Abstract
Over the past decade antiretroviral drugs have dramatically improved the prognosis for HIV-1 infected individuals, yet achieving better access to vulnerable populations remains a challenge. The principal obstacle to the CCR5-antagonist, maraviroc, from being more widely used in anti-HIV-1 therapy regimens is that the pre-treatment genotypic “tropism tests” to determine virus susceptibility to maraviroc have been developed primarily for HIV-1 subtype B strains, which account for only 10% of infections worldwide. We therefore developed PhenoSeq, a suite of HIV-1 genotypic tropism assays that are highly sensitive and specific for establishing the tropism of HIV-1 subtypes A, B, C, D and circulating recombinant forms of subtypes AE and AG, which together account for 95% of HIV-1 infections worldwide. The PhenoSeq platform will inform the appropriate use of maraviroc and future CCR5 blocking drugs in regions of the world where non-B HIV-1 predominates, which are burdened the most by the HIV-1 pandemic.
Since 2005, AIDS related deaths have fallen by 30%, largely due to the rollout of highly effective antiretroviral therapies. However, achieving adequate global access to antiretrovirals remains a major challenge, with approximately 66% of HIV-1 infected individuals considered eligible for therapy unable to access treatment1. One barrier to drug accessibility is the need for specialized prognostic laboratory tests to inform the appropriate use of anti-HIV-1 drugs.
Maraviroc is the first licensed drug in a relatively new class of HIV-1 entry inhibitors called CCR5 antagonists2. Entry of HIV-1 into cells of the immune system is initiated by primary engagement of the CD4 receptor on the cell surface, then secondary engagement of either of the chemokine coreceptors, CCR5 or CXCR4. Maraviroc blocks the ability of HIV-1 to engage CCR5, which inhibits viral entry into cells2, but does not block engagement of CXCR4 which is used by a substantial proportion of circulating virus, particularly at later stages of infection3,4,5,6. A specialized pre-treatment prognostic (or “tropism”) test is therefore mandatory to exclude patients with detectable CXCR4-using virus from being treated with CCR5 antagonists7,8,9.
Traditional “phenotypic” tropism tests for establishing HIV-1 coreceptor specificity use recombinant viruses pseudotyped with patient derived HIV-1 envelope proteins to infect cell lines expressing CD4 and either CCR5 or CXCR49,10,11,12,13. However, on a global scale the cost, lengthy turn-around time, and highly specialized nature of these tests have been obstacles to maraviroc being used more widely in HIV-1 treatment regimens.
In contrast, “genotypic” coreceptor usage prediction algorithms, trained on characteristic HIV-1 sequence alterations within the third variable region (V3) of the viral envelope gene (env) offer a comparatively inexpensive, rapid and accessible alternative to phenotypic tropism assays. However, while genotypic algorithms can be highly sensitive for predicting coreceptor usage and treatment outcome for patients receiving maraviroc9,10,11,13,14,15,16,17,18,19,20, the majority were trained on HIV-1 subtype B V3 sequences, and consequently they have limited capacity for establishing coreceptor usage of non-B HIV-1 subtypes that have distinctive V3 sequences, and which account for approximately 90% of infections worldwide13,14,21,22,23. This has particular implications for regions of the world where non-B HIV-1 strains predominate, and which have expanding economies and rapidly improving health care systems such as Eastern Europe and Russia where HIV-1 subtype A is endemic, India and China where HIV-1 subtype C predominates24, and Thailand, Indonesia and Vietnam where a circulating recombinant form of HIV-1 subtypes A and E predominates (referred to as HIV-1 CRF01_AE)25,26. Furthermore, HIV-1 CRF01_AE is becoming more prevalent in developed countries such as Japan and Singapore27,28. These are all populous regions with moderate to high HIV-1 burdens. These factors constitute an economic and clinical environment suitable for the introduction of CCR5 antagonists to HIV-1 treatment regimens, with significant potential to benefit large HIV-1 affected populations. The lack of genotypic algorithms designed specifically for non-B HIV-1 subtypes is presently a major barrier to informing the appropriate use of maraviroc for treatment of HIV-1 infected individuals from these regions, and will continue to be a barrier for new coreceptor blocking drugs as they are developed.
Here, we report the development and utility of PhenoSeq, a suite of new genotypic algorithms that are highly sensitive and specific for establishing coreceptor usage of HIV-1 subtypes A, B, C, D, CRF01_AE and of a circulating recombinant form of HIV-1 subtypes A and G (referred to as HIV-1 CRF02_AG), which together account for approximately 95% of HIV-1 strains worldwide (15% subtype A, 10% subtype B, 50% subtype C, 5% subtype D, 5% CRF01_AE and 10% CRF02_AG). Our free-to-use, automated online platform of prognostic tools (www.burnet.edu.au/phenoseq) will inform the appropriate use of maraviroc and future coreceptor blocking drugs in regions of the world where non-B HIV-1 strains predominate.
Results
Developing PhenoSeq algorithms
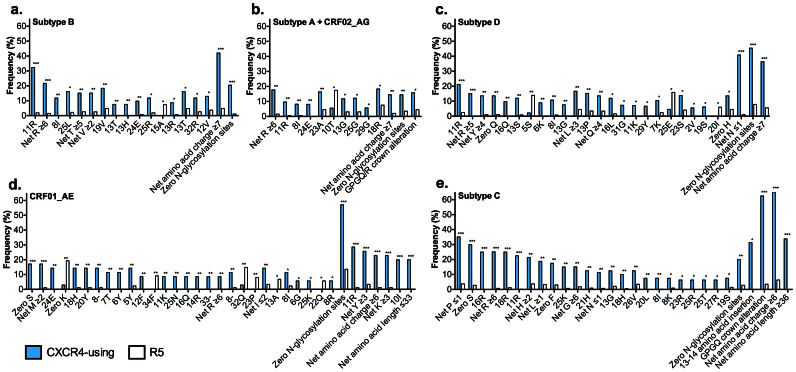
To develop the PhenoSeq algorithms, we first analyzed “training sets” comprising all available HIV-1 V3 sequences from the Los Alamos HIV-1 Database that had corresponding subtype and phenotypic tropism test results, to elucidate statistically significant alterations that distinguish CXCR4-using from CCR5-using (R5) viruses. Notably, we selected one sequence per phenotype per patient in order to avoid bias by resampling similar V3 sequences from a single HIV-1 infected individual. We focused on the V3 region since it is the major HIV-1 sequence determinant of coreceptor specificity29,30. For these analyses, we evaluated HIV-1 V3 amino acid length, net amino acid charge, number of N-linked glycosylation sites and the frequency of site-specific amino acid alterations. These sequence analysis results are summarized in Figure 1. Notably, because CRF02_AG strains contain a subtype A-like V3 region, we pooled and analyzed CRF02_AG and subtype A sequences together (Fig. 1b), and developed a single subtype A and CRF02_AG specific PhenoSeq algorithm (PhenoSeq-A/AG).
