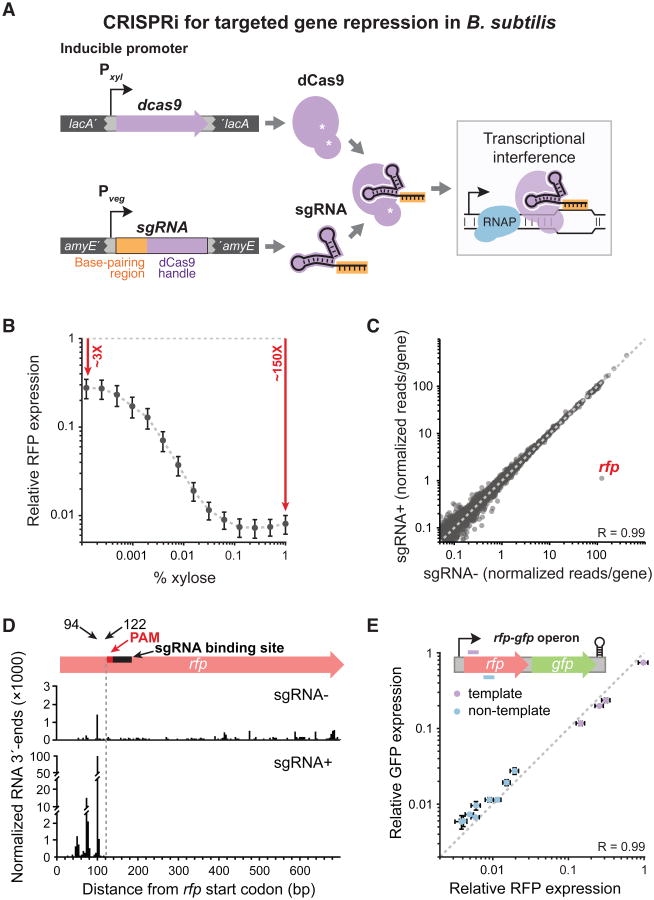
Figure 1. B. subtilis CRISPRi is Efficient, Titratable, and Specific.
A) B. subtilis xylose-inducible dCas9 is directed to specific DNA targets by constitutively expressed sgRNAs, where it represses transcription. dcas9 was stably integrated into the lacA locus, and sgRNAs into amyE or thrC.
B) Flow cytometry of cells that constitutively express rfp and an rfp-targeted sgRNA, in which dcas9 was induced by adding the specified concentration of xylose. Median RFP levels are relative to the no-sgRNA control (grey dashed line), and error bars are ±1 standard deviation.
C) RNA-seq of cells maximally induced for dcas9 (1% xylose) and constitutively expressing rfp and an rfp-targeted sgRNA versus cells without an sgRNA. Reads per gene were normalized by reads per kilobase per million reads. The dashed line is y = x.
D) NET-seq of cells maximally induced for dcas9 (1% xylose) and constitutively expressing rfp and an rfp-targeted sgRNA versus cells without an sgRNA. The y-axis is broken to accommodate the wide range of reads. The dashed line corresponds to the upstream boundary of the PAM sequence. We suggest that RNA 3′ end peaks upstream of the dCas9 block result from RNA polymerase queuing.
E) Flow cytometry of cells maximally induced for dcas9 (1% xylose) and constitutively expressing an rfp-gfp operon and various rfp-targeting sgRNAs. RFP and GFP levels are relative to the no-sgRNA control. The dashed line is y = x. Note that in this group of sgRNAs, only template strand-targeting sgRNAs exhibited low efficacy (violet).
Our Pxyl-based CRISPRi system is titratable, with unimodal rfp repression at the single-cell level across sub-saturating inducer concentrations (Figure 1B, S1C), and high specificity, repressing only rfp expression at saturating inducer concentrations (Figure 1C). We used NETseq to identify the genomic positions of transcribing RNA polymerase (Larson et al., 2014), showing that CRISPRi sterically blocks transcription in B. subtilis (Figure 1D, S1D), as in Escherichia coli (Qi et al., 2013). CRISPRi is polar (Peters et al., 2015), with all downstream genes in an operon showing equivalent knockdown (Figure 1E), and also reduces expression of upstream genes in the operon (Figure S1E). Thus, CRISPRi technology is suitable for examining gene function at the operon level.
A CRISPRi knockdown library of essential genes
We constructed an arrayed library of B. subtilis strains expressing computationally optimized sgRNAs (Extended Experimental Procedures) targeting the 289 known or proposed essential genes (Table S1). The sgRNAs targeted unique DNA sequences at the 5′ ends of genes, where CRISPRi is most effective (Qi et al., 2013). Nearly all sgRNAs (∼94%) targeting bona fide essential genes (258 genes total, Extended Experimental Procedures; Koo et al., in preparation) decreased colony size on agar plates with xylose (≥25% reduction in area compared to the control; Table S1; Extended Experimental Procedures). Control cells expressing only dcas9, or dcas9 and an sgRNA targeting an innocuous gene (rfp), had no growth defects (Figure S1F). We conclude that a single sgRNA is sufficient for effective knockdown of essential genes, simplifying CRISPRi library design in bacteria.
CRISPRi-based essential gene phenotyping and drug target discovery
The ∼3-fold repression of our knockdown library without induction (basal repression; Figure 1B, S1A) sensitized strains to various chemicals. This enabled us to define essential gene phenotypes via chemical-genomic analysis by measuring colony size against 35 unique compounds (Extended Experimental Procedures). We achieved high reproducibility, as measured by correlated colony sizes (R = 0.89, Figure S2A). We converted colony sizes to chemical-gene scores (Figure S2A-D; Table S2; Nichols et al., 2011), and identified significant chemical-gene phenotypes (false discovery rate ≤5%; Nichols et al., 2011). Most knockdowns of antibiotic targets were hypersensitized to their cognate drug (e.g. dfrA/folate biosynthesis to trimethoprim (Myoda et al., 1984), fabF/fatty acid metabolism to cerulenin (Moche et al., 1999); Table S2). Although fabI, the target of triclosan (Schujman et al., 2001), is not essential due to a gene duplication (Thomaides et al., 2007), another knockdown in the pathway (fabG) was sensitized (Table S2). We conclude that our CRISPRi platform effectively identifies known drug-gene interactions.
