Abstract
An enormous variety of biological redox reactions are accompanied by changes in proton content at enzyme active sites, in their associated cofactors, in substrates/products, and between protein interfaces. Understanding this breadth of reactivity is an ongoing chemical challenge. A great many workers have developed and investigated biomimetic model complexes to build new ways of thinking about the mechanistic underpinnings of such complex biological proton-coupled electron transfer (PCET) reactions. Of particular importance are those model reactions that involve transfer of one proton (H+) and one electron (e–), equivalently transfer of a hydrogen atom (H•). In this Current Topic Article we review key concepts in PCET reactivity, and describe important advances in biomimetic PCET chemistry, with special emphasis on research that has enhanced efforts to understand biological PCET reactions.
Graphical Abstract
1. Introduction and Historical Perspective
The importance of atom transfers in redox reactions (electron transfers) was established in the late 1800s, culminating most famously with the formulation of the Nernst equation.1 In a compendium published in 1963, Pourbaix applied the Nernst equation to an enormous variety of redox reactions that depend on proton concentration in widely used pH versus E diagrams that bear his name.2 Transfer of one electron (e–) and one proton (H+) together – often more easily described as hydrogen atom (H•) transfer (HAT) – has long been a focus of physical organic chemistry. HAT reactions of organic molecules were intensely investigated in the 1950s and 1960s,3 and in 1960, Wiberg showed that chromium(VI) compounds could oxidize organic molecules by HAT.4
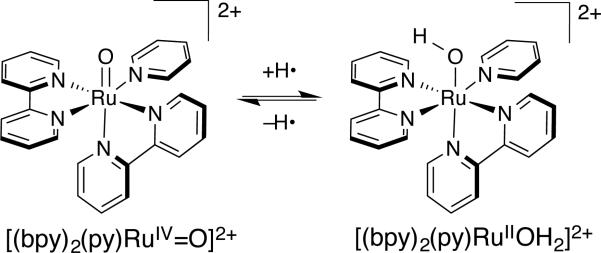
To our knowledge, the first paper explicitly invoking the coupling of H+ and e– transfers in a metalloenzyme was by Edward Stiefel in 1973, entitled “Proposed Molecular Mechanism for the Action of Molybdenum in Enzymes: Coupled Proton and Electron Transfer.” It is a particularly prescient essay, including the phrase “If both protons and electrons are delivered to the substrate in a concerted process, ....” In 1980, Eberson (likely independently) used the phrase “concerted electron/proton transfer” to describe the oxidation of toluene by tungsten polyoxometallate complexes,5 though the mechanism was later revised.6 Less than a year later, in 1981, Meyer coined the term “proton-coupled electron transfer” (PCET), to describe reactions of reactions between RuIV-oxo and RuII-aquo complexes (Figure 1).7
Figure 1.
Prototype for the first named PCET reaction between [(bpy)2(py)RuIIOH2]2+ to [(bpy)2(py)RuIV=O]2+ (bpy = 2,2′-bipyridyl, py = pyridine)
Researchers working on chemical and biological problems during the intervening 40 years have found that many, even most, electron transfer (ET) reactions are coupled in some way to proton transfer (PT) events.8 In many cases, biological “electron transport chains” could be better described as “electron-proton transport chains.” Sometimes the transfer of the e– and H+ is tightly coupled (e.g., hydride transfer from NAD(P)H or hydrogen abstraction from hydrocarbons by compound I of cytochrome P450s). At other times, the degree of coupling is less clear (e.g., pH dependent reduction potentials of redox proteins such as cytochromes c). The complexity of many of the biochemical processes, and the challenges of monitoring H+ in water, lead to the use of biomimetic small molecule complexes to provide understanding of such complex biological phenomena at a detailed (atomic) level. In many ways, this parallels the use of simpler chemical systems in the early work on pure ET reactions.9
The goal of this Current Topic article is to summarize key developments in biomimetic PCET chemistry and their relation to biological systems, rationalize different PCET reactivity patterns, and outline important frontiers in better understanding biological PCET reactions. We also touch upon important challenges associated with investigating and rationalizing PCET reactivity, both in small molecule models and more broadly in redox proteins.
1a. A view from the top: examples of diverse PCET in enzymes
PCET is recognized in many biological systems, but we think that it is underappreciated (or undiscovered) in an even greater number of biological processes and biomolecules. In our view, any system in which the reduction potential varies with pH is exhibiting PCET in some form. In this section, we develop a picture of PCET using two examples: class Ia ribonucleotide reductase (RNR) and cytochrome c oxidase (CcO). While no single system embodies all aspects of PCET, these two systems capture many salient points. In this section, we use these two examples to lay the groundwork for our subsequent discussion of the kinetics and thermodynamics of PCET reactions in small molecule models, underscoring the importance of those models in appreciating biological PCET.
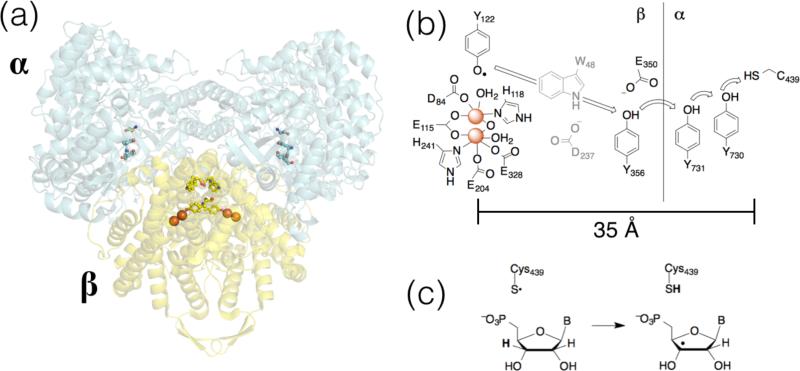
RNR enzymes are essential for DNA biosynthesis, converting ribonucleotides to deoxyribonucleotides. Perhaps the best studied RNR is the class Ia enzyme from E coli, but we note that detailed data for other enzymes recently became available.10 E. coli RNR (referred to as RNR hereafter, for convenience) is a heterotetrameric protein, consisting of homodimeric α and β subunits (Figure 2a).11 Each turnover of the catalytic cycle involves net transfer of e– about 35 Å from the substrate-binding site (on the α subunit) to a di-iron-tyrosyl site in the β subunit (Figure 2b); substrate- and effector-triggered conformational changes play key roles in the redox events.12 For these, and any PCET reaction, the pKa (describing the PT component) and E° (describing the ET component) values provide valuable insights. To that end, workers have incorporated modified tyrosine into the PCET chain of RNR (Figure 2b) to test for pKa perturbations13 and relative redox levels of the redox-active amino acids.14 Likewise, these studies also provide kinetics data that can be used to rationalize fundamental reactivity in a complex system involving conformational and redox changes.15
Figure 2.
(a) Docking model of E. coli RNR.11 (b) Schematic of the amino acid residues involved in PCET activation of Cys429. (c) H-abstraction from ribonucleotide substrate by Cys349.
The redox chemistry of tyrosine is central to RNR function. As outlined below, redox reactions of tyrosine are inherently proton-coupled because of the strong acidity of the oxidized (radical cation) form (Section 3c). By moving the electron and proton together, this high energy form can be avoided. Therefore, PCET likely is involved in the activation of the diiron-tyrosyl active oxidant to form the initial tyrosyl radical.16 Most notable is the long PCET cascade from the α to the β subunits in RNR, which also features two distinct types of PCET. The first involves 1H+/1e– oxidation of tyrosine where H+ and e– have distinct acceptors (Figure 2b); the second involves direct H• abstraction from a ribonucleotide C-H by a cysteinyl radical (Figure 2c). The former type of PCET highlights the disparity in distance scales for PT and ET, raising the interesting questions of mechanism (stepwise, with separate H+ and e– transfer steps or concerted, H• transfer) and PCET distance dependence.
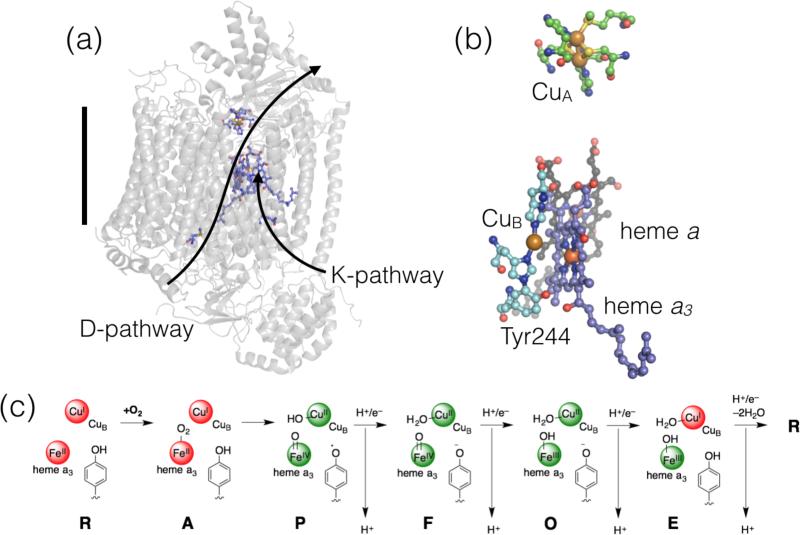
Cytochrome c oxidases (CcOs, Complex IV in the mitochondrial respiratory chain or, more broadly, heme-copper oxidases) are the terminal electron acceptors in the respiratory chain. In these transmembrane enzymes (e.g., Figure 3a), reduction of O2 to 2H2O is coupled to transmembrane H+ pumping.17 The mammalian enzyme contains 13 subunits, but studies on very similar and more easily obtained bacterial proteins have facilitated understanding of the remarkable mechanism of CcO enzymes.18 PCET is involved in every aspect of CcO function: reduction of O2 to H2O is a 4H+/4e– PCET reaction and for each input of an e– to the active site, H+ is translocated across the membrane (where the H+ stoichiometry is enzyme-dependent18a). In fact, PCET is critical throughout the mitochondrial and photosynthetic “electron-proton transport chains,” as highlighted above.
Figure 3.
Structure and mechanism of cytochrome c oxidase proteins. (a) Structure of CcO from bovine (PDB ID 2EIJ20) where the H+ loading pathways are labeled and the black bar indicates the approximate width of a lipid bilayer; (b) Close-up view of the redox cofactors in the active site; and (c) Mechanism of reduction of O2, including commonly used one-letter abbreviations for the intermediate species. The red colored spheres indicated reduced metal sites and the green spheres indicate oxidized sites.
The CcO active site contains one heme (heme a3), a Cu (CuB), and an tyrosine residue that is cross-linked with a nearby His (which ligates CuB) (Figure 3b). The protein also contains two redox cofactors: heme a and a purple copper (CuA) site. Theses sites serve as e– reservoirs for O2 reduction, and the latter is the primary redox partner with cytochromes c.19 Reduction of O2 starts from the fully reduced active site (R, Figure 3c). Binding of O2 gives state A. Four electron reduction of O2 substrate in an apparent single step yields the FeIV from of heme a3, CuIIB, and a tyrosyl radical. Subsequent additions of 1H+ and 1e– reduce the active site back to the resting state (R), release 2 equivalents of water, and each redox step is coupled to a H+ pumping step. This is only a bare overview of a complex process,17,18 but as for RNR, it demonstrates several aspects of PCET chemistry. First, the step going from state A to P is an example of a proton-coupled bond breaking (O=O and TyrO-H). Subsequent H+/e– additions are examples where e– are delivered to a metal site and H+ are delivered to a metal-ligated group. As for RNR, these latter steps (P to R) feature PCET reactions where e– and H+ have distinct donors/acceptors. Finally, each ET reaction at the active site is coupled to H+ translocation over a great distance, which is required for net transmembrane H+ pumping.