Figure 1. Training set V3 characteristics and amino acid mutations associated with coreceptor usage.
Sequence analysis results demonstrating amino acid alterations that differentiate phenotypically characterized CXCR4-using from R5 V3 sequences are shown for (a) HIV-1 subtype B (n = 93 CXCR4-using and n = 296 R5 sequences), (b) HIV-1 subtype A and CRF02_AG (n = 60 CXCR4-using and n = 172 R5 sequences), (c) HIV-1 CRF01_AE (n = 50 CXCR4-using and n = 128 R5 sequences), (d) HIV-1 subtype D (n = 57 CXCR4-using and n = 80 R5 sequences) and (e) HIV-1 subtype C (n = 80 CXCR4-using and n = 429 R5 sequences); “GPGQ/R crown alteration” refers to V3 sequences where residues 16–19 are not GPGQ or GPGR; “GPGQ crown alteration” refers to V3 sequences where residues 16–19 are not GPGQ; “13–14 amino acid insertion” refers to V3 sequences with a two amino acid insertion between residues 13 and 14. P-values for net amino acid length, net amino acid charge and number of glycosylation sites were calculated using a Mann Whitney U-test (two-tailed). P-values for amino acid alterations were calculated using a Fisher's exact t-test (two-tailed). *** p-value < 0.0001, ** p < 0.01, * p < 0.05. Notably, a number of these V3 amino acid alterations have been associated with HIV-1 coreceptor usage3,21,23,29,32,38,40,41,58,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76.
A unique feature of the PhenoSeq design method is the provision for continuous improvement and refinement of prediction criteria as new V3 sequences become available, to maintain maximal predictive accuracy. To illustrate this, here we re-evaluated and optimized our recently described HIV-1 subtype C specific algorithm CoRSeqV3-C21 (now re-named PhenoSeq-C), by adding 27 CXCR4-using and 35 R5 V3 sequences to the original CoRSeqV3-C training set, which have been made available since its development (Fig. 1e).
The coreceptor usage prediction criteria for each PhenoSeq algorithm was determined by testing all combinations of V3 sequence alterations and selecting the combinations with the fewest V3 sequence alterations that, when tested against their respective training set sequences, optimized their sensitivity for detecting CXCR4-using HIV-1, without compromising specificity for detecting R5 strains (Table 1). Notably, we selected the most accurate combination of prediction parameters that comprised the fewest V3 sequence alterations in order to minimize the potential for including parameters that were unique to the PhenoSeq training sets. For simplicity, only comparisons to the clinically validated Geno2Pheno (g2p) [at false positive rates (FPR) of 5.75% and 10%], WebPSSMX4R5 and WebPSSMSI/NSI algorithms are shown15,31,32. Extended comparisons to all the alternative algorithms and prediction rules that have been described in the literature are shown in Supplementary Tables 1–5. To demonstrate the optimization of the PhenoSeq prediction criteria, Table 1 shows that PhenoSeq-B, PhenoSeq-C, PhenoSeq-D, PhenoSeq-AE and PhenoSeq-A/AG are the most sensitive algorithms for detecting CXCR4-usage of the HIV-1 training set sequences without compromising specificity. The PhenoSeq-B, PhenoSeq-D, PhenoSeq-C, PhenoSeq-AE and PhenoSeq-A/AG prediction criteria are illustrated in Supplementary Figures 1–5.
Table 1. Sensitivity, specificity and AUROC of genotypic algorithms for predicting coreceptor usage of training set V3 sequences.
| PhenoSeq | g2p FPR 5.75% | g2p FPR 10% | WebPSSMX4R5 | WebPSSMSI/NSI* | ||
|---|---|---|---|---|---|---|
| PhenoSeq-B | Sens/Spec | 80.7/82.9 | 68.8/93.7 | 75.3/86.6 | 53.8/95.8 | 61.3/94.7 |
| Training Set | AUROC | 0.82 | 0.81 (p = 0.40) | 0.81 (p = 0.40) | 0.75 (p = 0.05) | 0.78 (p = 0.17) |
| 93 CXCR4-using, 269 R5 | ||||||
| PhenoSeq-C | Sens/Spec | 88.8/87.9 | 75/96.5 | 81.3/93.7 | - | 82.5/89 |
| Training Set | AUROC | 0.88 | 0.86 (p = 0.24) | 0.88 (p = 0.40) | - | 0.86 (p = 0.24) |
| 80 CXCR4-using, 429 R5 | ||||||
| PhenoSeq-D | Sens/Spec | 87.8/87.5 | 81.4/72.7 | 88.4/61.4 | 81.4/65.9 | 69.8/63.6 |
| Training Set | AUROC | 0.88 | 0.77 (p = 0.02) | 0.72 (p < 0.01) | 0.72 (p = 0.03) | 0.77 (p = 0.02) |
| 57 CXCR4-using, 80 R5 | ||||||
| PhenoSeq-AE | Sens/Spec | 88/92.2 | 82/76.6 | 88/56.3 | 78/78.9 | 80/81.3 |
| Training Set | AUROC | 0.90 | 0.79 (p = 0.02) | 0.72 (p < 0.01) | 0.79 (p < 0.01) | 0.81 (p = 0.03) |
| 50 CXCR4-using, 128 R5 | ||||||
| PhenoSeq-A/AG | Sens/Spec | 59.7/87.1 | 29/96 | 40.3/91.5 | 27.4/92 | 29/97 |
| Training Set | AUROC | 0.73 | 0.63 (p = 0.03) | 0.659 (p = 0.10) | 0.60 (p < 0.01) | 0.63 (p = 0.04) |
| 59 CXCR4-using, 172 R5 |
Sens, % sensitivity for correctly predicting CXCR4-usage (relative to phenotypic tropism assay results) was calculated by dividing the number of correctly predicted CXCR4-using sequences by the total number CXCR4-using sequences and multiplying by 100. Spec, % specificity for correctly predicting R5 strains (relative to phenotypic tropism assay results) was calculated by dividing the number of correctly predicted R5 sequences by the total number of R5 sequences and multiplying by 100. P-values (two-tailed) for comparing area under the receiver operator characteristic curves (AUROC) was performed according to Hanley et al33. P-values < 0.05 were considered significant and are highlighted in bold italicized text. FPR, false positive rate.
*The subtype C specific WebPSSMSI/NSI algorithm was used for subtype C HIV-1 predictions.
