Abstract
Background
Gastric carcinoma (GC) has one of the highest mortality rates of cancer diseases and has a high incidence rate in China. Palliative chemotherapy is the main treatment for advanced gastric cancer. It is necessary to compare the effectiveness and toxicities of different regimens. This study explores the possibility of methylation of DNA damage repair genes serving as a prognostic and chemo-sensitive marker in human gastric cancer.
Methods
The methylation status of five DNA damage repair genes (CHFR, FANCF, MGMT, MLH1, and RASSF1A) was detected by nested methylation-specific PCR in 102 paraffin-embedded gastric cancer samples. Chi-square or Fisher's exact tests were used to evaluate the association of methylation status and clinic-pathological factors. The Kaplan–Meier method and Cox proportional hazards models were employed to analyze the association of methylation status and chemo-sensitivity.
Results
The results indicate that CHFR, MLH1, RASSF1A, MGMT, and FANCF were methylated in 34.3 % (35/102), 21.6 % (22/102), 12.7 % (13/102), 9.8 % (10/102), and 0 % (0/102) of samples, respectively. No association was found between methylation of CHFR, MLH1, RASSF1A, MGMT, or FANCF with gender, age, tumor size, tumor differentiation, lymph node metastasis, and TNM stage. In docetaxel-treated gastric cancer patients, resistance to docetaxel was found in CHFR unmethylated patients by Cox proportional hazards model (HR 0.243, 95 % CI, 0.069–0.859, p = 0.028), and overall survival is longer in the CHFR methylated group compared with the CHFR unmethylated group (log-rank, p = 0.036). In oxaliplatin-treated gastric cancer patients, resistance to oxaliplatin was found in MLH1 methylated patients (HR 2.988, 95 % CI, 1.064–8.394, p = 0.038), and overall survival was longer in the MLH1 unmethylated group compared with the MLH1 methylated group (log-rank, p = 0.046).
Conclusions
CHFR is frequently methylated in human gastric cancer, and CHFR methylation may serve as a docetaxel-sensitive marker. MLH1 methylation was related to oxaliplatin resistance in gastric cancer patients.
Keywords: Gastric cancer, CHFR, MLH1, RASSF1A, MGMT, DNA methylation, DNA damage repair genes, Chemo-sensitive marker
Introduction
Gastric cancer is the fourth most commonly diagnosed cancer and the second leading cause of cancer-related death worldwide, with the highest incidence in Eastern Asian countries [1]. Most patients are diagnosed at an advanced stage, and chemotherapy becomes an important approach. However, prognosis remains unsatisfactory because of side effects and chemo-resistance. It is desirable to find a group of chemo-sensitive markers.
Methylation of the DNA damage repair gene was reported as a chemo-sensitive markers in a few cancers. To find chemo-sensitive markers in gastric cancer, we analyzed five DNA damage repair genes in this study.
Checkpoint with FHA and ring finger (CHFR) is a G2/M checkpoint inhibitor. Methylation of CHFR was reported to be sensitive to paclitaxel in endometrial and gastric cancer cell lines [2, 3]. The RAS association domain family 1A (RASSF1A) gene, a candidate tumor suppressor, may regulate microtubule and genomic stability [4]. RASSF1A hypermethylation mediated resistance to cisplatin and tamoxifen in male germ cell tumors and breast cancer [5, 6]. FANCF is a member of the Fanconi anemia gene family. Recently, it has been shown that promoter region hypermethylation disrupts the FA-BRCA pathway, resulting in cisplatin resistance in other malignant tumors [7, 8]. O6-Methylguanin DNA methyltransferase (MGMT) removes mutagenic alkyl groups from the O6 position of guanine, leading to G→A transitions after DNA damage [9]. MGMT is frequently lost because of methylation in brain tumors, and this methylation correlates with responsiveness to alkylator-based chemotherapy [10–12]. Demethylation of MLH1 (mutL homologue 1), a DNA mismatch repair gene, sensitizes the A2780 cell line to platinum [13]. In this study, we mainly focused on the methylation status of these five DNA repair genes and the sensitivity of oxaliplatin and docetaxel in gastric cancer.
Methods
Gastric cancer samples
We collected 102 cases of surgically resected gastric cancer tissue specimens from Chinese PLA General Hospital from April 2006 to June 2010. The patients, 30 women (29.4 %) and 72 men (70.6 %), were 26 to 76 years of age, with a median age of 53 years. None of the patients had received radiotherapy or chemotherapy before surgical resection. After surgery, 41 patients (40.2 %) received docetaxel therapy, 53 patients (52 %) received oxaliplatin treatment, and 8 patients (7.8 %) received other chemo-therapy. Docetaxel and oxaliplatin were administered intravenously on day 1 of every 21-day cycle, and capecitabine was administrated orally on week 1 and 2 of every 21-day cycle, for six cycles. Dosages of docetaxel, oxaliplatin, and capecitabine were 160 mg/m2, 130 mg/m2, and 2,000 mg/m2, respectively. Informed consent was obtained from all patients. The research protocol was approved by the institutional review board (IRB) of PLA General Hospital.
