Abstract
The dynamic reshaping of tissues during morphogenesis results from a combination of individual cell behaviours and collective cell rearrangements. However, a comprehensive framework to unambiguously measure and link cell behaviour to tissue morphogenesis is lacking. Here we introduce such a kinematic framework, bridging cell and tissue behaviours at an intermediate, mesoscopic, level of cell clusters or domains. By measuring domain deformation in terms of the relative motion of cell positions and the evolution of their shapes, we characterize the basic invariant quantities that measure fundamental classes of cell behaviour, namely tensorial rates of cell shape change and cell intercalation. In doing so we introduce an explicit definition of cell intercalation as a continuous process. We demonstrate how spatiotemporal mapping of strain rates in three models of tissue morphogenesis leads to new insight into morphogenetic mechanisms. Our quantitative approach has broad relevance for the precise characterisation and comparison of morphogenetic phenotypes.
Introduction
One of the most important challenges in developmental biology is to understand how molecular information leads to the individual and collective movement of cells that shape and form tissues1–4 via both cell-intrinsic stresses and cell responses to applied stress5–7. With the tremendous advances in molecular biology, genetics, imaging techniques and the automated tracking of many cells in parallel8,9, it is now possible to trace the evolution of morphogenetic phenotypes during development as a function of molecular perturbations as well as physical manipulation. This sets the stage for the identification and quantification of the geometry of shape change in terms of the resultant strain (deformation) rates, i.e. the amount of strain per unit of time (a glossary of the most commonly used terms can be found in Supplementary Note 1). The response of tissues to the sum of the external and cell-autonomous forces, constraints and stiffnesses is characterized by tissue strains and strain rates.
Current approaches to the statistical characterization of morphogenetic parameters rely on analogies of tissues to foams and granular materials, where topological methods that rely on connectivity are used10. Here, we deploy ideas from tensor analysis and continuum field theories11 and extend these methods for tissues composed of discrete cells to measure strain rates and rotations within biological tissues. This allows us to account for the continuous sliding process between cells regardless of their connectivity. In particular, we quantify the rates of individual cell shape change and collective cell intercalation (cell rearrangement), the two fundamental classes of mesoscopic cellular behaviour, combinations of which account for a wide range of tissue shaping behaviours. Cell shape change and cell intercalation can be active cell-autonomous behaviours, and also possibly passive deformations (elastic or plastic) in response to external forces or constraints. Patterns of strain rates allow modelling of the forces acting on and within tissues and reveal signatures of active and passive behaviour.
We deploy these measures on a variety of simulated and real deforming tissues, building dynamic maps of morphogenetic behavior. We show that we can determine the relative contribution of these two behaviours in shaping tissues. First, the Drosophila amnioserosa deforms predominantly by cell shape change during dorsal closure, and we show that there is little cell intercalation. Second, during mid-phase Drosophila germband extension we can resolve and distinguish different cellular modes underlying tissue extension. Lateral ectoderm extends by cell intercalation whereas ventral midline cells extend by stretching, without intercalation. Third, in the zebrafish trunk neuroectoderm, intercalation and cell shape change combine in an interesting way, the latter augmenting tissue convergence and reducing tissue extension. By following cell behaviour and coupling these to global movements of cellular domains we lay out a high-resolution quantitative framework for the geometric basis of morphogenesis.
Results
Computer-assisted cell tracking enables us to follow the behaviours of large numbers of epithelial cells within 2D surfaces that follow the shape of tissues within the embryo (Online Methods and Supplementary Videos 1-3). Morphogenetic deformations result from variation in the speed or direction of cell trajectories that change the relative positions of cells8. To quantify deformations at a multicellular scale, we follow, over a period +/-dt minutes, small groups of cells or domains. Domains are defined by a central cell and nc coronae of neighbouring cells at the central time point.
Tissue Strain Rates
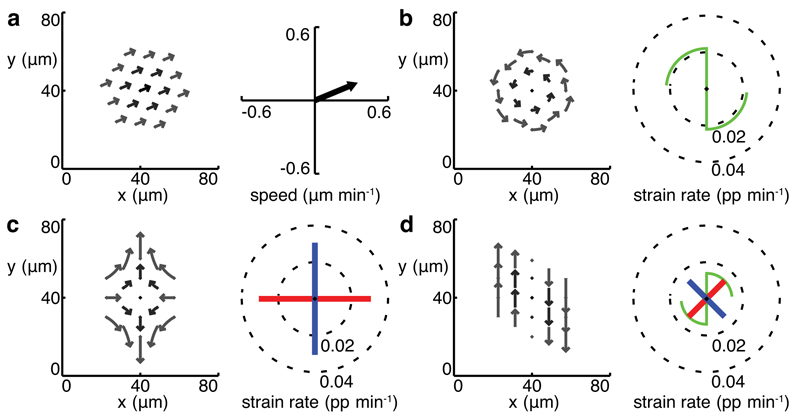
We treat each domain as a continuous field of tissue sampled at cell centroid locations, and calculate various domain metrics based on the movement of these centroids (see Online Methods). First, we calculate the average domain translation velocity, how fast the domain moves in space (Fig. 1a). Second, we calculate gradients of centroid velocities across the domain in two orthogonal orientations, and use these gradients to construct a tissue velocity gradient tensor Lt which characterizes the spatial variation of the local velocity field. Lt is then separated into a spin matrix Ωt, which yields the domain rotation (angular velocity) in radians per minute (Fig. 1b), and a deformation or strain rate tensor Ėt. An invariant description of domain strain rates (i.e. one that is independent of the chosen coordinate system) is given by the eigenvalues and associated eigenvectors of Ėt. These are the amplitudes (in proportion per minute) and orientations of the local principal strain rates associated with the velocity field (Fig. 1c). The eigenvalues of Ėt generalize the local convergence (negative) and extension (positive) rate constants described in Glickman et al.12.
Figure 1. Measuring tissue strain rates in simulated data.
