Abstract
In the U.S.A., cocaine is the second most abused illicit drug. Variants within the CHRNB3-A6 gene cluster have been associated with cigarette consumption in several GWAS. These receptors represent intriguing candidates for the study of cocaine dependence because nicotinic receptors are thought to be involved in generalized addiction pathways. Using genotypic data from a GWAS of the Study of Addiction: Genetics and Environment (SAGE) dataset, we tested for association of CHRNB3-A6 SNPs with DSM-5 cocaine use disorder. Multiple SNPs in the region were significantly associated with increased risk of cocaine use disorder. Inclusion of the most significant SNP as a covariate in a linear regression model provided evidence for an additional independent signal within this locus for cocaine use disorder. These results suggest that the CHRNB3-A6 locus contains multiple variants affecting risk for vulnerability to cocaine and nicotine dependence as well as bipolar disorder, suggesting that they have pleiotropic effects.
Drug addiction is a pervasive problem across cultures and is both an economic and psychological burden for the individuals and families involved. Illicit drugs have particularly devastating consequences: Cocaine users have six times the mortality rate of their age-matched peers1, and one study found that crack cocaine was the third most harmful drug overall2. Consequently, a greater understanding of the molecular mechanisms underlying the risk of developing cocaine dependence is urgently needed to aid in targeting prevention and treatment strategies.
Susceptibility to drug use, abuse, and dependence has been shown by several studies to have a moderate to high genetic component3,4. The one year point prevalence, or the proportion of people possessing a phenotype within a one year timeframe, for substance use disorders in the USA, excluding nicotine has been estimated to be 9.35%3. The heritability of cocaine dependence (CD) has been estimated from twin studies to be 63–79%5. There have been two genome-wide linkage studies. The first found suggestive linkage between CD phenotypes of cocaine induced paranoia, heavy use, and moderate use on chromosomes 9, 12, and 18 respectively (n = 986 individuals from 390 pedigrees)6. The second study reported genome-wide significant linkage to regions on chromosomes 5 and 7 with the phenotype of CD and its co-occurrence with major depressive disorder (n = 1896 individuals from 739 families)7. Candidate gene association studies have reported association between a SNP in pro-opiomelanocortin POMC (rs6713532) and CD in European-American families8, consistent with the role of the opioidergic system in addiction and reinforcement. Furthermore, several SNPs in the mannosidase, endo-alpha (MANEA) gene have also been associated with cocaine-induced paranoia9. Recently, Gelernter and colleagues completed a GWAS of cocaine dependence in over 4000 European and African-American subjects. They identified rs2329540 in the FAM53B (family with sequence similarity 53, member B) gene to be significantly associated with CD symptom count in both ethnic groups. Additional variants were also identified at genome-wide significant (GWS) levels for a diagnosis of CD as well as for cocaine-induced paranoia10.
In addition to the observed co-occurrence of cocaine dependence with other forms of substance disorders and psychopathology, numerous twin studies indicate a high degree of overlap among genetic factors influencing the liability to a variety of substance use disorders11,12. Genomic studies have also suggested that there are genetic loci that have substance-specific effects but also that loci exist that affect risk for the development of dependence on multiple substances (see4 for review). Loci that have been largely implicated to specifically influence a single substance use disorder include those that exert metabolic influence on the substance of abuse. For instance, SNPs in cytochrome P450 2A6 (CYP2A6), the gene encoding the major nicotine-metabolizing enzyme, affect cigarette consumption13,14 and a SNP in the Alcohol Dehydrogenase 1B (ADH1B) gene affects levels of alcohol consumption15 and risk for alcohol dependence4 via regulation of conversion of alcohol to acetaldehyde.
On the other hand, there are numerous examples of receptor encoding loci whose effects extend across multiple substance dependence phenotypes. One such example is the SNP rs16969968 (D398N) in the cholinergic nicotinic receptor subunit α5 (CHRNA5) that both increases nicotine dependence risk and decreases cocaine dependence risk5,16. The minor allele of this SNP is the most significant and widely replicated variant associated with cigarette consumption and is also associated with protection against cocaine dependence5,14,17. The protective effect of rs16969968 with CD has been replicated in both European and African-Americans16. The same study also found that another SNP in CHRNA5 (rs684513) is associated with risk for cocaine dependence in African-Americans (OR = 1.43, P = 0.0004).
In addition, a cluster of nicotinic receptors on chromosome 8 including CHRNB3-A6 was also previously shown to reduce risk for nicotine-related phenotypes in several GWAS of nicotine dependence and cigarettes smoked per day13,14,18,19,20,21,22,23. An overlapping set of SNPs in the CHRNB3-CHRNA6 region were also reported by Hartz et al. (2011) to be associated with risk of bipolar disorder24. Despite the positive correlation between nicotine dependence and bipolar disorder the associations are in opposite directions, i.e. SNPs in this locus are associated with reduced risk for nicotine dependence but increased risk for bipolar disorder. This suggests that the SNPs affect bipolar disorder independently of their role in nicotine dependence. The role of these specific SNPs in the etiology of CD remains unexplored but several rare variants in CHRNB3 have been associated with increased risk for both cocaine and alcohol dependence25. Together, these results suggest that nicotinic receptors are good candidate genes for susceptibility to nicotine and cocaine dependence vulnerability and that investigation of the role of common and low frequency variants within the CHRNB3-A6 locus in cocaine dependence is warranted.
In this study, we describe a novel association between DSM-5 cocaine use disorder and genotyped SNPs (~24 kb) upstream of the CHRNB3 transcription start site. We show that these SNPs remain significant after adjusting for genotype at the SNP previously reported to be associated with nicotine dependence in GWAS, suggesting that the cocaine association is not simply due to the nicotine association.