The above two examples set the groundwork for our ensuing discussion. A great many small-molecule model systems have been examined to provide atomic-level details to answer important questions about biological PCET. What are the factors that dictate H+ and e– and transfer between the same donors/acceptors or different donors acceptors? Are such reactions described by component theories of PT and ET, or is a new framework required? How can a cofactor (such as tyrosine) mediate different types of PCET? And importantly, how should these seemingly different reactions be rationalized, and what is the “best” experimental approach for answering such questions?
2. PCET reaction classes
The term PCET was initially used7 to distinguish concerted H+/e– transfer reactions of inorganic complexes from hydrogen atom transfer (HAT) reactions long studied by organic chemists. However, due to extensive and varied use, experimentalists describing H+/e– transfer reactions typically use the term “PCET,” regardless of stoichiometry or mechanism.21 In the literature of chemical theory, PCET is still used mostly to refer just to the concerted transfer of 1e– and 1H+.8a Because of this ambiguity, we encourage workers in the field use very clear descriptions of H+/e– transfer reactions.
Studies of small molecule models have made possible important mechanistic distinctions for PCET reactions. There are three limiting mechanisms for those PCET reactions where only one H+ and one e– transfer: stepwise ET, then PT; stepwise PT, then ET; and a concerted mechanism where H+ and e– transfer in a single kinetic step. We recommend following the Savéant group and calling the third mechanism concerted proton-electron transfer (CPET).22 Other workers term such reactions electron-proton transfer (EPT)21b or concerted electron-proton transfer (CEP).23
It has been suggested that HAT reactions might be considered as a special class of CPET where the transferring H+/e– come from the same chemical bond.21b The distinction is subtle, but the implications for reactivity can be profound. One example is H+/e– transfer from [(bpy)2(py)RuIIOH2]2+ to [(bpy)2(py)RuIV=O]2+ (Figure 1) The ground state products, from a CPET reaction, are 2 equivalents of [(bpy)2(py)RuIIIOH]2+. Using the above definition of HAT, the transferring H• from [(bpy)2(py)RuIIOH2]2+ would come from an O–H bond to give the high energy species [(bpy)2(py)RuII(•OH)]2+ as the initial product, before relaxing to the ground state. Here, and in a few other cases,24 there are clear energetic consequences for a CPET/HAT orbital distinction, but this is a rare example where a thorough mechanistic investigation can be used to distinguish two related, concerted mechanisms for net H• transfer. The above definition of HAT is also problematic because it often gives cases where a single reaction is called HAT in the forward direction, but not in the reverse. For instance, the net transfer of H• (e– + H+) from a tyrosine to a peroxyl radical would not be HAT, because the tyrosyl e– comes from the π orbitals and not from the O–H bond, but the reverse reaction would be classified as HAT. To further complicate matters, there are quite different theoretical arguments that concerted 1e–/1H+ transfers should be termed HAT when they are adiabatic, while those that are non-adiabatic (involve excited state energy surfaces) should be called CPET.25
Given the wide breadth of PCET reactivity, we think that restrictive and subtle distinctions such as HAT versus CPET can be easily misinterpreted and sometimes be (unintentionally) misleading. Furthermore, for the vast majority of PCET reactions, these distinctions are difficult, if not impossible, to draw from experimental results. This is an ongoing challenge in bridging concepts in new PCET theories with experimental results. For now we prefer to use the term PCET for any reaction involving overall H+/e– transfers, CPET for all concerted 1e–/1H+ transfers, and ET-PT or PT-ET for stepwise mechanisms.
In addition to the above examples where H• formally transfers from one donor molecule to one acceptor molecule, one of the most biologically diverse subsets of PCET reactions are those in which the transferring H+ and e– start from (or go to) distinct donors (or acceptors). This is shown schematically in equation 1, in which AH forms A• by delivering H+ to a Brønsted base B: and e– to an oxidant X+. These types of reactions are referred to as separated or multiple-site PCET (MS-CPET or sometimes MS-EPT or bidirectional PCET26).
| (1) |
The canonical biological example of MS-CPET is the reversible oxidation of YOHZ in photosystem II. YOHZ is oxidized by the chlorophyll P680 radical cation, likely concerted with transfer of H+ to Histidine191.27 The action of CcO, described above, is an even more complex example of MS-PCET.18 In that enzyme, all of the redox and H+ transfer processes must be coupled in some way, but they probably are not all CPET. Inspired by some of the issues in MS-PCET, we recently compiled and analyzed a more general series of oxidant + base combinations28
2a. Energetics and mechanisms
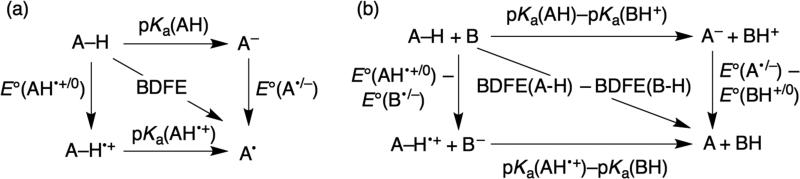
The energetics and mechanism of a PCET reaction are typically rationalized using thermodynamic square schemes. One version of such schemes describes the thermochemistry of a single reactant (Figure 4a). For a molecule AH, this includes two reduction potentials E(AH0/+) and E(A–/0) (E° in water or typically E1/2 in organic media), two pKas (for AH and AH+), and a homolytic bond strength for A–H. We strongly recommend the use of bond dissociation free energies (BDFEs) because they are directly related to the free energy changes associated with pKa and E°, as well as the equilibrium constants (Keq) for PCET reactions. The bond strength literature, however, has for many decades emphasized bond dissociation enthalpies (BDEs). The use of BDEs in PCET square schemes requires the assumption that any entropy change is negligible. While this is usually a reasonable assumption for reactions of organic molecules, it is not appropriate for redox reactions of transition metals.30 BDFEs are related to E° and pKa values by equation 2. The constant CG accounts for the free energy or formation and solvation of H• in a given solvent. A complete derivation is given elsewhere.31
| (2) |
Figure 4.
Thermodynamics square schemes (a) for a single PCET reagent (AH) and (b) for a PCET reaction (AH + B).
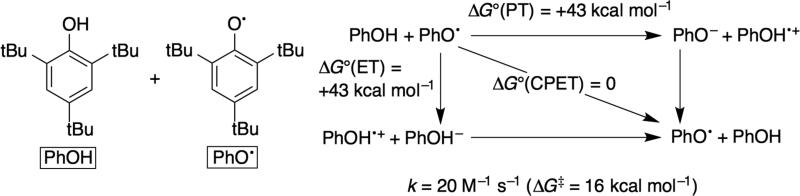
Information about the free energy landscape of a bimolecular PCET reaction AH + B is obtained by combining square schemes for two reactants (Figure 4b). For a separated MS-PCET (equation 1), the thermodynamics can be understood by combining the square scheme for AH with the reduction potential of the oxidant X+ and the basicity of B: (the pKa HB+).32 The overall driving force is given by the difference in BDFEs,33 and the free energy change for individual PT or ET steps is given by the appropriate differences in pKa or E°, respectively. Such information can provide mechanistic insight when kinetic data are available. In cases where kinetic barriers calculated from the Eyring equation (ΔG‡) are lower than the free energy change (ΔG°) for a stepwise ET or PT step, then a concerted mechanism is implicated. An example for the H• self-exchange reaction of 2,4,6-tri-tert-butylphenol in MeCN solvent is shown in Figure 5.21a The free energy required to produce intermediates required for stepwise ET-PT or PT-ET are much higher than the observed Eyring barrier; the self-exchange reaction must then proceed via a different pathway (CPET). In this case the large differences between the ΔG‡ and the ΔG°(ET) and ΔG°(PT) makes the mechanistic conclusion very convincing. Given the typical uncertainties in the pKa or E°, values, mechanistic conclusions require these differences to be at least a few kcal mol–1. Still, when the energy differences are large, even crude kinetics can be sufficient for strong mechanistic conclusions since ΔG‡ varies with the logarithm of the rate constant.
Figure 5.
Thermodynamic and kinetic parameters for PCET reactions of tBu3PhOH.21a
The unambiguous difference in energetics in Figure 5 does not hold for every PCET system. Some systems (e.g., ascorbate, see below) have a “flat” energetic landscape and are able to undergo facile CPET, ET, or PT reactions. A sense of the energetic preference of a reagent to undergo concerted versus stepwise PCET can be obtained from its square scheme, from the difference in the pKa of the oxidized and reduced forms. This is equivalent (in ΔG°) to the difference between the E° of the protonated and deprotonated forms. When the difference is large, as for toluene or for phenol, the reagent has a preference for concerted reactions, while it is small for ascorbate. Medium effects (e.g., protein active sites, solvent) can perturb such preferences for different PCET mechanisms; in this regard, the above is a rough guideline. Finally, note that “concerted” does not imply synchronous transfer of the e– and H+, only that the reaction does not proceed through any discrete intermediate.34
There are few systems where the kinetics and thermodynamics of all (or even most) of the relevant ET, PT, and CPET reactions are well defined. One example is the self-exchange reactions of FeII(H2bim)32+ + FeIII(H2bim)(Hbim)2+ (H2bim = 2,2′-biimidazoline) complexes that were developed as a model for C-H activations in non-heme iron enzymes such as lipoxygenase.35 For other selected organometallic examples see Ref. 36. In general, concerted transfer of H• avoids build-up of charge and other higher energy intermediates that can be associated with increased reorganization of the solvent. ET and PT reactions also can be facile, resulting in cases where both concerted and stepwise reactions are possible depending on reaction conditions; we return to this concept below.
2b. Medium Effects
The effects of the surrounding medium on the rates and efficiencies of ET reactions are well established.37 PCET reactions introduce another level of complexity because H+ (and H•) transfers must occur over short, well-defined reaction coordinates, in contrast to the more diffuse tunneling of e– through space or via multiple through-bond paths.38 Studies of medium effects on PCET reactions are limited, except for the extensive studies of organic HAT reactions.39 When H+ is transferred between two electronegative atoms, PCET is typically predicated by the presence of a hydrogen (H)-bond. The H-bonding properties of a solvent can markedly affect a PCET reaction X-H + Y by forming X-H•••solvent complexes that “block” formation of the reactive X-H•••Y hydrogen bonded precursor complex. Likewise, specific interactions supported by protein active sites can play key roles in modulating the H+/e– transfer reactivity of a given cofactor. We demonstrated that it is essential to account for solvent H-bonding properties for a series of solution CPET reactions of oxyl radicals using expressions that describe the H-bonding donating and accepting equilibria involving the solvent and the H• donors/acceptors.40 In a related study, we showed that the thermodynamics and kinetics of simple reactions of ascorbate are affected by its local environment; this likely influences how this ubiquitous PCET cofactor behaves within an enzyme active site or at a lipid interface, for instance.22b,41
To understand a biological PCET process or to build and study a model system, it is important to consider the role of the surrounding chemical environment. As described in the following sections, the reactivity patterns (thermodynamic and kinetic) of different cofactors are strongly influenced by solvents or other changes in the chemical environment. The mechanistic propensity of a give enzyme active site likely is steered by specific hydrophobic interactions (e.g., exclusion of water), salt bridges, H-bonding, and dielectric “tuning” by nearby amino acids. Such specific interactions can be cumbersome to reproduce in solution biomimetic studies, but progress has been made. For example, the effect of H-bonding additives to CPET reactions of ascorbate,42 the effect of different proton-accepting groups in oxidation of phenols with appended bases,43 and even a subset of PCET reactions in which other bonds (e.g., O-O) are broken.44 In addition, biomimetic interactions between ligands and metal centers can help stabilize reactive intermediates [e.g., Mn+=O45 or heme(Mn+-OO)46] that model biologically important PCET intermediates. Such rationally designed ligands are inspired by the discrete native interactions in protein active sites that stabilize the analogous intermediates.
