Abstract
Convincing epidemiological data suggest an inverse association between cancer and neurodegeneration, including Alzheimer's disease (AD). Since both AD and cancer are characterized by abnormal, but opposing cellular behavior, i.e., increased cell death in AD while excessive cell growth occurs in cancer, this motivates us to initiate the study into unraveling the shared genes and cell signaling pathways linking AD and glioblastoma multiform (GBM). In this study, a comprehensive bioinformatics analysis on clinical microarray datasets of 1,091 GBM and 524 AD cohorts was performed. Significant genes and pathways were identified from the bioinformatics analyses – in particular ERK/MAPK signaling, up-regulated in GBM and Angiopoietin Signaling pathway, reciprocally up-regulated in AD – connecting GBM and AD (P < 0.001), were investigated in details for their roles in GBM growth in an AD environment. Our results showed that suppression of GBM growth in an AD background was mediated by the ERK-AKT-p21-cell cycle pathway and anti-angiogenesis pathway.
The incidence of both cancer and Alzheimer's disease (AD) increases with age, but more and more studies have shown an inverse relationship of the two diseases; older persons with cancer have a reduced risk of AD and vice versa. For example, a recent nationwide population-based study of 6,960 patients with AD in Taiwan showed that patients with AD had a reduced risk of developing overall cancer [standardized incidence ratios (SIRs) = 0.88, 95% confidence interval (CI) = 0.80–0.97]1. Another case control analysis in a recent epidemiological study, based on the famed Framingham heart study consisting of 1,278 participants, concluded that patients with probable AD had a 61% decreased risk of incident cancer while cancer survivors (including GBM) had a 33% decreased risk of developing AD2. Similar findings of a significantly lower rate of hospitalization for cancer among AD patients [hazard ratio (HR) = 0.31, 95%CI = 0.12–0.86, p = 0.0237] and for a diagnosis of AD among white cancer patients [HR = 0.57, 95%CI = 0.36–0.90, p = 0.0155] have also been reported in two prospective studies by Roe and colleagues3,4. There was a recent cohort study within a population-based sample of adults aged 60 years and older in Northern Italy. They found that persons with incident AD (n = 2,832) were less likely to develop cancer in the future, and persons with incident cancer (n = 21,451) were less likely to develop AD later. Specifically, when compared to the general population of persons of the same age and sex, the risk of cancer in patients with AD dementia was decreased by 43%, while the risk of AD dementia in patients with cancer was reduced by 35%5. Furthermore, studies have shown that patients with Parkinson's disease, another neurodegenerative disease, also have reduced risk of cancer incidence6,7. Although these epidemiological studies indicate that cancer survivors may gain some protection from neurodegeneration, an in-depth understanding of the biological mechanism associated with this inverse relationship will help researchers gain more knowledge in understanding both of these complicated diseases.
Reports have shown that many molecular mechanisms are shared between the maintenance of neural function in neurodegenerative diseases and cell proliferation in oncogenic pathways8,9. Microarray correlation analyses revealed up-regulation of many tumor suppressors in AD with functions in phosphorylation, apoptosis, cell cycle, and other categories related with cancer (e.g., TGF-β, CDK2AP1)10. In addition, several molecules, like Pin111,12 and GSK313,14, have been found to be inversely proportional in expression between AD and GBM. Pin1 is necessary for cell division; its inhibition causes regression of tumors15, whereas in mouse models of AD its up-regulation in postnatal neurons reverses neurodegeneration16. GSK3, an up-regulated AD-related gene that exhibits tumor suppressor activity in the Wnt pathway17, also plays a role in the hyperphosphorylation of tau in AD models18. Taken together, these data suggest that there is a notable difference in the intrinsic genomic nature between these two diseases; thus, systemically studying the whole transcriptome may shed insight on their pathophysiological differences.
We performed a comprehensive bioinformatics analysis on clinical microarray datasets of GBM cohorts and AD cohorts in comparison to age-matched normal subjects in order to identify inversely regulated transcriptional processes and signaling pathways between GBM and AD. Furthermore, we elaborated in detail the specific roles of two significant signaling pathways in gliomagenesis and GBM tumor growth in an AD environment. One pathway the ERK/MAPK pathway is up-regulated in GBM, the other the Angiopoietin Signaling pathway is reciprocally up-regulated in AD. Since the Aβ peptide is naturally produced in brain and has been studied as an important biomolecule connecting AD and cancer19,20,21, we evaluated the effects of natural Aβ on GBM tumor cell migration, invasion, and GBM growth in APPswe transgenic mice. Our results showed that suppression of GBM growth in an AD background was mediated by the ERK-AKT-p21-cell cycle pathway and anti-angiogenesis pathway.
Results
Biological processes and cellular signaling pathways associated with AD and GBM inversion
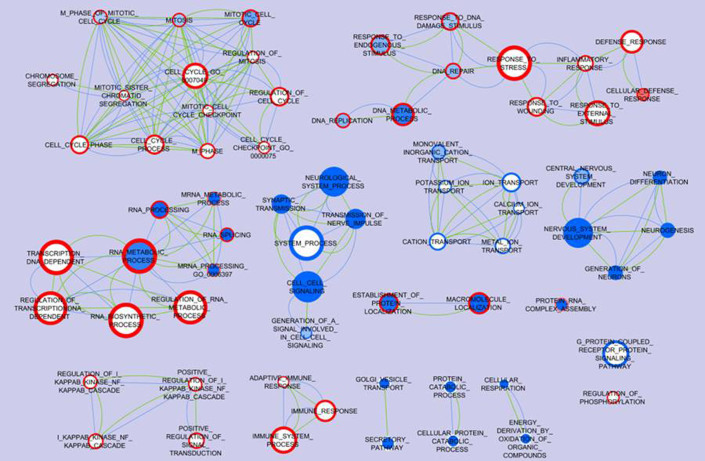
Through analyses of the clinical microarray datasets of AD and GBM cohorts, a total of 2,103 differential genes (827 up-regulated and 1,061 down-regulated) for AD patients and 1,384 differential genes (704 up-regulated and 634 down-regulated) for GBM patients were found to be significant (P < 0.01). Cytoscape pathway analyses of these genes determined the general biological processes that correlate with both diseases (Fig. 1). Gene sets involved in four biological processes were up-regulated in GBM and down-regulated in AD (Fig. 1): mitotic cell cycle, DNA metabolic process, RNA metabolic process, and protein localization. Gene sets involved in the immune system process IKB Kinase/NFKB cascade and regulation of phosphorylation were up-regulated in GBM and showed no significant changes in AD. Cell-cell signaling and central nervous system development gene sets showed down-regulation in both AD and GBM. Some other processes were either down-regulated in AD (vesicle transport, protein catabolic and cellular respiration) or in GBM (ion transport and G-protein coupled receptor protein signaling), but showed no significant changes in the other disease.
Figure 1. Biological processes associated with AD and GBM inversion.