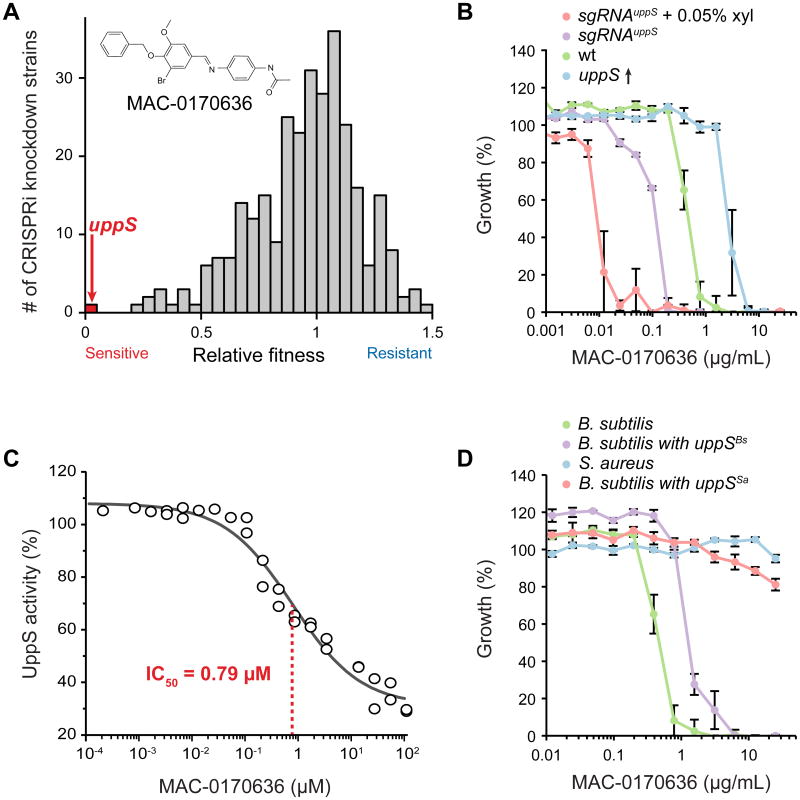
We tested our essential knockdown library as a platform for drug target discovery by screening against MAC-0170636, an antibiotic that upregulates the cell wall-damage responsive promoter PywaC (Czarny et al., 2014) by an unknown mechanism. Undecaprenyl pyrophosphate synthetase (uppS) was the most sensitized knockdown (Figure 2A; Table S2), and we confirmed its sensitivity in liquid (Figure 2B). Conversely, uppS overexpression increased MAC-0170636 resistance relative to wild-type (WT) cells (Figure 2B). Purified B. subtilis UppS activity was inhibited by MAC-0170636 with an IC50 of 0.79 μM (Figure 2C), indicating that UppS is the direct target of MAC-0170636. UppS purified from another Firmicute, Staphylococcus aureus, was completely resistant to MAC-017063 (Figure S2E), as was S. aureus itself. Likewise, B. subtilis expressing only S. aureus uppS was resistant (Figure 2D). These results highlight the utility of our knockdown library for identifying direct targets of uncharacterized compounds; CRISPRi portability suggests its utility in future organism-specific drug-discovery efforts.
Figure 2. CRISPRi Knockdowns of Essential Genes Enable Discovery of Direct Antibiotic Targets.
A) Relative fitness of CRISPRi essential gene knockdown strains (n = 289) with basal dcas9 expression (no xylose induction) grown on plates containing MAC-0170636, as determined by the ratio of normalized colony sizes on LB plates + DMSO versus LB + MAC-0170636.
B) Minimal inhibitory concentration (MIC) assay for strains over- or under-expressing uppS grown in liquid medium containing MAC-0170636. The MICs of these strains were (in μg/ml): sgRNAuppS + 0.05% xyl (∼0.012), sgRNAuppS (∼0.195), WT (∼0.78), and uppS overexpression (∼3.125). The values plotted are the means of at least three measurements, and error bars are ±1 standard deviation.
C) Concentration-dependent inhibition of purified B. subtilis UppS by MAC-0170636. Each point is an individual measurement.
D) MIC assay for strains expressing B. subtilis uppS (BsΔuppS/amyE∷Pspank-uppSBs) or S. aureus uppS (BsΔuppS/amyE∷Pspank-uppSSa) grown in liquid LB + MAC-0170636.
Functional analysis of the essential gene network
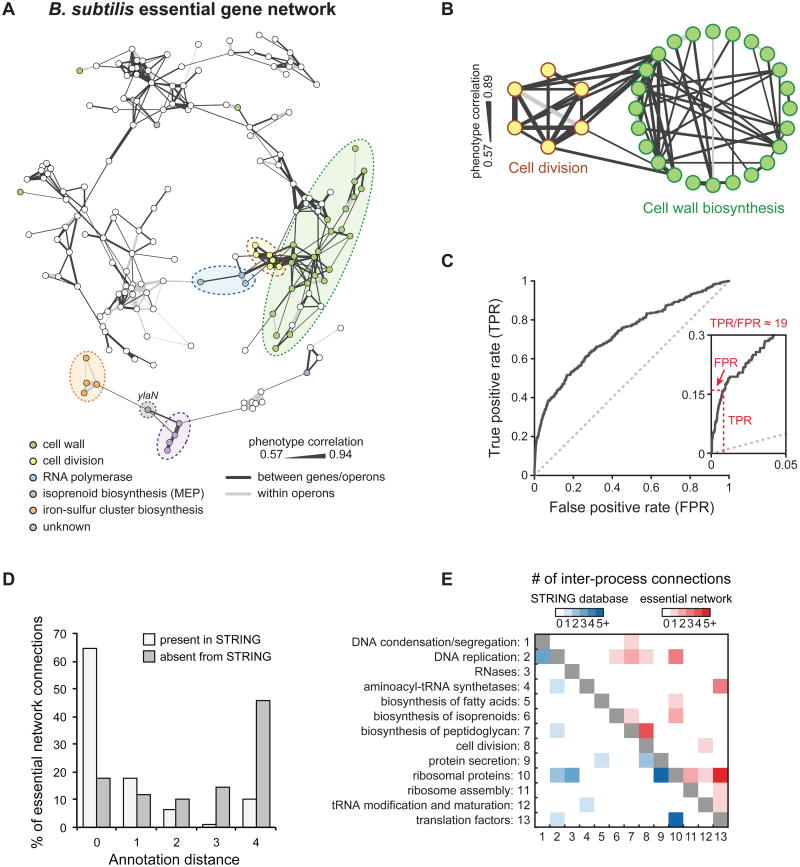
Highly correlated responses of gene knockdowns across chemical conditions (phenotypic signatures) indicate functional connections (Nichols et al., 2011). We established a network of gene-gene connections using statistically significant correlations among the phenotypic signatures of our essential gene knockdowns, based on both direct or indirect effects of drug-gene interactions (Figure 3A, S2B-D, S3; Table S3; Extended Experimental Procedures). The network was rich in known biological connections among genes in related processes such as cell-wall biosynthesis and cell division (Figure 3B).
Figure 3. An Essential Gene Network Reveals Numerous Intra- and Inter-process Connections.
A) Essential gene network based on correlations between chemical-gene phenotypes. Edge thickness is proportional to the extent of correlation. See also Figure S3 for network gene names.
B) Intra- and inter-process connections between cell division and cell wall-biosynthesis genes. Genes outside the main network or genes lacking intra-process connections were excluded.
C) ROC curve comparing connections between essential operons in our network to the STRING database. True-positive connections are those present in the high-confidence set of interactions from STRING, and false-positive connections are absent from STRING (these connections may be either truly false or novel).
D) Annotation distance for network connections between essential operons present or absent from the STRING database. Genes with an annotation distance of 0 are from the same functional group, while genes with an annotation distance of 4 are unconnected by annotation.
E) Functional annotations for intra-process connections between essential operons present in the STRING database or present in our essential network.
We quantitatively validated the network with a ROC curve. Because CRISPRi exhibits polarity, this analysis was based on operons containing essential genes (“essential operons”, n = 203), rather than on the individual genes themselves (Extended Experimental Procedures, Table S3). We compared high-confidence gene-gene connections in the STRING database (http://string-db.org/; Szklarczyk et al., 2015) with correlations between essential operons in our chemical-genomics dataset that met our correlation threshold (R=0.572; Figure 3C; Extended Experimental Procedures). Our network showed excellent agreement with STRING, with a ∼20-fold ratio of true to false positive rates (Figure 3C, inset).
We investigated the relationship between network connections and gene function by assigning essential operons to functional groups using the SubtiWiki online resource (http://subtiwiki.uni-goettingen.de/; Michna et al., 2014). Of the 61 non-overlapping functional groups, the 35 with two or more essential operons were assessed for connectivity within the group (intra-connectivity). Mean intra-connectivity was ∼2.4, compared with a background mean connectivity of ∼0.3 (p < 10−5), with 10 groups showing connectivity among ≥3 essential operons (Table S3). Indeed, the ROC analysis showed strong specificity and sensitivity for recapitulating existing intra-process connections in STRING (Figure S4A), with excellent recovery of connections within functional groups (Figure S4B). Connections within the peptidoglycan (PG) cell wall biosynthesis (18) and DNA replication (9) functional groups were most dense, likely reflecting convergence of PG precursor biosynthesis and localized cell wall assembly (Turner et al., 2014) and replisome protein-protein interactions (Sanders et al., 2010), respectively.