Results
The 47 SNPs within the CHRNB3-A6 region constitute 22 LD bins using an r2 cutoff ≥ 0.8, requiring a p-value of 0.002 after Bonferroni correction. Eleven SNPs, representing four LD bins met this cutoff and are associated with DSM-5 cocaine use disorder (Table 1). Overall, a total of thirty-one SNPs were nominally significant (2.34 × 10−4 < p < 4.66 × 10−2) in this single SNP analysis. Consistent with previous results in an overlapping dataset, we saw a protective effect of rs16969968 in CHRNA5 on risk for DSM-5 cocaine use disorder [5]. Inclusion of rs16969968 as a covariate had no effect on the association of the top SNP within CHRNB3-A6, rs9298626 with DSM-5 cocaine use disorder.
Table 1. Top association results for the linear models run for DSM-5 cocaine use disorder. Bolded SNPs passed multiple test correction (p > 0.002). Maximum FTND is the score from 0–10, L95 and U95 is the 95% confidence interval, and the frequency is in the SAGE dataset.
| European-Americans SAGE DSM-5 Cocaine Use Disorder with covariates age, sex, maximum FTND score, DSM-5 alcohol symptom count, study | ||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | LD Bin | bp | N | OR | L95 | U95 | Freq | P |
| rs9298626 | 1 | 42,647,165 | 1970 | 2.62 | 1.57 | 4.37 | 0.04 | 2.34E-04 |
| rs7844824 | 1 | 42,672,170 | 1970 | 2.65 | 1.58 | 4.46 | 0.04 | 2.43E-04 |
| rs4305884 | 2 | 42,637,880 | 1966 | 2.13 | 1.42 | 3.21 | 0.06 | 2.69E-04 |
| rs7824160 | 1 | 42,705,413 | 1969 | 2.50 | 1.49 | 4.19 | 0.04 | 4.88E-04 |
| rs11986893 | 4 | 42,772,016 | 1971 | 1.56 | 1.22 | 2.01 | 0.2 | 4.92E-04 |
| rs7002907 | 1 | 42,702,998 | 1970 | 2.49 | 1.49 | 4.17 | 0.04 | 5.03E-04 |
| rs6997994 | 1 | 42,702,328 | 1971 | 2.49 | 1.49 | 4.18 | 0.04 | 5.04E-04 |
| rs7815274 | 1 | 42,701,740 | 1967 | 2.47 | 1.47 | 4.15 | 0.04 | 6.41E-04 |
| rs4952 | 1 | 42,706,222 | 1971 | 2.43 | 1.44 | 4.08 | 0.04 | 8.12E-04 |
| rs10107450 | 5 | 42,749,052 | 1969 | 1.50 | 1.18 | 1.92 | 0.22 | 1.04E-03 |
| rs1868859 | 2 | 42,634,958 | 1971 | 1.85 | 1.27 | 2.69 | 0.07 | 1.36E-03 |
| rs892413 | 3 | 42,733,535 | 1971 | 1.48 | 1.15 | 1.89 | 0.2 | 2.11E-03 |
| rs4950 | 6 | 42,671,790 | 1957 | 1.42 | 1.11 | 1.83 | 0.22 | 5.93E-03 |
| rs13280604 | 6 | 42,678,743 | 1971 | 1.41 | 1.10 | 1.80 | 0.22 | 7.49E-03 |
| rs1530848 | 6 | 42,672,065 | 1964 | 1.40 | 1.09 | 1.79 | 0.22 | 7.53E-03 |
| rs2196128 | 3 | 42,737,443 | 1971 | 1.37 | 1.08 | 1.74 | 0.23 | 8.83E-03 |
| rs6997909 | 6 | 42,679,406 | 1971 | 1.39 | 1.08 | 1.78 | 0.22 | 9.67E-03 |
| rs6474414 | 6 | 42,679,493 | 1971 | 1.39 | 1.08 | 1.78 | 0.22 | 9.67E-03 |
| rs4736835 | 6 | 42,666,190 | 1971 | 1.39 | 1.08 | 1.78 | 0.22 | 1.03E-02 |
| rs9298628 | 3 | 42,725,148 | 1968 | 1.38 | 1.08 | 1.76 | 0.21 | 1.04E-02 |
| rs6474415 | 6 | 42,682,095 | 1970 | 1.38 | 1.08 | 1.77 | 0.22 | 1.05E-02 |
| rs1451240 | 6 | 42,665,868 | 1970 | 1.38 | 1.08 | 1.77 | 0.22 | 1.14E-02 |
| rs7004381 | 6 | 42,670,318 | 1971 | 1.38 | 1.07 | 1.77 | 0.22 | 1.24E-02 |
| rs13273442 | 6 | 42,663,174 | 1970 | 1.38 | 1.07 | 1.77 | 0.22 | 1.25E-02 |
| rs1955185 | 6 | 42,668,804 | 1971 | 1.37 | 1.07 | 1.76 | 0.22 | 1.30E-02 |
| rs6474413 | 6 | 42,670,221 | 1971 | 1.37 | 1.07 | 1.76 | 0.22 | 1.31E-02 |
| rs16891620 | 7 | 42,744,820 | 1970 | 1.42 | 1.06 | 1.92 | 0.13 | 2.00E-02 |
| rs10958726 | 6 | 42,655,066 | 1971 | 1.34 | 1.04 | 1.73 | 0.21 | 2.20E-02 |
| rs6474421 | 12 | 42,776,255 | 1969 | 1.50 | 1.05 | 2.14 | 0.07 | 2.62E-02 |
| rs10958725 | 6 | 42,643,741 | 1968 | 1.31 | 1.02 | 1.68 | 0.22 | 3.37E-02 |
| rs7012713 | 13 | 42,711,460 | 1970 | 1.65 | 1.01 | 2.70 | 0.04 | 4.66E-02 |
Conditional analyses
To determine whether there was evidence for multiple independently associated variants at this locus contributing to risk for DSM-5 cocaine use disorder, the most significant SNP in the region (rs9298626) was added to the model as a covariate. Conditioning on rs9298626 eliminated the association with SNPs in LD bins 1 and 2, but the association remained for SNPs in bins 4 and 5. After including rs9298626 as a covariate, the top SNP associated with this phenotype was rs892413 (p = 3.57 × 10−4, OR = 1.58, CI = 1.23–2.04). The p-value for bin 4 tagged here by rs11986892 is 0.001, OR = 1.51, CI = 1.17–1.94. Similarly the p-value for bin 5 tagged by rs10107450 is 0.003, OR = 1.45, CI = 1.14–1.85. This means that after conditioning on rs9298626, rs892413 and rs11986892 still pass the multiple test correction threshold of 0.002, whereas rs10107450 does not, although it does come close.