3. PCET Theories
A number of different theoretical approaches have been developed to understand PCET reactivity. Many studies use traditional transition state theory, typically calculating barrier heights and energy surfaces with density functional theory (DFT).47 However, ET cannot be is not well treated in this fashion, so a number of groups have developed more sophisticated approaches,48 drawing heavily on the framework of semiclassical ET theory.49 A full treatment is not possible in this article and interested readers are pointed to the above reviews. One of the most important features of PCET theories include explicit terms that account for vibronic coupling between reactant and product states, emphasizing that the behavior of H+ is as important as that of e–. The notation used in PCET and ET theories is very similar, but the parameters are not interchangeable. For example, the driving force (–ΔG°) and reorganization energy (λ) for a given ET or PCET process correspond to different chemical reactions.
4. PCET in organic molecules
Organic cofactors are the most widely studied class of PCET molecules. This is largely because of the enormous breadth of biological systems that use organic PCET cofactors [e.g., NAD(P)H and flavins], as well as the ready availability of many reasonable model compounds (e.g., phenol as a model for tyrosine). The biological ubiquity of organic redox cofactors, and their frequent reactions with metal-containing cofactors, makes them a good starting point. Furthermore, the reactions carried out at biological metal sites can be predicated by the reactivity of the molecule being acted upon (e.g., C-H activations). In this section we describe important general features of the PCET reactivity of different classes organic compounds.
4b. Carbon-hydrogen bonds
Carbon-hydrogen bonds are among the most intrinsically unreactive PCET functional groups. This is likely in part because they do not form strong hydrogen bonds. One way of demonstrating this is the hydrogen atom self-exchange rate constants reactions of C-H bonds, which are sluggish compared to sterically congested phenols. For instance, the kself-exchange for toluene + benzyl radical50 has ks.e. ~ 8 × 10–5 M–1 s–1 compared to 20 M–1 s–1 for the sterically congested 2,4,6-tri-t-butyl-phenol (Figure 5)21a and ca. 106 M–1 s–1 for phenol.51 In addition, the thermodynamics of stepwise ET-PT activation of C-H bonds are typically quite unfavorable. For example, in MeCN solvent, toluene has pKa(C6H5CH2-H) ~ 54 and E1/2(C6H5-CH3•+/0 ~ 1.87 V versus Cp Fe+/0 (Cp = cyclopentadienyl).21a
Linear free-energy relationships often are used to correlate observed reactivity of a series of C-H bonds due to the propensity of C-H bonds to react predominantly3,52 by concerted H• transfer. The energetics of net C-H bond activations also are less sensitive to solvent composition since, as noted above, C-H groups do not form strong H-bonds. C-H BDFEs are slightly solvent dependent due to differences in the free energy of solvation of H•.31 As noted above, medium effects can modulate specific reactivity (and energetics) of different substrates.
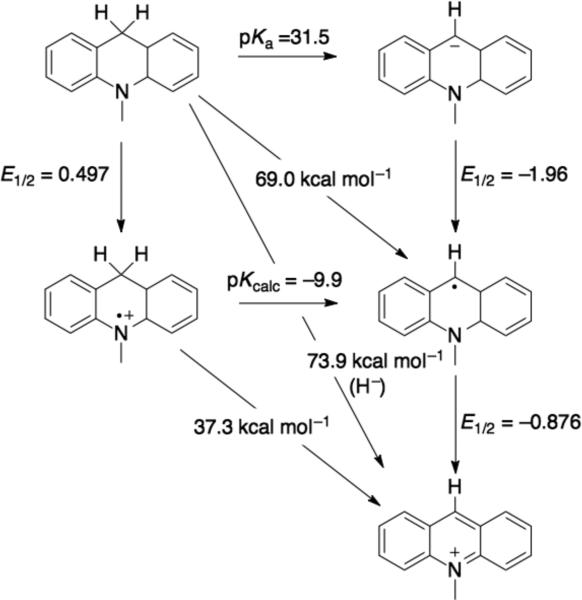
One exception the above reactivity pattern for C-H bonds is the reactivity of nicotinamide-containing molecules, which are net hydride (H–) donors. Reactions of model complexes can proceed via initial H• followed by e– transfer,53 but biological reactions, such as in the mitochondrial respiratory Complex I,54 are through to proceed via direct H– transfer mechanisms. Hydride transfer is another kind of PCET, although not always grouped that way. Like other PCET reactions, H– transfer reactions can be rationalized using thermodynamic cycles55 (e.g., Figure 656). The thermodynamics of the discrete H+, H• or e– steps are often better known in aprotic media, though some data are available in aqueous solutions for NADH models.57 To the best of our knowledge, H• transfer reactions of nicotinamides in biological systems are not well established. However, biomimetic solution reactivity of nicotinamide models suggests that, under appropriate conditions, single e–, single H+, H•, or H– reactivity are all possible modes of reactivity.58
Figure 6.
Thermodynamic square scheme for 10-methyl-9,10-dihydroacridine in DMSO.56
3c. Phenols and quinones
PCET reactions of phenol have been the subject of intense research, and some controversy, over the past 30 years. As such, it is impossible to even highlight this work in any detail. Research on phenol PCET chemistry is a result of the recognition that redox reactions of the amino acid tyrosine (YOH) are integral to the function of many proteins, including ribonucleotide reductase,59 photosystem II,60 prostaglandin-H-synthase/cyclooxygenase (PGSH/COX),61 and cytochrome c oxidase.17,18 Furthermore, products of radical reactions of YOH are markers for oxidative stress, which is implicated the pathology of many disease states, such as atherosclerosis, asthma, some cancers, and neurodegenerative diseases.62 Note that we prefer the unusual notation “YOH” for tyrosine because it keeps track of H+ and e– in deprotonated/oxidized species, using YO– for tyrosinate and YO• for tyrosyl radical.
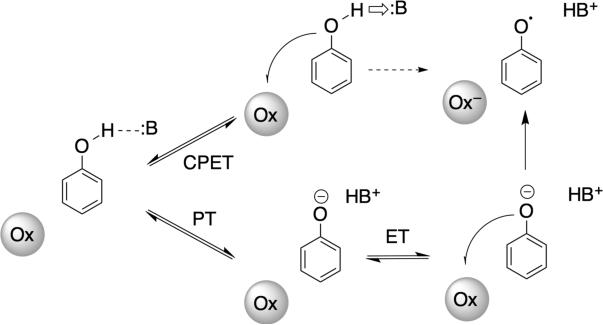
The solution properties of many phenol (PhOH) derivatives are known in many different solvents,21a with the most extensive data available in DMSO63 and in water.64 In all cases, PhO– is more easily oxidized than PhOH by hundreds of millivolts, and the PhO– is a good Brønsted base. In aprotic media, concerted reactions (Figure 7) are preferred in order to avoid formation of the high-energy species PhO– (from initial PT) or PhOH•+ (from initial ET). While the same arguments can hold in water, PT is much more facile. The ratio of rate constants for forward and reverse PT, KPT, is dictated by the pKas of PhOH and the H+-acceptor. Exoergic PT between electronegative atoms, as in this case, occurs without barrier at the diffusion limit, so the rate constant for the reverse reaction is simply the diffusion limit times. The PhO–/PhO• electron self-exchange reaction is also very rapid65 (1.9 × 108 M–1 s–1 in water, pH 12), so ET can be very rapid, even the in the low ET driving force regime typical for biological systems [where |ΔG°|<< λ (Marcus reorganization energy). Therefore tyrosine and phenols are known to react by PTET, CPET and sometimes ET-PT mechanisms66 (Figure 7), depending on the other reactant and the medium. Among the most detailed analysis of phenol oxidations are reactions with outer-sphere ET partners,67 electrochemical oxidations,68 and laser flash-quench oxidations,69 including cases where the modified phenol is covalently linked to the photosensitizer.70 Finally, we highlight recent work on PCET to electronically excited oxidants, 71 a mode of reactivity that is receiving new interest in H+/e– transfer model chemistry.
Figure 7.
Stepwise and concerted PCET mechanisms accessible by phenols using separate oxidants (Ox) and Brønsted bases (:B). Initial ET is rare because of the high PhOH+•/0 reduction potential, but such a pathway has been suggested is a few systems.66
In path breaking work, Linschitz et al. surveyed MS-PCET reactions of phenols using combinations of a photo-oxidant and a large excess of Brønsted bases.72 To address the issue of proton release/return on oxidation/reduction, workers developed a variety of small molecule model compounds that incorporate a Brønsted base proximal to a phenol-OH. Some of the molecules we have used in our laboratories are shown in Figure 8, based in part on related systems developed by other groups (see below).73 In these molecules, 1e– oxidation results in production of the corresponding distonic radical cation, where H+ has migrated to the pendant base (equation 3). In many cases, electrochemical oxidation-reduction is reversible, in contrast to the parent phenol radicals, which undergo H+ loss and dehydrodimerization at nearly the diffusion limit. The electrochemical reversibility indicates that the H+ transfer steps associated with oxidation and reduction are reversible, making these excellent small molecule models with which to explore the details of PCET reactivity, most notably the tyrosine-histidine pair in Photosystem II mentioned above. A remarkable example of reversible tyrosine/tyrosyl radical redox cycling occurs in RNR (see above), in which a hole is transferred along a series of tyrosine residues to ultimately initiate catalysis at a cysteine in another subunit.74 This long-range PCET appears to occur by the redox equivalent hopping across multiple residues, including at least two intermediate tyrosines.59
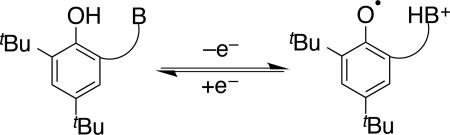 |
(3) |
Figure 8.
Phenol PCET model complexes that incorporate a pendant Brønsted base.
The phenol-base systems are examples of MS-PCET because the oxidant is distinct from the Brønsted base. This is the situation for a great many biological PCET reactions, especially in oxidations of amino acids (tyrosine, tryptophan, cysteine). The small molecule models have allowed for many tests of PCET parameters that are very difficult to probe in complex biomolecules. These parameters include: driving force dependence,68b,69,73d,75 reactivity modulation due to the pKa of the base,68b,70c,73h,76,77 the role of conjugation between the phenol and the base,78 and the effect of PT distance.79 Furthermore, because of their small size and a variety of well-defined experimental observables (e.g., IR and NMR characterization, X-ray structures), these systems have proven to be valuable tests of modern PCET theories.48b,80 The important lesson from these molecules is that the nature of the phenol•••base hydrogen bond plays a central role in reactivity. Many systems that feature such strong H-bonds react primarily through concerted processes.
The demonstration of different regimes of phenol PCET (stepwise PT-ET versus concerted) can be used to rationalize many protein redox reactions. Solution models can sometimes be misleading because a “bulk” model poorly describes protein active sites, which use carefully arranged amino acids to promote a given function. For example, ready return of H+ lost upon oxidation of YOHZ in photosystem II (PSII) is imperative for maintaining the high energy required to oxidize water. By taking advantage of a reversible bond homolysis step, PSII is able to maintain ≥ 1 eV in energy,81 which is necessary to oxidize water. Loss of H+ from the vicinity of YO•Z would shift the redox couple to that of YO•/– (E° = 0.71 V in H2O), which is not sufficient to oxidize water. Conversely, for H-abstraction from arachidonic acid by YO• in PGHS, the fate of H+ lost upon oxidative activation of YOH should not affect function in the following C-H abstraction step.