Developing the bioinformatics tool bulk2clonal
Validating the predictive accuracy of the PhenoSeq algorithms in a clinically relevant setting requires testing their performance against sets of phenotypically characterized V3 sequences derived from plasma of individuals infected with HIV-1 subtypes A, B, C, D, CRF01_AE or CRF02_AG that are independent of the training sets of Los Alamos HIV Database derived V3 sequences. However, to do this we first needed to develop a new program that would make the PhenoSeq algorithms compatible with V3 sequences generated by routine diagnostic laboratories.
This is because, when selecting our PhenoSeq training set sequences from the Los Alamos HIV Database, it was necessary to exclude V3 sequences that contained sites of base-call ambiguity, since this would impede analyses of V3 characteristics. Consequently, at this point of development, the predictive accuracy of PhenoSeq algorithms is dependent on unambiguous query V3 sequences. Unambiguous V3 sequences can be isolated from patient blood samples using contemporary clonal and next-generation deep sequencing techniques, however in the clinical setting, the majority of V3 sequences are isolated by much less expensive Sanger (or “bulk”) sequencing techniques. Bulk V3 sequencing produces a consensus sequence whereby each nucleotide represents the most frequent base at a given position within the sampled HIV-1 quasispecies. Consequently, bulk sequencing often produces sequences with sites of ambiguity whereby two or more nucleotides occur with equal frequency. Therefore, to enable compatibility with bulk V3 sequencing techniques we interfaced each of the PhenoSeq algorithms with a new bioinformatic tool that we developed, called “bulk2clonal”.
Briefly, bulk2clonal converts nucleotide V3 sequences containing ambiguity into multiple, unambiguous amino acid sequences, by generating and translating all possible nucleotide combinations, as described in the Methods (Supplementary Fig. 6). We configured PhenoSeq algorithms to predict a bulk V3 sequence containing ambiguity to be CXCR4-using if ≥10% of the protein sequences generated by bulk2clonal were predicted to be CXCR4-using. PhenoSeq then predicts a patient to harbour CXCR4-using HIV-1 if one or more bulk V3 nucleotide sequences isolated from that patient are predicted to be CXCR4-using.
Validating PhenoSeq algorithms integrated with bulk2clonal
To validate the predictive accuracy of each PhenoSeq algorithm (now integrated with bulk2clonal) in a clinically relevant setting, we next compared their sensitivity, specificity and area under the receiver operating characteristic curve (AUROC) to several alternate algorithms for predicting the coreceptor usage of panels of bulk V3 sequences isolated from patient plasma samples that are unavailable on the Los Alamos HIV Database and thus independent of PhenoSeq training sets, relative to known phenotypic tropism assay results. Statistical comparison of AUROC scores was performed to assess whether the PhenoSeq algorithms were statistically more accurate than alternative genotypic algorithms, as described by Hanley et al33.
PhenoSeq-B was tested against 12 CXCR4-using and 41 R5 bulk V3 sequences from patients of a study by Mulinge et al13 that were phenotyped by an in-house recombinant virus phenotypic tropism assay (RVA) (Table 2; PhenoSeq-B Test Set 1). PhenoSeq-B demonstrated the highest possible sensitivity (100%), without compromising specificity (87.8%), and an AUROC that was statistically greater than WebPSSMX4R5 (p < 0.01) and WebPSSMSI/NSI (p < 0.01), and statistically similar to g2p at FPRs of 5.75% and 10%. Considering the relatively low number of CXCR4-using V3 sequences used here, we also tested PhenoSeq-B against 92 CXCR4-using and 269 R5 independent V3 sequences from the Los Alamos HIV Database (Table 2; Test Set 2). Here, PhenoSeq demonstrated the highest sensitivity (78.4%), without compromising specificity (80.3%), and an AUROC that was statistically similar to g2p at FPRs of 5.75% and 10%, and WebPSSMX4R5 and WebPSSMSI/NSI.
Table 2. Sensitivity, specificity and AUROC of genotypic algorithms for predicting coreceptor usage of test set V3 sequences.
| PhenoSeq | g2p FPR 5.75% | g2p FPR 10% | WebPSSMX4R5 | WebPSSMSI/NSI* | ||
|---|---|---|---|---|---|---|
| PhenoSeq-B Test Set 1 | Sens/Spec | 100/87.8 | 100/97.6 | 100/90.2 | 41.7/95.1 | 16.7/87.8 |
| Mulinge et al | AUROC | 0.94 | 0.99 (p = 0.17) | 0.95 (p = 0.44) | 0.68 (p < 0.01) | 0.52 (p < 0.01) |
| 12 CXCR4-using, 41 R5 | ||||||
| PhenoSeq-B Test Set 2 | Sens/Spec | 78.4/80.3 | 70.7/92.2 | 72.8/84 | 55.4/96.3 | 62/94.1 |
| Los Alamos HIV Database | AUROC | 0.79 | 0.82 (p = 0.32) | 0.78 (p = 0.41) | 0.76 (p = 0.24) | 0.78 (p = 0.41) |
| 92 CXCR4-using, 269 R5 | ||||||
| PhenoSeq-C Test Set 1 | Sens/Spec | 83.6/92.5 | 78.2/95 | 81.8/95 | - | 85.5/77.5 |
| A4001064 | AUROC | 0.88 | 0.87 (p = 0.22) | 0.88 (p = 0.30) | - | 0.82 (p = 0.22) |
| 55 CXCR4-using, 40 R5 | ||||||
| PhenoSeq-C Test Set 2 | Sens/Spec | 77.8/75.4 | 11.1/93 | 50/82.4 | - | 61.1/81.3 |
| MERIT | AUROC | 0.77 | 0.52 (p < 0.01) | 0.66 (p = 0.15) | - | 0.71 (p = 0.29) |
| 18 CXCR4-using, 187 R5 | ||||||
| PhenoSeq-D Test Set | Sens/Spec | 80.5/77.3 | 81.4/72.7 | 88.4/61.4 | 81.4/65.9 | 69.8/63.6 |
| A4001064 | AUROC | 0.79 | 0.77 (p = 0.40) | 0.75 (p = 0.29) | 0.74 (p = 0.24) | 0.67 (p = 0.05) |
| 43 CXCR4-using, 44 R5 | ||||||
| PhenoSeq-AE Test Set | Sens/Spec | 85.7/96 | 78.6/68 | 85.7/56 | 85.7/72 | 85.7/76 |
| A4001064 | AUROC | 0.91 | 0.73 (p = 0.046) | 0.71 (p = 0.03) | 0.79 (p = 0.11) | 0.81 (p = 0.15) |
| 14 CXCR4-using, 25 R5 | ||||||
| PhenoSeq-A/AG Test Set 1 | Sens/Spec | 88.9/76.8 | 44.4/97.1 | 55.6/94.2 | 44.4/87 | 66.7/84 |
| A4001064 | AUROC | 0.83 | 0.71 (p = 0.18) | 0.75 (p = 0.27) | 0.66 (p = 0.10) | 0.75 (p = 0.28) |
| 9 CXCR4-using, 69 R5 | ||||||
| PhenoSeq-A/AG Test Set 2 | Sens/Spec | 62.5/97.2 | 50/94.4 | 62.5/80.6 | 37.5/88.9 | 37.5/91.7 |
| Mulinge et al | AUROC | 0.80 | 0.72 (p = 0.30) | 0.72 (p = 0.29) | 0.63 (p = 0.14) | 0.65 (p = 0.16) |
| 8 CXCR4-using, 36 R5 |
Sens (% sensitivity) and Spec (% specificity) was calculated as described in the Table 1 legend. P-values (two-tailed) for comparisons of area under the receiver operator characteristic curve (AUROC) were performed according to Hanley et al33. P-values < 0.05 were considered significant and are highlighted in bold italicized text. FPR, false positive rate.