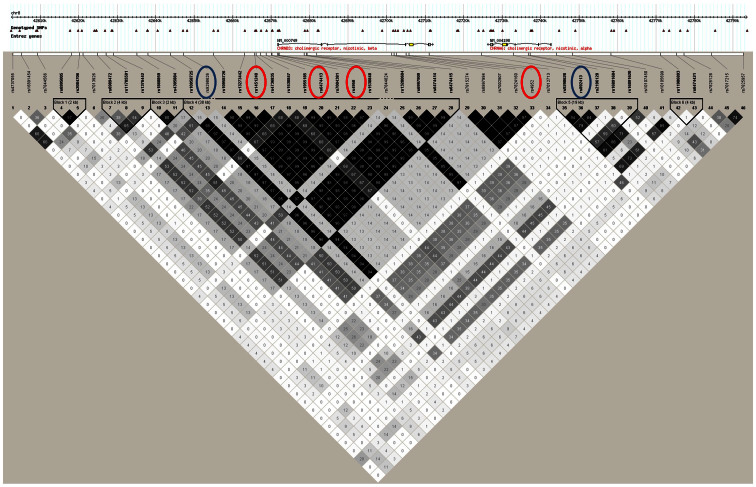
Examination of LD shows that the r2 between rs9298626 and rs892413 is low, suggesting that these SNPs represent independent association signals (r2 = 0.01; D′ = 0.85). Because the minor allele frequency for rs9298626 is low, the r2 will never be high but the D′ indicates that the minor allele of this SNP is usually but not always on the background of one allele of rs892413. Neither of these SNPs is in significant LD with the previously identified genome-wide significant signal (rs1451240) associated with cigarette consumption and nicotine dependence14,23 in this region (r2 = 0.14 between rs9298626 and rs1451240, r2 = 0.35 between rs892413 and rs1451240; Figure 1), although rs9298626 is in high LD (r2 = 0.94) with rs4952, another SNP previously reported to be associated with nicotine dependence18. We find that rs9298626, rs892413 and rs1451240 correspond to different LD bins using r2 = 0.8 as the threshold for defining the bins (Table 1). This is consistent with recent results from our group showing, in an overlapping dataset, that the genome-wide significant signal in this region, tagged by rs1451240, is solely associated with nicotine dependence. Taken together, this suggests that these three SNPs represent different association signals in this region.
Figure 1. LD plot of the region.
Values and gray-scale represent r-squared. Importantly, rs9298626, the top SNP associated with DSM-5 cocaine use disorder and the shared SNP that becomes significant when putting this SNP into the respective model as a covariate (rs892413) are circled in blue, and SNPs representing the previously discovered association with nicotine dependence (rs1451240, rs6474413, and rs4952) are circled in red.
Minor allele frequency examination
To determine whether there is a significant effect of lifetime cocaine exposure on these associations, we compared the frequency of the minor allele of rs9298626 with DSM-5 cocaine use disorder, exposed non-dependent individuals and non-exposed individuals. Control individuals used in our initial analyses included both those who were exposed to cocaine but were unaffected (N = 393 with 0–1 DSM-5 criteria), as well as those who were never exposed (N = 1077). The minor allele frequency of rs9298626 was 9.9% among those with DSM-5 cocaine use disorder and 5.3% among subjects who have been exposed to cocaine but did not progress to cocaine use disorder. Non-exposed controls have an intermediate minor allele frequency of 7.7%, suggesting that both those with DSM-5 cocaine use disorder and those who were cocaine-exposed non-dependent controls show allele frequency differences from unselected controls (Table 2), although the difference in frequency of rs9298626 between exposed but unaffected controls and unexposed controls was not significant. When control subjects were restricted to those who had been exposed to cocaine but were unaffected (n = 899 vs. 1976), the association between rs9298626 and DSM-5 cocaine use disorder was reduced but the odds ratio was unchanged (p = 3.12 × 10−3, OR = 2.68, CI = 1.40–5.16), supporting the role of this SNP, or another SNP in LD with rs9298626, in risk for DSM-5 cocaine use disorder, even after accounting for cocaine exposure. While the exposed and unaffected individuals have higher rates of nicotine and alcohol dependence we conclude that this is likely because exposure to cocaine, regardless of dependence, is correlated with other substance use. We conclude that the minor allele of rs9298626 is correlated with cocaine use disorder, which is strengthened by the fact that there remains an association even when not considering unexposed individuals in the analysis.