DNA preparation
Paraffin-embedded samples were sliced to 8 lm and deparaffinized by xylene. DNA was prepared by the proteinase K method. DNA samples were stored at −20 °C in low TE buffer.
Nested methylation-specific polymerase chain reaction (PCR)
Nested PCR was performed to facilitate the examination of the methylation of CHFR, MLH1, MGMT, RASSF1A and FANCF on paraffin-embedded gastric cancer samples. The bisulfite-modified DNA was subjected to a first-stage PCR incorporating external primer sets. The PCR reaction was carried out in a total volume of 25 μl with 1 U Taq Polymerase (Invitrogen, Carlsbad, CA, USA), 25 pmol external primer, 100 pmol dNTPs, 2.5 μl 10× PCR buffer, and 2 μl bisulfite-modified DNA. External primer sequences are shown in Table 1. PCR conditions included an initial denaturation at 95 °C for 5 min, followed by 35 cycles of amplification (95 °C × 30 s, 55 °C × 30 s, 72 °C × 40 s), and a final elongation step at 72 °C for 5 min. PCR products were analyzed on a 2 % agarose gel to confirm adequate template for subsequent second-stage internal methylation-specific PCR (MSP).
Table 1.
Methylation-specific polymerase chain reaction (PCR) (MSP) primers used in the present study
| Genes | Primer sequence (5′–3′) | Products size |
|---|---|---|
| CHFR Flank Up | AAAAATAAGGAAAAGATGTAGAYG | 167 |
| CHFR Flank Down | TAAAAATTTCCRATTAAAAAAACCC | |
| CHFR Me-Sense | AGATGTAGACGTTTTTTTTTGGAGGC | 100 |
| CHFR Me-Anti-Sense | ATTTCCGATTAAAAAAACCCCTTAACG | |
| CHFR Un-Sense | AGATGTAGATGTTTTTTTTTGGAGGT | 115 |
| CHFR Un-Anti-Sense | AAATTTCCAATTAAAAAAACCCCTTAACA | |
| MGMT Flank Up | GYGTTTYGGATATGTTGGGATAGTT | 135 |
| MGMT Flank Down | AAACTCCRCACTCTTCCRAAAAC | |
| MGMT Me-Sense | TTTCGACGTTCGTAGGTTTTCGC | 81 |
| MGMT Me-Anti-Sense | GCACTCTTCCGAAAACGAAACG | |
| MGMT Un-Sense | TTTGTGTTTTGATGTTTGTAGGTTTTTGT | 93 |
| MGMT Un-Anti-Sense | AACTCCACACTCTTCCAAAAACAAAACA | |
| FANCF Flank Up | GGTTTTAAGTATTATTTAYGTTAGTATTT | 165 |
| FANCF Flank Down | CCTCTTACCTCCACTAATTATACAAC | |
| FANCF Me-Sense | AGTATTTGGGATTTCGTTATCGTGC | 99 |
| FANCF Me-Anti-Sense | GAATAAAACCATACCGACCAAAACG | |
| FANCF Un-Sense | TATGTTAGTATTTGGGATTTTGTTATTGTGT | 106 |
| FANCF Un-Anti-Sense | ACAAATAAAACCATACCAACCAAAACA | |
| MLH1 Flank Up | GGAGTGAAGGAGGTTAYGGGTAAGT | 182 |
| MLH1 Flank Down | AAAAACRATAAAACCCTATACCTAATCTATC | |
| MLH1 Me-Sense | ACGTAGACGTTTTATTAGGGTCGC | 115 |
| MLH1 Me-Anti-Sense | CCTCATCGTAACTACCCGCG | |
| MLH1 Un-Sense | TTTTGATGTAGATGTTTTATTAGGGTTGT | 124 |
| MLH1 Un-Anti-Sense | ACCACCTCATCATAACTACCCACA | |
| RASSf1 Flank Up | GTTTAGTTTGGATTTTGGGGGAG | 144 |
| RASSf1 Flank Down | CCCRCAACTCAATAAACTCAAACTC | |
| RASSf1 Me-Sense | GGGTTCGTTTTGTGGTTTCGTTC | 76 |
| RASSf1 Me-Anti-Sense | TAACCCGATTAAACCCGTACTTCG | |
| RASSf1 Un-Sense | GGGGTTTGTTTTGTGGTTTTGTTT | 81 |
| RASSf1 Un-Anti-Sense | AACATAACCCAATTAAACCCATACTTCA |
Methylation-specific PCR
MSP primers were designed according to genomic sequences flanking the presumed transcriptional start sites. Primer sequences were oligo-synthesized (Invitrogen) to detect bisulfite-induced changes affecting unmethylated (U) and methylated (M) alleles. The MSP of 5 genes was carried out using primers showed in Table 1. A 1:1,000 dilution of external PCR product was used as internal MSP template. For each reaction, 2 μl diluted external PCR product was added. The composition of the PCR reaction is as same as the external PCR. Cycle conditions were 95 °C × 5 min for 1 cycle; 28 cycles × (95 °C × 30 s, 60 °C × 30 s, 72 °C × 30 s); and 72 °C × 5 min for 1 cycle. Each PCR assay included a positive control, using DNA treated in vitro with SssI methyltransferase (New England Biolabs, Ipswich, MA, USA). MSP products were analyzed using 2 % agarose gel electrophoresis.