*The subtype C specific WebPSSMSI/NSI algorithm was used for subtype C HIV-1 predictions.
We next tested the newly-optimized PhenoSeq-C algorithm against 55 CXCR4-using and 40 R5 bulk V3 sequences from 95 participants of the Pfizer epidemiology trial A4001064 that were phenotyped by the original Trofile™ phenotypic tropism assay (OTA)14,34, and against 18 CXCR4-using and 187 R5 bulk V3 sequences from 205 participants of the Pfizer phase III Maraviroc versus Efavirenz in Treatment-Naïve Patients (MERIT) trial that were phenotyped by the enhanced sensitivity Trofile™ phenotypic tropism assay (ESTA)35 (Table 2; PhenoSeq-C Test Sets 1 and 2, respectively). For the A4001064 patients, PhenoSeq-C demonstrated the highest sensitivity (83.6%), without compromising specificity (92.5%), and an AUROC that was statistically similar to g2p at FPRs of 5.75% and 10%, and WebPSSMSI/NSI-C. For the MERIT patients, PhenoSeq-C demonstrated the highest sensitivity (77.8%), without compromising specificity (75.4%), and an AUROC that was statistically greater than g2p at a FPR of 5.75% (p < 0.01), and which was similar to g2p at a FPR of 10% and WebPSSMSI/NSI-C.
PhenoSeq-D was tested against 43 CXCR4-using and 44 R5 bulk V3 sequences from 87 A4001064 participants that were phenotyped by OTA14,34 (Table 2; PhenoSeq-D Test Set). Our results show that PhenoSeq-D had the most favorable sensitivity (80.5%) and specificity (77.3%) profile and the highest AUROC, which was statistically similar to g2p at FPRs of 5.75% and 10%, WebPSSMX4R5 and WebPSSMSI/NSI.
PhenoSeq-AE was tested against 14 CXCR4-using and 25 R5 bulk V3 sequences from 39 patients phenotyped by RVA13 (Table 2; PhenoSeq-AE Test Set). Our results show that PhenoSeq-AE had the highest sensitivity (85.7%) without compromising specificity (96%), and an AUROC that was statistically greater than g2p at FPRs of 5.75% (p = 0.046) and 10% (p = 0.03).
Finally, PhenoSeq-A/AG was tested against 9 CXCR4-using and 69 R5 bulk V3 sequences from 78 A4001064 participants that were phenotyped by OTA14,34 (Table 1; PhenoSeq-A/AG Test Set 1), and 8 CXCR4-using and 36 R5 bulk V3 sequences from 44 patients that were phenotyped by RVA13 (Table 2; PhenoSeq-A/AG Test Set 2). For both test sequence sets, PhenoSeq-A/AG demonstrated the most favorable sensitivity (88.9% and 62.5%, respectively) and specificity (76.8% and 97.2%, respectively) profiles, and had the highest AUROC.
Discussion
Maraviroc is the first licensed drug in a relatively new class of anti-HIV-1 therapeutics called CCR5 antagonists, which bind to CCR5 and block CCR5-mediated HIV-1 entry into cells. Since maraviroc is ineffective against CXCR4-using HIV-1, a coreceptor usage (or “tropism”) test is required before its administration. The most frequently used phenotypic tropism test is the enhanced sensitivity Trofile™ assay (ESTA)35,36,37, yet several factors such as cost and turn-around time have limited the widespread clinical use of ESTA and other phenotypic tropism assays, which in turn has limited the access of maraviroc to many eligible patients. On the other hand, genotypic algorithms enable most diagnostic laboratories to establish HIV-1 coreceptor usage by amplifying and sequencing the relatively short V3 region of env from patient blood samples, which compared to phenotypic tropism assays is a relatively inexpensive, rapid and straightforward process. Unfortunately, the majority of the currently available genotypic algorithms have been developed against HIV-1 subtype B V3 sequences and consequently they lack optimal predictive accuracy against non-B HIV-1 V3 sequences, as many of the V3 loop determinants of coreceptor specificity are subtype specific3,13,22,23,29,38,39,40,41,42,43,44,45,46,47. The lack of reliable genotypic algorithms that have been designed specifically for non-B HIV-1 subtypes is presently a major barrier to informing the appropriate use of maraviroc and future HIV-1 coreceptor blocking drugs in subjects infected with non-B HIV-1, which comprise approximately 90% of infections worldwide.
Here, we have conducted the most extensive and comprehensive analysis of phenotypically characterized HIV-1 subtype A, B, C, D, CRF01_AE and CRF02_AG V3 sequences to date, and developed subtype specific genotypic algorithms that are highly sensitive for predicting CXCR4-usage of HIV-1 in a clinical setting, without compromising specificity. Furthermore, we report the development and utility of a novel bioinformatic tool termed bulk2clonal, which computes and translates every possible amino acid sequence from nucleotide V3 sequences containing sites of base-call ambiguity. We showed that each of the PhenoSeq algorithms, when interfaced with bulk2clonal, are highly sensitive and specific for predicting CXCR4-usage of clinically relevant independent plasma-derived bulk V3 sequences that were generated by routine diagnostic laboratories. The performance of PhenoSeq-C against the MERIT clinical trial samples was particularly revealing. Of the 205 C-HIV infected individuals previously enrolled in MERIT, 18 belonged to a unique subset that was initially determined to harbor only R5 viruses by OTA, but then after failing maraviroc therapy were retrospectively shown to have harbored low frequency CXCR4-using strains by ESTA10,35,48,49. PhenoSeq-C detected minor CXCR4-using variants in 14 of these 18 subjects (accuracy 77.8%), thus correctly predicting their maraviroc treatment failure. These findings further demonstrate that our novel approach to genotypic tropism testing is highly sensitive and clinically valuable.