Differentially expressed genes identified from each AD or GBM microarray dataset compared to the normal subjects control were performed gene set enrichment analysis (GSEA) for biological processes. Enrichment Map software was used to visualize the biological processes association between AD and GBM. Each node represents a gene ontology (GO) term, and all of the nodes pose GSEA p-value < 0.01 and FDR q-value < 0.25. The color tone in the face of the node represents the regulation in AD datasets: red means the genes are up-regulated while blue means the genes are down-regulated; meanwhile the color tone in the boundary of the node represents the regulation in GBM datasets. The actual color corresponds to the GSEA enrichment p-value: darker red or blue means this GO term is more enriched with differentially expressed genes, while white means the corresponding GO term is not enriched; e.g. the node Mitosis has a blue face color and dark red boundary color indicating high enrichment of down-regulated genes in the AD datasets and up-regulated genes in the GBM datasets. The edges indicate that the connected two nodes share at least 30% of genes, and the thicker edge represents a larger ratio of shared genes. The green edges represent the shared genes based on the annotation of AD datasets and the blue edges represent the shared genes based on the annotation of GBM datasets; e.g. DNA Metabolic process and DNA Replication share more than 30% genes in both the GBM and AD datasets.
In the aforementioned example, Pin1 is prevalently up-regulated in human cancers, and opposite regulation of this gene contributes to hallmark symptoms in neurodegeneration. To analyze and find such genes systematically, our research focuses on the set of genes that are inversely regulated between AD and GBM. Cross-comparison of significantly reversely expressed genes, genes up-regulated in GBM and down-regulated in AD (GBM+/AD−) and genes up-regulated in AD and down-regulated in GBM (AD+/GBM−), were further explored. We identified 184 genes in GBM+ & AD− and 69 genes in GBM− & AD+ (Supplementary Table S1, S2). Ingenuity Pathway Analysis (IPA) software was used to analyze cellular signaling pathways for each group of genes. For GBM+ & AD− genes, significant pathways found include: cAMP signaling, IL-3 signaling, 14-3-3 mediated signaling, axonal guidance signaling, IGF-1 signaling, ERK/MAPK signaling, and inhibition of angiogenesis by TSP1 (Table 1). For GBM−&AD+ genes, significant pathways include: interleukin signaling, PDGF signaling, PPAR signaling, TNFR1 signaling, angiopoietin signaling, and NF-κB signaling (Table 1).
Table 1. IPA analysis of significant genes and pathways up-regulated in GBM+&AD− and GBM−&AD+ gene sets.
| GBM+&AD− | ||
|---|---|---|
| Significant Pathways | P-Value | Ratio |
| cAMP-mediated signaling | 4.07E-05 | 5.16E-02 |
| IL-3 Signaling | 1.86E-04 | 8.22E-02 |
| 14-3-3-mediated Signaling | 3.80E-04 | 6.09E-02 |
| Axonal Guidance Signaling | 7.08E-04 | 3.08E-02 |
| Rac Signaling | 1.38E-03 | 5.13E-02 |
| Protein Kinase A Signaling | 2.82E-03 | 3.27E-02 |
| Inositol Phosphate Metabolism | 4.90E-03 | 4.44E-02 |
| IGF-1 Signaling | 5.75E-03 | 4.90E-02 |
| ERK/MAPK Signaling | 6.17E-03 | 3.54E-02 |
| Inhibition of Angiogenesis by TSP1 | 6.76E-03 | 9.09E-02 |
| Glutamate Metabolism | 7.94E-03 | 8.57E-02 |
| Chemokine Signaling | 8.91E-03 | 5.80E-02 |
Aβ mediates the ERK/MAPK pathway differentiation in AD versus GBM
From the clinical microarray data analysis, we observed that ERK/MAPK signaling genes exhibited inverse expression between AD and GBM patients (up-regulated in GBM and down-regulated in AD). Specifically, in the GBM U133A and Agilent datasets, the Erk1 gene shows 1.77- (P = 1.69E-11) and 1.78- (P = 8.90E-19) fold increase, and the Erk2 gene shows 2.37- (P = 6.79E-08) and 1.52- (P = 7.29E-09) fold increase, in comparison with the normal controls in the two datasets, respectively; in the AD GSE5281 and GSE15222 datasets, Erk1 gene shows 1.28- (P = 6.29E-04) and 1.22-(P = 1.18E-02) fold decrease, and Erk2 gene shows 2.26- (P = 1.99E-10) and 1.52- (P = 3.36E-09) fold decrease, in comparison with the normal controls. Although elevated Erk1/2 as a critical signaling pathway in GBM is well-appreciated22, what molecular determinants trigger the ERK/MAPK pathway differentiation in AD versus GBM has rare been explored.
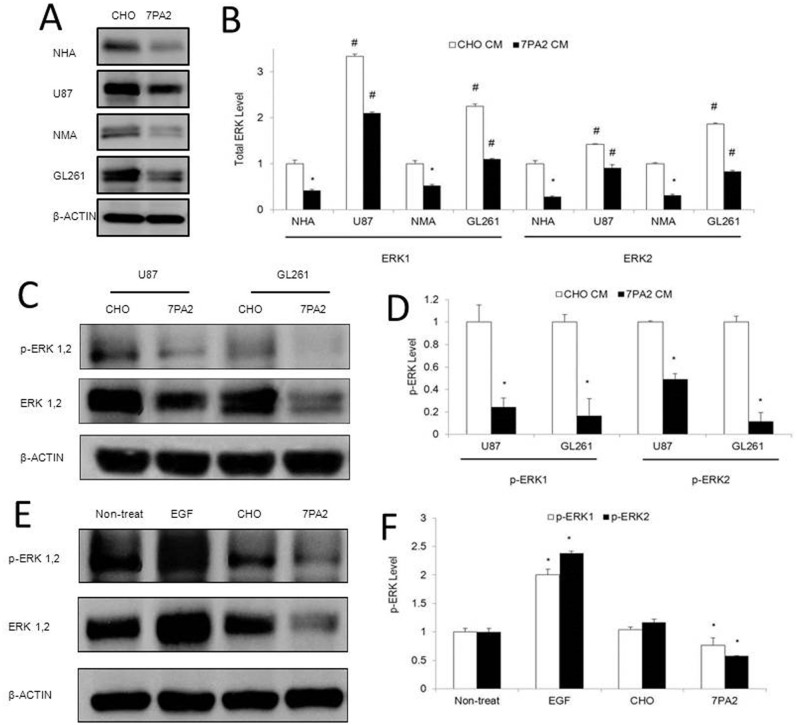
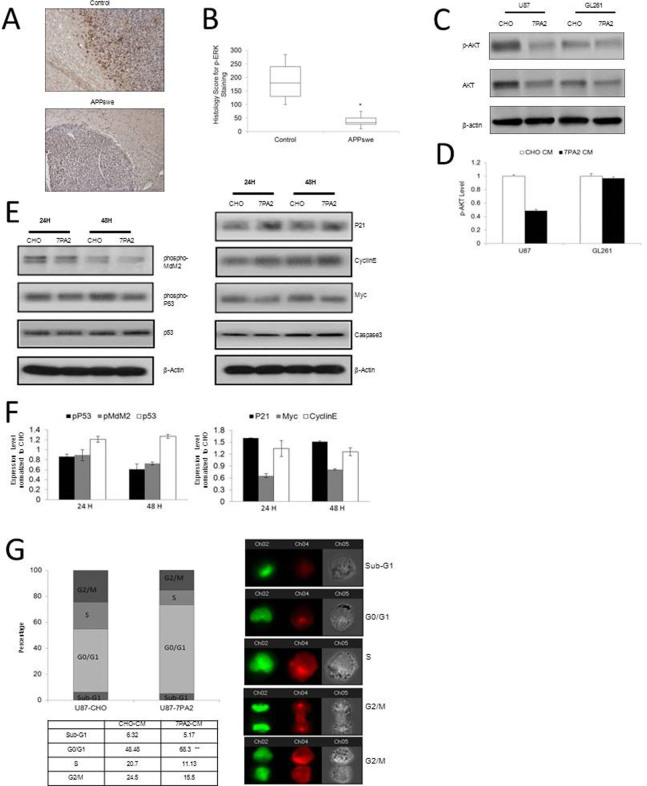
Aβ peptide is naturally produced in brain, and accumulates extensively in brains of AD patients23. Studies also hinted Aβ as an important biomolecule connecting AD and cancer. For example, inflammation responses to Aβ mediated by glial cells in the brain24, can be tumor-suppressive20,21. Aβ leads to abnormal Ca2+ signaling25, which can inhibit tumor growth and progression26. We thus investigated how Aβ would trigger the inverse activation of ERK1/2 signaling in AD and GBM conditions. To mimic the AD condition in vitro, as in our previous studies, we took advantage of conditioning media (CM) from Aβ precursor protein (APP)-expressing cells27, which contains natural forms of Aβ monomers and oligomers28. Because astrocytes are the most abundant cell of the human brain, we first applied the Aβ-CM to normal human astrocytes and found that the ERK1 and 2 protein expression decreased significantly in comparison to the cells cultured in the non Aβ-CM (Fig. 2A, B).