Statistical analysis
Statistical analysis was performed using SPSS 16.0 software (SPSS, Chicago, IL, USA). Chi-square or Fisher's exact tests were used to evaluate the relationship between methylation status and clinicopathological characteristics. Survival rates were calculated by the Kaplan–Meier method, and differences in survival curve were evaluated using the log-rank test. Cox proportional hazards models were fit to determine independent associations of CHFR or MLH1 methylation with overall survival outcome. Two-sided tests were used to determine significance, and p < 0.05 was considered statistically significant.
Results
Methylation status of DNA repair genes and the association of methylation with clinicopathological features in gastric cancer
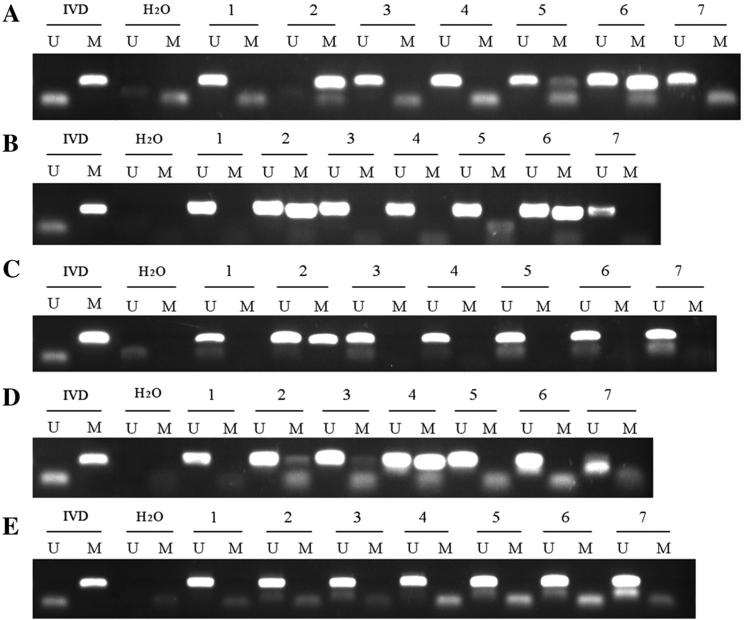
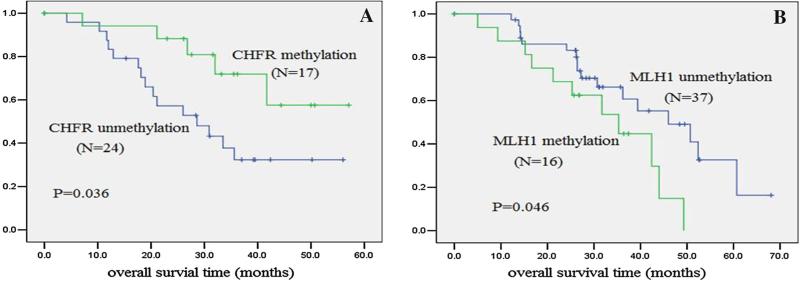
Methylation status of CHFR, FANCF, MGMT, MLH1, and RASSF1A was examined by MSP in 102 cases of human primary gastric cancer. No methylation was found in the FANCF gene; 35 cases (34.4 %) were methylated in the CHFR gene, 22 cases (21.6 %) were methylated in the MLH1 gene, 10 cases (9.8 %) were methylated in the MGMT gene, and 13 cases (12.7 %) were methylated in the RASSF1A gene (Fig. 1a–e). No association was found among CHFR, MLH1, MGMT, and RASSF1A methylation with gender, age, tumor size, degree of differentiation, lymph node metastasis, and TNM stage (Table 2). In 41 cases of docetaxel-treated patients, CHFR methylation was found in 17 cases (41.5 %). In 53 cases of oxaliplatin-treated patients, MLH1 methylation was found in 16 cases (30.2 %). In docetaxel-treated gastric cancer patients, overall survival was longer in the CHFR methylated group than in the CHFR unmethylated group (log-rank, p = 0.036; Fig. 2a). In oxaliplatin-treated gastric cancer patients, overall survival was longer in the MLH1 unmethylated group than in the MLH1 methylated group (log-rank, p = 0.046; Fig. 2b).
Fig. 1.
Methylation of five DNA damage repair genes in gastric cancer tissues. a Methylation-specific PCR (MSP) results of CHFR. b MSP results of MLH1. c MSP results of RASSF1A. d MSP results of MGMT. e MSP results of FANCF. H2O negative control to confirm the specificity of MSP, IVD positive control to confirm the specificity of MSP, M presence of methylated alleles, U presence of unmethylated alleles, 1–7 gastric cancer samples
Table 2.