Table 2. Characteristics of the sample broken down by: DSM-5 cocaine use disorder, exposed but unaffected, and non-exposed unaffected.
| DSM-5 Cocaine Use Disorder EAs | Exposed Unaffected EAs | Non-exposed Unaffected EAs | Total | |
|---|---|---|---|---|
| N = 506 | N = 393 | N = 1077 | N = 1976 | |
| Age (years) | ||||
| mean age | 36 | 38 | 39 | 38 |
| <35 | 37% | 26% | 35% | 34% |
| 35–39 | 28% | 27% | 22% | 25% |
| 40–44 | 23% | 35% | 25% | 26% |
| ≥45 | 12% | 12% | 18% | 13% |
| Male | 62% | 50% | 34% | 44% |
| Female | 38% | 50% | 66% | 56% |
| Nicotine Dependence | ||||
| FTND 0,1 | 10% | 47% | 68% | 49% |
| FTND 2,3 | 12% | 9% | 9% | 10% |
| FTND ≥ 4 | 78% | 44% | 23% | 41% |
| DSM-5 Alcohol Use Disorder | ||||
| Case | 99% | 88% | 37% | 42% |
| Control | 1% | 12% | 63% | 58% |
| rs9298626 minor allele carrier | ||||
| Yes | 9.9% | 5.3% | 7.7% | 7.8% |
| No | 90.1% | 94.7% | 92.3% | 92.2% |
Stratified analysis
Because SAGE is composed of individuals from three independent studies, each ascertained for a different substance dependence, we performed stratified analyses both by study and by nicotine dependence and alcohol dependence to determine if there existed a subset of subjects in which the association was most pronounced. The top SNP associated with DSM-5 cocaine use disorder (rs9298626) in the whole SAGE dataset was significantly associated with DSM5 cocaine use disorder in the COGA subset and showed a trend in the same direction in the FSCD and COGEND subsets. Furthermore, when individuals from the whole dataset were stratified by DSM-5 alcohol use disorder, or FTND nicotine dependence there was evidence of association between rs9298626 and cocaine use disorder in both groups (Table 3). This suggests that the observed associations are not an artifact of ascertainment. This analysis suggests that this SNP is associated with DSM-5 cocaine use disorder and that the CHRNB3-A6 locus is robustly associated with DSM-5 cocaine use disorder, regardless of comorbidity or ascertainment.
Table 3. Results of stratified analyses in these groups for rs9298626 – controls included here are both those who are exposed and unaffected, as well as those who are unexposed.
| European-American DSM-5 Cocaine Use Disorder | ||||
|---|---|---|---|---|
| N | OR | 95% CI | P-Value for rs9298626 | |
| Study | ||||
| COGEND | 1077 | 1.77 | 0.67–4.62 | 0.25 |
| COGA | 599 | 2.62 | 1.25–5.52 | 0.01 |
| FSCD | 294 | 6.35 | 0.67–60.43 | 0.11 |
| Smoking Status | ||||
| FTND Cases | 814 | 2.31 | 1.06–5.02 | 0.03 |
| FTND Controls | 1156 | 2.59 | 1.22–5.50 | 0.01 |
| Alcohol Status | ||||
| DSM-5 Cases | 1522 | 2.41 | 1.46–3.98 | 0.0006 |
| DSM-5 Controls | 448 | 7.05 | 0.52–94.60 | 0.14 |
Haplotype analysis
To further examine the relationship between our top variant identified for DSM-5 cocaine use disorder and the group of variants known to be associated with smoking tagged by rs1451240, we performed haplotype-based association testing using rs9298626 and rs1451240. These two SNPs occur on three haplotypes that occur with a frequency >1% (Table 4). We chose rs1451240 because it was found to be genome-wide significant for nicotine dependence in a previous study using the SAGE GWAS data23. The most common haplotype, composed of the major alleles of both SNPs, has a frequency of 78%. The haplotype associated with the highest risk for DSM-5 cocaine use disorder, has a frequency of 4% and is composed of the minor alleles at both rs9298626 and rs1451240 (OR = 3.19 p = 1.35 × 10−4, 95% CI = 1.64–4.73). A haplotype composed of the major allele at rs9298626 and the minor allele at rs1451240 has a frequency of 18% but was not associated with DSM-5 cocaine use disorder (p = 0.14). Because the frequencies and odds ratio of the haplotype with both minor alleles is nearly identical to that of the single SNP analysis for rs9298626 and the fact that the other haplotype containing the minor allele of rs1451240 is not associated with DSM-5 cocaine use disorder, we conclude that the functional allele responsible for this association is in high LD with the low frequency variant, rs9298626.
Table 4. Haplotypes observed in the SAGE GWAS European-American sample for DSM-5 cocaine use disorder. The grey box indicates the major allele for that SNP. P-values are denoting significance of that haplotype relative to the reference haplotype. SNPs are arranged in the order they occur on the chromosome. Covariates used are age, sex, study, DSM-5 alcohol symptom count, FTND total.
| Haplotypes | DSM-5 Cocaine Use Disorder | |||
|---|---|---|---|---|
| rs9298626 | rs1451240 | Frequency | Odds Ratio | P-Value |
| A | A | 0.04 | 3.19 | 1.35 × 10−4 |
| A | G | <0.01 | 9.89 | 0.89 |
| C | A | 0.18 | 1.23 | 0.14 |
| C | G | 0.78 | - | - |
Cocaine use disorder and nicotine dependence
To further examine the relationship between nicotine dependence and DSM-5 cocaine use disorder associations in this region, we performed additional conditional analyses. In a linear regression model using age, sex, study, DSM-5 alcohol symptom count, total FTND score and rs1451240 genotype as covariates, the association with DSM-5 cocaine use disorder remained significant (Table 5). Lastly, the DSM-5 cocaine use disorder signal remains significant when conditioning on the two rare variants (rs35327613 and rs149775276) recently identified by our group to be associated with DSM-IV alcohol and cocaine dependence symptom count26. This is not surprising given that these rare missense variants are present on the haplotypes containing the major allele of rs9298626, whereas the association reported here is with the minor allele. The fact that the association with cocaine use disorder remains when conditioning on the genome-wide significant signal with nicotine dependence in the region, suggests that the association is independent and not acting through nicotine dependence.