We next compared connectivity between functional groups (inter-connectivity) in our network with those in STRING, finding common connections and important distinctions (Figure 3D, 3E). First, the balance of intra- and inter-process connections differed; 46.2% of connections in STRING were inter-process, versus 59.0% in our network (p < 10−4). Second, STRING inter-process connections were biased toward extensively studied processes, e.g. 39% of STRING but only 4.3% of our inter-process connections were between ribosomal proteins and translation factors. Moreover, 84% (113/134) of connections unique to our network were between processes (Table S3), highlighting the ability of our open-ended approach to detect such connections. Finally, our network revealed many connections between operons in distant functional groups. Using the hierarchical annotation levels from SubtiWiki (Michna et al., 2014) as an approximation of “annotation distance”, we found that novel connections in our network were skewed toward processes furthest apart in annotation, whereas those in STRING were predominantly between related processes (Figure 3D). Some distant connections were intuitive, such as DNA replication (holB) and folate biosynthesis (folC and the sul-folB-folK operon), likely reflecting the folate requirement for dTTP production (Hardy et al., 1987). Other connections were unexpected, such as those between peptidoglycan (PG) biosynthesis/cell division and DNA replication/modification (e.g., ftsL and dnaX, murC and gyrB, ddl-murF and ydiO); these connections may be involved in failsafe mechanisms that link division and DNA replication (Arjes et al., 2014).
We explored the unexpected connection between transcription (rpoB) and cell division (ftsL), which is based on shared sensitivities to DNA intercalators and cell-wall antibiotics (Figure S2D). Using RNAseq, we found that basal knockdown of rpoB reduced the rpoB-rpoC mRNA level by two-fold. Among the few other substantial expression changes (Figure S4D), we found several down-regulated envelope genes including manA (4-fold), which is involved in cell-wall integrity (Elbaz and Ben-Yehuda, 2010), and sigW (1.5-fold to 2-fold), a master regulator of envelope stress (Cao et al., 2002) (Table S3). Selective reduction of several envelope functions due to rpoB knockdown could result in cell-wall defects that mimic those caused by knockdown of late-acting cell-division genes.
We also identified novel connections to essential genes of unknown function (Figure 3A, S3, S4E). For example, resistance to cell wall-targeting antibiotics drove strong correlations among the largely uncharacterized gene ylaN (Figure S2F; Hunt et al., 2006; Xu et al., 2007), iron-sulfur cluster biogenesis, and isoprenoid biosynthesis, the latter of which depends on the iron-sulfur cluster enzyme IspH (Gräwert et al., 2004; Wolff et al., 2003). These results suggest that defects in cellular iron homeostasis underlie the connections to ylaN. Indeed, in follow up experiments, we determined that ylaN is non-essential with added iron(III) (Figure S4F), further highlighting the ability of our unbiased approach to identify novel connections among essential processes.
Growth characteristics of the essential gene knockdown library
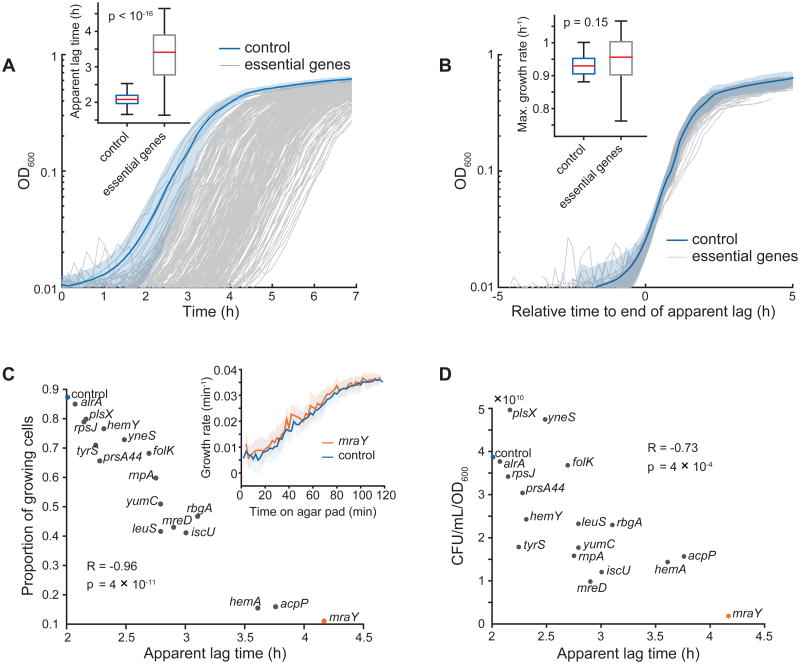
We measured apparent lag, maximum growth rate, and saturating density of growth curves for all basal knockdowns in LB medium (Figure 4A, 4B; Extended Experimental Procedures). Almost all knockdowns (∼80%) had a maximal growth rate equivalent to the control (Figure 4B and inset); ribosomal proteins were the only functional category with slower growth rates relative to other knockdowns (p = 0.0008, t-test). However, most knockdowns (95%) had longer apparent lag times (Figure 4A, inset), and cell wall-synthesis genes were enriched for the longest lags (p = 0.0013, Mann–Whitney U test, Table S4). CRISPRi repression was maintained in stationary phase (Figure S4G).
Figure 4. High-resolution Growth Profiles of Essential Gene Knockdowns Reveal Widespread Defects in Stationary-phase Survival.
A) Microplate reader growth curves of essential gene knockdown library strains. We grew cells for 18 h in LB before back-diluting into fresh LB (t = 0). Shaded area is mean ± standard deviation (S.D.) for the no-sgRNA control, n = 23.
B) Growth curves from A with the apparent lag time of each strain shifted to zero. Shaded area is mean ± S.D. for the no-sgRNA control, n = 23.
C) Data extracted from single-cell, time-lapse microscopy of selected essential gene knockdown strains grown on agarose pads with fresh LB after liquid growth in LB for 18 h. Although fewer mraY knockdown cells grow on LB pads after 18 h (11% versus 87% for the no-sgRNA control), the growth rate of elongating cells quantitatively matches that of the control (inset).
D) Viable cell plating of selected essential gene knockdown strains grown in LB for 18 h. CFU: colony-forming units.
We determined whether longer apparent lag times reflected strain-specific differences or decreased viability. Both the fraction of growing cells on agarose pads (Figure 4C) and plate viability measurements (Figure 4D) of strains spanning the apparent lag times negatively correlated with apparent lag. Apparent lag times also tracked with expectations from the number of live cells present. For example, using the 45-min doubling time in batch culture (Figure 4B inset), the ∼8-fold reduction in mraY knockdown viable cells (Figure 4C, 4D) requires ∼3 additional divisions, accounting for the 2.1-h increase in apparent lag. Moreover, viable mraY knockdown cells and the control had equivalent growth rates after transfer to an agarose pad with fresh LB (Figure 4C, inset). Together, these surprising results suggest that slight reductions in essential gene products affect outgrowth from stationary phase, creating a mixed population in which some cells exhibit WT outgrowth and others are either dead or non-growing.