Figure 2. Effects of the Aβ-containing conditioned medium on basal and activated ERK expression in astrocytes and brain tumor cells.
(A–B). Normal human (NHA) and mouse (NMA) primary astrocytes, and human GBM (U87) and mouse GBM (GL261) cells were exposed to Aβ-containing conditioned medium (7PA2-CM) for 72 hours, and total ERK expression was detected by Western blot. (C–D). U87 and GL261 cells exposed to 7PA2-CM for 30 minutes were detected for phosphorylated ERK1/2 (pERK1/2). (E–F). U87VIII cells exposed to EGF and 7PA2-CM for 1 hour were detected for pERK1/2. *P < 0.05 vs. CHO-CM. The quantification data were obtained from three independent experiments under the same condition. In all panels, western blots images shown are cropped to show the protein of interest, and all blots were performed under the same experimental conditions.
In clinical samples, the total ERK1 and 2 protein expression showed decrease in the prefrontal neocortices of AD patients that correlates with the high level of Aβ29. Secondly, we detected a strong inhibition on the highly expressed total ERK1/2 in GBM cell lines by the Aβ-CM (Fig. 2A, B). Moreover, we examined the effect of CM on the activation of ERK1/2 in GBM cell lines. In basal conditions, ERK1/2 phosphorylation in U87 and GL261 cells decreased 30–40% after 30 min-treatment with Aβ-CM (Fig. 2C, D). Treatment of U87VIII cells (a U87 cell line overexpressing the epidermal growth factor (EGF) receptor) with EGF, an activator of ERK1/2 signaling pathway, produced a 2-fold increase of ERK1/2 phosphorylation. Aβ-CM almost completely abolished EGF-induced activation of ERK1/2 (Fig. 2E, F). These results suggest that the elevated Aβ may lead to the decreased ERK1/2 signaling in AD. While in GBM, the ERK1/2 signaling is intrinsically activated, and the signaling activity could be suppressed in an AD environment.
High level Aβ suppresses gliomagenesis and GBM tumor growth through negatively regulate ERK1/2 signaling
To further explore how Aβ-CM or an AD environment would affect the tumorigenesis or GBM tumor development, we collected two lines of evidence. First, abolished ERK1/2 phosphorylation by a combined Mek1/2 knockout (the only known MEK1/2 targets are ERK1 and ERK2) in development or adulthood mice led to a diminished oncogenic RAS-induced hyperplasia30, which is the driving force for many human GBMs31. Since both the basal and growth factor stimulated-ERK1/2 phosphorylation could be suppressed significantly by the Aβ-CM, we postulate that such suppression might lead to the extinction of gliomagenesis in the AD background.
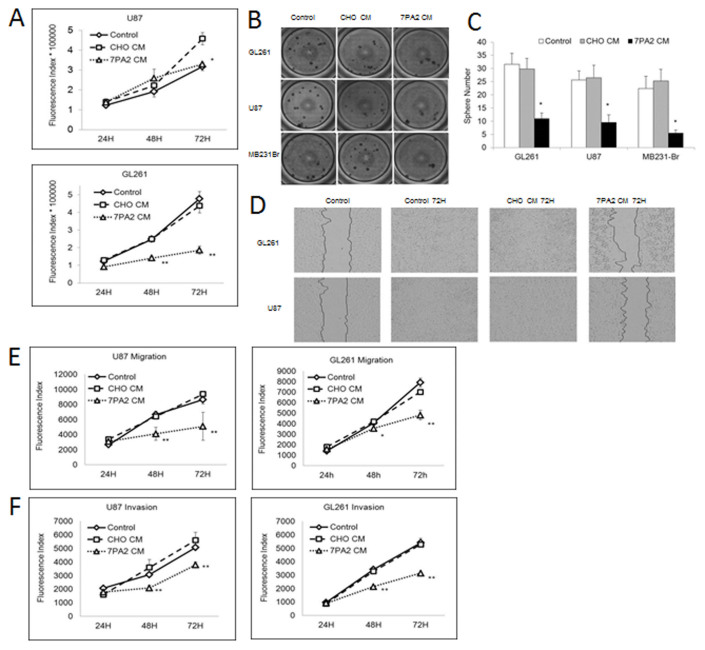
Second, GBM tumor development was retarded in the AD environment. In specific, two GBM cell lines were studied in vitro. The proliferation of GL261 cells was inhibited early from 24 h Aβ-CM culturing (P < 0.01) (Fig. 3A), and U87 cells exhibited decreased proliferation until 72 h Aβ-CM culturing (P < 0.05). For the three-dimensional proliferation determined by the anchorage-independent growth in soft agar, tumor cells cultured in Aβ-CM exhibited 60–70% reduced colony formation in GL261 and U87 cell lines (Fig. 3B, C). Aβ-CM also reduced the cell migration by both the wound healing migration assay and Boyden chamber assay (Fig. 3D, E). In the invasion assay, U87 cells exposed to the Aβ-CM exhibited at least 32% (P < 0.01) and 25.3% (P < 0.01) fewer penetrating cells in comparison to other control groups at 48 h and 72 h respectively. Similar results were found for GL261 (Fig. 3F).