(a)-(d) left panels trajectories, right panels domain translation (a) and tissue strain rates (b)-(d) for simulated domains (nc = 2, dt = 9 min). (a) the domain behaves as a rigid block translating with velocities uniform within the domain. (b) pure rotation. (c) balanced convergence and extension, or pure shear. (d) an equal combination of pure shear and rotation, or simple shear. Principal strain rates are represented by orthogonal line segments with length equal to strain rate amplitude (blue positive, red negative). Rotation is represented by a green scythe motif, with radius indicating radians per minute on the same scale as the strain rates. Blades point in the direction of rotation (anticlockwise in (b), clockwise in (d)).
Domain translation and rotation measure how the domain moves as a whole in space, while the strain rate tensor measures how the domain itself deforms, i.e. how distances and angles between cells vary locally. The rotation component nevertheless remains important for interpreting simple classes of local deformations. For instance, simple shear arises from a combination of rotation and a pure shear deformation (see Supplementary Note 1 and Fig. 1d). However, a mechanistic interpretation remains ambiguous at the level of the single domain since it is not possible to distinguish between local rotation that is 1) due to an advective motion resulting from activity extraneous to the domain or because of embryo movement, and 2) part of a simple shear deformation.
Cell Shape and Cell Intercalation Strain Rates
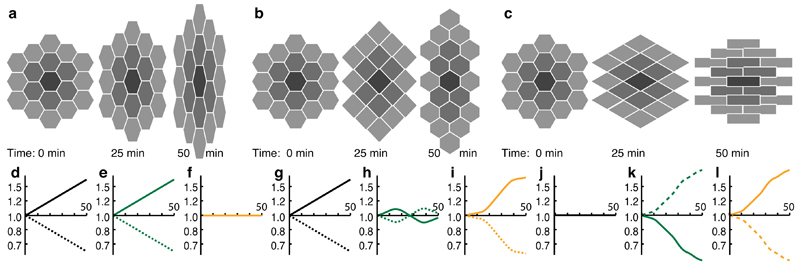
To interpret tissue deformation in terms of the intrinsic cell behaviour within each domain, we identify two complementary components: cell shape change and cell intercalation. We illustrate their distinction with two simulated scenarios (Supplementary Videos 4,5) wherein tissue convergence and extension are equal but are accounted for by cell shape change or cell intercalation respectively (Fig. 2a,b). In a further example (Fig. 2c) mutually cancelling cell shape change and intercalation cancel resulting in no tissue deformation. In vivo, we expect morphogenesis to involve combinations of cell shape change and cell intercalation that differ with tissue, time and location. We estimate a cell shape strain rate tensor Lc from traced cell outlines based upon their best-fit ellipses (see Online Methods). We assume that average cell spin Ωc equals Ωt, that is cells rotate only with the domain. We therefore eliminate the effects of the rotation Ωt before calculating a symmetrical cell shape strain rate tensor Ėc, which represents the average rate of change in the shape of cells within a domain. The average rate of change in cell area is given by the Trace of Ėc, as a result of movement of cell volume into or out of the plane13.
Figure 2. Cellular simulations of tissue morphogenesis.
Three cellular scenarios are simulated in (a)-(c) demonstrating tissue outcomes for different combinations of cell shape change and cell intercalation. Focal cell is black, with first and second coronae of neighbours in dark and light gray, respectively. (d)-(f), (g)-(i) and (j)-(l) show cumulative stretch ratios on a log scale versus time in minutes for examples (a), (b) and (c) respectively. Cumulative stretch ratios in vertical (solid) and horizontal (dotted) orientations are plotted for tissue (d), (g), (j) in black, cell shape (e), (h), (k) in dark green and cell intercalation (f), (i), (l) in orange.
To characterize cell intercalation in terms of these tensorial quantities, we propose that the principal characteristic of cell intercalation is the continuous sliding of the bulk of neighbouring cells past each other. We show that the local rate and principal directions of sliding derive directly from what we call the cell intercalation strain rate tensor Li defined by (see Online Methods and Supplementary Fig. 1):
| (1) |
As one would expect, Li is intimately related with the mismatch between the cell shape and tissue strain rates. This explicitly defines cell intercalation as a measure that bridges spatial scales: it represents the residual difference in bulk movement between Lt, the redistribution of the bulk of the domain relative to its centre, and Lc, the average redistribution of each cell’s bulk relative to its own centroid. Since both Lc and Lt capture the change in area of the cells in the domain (volume change in 3D), Li has zero dilatation and is a pure shear deformation. This definition is valid in both two and three dimensions. Thus intercalation is a process in which convergence rate in one orientation is balanced by an equal rate of extension in the orthogonal orientation: it preserves area (in two dimensions) and volume (in three dimensions).
To get an intuitive understanding of this simple measure, we apply our methods to simulated data. In our first example, net cell shape strain is identical to tissue strain and there is no cell intercalation (Fig. 2d-f). In the second example, cell intercalation accounts for virtually all of the tissue strain (Fig. 2g-i) and is continuous, unlike the synchronous and discrete neighbour-exchange events (Supplementary Video 5, frame 25). Net cell shape strain is minimal, fluctuating due to transient packing changes required to exchange neighbours by the T1 process14, as in biological tissues undergoing cell intercalation2,12. In our third example, there is zero tissue strain and dramatic cell shape strain is balanced by intercalation strain in opposite orientations (Fig. 2j-l). These examples highlight the differences between our continuous measures based on cell shape measurements and topological measures which are based upon discrete changes in cell-cell connectivity8,15,16. We consider the more fundamental and continuous sliding process that our methods capture, regardless of cell topology, to be better suited to precise spatiotemporal mapping. In each of these examples, we have the ability to track four related measures; cell shape, cell intercalation and tissue strain rates, and the tissue rotation rate.