Correlation between hypermethylation in DNA repair genes and clinicopathological characteristics of 102 gastric cancer cases
| Clinicopathological parameters (n) | Frequency of methylation, n (%) |
|||
|---|---|---|---|---|
| CHFR | MLH1 | MGMT | RASSFA1 | |
| Gender | ||||
| Male (72) | 24 (33.3) | 19 (26.4) | 4 (5.6) | 9 (12.5) |
| Female (30) | 11 (36.7) | 3 (10.0) | 6 (20.0) | 4 (13.3) |
| p | 0.747 | 0.067 | 0.061 | 1.000 |
| Age (years) | ||||
| n < 60 (73) | 26 (35.6) | 14 (19.2) | 9 (12.3) | 9 (12.3) |
| n ≥ 60 (29) | 9 (31.0) | 8 (27.6) | 1 (3.4) | 4 (13.8) |
| p | 0.660 | 0.352 | 0.321 | 1.000 |
| Tumor histology | ||||
| Intestinal type (64) | 22 (34.3) | 13 (20.3) | 7 (10.9) | 7 (10.9) |
| Diffused and mixed type (33) | 13 (34.2) | 9 (23.6) | 3 (7.9) | 6 (15.8) |
| p | 0.938 | 0.638 | 0.646 | 1.445 |
| Differentiation | ||||
| Well and moderately differentiated (29) | 9 (31.0) | 3 (10.3) | 3 (17.6) | 1 (5.9) |
| Poorly differentiated (73) | 26 (35.6) | 19 (26.0) | 7 (8.2) | 12 (14.1) |
| p | 0.660 | 0.082 | 0.457 | 0.595 |
| Tumor size | ||||
| d < 5 (45) | 13 (28.9) | 12 (26.7) | 5 (11.1) | 4 (8.9) |
| d ≥ 5 (57) | 22 (38.6) | 10 (17.5) | 5 (8.8) | 9 (15.8) |
| p | 0.305 | 0.266 | 0.953 | 0.299 |
| Tumor invasion | ||||
| T1–T2 (13) | 7 (53.8) | 3 (23.1) | 2 (15.4) | 1 (7.7) |
| T3–T4 (89) | 28 (31.5) | 19 (21.3) | 8 (9.0) | 12 (13.5) |
| p | 0.112 | 0.887 | 0.822 | 0.889 |
| Lymph node metastasis | ||||
| N0–N1 (36) | 10 (27.8) | 10 (27.8) | 2 (5.6) | 4 (11.1) |
| N1–N3 (66) | 25 (37.9) | 12 (18.2) | 8 (12.5) | 9 (14.3) |
| p | 0.304 | 0.260 | 0.473 | 0.956 |
| Peritoneal or distant metastasis | ||||
| M0 (94) | 30 (31.9) | 21 (22.3) | 8 (8.5) | 11 (11.7) |
| M1 (8) | 5 (62.5) | 1 (12.5) | 2 (25) | 2 (25) |
| p | 0.080 | 0.516 | 0.375 | 0.596 |
| Tumor stage | ||||
| I–II (19) | 6 (31.6) | 6 (31.6) | 2 (10.5) | 3 (15.8) |
| III–IV (83) | 29 (34.9) | 16 (19.3) | 8 (9.6) | 10 (12.0) |
| p | 0.781 | 0.240 | 0.100 | 0.952 |
| Vessel invasion | ||||
| Negative (72) | 25 (34.7) | 15 (20.8) | 9 (12.5) | 9 (12.5) |
| Positive (30) | 10 (33.3) | 7 (23.3) | 1 (3.3) | 4 (13.3) |
| p | 0.893 | 0.780 | 0.292 | 1.000 |
Pathological tumor invasion, lymph node status, distant metastasis status, and TNM stage of gastric cancer were evaluated according to American Joint Committee on Cancer (AJCC), 7th edition, 2009. Statistical analysis was performed by χ2 or Fisher exact test
p < 0.05 was considered statistically significant
Fig. 2.
Association of gene methylation and overall survival in gastric cancer patients. Kaplan–Meier survival curves were constructed according to the presence of gene methylation in gastric cancer. a In the docetaxel-treated group, overall survival is longer in the CHFR methylated group than in the CHFR unmethylated group. b In the oxaliplatin-treated group, overall survival is longer in the MLH1 unmethylated group than in the MLH1 methylated group
Resistance to docetaxel treatment was found in CHFR unmethylated patients by the Cox proportional hazards model (HR 0.243, 95 % CI, 0.069–0.859, p = 0.028; Table 3), and resistance to oxaliplatin treatment was found in MLH1 methylated patients (HR 2.988, 95 % CI, 1.064–8.394, p = 0.038; Table 4).
Table 3.