Figure 3. Aβ-containing conditioned medium on tumor cell proliferation, migration and invasion.
(A). Proliferation of GL261 and U87 cells grown in 7PA2-CM and control medium. (B–C). Anchorage-independent growth of GL261 and U87 cells grown in 7PA2-CM and control medium. (D–E). Migration abilities of GL261 and U87 cells grown in 7PA2-CM and control medium. (F). Invasion abilities of GL261 and U87 cells grown in 7PA2-CM and control medium. *P < 0.05, **P < 0.01 vs. CHO-CM. The quantification data were obtained from three independent experiments under the same condition.
In addition to Aβ, APP over-expressing cells also secrete high levels of soluble APP, the majority of which is APPsα, which is derived from α-secretase cleavage of APP, with very low levels of APPsβ (derived from β-secretase cleavage of APP). We thus utilized the conditioned media containing a high level of APPsα but only endogenous levels of Aβ (undetectable by our ELISA) from a stable HEK293 cell line overexpressing soluble APPsα. In previous work on multiple tumor cell lines and under different culturing periods, we did not observe any inhibitory effects on tumor cell proliferation27. Moreover, we identified that the inhibitory effect on tumor cell proliferation correlates with Aβ concentrations in the Aβ-CM27. These previous results ruled out the involvement of APPsα in inhibiting GBM tumor cell proliferation in the current same setting. We thus assert that Aβ inhibited the GBM tumor cell proliferation, migration and invasion.
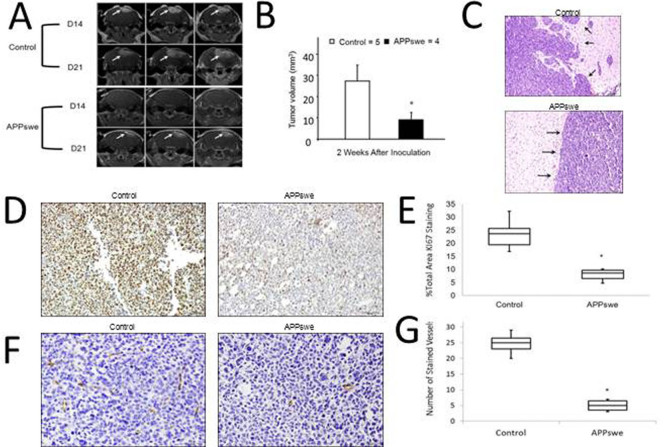
Our in vitro findings were further corroborated by a subsequent in vivo study, in which we explored GBM growth in APPswe transgenic mice. GL261 murine GBM cells were intracranially injected into 12-month old APPswe transgenic mice and aged-matched littermate control normal mice. The Aβ level in the 12-month old APPswe mice brains is high32 and the plaque deposition is visible by microscope (Fig S1A). The results indicated that average GBM tumor volume and growth had been significantly suppressed in the APPswe group in comparison to the normal control group (P < 0.05, student's t test) (Fig. 4A, B). Furthermore, histological studies revealed decreased Ki67-positive proliferative tumor cells in the brain tumors of APPswe mice (Fig. 4D, E). APPswe transgenic mice also showed more confined tumor boundaries than those in the control group, which indicates less migration and invasion of tumor cells (Fig. 4C). Apoptosis, examined by TUNEL staining, was undetected in all mice (data not shown). APPswe transgenic mice have been reported with astrocytosis and gliosis in brains, which may affect the injected GL261 cells. However, previous studies in injecting Aβ directly into the GBM tumors of young mice without astrocytosis also reported similar findings of reduced tumor volume. Thus, we conclude that Aβ played a major role in inhibiting the GBM tumor growth.
Figure 4. GBM growth in AD transgenic mice.
GL261 murine GBM cells were intracranially injected into 12-month old APPswe transgenic and litter-match wild-type mice. (A). Representative brain MRI images for 3 slices of each brain of the AD and control mice. Arrows indicate the tumor. (B). Tumor volume from week 2 was calculated from the MRI whole brain slices and data was presented as mean with SD. (C). Representative H&E images of tumor boundary in AD and control mice. Arrows indicate the tumor boundary regions. (D). Representative images of Ki67-positive proliferative cells in the brain tumors of AD and control mice (100×). (E). Ki67-positive cells (brown staining) were counted in 5 fields of 5 sections/mouse brain under 20× objective, and data was presented as % of Ki67-positive cells to the total cells. (F). Representative images of CD34-positive microvessels (brown staining) in the non-tumor area and tumor area of AD mice (200×). (G). CD34-positive microvessels were counted in 5 fields of 5 sections/mouse brain under 20× objective, and data was presented as mean with SD. *P < 0.05 vs. wild-type control. n = 4 for APPswe transgenic mice and n = 5 for litter-match wild-type mice.
In addition to the in vitro data that Aβ-CM decreased the expression and activation of ERK1/2 in GBM cell lines (Fig. 2), we also observed lower p-ERK1/2 immunostaining in the tumor regions of the APPswe transgenic mice in contrast to the normal control mice (Fig. 5A, B). Especially in the tumor margin area, the invasive tumor cells forming metastatic satellites show strong p-ERK1/2 expression in the normal control mice, while the high expression of p-ERK1/2 was significantly suppressed in the APPswe transgenic mice. These results suggest that a high level of Aβ suppresses GBM growth through negatively regulating the ERK1/2 activation.
Figure 5. ERK1/2 and AKT signaling connecting AD and GBM.
(A). Representative images of p-ERK1/2 immuno-staining in the brain tumor margin area of AD and wild-type control mice (100×). (B). Quantification of the H scores for p-ERK1/2 immuno-reactivity. *P < 0.05 vs. wild-type control. n = 4 for APPswe transgenic mice and n = 5 for litter-match wild-type mice. (C–D). U87 and GL261 cells exposed to 7PA2-CM for 30 minutes were detected for pAKT expression. (E–F). Signaling proteins downstream to ERK1/2 and AKT in the U87 cells exposed to 7PA2-CM for 24 and 48 hours. (G). Cell cycle distribution of U87 cells exposed to 7PA2-CM for 24 hours. The in vitro quantification data were obtained from three independent experiments under the same condition. *P < 0.05 vs. CHO-CM. In all panels, western blots images shown are cropped to show the protein of interest, and all blots were performed under the same experimental conditions.
ERK-AKT crosstalk and p21-cell cycle mechanism dominates the Aβ-mediated GBM suppression
The ERK/MAPK pathway is known to have crosstalk with the PI3K/AKT pathway33, and specifically from our AD and GBM clinical microarray data analysis, the Akt1 gene was examined to be consistently down-regulated in both GBM datasets (1.8- and 1.24-fold decrease in the U133A and Agilent data, respectively) and up-regulated in both AD datasets (1.1- and 1.26-fold increase in the GSE5281 and GSE15222 datasets, respectively).