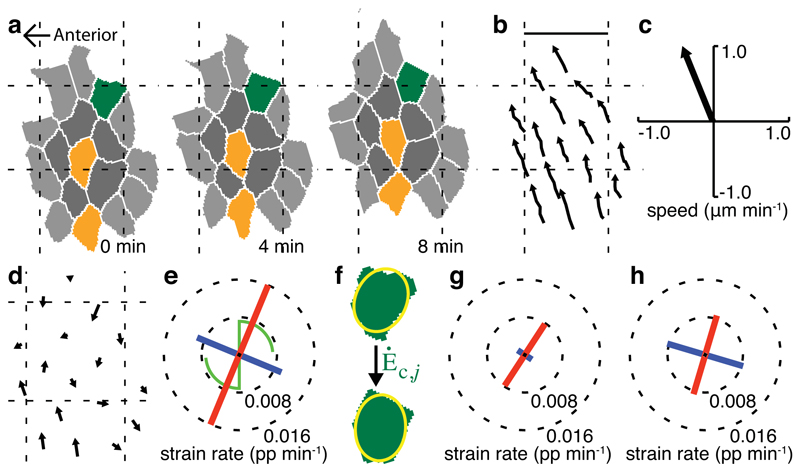
We deploy these measures to quantify convergence and extension in a single example domain of zebrafish trunk neuroectoderm (Fig. 3). Tissue strain rate (Fig. 3e) shows the domain to exhibit unequal convergence and extension, with extension oriented close to the anterior-posterior (AP) axis. A clockwise rotation, of similar magnitude to the strain rates, shows that a simple shear has taken place. Both cell shape change (Fig. 3g) and cell intercalation (Fig. 3h) contribute to the net domain deformation, with the convergence orientation of intercalation aligned with the orientation of cell shape contraction. We also calculate confidence intervals associated with each strain rate and use these to establish an appropriate domain size for a tissue (see Online Methods and Supplementary Fig. 2). An outline of the sequence of algorithms used to calculate strain rates from 4D movies of tissues with membranes marked with ubiquitous fluorescence is provided (Supplementary Fig. 3), and full code is provided for calculating strain rates and associated information for a domain of cells (Supplementary Software).
Figure 3. Measuring strain rates for a domain of zebrafish ectoderm.
(a) cell shapes and (b) cell centroid trajectories for a domain (nc = 2, dt = 4 min) are used to calculate strain rates. Cell colors in (a) show first (dark grey) and second (light grey) coronae of neighbours around the focal central cell, with examples of cell shape change (dark green) and intercalation (orange). Scale bar in (b) 25 µm. (c) average domain translation. (d) velocity field. (e) tissue strain and rotation rates. Strain rate line-segments and rotation rates are drawn as in Figure 1. (f) cell shapes are approximated to their best-fit ellipses and strain rates that must be applied to account for a cell’s shape evolution from t-dt to t+dt are calculated for all cells of the domain. (g) area-weighted average cell shape strain rates. (h) cell intercalation strain rates.
Biological applications
We have quantitatively mapped the morphogenesis of three diverse embryonic epithelial tissues in which we expect different combinations of cell shape change and intercalation. To create maps of developing tissues, we calculated strain rates for domains surrounding each cell and for each time point. Summary measures of strain rates are averaged over space and integrated over time from average or cumulative tensors, respectively. In all three tissues, the dominant orientations of deformation are strongly aligned with the body axes so we present cumulative stretch ratios (the exponent of the cumulative strains) projected onto these axes.
Drosophila amnioserosa
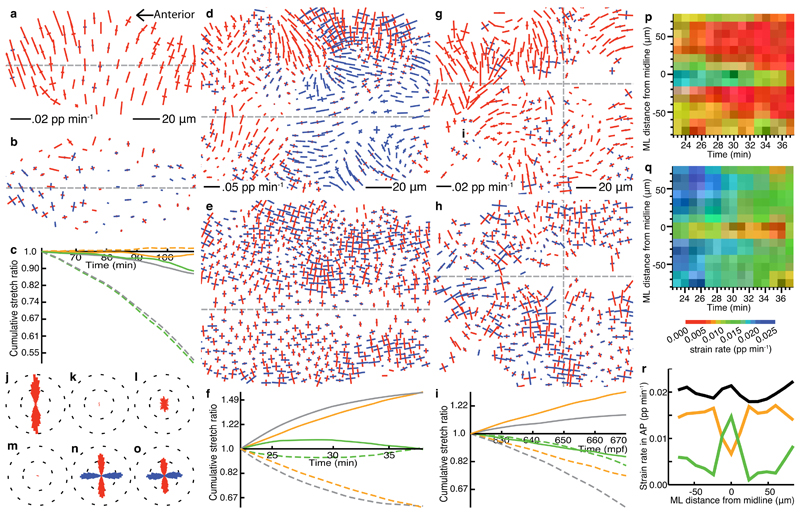
The Drosophila amnioserosa is an ectodermal tissue eclipsed by the dorsal closure of lateral epidermis17 (Fig. 4a, Supplementary Video 1). The forces acting at the tissue margins are well understood5 and the amnioserosa tissue is thought to contract actively in the medio-lateral (ML) orientation18,19. However, little is known about the behaviour of individual cells during this process. Tissue deformation shows strong ML-oriented convergence across the tissue but also some weaker AP-oriented convergence (Fig. 4b,c,j, Supplementary Video 6). Highly asymmetric tissue deformation results almost exclusively from equally asymmetric cell shape changes, by narrowing cells predominantly in the ML orientation at a rate that accelerates with time (Fig. 5a,c,j, Supplementary Video 7). Cells contract to nearly 50% of their original ML size (average -0.013 pp/min), and to 88% of their AP size in the mid-zippering phase of dorsal closure analyzed. There is only weak and late cell intercalation that sums to a minor net contribution (Fig. 5b,c,m, Supplementary Video 8).
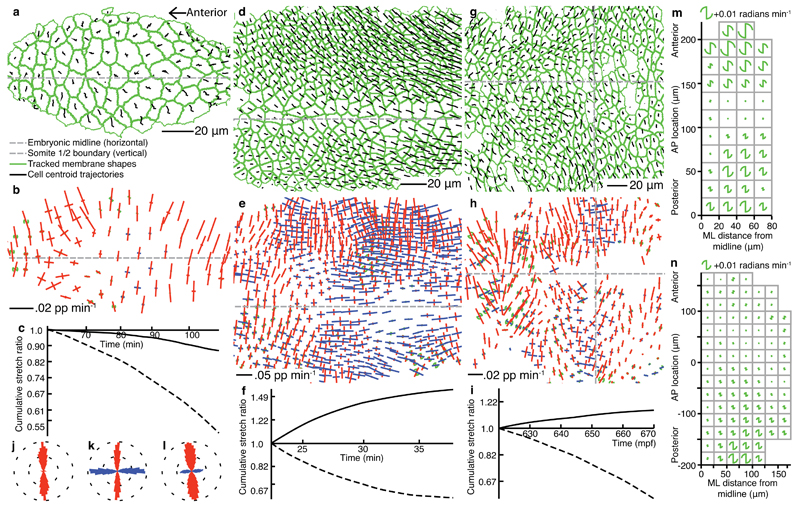
Figure 4. Tissue strain rate patterns in Drosophila and zebrafish ectoderm.