Basal knockdown of essential genes results in changes in cellular dimensions that reveal shape actuators and modulators
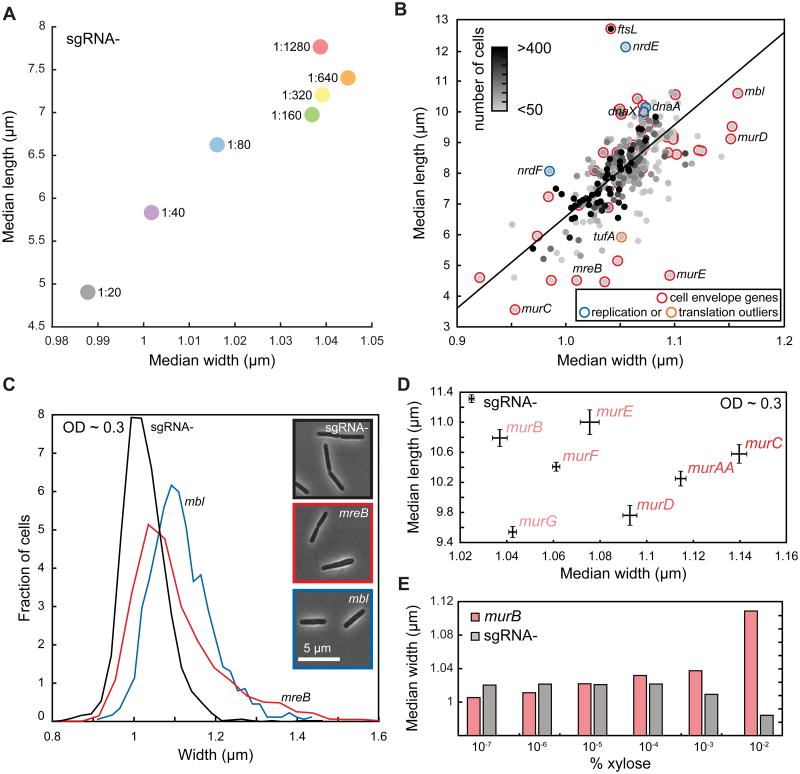
Our library offers an unprecedented opportunity to examine whether basal knockdown of essential genes affects cell morphology. To provide baseline values, we quantified the dimensions of WT cells. Cells were imaged 3.5 h after different degrees of dilution into fresh medium (Extended Experimental Procedures), and therefore had different culture optical densities (OD). We found that median cell length and width varied systematically with extent of dilution (Figure 5A), likely reflecting growth phase differences as cells cycle through nutrients in rich medium (LB). Cells diluted less were smaller, reflecting their slower growth rate at the time of imaging, similar to previous reports linking cell size and steady-state growth rate (Schaechter et al., 1958) and consistent with known shortening in stationary phase (Overkamp et al., 2015).
Figure 5. Partial Knockdown of Essential Genes Identifies Potential Morphological Regulators, with Envelope Gene Knockdown Leading to Changes in Cell Width.
A) Cell volume of the no-sgRNA control varies substantially after 3.5 h of growth across dilutions of the same overnight culture into fresh LB. Cell length and width were highly correlated (Pearson's R = 0.96, p<0.001).
B) Median cell length and width of essential knockdown strains are highly correlated after 3.5 h of growth (Pearson's R = 0.66, p<10−38). “Cell envelope” is the only functional category that is enriched in the outliers to the best-fit line (black) between length and width. Selected non-cell envelope outliers in DNA replication (blue) and translation (orange) are also shown.
C) Distribution of cell widths of mbl and mreB knockdown strains at OD600∼0.3 reveals subtle distinctions between the two actin homologs, with mbl cells wider than no-sgRNA control cells and mreB cells adopting a broader range of widths. n > 2000 cells for each histogram.
D) Knockdown strains in the mur pathway have similar median cell length at OD600∼0.3, while cell width varies over a wide range. Bars represent standard error of the mean.
E) Cell width of the murB knockdown strain increases monotonically with the degree of dcas9 induction, in contrast to the no-sgRNA control.
We next determined morphological parameters of the essential gene basal knockdowns to pinpoint proteins for which a small expression change results in altered shape, using high-throughput imaging at 3.5 h after dilution (Extended Experimental Methods). Because of variable stationary-phase outgrowth (Figure 4C, 4D), culture OD varied widely at the time of imaging. We found a general relationship between growth rate and OD for virtually all knockdowns (Figure S4H), indicating a large range of instantaneous growth rates at imaging. Nonetheless, the length and width of the knockdowns were highly correlated (Figure 5B), similar to WT cells across dilutions (Figure 5A). The fact that cells reach their maximum growth rate at a fixed OD (Figure S4H), rather than at a fixed number of doublings after growth resumption, underscores the importance of cell density and the extracellular milieu in growth rate control.
Basal knockdowns that deviated from the length/width trendline (Figure 5B; Table S5) could represent proteins involved in either actuating or regulating growth of the shape-determining cell wall. Only cell-envelope genes exhibited significant enrichment of outliers (average median length deviation = 0.99 μm, p < 0.0005 by bootstrapping, Extended Experimental Procedures; Figure 5B), as expected because they are largely PG synthesis-related and are therefore actuators of cell shape. To validate our identification of outliers and ensure that cell chaining was rare, we used the membrane stain, FM4-64, on a subset of strains with different median cell lengths, finding that length measurements from phase images and peripheral fluorescence were highly correlated (Figure S5A).
Several outliers in other functional groups were intriguing, as they could identify cell shape regulators. Basal knockdown of tufA, encoding the translation elongation factor Tu, resulted in cells that were substantially shorter than expected (Figure 5B, Table S5); Tu interacts with the bacterial cytoskeleton protein and rod-shape determinant MreB (Defeu Soufo et al., 2010). Knockdowns of several DNA replication genes (dnaX, dnaA, nrdE/F) resulted in longer cells (Figure 5B, Table S5). Moreover, dnaX and ftsL, both of which are large mophological outliers, were also significantly correlated in our chemical screen, suggesting a functional connection between replication and division corroborated by multiple independent data types.
As cell size is dependent on the degree of dilution prior to regrowth (Figure 5A), direct comparisons of cellular dimensions among different strains require cells to be at the same OD. Large variation in lag times prevented us from measuring the entire library by this method. Instead, we examined two actin homologs (mreB and mbl) involved in coordinating PG synthesis (Scheffers and Pinho, 2005) and the mur genes responsible for PG precursor synthesis at OD 0.3±0.05. Both actin homologs were wider than WT on average, with mreB exhibiting larger standard deviation, suggesting that cell width is particularly sensitive to MreB levels (Figure 5C). The median cell lengths of mur strains were similar to WT (Figure 5D), but widths (population median between 1.04-1.14 μm) were larger than no-sgRNA cells (1.02 μm), validating their classification as outliers (Figure 5D). The murB cell width increased monotonically with dcas9 induction (Figure 5E), indicating that cell width is responsive to the cellular levels of PG precursors.
In summary, our systematic screen for the essential gene products most intimately tied to cell shape identified genes with known ties to morphology (e.g. mreB/mbl) and cell wall synthesis, revealed a quantitative relationship between PG precursor gene expression and cell width, and uncovered potential new regulators of cell morphology.