Associations between clinicopathological factors and overall survival among docetaxel-treated gastric cancer patients
| Clinicopathological factors | Short time survival, hazard ratio | 95 % confidence interval | p value |
|---|---|---|---|
| CHFR | |||
| CHFR unmethylation | 1.000 | ||
| CHFR methylation | 0.243 | 0.069–0.859 | 0.028* |
| Gender | |||
| Male | 1.000 | ||
| Female | 2.115 | 0.555–8.062 | 0.272 |
| Age (years) | |||
| n < 60 | 1.000 | ||
| n ≥ 60 | 1.215 | 0.289–5.113 | 0.790 |
| Tumor histology | |||
| Intestinal type | 1.000 | ||
| Diffused and mixed type | 0.815 | 0.391–1.701 | 0.586 |
| Differentiation | |||
| Well and moderately differentiated | 1.000 | ||
| Poorly differentiated | 1.997 | 0.440–9.061 | 0.370 |
| Tumor size | |||
| d < 5 | 1.000 | ||
| d ≥ 5 | 0.351 | 0.107–1.149 | 0.084 |
| Tumor invasion | |||
| T1–T2 | 1.000 | ||
| T3–T4 | 2.729 | 0.527–14.121 | 0.231 |
| Lymph node metastasis | |||
| N0–N1 | 1.000 | ||
| N2–N3 | 1.550 | 0.785–3.060 | 0.206 |
| Peritoneal or distant metastasis | |||
| M0 | 1.000 | ||
| M1 | 4.048 | 0.256–64.064 | 0.321 |
| Tumor stage | |||
| I–II | 1.000 | ||
| II–IV | 0.300 | 0.038–2.353 | 0.252 |
| Vessel invasion | |||
| Negative | 1.000 | ||
| Positive | 0.837 | 0.200–3.508 | 0.808 |
The multivariate Cox regression model showed that CHFR methylation was associated with a decreased (75.7 %) risk of shorter overall survival compared to CHFR unmethylation (HR 0.243, 95 % CI, 0.069–0.859, p = 0.028; Table 3) in docetaxel-treated gastric cancer patients. No significant association between the other clinicopathological factors and overall survival was observed
Table 4.
Associations between clinicopathological factors and overall survival among oxaliplatin-treated gastric cancer patients
| Clinicopathological factors | Hazard ratio | 95 % confidence interval (CI) | p value |
|---|---|---|---|
| MLH1 | |||
| MLH1 unmethylation | 1.000 | ||
| MLH1 methylation | 2.988 | 1.064–8.394 | 0.038* |
| Gender | |||
| Male | 1.000 | ||
| Female | 0.553 | 0.143–2.131 | 0.389 |
| Age (years) | |||
| n < 60 | 1.000 | ||
| n ≥ 60 | 0.398 | 0.142–1.116 | 0.080 |
| Tumor histology | |||
| Intestinal | 1.000 | ||
| Mixed | 1.173 | 0.704–1.955 | 0.539 |
| Differentiation | |||
| Well and moderately differentiated | 1.000 | ||
| Poorly differentiated | 0.653 | 0.184–2.319 | 0.510 |
| Tumor size | |||
| d < 5 | 1.000 | ||
| d ≥ 5 | 1.999 | 0.794–5.036 | 0.142 |
| Tumor invasion | |||
| T1–T2 | 1.000 | ||
| T3–T4 | 0.596 | 0.158–2.242 | 0.444 |
| Lymph node metastasis | |||
| N0–N1 | 1.000 | ||
| N2–N3 | 0.727 | 0.344–1.535 | 0.403 |
| Peritoneal or distant metastasis | |||
| M0 | 1.000 | ||
| M1 | 0.000 | 0.000 | 0.980 |
| Tumor stage | |||
| I–II | 1.000 | ||
| III–IV | 1.544 | 0.206–11.592 | 0.673 |
| Vessel invasion | |||
| Negative | 1.000 | ||
| Positive | 0.533 | 0.183–1.556 | 0.250 |
The multivariate Cox regression model showed that MLH1 methylation was associated with an increased (198.8 %) risk of shorter overall survival compared to MLH1 (HR 2.988, 95 % CI, 1.064–8.394, p = 0.038; Table 4) in oxaliplatin-treated gastric cancer patients. No significant association between the other clinicopathological factors and overall survival was observed. Statistical analysis was performed by the Cox proportional hazards model
p < 0.05 was considered statistically significant
p < 0.05
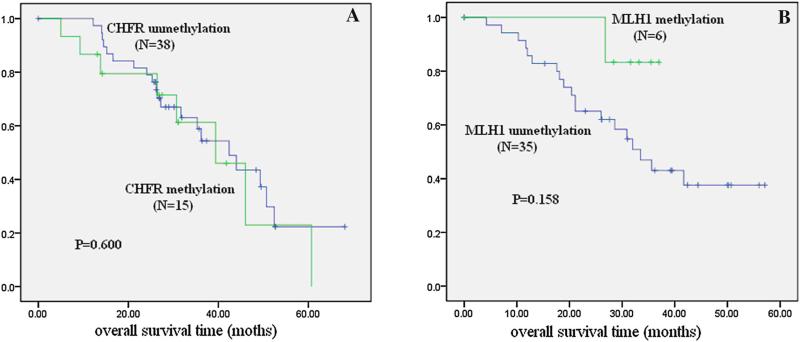
In the oxaliplatin-treated group, there was no significant difference in overall survival between the CHFR methylated group and unmethylated group (p > 0.05; Fig. 3a). In the docetaxel-treated group, there was no statistical difference in overall survival between the MLH1 methylation group and the unmethylated group (p > 0.05; Fig. 3b). These results further suggest that CHFR methylation is a docetaxel-sensitive marker, and that MLH1 methylation is a oxaliplatin-resistant marker in gastric cancer.