Table 5. All SNPs remain associated with DSM-5 cocaine use disorder after conditioning on rs1451240. P-values for all covariates are also shown.
| DSM-5 Cocaine Use Disorder with Age, Sex, DSM-5 Alcohol Symptom Count, Study, FTND Total and rs1451240 as covariates | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | N | OR | 95% CI | P | P(age) | P(sex) | P(FTND Total) | P(DSM-5 Alc Sx) | P(COGEND) | P(COGA) | P(rs1451240) |
| rs9298626 | 1969 | 2.28 | (1.32,3.95) | 3.22 × 10−3 | 2.56 × 10−10 | 8.07 × 10−3 | 2.12 × 10−9 | 1.55 × 10−27 | 9.31 × 10−41 | 1.43 × 10−21 | 0.17 |
| rs4952 | 1970 | 2.10 | (1.21,3.64) | 8.24 × 10−3 | 1.71 × 10−10 | 0.01 | 4.10 × 10−9 | 7.67 × 10−28 | 1.35 × 10−40 | 2.98 × 10−21 | 0.13 |
| rs892413 | 1970 | 1.35 | (0.99,1.83) | 5.26 × 10−2 | 8.69 × 10−11 | 8.83 × 10−3 | 6.43 × 10−9 | 6.39 × 10−28 | 4.28 × 10−40 | 7.46 × 10−21 | 0.34 |
Discussion
We have shown, in genotyped data from European-Americans in the SAGE dataset, that there are at least two statistically independent signals associated with increased risk for DSM-5 cocaine use disorder in the region of the CHRNB3-A6 nicotinic receptors on chromosome 8. Several SNPs representing the rs9298626 LD bin surpass the multiple test correction for the region with DSM-5 cocaine use disorder (p = 0.002). rs9298626 is also associated with reduced risk for nicotine dependence (OR = 0.47, 95% CI = 0.30–0.76, p = 1.80 × 10−3) in a univariate genetic analysis. This may be due in part to the fact that, in European ancestry populations, the minor allele of rs9298626 (MAF = 0.04) occurs almost exclusively on the background of the more frequent minor allele (MAF = 0.22) for rs1451240 previously reported to be genome-wide significantly associated with nicotine dependence (Table 3). Conditioning on rs1451240 had no effect on the association with DSM-5 cocaine use disorder (Table 5). This is not surprising because rs1451240 is not associated with DSM-5 cocaine use disorder in our data (Table 1).
LD bins tagged by rs9298626 and rs892413 each show association with DSM-5 cocaine use disorder in joint SNP analysis. Analyses conditioning on rs9298626 reveal that rs892413 is independently associated with DSM-5 cocaine use disorder. rs892413 is also associated with DSM-5 cocaine use disorder independent of the previously identified genome-wide significant association in the region with nicotine dependence (represented by rs1451240), providing support for a direct effect of this SNP on higher DSM-5 cocaine use disorder risk, as opposed to acting through nicotine dependence risk (Tables 3 and 5).
The LD bin containing rs9298626 also contains rs4952 and rs4953, two low frequency synonymous variants in CHRNB3 that have previously been reported to be associated with reduced risk for nicotine dependence18 and increased risk for bipolar disorder (OR = 1.7, 95%, CI = 1.2–2.4, p = 0.001)24. Interestingly the association of rs4952/rs4953 with cocaine use disorder is in the same direction as the association with bipolar disorder (risk) but in the opposite direction to the association with nicotine dependence (protective) suggesting that CHRNB3 variants have pleiotropic effects on substance use disorders and other psychiatric diseases. Many epidemiological studies have reported the common co-occurrence of bipolar disorder and substance dependence27,28,29. Studies have also implicated shared genes with substance dependence and bipolar disorder29,30. It is therefore possible that the high frequency of bipolar disorder and substance dependence comorbidity is in part due to common underlying genetic risk factors such as the risk alleles in the CHRNB3-A6 locus reported here.
Our group has previously reported that rare missense variants in CHRNB3 increase risk for cocaine dependence26. The results reported here demonstrate that low frequency and common alleles within the CHRNB3 locus are also associated with increased risk of DSM-5 cocaine use disorder. Cocaine dependence has now been associated with SNPs in two different nicotinic receptor gene clusters, on chromosomes 8 and 15 5,26. It is interesting, however, that the variant on chromosome 15, within CHRNA5 is associated with decreased risk for cocaine dependence, while rs9298626 and other variants in the CHRNB3-A6 region are associated with higher (OR = 2.62) risk for DSM-5 cocaine use disorder.
Furthermore, similar to the observation on chromosome 15, the chromosome 8 locus is associated with opposing effects on the risk for cocaine dependence and nicotine dependence. The CHRNB3-A6 locus is associated with decreased risk for nicotine dependence and increased risk for DSM-5 cocaine use disorder. In contrast, in CHRNA5, the same variant, D398N (rs16969968), increases risk for nicotine dependence and decreases risk for cocaine dependence. Furthermore, our data suggest that different but overlapping SNPs may explain the cocaine and nicotine dependence associations in CHRNB3-A6 rather than a single SNP causing opposing effects as was seen on chromosome 15. These results suggest that CHRNA5 and CHRNB3-A6 demonstrate pleiotropic effects on substance dependence risk.