Single-cell characterization of terminal phenotypes provides a novel view of essential gene function
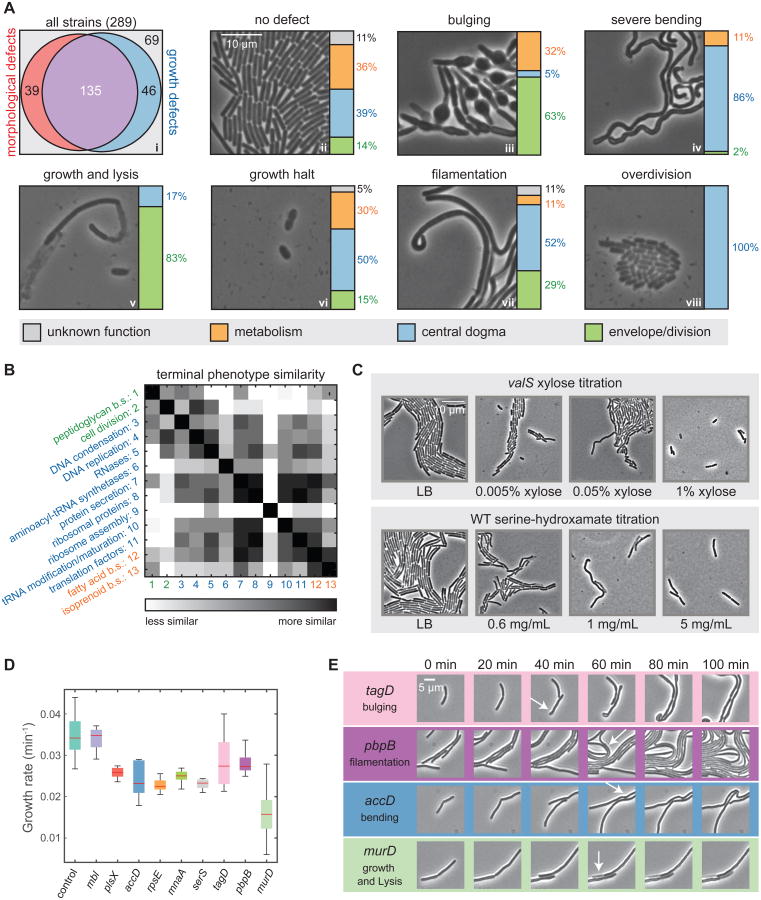
We examined whether substantial depletion results in more extreme morphological changes than basal knockdown by imaging the entire library after prolonged (∼24 h) induction of dcas9 (“terminal phenotypes”; Extended Experimental Procedures). Remarkably, >60% of the strains (174/289, or 166/258 bona-fide essentials) exhibited morphological phenotypes, even excluding strains with growth defects only (over-division or growth halting; Figure 6Ai), a number far exceeding the 48 known cell envelope-related strains. Knockdowns displayed several predominant morphologies (Figure 6Aii-viii, S5B), whereas the control formed a uniform lawn of normal, rod-shaped cells (Figure 6Aii).
Figure 6. Essential Gene Depletion Reveals a Diversity of Terminal Phenotypes.
A) (i) Area-proportional graph of the fraction of essential gene knockdowns that give rise to morphological and growth terminal phenotypes. (ii-viii) Single-cell imaging of common terminal phenotypes of essential-gene knockdowns, with bar graphs depicting the broad functional categories underlying each terminal phenotype.
B) Matrix of the similarity of terminal phenotypes achieved by genes belonging to the most common essential-gene functional groups. b.s., biosynthesis.
C) Titrated depletion of the valS knockdown and inhibition of WT by serine hydroxamate led to similar trends in terminal phenotypes, from over-division to filamenting and bending, and finally growth halting.
D) Post-induction growth rates of selected strains with different terminal phenotypes. Most strains reduced growth rates by 20-30% in the first hour, except for murD and mbl. murD had a larger decrease in growth rate, whereas mbl had a similar growth rate as the no-sgRNA control.
E) Morphological phenotypes during depletion post-CRISPRi induction. Phenotypes were observable by 40-60 min post-induction.
We quantified terminal morphologies by manually classifying phenotypes with an eight-dimensional phenotype vector including lysis, bulging, uniform shape loss, bending, filamentation, over-division (shorter cells), and extent of growth. We assessed whether functionally related genes mapped to particular phenotypes. Many cell-envelope genes bulged (e.g. tagA/B/F/G/O (teichoic acid), mreC (cell shape/PG biosynthesis); Figure 6Aiii), while many ribosomal genes displayed severe cell bending (Figure 6Aiv). However, in some cases genes in different processes had similar terminal phenotypes, such as growth halting (Figure 6Avi) and bending (Figure 6Aiv). Several metabolic genes (e.g. metK, dxs) exhibited bulging (Table S5), consistent with network connections between these processes and cell envelope synthesis/division (Figure 3A). Finally, some strains displayed both bulging and lysis (Table S5), indicating that depletions in one process can lead to more than one terminal phenotype.
Phenotypic variability within functional groups and common phenotypes of different groups made it challenging to map phenotype to process with simple visual comparison. Instead, we evaluated the similarities between terminal phenotype vectors for all SubtiWiki annotation pairs (Figure 6B, Extended Experimental Procedures), a process conceptually similar to the one we used to identify inter-process connections in the chemical-genomics dataset. Our similarity matrix recapitulated known connections (e.g. PG synthesis/cell division) while providing support for novel network connections (e.g. fatty acid metabolism with both isoprenoid biosynthesis and several central-dogma annotations; Figure 6B, 3A, and 3B). Terminal-phenotype links are consistent with and complement the high level of inter-process connectivity of the essential gene network (Figure 3E).
Variable terminal phenotypes within a group might reflect the rate of gene-product depletion. We tested this hypothesis by inhibiting tRNA charging (Figure 6C) either by valS (ValS aminoacyl-tRNA synthetase) knockdown, or by addition of serine hydroxymate, (tRNAser aminoacylation inhibitor). We observed dose-dependent phenotypes (Figure 6C): shorter cells with slight depletion/inhibition (Figure S5C, S5D), filamentation and bending with intermediate levels, and growth halting at high levels; these phenotypes were also exhibited by other aminoacyl-tRNA synthetase knockdowns (Figure 6Aiv, vi, viii). Thus, the rate of inhibition of essential processes can qualitatively affect their terminal phenotypes.
In summary, the majority of essential genes knockdowns display an altered terminal morphological phenotype. Phenotypes group both within and across functional processes, demonstrating the utility of morphology for revealing interactions, and can vary with the kinetics and extent of gene knockdown.
Cellular behavior during the depletion of essential proteins
To determine how rapidly terminal phenotypes are established, we performed timelapse imaging on nine representative knockdown strains in the presence of inducer. We placed exponentially growing cells on agarose pads of LB+xylose to initiate depletion, and imaged every 5 min to determine microcolony growth rates (Figure 6D, S5E). During the first 60 min post-induction, WT cells maintained a constant elongation rate. The mbl knockdown maintained the WT growth rate, murD had a ∼50% reduction, and the rest had a 20-30% reduction. Bulging (tagD) was the first prominent morphological phenotype observed (∼40 min; 1-2 cell doublings on agarose pads) with filamentation (pbpB), bending (accD), and lysis (murD) occurring after ∼60 min (2-3 doublings; Figure 6E). As depletion is primarily by dilution, these data indicate that reduction of at least 50-75% below basal knockdown is required to substantially affect growth rate and morphology. Many factors prevent precise quantification, including differential protein stabilities and polar effects on transcription.
We examined growth of the entire library during dcas9 induction by diluting stationary-phase cultures into liquid LB+xylose. Approximately 1/3 of the strains never emerged from stationary phase (OD600<0.06 at 7 h; Figure S6A). These strains covered the entire range of lags, hence lack of growth could not be explained by the variable stationary-phase outgrowth exhibited under basal knockdown conditions (Figure S6B). Instead, these strains identify the subset of essential proteins present in limiting amounts such that additional depletion beyond basal knockdown results in growth cessation or lysis (Table S4).