Fig. 3.
Association of gene methylation and overall survival in gastric cancer patients. Kaplan–Meier survival curves were constructed according to the presence of gene methylation in gastric cancer. a In the oxaliplatin-treated group, there is no significant difference in overall survival between the CHFR methylated group and the unmethylated group. b In the docetaxel-treated group, there is no statistical difference in overall survival between the MLH1 methylation group and the unmethylated group
Discussion
Hypermethylation of DNA repair genes is a frequent event in the carcinogenesis of gastric cancer. It is reasonable to explore the possibility of DNA methylation serving as prognostic and chemo-sensitive markers in gastric cancer. In 102 cases of primary gastric cancer, no FANCF was methylated; 34.4 % of CHFR was methylated, 21.6 % of MLH1 was methylated, 9.8 % of MGMT was methylated, and 12.7 % of RASSFA1 was methylated. Further analysis was only focused on CHFR and MLH1 because of the limited number of methylation cases.
Cell-cycle checkpoint dysfunction is often associated with sensitivity of chemotherapeutic agents [14, 15], but mutation of mitotic checkpoint genes is rare in gastric cancer [16, 17]. Satoh et al. [3] found that CHFR is frequently methylated in gastric cancers, and other mitotic checkpoint genes (BUB1, BUB1B, BUB3, MAD2L1, MAD2L2, CENP-E, and EB1) are not inactivated. Further studies indicated that CHFR methylation is a sensitive molecular marker of microtubule inhibitors (docetaxel and paclitaxel) in gastric cancer cells. However, the relationship of CHFR methylation status and responsiveness to microtubule inhibitors in primary gastric cancer remains unclear. In our study, the methylation rate of CHFR gene was similar to previous reports in gastric cancer [18–20]. Resistance to docetaxel treatment was found in CHFR unmethylated patients by the Cox proportional hazards model (HR 0.243, 95 % CI, 0.069–0.859, p = 0.028; Table 3), and the overall survival was longer in the CHFR methylated group than in the CHFR unmethylated group in docetaxel-treated gastric cancer patients (log-rank, p = 0.036; Fig. 2a). The results suggest that CHFR methylation is a sensitive marker for docetaxel in gastric cancer patients.
MLH1 is a major mismatch repair gene that has a role in maintaining the stability of the genome. Compared with colorectal cancer, mutations of DNA mismatch repair (hMLH1) are rare, despite the finding of high-frequency microsatellite instability (MSI) in gastric cancer [21, 22]. MSI is reported to be mainly caused by hypermethylation of the MLH1 promoter [23–25]. Our data demonstrated that MLH1 was methylated in 21.6 % of Chinese primary gastric cancer cases. The methylation rate is similar to other reports [26, 27]. DNA mismatch repair plays a critical role in maintaining genomic integrity. Loss of DNA mismatch repair occurs in many types of tumors. The ability of mismatch repair proteins to correct DNA mismatches that occur during DNA replication, repair, and recombination was considered to be the major mechanism for maintaining genomic stability, but increasing evidence supports the suggestion that the mismatch repair system also contributes to genomic stability by stimulating DNA damage-induced apoptosis as part of the cytotoxic response to chemical agents [28]. It was reported that loss of MLH1 expression results in resistance to both cisplatin and carboplatin in a colorectal cancer cell line [29]. In an ovarian tumor cell line, development of cisplatin resistance is associated with MLH1 promoter hypermethylation [30]. Previous studies in breast and ovarian cancer have shown that MLH1 methylation significantly correlates with poor survival [31, 32]. There is no report about MLH1 methylation status and oxaliplatin sensitivity in gastric cancer. Resistance to oxaliplatin treatment was found in MLH1 methylated gastric cancer patients in our study (HR 2.988, 95 % CI, 1.064–8.394, p = 0.038; Table 4), and the overall survival was longer in the MLH1 unmethylated group compared with the MLH1 methylated group in oxaliplatin-treated gastric cancer patients (log-rank, p = 0.046; Fig. 2b). However, there is no significant difference of overall survival between the CHFR methylation group and unmethylated group in the oxaliplatin-treated group, and no statistical difference of overall survival between the MLH1 methylation group and unmethylated group in the docetaxel-treated group in gastric cancer. These results suggest that MLH1 methylation may serve as an oxaliplatin resistance marker in gastric cancer. Some have tried to find methylation changes induced by chemotherapeutic reagents, but no strong evidence was found [33–35].
In conclusion, CHFR is frequently methylated in human gastric cancer, and CHFR methylation may serve as a marker of docetaxel sensitivity. MLH1 methylation was related to oxaliplatin-resistant gastric cancer patients.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (973 Program No. 2012CB934002, 2010CB912802); National High-tech R&D Program of China (863 Program No. SS2012AA020314, SS2012AA020821, SS2012AA020303); National Key Scientific instrument Special Programme of China (Grant No. 2011YQ03013405); and National Science Foundation of China (Grant No. 81121004, 81071953, 81161120432).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10120-014-0370-2) contains supplementary material, which is available to authorized users.