Nicotinic acetylcholine receptors (nAChR) are expressed in multiple types of neurons, and have been shown to modulate reward response for several substances5. For example, work in animals suggests that activation of α3β4 nAChR can increase cocaine self-administration31. Because comorbidity between substance dependencies is so high, it is plausible that these receptors could play a role in addiction to multiple substances. Most drugs of abuse act on the mesolimbic dopamine-containing receptors in the brains of humans and many other mammals. Among other functions, this system is known to regulate motivation32,33 and has similar effects across mammalian species34. Activation and reinforcement of this system is a necessary part of drug abuse32. The dopaminergic system is therefore crucial to addiction, however other neurotransmitters besides dopamine affect the mesolimbic system, especially acetylcholine31. The biological connection between these two systems could be related to the reversal of the odds ratio for rs16969968, which is protective for cocaine dependence but a risk factor for nicotine dependence, as well as our observation that in the CHRNB3-A6 locus on chromosome 8, there are variants associated with protection against nicotine dependence, in addition to variants associated with risk for DSM-5 cocaine use disorder and bipolar disorder. Since the finding in CHRNA5 on chromosome 15 is the only association with cocaine dependence to be successfully replicated, it would be interesting to examine the CHRNB3-A6 region in other datasets that have assessed cocaine dependence phenotypes, as well as to analyze datasets of other ethnicities. Currently, there is no evidence that either of the variants reported here are correlated with SNPs that have known functional consequences. However, rs4952 and rs4953 are both synonymous variants in CHRNB3 and may therefore have some, as yet unknown effect on transcription or translation of CHRNB3 mRNA. Overall, our findings underscore the comorbidity among drug dependencies and corroborate the role of nicotinic receptors in cocaine-related phenotypes. This study represents one of only a few to implicate specific variants in cocaine dependence phenotypes and the first to implicate low frequency variants within the CHRNB3-A6 locus in risk for DSM-5 cocaine use disorder.
Methods
Samples
Subjects were members of the Study of Addiction: Genetics and Environment (SAGE) dataset, part of the Gene Environment Association Studies (GENEVA) program of the National Institutes of Health (NIH) Genes, Environment, and Health Initiative35. SAGE was designed to study alcohol dependence, and as a result is composed largely of unrelated alcohol-dependent cases (n = 1048) and non-alcohol-dependent control subjects (n = 928). The SAGE dataset was ascertained from 3 large substance dependence datasets: the Collaborative Genetic Study of Nicotine Dependence (COGEND), the Collaborative Study on the Genetics of Alcoholism (COGA) and the Family Study of Cocaine Dependence (FSCD)36. For the purpose of the current analyses, there were 1976 European-Americans as defined both by self-report and principal components from the GWAS data, who met criteria for cocaine use disorder or unaffected controls.
The DSM-5 was published on May 18, 2013, and supersedes the DSM-IV text revision (DSM-IV-TR) published in 2000. In the DSM-5, the DSM-IV criteria cocaine abuse and cocaine dependence have been combined into a single cocaine use disorder. Cocaine use disorder is now divided into mild (2–3 criteria), moderate (4–5 criteria) and severe (6 or more criteria). A further difference is that whereas in the DSM-IV, cocaine abuse diagnostic criteria required only one symptom, in the DSM-5 diagnostic criteria, a diagnosis of mild cocaine use disorder requires at least two criteria to be met. Lastly, the DSM-IV recurrent legal problems criterion for cocaine abuse was replaced with the new criterion of craving37.We recoded DSM-IV values in SAGE to DSM-5 for both cocaine and alcohol use, since alcohol use is used as a covariate in our analyses.
COGEND sample
COGEND was designed as a community based case–control study of nicotine dependence. COGEND contains current smokers with nicotine dependence defined by a Fagerstrom Test for Nicotine Dependence (FTND) score ≥ 4 (maximum score of 10) and non-nicotine dependent subjects who had smoked at least 100 cigarettes and had a lifetime FTND score of zero or one. All subjects were ascertained from Detroit and St Louis. Out of the 53,000 subjects who were screened by telephone, 2,800 were interviewed in person and approximately 2,700 donated blood samples for genetic studies23. COGEND contributed 76 individuals with cocaine use disorder (54 nicotine dependent cases based on the FTND, 22 non-dependent smokers) and 1005 individuals without cocaine use disorder (135 nicotine dependent cases based on the FTND, 870 non-dependent smokers).
COGA sample
Out of more than 11,000 subjects who participated in COGA, a case-control series of unrelated individuals was selected for SAGE. COGA recruited subjects in Hartford, Connecticut; Indianapolis, Indiana; Iowa City, Iowa; New York City, New York; San Diego, California; St Louis, Missouri; and Washington, DC. For inclusion in SAGE, cases had to meet life-time criteria for DSM-IV alcohol dependence, the majority of cases were recruited from alcoholism treatment centers. Control subjects, were both biologically unrelated to cases, and had consumed alcohol but never experienced any significant alcohol or drug-related problems, according to the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA)23. COGA contributed 212 individuals with DSM-5 cocaine use disorder (all 212 of which also had DSM-5 alcohol use disorder) and 389 individuals without cocaine use disorder (271 with DSM-5 alcohol use disorder, 118 alcohol drinkers without DSM-5 alcohol use disorder).
FSCD sample
Subjects in the FSCD were specifically recruited for cocaine use from chemical dependency treatment units in the greater St Louis metropolitan area. The Missouri Family Registry identified community-based control subjects and matched them by age, race, gender and residential zip code. Controls were biologically unrelated individuals from the same communities who consumed alcohol, but had no lifetime history of dependence on any substance23. FSCD contributed 218 individuals with DSM-5 cocaine use disorder and 76 individuals without DSM-5 cocaine use disorder (51 non-exposed controls and 25 exposed but without DSM-5 cocaine use disorder).
Genotyping and Q/C
All DNA samples were genotyped on the Illumina Human 1M-Duo beadchip by the Center for Inherited Disease Research (CIDR) at Johns Hopkins University. After thorough genotype quality control process, 948,758 of the 1,049,008 genotyped SNPs were available for genetic analysis. Sixty-five of these genotyped SNPs fell within the region containing the CHRNA6 and CHRNB3 genes on chromosome 8. Of the 65, only SNPs with a minor allele frequency (MAF) >1% and a genotyping call rate >0.98 were considered (47 SNPs). Full details regarding the quality control procedures are provided in the data cleaning report posted on the GENEVA website (http://www.genevastudy.org/docs/GENEVA_Alcohol_QC_report_8Oct2008.pdf) and in related publications35,36.