For determining coreceptor usage of HIV-1 subtype A and CRF02_AG, although PhenoSeq-A/AG exhibited a more favorable sensitivity and specificity profile than the clinically validated g2p at FPRs of 5.75% and 10%, WebPSSMX4R5 and WebPSSMSI/NSI, the recently developed HIVcoPRED (SAAC) and HIVcoPRED (SAAC + BLAST) algorithms exhibited the most favorable sensitivity (88% and 90%, respectively) and specificity (both 85.2%) profiles when tested against the PhenoSeq-A/AG training set sequences that were obtained from the Los Alamos HIV database (Supplementary Table 4). However, the performance of both of the HIVcoPRED algorithms was relatively poor compared to PhenoSeq-A/AG when tested against clinically relevant patient-derived bulk V3 sequences, even when they were coupled with bulk2clonal (Supplementary Table 4). These findings may be explained by the fact that HIVcoPRED (SAAC) and HIVcoPRED (SAAC + BLAST) training sets consisted of HIV-1 subtype A and CRF02_AG V3 sequences sampled from the Los Alamos HIV Database50, many of which were likely used here to test these algorithms.
Given that the PhenoSeq platform is highly sensitive and specific for predicting CXCR4-using HIV-1 strains, it is likely to be clinically useful for predicting treatment outcome for patients receiving maraviroc or indeed future CCR5 antagonists as they are developed. However, we acknowledge that the sensitivity and specificity of PhenoSeq for correctly determining HIV-1 coreceptor usage was measured against the results of phenotypic tropism assays rather than against maraviroc treatment outcome. In future studies we plan to determine the ability of the PhenoSeq platform to retrospectively predict virological outcome in patients who received maraviroc in the Pfizer phase III clinical trials MOTIVATE, MERIT and A4001029 using plasma-derived V3 sequences isolated by bulk and deep sequencing techniques7,35,49,51,52,53. For this study, we arbitrarily assigned a liberal cut-off of ≥10% for the analysis of amino acid sequences generated by bulk2clonal in order to maintain high sensitivity for predicting CXCR4-usage without compromising specificity. Through these planned studies we will more precisely determine the clinically relevant bulk2clonal cut-off required to accurately predict virological outcome in patients receiving maraviroc.
PhenoSeq is the first open access, online suite of genotypic algorithms to offer coreceptor usage analyses specifically designed for the major HIV-1 subtypes, which together account for approximately 95% of circulating viruses worldwide. Furthermore, the provision for constant revision and optimization of the PhenoSeq predictive criteria will ensure sustained high predictive accuracy. At an operational level, the online interface allows users to select different functions depending on how well the sequences have been characterized. For example, if the subtype is unknown we have provided an option to select an “unknown” PhenoSeq algorithm that first performs a BLAST align/search using the Los Alamos HIV BLAST tool (default settings) to determine the HIV-1 subtype, and then automatically selects and reports the subtype specific PhenoSeq algorithm used to determine coreceptor usage. To assess the performance of our PhenoSeq BLAST HIV-1 subtyping tool, we tested its accuracy for correctly predicting the HIV-1 subtype of three data sets comprising V3 sequences downloaded from the Los Alamos HIV Database, namely data sets 1, 2 and 3. Each data set consisted of 10 CXCR4-using and 10 R5 V3 sequences from HIV-1 subtypes B, C, D, CRF01_AE, and A or CRF02_AG, totaling 100 discrete V3 sequences per data set. The PhenoSeq “unknown” algorithm correctly predicted the HIV-1 subtype of 93%, 83% and 90% of data sets 1, 2 and 3, respectively, relative to the HIV-1 subtype reported on Los Alamos HIV Database. Currently, PhenoSeq can process up to 10,000 V3 sequences at a time and has been fully integrated with the bulk2clonal software.
AIDS-related deaths have fallen by 30% since 2005 largely due to increased accessibility of antiretroviral therapies to vulnerable HIV-1 affected populations1. As a new prognostic tool, PhenoSeq may improve access to maraviroc and future CCR5 blocking drugs, particularly for patients infected with non-B HIV-1 strains who comprise the vast majority of HIV-1 infected individuals worldwide. Furthermore, PhenoSeq may be a valuable tool for the monitoring of novel maraviroc therapies that have advanced to clinical trials, such as its use in intensification therapies to purge viral reservoirs54, and as a pre-exposure prophylaxis for the prevention of HIV-1 transmission55. In order to maximize the reach and potential public health benefit of PhenoSeq, we have made the platform freely available online at www.burnet.edu.au/phenoseq.
Methods
Assembly of phenotypically characterized HIV-1 V3 amino acid sequences
Previously published V3 sequences were obtained from the Los Alamos HIV Database (LANL) (http://www.hiv.lanl.gov/). HIV-1 subtypes were assigned as reported in LANL. Plasma derived bulk V3 sequences were obtained from participants of the Pfizer epidemiology study A400106414,34, the Pfizer maraviroc clinical trial MERIT35, and from Mulinge et al13. V3 sequences were defined as “CXCR4-using” if they were documented to solely use CXCR4 (X4) or to use CXCR4 together with CCR5 (R5X4) in phenotypic tropism assays, cause syncytia in MT2 cells or were isolated from plasma virus phenotyped by OTA or ESTA as X4 or dual-mixed. Alternatively, V3 sequences were defined as “R5” if they used CCR5 solely in phenotypic tropism assays, did not cause syncytia in MT2 cells or were isolated from plasma virus phenotyped by OTA or ESTA as R5. We selected one sequence per phenotype per patient using a random number generator to avoid biasing sequence analysis results by resampling related sequences.
In total, from LANL we collected 185 CXCR4-using and 538 R5 HIV-1 subtype B sequences, 80 CXCR4-using and 429 R5 HIV-1 subtype C sequences, 57 CXCR4-using and 80 R5 HIV-1 subtype D sequences, 18 CXCR4-using and 118 R5 HIV-1 subtype A sequences, 41 CXCR4-using and 54 R5 HIV-1 CRF02_AG sequences, and 50 CXCR4-using and 128 R5 HIV-1 CRF01_AE sequences. From the A4001064 study, we obtained bulk V3 nucleotide sequences (one per patient) from 78 subtype A infected individuals (9 CXCR4-using and 69 R5), 95 subtype C infected individuals (55 CXCR4-using and 40 R5) and 87 subtype D infected individuals (43 CXCR4-using and 44 R5), all of which were phenotyping by OTA (Monogram Biosciences). From the MERIT trial, we obtained 615 bulk V3 nucleotide sequences from pre-treatment plasma samples of 205 HIV-1 subtype C infected individuals (18 CXCR4-using and 187 R5), which were phenotyped by ESTA (Monogram Biosciences). From the Mulinge et al study13, we obtained bulk V3 nucleotide sequences (one per patient) from 53 subtype B infected individuals (12 CXCR4-using and 41 R5), 15 HIV-1 subtype A infected individuals (2 CXCR4-using and 13 R5), 30 CRF02_AG infected individuals (7 CXCR4-using and 23 R5) and 39 CRF01_AE infected individuals (14 CXCR4-using and 25 R5), all of which were phenotyped by an in-house recombinant virus phenotypic tropism assay.