Conflict of interest J.G.H. is a consultant to MDxHealth. The other authors declare no conflict of interest.
Contributor Information
Yazhuo Li, Department of Pathology, Chinese PLA General Hospital, Haitangwan Town, Sanya 572000, Hainan, China; Department of Gastroenterology and Hepatology, Chinese PLA General Hospital, #28 Fuxing Road, Beijing 100853, China.
Yunsheng Yang, Department of Gastroenterology and Hepatology, Chinese PLA General Hospital, #28 Fuxing Road, Beijing 100853, China.
Youyong Lu, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Laboratory of Molecular Oncology, Peking University Cancer Hospital and Institute, Beijing 10000, China.
James G. Herman, Oncology Center, Johns Hopkins University, 1650 Orleans Street, Baltimore, MD 21231, USA
Malcolm V. Brock, Oncology Center, Johns Hopkins University, 1650 Orleans Street, Baltimore, MD 21231, USA
Po Zhao, Department of Pathology, Chinese PLA General Hospital, #28 Fuxing Road, Beijing 100853, China.
Mingzhou Guo, Department of Gastroenterology and Hepatology, Chinese PLA General Hospital, #28 Fuxing Road, Beijing 100853, China.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Yang Y, Xu C, Xiao L, Shen H, Zhang X, et al. CHFR suppression by hypermethylation sensitizes endometrial cancer cells to paclitaxel. Int J Gynecol Cancer. 2011;21:996–1003. doi: 10.1097/IGC.0b013e31821e05e8. [DOI] [PubMed] [Google Scholar]
- 3.Satoh A, Toyota M, Itoh F, Sasaki Y, Suzuki H, Ogi K, et al. Epigenetic inactivation of CHFR and sensitivity to microtubule inhibitors in gastric cancer. Cancer Res. 2003;63:8606–13. [PubMed] [Google Scholar]
- 4.Vos MD, Martinez A, Elam C, Dallol A, Taylor BJ, Latif F, et al. A role for the RASSF1A tumor suppressor in the regulation of tubulin polymerization and genomic stability. Cancer Res. 2004;64:4244–50. doi: 10.1158/0008-5472.CAN-04-0339. [DOI] [PubMed] [Google Scholar]
- 5.Koul S, McKiernan JM, Narayan G, Houldsworth J, Bacik J, Dobrzynski DL, et al. Role of promoter hypermethylation in cisplatin treatment response of male germ cell tumors. Mol Cancer. 2004;3:16. doi: 10.1186/1476-4598-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiegl H, Millinger S, Mueller-Holzner E, Marth C, Ensinger C, Berger A, et al. Circulating tumor-specific DNA: a marker for monitoring efficacy of adjuvant therapy in cancer patients. Cancer Res. 2005;65:1141–5. doi: 10.1158/0008-5472.CAN-04-2438. [DOI] [PubMed] [Google Scholar]
- 7.Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, Joenje H, et al. Disruption of the Fanconi anemia–BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–74. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 8.Swisher EM, Gonzalez RM, Taniguchi T, Garcia RL, Walsh T, Goff BA, et al. Methylation and protein expression of DNA repair genes: association with chemotherapy exposure and survival in sporadic ovarian and peritoneal carcinomas. Mol Cancer. 2009;8:48. doi: 10.1186/1476-4598-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4(4):296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 10.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–4. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 11.Paz MF, Yaya-Tur R, Rojas-Marcos I, Reynes G, Pollan M, Aguirre-Cruz L, et al. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10:4933–8. doi: 10.1158/1078-0432.CCR-04-0392. [DOI] [PubMed] [Google Scholar]
- 12.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 13.Zeller C, Dai W, Steele NL, Siddiq A, Walley AJ, Wilhelm-Benartzi CS, et al. Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene. 2012;31:4567–76. doi: 10.1038/onc.2011.611. [DOI] [PubMed] [Google Scholar]
- 14.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–9. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki H, Itoh F, Toyota M, Kikuchi T, Kakiuchi H, Imai K. Inactivation of the 14–3-3 sigma gene is associated with 5′-CpG island hypermethylation in human cancers. Cancer Res. 2000;60:4353–7. [PubMed] [Google Scholar]
- 16.Tanaka K, Nishioka J, Kato K, Nakamura A, Mouri T, Miki C, et al. Mitotic checkpoint protein hsMAD2 as a marker predicting liver metastasis of human gastric cancers. Jpn J Cancer Res. 2001;92:952–8. doi: 10.1111/j.1349-7006.2001.tb01186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shigeishi H, Yokozaki H, Kuniyasu H, Nakagawa H, Ishikawa T, Tahara E, et al. No mutations of the Bub1 gene in human gastric carcinomas. Oncol Rep. 2001;8:791–4. doi: 10.3892/or.8.4.791. [DOI] [PubMed] [Google Scholar]