Phenotype
Cocaine use disorder for all members of the SAGE dataset was measured using the DSM-5 criteria37. As outlined in the manual, 11 criteria (3 abuse, 7 dependence and craving) were combined and cocaine use disorder was scored as the endorsement of 2 or more of these 11 criteria (N = 506). Unaffected individuals met zero or one of the DSM-5 criteria (N = 1470).
Statistical analyses
All analyses were performed on genotyped data. Association analyses were conducted in PLINK38 for SNPs in the region on chromosome 8 encompassing the α6 and β3 subunits of nicotinic receptors (42600000 kb to 42800000 kb). Logistic regression with DSM-5 cocaine use disorder as the dependent variable was performed. Covariates included were age at interview as a continuous variable, gender, study, maximum lifetime FTND score (0–10, based on the Fagerström Test of Nicotine Dependence) to control for smoking status, and DSM-5 alcohol use disorder. Study was coded using two dummy variables (yes/no for two of the three studies) in order to control for differences in ascertainment. Haploview was run using the genotypes of the study population to determine the number of independent linkage disequilibrium (LD) bins in the region using a threshold of r2 ≥ 0.8. The Bonferroni correction used in this study is p = 0.002 (0.05/22), as the number of LD bins in the region examined is 22. A conditional analysis was conducted including allele dosage for the top associated SNP as a covariate in the logistic model.
In a case/control division of subjects based on presence or absence of cocaine use disorder, logistic regressions were run both using as controls only those who had been exposed to cocaine but had not become dependent (i.e. have used cocaine at least once in their lifetime) and all non-cocaine-dependent individuals in the sample, regardless of exposure status.
To improve our understanding of observed associations, the top SNPs identified in the whole SAGE dataset were examined using the same models described above in strata of the data defined by study (COGEND, COGA, FSCD), smoking status (FTND cases and FTND controls), and alcohol use disorder (DSM-5 cases and DSM-5 controls). A two-SNP haplotype analysis was run in R using the top SNP and the SNP tagging the bin previously found to be genome-wide significant with nicotine dependence (rs1451240)23. This model included the covariates age, sex, study, DSM-5 alcohol symptom count, FTND total (in the cocaine haplotypes) and DSM-5 cocaine use disorder (in the FTND haplotypes), and examined the association with each haplotype with the phenotype compared to homozygotes for the reference allele at both SNPs. Finally, we used conditional analyses to examine the extent of independence between these cocaine-associated SNPs and the previous association in the region with nicotine dependence tagged by rs1451240.
Acknowledgments
The Collaborative Study on the Genetics of Alcoholism (COGA): COGA, Principal Investigators B Porjesz, V Hesselbrock, H Edenberg, L Bierut includes ten different centers: University of Connecticut (V Hesselbrock); Indiana University (HJ Edenberg, J Nurnberger Jr, T Foroud); University of Iowa (S Kuperman, J Kramer); SUNY Downstate (B Porjesz); Washington University in Saint Louis (L Bierut, A Goate, J Rice, K Bucholz); University of California at San Diego (M Schuckit); Rutgers University (J Tischfield); Southwest Foundation (L Almasy), Howard University (R Taylor) and Virginia Commonwealth University (D Dick). A Parsian and M Reilly are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P Michael Conneally, Raymond Crowe and Wendy Reich, for their critical contributions. In memory of Theodore Reich, founding Principal Investigator of COGEND, we are indebted to his leadership in the establishment and nurturing of COGEND and acknowledge with great admiration his seminal scientific contributions to the field. Lead investigators directing data collection for COGEND are Laura Bierut, Naomi Breslau, Dorothy Hatsukami, and Eric Johnson. The authors thank Heidi Kromrei and Tracey Richmond for their assistance in COGEND data collection. COGA is a national collaborative study supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). COGEND is a collaborative research group and part of the National Institute on Drug Abuse (NIDA) Genetics Consortium. Subject collection was supported by NIH grant P01 CA89392 (PI - L Bierut) from the NCI. Phenotypic and genotypic data are stored in the NIDA Center for Genetic Studies (NCGS) at http://zork.wustl.edu/ under NIDA Contract HHSN271200477451C (PIs J Tischfield and J Rice).
Footnotes
Drs. LJ Bierut and AM Goate, are listed as inventors on the patent “Markers for Addiction” (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. Dr. N. Saccone is the spouse of Dr. S. Saccone, who is also listed as an inventor on the above patent.
Author Contributions B.S., G.H., A.A., R.C., K.B., A.B., J.T., E.J., H.E., M.S., N.S., L.B. and A.G. critically reviewed and approved the manuscript. B.S. ran all analyses and wrote the manuscript, G.H. provided crucial technical support, A.A. helped compile the DSM-5 phenotype, N.S. supervised data management for COGEND and FSCD, E.J. supervised data collection for COGEND, L.B. supervised data collection for all three components of SAGE and designed the SAGE study, A.G. critically reviewed all data and data analysis.