(a), (d), (g) tracked cell trajectories and cell shapes within curved layers taken through the apices of three different epithelial tissues imaged by 3D time-lapse confocal microscopy. (a)-(c),(j) Drosophila amnioserosa mid-way through dorsal closure (time is from the start of dorsal closure in (c)). (d)-(f),(k) Drosophila germband mid-way through germband extension (time is from the start of germband extension in (f)), with the left margin of the field of view approximately four cell diameters posterior to the cephalic furrow. (g)-(i),(l) zebrafish trunk neuroectoderm just prior to the onset of neurulation, starting at 10.2 hpf. Gaps indicate tracking ambiguities. (b),(e),(h), tissue strain rates. Colors of strain rates and rotations are as in Figure 1. Strain rate scale bars are shown. (c),(f),(i) cumulative tissue stretch ratios, integrated across AP (solid) and ML (dotted) orientations. (j)-(l) radial histograms (east-west is AP) of whole movie pooled strain rate orientations, weighted by their absolute magnitude. (m),(n) pooled rotation data from multiple aligned embryos (see Online Methods), summarized for squares of tissue for Drosophila germband (average over 5-20 min after the start of germband extension) and zebrafish trunk ectoderm (average over 560-610 mpf) respectively. Data on the left of each embryo has been mirrored onto the right-hand side. AP location of ~220 µm is the cephalic furrow in (m) and 0 µm is the somite 1/2 boundary in (n).
Figure 5. Cell shape and cell intercalation strain rate patterns in Drosophila and zebrafish ectoderm.
(a)-(o) are from the same movies represented in Fig. 4a-l, but here the first two panels of each column represent cell shape and cell intercalation strain rates respectively. (a)-(c),(j),(m) Drosophila amnioserosa, (d)-(f),(k),(n) Drosophila germband, (g)-(i),(l),(o) zerbrafish trunk neuroectoderm. (c), (f), (i), cumulative stretch ratios of cell shape (green), cell intercalation (orange) and total tissue (gray) plotted as in Figure 2. Whole movie pooled strain rate orientation histograms of cell shape (j)-(l) and intercalation (m)-(o). (p)-(r) average strain rates projected onto the AP (extension) axis for the epoch of germband extension shown in (l). (p) cell shape, (q) intercalation strain rates. (r) colors are as for cumulative stretch ratios.
Drosophila germband
The Drosophila post-gastrulation germband extends to the posterior as cells converge towards the ventral midline20(Fig. 4d, Supplementary Video 2). Cell intercalation is known to contribute to this process2,21 but how much this alone can account for tissue deformation is unknown. In the mid-phase, strong tissue convergence in ML is matched by extension in AP (Fig. 4e,f,k, Supplementary Video 9). Ectodermal tissue deformation is predominantly due to strong cell intercalation co-oriented with the body axes (Fig. 5e,f,n, Supplementary Video 10). In contrast to the amnioserosa, strong cell shape changes are heterogeneous across the tissue and cancel to give minimal net contribution over time (Fig. 5d,f,k, Supplementary Video 11). Further analysis of cell shape changes are presented elsewhere22. Average tissue rotations are shown for data pooled from 5 embryos (see Online Methods) during the fast phase, 5-20 min after the start of germband extension. The rotation rate varies from clockwise anteriorly (for the right hand ventral ectoderm) to anticlockwise in the posterior, passing through a region with little rotation in between (Fig. 4m).
Zebrafish trunk neuroectoderm
Zebrafish trunk neural ectoderm converges in ML while extending along AP, prior to and during neurulation3,23 (Fig. 4g, Supplementary Video 3). The temporal and spatial contributions of cell intercalation and cell shape change in the neural plate are poorly understood3,24. The neural plate has strong ML convergence in tissue strain rate but heterogeneous AP extension (Fig. 4h,i,l, Supplementary Video 12). The rate of tissue deformation increases over time, but AP-oriented extension never matches ML-oriented convergence. Cell shape strain rate within the neural plate is mostly negative (Fig. 5g,i,l, Supplementary Video 13), both in ML and AP orientations, as cells get smaller in the plane and lengthen in depth. Strikingly, cell intercalation across the tissue is strongly aligned with the body axes (Fig. 5h,i,o, Supplementary Video 14). The average magnitude of cell shape change is about half that of cell intercalation. The imbalance between tissue convergence and tissue extension can now be correctly interpreted: The combination of cell shape change and cell intercalation are additive in the ML orientation but cancel in the AP orientation. Similar to the germband, there is an AP gradient of the rotation rate (Fig. 4n).
The high spatiotemporal resolution of our approach allows us to detect local variation in cell behaviour. The patterns of cell shape change and the strength of intercalation, in particular in the germband and zebrafish trunk, are remarkably complex in space (Fig. 5d,e,g,h) and in time (Supplementary Videos 10,11,13,14), presumably reflecting variation in combinations of gene expression, cell signaling, cell-intrinsic fluctuations, local stresses21, and physical constraints. For cell shape change in the amnioserosa, and intercalation in the germband and zebrafish trunk, the orientations of domain deformation are found to be aligned predominantly with the body axes (Fig. 5j-o). The orientation of intercalation is correlated with cell polarization in the germband and the zebrafish trunk1,20 with characteristic average rates of ~0.03 pp min-1 and ~0.006 pp min-1, respectively. We identified a consistent spatial heterogeneity during germband extension, along a narrow band of cells at the ventral midline, so we further explored the ventral midline of Drosophila and the dorsal midline of zebrafish using data pooled from several embryos (see Online Methods). In the germband, while AP-oriented tissue extension is constant across ML, cell shape change is weak laterally but strong along the ventral midline (Fig. 5d,p). Conversely, cell intercalation is strong laterally but weak medially (Fig. 5e,q). A 15 µm region either side of the ventral midline comprises two lines of ventral midline neural precursor cells (VML). Thus, during germband extension the VML cells extend in AP not by intercalation but by shape stretch, while the lateral cells intercalate. The same analysis of the dorsal midline of zebrafish embryos shows that there is no such inversion of cellular mechanisms at the midline of the fish trunk neuroectoderm at ~10-11hpf (Supplementary Fig. 4). Indeed, at this pre-neurulation stage, intercalation is strong across the midline.