We further explored how the Aβ-CM would affect AKT and the downstream signals behind ERK1/2-AKT crosstalk in suppressing GBM. Although less noticeable than p-ERK, GBM cells exposed to the Aβ-CM also had reduced expression of p-AKT compared to the cells exposed to the non-Aβ-CM (Fig. 5C, D). Proteins located downstream of the ERK/MAPK and PI3K/AKT pathways were tested, and the results showed increased expression of p21, cyclin E, and p53, and reduced expression of Myc, p-MDM2, and p-p53 in GBM cells treated with Aβ-CM, but no changes of caspase-3 were detected by the Aβ-CM treatment (Fig. 5E, F). Together with the data from cell cycle analyses (Fig. 5G), the results suggest that negative regulation on cell cycle (G1 arrest) rather than cell survival or apoptosis by Aβ was the dominant mechanism behind the ERK1/2-AKT crosstalk in suppressing GBM (Fig. 6).
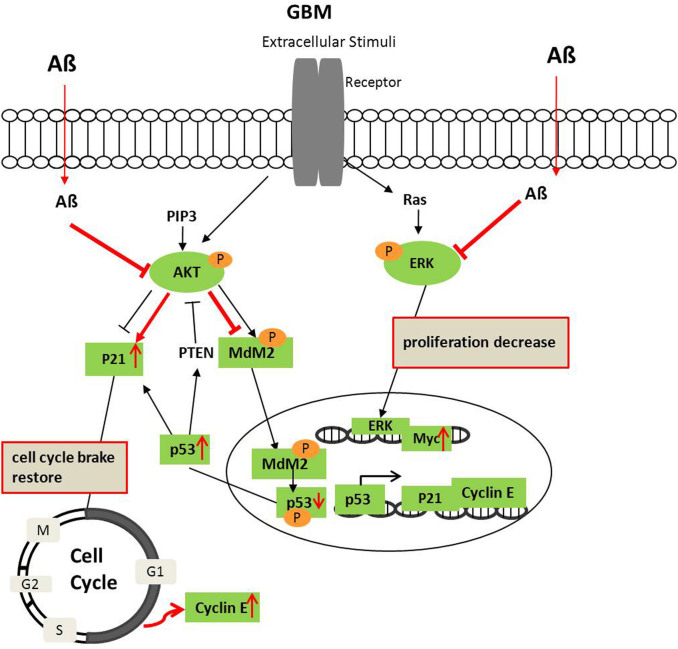
Figure 6. Diagram of Aβ affecting the ERK1/2 and AKT signaling crosstalk in GBM.
AKT and ERK are de-phosphorylated in GBM after Aβ treatment. AKT deactivation decreases MDM2 and p53 phosphorylation and increases stable p53, which trigger its downstream p21 and cyclin E expression. Simultaneously, p53 may increase PTEN to suppress AKT activation further. Red labels indicate the effects of Aβ on GBM.
Distinction of angiogenesis signaling in GBM and AD
In addition to the up-regulated ERK1/2 signaling in GBM, we found another important signaling mechanism that is reciprocally up-regulated in AD and down-regulated in GBM angiopoietin signaling (Table 1). Specifically, in the AD GSE5281 and GSE15222 datasets, the ANGPT1 gene shows 1.68- (P = 3.04E-08) and 1.59- (P = 4.08E-011) fold increase, and the ANGPNT2 gene shows 1.76- (P = 2.76E-14) and 1.51- (P = 5.12E-09) fold increase, in comparison with the normal controls. Angiopoietins are proteins binding with an endothelial cell-specific tyrosine-protein kinase receptor to stimulate sprouting angiogenesis. In AD brains, large populations of endothelial cells are activated by angiopoietins due to brain hypoxia and inflammation, and secrete the precursor substrate as well as the neurotoxic Aβ peptide to kill neurons34. Although GBM is highly angiogenic35, we observed the deactivation of angiopoietin signaling and activation of inhibition of angiogenesis by TSP1 signaling (Table 1). Specifically, in the GBM U133A and Agilent datasets, Angpt1 and Angpt2 genes show 2.23- and 5.86- (P = 0.004 and 1.95E-10), and 1.38- and 4.74- (P = 0.023 and 3.56E-31) fold decrease while Tsp1 gene shows 1.85- (P = 0.005) and 1.72- (P = 1.51E-05) fold increase, in comparison with the normal controls in the two datasets, respectively. These results suggest that distinctive angiogenic signaling mechanisms might be involved in GBM and AD although hyper-angiogenesis is a common pathophysiological character for both diseases.
Clinically, the angiogenesis inhibitor bevacizumab has been shown to increase progression-free survival in GBM patients majorly through inhibiting the vascular endothelial growth factor (VEGF)-dependent vessels formation35, which confirmed our finding that angiopoietin-mediated angiogenesis may not be a major mechanism in GBM. Nevertheless, in our animal study, we found a decreased level of CD34-positive microvessels in the GBM tumor area in the APPswe mice compared with the non-tumor area (Fig. 4F, G). A previous study also showed that intra-tumoral injection of Aβ potently inhibits the angiogenesis of human GBM and thus inhibits the growth of the tumor19. Although how Aβ would impact angiogenesic signaling is not clear; taken together, the facts indicate that our bioinformatics analysis was able to identify the detailed distinction of angiogenesis signaling in GBM and AD.
Discussion
The inverse trend of brain cancer patients rarely developing AD and AD patients rarely developing brain cancer presents an enticing entry point in the study of these two diseases. Many links have been found between cancer and aging36. For example, genetic mutations in the DNA damage repair pathway lead to fast-aging and tumorigenesis. Impaired autophagy associates with tumor formation and age-related neuronal degeneration. Also, extrinsic factors37 and protein translational factors38 could affect the inverse trend exhibited between AD and GBM. The present study, however, focused on a different angle in studying patterns in the respective transcriptome that could link the inverse relation of AD and GBM. Overall, our bioinformatics results reveal widespread changes in transcriptional regulation of multiple cellular pathways in AD and GBM. Importantly, our study is the first to elucidate GBM mechanisms using AD as a reference in a multidisciplinary disease systems approach using a combination of in vitro, in vivo, and computational methods.
Our analysis on the clinical microarray datasets revealed biological processes that inversely correlate with the two diseases. Not surprisingly, we found some biological processes are up-regulated in GBM and down-regulated in AD, such as in mitotic cell cycle, DNA replication, metabolism and repair, and RNA metabolism and processing into proteins. This coincides with alternative cell cycle hypotheses in explaining the causes of many degenerative diseases, AD included39. This explanation is based on the post-mitotic nature of neurons, which normally do not proliferate. Anomalies in cell cycle regulators and checkpoint proteins spur neurons resting in G0 phase back into cell-cycle-initiating G1 and S phases. While DNA is replicated, the inability to complete cell mitotic division and aneuploidistic chromosome numbers lead to neuronal cell death and degeneration40,41. GBM and all other cancers are also characterized by anomalies in cell cycle regulators and DNA damage-related factors42,43, albeit a different nature from trends seen in neurodegenerative diseases. In juxtaposition, this presents an interesting cell cycle connection between cancer and AD; cancer is characterized by the uncontrolled over-proliferation of cell division, whereas AD exhibits forced re-entry and inability to divide in the cell cycle.