Sequence analysis parameters
Parameters were used to limit V3 sequence analysis results to alterations most likely to maximize sensitivity for correctly predicting CXCR4-usage, without compromising specificity for correctly predicting R5-tropism. Specifically, cutoffs used to predict CXCR4-usage based on V3 length, charge and/or the number of potential N-linked glycosylation sites were assigned where the frequency in R5 sequences was <5% and the frequency in CXCR4-using sequences was ≥10%. Specific amino acid alterations were considered to have predictive value if the difference in frequency of the alteration between CXCR4-using and R5 sequences was statistically significantly (p < 0.05; two-tailed Fisher's exact t-test) and occurred in ≤10% of CXCR4-using or R5 sequences.
V3 sequence numbering
Throughout this study V3 sequence amino acids were numbered according to a modified version of the HXB2 V3 sequence numbering system56; Cys1, Thr2, Arg3, Pro4, Asn5, Asn6, Asn7, Thr8, Arg9, Lys10, Arg11, Ile12, Arg13, Ile13-14 insert, Gln13-14 insert, Arg14, Gly15, Pro16, Gly17, Arg18, Ala19, Phe20, Val21, Thr22, Ile23, Gly24, Lys25, -26, Ile27, Gly28, Asn29, Met30, Arg31, Gln32, Ala33, His34, Cys35.
Bulk2clonal
At sites of sequence ambiguity, i.e. ≥2 nucleotide variants, bulk2clonal generates all possible nucleotide combinations, based on the International Union of Pure and Applied Chemistry (IUPAC) nomenclature, and translates each possible nucleotide sequence to amino acids. IUPAC nomenclature states that within a nucleotide sequence; R represents the nucleotides A or G, Y represents C or T, S represents G or C, W represents A or T, K represents G or T, M represents A or C, B represents C or G or T, D represents A or G or T, H represents A or C or T, V represents A or C or G and N represents any base.
Alternative genotypic algorithms
Genotypic algorithms used for comparison were g2p at false positive rates of 1%, 2.5%, 5%, 5.75%, 10%, 15% and 20% (http://coreceptor.bioinf.mpi-inf.mpg.de/)15, WebPSSMX4R5, WebPSSMSINSI and subtype C specific WebPSSMSINSI-C, using default cutoff settings (http://indra.mullins.microbiol.washington.edu/webpssm/)31,32, HIVcoPRED split amino acid compositions (SAAC) and SAAC + Basic Local Alignment Search Tool (SAAC + BLAST) algorithms (http://www.imtech.res.in/raghava/hivcopred/submit.html) at default thresholds50, dsKernel (http://www.webcitation.org/query.php?url=http://genome.ulaval.ca/hiv-dskernel&refdoi=10.1186/1742-4690-5-110)57, the 11/25 rule, the 11 and/or 25 rule, the 11/24/25 rule30,58, the Raymond et al 11/25 + V3 charge rule22,43, the Lin et al rule59, the Raymond et al subtype D specific rules23, the Raymond et al CRF01_AE specific rules39 and the Esbjömsson et al subtype A and CRF02_AG specific rules60.
Supplementary Material
Supplementary material
Acknowledgments
We thank Rohan Gray for assistance with writing the code for the PhenoSeq algorithms and the bulk2clonal software, and Brendan Crabb for helpful comments. This study was supported by grants from (i) the Australian National Health and Medical Research Council (NHMRC) to P.R.G. and M.J.C. (1022066), (ii) the National Institutes of Health (NIH) to P.R.G. and M.J.C. (R21 MH100594), and (iii) the Delaney AIDS Research Enterprise (DARE) to M.J.C. and L.R.G. (U19 AI096109). K.Y.C. and K.L.H. are supported by Australian Postgraduate Awards from the University of Melbourne. P.R.G. is supported by an Australian Research Council (ARC) Future Fellowship (FT2). L.R.G. was supported by an Australian NHMRC Postdoctoral Training Fellowship. The authors gratefully acknowledge the contribution to this work of the Victorian Operational Infrastructure Support Program received by the Burnet Institute.
Footnotes
JFD and FD are employees of ViiV Healthcare. PRG is a former member of the ViiV Australia scientific advisory board and has received honoraria. KC and PRG have received honoraria from ViiV Healthcare Australia for conference travel. PRH is supported by CIHR/GSK Research Chair in Clinical Virology and has consulted and/or received grant funding from a variety pharmaceutical diagnostic companies and has received grants from, served as an ad hoc advisor to, or spoke at various events sponsored by: Pfizer, Glaxo-Smith Kline, Abbott, Merck, Selah, Tobira, Virco and Quest Diagnostics. None of the other authors have any competing interests to declare.
Author Contributions K.C. performed the sequence and statistical analyses and designed the algorithms. L.R.G. assisted with the design of the algorithms and of the bulk2clonal software. K.L.H., J.S., M.R., M.J.C. and P.R.G. contributed intellectually and helped analyze the data. D.P.B., G.Q.L. and P.R.H. provided published and unpublished V3 sequences and helped analyze the data. K.C., P.R.H., M.J.C. and P.R.G. interpreted the results. J.F.D. and F.D. provided clinical trial data. K.C., K.L.H. and P.R.G. wrote the manuscript. All authors have read and helped edit the manuscript.