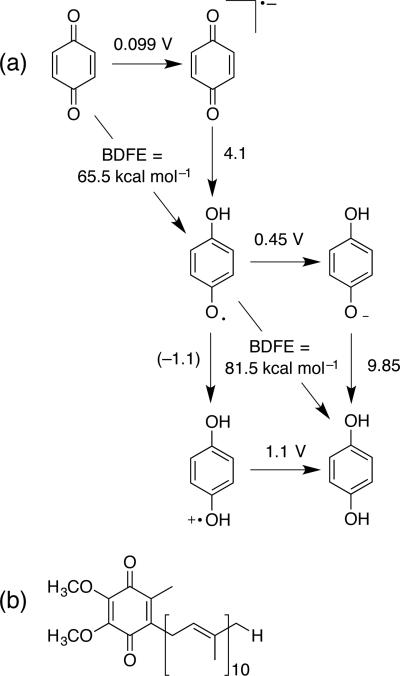
Many of the lessons from phenol chemistry apply to quinone/semiquinone/hydroquinone PCET. Quinones are ubiquitous redox cofactors. The quinone/quinol cycle is a 2H+/2e– cycle with mechanistic preferences that are sensitive to solvent and hydrogen bonding.82 The fully reduced (quinol) form displays reactivity patterns like those of phenols; 1 e– oxidation or deprotonation are unfavorable pathways in most cases (e.g., Figure 9a), especially in polar, aprotic solvents. On the other hand, stepwise PT-ET can be facile in water. The fully oxidized quinones have mild reduction potentials for single ET (e.g., 0.1 V83 for benzoquinone0/–) that allow facile 1e– reduction to the semiquinone radical anion (whose protonated form HQ• also is not too acidic, pKa = 4.1). Meyer and co-workers recently evaluated the many PCET pathways that are possible for quinones.84 We note that the relative stability of these odd electron intermediates, and the “tunability” of reactivity are important features that underlie function in biological quinone/quinol cycles, as for coenzyme Q (Figure 9b). The average BDFEs (corresponding to net 2H+/2e– conversions) cover a narrow range from about 74 (hydroquinone) to 69 (tetramethylhydroquinone) kcal mol–1, and the individual E° and pKa values follow Hammett-type trends.21a Finally, some work suggests that H– transfer to quinones is possible, but stepwise mechanisms (e.g., H• + e– or e– + H+ + e–) are likely preferred.85
Figure 9.
(a) Partial square scheme for the benzoquinone/hydroquinone thermochemistry.21a (b) Coenzyme Q10.
3d. Antioxidants
Historically, antioxidant chemistry places emphasis on “free radicals,” but it was a comparatively recent notion that biological antioxidants can formally donate H•. In 1981, Creutz outlined the H+/e– reactivity of ascorbate (HAsc–) with model complexes, even before the term PCET was coined,86 and in 1991, Njus and co-workers highlighted the CPET reactivity of HAsc– in vivo.22 As another example, oxidation of 2•glutatinone (GSH) gives glutathione disulfide (GSSG), a process that involves loss of H+ + e– from each GSH. Vitamin E (α-tocopherol) also is a good example of antioxidant H• donors. Indeed, a great many of the net reactions carried out by antioxidants are inherently PCET processes, including reactions with reactive oxygen species and reactions with alkyl radicals.
Ascorbate (HAsc–) redox chemistry has often been thought about in the context of outer-sphere ET, but H+ have profound effects on reactivity, even with 1e– oxidants.87 Oxidation under biological conditions always is accompanied by loss of H+ since pKa(HAsc•) = –0.45. The O–H bond in ascorbate is weak, so HAsc– readily transfers H• to other radical species. We demonstrated that derivatives of HAsc– can participate in CPET reactions with organic radicals88 and with iron-porphyrin model complexes,89 and that the CPET reactivity of ascorbate is sensitive to its chemical (H-bonding) environment.42 One biological example of where this sort of thermodynamic tuning would be important is for redox reactions at membrane interfaces.41 In sum, the weak O–H bond, the mild reduction potentials for different HAsc– species, and the facile PT in water, mean that ascorbate likely can react by CPET, PT-ET, or ET-PT, depending on the reacting partners and biological environment.
The PCET reactivity tocopherols and related phenolic antioxidants, resembles that of phenols and YOH, with a few caveats. Tocopherols have a lipid tail that confers membrane solubility,90 and it is established that tocopherols readily react with lipid alkyl (R•) or peroxyl (ROO•) radicals produced from reactive oxygen species.91 It is thought that such reactions proceed via H• transfer from the tocopherol-OH to the radical (HAT or CPET in the terminology used here), and that this is the primary in vivo function.90,91 This is a reasonable proposal (in our view) as the hydrophobic membrane environment will disfavor formation of the charged intermediates required for a stepwise mechanism. In some cases, tocopherol radicals can be reduced at membrane interfaces by PCET from HAsc–, which likely also follows an HAT/CPET mechanism.22,92
3d. Flavins
Flavins are 2H+/2e– PCET reagents that serve as important “switches” between 2e– and 1e– cofactors (e.g., NADH and hemes in cytochromes P450). One important biological example is the 2e– reduction of the oxidized (quinonoid) form of flavins by NADH, which is followed by two sequential 1 e– oxidations, where the electrons are usually transferred to a heme or [FeS] cluster. Flavins also are involved with O2 activation (e.g., phenylalanine hydroxylase),93 which is an inherently PCET process as O2 is converted to H2O or H2O2. The fundamental data required to construct thermochemical square schemes for flavins are available, but these data are probably only a starting point for thinking about PCET in flavins94 because of the breadth of reactions that they carry out.95 The variety of 2e– transformations of flavins is aided by the relative stability and mild acid/base properties (pKa ~ 8.5) of the flavosemiquinones.
While the PCET chemistry of flavins is widely appreciated, relatively few model studies have been reported. However, PCET reactivity in a variety of natural flavoenzymes has been described in a variety of contexts because of their remarkable range of PCET and non-PCET reactivity.96 Flavins also are involved with the PCET reduction of O2 to H2O2 by glucose oxidase, the process of which has been investigated in some detail.97 In these examples the extended structure of the protein active sites is very important, so they remain a significant challenge to fully model using small molecules. In these cases, careful kinetics and thermodynamics analyses of the natural enzymes have proven more fruitful.
3e. Hydroxylamines
Hydroxylamines are not typical biological PCET substrates (with the exception of the nitrogen cycle), but they are invaluable reagents for studying biomimetic PCET reactions. Like most hydrocarbons and their C-H bonds, alkyl hydroxylamines (R2NO-H) are very poor Brønsted acids and very poor reductants, so they strongly prefer to react via CPET. In many cases, the resulting aminoxyl radical is stable – the most prevalent example being 2,2′-6,6′-tetramethylpiperidinoxyl (TEMPO) and its relatives. The radicals also are poor bases and have very low reduction potentials, leading them to also prefer a CPET mechanism. Finally, the O-H bond in hydroxylamines is fairly weak [BDFE(TEMPO-H) = 71 kcal mol–1 in water], facilitating investigations of metal complexes and active sites with BDFEs from 60 to >80 kcal mol–1.
4. Inorganic PCET Cofactors and Reactivity
The idea that transition metal sites can participate in reactions involving e– and H+ gained momentum in the 1980's, well in advance of the focus on this for organic cofactors. Investigations of a remarkable breadth of inorganic PCET model complexes provide a basis for understanding function in many metalloproteins. It is not possible here to provide a comprehensive picture of this work; we restrict ourselves to the prevalent and important patterns of reactivity. In the concluding section we place special emphasis on new challenges for biomimetic inorganic chemistries.
3b. Small gaseous molecules
The PCET chemistries of H2, O2, N2 and CO2 are at the heart of life, and the PCET thermodynamics of these building blocks have long been understood in isolation.21a,98 The PCET reactivity of the above small molecules at transition metal sites has recently received a great deal of attention, especially for the respective reduction reactions: 2H+ + 2e– → H2; CO2 + 2H+ + 2e– → CO + H2O;99 N2 + 6H+ + 6e– → 2NH3;100 and O2 + 4H+ + 4e– → 2H2O.101 Understanding the details of how enzymes use metal centers to carry out respiration, nitrogen fixation, and photosynthesis could provide pivotal insights across diverse areas, such as understanding biological functions and addressing emerging energy challenges across the planet.
4b. Porphyrin-based models
One of the earliest and important heme model compounds was Groves’ TMP•+FeIV=O complex (TMP = 5,10,15,20-tetramesitylporphyrin).102 This complex exhibited diverse PCET and oxygen atom transfer reactivity. It led to the proposal of the rebound mechanism that is now widely accepted for the widespread net oxygen insertion reaction accomplished by cysteineligated heme oxygenases.103 Conceptually related porphyrin models with very bulky substituents in the periphery are models for oxygen binding and activating enzymes. Hydrogen bonding moieties can stabilize bound superoxide [FeIII-O2], facilitating the intramolecular ET, as in oxygen binding heme enzymes.101a
One of the final pieces of the P450 cycle was demonstrated recently, with Green's direct observation of the O=FeIV(P•+) intermediate (Compound I, P = protoporphyrin IX), the observation of its abstraction of H• from R-H to give FeIII(P) and R-OH, and the determination of the pKa value of HO=FeIV(P) (Compound II).104 Though the substrate preference or reactivity of sub-classes of P450 enzymes are distinct,105 the C-H activating moieties share common features: all are both oxidizing and their reduced form is basic. In accord with the above outline of C-H thermochemistry, activation of those bonds almost never proceeds by initial ET because that would require a very strong oxidant, which would be non-selective. Similarly, an unreasonably strong base would be required for initial deprotonation of most C-H bonds. Therefore, the enzymes use a less demanding combination of a milder oxidant and a moderate base for PCET activation. The activation of C–H bonds by Compound I is always described as a hydrogen atom transfer, but it is probably better described as MS-CPET: the H+ removed from the C–H bonds adds to the oxo group, but the e– is transferred to a half-occupied orbital located on the porphyrin and thiolate.
PCET reactions of hemes are not necessarily mediated by high-valent intermediates. Another early use of model porphyrin complexes used to study PCET are the photochemical studies of Nocera et al., in which photoinduced ET is modulated by H+ in a hydrogen-bonded bridge.106 Not only are these models useful for investigations of light-induced PCET, they also serve as scaffolds for investigation the importance of hydrogen bonded bridges in mediating PCET in ways related to the pathway model for biological ET.38
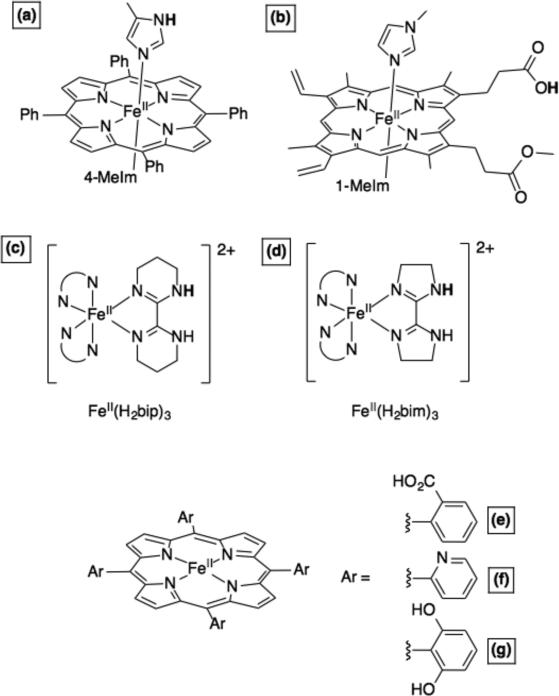
Later, we demonstrated that imidazolate-ligated complexes of FeIII-5,10,15,20-tetraphenylporphyrin (Figure 10a) can act as H• acceptors, reduction of the iron and protonation of the imidazolate. TEMPOH, hydroquinone, or 5,6,-isopropylidene ascorbate are all effective donors.107 Analogous complexes of FeIII-protoporphyin IX dimethyl ester display very similar reactivity patterns.108 The mono-methyl ester complexes (Figure 10b) can undergo PCET coupling redox activity at the iron with protonation/deprotonation of the distant carboxylate, for instance with ascorbate.89b These compounds are models for 6-coordinate heme proteins that have pH-dependent reduction potentials.109 Coordinatively saturated hemes are typically through of as electron carriers (e.g., cytochrome c) but our model systems – and the pH dependent potentials – suggest that bis(His)-ligated hemes can participate in PCET reactions. This is consistent with some biochemical investigations where alkylation of a ligating His destroys enzyme function.41
Figure 10.