In comparison to the general biological process, cellular signaling pathways were identified underlying the significantly inversely expressed genes between AD and GBM. We identified the transcriptional responses of ERK/MAPK signaling are significantly up-regulated in GBM and down-regulated in AD, and reversely, PI3K/Akt signaling are up-regulated in AD and down-regulated in GBM. Crosstalk between them has been implicated in the regulation of both G1/S cell cycle progression and G2/M transition44. Akt can negatively regulate the Raf/MEK/ERK pathway through specifically phosphorylating Raf45, which may switch the biological response from growth arrest to proliferation in cancer cells46. Thus, in the scenario of GBM, the decreased Akt and increased ERK1/2 expression favor the tumor cell proliferation while in the scenario of AD, the increased Akt and decreased ERK1/2 mediate the down-regulation of cell cycle progress, especially the mitotic cell cycle (Fig. 1). Furthermore, our data showed that Aβ containing medium strongly inhibited the activation of ERK1/2 and moderately inhibited the activity of AKT in the GBM cells, suggesting that the effect of Aβ on endogenous MEK/ERK activation in regulating cell cycle is more potent than on the AKT activation in protecting GBM cells from apoptosis. Our data on examining the apoptosis signals, including caspase 3 and Bcl-2, were not changed, but the G1-S cell cycle regulators, including p21 and cyclin E were remarkably increased by the treatment of Aβ containing medium, provided direct evidence to support this conclusion. In addition, AKT deactivation decreased MDM2 and p53 phosphorylation and increased stable p53, which may trigger its downstream p21 and cyclin E expression. Simultaneously, stable p53 may lead to the increase of PTEN to suppress AKT activation further (Fig. 6).
ERK1/2 can regulate processing of Aβ and phosphorylation of the microtubule-associated protein tau, both events critical to the pathophysiology of AD29,47,48. It has been reported that the increased ERK1/2 in cerebrospinal fluid (CSF) may serve as a biomarker for AD49. In our study, we found the expression of ERK1/2 genes was decreased in the brain tissue samples of AD patients. We reasoned that both the decreased expression of ERK1/2 in brain tissue and increased ERK1/2 in CSF occur at a later disease stage. ERK1/2 acts as a restraint to the processing of APP and production of Aβ42, through negative regulation of β-secretase (BACE1) and γ-secretase, the endo-proteases essential for the production of Aβ peptides29,47. In the late stage disease condition, the overwhelming Aβ production causes the excessive negative feedback to ERK1/2 expression and activity, and the vulnerable neurons or other types of cells in the brain may release the cellular proteins, such as ERK1/2.
Besides the ERK/MAPK pathway, many other significant pathways from our analysis are related to the signature pathophysiology found in AD and GBM, such as the angiopoietin signaling and inhibition of angiogenesis by TSP1 signaling. Clinical data favors our conclusion that distinctive angiogenic signaling mechanisms might be involved in GBM and AD. Further validation on these pathway signals in driving the exclusive disease patterns between AD and GBM may offer insights into alternative therapeutic strategies. Nevertheless, the current study of an in-depth bioinformatics analysis opens a new vista in discovering insights connecting these two complicated diseases.
Methods
Microarray data and pathway analysis
Four age-matched clinical microarray datasets were used to explore the inversely expressed genes between the two diseases, 2 for GBM (TCGA) and 2 for AD (GSE 5281, GSE 15222). The 2 GBM microarray datasets contain 555 samples (545 GBM patients, 10 controls) and 536 samples (517 GBM patients, 19 controls), and were analyzed with Affymetrix U133A and Agilent platforms respectively. The 2 AD microarray datasets contain 161 samples (87 AD patients, 74 controls) and 363 samples (176 AD patients, 187 controls), and were analyzed with Affymetrix HG-U133_Plus_2 and Sentrix Human Ref-8 Expression BeadChip platforms respectively. Using 2 independent microarray platforms for each disease alleviates errors caused by data noise from each microarray platform itself and improves confidence of results. Partek and R software packages were used to process the raw data, and student's t-test was applied to identify differentially expressed genes (G) for each disease compared with its respective controls. Then, oppositely up-regulated (+) and down-regulated (−) genes for GBM and AD were compared to one another (GBM+&AD− and GBM−&AD+). Gene Set Enrichment Analysis (GSEA), Cytoscape and Ingenuity Pathway Analysis (IPA) software packages were used for analyzing and visualizing gene set enriched biological processes and signaling pathways for the genes in GBM+&AD− and GBM−&AD+ groups.
Cell lines and Aβ-conditioning medium
Brain tumor cell lines U87 (human glioblastoma, from ATCC), U87VIII (a U87 cell line overexpressing the epidermal growth factor (EGF) receptor), GL261 (murine glioblastoma), primary normal human astrocytes (NHA, from Cell Systems Corporation), and primary normal mouse astrocytes (NMA) were used. The Chinese hamster ovary (CHO) cell line stably transfected with the V717F Aβ precursor protein (APP) mutation (referred to as 7PA2 cells) was characterized previously28. 7PA2 cells naturally secrete Aβ, so 7PA2 and CHO cells were grown to near confluence and allowed to condition DMEM for ~16 h. The 7PA2 conditioned medium (CM) used in this study had a total Aβ concentration of ~2.6 ng/ml, of which ~15% is known to be Aβ4228. Conditioned medium was cleared of cells (200 g for 10 min, 4°C) before utilization. All cells were grown in DMEM (InVitrogen) medium supplemented with 10% fetal bovine serum (Sigma), 2 mmol/L L-glutamine (InVitrogen), and 100 units/ml penicillin/streptomycin (InVitrogen), in a humidified atmosphere at 37°C in 5% CO2.
Cell proliferation assay
Cells were plated at a density of 1 × 104 cells per well in 96-well plates. Following serum starvation overnight, the cells were cultured in the conditioned medium. The number of viable cells was followed for 3 days after cell seeding by adding CellTiter-Glo® Luminescent Cell Viability reagent (Promega). Images were taken using IVIS imaging System (Xenogen), and luminescence data was analyzed using Living Image Software (Xenogen).
Wound healing assay
Cells were plated in triplicate sets of 24-well plates and grown to confluence. After serum starvation overnight, cells were scraped using a sterile tip to create a “wound”. The scraped off cells were washed out and remaining cells were cultured in fresh conditioned medium. Photographs were taken at the time of “wound” creation and at 24, 48, and 72 h after its induction using X81 Inverted Microscope (Olympus).
Invasion and migration assays
Cell invasion and migration assays were performed as described50 in 24-well BD Chambers (Becton Dickinson, Bedford, MA). The number of cells invading or migrating on the underside of the membrane was determined by using Olympus IX81 automated microscope system and Slidebook software (Olympus) to take montage images of the whole membranes. The number of total cells throughout the whole membrane was counted with Simple PCI software (Olympus).
Anchorage-independent cell proliferation
Culture medium (0.5 ml) containing 0.4% agarose, cells (10,000/well) and 10% FBS were overlaid onto the bottom layer containing 0.6% agarose in the same culture medium (0.5 ml) in 24-well plates. The plates were cultured in the conditioned medium for 2 weeks. At endpoint, colonies larger than 100 μm in diameter were counted.