Among strains that grew, we identified two distinct phenotypes (Figure S6C, Table S4, Extended Experimental Procedures). Thirteen strains showed nearly linear growth; 9/13 affected cofactor biosynthesis (e.g. heme biosynthesis) or electron transport (Figure S6D), and may reflect cofactor availability. Fourteen strains enriched in either cell-envelope synthesis (5/14) or DNA replication (7/14) (Table S4) stalled after initial growth and then exhibited a marked decrease in OD, indicative of lysis (Figure S6E). This common phenotype may reflect the close network and morphological connections observed between DNA replication and envelope synthesis (Figure 3A).
Dissecting a highly redundant gene network with multiplexed CRISPRi
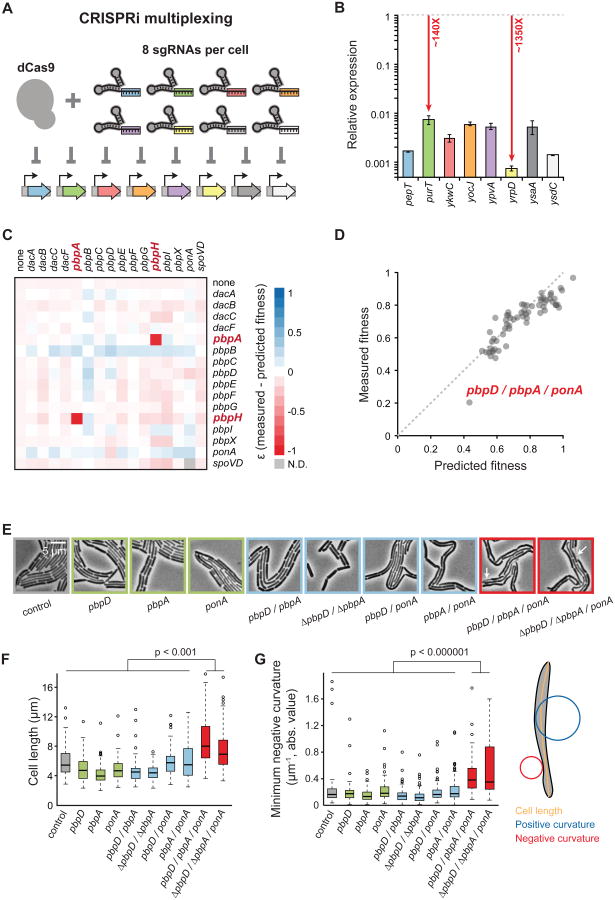
Genetic redundancy, prevalent in complex processes (e.g. construction of PG; Meeske et al., 2015) can mask the contributions of individual genes to essential processes. CRISPRi can simultaneously knockdown two genes (Qi et al., 2013); we show that CRISPRi can simultaneously knockdown eight unrelated, nonessential genes effectively (Figure 7A and 7B). Using this multiplexing capability, we examined redundancy for the 16 penicillin binding protein (PBP)-encoding genes involved in PG synthesis. Simultaneous knockdown of pairwise pbp genes were viable at full dcas9 induction except pbpA/pbpH, a known synthetic lethal pair, and those including pbpB, the only essential PBP (Figure 7C, S7A, S7B, and S7C; Table S6; Wei et al., 2003). The observed fitness of double knockdowns was largely predicted by multiplying the fitness of single mutants, a formalism developed for null mutations in parallel pathways (Figure S7A, Extended Experimental Procedures).
Figure 7. Multiplexed CRISPRi Knockdowns Facilitate Genetic Analysis of Complex Pathways.
A) Schematic of knockdown of eight genes in a single cell. Knockdowns correspond to the genes in (B).
B) Quantitative PCR of RNA levels in the eight-gene knockdown strain at maximal dcas9 induction (1% xylose). Expression is relative to the no-sgRNA control. Points are the means of at least three measurements, and error bars are ±1 standard deviation.
C) The difference between measured and predicted fitness (ε, Extended Experimental Procedures) for each pbp double knockdown based on normalized colony size at maximal dcas9 induction (1% xylose). Negative and positive ε represent reduced or improved fitness, respectively.
D) Measured versus predicted fitness of pbp triple knockdowns based on normalized colony size at maximal dcas9 induction (1% xylose).
E) Terminal phenotypes of pbp knockdown strains. Arrows indicate lysed cells.
F) Box plots of cell length from pbp knockdowns in E. n > 100 cells for each strain.
G) Box plots of cell curvature from pbp knockdowns in E. n > 100 cells for each strain.
We identified double knockdowns that were hypersensitive to PBP inhibitors (mecillinam, cefoxitin, and aztreonam; Figure S7D, E), as these suggest possible triple synthetic gene combinations. Fitness of the pbpA/ponA/pbpD triple knockdown was lower than predicted (Figure 7D) and we could not construct a ΔpbpA/ΔponΔ/ΔpbpD triple deletion, although we could introduce an innocuous deletion (ΔtrpC) or an unrelated pbp deletion (ΔpbpC) into ΔpbpA/ΔponA. Thus, the triple deletion is either extremely sick or synthetic lethal.
PBP4 (pbpD), PBP2a (pbpA), and PBP1a/b (ponA) have transpeptidase activity and are septally localized (Scheffers et al., 2004). Reduced septal PG transpeptidation during cell division may underlie the severe growth defect of the triple knockdown. Control and single- and double-knockdown cells exhibited the expected rod-like shape, albeit with reduced cell length (Figure 7E and 7F), potentially implicating these three genes in the regulation of cell division. By contrast, the triple mutant had distinct phenotypes including filamentation, cell lysis (Figure 7E, white arrows), and bending along the cell contour (Figure 7G, p<10−6), whether constructed with complete knockdown of all three genes or ponA knockdown in the ΔpbpA/ΔpbpD double. This phenotype is consistent with lethality caused by reduced septal transpeptidation, and showcases the ease of dissecting redundant pathways with CRISPRi.
Discussion
Bacteria typically have several hundred essential genes that encode the core reactions central to viability, together constituting ∼10% of their total genetic complement. Lacking a facile way to reduce their expression, we had little understanding of in vivo relationships among essential gene processes, or how subtle imbalances in essential pathways impact cellular homeostasis. This work is a major advance in the study of bacterial essential genes, providing a systematic, unbiased study of their phenotypes in vivo using CRISPRi (Figure 1) to obtain facile and precise down-regulation. Using chemical genomics profiling and high-throughput microscopy, we identified complex phenotypes including chemical vulnerabilities (Figure 3), growth and shape phenotypes (Figure 4, 5), and terminal death phenotypes (Figure 6). Together, these revealed a complex web of connections among essential processes. Given that CRISPR systems are broadly active in bacteria, our approach can be readily extended to other bacterial species including pathogenic and non-culturable species.
Our essential gene network (Figure 3) reveals numerous inter-process connections not previously annotated, and is highly enriched in novel connections among distant processes. Process inter-connectivity may provide the cell with mechanisms for restoring cellular homeostasis in response to transient imbalances. Some connections make intuitive sense. For example, folate is likely linked to replication via its necessity for dTTP synthesis. Such correlations readily suggest specific hypotheses that can be directly tested experimentally; e.g. a particular DNA polymerase subunit senses dTTP levels. Other connections have no facile explanation, and underscore the fact that connections between core processes are abundant and understudied. Very likely, our network significantly underestimates connections as it is based only on the highest confidence interactions to avoid false positives. Comparable datasets in other organisms will provide the basis for evolutionary studies to investigate the extent to which the logic of these functional circuits is conserved across organisms.
Our studies suggest that the levels of essential B. subtilis proteins are higher than necessary to maintain optimal growth, as the vast majority of basal knockdown strains grew indistinguishably from WT in exponential phase (Figure 4B). Thus, either protein levels are set high enough to be robust to small (1.5-3-fold) decreases in expression or their levels are maintained by an elaborate posttranscriptional regulatory system. Moreover, there is a sufficient excess of essential proteins that most strains (70%) emerge from stationary phase even when their gene products are depleted during regrowth. The 30% of strains unable to exit stationary phase (Figure S6A) are those whose protein levels are closest to the levels necessary for normal growth.