Iron heme and non-heme model complexes for PCET.89b,101d,e,107,110,113,117 The transferring H+ are shown in bold.
Catalysis at heme sites often relies upon ready delivery or extraction of H+. To that end, we101d,e and others99b,110 illustrated the importance of Brønsted acids/bases positions around the metal (Figure 10e-g); the delivery of H+ was suggested to play a key role in the PCET reduction of O2 and CO2, respectively. As noted above, the distinct mechanism of these transformations could involve stepwise or concreted steps, but it is the controlled delivery of both e– and H+ that is central to reactivity and function.
4b. Non-heme iron
Non-heme iron enzymes undergo a wide variety of PCET processes. For instance, the oxidation of a YOH to YO• by a di-iron site RNR was mentioned above. This field has grown enormously in recent years with the isolation of FeIV-oxo (ferryl) compounds and their identification in enzyme catalytic cycles. The interested reader is referred to relevant reviews,111 and those highlighted below. In addition to the ferryl intermediates, we and others set out to provide understanding of lipoxygenase enzymes, which abstract H• from the weak C–H bonds in polyunsaturated fatty acids using an FeIII–OH active site, i.e., R–H + FeIII–OH → R• + FeII–OH2. The first model for this reaction is likely the report by Stack et al., using an FeIII-methoxide complex.112 We were concurrently working on less biomimetic iron complexes with three biimidazoline or three bipyrimidine ligands (Figure 10c,d).113 The oxidized/deprotonated forms of these complexes can activate weak C–H and O–H bonds, and the reduced/protonated complexes will react with phenoxyl radicals (e.g., 2,4,6-tri-tert-butylphenoxyl114) to give the corresponding phenols and the oxidized/deprotonated congener. These FeIII systems are very mild oxidants, and are able to oxidize weak C-H bonds only because they are fairly strong bases. The discoveries that C-H bonds are oxidized by FeIII active sites, as well as ferryl cofactors, reinforces the PCET perspective that both redox potential and basicity are required.
The iron systems with N-heterocyclic ligands proved to be robust and powerful systems to test fundamental properties of CPET reactions. First, our investigation of CPET reactions of Fe(bip) and Fe(bim) complexes (Figure 10c,d), as well as other systems, showed that the Marcus Cross Relation could be applied to reactions where H• is transferred (as opposed its original derivation for outer-sphere ET).115 We later expanded upon this idea to include a more comprehensive range of CPET reagents, including many of biochemical interest.40,116 This demonstration that a version of Marcus Theory holds for many reactions has some overlap with current, more sophisticated theories of CPET. In addition, some of these non-heme metal couples were found to have very large ground state entropic changes, in contrast to related organic reagents.117 These studies demonstrated that the historical use of bond dissociation enthalpies (BDEs) in rationalizing hydrogen atom transfer reactivity was not appropriate for closely related reactions of transition metal complexes. We strongly encourage workers to employ bond dissociation free energies (BDFEs) when discussing PCET reaction driving forces. This follows from the use of free energies in Marcus theory and current theories of PCET, and is consistent with the dominant use of linear free energy relationships in physical organic chemistry. Not only are BDFEs more appropriate, they also are directly related to the commonly reported PT and ET thermodynamic values, the pKa and E°.
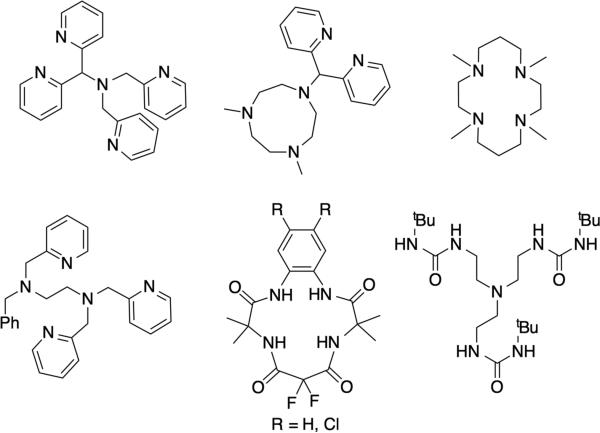
Proton-coupled redox transformations at biological iron sites are very widespread and varied, including enzymatic118 and non-enzymatic sites.119 One major area is the activation of O2 for substrate functionalization by a great many non-heme iron enzymes,120 and such processes have been extensively modeled using a diverse array of ligands.121 Modeling such reactions is challenging because of the fleeting nature of the intermediates, and the lack of a strong chromophore. In terms of PCET reactivity, among the most comprehensively investigated systems are those of Borovik,45 Collins,122 Nam123 and Que124 (examples given in Figure 11). In addition, the design criteria and reaction chemistry from Collins and co-workers provide some insight into important enzymatic features that protect proteins from oxidation by their own cofactors.111b The number of PCET reactions carried out by these model systems (Figure 11) is too large to elaborate here. Each system also can carry out a range of oxo transfer reactions and hydroxylations, depending on the substrate. Importantly, the great many model systems provide a broad foundation for the spectroscopy and reactivity of difficult to study non-heme iron in proteins.
Figure 11.
Examples of ligands that support formation of high valent (FeIV/FeV) iron-oxo moieties.
Iron-sulfur cluster and related compounds have long been of interest for their range of ET properties and rich spectroscopy.125 Of special interest to modern energy challenges are the hydrogenase enzymes, which catalyze perhaps the simplest PCET process, 2H+ + 2e– = H2. Early hydrogenase models roughly reproduced features of the enzyme active sites and reactivity, but a new generation of models provides a great deal of insight into reactivity and potential catalytic intermediates. Several groups demonstrated PCET reactivity in such models, including production of hydrides using combinations of reductants and acids.126 An extremely important feature of some of the new models is the incorporation of a “proton relay” in the second (or outer) coordination sphere.127 This idea was most clearly demonstrated in Ni catalysts,128 but was recently extended to those that contain Fe.129 The detailed mechanisms of H+ reduction and H2 oxidation are beginning to become clear.130 However as emphasized above, the key feature required for function is facile ET chemistry and the ready availability of a H+ donor/acceptor. The situation is recapitulated on a much larger scale in enzymes, notably the water channels (PT pathways) in laccase enzymes131 or cytochrome c oxidase.17,18
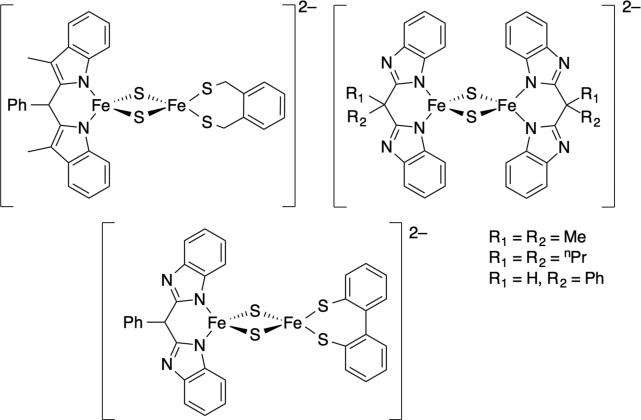
More recently, we132 and others133 reported the synthesis and PCET reactivity of small molecule models for Rieske iron-sulfur clusters (Figure 12). These models are inspired by the first structural and spectroscopic model133a (top left, Figure 12). Rieske proteins are involved with respiration and contain an unusual [2Fe-2S] cluster where one iron is ligated by two histidine ligands and the other by two cysteine ligands. The markedly pH dependent reduction potentials of the protein suggest that it can be involved in PCET reactions, where PT is mediated by the ligated histidine(s).134
Figure 12.
These Rieske models undergo CPET reactions with hydroxylamines and quinones (as models for natural redox partners). Both the homoleptic asymmetric models have about the same BDFE (60.2 kcal mol–1), which is similar to those derived from thermodynamics data for the native protein.134 However, the homoleptic complex undergoes CPET with TEMPO almost 50 times slower than the asymmetric complex, highlighting the importance of the contribution of ligands to differences in intrinsic reactivity. The facile ET and CPET reactivity of these clusters suggests that they have little bias for stepwise or concerted reactions.132b Such an observation is perfectly reasonable from a biological perspective; Rieske clusters react with quinones and quinols, the former usually preferring ET-PT and the latter favoring CPET, as discussed above.
4c. Manganese-containing models
The PCET chemistry of PSII and Mn-superoxide dismutase (MnSOD) were historical drivers for the awareness of the importance of PCET in enzyme active sites, and it has stimulated the development of many model systems. The model chemistry of Mn-oxo cores was especially prevalent during the time where PSII X-ray structures were disclosed with increasingly high resolution and different interpretations of the Mn4Ca Oxygen Evolving Complex (OEC). Even with an atomic resolution structure available,135 researchers are still trying to understand the discrete mechanism of water oxidation at the OEC, with improving X-ray methods,136 and correlating structural models with spectroscopy and reactivity.137
The simplest Mn-O containing PCET reagent is the permanganate anion (MnO4–), which has long been employed in organic oxidations.138 The ability of MnO4– to abstract H• from X-H bonds is a result of the combination of the MnO4–/2– reduction potential (0.564 V versus NHE) and the basicity of MnO42– (pKa = 7.4), yielding an aqueous BDFE of 80.7 kcal mol–1. The above thermodynamic parameters are both reasonably mild, but together they yield a PCET reagent with a strong affinity for H•.139 The late Jerry Babcock, who was a very early proponent of the importance of PCET in biological water oxidation and O2 reduction, was excited that this value was slightly below that for tyrosine (87.8 in water), supporting his hypothesis that YZO• was abstracting H• from a MnOH group.140 While this direct CPET mechanism is not supported by the high-resolution structure and other data, the focus on PCET thermochemistry that he and others emphasized is now a cornerstone of the field.
A great many SOD mimics are available, with many different metal sites (and even purely organic systems).141 Many of these compounds are of interest as drug candidates that can be used to address health problems related to oxidative stress and the associate production of reaction oxygen species.142 The classical SOD mechanism involves initial ET from MnII to O2– to give peroxide (O22–) bound to MnIII. Subsequently, MnIII reacts with a second equivalent of O2– to give MnII and O2. Studies of model complexes suggest that the first reaction could occur via PCET from MnII-OH2 or via an inner-sphere mechanism involving displacement of water by O2–.143 Detailed work on the native SOD enzymes suggests that PT and ET are closely related and are tightly controlled during turnover, and that diseases such as amyotrophic lateral sclerosis (ALS) can result if they are not tightly controlled.144 These observations, in combination with PCET models involving phenol and hydrogenase mimics (see above), suggest that more efficacious artificial SOD systems (e.g., drug candidates) would incorporate and Brønsted acid proximal to the active site.
4d. Other transition metal systems
The PCET chemistry of a wide variety of other metal complexes have been studied in detail. Ruthenium model complexes play a pre-eminent role, starting from the path breaking studies of Meyer and co-workers starting in the early 1980s and continuing to this day.21b Many systems with first-row transition metals have been developed, many of them quite biomimetic and directly relevant to specific metalloenzyme systems. The elegant work on copper systems in is particularly notable in this context.145
Workers have marched across the periodic table in their investigations of transition metal PCET. From vanadium-oxo complexes,146 Cr-peroxide complexes147 to second/third row metal hydrides,31 (to name only a few examples!) the basis PCET reactivity has strong parallels to biochemical and biomimetic systems: good-H-atom abstractors display both moderate reduction potentials and moderate acidity/basicity; observed PCET reactivity often depends strongly on the substrate (e.g., CPET preference for C-H bonds); and intrinsic barriers and reorganization associated with redox change are central to rationalizing reactivity.