- 18.Morioka Y, Hibi K, Sakai M, Koike M, Fujiwara M, Kodera Y, et al. Aberrant methylation of the CHFR gene in digestive tract cancer. Anticancer Res. 2006;26:1791–5. [PubMed] [Google Scholar]
- 19.Koga Y, Kitajima Y, Miyoshi A, Sato K, Sato S, Miyazaki K. The significance of aberrant CHFR methylation for clinical response to microtubule inhibitors in gastric cancer. J Gastroenterol. 2006;41:133–9. doi: 10.1007/s00535-005-1732-7. [DOI] [PubMed] [Google Scholar]
- 20.Gao YJ, Xin Y, Zhang JJ, Zhou J. Mechanism and pathobiologic implications of CHFR promoter methylation in gastric carcinoma. World J Gastroenterol. 2008;14:5000–7. doi: 10.3748/wjg.14.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiyama Y, Nakasaki H, Nihei Z, Iwama T, Nomizu T, Utsunomiya J, et al. Frequent microsatellite instabilities and analyses of the related genes in familial gastric cancers. Jpn J Cancer Res. 1996;87:595–601. doi: 10.1111/j.1349-7006.1996.tb00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura G, Sakata K, Maesawa C, Suzuki Y, Terashima M, Satoh K, et al. Microsatellite alterations in adenoma and differentiated adenocarcinoma of the stomach. Cancer Res. 1995;55:1933–6. [PubMed] [Google Scholar]
- 23.Fleisher AS, Esteller M, Wang S, Tamura G, Suzuki H, Yin J, et al. Hypermethylation of the hMLH1 gene promoter in human gastric cancers with microsatellite instability. Cancer Res. 1999;59:1090–5. [PubMed] [Google Scholar]
- 24.Ottini L, Falchetti M, Lupi R, Rizzolo P, Agnese V, Colucci G, et al. Patterns of genomic instability in gastric cancer: clinical implications and perspectives. Ann Oncol. 2006;17(suppl 7):97–102. doi: 10.1093/annonc/mdl960. [DOI] [PubMed] [Google Scholar]
- 25.Endoh Y, Tamura G, Ajioka Y, Watanabe H, Motoyama T. Frequent hypermethylation of the hMLH1 gene promoter in differentiated-type tumors of the stomach with the gastric foveolar phenotype. Am J Pathol. 2000;157:717–22. doi: 10.1016/S0002-9440(10)64584-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roa JC, Anabalón L, Roa I, Tapia O, Melo A, Villaseca M, et al. Promoter methylation profile in gastric cancer. Rev Med Chil. 2005;133:874–80. [PubMed] [Google Scholar]
- 27.Moura Lima E, Ferreira Leal M, Cardoso Smith Mde A, Rodríguez Burbano R, Pimentel de Assumpção P, Bello MJ, et al. DNA mismatch repair gene methylation in gastric cancer in individuals from northern Brazil. Biocell. 2008;32:237–43. [PubMed] [Google Scholar]
- 28.Li GM. The role of mismatch repair in DNA damage-induced apoptosis. Oncol Res. 1999;11:393–400. [PubMed] [Google Scholar]
- 29.Fink D, Nebel S, Aebi S, Zheng H, Cenni B, Nehmé A, et al. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881–6. [PubMed] [Google Scholar]
- 30.Strathdee G, MacKean MJ, Illand M, Brown R. A role for methylation of the hMLH1 promoter in loss of hMLH1 expression and drug resistance in ovarian cancer. Oncogene. 1999;18:2335–41. doi: 10.1038/sj.onc.1202540. [DOI] [PubMed] [Google Scholar]
- 31.Mackay HJ, Cameron D, Rahilly M, Mackean MJ, Paul J, Kaye SB, et al. Reduced MLH1 expression in breast tumours after primary chemotherapy predicts disease-free survival. J Clin Oncol. 2000;18:87–93. doi: 10.1200/JCO.2000.18.1.87. [DOI] [PubMed] [Google Scholar]
- 32.Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res. 2004;10:4420–6. doi: 10.1158/1078-0432.CCR-03-0732. [DOI] [PubMed] [Google Scholar]
- 33.Jung TY, Jung S, Moon KS, Kim IY, Kang SS, et al. Changes of the O6-methylguanine-DNA methyltransferase promoter methylation and MGMT protein expression after adjuvant treatment in glioblastoma. Oncol Rep. 2010;23(5):1269–76. doi: 10.3892/or_00000760. [DOI] [PubMed] [Google Scholar]
- 34.Lin R, Li X, Li J, Zhang L, Xu F, Chu Y, Li J. Long-term cisplatin exposure promotes methylation of the OCT1 gene in human esophageal cancer cells. Dig Dis Sci. 2013;58(3):694–8. doi: 10.1007/s10620-012-2424-9. [DOI] [PubMed] [Google Scholar]
- 35.Nyce JW. Drug-induced DNA hypermethylation: a potential mediator of acquired drug resistance during cancer chemotherapy. Mutat Res. 1997;386(2):153–61. doi: 10.1016/s1383-5742(96)00051-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.