In stark contrast, almost all basal knockdowns strains exhibited increased cell inviability during the exit from stationary phase. Thus, even a small decrease in protein product increases the vulnerability of cell regrowth (Figure 4A). Importantly, there is little overlap between strains ceasing growth soon after depletion and those most vulnerable to inhibition of outgrowth from stationary phase (Figure S6B). This discordance suggests that the set of essential genes whose expression level is close to that necessary for rapid cell growth in ideal conditions is distinct from the set necessary for survival during stationary phase. Speculatively, protein levels may reflect the ecological niche of B. subtilis in soil, where cells spend much time in a non-growing state and must survive long enough to enter the sporulation program with high efficiency.
Classically, genetic perturbations resulting in morphological variation, such as homeotic mutants and variegated maize, provided critical insights into fundamental cellular circuits. However, the essentiality or redundancy of most cell wall-synthesis proteins has made it difficult to uncover the molecular mechanisms underlying bacterial cell shape and size determination. Our CRISPRi knockdown approach allowed us to probe this relationship both under conditions of partial knockdown (1.5 to 3-fold) and during complete depletion. Partial knockdown identifies outliers exceptionally sensitive to depletion. As a consequence, we identified the critical envelope gene actuators of the response, showed that cell width as well as length is controlled (Figure 5), and determined that cell width varies montonically with extent of depletion of mur genes (Figure 5D, 5E). The few outliers involved in other processes identify putative regulators of cell wall synthesis. The translation and DNA replication-related outliers may tie the rate of protein synthesis and DNA replication to cell-wall growth, possibly using previously unrecognized moonlighting functions of these proteins. Moreover, since maximal growth rate is not substantially affected across the basal knockdown strains, our findings indicate that cell size and growth rate can be at least partially decoupled. Additionally, a graded morphological response to the levels of essential proteins may drive physiological heterogeneity enabling population adaptation to dynamic environments.
In contrast, complete depletion probes the intimate relationship between growth and morphology. Here, cells exhibit a wide array of terminal phenotypes with alterations in both growth and morphology (Figure 6A). By comparison, only a single nonessential E. coli gene deletion (rodZ) has a morphological phenotype (Shiomi et al., 2008). Some phenotypes result from gradual depletion of function (Figure 6C), which may mimic imbalances that occur transiently due to stochasticity in gene expression. Tracing changes at different rates of depletion may provide clues to the cellular program for morphology. Moreover, the dynamics of removing an essential function may be as important to growth and viability as the presence or absence of such a protein.
The evolution of essential processes has remained largely mysterious. Long-term evolution experiments have detected many mutational events in the RNA polymerase complex, cell-wall synthesis, and cell-shape determination (Tenaillon et al., 2012), suggesting a great diversity of molecular adaptations, including to essential processes. The apparent excess abundance of most essential proteins (Figure 6D, 6E) suggests that new functions could rapidly develop from the existing repertoire of proteins without compromising growth, possibly explaining the prevalence of moonlighting proteins (Huberts and van der Klei, 2010). For example, actin and tubulin homologs have diverse roles in bacteria and eukaryotes (Busiek and Margolin, 2015). Phenotypic heterogeneity and variable survival during stationary phase, as observed here (Figure 4), may indicate that stressful environments have shaped the evolutionary history of essential genes. The complexity of the network (Figure 3A) suggests context dependence that may be critical for the evolution of essential genes and their interactions; perhaps certain essential genes can be rendered non-essential in environments such as biofilms. Expansion of our study to varied environments may provide a more nuanced view of the instances when a particular process is limiting for growth.
Experimental Procedures
CRISPRi Library Design, Cloning, Chemical Screens, and Growth Analysis
sgRNAs targeted all putative essential genes (subtiwiki.uni-goettingen.de) and those recently identified in our B. subtilis gene knockout library (Koo et al., unpublished). sgRNAs were designed to target within the gene body near the 5′ end of the gene on the non-template strand. sgRNA libraries were cloned via inverse PCR as previously described (Larson et al., 2013) and strains were constructed using natural competence transformation. Chemical screening was performed and chemical-gene scores were calculated as previously described (Nichols et al., 2011). B. subtilis and S. aureus UppS proteins were purified using nickel-affinity chromatography, and assayed using a Kinetic EnzCheck pyrophosphate assay (Life Technologies). The B. subtilis essential gene network was constructed by calculating all pairwise Pearson correlations between sgRNA knockdown strains, randomly permuting gene identity relative to chemical-gene scores to generate a background distribution, then applying a significance cutoff to the correlations based on the 95% confidence interval of the background distribution. Population growth curves were obtained from a microplate reader, and growth information was extracted by fitting the curves to a Gompertz equation (Zwietering et al., 1990).
High-throughput Microscopy
Images of single cells were acquired after transferring cells from a 96-well plate onto a large-format agarose pad. Analysis of cellular morphologies was performed using custom MATLAB code. A minimum of 100 cells were analyzed for quantitative descriptions of single-cell phenotypes. Terminal phenotypes were examined by spotting cultures outgrown from stationary phase for two hours in liquid LB medium, transferred to agarose pads containing LB + 1% xylose, then imaged after overnight incubation. Time-lapse images were taken in an active- control environmental chamber at 37 °C (HaisonTech). Further details of methods are in the Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We thank D. Rudner for strains/plasmids, O. Rosenberg for S. aureus DNA, G. Kritikos and N. Typas for colony-sizing software, and A. Grossman, G. W. Li, N. Typas, and anonymous reviewers for helpful comments. This work was supported by the UCSF Center for Systems and Synthetic Biology (to L.S.Q.), a Stanford Graduate Fellowship (to A.C.), a Stanford Agilent Fellowship (to H.S.), a Foundation Grant (from the Canadian Institutes of Health Research) and a Canada Research Chair to (E.D.B.), NIH F32 GM108222 (to J.M.P.), NIH DA036858 and HHMI funding (to J.S.W.), NIH OD017887 (to L.S.Q.), NIH R01 DA036858 (to L.S.Q.), NIH DP2-OD006466 (to K.C.H.), NSF MCB-1149328 (to K.C.H.), and NIH R01 GM102790 (to C.A.G.).
Footnotes
Author Contributions: Conceptualization, J.M.P., A.C., H.S., L.S.Q., K.C.H., and C.A.G.; Methodology, J.M.P., A.C., H.S., T.L.C., M.H.L., B.M.K., J.S.W., E.D.B., L.S.Q., K.C.H., and C.A.G.; Investigation, J.M.P., A.C., H.S., T.L.C., M.H.L., S.W., C.H.S.L., E.M., and E.H.W.; Formal Analysis, J.M.P., A.C., H.S., T.L.C., M.H.L., J.S.H., A.L.S., J.S.W., E.D.B., L.S.Q., K.C.H., and C.A.G.; Writing-Original Draft, J.M.P., A.C., H.S., L.S.Q., K.C.H., and C.A.G.; Writing-Review & Editing, J.M.P., A.C., H.S., L.S.Q., K.C.H., and C.A.G.; Funding Acquisition, J.M.P., J.S.W., E.D.B., L.S.Q., K.C.H., and C.A.G.; Supervision, J.M.P., J.S.W., E.D.B., L.S.Q., K.C.H. and C.A.G.