Discussion
Our quantitative measurements go well beyond qualitative comparisons of dominant behaviours in three example tissues and reveal detailed spatial and temporal patterns in morphogenetic processes. We have identified novel and specific features in each tissue. Cell intercalation in the Drosophila amnioserosa makes only a weak contribution to late dorsal closure. We reveal cell shape contraction in AP, in addition to the stronger ML contraction, which suggests that isometric apical contraction encounters greater resistance to deformation in AP, since amnioserosa cells are not known to be polarized25. Further investigation of the dynamics in dorsal closure mutants will help elucidate the relative contribution of the various forces and constraints26. In the Drosophila germband, but not in the zebrafish trunk, there is an inversion of mechanisms at the midline, with VML cells undergoing cell shape extension in AP whereas the lateral ectoderm extends by intercalation. A mechanistic understanding of intercalation in the germband must therefore be able to explain why there is no intercalation across the midline, and how the lateral ectoderm cells can make new connections with but not through VML cells. In the zebrafish trunk, cell shapes contract in AP and ML as cells elongate in depth, reducing tissue AP extension and augmenting ML convergence. Our strain breakdown clarifies the sorts of planar combinations that are possible. Cell intercalation is by definition balanced orthogonal convergence and extension. It is only the addition of unbalanced cell shape (i.e. when there is a change in area) that leads to an imbalance in tissue convergence and extension.
The rotation measure, in addition to the tissue deformation field, provides a key component to test mechanical models of tissue tectonics, as used in plate tectonics27. Such patterns signify deformation gradients, here predominantly due to an increasing rate of AP extension towards the midline in both the germband22 and the zebrafish trunk (N.L.S., G.B.B., A.J.K., L.M. & R.J.A. in preparation). Where intercalation and rotation happen together, two interpretations are possible. If the tissue rotation is a result of cell rotation then the tissue rotation is not associated with slippage. However, if there is no cell rotation then a new type of behaviour, intercalation simple shear, is identified, in which lines of fracture are a result of slippage in one orientation only. However, we cannot currently unambiguously separate intercalation rotation from cell shape rotation, so further analysis of the shear structure and planes of shear at the cellular and sub-cellular scale will reveal insights about the cellular organisation of tissues.
Our biological examples confirm that two fundamental and complementary classes of cellular events can now be faithfully distinguished and exquisite patterns revealed. Our quantitative approaches allow us to assay morphogenetic phenotypes with unprecedented detail and rigor, and facilitate comparisons between individuals, manipulations, genotypes and species to probe the variability of developmental programs. We suspect that many genetic phenotypes are being missed or misinterpreted through lack of suitable methods, and because similar final phenotypes can be achieved through different developmental mechanisms. An immediate application of our methods would therefore be quantitative phenotypic screening based on comparisons of rates of cell shape change and intercalation.
Online Methods
Tissue strain rate
For each cell j in a domain, we know its location, Rj (xj,yj), and its velocity, Uj (uj,vj). We typically use domains of nc = 1-3 coronae, which are sufficiently small to be treated as flat planes, once corrected for surface tilt. The local velocity field can therefore be treated as linear and a planar regression used to characterise the domain behaviour in terms of the local strain rates using the form:
| (2) |
| (3) |
which can be written in matrix notation as:
| (4) |
Here the average domain translation velocity is and the tissue velocity gradient tensor is Lt. We choose the time interval dt such that the tissue domain deformation is small (each coefficient of Lt < 0.1), permitting a simple decomposition of Lt into an antisymmetric spin Ωt = (Lt + Lt T) / 2 and a symmetric strain rate tensor, Ėt = (Lt + Lt T) / 2. The trace of Lt (Tr Lt = Tr Ėt) gives the rate of change in area of the domain.
Cell shape strain rate
Without independent fiducial (registration) marks within or around cells that can be followed over time, cell rotation cannot be determined unambiguously. Movements of individual vertices at the junctions between cells have no direct relationship with the overall changes in cell shape; the latter would therefore not be properly measured by registration methods that track details of the cell contour. We instead use the traced outlines of tracked cells to extract a statistical representation of their deformation. We find the deformation that minimises the squared area difference between the starting cell shape, deformed by the estimated Ėc,j, and the final cell shape. Tr Ėc,j is constrained to be ln(Area(t+dt), j/Area(t-dt), j) to conserve area change. Eigenvector and eigenvalue space is searched for the optimal Ėc,j using the Nelder-Mead minimization method28. Ėc is then the area-weighted average of Ėc,j for all cells of the domain. In practice, we find that simplifying real cell shapes to their best-fit ellipses dramatically speeds up calculations with minimal change to the resulting Ėc values. The fit of an ellipse (see Fig. 3f) to a cell shape is optimized by maximising the area overlap, with the area of the ellipse constrained to be identical to that of the cell shape. An alternative approach, that will not developed here, is to use methods derived from the statistical strain tensors introduced by Aubouy et al.29.
Intercalation strain rate
Lt characterizes the relative velocities of cell centroids and Lc represents the average redistribution of each cell’s bulk relative to its centroid, i.e. the relative velocities of points within a cell. Any mismatch between them can be conveniently represented by a slippage velocity. We use slippage in an abstract sense of a relative movement of cell bulk without being explicit about what biological mechanism mediates this at the interfaces between cells. Consider a point P located between two neighbouring cells 1 and 2 (Supplementary Fig. 1). If we estimate the displacement of this point with respect to the centre of cell 1, we obtain Similarly, for cell 2, , and the centre of cell 2 moves itself at a speed The local rate of slippage on the point A is then:
| (5) |
This construction shows that the knowledge of the displacement and shape change of two cells is a priori sufficient to give an estimate of the continuous process of planar intercalation. Our general method provides however the averaging that is required for accurate and robust measurements at the mesoscopic scale.