Western blot analysis
Cell lysates were collected after treatment with conditioned medium for 30 min. Immunoblots were performed as described50. Antibodies used included p-ERK 1/2 (Sigma), ERK1/2(Santa Cruz Biotechnology), p-AKT S473 (Cell Signaling), and pan AKT (Cell Signaling), Ki67 (Cell Signaling Technology) and CD34 (Santa Cruz Biotechnology).
Imaging flow cytometry cell cycle analysis
Cells were harvested, washed, and then gently resuspended for DAPI and anti-tubulin staining. The cell cycle distribution based on DNA content and tubulin was analyzed on the ImageStreamX high content screening system (Amnis) and analysis of the data using the IDEAS application.
In Vivo animal experiments
APPswe transgenic mice B6;SJL-Tg(APPSWE)2576Kha (Taconic, Hudson, NY) and litter-match control group mice were used to study tumor growth in an AD model in accordance with approvals and the guidelines of Institutional Animal Care and Use Committee (IACUC) of Brigham and Women's Hospital at Harvard Medical School. 12-month old female mice were intracranially injected with GL261 cells, as described before51. Tumor growth was monitored once per week by gadolinium-enhanced MRI with a 4.7-T small-animal scanner (Bruker Biospec, Ettlingen, Germany). The scanner is equipped with a volume-based transmitter and receiver coil with an inner diameter of 4 cm. T2-weighted images (TR/TEeff = 2000 ms/72 ms) were obtained at 1 mm slice thickness with 80 μm × 130 μm in-plane resolution. MagnevistTm Gd-DTPA was injected i.p. at 0.7 mmol/kg, and T1-weighted images (TR/TEeff = 417 ms/24.5 ms) were acquired after a 12 min delay to allow for optimal contrast enhancement. Regions of Interest (ROIs) were drawn manually on the T1-weighted images with reference to tumor size and location from T2-weighted images. After the third MRI imaging, mice were sacrificed and brains were harvested for immunochemistry staining. Images were obtained using Olympus BX61 automated microscopy system. Five fields of 5 sections/mouse brain were captured using a 20× microscope objective. Ki67-positive and total cells in each field were counted with Simple PCI software (Olympus). For evaluating ERK immuno-staining, H scores were calculated by multiplying the fraction of positively stained tumor (percentage) by staining intensity (0, 1+, 2+ or 3+)52.
Statistical analysis
Microarray data were processed as described53. All other experiments were analyzed by one-way ANOVA followed by Fisher's post-hoc test using statistical software (SPSS, version 10.0). A p-value of less than 0.05 is considered to be statistically significant. Data were presented as a mean with standard deviation (SD).
Supplementary Material
Supplementary Information
Acknowledgments
This research is funded by NIH U54 CA149196, NIH R01 CA121225, TT & WF Chao Foundation grant and John S. Dunn Research Foundation grant to STCW. We thank Dr. Neal Copeland for providing comments on the manuscript, and Drs. James Mancuso and Rebecca Danforth for proofreading, Drs. Dennis Selkoe (Harvard Medical School) and Santosh Kesari (UC San Diego School of Medicine) for providing the 7PA2, U87VIII, and GL261 cell lines. All the microscopic imaging and ImageStream analysis were performed at Houston Methodist Research Institute's Advanced Cellular and Tissue Microscope Core Facility. We also acknowledge the computational time funding support from the Texas Advanced Computing Center (TACC; Project ID: TG-MCB110130) at the University of Texas in Austin and BlueBioU (IBM POWER 7 Bioscience Computing Core at Rice University) to access super-computing resources.
Footnotes
Author Contributions H.Z. and S.T.C.W. conceived the question and initiated the study. T.L., D.R., H.Z. and S.T.C.W. participated in the design, analysis of experiments and drafted the manuscript. T.L., D.R., X.Z., Z.Z., X.L., K.W., K.C. and H.Z. participated in the execution and analysis of wet lab studies. Z.Y., G.J., P.C., D.R., H.Z. and S.T.C.W. participated in the analysis of dry lab studies.
References
- Ou S.-M., Lee Y.-J., H Y.-W., Liu C.-J., Chen T.-J., Fuh J.-L. & Wang S.-J. Does Alzheimer's Disease Protect against Cancers? A Nationwide Population-Based Study. Neuroepidemiology 40, 42–49 (2013). [DOI] [PubMed] [Google Scholar]
- Driver J. A. et al. Inverse association between cancer and Alzheimer's disease: results from the Framingham Heart Study. BMJ 344, e1442 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe C. M., Behrens M. I., Xiong C., Miller J. P. & Morris J. C. Alzheimer disease and cancer. Neurology 64, 895–898 (2005). [DOI] [PubMed] [Google Scholar]
- Roe C. M. et al. Cancer linked to Alzheimer disease but not vascular dementia. Neurology 74, 106–112 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musicco M. et al. Inverse occurrence of cancer and Alzheimer disease: a population-based incidence study. Neurology 81, 322–328 (2013). [DOI] [PubMed] [Google Scholar]
- Driver J. A., Kurth T., Buring J. E., Gaziano J. M. & Logroscino G. Prospective case-control study of nonfatal cancer preceding the diagnosis of Parkinson's disease. Cancer Causes Control 18, 705–711 (2007). [DOI] [PubMed] [Google Scholar]
- Driver J. A., Logroscino G., Buring J. E., Gaziano J. M. & Kurth T. A prospective cohort study of cancer incidence following the diagnosis of Parkinson's disease. Cancer Epidemiol Biomarkers Prev 16, 1260–1265 (2007). [DOI] [PubMed] [Google Scholar]
- Behrens M. I., Lendon C. & Roe C. M. A common biological mechanism in cancer and Alzheimer's disease? Curr Alzheimer Res 6, 196–204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver J. A. & Lu K. P. Pin1: a new genetic link between Alzheimer's disease, cancer and aging. Curr Aging Sci 3, 158–165 (2010). [DOI] [PubMed] [Google Scholar]
- Blalock E. M. et al. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A 101, 2173–2178 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf G. M. et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J 20, 3459–3472 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino L. et al. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature 440, 528–534 (2006). [DOI] [PubMed] [Google Scholar]
- Manoukian A. S. & Woodgett J. R. Role of glycogen synthase kinase-3 in cancer: regulation by Wnts and other signaling pathways. Adv Cancer Res 84, 203–229 (2002). [DOI] [PubMed] [Google Scholar]
- Fuentealba R. A. et al. Signal transduction during amyloid-beta-peptide neurotoxicity: role in Alzheimer disease. Brain Res Brain Res Rev 47, 275–289 (2004). [DOI] [PubMed] [Google Scholar]