5. Long-range PCET and Separated PCET
Electrons can tunnel many Ångstroms between cofactors. In contrast, H+ movement is limited to very short distances, < ~0.5 Å, often between donor and acceptor atoms separated by less than 3 Å in a well-defined hydrogen bond. To our knowledge, there are two reports of the PT distance dependence in PCET reactions that offer contrasting views;79 more work is needed to provide a comprehensive picture. The mismatch between ET and PT distance scales raises interesting questions about the nature of long-range PCET processes. For example, catalysis in RNR, PSII, and COX all involve transformation of YOH to YO• radical; the requisites ET reactions have at least 6 Å (edge-to-edge distances), while the distances between the H+ donor and acceptor atoms are 3 Å or less. Long-range ET in biological systems is understood using semi-classical theory,148 but it is still not clear what modifications are needed to rationalize PCET reactions where long-range ET is coupled to PT. We focus on the ET component of PCET distance dependence in this section as is an area of active research in many laboratories, on both biochemical and model systems.
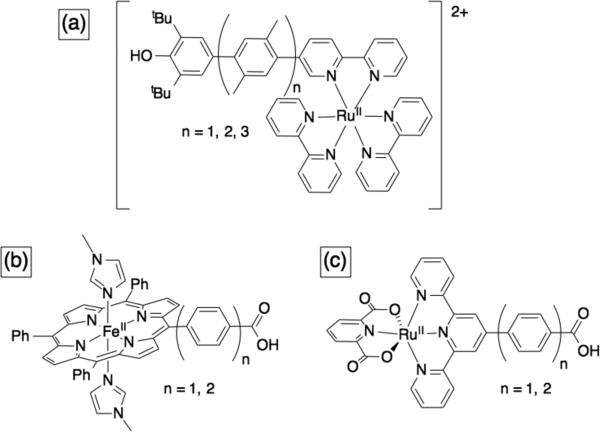
One example of a model system where long-range ET is coupled to short range PT are the indole and phenol appended Ru or Re complexes investigated by Hammarström and Nocera.70 Wenger and co-workers recently reported on the distance dependence of PCET reactions in Ru-phenol complexes;149 they used rigid xylene linkers to select defined separations between the Ru and the phenol (Figure 13a). They found that the distance decay constant (β) for CPET reactions involving phenol is similar to that for analogous ET reactions (β ~ 0.5 - 0.8 Å).150
Figure 13.
Examples of models used to investigate long-range PCET.150-151
We also investigated the ET distance dependence of CPET reactions, but using bimolecular reactions and organic H-donors with Fe-porphyrin151 or Ru-terpyridine152 complexes (Figure 13b and 13c respectively). We were not able to evaluate the distance decay constant (β) in the Ru complexes, but the observed CPET rates constants were slower with longer linkers, in accord with observations for ET reactions. With the help of computations, we evaluated β for CPET reactions of Fe-porphyrin-benzoates; β = 0.23 Å–1, which is in accord with β for ET though phenyl bridges (0.2 to 0.5 Å–1).
The above models start to bridge the conceptual gap between “inner-sphere” PCET (e.g., X-H activation by metal oxos) and purely separated or multiple-site PCET, where the redox and acid base moieties are distinct.32 In the stretched model systems above, the protonation state of the acid/base group still affects the reduction potential of the metal site, although only slightly. In the porphyrin models, for example, the protonation state of the benzoate shifts the FeIII/II potential by 30 mV (n = 1) or 15 mV (n = 2).151 This is an example of a “flat” square scheme, described above. In contrast, the protonation state of imidazole in related porphyrin models (Figure 11a), changes E1/2(FeIII/II) by 365 mV.89a
The coupling of thermodynamic parameters (E and pKa) is of central importance to PCET reactivity, but the biological mechanistic implications are not yet clear. As noted above, activation of C-H bonds requires a combination of oxidant and base (a strongly coupled system). Conversely, long-range PCET in enzymes, (e.g., RNR) are not as strongly coupled, yet holes are efficiently steered through several distinct steps involving the PCET cofactor YOH. An important challenge is understanding how these distant steps are coupled.
6. Frontiers for biomimetic and biochemical PCET
Having started this essay with Stiefel's 1973 paper on molybdenum enzymes, it is fitting to close with a portion of his concluding section:
“The coupled proton-electron transfer may be involved in several chemical processes, e.g., .... However, the chemical mechanism may be inferior to the enzymatic process due to the role of the protein. Thus, the protein will have the capacity to precisely orient the substrate and metal so that the coupled transfer can facilely occur.”
More than 40 years later this still is an excellent blueprint and challenge for the field. Much is understood about proton-coupled electron transfer processes. It is exciting to see the increasingly close interplay of biochemical, biomimetic and theoretical studies.
Synthetic models for metalloenzymes are increasingly controlling not only the coordination environment around the metal but also the second coordination sphere, including groups to position protons and facilitate their transfer.153 With substantial advances in the production of peptide-based models and modified metalloproteins, future biomimetic systems likely will more closely resemble actual biological systems. Recent examples include tyrosine-containing synthetic peptides,154 proteins,155 and peptide scaffolds on synthetic hydrogenase catalysts.156 Likewise, investigations of intermediates in proteins can draw a great deal of insight from small molecule models. For instance, the comprehensive characterization of cytochrome P450 Compound I and measurement of the pKa of Compound II was in part inspired by model systems showing the importance of the basicity of the reduced species for C–H bond activation.157 Remarkably, the distinction between biomimetic and biochemical has recently been blurred by the report that a biomimetic iron complex is taken up by apo-hydrogenase protein scaffold and the resulting protein is an active catalyst.158 Finally, PCET theory is showing how control of substrate orientation within an active site facilitates catalysis, and that both structure and dynamics of the active site are important.159 We are optimistic that the confluence of these thrusts will not only enrich our understanding of how biology efficiently accomplishes complex PCET processes, but also how valuable synthetic catalysts might be designed.
Acknowledgments
Funding Information
Work at the University of Washington and at Yale University has been supported primarily by a grant (to J.M.M.) from the U.S. National Institutes of Health (GM50422), with additional support from the U.S. National Science Foundation, and the two Universities listed above. This work was also in part supported as part of the Center for Molecular Electrocatalysis, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Basic Energy Sciences. Work at Simon Fraser University is supported by the Natural Sciences and Engineering Research Council of Canada (RGPIN05559) and the SFU President's Research Startup Fund (to J.J.W.).
Contributor Information
Jeffrey J. Warren, Simon Fraser University, Department of Chemistry, 8888 University Drive, Burnaby BC, Canada V5A 1S6.
James M. Mayer, Yale University, Department of Chemistry, P.O. Box 208107, 225 Prospect Street, New Haven, CT 06520-8107.
References
- 1.Orna Mary Virginia, Stock John. Electrochemistry, past and present. American Chemical Society; Columbus, Ohio: 1989. [Google Scholar]
- 2.Pourbaix M. Atlas of Electrochemical Equilibria in AqueousSolutions. 2nd English ed. National Association of Corrosion Engineers; Houston, TX: 1974. [Google Scholar]
- 3.a Kochi JK, editor. Free Radicals. Wiley; New York: 1973. [Google Scholar]; b Anslyn EV, Dougherty DA. Modern Physical Organic Chemistry University Science Books. Sausalito CA: 2006. [Google Scholar]
- 4.Wiberg KB, Evans RJ. Tetrahedron. 1960;8:313. [Google Scholar]
- 5.Eberson L, Wistrand L-G. Acta. Chem. Scand. 1980;B34:349–357. [Google Scholar]
- 6.Eberson L. J. Am.Chem. Soc. 1983;105:3192–3199. [Google Scholar]
- 7.Binstead RA, Moyer BA, Samuels GJ, Meyer TJ. J. Am. Chem. Soc. 1981;103:2897–2899. [Google Scholar]
- 8.For example: Proton-Coupled Electron Transfer Themed Issue, Chem. Rev. 2010;110:6937–7100. doi: 10.1021/cr100367q.Formosinho S, Barroso M, editors. Proton-coupled Electron Transfer: A Carrefour for Chemical Reactivity Traditions. RSC Publishing; Cambridge UK: 2012. Dempsey JL, Winkler JR, Gray HB. Chem. Rev. 2010;110:7024–7039. doi: 10.1021/cr100182b.
- 9.Marcus RA, Sutin N. Biochim. Biophys. Acta. 1985;811:265–322. [Google Scholar]
- 10.a Jiang W, Yun D, Saleh L, Barr EW, Xing G, Hoffart LM, Maslak M-A, Krebs C, Bollinger JM. Science. 2007;316:1188–1191. doi: 10.1126/science.1141179. [DOI] [PubMed] [Google Scholar]; b Jiang W, Saleh L, Barr EW, Xie J, Gardner MM, Krebs C, Bollinger JM. Biochemistry. 2008;47:8477–8484. doi: 10.1021/bi800881m. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Kwak Y, Jiang W, Dassama LMK, Park K, Bell CB, Liu LV, Wong SD, Saito M, Kobayashi Y, Kitao S, Seto M, Yoda Y, Alp EE, Zhao J, Bollinger JM, Krebs C, Solomon EI. J. Am. Chem. Soc. 2013;135:17573–17584. doi: 10.1021/ja409510d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uhlin U, Eklund H. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama K, Uhlin U, Stubbe J. J. Am. Chem. Soc. 2010;132:15368–15379. doi: 10.1021/ja1069344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoyama K, Uhlin U, Stubbe J. J. Am. Chem. Soc. 2010;132:8385–8397. doi: 10.1021/ja101097p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokoyama K, Smith AA, Corzilius B, Griffin RG, Stubbe J. J. Am. Chem. Soc. 2011;133:18420–18432. doi: 10.1021/ja207455k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minnihan EC, Seyedsayamdost MR, Uhlin U, Stubbe J. J. Am. Chem. Soc. 2011;133:9430–9440. doi: 10.1021/ja201640n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stubbe J. Curr. Opin. Chem. Biol. 2003;7:183–188. doi: 10.1016/s1367-5931(03)00025-5. [DOI] [PubMed] [Google Scholar]
- 17.Hosler JP, Ferguson-Miller S, Mills DA. Annu. Rev. Biochem. 2006;75:165–187. doi: 10.1146/annurev.biochem.75.062003.101730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a von Ballmoos C, Ädelroth P, Gennis RB, Brzezinski P. Biochim. Biophys. Acta. 2012;1817:650–657. doi: 10.1016/j.bbabio.2011.11.015. [DOI] [PubMed] [Google Scholar]; b Kaila VRI, Verkhovsky MI, Wikström M. Chem. Rev. 2010;110:7062–7081. doi: 10.1021/cr1002003. [DOI] [PubMed] [Google Scholar]; c Ishigami I, Hikita M, Egawa T, Yeh S-R, Rousseau DL. Biochim. Biophys. Acta. 2015;1847:98–108. doi: 10.1016/j.bbabio.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muresanu L, Pristovsek P, Löhr F, Maneg O, Mukrasch MD, Rüterjans H, Ludwig B, Lücke C. J. Biol. Chem. 2006;281:14503–14513. doi: 10.1074/jbc.M601108200. [DOI] [PubMed] [Google Scholar]
- 20.Muramoto K, Hirata K, Shinzawa-Itoh K, Yoko-o S, Yamashita E, Aoyama H, Tsukihara T, Yoshikawa S. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7881–7886. doi: 10.1073/pnas.0610031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a Warren JJ, Tronic TA, Mayer JM. Chem. Rev. 2010;110:6961–7001. doi: 10.1021/cr100085k. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Weinberg DR, Gagliardi CJ, Hull JF, Murphy CF, Kent CA, Westlake BC, Paul A, Ess DH, McCafferty DG, Meyer T. J. Chem. Rev. 2012;112:4016–4093. doi: 10.1021/cr200177j. [DOI] [PubMed] [Google Scholar]
- 22.The term CPET also has been used in a biochemical context. See: Njus D, Jalukar V, Zu J, Kelley PM. Am. J. Clin. Nutr. 1991;54:1179S–1183S. doi: 10.1093/ajcn/54.6.1179s.Jalukar V, Kelley PM, Njus D. J. Biol. Chem. 1991;11:6878–6882.Njus D, Kelley PM. Biochim. Biophys. Acta. 1993;1144:235–248. doi: 10.1016/0005-2728(93)90108-r.Njus D, Wigle M, Kelley PM, Kipp BH, Schlegel HB. Biochemistry. 2001;40:11905–11911. doi: 10.1021/bi010403r.