Strain rate errors
Practical considerations in calculating tissue deformations and spins include the choice of domain size and time interval, the requirements for which may vary between tissues. We measure the consensus tissue deformation in each domain using regression to minimize residual cell centroid movement (Eqns. 2,3). The sources and magnitudes of residuals will depend both on cell behaviour inhomogeneity and measurement error. Biological sources of inhomogeneity include non-linear patterns of behaviour within domains, domains spanning boundaries between differing tissues or behaviours, and non-uniform cell behaviour, such as cell division or isolated cells moving into or from the plane. In these analyses, we treat a cell going through cell division as one cell for the duration of a domain even if it splits fully into daughters, and hence this is measured as a cell shape change. Measurement errors will arise from noise due to pixelation of cell shapes and remaining errors in cell-tracking. It is difficult to tease apart these potential sources so we combine them into a total domain error, e. Irrespective of its source, we want to be able to quantify the error and relate it to the signal, measured as a composite mean magnitude of domain deformation and spin, µ.
We calculate confidence intervals32 associated with the linear regressions for each of the elements of Lt. We summarize the mean absolute value of these as a single confidence value et, which has the same units of proportion per minute as Lt. We similarly summarize the amount of deformation and spin in Lt as µt, the mean absolute value of its four elements. In our example domain (Fig. 3e), µt = 0.0079 and et = 0.0053, showing that there is both strong signal and significant inhomogeneity within the domain, which can be seen in the velocity field (Fig. 3d).
We explore the effect of varying nc and dt on µt and et in our three example tissues (Supplementary Fig. 2a-c). We require domain sizes small enough to capture morphogenetic detail, but large enough to have good signal to noise ratios. Increasing either nc or dt reduces µt and et in all three tissues. There is greatest reduction of et from nc = 1 to nc = 2, while µt reduces from nc = 1 to nc = 2 but is then stable. We therefore select nc = 2 for all three example tissues. Similarly, µt and et stabilize at dt = 1 min for the Drosophila germband, and dt = 4 mins for the other two examples and we use these settings throughout the paper.
We calculate a standard error, ec, for the distribution of individual cell shape deformations within each domain, of which Ėc is the mean. We also calculate the strength of cell shape deformation, µc, as the mean absolute value of the elements of Ėc. In our example domain (Fig. 3g), µc = 0.0039 and ec = 0.0031 showing a weaker cell shape signal compared to the tissue measure, while the error is proportionately larger. The distribution of ec for each example tissue is shown (Supplementary Fig. 2d-f, and summary in g).
Li is the most derived of our three strain rate measures, dependent on both cell centroid and cell shape measures being accurate and consistent (Eqn. 1). The dilatation rate (rate of area change) of Li should be zero but in practice this is not guaranteed. Differences in the dilatation rates of Lt and Lc cause the dilatation rate of Li to be non-zero. This can occur when gaps within domains, caused by missing cells, behave differently to included cells. For example, gaps may change shape, influencing Lt, but not influencing Lc. We use ei = |Tr Li| / 2 as a measure of intercalation error and define intercalation signal, µi, as half the absolute difference between the eigenvalues derived from Li. In our example domain (Fig. 3h), µi = 0.0085 and ei = 0.00016. The error is very low because the traces of Lt and Lc are very similar. The means of the distributions of ei are small compared to those of µi (Supplementary Fig. 2g). We set an intercalation error threshold of 0.009 pp min-1 above which domains are classified as being based on inconsistent Lc and Li measures and are rejected from the analyses. This removes 0.1%, 3.2% and 5.5% of domains from our three example tissues respectively.
Imaging morphogenesis in living tissues
Wild-type zebrafish (Danio rerio) and Drosophila melanogaster embryos with fluorescently-labelled cell membranes were imaged using 3D time-lapse confocal microscopy. Transgenic WT Drosophila embryos carried an ubiDECadherinGFP construct30 that expresses a fusion between DE-Cadherin and GFP, thus labelling the apical cell outlines. Dechorionated embryos were covered with Voltalef oil 10S (Attachem) and viewed with a 40X oil immersion Plan/Fluor (NA=1.3) objective. For imaging the Drosophila amnioserosa during dorsal closure, stage 1431 embryos were mounted on a coverslip with the dorsal side glued to the glass. A 50 minute time-lapse movie, starting at the beginning of the zippering stage16, was taken using an inverted LSM 510 Meta laser scanning microscope. 50 z-sections 1µm apart were collected every two minutes, with the whole of the amnioserosa in view (Fig. 4a and Supplementary Video 1). For the Drosophila germband, cellularised embryos were mounted ventral side up between an 02-permeable membrane (Sartorius) and a coverslip using a custom-made frame for imaging on an upright Nikon Eclipse E800 microscope coupled to a MRC1024 Biorad confocal. 10 z-sections 1µm apart starting from the apical surface of the cells were taken every 30s with the ventral and ventral-lateral cells of the extending germband in view (Fig. 4d and Supplementary Video 2).
For visualizing morphogenesis in the zebrafish, all cell membranes were rendered fluorescent by injecting a one-cell stage embryo with 230 pg of capped membrane-targeted mRNA generated from pCS2-fGFP (subcloned from Clontech pEGFP-F), linearised by NotI, and transcribed using the MEGAscriptTM SP6 in vitro transcription kit (Ambion, Inc.). Live gastrulating embryos were mounted in a custom-made chamber in 0.3% low-gelling agarose and imaged dorsal side down on an inverted Leica Microsystems TSC-SP2-MP confocal microscope, using a long distance 40x/0.8 water immersion objective. 50 z-sections every 2µm were collected every two minutes (Fig. 4g and Supplementary Video 3).
Multiple embryo alignment
We aligned 5 Drosophila and 9 zebrafish wild-type embryo movies in space and time, by co-aligning the embryonic midlines and locating a common developmental staging point along AP in each movie. For the fly this is the location of the cephalic furrow 5 minutes after the start of germband extension, and for the fish trunk ectoderm it is the location of the boundary between somites 1 and 2 when it first becomes visible (10.5 hpf). The pooled embryos were broken down into 20 and 25 µm grid squares, in fly and fish respectively, and average strains and rotation in each square were calculated.