Supplemental Information: Supplemental information includes Extended Experimental Procedures, 7 Figures, and 6 tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arjes HA, Kriel A, Sorto NA, Shaw JT, Wang JD, Levin PA. Failsafe mechanisms couple division and DNA replication in bacteria. Curr Biol CB. 2014;24:2149–2155. doi: 10.1016/j.cub.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen VA, Májek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5:711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busiek KK, Margolin W. Bacterial actin and tubulin homologs in cell growth and division. Curr Biol CB. 2015;25:R243–R254. doi: 10.1016/j.cub.2015.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Wang T, Ye R, Helmann JD. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons. Mol Microbiol. 2002;45:1267–1276. doi: 10.1046/j.1365-2958.2002.03050.x. [DOI] [PubMed] [Google Scholar]
- Choudhary E, Thakur P, Pareek M, Agarwal N. Gene silencing by CRISPR interference in mycobacteria. Nat Commun. 2015;6 doi: 10.1038/ncomms7267. [DOI] [PubMed] [Google Scholar]
- Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, Coller JA, Fero MJ, McAdams HH, Shapiro L. The essential genome of a bacterium. Mol Syst Biol. 2011;7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarny TL, Perri AL, French S, Brown ED. Discovery of novel cell wall-active compounds using P ywaC, a sensitive reporter of cell wall stress, in the model gram-positive bacterium Bacillus subtilis. Antimicrob Agents Chemother. 2014;58:3261–3269. doi: 10.1128/AAC.02352-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Reimold C, Linne U, Knust T, Gescher J, Graumann PL. Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein. Proc Natl Acad Sci U S A. 2010;107:3163–3168. doi: 10.1073/pnas.0911979107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz M, Ben-Yehuda S. The metabolic enzyme ManA reveals a link between cell wall integrity and chromosome morphology. PLoS Genet. 2010;6:e1001119. doi: 10.1371/journal.pgen.1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräwert T, Kaiser J, Zepeck F, Laupitz R, Hecht S, Amslinger S, Schramek N, Schleicher E, Weber S, Haslbeck M, et al. IspH protein of Escherichia coli: studies on iron-sulfur cluster implementation and catalysis. J Am Chem Soc. 2004;126:12847–12855. doi: 10.1021/ja0471727. [DOI] [PubMed] [Google Scholar]
- Hardy LW, Finer-Moore JS, Montfort WR, Jones MO, Santi DV, Stroud RM. Atomic structure of thymidylate synthase: target for rational drug design. Science. 1987;235:448–455. doi: 10.1126/science.3099389. [DOI] [PubMed] [Google Scholar]
- Huberts DHEW, van der Klei IJ. Moonlighting proteins: an intriguing mode of multitasking. Biochim Biophys Acta. 2010;1803:520–525. doi: 10.1016/j.bbamcr.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Hunt A, Rawlins JP, Thomaides HB, Errington J. Functional analysis of 11 putative essential genes in Bacillus subtilis. Microbiol Read Engl. 2006;152:2895–2907. doi: 10.1099/mic.0.29152-0. [DOI] [PubMed] [Google Scholar]
- Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Sham LT, Kimsey H, Koo BM, Gross CA, Bernhardt TG, Rudner DZ. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci U S A. 2015;112:6437–6442. doi: 10.1073/pnas.1504967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michna RH, Commichau FM, Tödter D, Zschiedrich CP, Stülke J. SubtiWiki-a database for the model organism Bacillus subtilis that links pathway, interaction and expression information. Nucleic Acids Res. 2014;42:D692–D698. doi: 10.1093/nar/gkt1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimee M, Tucker AC, Voigt CA, Lu TK. Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell Syst. 2015;1:62–71. doi: 10.1016/j.cels.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moche M, Schneider G, Edwards P, Dehesh K, Lindqvist Y. Structure of the complex between the antibiotic cerulenin and its target, beta-ketoacyl-acyl carrier protein synthase. J Biol Chem. 1999;274:6031–6034. doi: 10.1074/jbc.274.10.6031. [DOI] [PubMed] [Google Scholar]
- Myoda TT, Lowther SV, Funanage VL, Young FE. Cloning and mapping of the dihydrofolate reductase gene of Bacillus subtilis. Gene. 1984;29:135–143. doi: 10.1016/0378-1119(84)90174-4. [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overkamp W, Ercan O, Herber M, van Maris AJA, Kleerebezem M, Kuipers OP. Physiological and cell morphology adaptation of Bacillus subtilis at near-zero specific growth rates: a transcriptome analysis. Environ Microbiol. 2015;17:346–363. doi: 10.1111/1462-2920.12676. [DOI] [PubMed] [Google Scholar]
- Peters JM, Silvis MR, Zhao D, Hawkins JS, Gross CA, Qi LS. Bacterial CRISPR: accomplishments and prospects. Curr Opin Microbiol. 2015;27:121–126. doi: 10.1016/j.mib.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders GM, Dallmann HG, McHenry CS. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol Cell. 2010;37:273–281. doi: 10.1016/j.molcel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- Schaechter M, Maaloe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev MMBR. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers DJ, Jones LJF, Errington J. Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis. Mol Microbiol. 2004;51:749–764. doi: 10.1046/j.1365-2958.2003.03854.x. [DOI] [PubMed] [Google Scholar]
- Schujman GE, Choi KH, Altabe S, Rock CO, de Mendoza D. Response of Bacillus subtilis to cerulenin and acquisition of resistance. J Bacteriol. 2001;183:3032–3040. doi: 10.1128/JB.183.10.3032-3040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Sakai M, Niki H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J. 2008;27:3081–3091. doi: 10.1038/emboj.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Rodríguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. The molecular diversity of adaptive convergence. Science. 2012;335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- Thomaides HB, Davison EJ, Burston L, Johnson H, Brown DR, Hunt AC, Errington J, Czaplewski L. Essential bacterial functions encoded by gene pairs. J Bacteriol. 2007;189:591–602. doi: 10.1128/JB.01381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RD, Vollmer W, Foster SJ. Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol. 2014;91:862–874. doi: 10.1111/mmi.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weart RB, Lee AH, Chien AC, Haeusser DP, Hill NS, Levin PA. A metabolic sensor governing cell size in bacteria. Cell. 2007;130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Havasy T, McPherson DC, Popham DL. Rod shape determination by the Bacillus subtilis class B penicillin-binding proteins encoded by pbpA and pbpH. J Bacteriol. 2003;185:4717–4726. doi: 10.1128/JB.185.16.4717-4726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wolff M, Seemann M, Tse Sum Bui B, Frapart Y, Tritsch D, Garcia Estrabot A, Rodríguez-Concepción M, Boronat A, Marquet A, Rohmer M. Isoprenoid biosynthesis via the methylerythritol phosphate pathway: the (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase (LytB/IspH) from Escherichia coli is a [4Fe-4S] protein. FEBS Lett. 2003;541:115–120. doi: 10.1016/s0014-5793(03)00317-x. [DOI] [PubMed] [Google Scholar]
- Xu HH, Trawick JD, Haselbeck RJ, Forsyth RA, Yamamoto RT, Archer R, Patterson J, Allen M, Froelich JM, Taylor I, et al. Staphylococcus aureus TargetArray: comprehensive differential essential gene expression as a mechanistic tool to profile antibacterials. Antimicrob Agents Chemother. 2010;54:3659–3670. doi: 10.1128/AAC.00308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Sedelnikova SE, Baker PJ, Hunt A, Errington J, Rice DW. Crystal structure of S. aureus YlaN, an essential leucine rich protein involved in the control of cell shape. Proteins. 2007;68:438–445. doi: 10.1002/prot.21377. [DOI] [PubMed] [Google Scholar]
- Zwietering MH, Jongenburger I, Rombouts FM, van 't Riet K. Modeling of the bacterial growth curve. Appl Environ Microbiol. 1990;56:1875–1881. doi: 10.1128/aem.56.6.1875-1881.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.