- 23.Sjödin M, Styring S, Wolpher H, Xu Y, Sun L, Hammarström L. J. Am. Chem. Soc. 2005;127:3855–3863. doi: 10.1021/ja044395o. [DOI] [PubMed] [Google Scholar]
- 24.a Mayer JM, Hrovat DA, Thomas JL, Borden WT. J. Am. Chem. Soc. 2002;124:11142–11147. doi: 10.1021/ja012732c. [DOI] [PubMed] [Google Scholar]; b Isborn C, Hrovat DA, Borden WT, Mayer JM, Carpenter BK. J. Am. Chem. Soc. 2005;127:5794–5795. doi: 10.1021/ja050024b. [DOI] [PubMed] [Google Scholar]; c Lingwood M, Hammond JR, Hrovat DA, Mayer JM, Borden WT. J. Chem. Theory Comput. 2006;2:740–745. doi: 10.1021/ct050282z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.a Tishchenko O, Truhlar DG, Ceulemans A, Nguyen MT. J. Am. Chem. Soc. 2008;130:7000–7010. doi: 10.1021/ja7102907. [DOI] [PubMed] [Google Scholar]; b Cembran A, Provorse MR, Wang C, Wu W, Gao J. J. Chem. Theory Comput. 2012;8:4347–4358. doi: 10.1021/ct3004595. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Hammes-Schiffer S. J. Phys. Chem. Lett. 2011;2:1410–1416. doi: 10.1021/jz101532g. [DOI] [PubMed] [Google Scholar]
- 26.Reece SY, Nocera DG. Annu. Rev. Biochem. 2009;78:673–699. doi: 10.1146/annurev.biochem.78.080207.092132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umena Y, Kawakami K, Shen J-R, Kamiya N. Nature. 2011;473:55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- 28.Waidmann CR, Miller AJM, Ng C-WA, Scheuermann ML, Porter TR, Tronic TA, Mayer JM. Energy Environ. Sci. 2012;5:7771–7780. [Google Scholar]
- 29.Bordwell FG, Bausch MJ. J. Am. Chem. Soc. 1986;108:1979–1985. doi: 10.1021/ja00269a071. [DOI] [PubMed] [Google Scholar]
- 30.a Mader EA, Manner VW, Markle TF, Wu A, Franz JA, Mayer JM. J. Am. Chem. Soc. 2009;131:4335–4345. doi: 10.1021/ja8081846. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Mader EA, Davidson ER, Mayer JM. J. Am. Chem. Soc. 2007;129:5153–5166. doi: 10.1021/ja0686918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tilset M. In: Electron Transfer in Chemistry. Balzani V, editor. Wiley-VCH; Weinheim: 2001. pp. 677–713. [Google Scholar]
- 32.Waidmann CR, Miller AJM, Ng C-WA, Scheuermann ML, Porter TR, Tronic TA, Mayer JM. Energy Environ. Sci. 2012;5:7771–7780. [Google Scholar]
- 33.An effective BDFE can be defined for the separated reagents of an MS-PCET reaction, even though no E-H bond is homolytically broken. This is the ΔG° for BH+ + X → B: + X+ + H•, and can be evaluated given the same equation (2).
- 34.An intermediate is typically defined as a species that is a minimum on the free energy surface and that lives for multiple vibrational periods.
- 35.a Roth JP, Lovell S, Mayer JM. J. Am. Chem. Soc. 2000;122:5486–5498. [Google Scholar]; b Yoder JC, Roth JP, Gussenhoven EM, Larsen AS, Mayer JM. J. Am. Chem. Soc. 2003;125:2629–2640. doi: 10.1021/ja0273905. [DOI] [PubMed] [Google Scholar]
- 36.a Song JS, Bullock RM, Creutz C. J. Am. Chem. Soc. 1991;113:9862–9864. [Google Scholar]; b Protasiewicz JD, Theopold KH. J. Am. Chem. Soc. 1993;115:5559–5569. [Google Scholar]; c Bullock RM. Comments Inorg. Chem. 1991;12:1–33. [Google Scholar]
- 37.Newton MD, Sutin N. Annu. Rev. Phys. Chem. 1984;35:437–480. [Google Scholar]
- 38.Beratan DN, Skourtis SS, Balabin IA, Balaeff A, Keinan S, Venkatramani R, Xiao D. Acc. Chem. Res. 2009;42:1669–1678. doi: 10.1021/ar900123t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.a Litwinienko G, Ingold KU. J. Org. Chem. 2003;68:3433–3438. doi: 10.1021/jo026917t. [DOI] [PubMed] [Google Scholar]; b Litwinienko G, Ingold KU. J. Org. Chem. 2004;69:5888–5896. doi: 10.1021/jo049254j. [DOI] [PubMed] [Google Scholar]; c Litwinienko G, Ingold KU. J. Org. Chem. 2005;70:8982–8990. doi: 10.1021/jo051474p. [DOI] [PubMed] [Google Scholar]; d Litwinienko G, Ingold KU. Acc. Chem. Res. 2007;40:222–230. doi: 10.1021/ar0682029. [DOI] [PubMed] [Google Scholar]
- 40.Warren JJ, Mayer JM. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5282–5287. doi: 10.1073/pnas.0910347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakanishi N, Takeuchi F, Tsubaki MJ. Biochem. 2007;142:553–560. doi: 10.1093/jb/mvm181. [DOI] [PubMed] [Google Scholar]
- 42.Warren JJ, Mayer JM. J. Am. Chem. Soc. 2010;132:7784–7793. doi: 10.1021/ja102337n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.a Rhile IJ, Markle TF, Nagao H, DiPasquale AG, Lam OP, Lockwood MA, Rotter K, Mayer JM. J. Am. Chem. Soc. 2006;128:6075–6088. doi: 10.1021/ja054167+. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Markle TF, Rhile IJ, DiPasquale AG, Mayer JM. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8185–8190. doi: 10.1073/pnas.0708967105. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Markle TF, Tronic TA, DiPasquale AG, Kaminsky W, Mayer JM. J. Phys. Chem. A. 2012;116:12249–12259. doi: 10.1021/jp311388n. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Sjödin M, Irebo T, Utas JE, Lind J, Merényi G, Åkermark B, Hammarström L. J. Am. Chem. Soc. 2006;128:13076–13083. doi: 10.1021/ja063264f. [DOI] [PubMed] [Google Scholar]; e Zhang M-T, Irebo T, Johansson O, Hammarström L. J. Am. Chem. Soc. 2011;133:13224–13227. doi: 10.1021/ja203483j. [DOI] [PubMed] [Google Scholar]; f Costentin C, Robert M, Savéant J-M. J. Am. Chem. Soc. 2007;129:9953–9963. doi: 10.1021/ja071150d. [DOI] [PubMed] [Google Scholar]
- 44.Costentin C, Hajj V, Robert M, Savéant J-M, Tard C. Proc. Natl. Acad. Sci. U.S.A. 2011;108:8559–8564. doi: 10.1073/pnas.1104952108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borovik AS. Acc. Chem. Res. 2005;38:54–61. doi: 10.1021/ar030160q. [DOI] [PubMed] [Google Scholar]
- 46.Collman JP, Fu L. Acc. Chem. Res. 1999;32:455–463. [Google Scholar]
- 47.Siegbahn PEM, Margareta RA, Blomberg MRA. Chem. Rev. 2010;110:7040–7061. doi: 10.1021/cr100070p. [DOI] [PubMed] [Google Scholar]
- 48.For reviews see [Google Scholar]; a Hammes-Schiffer S, Soudackov AV. J. Phys. Chem. B. 2008;112:14108–14123. doi: 10.1021/jp805876e. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hammes-Schiffer S, Stuchebrukhov AA. Chem. Rev. 2010;110:6939–6960. doi: 10.1021/cr1001436. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Tishchenko O, Truhlar DG, Ceulemans A, Nguyen MT. J. Am. Chem. Soc. 2008;130:7000–7010. doi: 10.1021/ja7102907. [DOI] [PubMed] [Google Scholar]
- 49.Cukier RI, Nocera DG. Annu. Rev. Phys. Chem. 1998;49:337–369. doi: 10.1146/annurev.physchem.49.1.337. [DOI] [PubMed] [Google Scholar]
- 50.Jackson RA, O'Neill DW. J. Chem. Soc., Chem. Comm. 1969:1210–1211. [Google Scholar]
- 51.Foti M, Ingold KU, Lusztyk J. J. Am. Chem. Soc. 1994;116:9440–9447. This paper reports k = 4.5 × 106 M−1 s−1 for phenoxyl + β-naphthol, which is a little more exoergic than the self-exchange PhO• + PhOH.
- 52.Kochi JK. Electron-Transfer Oxidation. In: Trost BM, editor. Comprehensive Organic Synthesis. Pergamon; New York: 1991. [Google Scholar]
- 53.Matsuo T, Mayer JM. Inorg. Chem. 2005;44:2150–2158. doi: 10.1021/ic048170q. [DOI] [PubMed] [Google Scholar]
- 54.Hirst J. Annu. Rev. Biochem. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- 55.a DuBois DL, Berning DE. Appl. Organometal. Chem. 2000;14:860–862. [Google Scholar]; b Ellis WW, Raebiger JW, Curtis CJ, Bruno JW, DuBois DL. J. Am. Chem. Soc. 2004;126:2738–2743. doi: 10.1021/ja038567d. [DOI] [PubMed] [Google Scholar]
- 56.a Cheng J-P, Lu Y. J. Phys. Org. Chem. 1997;10:577–584. [Google Scholar]; b Cheng J-P, Lu Y, Zhu X, Mu L. J. Org. Chem. 1998;63:6108–6114. doi: 10.1021/jo9715985. [DOI] [PubMed] [Google Scholar]
- 57.a Farrington JA, Land EJ, Swallow A. J. Biochim. Biophys. Acta. 1980;590:273. doi: 10.1016/0005-2728(80)90031-6. [DOI] [PubMed] [Google Scholar]; b Carlson BW, Miller LL, Neta P, Grodkowski J. J. Am. Chem. Soc. 1984;106:7233. [Google Scholar]; c Zhu X-C, Yang Y, Zhang M, Cheng J-P. J. Am. Chem. Soc. 2003;125:15298. doi: 10.1021/ja0385179. [DOI] [PubMed] [Google Scholar]
- 58.Matsuo T, Mayer JM. Inorg. Chem. 2005;44:2150–2158. doi: 10.1021/ic048170q. references therein. [DOI] [PubMed] [Google Scholar]
- 59.Minnihan EC, Nocera DG, Stubbe J. Acc. Chem. Res. 2013;46:2524–2535. doi: 10.1021/ar4000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.a McEvoy JP, Brudvig GW. Chem. Rev. 2006;106:4455–4483. doi: 10.1021/cr0204294. [DOI] [PubMed] [Google Scholar]; b Siegbahn PEM. Dalton Trans. 2009;45:10063–10068. doi: 10.1039/b909470a. [DOI] [PubMed] [Google Scholar]
- 61.Smith WL, Urade Y, Jakobsson P-J. Chem. Rev. 2011;111:5821–5865. doi: 10.1021/cr2002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.a Giulivi C, Traaseth NJ, Davies KJA. Amino Acids. 2003;25:227–232. doi: 10.1007/s00726-003-0013-0. [DOI] [PubMed] [Google Scholar]; b Heinecke JW. Free Radical Bio. Med. 2002;32:1090–1101. doi: 10.1016/s0891-5849(02)00792-x. [DOI] [PubMed] [Google Scholar]
- 63.Bordwell FG, Cheng J. J. Am. Chem. Soc. 1991;113:1736–1743. [Google Scholar]
- 64.a Lind J, Shen X, Eriksen TE, Merenyi G. J. Am. Chem. Soc. 1990;112:479–482. [Google Scholar]; b Steenken S, Neta P. J. Phys. Chem. 1982;86:3661–3667. [Google Scholar]
- 65.For an extensive tabulation of reaction rates for phenoxyl radicals see: Neta P, Grodkowski J. J. Phys. Chem. Ref. Data. 2005;34:109–199.