References
- Joint United Nations Programme on HIV/AIDS report on the global AIDS epidemic. UNAIDS 1, 1–198 (2013). [Google Scholar]
- Dorr P. et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother 49, 4721–4732 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashin K. et al. Linkages between HIV-1 specificity for CCR5 or CXCR4 and in vitro usage of alternative coreceptors during progressive HIV-1 subtype C infection. Retrovirology 10, 98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor R. I., Sheridan K. E., Ceradini D., Choe S. & Landau N. R. Change in coreceptor use correlates with disease progression in HIV-1--infected individuals. J Exp Med 185, 621–628 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell B. J. et al. Emergence of X4 usage among HIV-1 subtype C: evidence for an evolving epidemic in South Africa. AIDS 22, 896–899 (2008). [DOI] [PubMed] [Google Scholar]
- Michler K. et al. Genotypic characterization and comparison of full-length envelope glycoproteins from South African HIV type 1 subtype C primary isolates that utilize CCR5 and/or CXCR4. AIDS Res Hum Retroviruses 24, 743–751 (2008). [DOI] [PubMed] [Google Scholar]
- Gulick R. M. et al. Maraviroc for previously treated patients with R5 HIV-1 infection. N Engl J Med 359, 1429–1441 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekerckhove L., Verhofstede C. & Vogelaers D. Maraviroc: integration of a new antiretroviral drug class into clinical practice. J Antimicrob Chemother 61, 1187–1190 (2008). [DOI] [PubMed] [Google Scholar]
- Whitcomb J. M. et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother 51, 566–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkin T. J. et al. Reanalysis of coreceptor tropism in HIV-1-infected adults using a phenotypic assay with enhanced sensitivity. Clin Infect Dis 52, 925–928 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond S. et al. Development and performance of a new recombinant virus phenotypic entry assay to determine HIV-1 coreceptor usage. J Clin Virol 47, 126–130 (2010). [DOI] [PubMed] [Google Scholar]
- Braun P. & Wiesmann F. Phenotypic assays for the determination of coreceptor tropism in HIV-1 infected individuals. Eur J Med Res 12, 463–472 (2007). [PubMed] [Google Scholar]
- Mulinge M. et al. HIV-1 tropism determination using a phenotypic Env recombinant viral assay highlights overestimation of CXCR4-usage by genotypic prediction algorithms for CRRF01_AE and CRF02_AG. PloS one 8, e60566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G. Q. et al. Comparison of population and 454 “deep” sequence analysis for HIV type 1 tropism versus the original trofile assay in non-B subtypes. AIDS Res Hum Retroviruses 29, 979–984 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer T., Sander O., Sierra S., Thielen A. & Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol 25, 1407–1410 (2007). [DOI] [PubMed] [Google Scholar]
- Swenson L. C. et al. Use of cellular HIV DNA to predict virologic response to maraviroc: performance of population-based and deep sequencing. Clin Infect Dis 56, 1659–1666 (2013). [DOI] [PubMed] [Google Scholar]
- Swenson L. C. et al. Deep sequencing to infer HIV-1 co-receptor usage: application to three clinical trials of maraviroc in treatment-experienced patients. J Infect Dis 203, 237–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson L. C. et al. Deep V3 sequencing for HIV type 1 tropism in treatment-naive patients: a reanalysis of the MERIT trial of maraviroc. Clin Infect Dis 53, 732–742 (2011). [DOI] [PubMed] [Google Scholar]
- Diez-Fuertes F. et al. Improvement of HIV-1 coreceptor tropism prediction by employing selected nucleotide positions of the env gene in a Bayesian network classifier. J Antimicrob Chemother 68, 1471–1485 (2013). [DOI] [PubMed] [Google Scholar]
- Pillai S., Good B., Richman D. & Corbeil J. A new perspective on V3 phenotype prediction. AIDS Res Hum Retroviruses 19, 145–149 (2003). [DOI] [PubMed] [Google Scholar]
- Cashin K. et al. CoRSeqV3-C: a novel HIV-1 subtype C specific V3 sequence based coreceptor usage prediction algorithm. Retrovirology 10, 24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond S. et al. Genotypic prediction of human immunodeficiency virus type 1 CRF02-AG tropism. J Clin Microbiol 47, 2292–2294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond S. et al. Genotypic prediction of HIV-1 subtype D tropism. Retrovirology 8, 56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen M. R., Ellett A., Churchill M. J. & Gorry P. R. Viral tropism, fitness and pathogenicity of HIV-1 subtype C. Future Virology 5, 219–231 (2010). [Google Scholar]
- Hemelaar J., Gouws E., Ghys P. D. & Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 20, W13–23 (2006). [DOI] [PubMed] [Google Scholar]
- Hemelaar J., Gouws E., Ghys P. D. & Osmanov S. Isolation W-UNfH, Characterisation. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 25, 679–689 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K. Y. et al. High prevalence of CXCR4 usage among treatment-naive CRF01_AE and CRF51_01B-infected HIV-1 subjects in Singapore. BMC Infect Dis 13, 90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S. et al. Differential prevalence of HIV type 1 subtype B and CRF01_AE among different sexual transmission groups in Tokyo, Japan, as revealed by subtype-specific PCR. AIDS Res Hum Retroviruses 19, 1057–1063 (2003). [DOI] [PubMed] [Google Scholar]
- De Jong J. J., De Ronde A., Keulen W., Tersmette M. & Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol 66, 6777–6780 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda T., Levy J. A. & Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 89, 9434–9438 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. A., Coetzer M., van 't Wout A. B., Morris L. & Mullins J. I. A reliable phenotype predictor for human immunodeficiency virus type 1 subtype C based on envelope V3 sequences. J Virol 80, 4698–4704 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. A. et al. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J Virol 77, 13376–13388 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley J. A. & McNeil B. J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143, 29–36 (1982). [DOI] [PubMed] [Google Scholar]
- Ataher Q. et al. The epidemiology and clinical correlates of HIV-1 co-receptor tropism in non-subtype B infections from India, Uganda and South Africa. J Int AIDS Soc 15, 2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. A. et al. Maraviroc versus efavirenz, both in combination with zidovudine-lamivudine, for the treatment of antiretroviral-naive subjects with CCR5-tropic HIV-1 infection. J Infect Dis 201, 803–813 (2010). [DOI] [PubMed] [Google Scholar]
- Reeves J. D., Coakley D. E., Petropoulos C. J. & Whitcomb J. M. An Enhanced-Sensitivity Trofile™ HIV Coreceptor Tropism Assay for Selecting Patients for Therapy with Entry Inhibitors Targeting CCR5: A Review of Analytical and Clinical Studies. The Journal of Viral Entry 3 (2009). [Google Scholar]
- Trinh L. et al. Validation of an enhanced sensitivity Trofile™ HIV-1 co-receptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5. Journal of the International AIDS Society 11, 197 (2008). [Google Scholar]
- Jakobsen M. R. et al. Longitudinal Analysis of CCR5 and CXCR4 Usage in a Cohort of Antiretroviral Therapy-Naive Subjects with Progressive HIV-1 Subtype C Infection. PLoS One 8, e65950 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond S. et al. Genotypic prediction of HIV-1 CRF01-AE tropism. J Clin Microbiol 51, 564–570 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman N. G., Seillier-Moiseiwitsch F., Ahn J., Walker J. M. & Swanstrom R. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J Virol 76, 3852–3864 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S. S., Boyle T. J., Lyerly H. K. & Cullen B. R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 253, 71–74 (1991). [DOI] [PubMed] [Google Scholar]