Embryo layers and cell tracking
We sense the shape of the surface of the embryo in each 3D image stack8, and use these shapes to extract curved image layers of constant radial depth within the embryo. Our methods for quantifying strain rates use distances and velocities calculated across the surface of such curved layers, taking into account the local inclination of the embryo. For Drosophila tissues we extracted layers through the apical zonula adherens of the ectoderm, the site at which much cell-cell interaction is controlled, and for zebrafish tissues we took a surface cutting through the middle of the outermost layer of epiblast cells. Repeating the analysis for different depths of tissues will identify whether the tissue is behaving homogenously in depth, or which layers deform first. The approximate angular curvature spanning an average domain of cells (nc = 2) are 26.9, 13.3 and 9.0 degrees for our three example tissues respectively. We wrote software to track all cells in these pseudo-2D layers over time (G.B.B. & R.J.A., unpublished), based on the identification of the cell membranes (Supplementary Videos 1-3 and Figure 4a,d,g). The tracking software records for each valid cell, at each time point: the pixelated shape described by the fluorescent cell membrane and hence the cell area; the location of the cell centroid (centre of mass); cell identity; the identity of all touching neighbours. We filter out parts of tracked cell lineages that do not meet criteria for reasonable behaviour, such as anomalously high rates of volume change or cell displacement compared to the immediate cell neighbourhood. This removes 2.5%, 3.9% and 20.9% from our three example tissues respectively. Our analyses are designed to have the essential quality of being robust to occasional missing cells. All cells that touch the edge of the field of view are excluded from the analyses because they may be incomplete. The average number of cells per time point used for strain rate analysis is 87, 522 and 503 respectively.
Supplementary Material
| Supplementary File | Title |
|---|---|
| Supplementary Note | Glossary |
| Supplementary Figures | Supplementary figures 1-3 |
| Software | Zip file of code written in IDL for calculating strain rates in a domain of tissue |
| Supplementary Video 1 |
Tracked cell trajectories and membrane shapes in Drosophila amnioserosa. Video to accompany Figure 4a. Video duration is 50 mins, with frames at 2 min intervals. Details are as in Figure 4. |
| Supplementary Video 2 |
Tracked cell trajectories and membrane shapes in Drosophila germband. Video to accompany Figure 4d. Video duration is 15 mins, with frames at 30s intervals. Details are as in Figure 4. |
| Supplementary Video 3 |
Tracked cell trajectories and membrane shapes in zebrafish trunk ectoderm. Video to accompany Figure 4g. Video duration is 50 mins, with frames at 2 min intervals. Details are as in Figure 4. |
| Supplementary Video 4 |
Cellular simulation of a domain dominated by cell shape change. Video to accompany Figure 2a simulation. Video duration is 50 mins, with frames every minute. |
| Supplementary Video 5 |
Cellular simulation of a domain dominated by cell intercalation. Video to accompany Figure 2b simulation. Video duration is 50 mins, with frames every minute. |
| Supplementary Video 6 |
Tissue deformations and rotations in Drosophila amnioserosa. Video to accompany Figure 4b. Video duration is 50 mins, with frames at 2 min intervals. Details are as in Figure 4. |
| Supplementary Video 7 |
Cell shape deformations in Drosophila amnioserosa. Video to accompany Figure 5a. Video duration is 50 mins, with frames at 2 min intervals. Details are as in Figure 5. |
| Supplementary Video 8 |
Cell intercalation deformations in Drosophila amnioserosa. Video to accompany Figure 5b. Video duration is 50 mins, with frames at 2 min intervals. Details are as in Figure 5. |
| Supplementary Video 9 |
Tissue deformations and rotations in Drosophila germband. Video to accompany Figure 4e. Video duration is 15 mins, with frames at 30s intervals. Details are as in Figure 4. |
| Supplementary Video 10 |
Cell shape deformations in Drosophila germband. Video to accompany Figure 5d. Video duration is 15 mins, with frames at 30s intervals. Details are as in Figure 5. |
| Supplementary Video 11 |
Cell intercalation deformations in Drosophila germband. Video to accompany Figure 5e. Video duration is 15 mins, with frames at 30s intervals. Details are as in Figure 5. |
| Supplementary Video 12 |
Tissue deformations and rotations in zebrafish trunk ectoderm. Video to accompany Figure 4h. Video duration is 50 mins, with frames at 2 min intervals. Details are as in Figure 4. |
| Supplementary Video 13 |
Cell shape deformations in zebrafish trunk ectoderm. Video to accompany Figure 5g. Video duration is 50 mins, with frames at 2 min intervals. Details are as in Figure 5. |
| Supplementary Video 14 |
Cell intercalation deformations in zebrafish trunk ectoderm. Video to accompany Figure 5h. Video duration is 50 mins, with frames at 2 min intervals. Details are as in Figure 5. |
Acknowledgements
We acknowledge financial support from the MRC (R.A.) and the Harvard-MRSEC (L.M.). Additional financial support: for the zebrafish trunk, a Wellcome Trust Studentship to N.L.S.; for Drosophila germband extension, a HFSP grant to B.S. and a Wellcome Trust studentship to L.C.B.; for Drosophila dorsal closure, a BBSRC grant to Alfonso Martinez Arias and N.G‥ We thank N. J. Lawrence who initiated Drosophila germband extension imaging, and thank S.J. England, S.R. Young and other members of our laboratories for fruitful discussions. This paper is dedicated to the memory of Locke G.N.B.
Footnotes
Author contributions: The project was conceived and developed and the manuscript written by G.B.B., A.J.K., L.M. and R.J.A‥ Data analysis and code development was performed by G.B.B. and A.J.K‥ Analyses were developed in collaboration with N.L.S. for the zebrafish trunk, L.C.B. and B.S. for Drosophila germband extension and N.G. for Drosophila dorsal closure, who all contributed time-lapse movies and expertise on their models.