- Wulf G., Garg P., Liou Y. C., Iglehart D. & Lu K. P. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J 23, 3397–3407 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. et al. Pin1 has opposite effects on wild-type and P301L tau stability and tauopathy. J Clin Invest 118, 1877–1889 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble W. et al. Cdk5 is a key factor in tau aggregation and tangle formation in vivo. Neuron 38, 555–565 (2003). [DOI] [PubMed] [Google Scholar]
- Wang J. Z., Wu Q., Smith A., Grundke-Iqbal I. & Iqbal K. Tau is phosphorylated by GSK-3 at several sites found in Alzheimer disease and its biological activity markedly inhibited only after it is prephosphorylated by A-kinase. FEBS Lett 436, 28–34 (1998). [DOI] [PubMed] [Google Scholar]
- Paris D. et al. Inhibition of angiogenesis by Abeta peptides. Angiogenesis 7, 75–85 (2004). [DOI] [PubMed] [Google Scholar]
- Xie R. L. et al. The tumor suppressor interferon regulatory factor 1 interferes with SP1 activation to repress the human CDK2 promoter. J Biol Chem 278, 26589–26596 (2003). [DOI] [PubMed] [Google Scholar]
- Bordin S. & Tan X. C1q arrests the cell cycle progression of fibroblasts in G(1) phase: role of the cAMP/PKA-I pathway. Cell Signal 13, 119–123 (2001). [DOI] [PubMed] [Google Scholar]
- Lama G. et al. Activated ERK1/2 expression in glioblastoma multiforme and in peritumor tissue. Int J Oncol 30, 1333–1342 (2007). [PubMed] [Google Scholar]
- Hardy J. & Selkoe D. J. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 297, 353–356 (2002). [DOI] [PubMed] [Google Scholar]
- Mrak R. E. & Griffin W. S. Interleukin-1, neuroinflammation, and Alzheimer's disease. Neurobiol Aging 22, 903–908 (2001). [DOI] [PubMed] [Google Scholar]
- Grolla A. A. et al. Abeta leads to Ca(2+) signaling alterations and transcriptional changes in glial cells. Neurobiol Aging 34, 511–522 (2013). [DOI] [PubMed] [Google Scholar]
- Capiod T. S. Y., Skryma R. & Prevarskaya N. in Subcell Biochem, Vol. 45, 405–427 (2007). [DOI] [PubMed] [Google Scholar]
- Zhao H. et al. Bioluminescence imaging reveals inhibition of tumor cell proliferation by Alzheimer's amyloid beta protein. Cancer Cell Int 9, 15 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D. M. et al. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416, 535–539 (2002). [DOI] [PubMed] [Google Scholar]
- Tamagno E. et al. JNK and ERK1/2 pathways have a dual opposite effect on the expression of BACE1. Neurobiol Aging 30, 1563–1573 (2009). [DOI] [PubMed] [Google Scholar]
- Scholl F. A. et al. Mek1/2 MAPK kinases are essential for Mammalian development, homeostasis, and Raf-induced hyperplasia. Dev Cell 12, 615–629 (2007). [DOI] [PubMed] [Google Scholar]
- Holland E. C. et al. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet 25, 55–57 (2000). [DOI] [PubMed] [Google Scholar]
- Hsiao K. et al. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science 274, 99–102 (1996). [DOI] [PubMed] [Google Scholar]
- Sunayama J. et al. Crosstalk between the PI3K/mTOR and MEK/ERK pathways involved in the maintenance of self-renewal and tumorigenicity of glioblastoma stem-like cells. Stem Cells 28, 1930–1939 (2010). [DOI] [PubMed] [Google Scholar]
- Vagnucci A. H. Jr & Li W. W. Alzheimer's disease and angiogenesis. Lancet 361, 605–608 (2003). [DOI] [PubMed] [Google Scholar]
- Das S. & Marsden P. A. Angiogenesis in glioblastoma. N Engl J Med 369, 1561–1563 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T., Serrano M. & Blasco M. A. The common biology of cancer and ageing. Nature 448, 767–774 (2007). [DOI] [PubMed] [Google Scholar]
- Park L. et al. Innate immunity receptor CD36 promotes cerebral amyloid angiopathy. Proc Natl Acad Sci U S A 110, 3089–3094 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. N. et al. Dual modification of Alzheimer's disease PHF-tau protein by lysine methylation and ubiquitylation: a mass spectrometry approach. Acta Neuropathol 123, 105–117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K. & Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci 8, 368–378 (2007). [DOI] [PubMed] [Google Scholar]
- Ye W. & Blain S. W. S phase entry causes homocysteine-induced death while ataxia telangiectasia and Rad3 related protein functions anti-apoptotically to protect neurons. Brain 133, 2295–2312 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt T., Bruckner M. K., Mosch B. & Losche A. Selective cell death of hyperploid neurons in Alzheimer's disease. Am J Pathol 177, 15–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester A. C. et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145, 435–446 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. S. et al. Cell cycle-related kinase: a novel candidate oncogene in human glioblastoma. J Natl Cancer Inst 99, 936–948 (2007). [DOI] [PubMed] [Google Scholar]
- Dai R., Chen R. & Li H. Cross-talk between PI3K/Akt and MEK/ERK pathways mediates endoplasmic reticulum stress-induced cell cycle progression and cell death in human hepatocellular carcinoma cells. Int J Oncol 34, 1749–1757 (2009). [DOI] [PubMed] [Google Scholar]
- Rommel C. et al. Differentiation stage-specific inhibition of the Raf-MEK-ERK pathway by Akt. Science 286, 1738–1741 (1999). [DOI] [PubMed] [Google Scholar]
- Liu J. F. et al. Functional Rac-1 and Nck signaling networks are required for FGF-2-induced DNA synthesis in MCF-7 cells. Oncogene 18, 6425–6433 (1999). [DOI] [PubMed] [Google Scholar]
- Kim S. K. et al. ERK1/2 is an endogenous negative regulator of the gamma-secretase activity. FASEB J 20, 157–159 (2006). [DOI] [PubMed] [Google Scholar]
- Mei M. et al. Distribution, levels and phosphorylation of Raf-1 in Alzheimer's disease. J Neurochem 99, 1377–1388 (2006). [DOI] [PubMed] [Google Scholar]
- Spitzer P. et al. Evidence for Elevated Cerebrospinal Fluid ERK1/2 Levels in Alzheimer Dementia. Int J Alzheimers Dis 2011, 739847 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. et al. The effect of mTOR inhibition alone or combined with MEK inhibitors on brain metastasis: an in vivo analysis in triple-negative breast cancer models. Breast Cancer Res Treat 131, 425–436 (2012). [DOI] [PubMed] [Google Scholar]
- Wang L. et al. Differential effects of low- and high-dose GW2974, a dual epidermal growth factor receptor and HER2 kinase inhibitor, on glioblastoma multiforme invasion. J Neurosci Res 91, 128–137 (2013). [DOI] [PubMed] [Google Scholar]
- Andersen J. N. et al. Pathway-based identification of biomarkers for targeted therapeutics: personalized oncology with PI3K pathway inhibitors. Science translational medicine 2, 43ra55. [DOI] [PubMed] [Google Scholar]
- Jin G. et al. A novel method of transcriptional response analysis to facilitate drug repositioning for cancer therapy. Cancer Res 72, 33–44 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information