- Schuitemaker H., van 't Wout A. B. & Lusso P. Clinical significance of HIV-1 coreceptor usage. J Transl Med 9 Suppl 1, S5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond S. et al. Prediction of HIV type 1 subtype C tropism by genotypic algorithms built from subtype B viruses. J Acquir Immune Defic Syndr 53, 167–175 (2010). [DOI] [PubMed] [Google Scholar]
- Dimonte S. et al. Selected amino acid changes in HIV-1 subtype-C gp41 are associated with specific gp120(V3) signatures in the regulation of co-receptor usage. Virus Res 168, 73–83 (2012). [DOI] [PubMed] [Google Scholar]
- Dina J. et al. Algorithm-based prediction of HIV-1 subtype D coreceptor use. J Clin Microbiol 51, 3087–3089 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido C. et al. Evaluation of eight different bioinformatics tools to predict viral tropism in different human immunodeficiency virus type 1 subtypes. J Clin Microbiol 46, 887–891 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recordon-Pinson P. et al. Evaluation of the genotypic prediction of HIV-1 coreceptor use versus a phenotypic assay and correlation with the virological response to maraviroc: the ANRS GenoTropism study. Antimicrob Agents Chemother 54, 3335–3340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern R. A. et al. Population-based sequencing of the V3-loop can predict the virological response to maraviroc in treatment-naive patients of the MERIT trial. J Acquir Immune Defic Syndr 61, 279–286 (2012). [DOI] [PubMed] [Google Scholar]
- Saag M. et al. A double-blind, placebo-controlled trial of maraviroc in treatment-experienced patients infected with non-R5 HIV-1. J Infect Dis 199, 1638–1647 (2009). [DOI] [PubMed] [Google Scholar]
- Kumar R. & Raghava G. P. Hybrid approach for predicting coreceptor used by HIV-1 from its V3 loop amino acid sequence. PLoS One 8, e61437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern R. A. et al. Population-based V3 genotypic tropism assay: a retrospective analysis using screening samples from the A4001029 and MOTIVATE studies. AIDS 24, 2517–2525 (2010). [DOI] [PubMed] [Google Scholar]
- Fatkenheuer G. et al. Subgroup analyses of maraviroc in previously treated R5 HIV-1 infection. N Engl J Med 359, 1442–1455 (2008). [DOI] [PubMed] [Google Scholar]
- Kagan R. M. et al. A genotypic test for HIV-1 tropism combining Sanger sequencing with ultradeep sequencing predicts virologic response in treatment-experienced patients. PLoS One 7, e46334 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. et al. Intensification of antiretroviral therapy with a CCR5 antagonist in patients with chronic HIV-1 infection: effect on T cells latently infected. PLoS One 6, e27864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff C. P., Ndolo T., Tandon A., Habu Y. & Akkina R. Oral pre-exposure prophylaxis by anti-retrovirals raltegravir and maraviroc protects against HIV-1 vaginal transmission in a humanized mouse model. PLoS One 5, e15257 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner L. et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313, 277–284 (1985). [DOI] [PubMed] [Google Scholar]
- Boisvert S., Marchand M., Laviolette F. & Corbeil J. HIV-1 coreceptor usage prediction without multiple alignments: an application of string kernels. Retrovirology 5, 110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R. A. et al. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol 66, 3183–3187 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N. H. et al. Env sequence determinants in CXCR4-using human immunodeficiency virus type-1 subtype C. Virology 433, 296–307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esbjornsson J. et al. Frequent CXCR4 tropism of HIV-1 subtype A and CRF02_AG during late-stage disease--indication of an evolving epidemic in West Africa. Retrovirology 7, 23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashin K. et al. Differences in coreceptor specificity contribute to alternative tropism of HIV-1 subtype C for CD4 + T-cell subsets, including stem cell memory T-cells. Retrovirology 11, 97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashin K. et al. Alternative coreceptor requirements for efficient CCR5- and CXCR4-mediated HIV-1 entry into macrophages. J Virol 85, 10699–10709 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashin K. et al. Covariance of charged amino acids at positions 322 and 440 of HIV-1 Env contributes to coreceptor specificity of subtype B viruses, and can be used to improve the performance of V3 sequence-based coreceptor usage prediction algorithms. PLoS One 9, e109771 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorry P. R. et al. Changes in the V3 region of gp120 contribute to unusually broad coreceptor usage of an HIV-1 isolate from a CCR5 Delta32 heterozygote. Virology 362, 163–178 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. et al. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol 81, 7885–7893 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakso M. M. et al. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS pathogens 3, e117 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milich L., Margolin B. H. & Swanstrom R. Patterns of amino acid variability in NSI-like and SI-like V3 sequences and a linked change in the CD4-binding domain of the HIV-1 Env protein. Virology 239, 108–118 (1997). [DOI] [PubMed] [Google Scholar]
- Nabatov A. A. et al. Intrapatient alterations in the human immunodeficiency virus type 1 gp120 V1V2 and V3 regions differentially modulate coreceptor usage, virus inhibition by CC/CXC chemokines, soluble CD4, and the b12 and 2G12 monoclonal antibodies. J Virol 78, 524–530 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neogi U. et al. Genetic analysis of HIV-1 Circulating Recombinant Form 02_AG, B and C subtype-specific envelope sequences from Northern India and their predicted co-receptor usage. AIDS research and therapy 6, 28 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrgren H., Bamba S., Da Silva Z. J., Koivula T. & Andersson S. Higher mortality in HIV-2/HTLV-1 co-infected patients with pulmonary tuberculosis in Guinea-Bissau, West Africa, compared to HIV-2-positive HTLV-1-negative patients. International journal of infectious diseases: IJID: official publication of the International Society for Infectious Diseases 14 Suppl 3, e142–147 (2010). [DOI] [PubMed] [Google Scholar]
- Resch W., Hoffman N. & Swanstrom R. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology 288, 51–62 (2001). [DOI] [PubMed] [Google Scholar]
- Rosen O., Samson A. O. & Anglister J. Correlated mutations at gp120 positions 322 and 440: implications for gp120 structure. Proteins 71, 1066–1070 (2008). [DOI] [PubMed] [Google Scholar]
- Rosen O., Sharon M., Quadt-Akabayov S. R. & Anglister J. Molecular switch for alternative conformations of the HIV-1 V3 region: implications for phenotype conversion. Proc Natl Acad Sci U S A 103, 13950–13955 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suphaphiphat P., Essex M. & Lee T. H. Mutations in the V3 stem versus the V3 crown and C4 region have different effects on the binding and fusion steps of human immunodeficiency virus type 1 gp120 interaction with the CCR5 coreceptor. Virology 360, 182–190 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. Molecular determinants of HIV-1 subtype C coreceptor transition from R5 to R5X4. Virology 407, 68–79 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajoge H. O. et al. Genetic characteristics, coreceptor usage potential and evolution of Nigerian HIV-1 subtype G and CRF02_AG isolates. PLoS One 6, e17865 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material