References
- 1.Keller R, et al. Mechanisms of convergence and extension by cell intercalation. Phil Trans Roy Soc B. 2000;355:897–922. doi: 10.1098/rstb.2000.0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertet C, Sulak L, Lecuit T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 2004;429:667–671. doi: 10.1038/nature02590. [DOI] [PubMed] [Google Scholar]
- 3.Concha M, Adams RJ. Oriented cell divisions and cellular morphogenesis in the zebrafish gastrula and neurula: a time-lapse analysis. Development. 1998;125:983–994. doi: 10.1242/dev.125.6.983. [DOI] [PubMed] [Google Scholar]
- 4.Neumann M, Affolter M. Remodelling epithelial tubes through cell rearrangements: from cells to molecules. EMBO reports. 2006;7:36–40. doi: 10.1038/sj.embor.7400597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutson MS, et al. Forces for morphogenesis investigated with laser microsurgery and quantitative modelling. Science. 2003;300:145–149. doi: 10.1126/science.1079552. [DOI] [PubMed] [Google Scholar]
- 6.Moore SW, Keller RE, Koehl MAR. The dorsal involuting marginal zone stiffens anisotropically during its convergent extension in the gastrula of Xenopus laevis. Development. 1995;121:3131–3140. doi: 10.1242/dev.121.10.3131. [DOI] [PubMed] [Google Scholar]
- 7.Keller R, Shook D, Skogland P. The forces that shape embryos: physical aspects of convergent extension by cell intercalation. Phys Biol. 2008;5:15007. doi: 10.1088/1478-3975/5/1/015007. [DOI] [PubMed] [Google Scholar]
- 8.England SJ, Blanchard GB, Mahadevan L, Adams RJ. A dynamic fate map of the forebrain shows how vertebrate eyes form and explains two causes of cyclopia. Development. 2006;133:4613–4617. doi: 10.1242/dev.02678. [DOI] [PubMed] [Google Scholar]
- 9.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. doi: 10.1126/science.1162493. in press. [DOI] [PubMed] [Google Scholar]
- 10.Graner F, Dollet B, Raufaste C, Marmottant P. Discrete rearranging disordered patterns, part I: Robust statistical tools in two or three dimensions. Eur Phys JE. 2008;25:349–369. doi: 10.1140/epje/i2007-10298-8. [DOI] [PubMed] [Google Scholar]
- 11.Fung YC, Tong P. Classical and Computational Solid Mechanics. World Scientific; 2001. [Google Scholar]
- 12.Glickman NS, Kimmel CB, Jones MA, Adams RJ. Shaping the zebrafish notochord. Development. 2003;130:873–887. doi: 10.1242/dev.00314. [DOI] [PubMed] [Google Scholar]
- 13.Yin C, et al. Cooperation of polarized cell intercalations drives convergence and extension of presomitic mesoderm during zebrafish gastrulation. J Cell Biol. 2008;180:221–232. doi: 10.1083/jcb.200704150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weaire D, Hutzler S. The Physics of Foams. Oxford University Press; 2001. [Google Scholar]
- 15.Farhadifar R, et al. The influence of cell mechanics, cell-cell interactions, and proliferation on epithelial packing. Curr Biol. 2007;17:2095–2104. doi: 10.1016/j.cub.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 16.Hilgenfeldt S, Erisken S, Carthew RW. Physical modeling of cell geometric order in an epithelial tissue. PNAS. 2008;105:907–911. doi: 10.1073/pnas.0711077105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacinto A, Woolner S, Martin P. Dynamic analysis of dorsal closure in Drosophila: from genetics to cell biology. Dev Cell. 2002;3:9–19. doi: 10.1016/s1534-5807(02)00208-3. [DOI] [PubMed] [Google Scholar]
- 18.Kiehart DP, et al. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J Cell Biol. 2000;149:471–490. doi: 10.1083/jcb.149.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez BG, Arias AM, Jacinto A. Dpp signalling orchestrates dorsal closure by regulating cell shape changes both in the amnioserosa and in the epidermis. Mech Dev. 2007;124:884–897. doi: 10.1016/j.mod.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Irvine KD, Weischaus E. Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development. 1994;120:827–841. doi: 10.1242/dev.120.4.827. [DOI] [PubMed] [Google Scholar]
- 21.Blankenship JT, et al. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev Cell. 2006;11:459–470. doi: 10.1016/j.devcel.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 22.Butler LC, et al. Cell shape changes indicate a role for extrinsic tensile forces in Drosophila germband extension. Nat Cell Biol. doi: 10.1038/ncb1894. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller R, Shih J, Sater A. The cellular basis of the convergence and extension of the Xenopus neural plate. Dev Dynam. 1992;193:199–217. doi: 10.1002/aja.1001930302. [DOI] [PubMed] [Google Scholar]
- 24.Hong E, Brewster R. N-cadherin is required for the polarized cell behaviors that drive neurulation in the zebrafish. Development. 2006;133:3895–3905. doi: 10.1242/dev.02560. [DOI] [PubMed] [Google Scholar]
- 25.Pope KL, Harris TJ. Control of cell flattening and junctional remodelling during squamous epithelial morphogenesis in Drosophila. Development. 2008;135:2227–2238. doi: 10.1242/dev.019802. [DOI] [PubMed] [Google Scholar]
- 26.Gorfinkiel N, Blanchard GB, Adams RJ, Arias AM. Mechanical control of global cell behaviour during Dorsal Closure in Drosophila. Development. doi: 10.1242/dev.030866. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allmendinger RW, Relinger R, Loveless J. Strain and rotation rate from GPS in Tibet, Anatolia, and the Altiplano. Tectonics. 2007;26:TC3013. [Google Scholar]
- 28.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes in C. Cambridge University Press; 1988. [Google Scholar]
- 29.Aubouy M, Jiang Y, Glazier JA, Graner F. A texture tensor to quantify deformations. Granular Matter. 2003;5:67–70. [Google Scholar]
- 30.Oda H, Tsukita S. Real-time imaging of cell-cell adherens junctions reveals that Drosophila mesoderm invagination begins with two phases of apical constriction of cells. J Cell Sci. 2001;114:493–501. doi: 10.1242/jcs.114.3.493. [DOI] [PubMed] [Google Scholar]
- 31.Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Springer-Verlag; 1997. [Google Scholar]
- 32.Draper NR, Smith H. Applied Regression Analysis. 3rd Edition. John Wiley & Sons Inc; New York: 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.