References
- Gossop M., Stewart D., Treacy S. & Marsden J. A prospective study of mortality among drug misusers during a 4-year period after seeking treatment. Addiction 97, 39–47 (2002). [DOI] [PubMed] [Google Scholar]
- Nutt D. J., King L. A. & Phillips L. D. Drug harms in the UK: a multicriteria decision analysis. Lancet 376, 1558–1565, 10.1016/S0140-6736(10)61462-6 (2010). [DOI] [PubMed] [Google Scholar]
- Goldman D., Oroszi G. & Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet 6, 521–532, 10.1038/nrg1635 (2005). [DOI] [PubMed] [Google Scholar]
- Wang J. C., Kapoor M. & Goate A. M. The genetics of substance dependence. Annu Rev Genomics Hum Genet 13, 241–261, 10.1146/annurev-genom-090711-163844 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grucza R. A. et al. A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol Psychiatry 64, 922–929, 10.1016/j.biopsych.2008.04.018 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J. et al. Genomewide linkage scan for nicotine dependence: identification of a chromosome 5 risk locus. Biol Psychiatry 61, 119–126, 10.1016/j.biopsych.2006.08.023 (2007). [DOI] [PubMed] [Google Scholar]
- Yang B. Z., Han S., Kranzler H. R., Farrer L. A. & Gelernter J. A genomewide linkage scan of cocaine dependence and major depressive episode in two populations. Neuropsychopharmacology 36, 2422–2430, 10.1038/npp.2011.122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. Pro-opiomelanocortin gene variation related to alcohol or drug dependence: evidence and replications across family- and population-based studies. Biol Psychiatry 66, 128–136, 10.1016/j.biopsych.2008.12.021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer L. A. et al. Association of variants in MANEA with cocaine-related behaviors. Arch Gen Psychiatry 66, 267–274, 10.1001/archgenpsychiatry.2008.538 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J. et al. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry, 10.1038/mp.2013.99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler K. S., Myers J. & Prescott C. A. Specificity of genetic and environmental risk factors for symptoms of cannabis, cocaine, alcohol, caffeine, and nicotine dependence. Arch Gen Psychiatry 64, 1313–1320, 10.1001/archpsyc.64.11.1313 (2007). [DOI] [PubMed] [Google Scholar]
- Tsuang M. T., Bar J. L., Harley R. M. & Lyons M. J. The Harvard Twin Study of Substance Abuse: what we have learned. Harv Rev Psychiatry 9, 267–279 (2001). [PubMed] [Google Scholar]
- Mwenifumbo J. C. & Tyndale R. F. Molecular genetics of nicotine metabolism. Handb Exp Pharmacol, 235–259, 10.1007/978-3-540-69248-5_9 (2009). [DOI] [PubMed] [Google Scholar]
- Thorgeirsson T. E. et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet 42, 448–453, 10.1038/ng.573 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut L. J. et al. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry 17, 445–450, 10.1038/mp.2011.124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R. et al. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology 35, 1921–1931, 10.1038/npp.2010.64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Gen 42, 441–447, 10.1038/ng.571 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone S. F. et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet 16, 36–49, 10.1093/hmg/ddl438 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiger J. S. et al. The neuronal nicotinic receptor subunit genes (CHRNA6 and CHRNB3) are associated with subjective responses to tobacco. Hum Mol Genet 17, 724–734, 10.1093/hmg/ddm344 (2008). [DOI] [PubMed] [Google Scholar]
- Hoft N. R. et al. Genetic association of the CHRNA6 and CHRNB3 genes with tobacco dependence in a nationally representative sample. Neuropsychopharmacology 34, 698–706, 10.1038/npp.2008.122 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone N. L. et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res 69, 6848–6856, 10.1158/0008-5472.CAN-09-0786 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone N. L. et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet 6, 10.1371/journal.pgen.1001053 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice J. P. et al. CHRNB3 is more strongly associated with Fagerstrom test for cigarette dependence-based nicotine dependence than cigarettes per day: phenotype definition changes genome-wide association studies results. Addiction 107, 2019–2028, 10.1111/j.1360-0443.2012.03922.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz S. M. et al. Genetic association of bipolar disorder with the beta(3) nicotinic receptor subunit gene. Psychiatr Genet 21, 77–84, 10.1097/YPG.0b013e32834135eb (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller G. et al. Rare missense variants in CHRNB3 and CHRNA3 are associated with risk of alcohol and cocaine dependence (accepted). Hum Mol Genet, 10.1093/hmg/ddt463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin R. D., Zvolensky M. J., Keyes K. M. & Hasin D. S. Mental disorders and cigarette use among adults in the United States. Am J Addic 21, 416–423, 10.1111/j.1521-0391.2012.00263.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenneson A., Funderburk J. S. & Maisto S. A. Substance use disorders increase the odds of subsequent mood disorders. Drug Alcohol Depend, 10.1016/j.drugalcdep.2013.06.011 (2013). [DOI] [PubMed] [Google Scholar]
- Post R. M. & Kalivas P. Bipolar disorder and substance misuse: pathological and therapeutic implications of their comorbidity and cross-sensitisation. Br J Psychiatry 202, 172–176, 10.1192/bjp.bp.112.116855 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P. I. et al. Clinical correlates and familial aggregation of age at onset in bipolar disorder. Am J Psychiatry 163, 240–246, 10.1176/appi.ajp.163.2.240 (2006). [DOI] [PubMed] [Google Scholar]
- Hansen S. T. & Mark G. P. The nicotinic acetylcholine receptor antagonist mecamylamine prevents escalation of cocaine self-administration in rats with extended daily access. Psychopharmacology (Berl) 194, 53–61, 10.1007/s00213-007-0822-z (2007). [DOI] [PubMed] [Google Scholar]
- Koob G. F. Drug addiction: the yin and yang of hedonic homeostasis. Neuron 16, 893–896 (1996). [DOI] [PubMed] [Google Scholar]
- Wise R. A. Addictive drugs and brain stimulation reward. Annu Rev Neurosci 19, 319–340, 10.1146/annurev.ne.19.030196.001535 (1996). [DOI] [PubMed] [Google Scholar]
- Tanda G., Pontieri F. E. & Di Chiara G. Cannabinoid and heroin activation of mesolimbic dopamine transmission by a common mu1 opioid receptor mechanism. Science 276, 2048–2050 (1997). [DOI] [PubMed] [Google Scholar]
- Laurie C. C. et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol 34, 591–602, 10.1002/gepi.20516 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut L. J. et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A 107, 5082–5087, 10.1073/pnas.0911109107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin D. S. et al. DSM-5 Criteria for Substance Use Disorders: Recommendations and Rationale. Am J Psychiatry 170, 834–851, 10.1176/appi.ajp.2013.12060782 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575, 10.1086/519795 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81, 559–75 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]