- 66.a Bronner C, Wenger OS. Phys. Chem. Chem. Phys. 2014;16:3617–3622. doi: 10.1039/c3cp55071k. [DOI] [PubMed] [Google Scholar]; b Irebo T, Zhang M-T, Markle TF, Scott AM, Hammarström L. J. Am. Chem. Soc. 2012;134:16247–16254. doi: 10.1021/ja3053859. [DOI] [PubMed] [Google Scholar]
- 67.a Song N, Stanbury DM. Inorg. Chem. 2008;47:11458–11460. doi: 10.1021/ic8015595. [DOI] [PubMed] [Google Scholar]; b Song N, Stanbury DM. Inorg. Chem. 2012;51:4909–4911. doi: 10.1021/ic300562u. [DOI] [PubMed] [Google Scholar]; c Osako T, Ohkubo K, Taki M, Tachi Y, Fukuzumi S, Itoh S. J. Am. Chem. Soc. 2003;125:11027–11033. doi: 10.1021/ja029380+. [DOI] [PubMed] [Google Scholar]
- 68.a Costentin C, Louault C, Robert M, Savéant J-M. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18143–18148. doi: 10.1073/pnas.0910065106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Fecenko CJ, Thorp HH, Meyer TJ. J. Am. Chem. Soc. 2007;129:15098–15099. doi: 10.1021/ja072558d. [DOI] [PubMed] [Google Scholar]; c Fecenko CJ, Meyer TJ, Thorp HH. J. Am. Chem. Soc. 2006;128:11020–11021. doi: 10.1021/ja061931z. [DOI] [PubMed] [Google Scholar]
- 69.a Bonin J, Costentin C, Louault C, Robert M, Routier M, Savéant J-M. Proc. Natl. Acad. Sci. U.S.A. 2010;107:3367–3372. doi: 10.1073/pnas.0914693107. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Glover SD, Jorge C, Liang L, Valentine KG, Hammarström L, Tommos C. J. Am. Chem. Soc. 2014;136:14039–14051. doi: 10.1021/ja503348d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.a Reece SY, Nocera DG. J. Am. Chem. Soc. 2005;127:9448–9458. doi: 10.1021/ja0510360. [DOI] [PubMed] [Google Scholar]; b Sjödin M, Styring S, Kermark B, Sun L, Hammarström L. J. Am. Chem. Soc. 2000;122:3932–3936. [Google Scholar]; c Irebo T, Reece SY, Sjoedin M, Nocera DG, Hammarström L. J. Am. Chem. Soc. 2007;129:15462–15464. doi: 10.1021/ja073012u. [DOI] [PubMed] [Google Scholar]
- 71.a Cape JL, Bowman MK, Kramer DM. J. Am. Chem. Soc. 2005;127:4208–4215. doi: 10.1021/ja043955g. [DOI] [PubMed] [Google Scholar]; b Westlake BC, Brennaman MK, Concepcion JJ, Paul JJ, Bettis SE, Hampton SD, Miller SA, Lebedeva NV, Forbes MDE, Moran AM, Meyer TJ, Papanikolas JM. Proc. Natl. Acad. Sci. U.S.A. 2011;108:8554–8558. doi: 10.1073/pnas.1104811108. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Eisenhart TT, Dempsey JL. J. Am. Chem. Soc. 2014;136:12221–12224. doi: 10.1021/ja505755k. [DOI] [PubMed] [Google Scholar]; d Wenger OS. Coord. Chem. Rev. 2015;282-283:150–158. [Google Scholar]; e Chen J, Kuss-Petermann M, Wenger OS. J. Phys. Chem. B, Article ASAP. DOI: 10.1021/jp506087t. [Google Scholar]; f Gagliardi CJ, Westlake BC, Kent CA, Paul JJ, Papanikolas JM, Meyer TJ. Coord. Chem. Rev. 2010;254:2459–2471. [Google Scholar]; g Lebedeva NV, Schmidt RD, Concepcion JJ, Brennaman MK, Stanton IN, Therien MJ, Meyer TJ, Forbes MDE. J. Phys. Chem. A. 2011;115:3346–3356. doi: 10.1021/jp200381n. [DOI] [PubMed] [Google Scholar]
- 72.a Gupta N, Linschitz H, Biczok L. Fullerene Sci. Technol. 1997;5:343–353. [Google Scholar]; b Biczok L, Gupta N, Linschitz H. J. Am. Chem. Soc. 1997;119:12601–12609. [Google Scholar]
- 73.Maki T, Araki Y, Ishida Y, Onomura O, Matsumura Y. J. Am. Chem. Soc. 2001;123:3371–3372. doi: 10.1021/ja002453+.Benisvy L, Blake AJ, Collison D, Davies ES, Garner CD, McInnes EJL, McMaster J, Whittaker G, Wilson C. J. Chem. Soc., Dalton Trans. 2003:1975–1985.Benisvy L, Bill E, Blake AJ, Collison D, Davies ES, Garner CD, Guindy CI, McInnes EJL, McArdle G, McMaster J, Wilson C, Wolowska J. Dalton Trans. 2004:3647–3653. doi: 10.1039/b410934a. d References 75,76,78,79.
- 74.Stubbe J, Nocera DG, Yee CS, Chang MCY. Chem. Rev. 2003;103:2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 75.a Rhile IJ, Markle TF, Nagao H, DiPasquale AG, Lam OP, Lockwood MA, Rotter K, Mayer JM. J. Am. Chem. Soc. 2006;128:6075–6088. doi: 10.1021/ja054167+. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Schrauben JN, Cattaneo M, Day TC, Tenderholt AL, Mayer JM. J. Am. Chem. Soc. 2012;134:16635–16645. doi: 10.1021/ja305668h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Markle TF, Tronic TA, DiPasquale AG, Kaminsky W, Mayer JM. J. Phys. Chem. A. 2012;116:12249–12259. doi: 10.1021/jp311388n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bonin J, Costentin C, Robert M, Savéant J-M. Org. Biomol. Chem. 2011;9:4064–4069. doi: 10.1039/c1ob05090g. [DOI] [PubMed] [Google Scholar]
- 78.a Markle TF, Rhile IJ, DiPasquale AG, Mayer JM. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8185–8190. doi: 10.1073/pnas.0708967105. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Markle TF, Mayer JM. Angew. Chem. (Int. Ed.) 2008;47:738–740. doi: 10.1002/anie.200702486. [DOI] [PubMed] [Google Scholar]
- 79.a Markle TF, Rhile IJ, Mayer JM. J. Am. Chem. Soc. 2011;133:17341–17352. doi: 10.1021/ja2056853. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Zhang M-T, Irebo T, Johansson O, Hammarström L. J. Am. Chem. Soc. 2011;133:13224–13227. doi: 10.1021/ja203483j. [DOI] [PubMed] [Google Scholar]
- 80.a Johannissen LO, Irebo T, Sjödin M, Johansson O, Hammarström L. J. Phys. Chem. B. 2009;113:16214–16225. doi: 10.1021/jp9048633. [DOI] [PubMed] [Google Scholar]; b Markle TF, Tenderholt AL, Mayer JM. J. Phys. Chem. B. 2011;116:571–584. doi: 10.1021/jp2091736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rappaport F, Diner BA. Coord. Chem. Rev. 2008;252:259–272. [Google Scholar]
- 82.a Costentin C. Chem. Rev. 2008;108:2145–2179. doi: 10.1021/cr068065t. [DOI] [PubMed] [Google Scholar]; b Costentin C, Robert M, Savéant J-M. Chem. Rev. 2010;110:PR1–PR40. doi: 10.1021/cr100038y. [DOI] [PubMed] [Google Scholar]
- 83.Wardman P. J. Phys. Chem. Ref. Data. 1989;18:1637–1755. [Google Scholar]
- 84.Song N, Gagliardi CJ, Binstead RA, Zhang M-T, Thorp H, Meyer TJ. J. Am. Chem. Soc. 2012;134:18538–18541. doi: 10.1021/ja308700t. [DOI] [PubMed] [Google Scholar]
- 85.Cheng J-P, Lu Y, Zhu X, Mu L. J. Org. Chem. 1998;63:6108–6114. doi: 10.1021/jo9715985. [DOI] [PubMed] [Google Scholar]
- 86.Creutz C. Inorg. Chem. 1981;20:4449–4452. [Google Scholar]
- 87.Macartney DH, Sutin N. Inorg. Chim. Acta. 1983;74:221–228. [Google Scholar]
- 88.Warren JJ, Mayer JM. J. Am. Chem. Soc. 2008;130:7546–7547. doi: 10.1021/ja802055t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.a Warren JJ, Mayer JM. J. Am. Chem. Soc. 2008;130:2774–2776. doi: 10.1021/ja711057t. [DOI] [PubMed] [Google Scholar]; b Warren JJ, Mayer JM. J. Am. Chem. Soc. 2011;133:8544–8551. doi: 10.1021/ja201663p. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Warren JJ, Menzeleev AR, Kretchmer JS, Miller TF, Gray HB, Mayer JM. J. Phys. Chem. Lett. 2013;4:519–523. doi: 10.1021/jz400029w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.a Brigelius-Flohé R, Traber MG. FASEB J. 1999;13:1145–1155. [PubMed] [Google Scholar]; b Traber MG, Atkinson J. Free Radical Bio. Med. 2007;43:4–15. doi: 10.1016/j.freeradbiomed.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burton GW, Ingold KU. Acc. Chem. Res. 1986;19:194–201. [Google Scholar]
- 92.Bisby RH, Parker AW. Arch. Biochem. Biophys. 1995;317:170–178. doi: 10.1006/abbi.1995.1150. [DOI] [PubMed] [Google Scholar]
- 93.a Fitzpatrick PF. Annu. Rev. Biochem. 1999;68:355–381. doi: 10.1146/annurev.biochem.68.1.355. [DOI] [PubMed] [Google Scholar]; b Massey V. J. Biol. Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- 94.a Mayhew SG. Eur. J. Biochem. 1999;265:698–702. doi: 10.1046/j.1432-1327.1999.00767.x. [DOI] [PubMed] [Google Scholar]; b Anderson RF. Biochim. Biophys. Acta. 1983;722:158–162. doi: 10.1016/0005-2728(83)90169-x. [DOI] [PubMed] [Google Scholar]
- 95.e.g., for the Old Yellow enzyme. Toogood HS, Gardiner JM, Scrutton NS. ChemCatChem. 2010;2:892–914.
- 96.a Massey V. J. Biol. Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]; b Fitzpatrick PF. Annu. Rev. Biochem. 1999;68:355–381. doi: 10.1146/annurev.biochem.68.1.355. [DOI] [PubMed] [Google Scholar]
- 97.a Roth JP, Klinman JP. Proc. Natl. Acad. Sci. U.S.A. 2003;100:62–67. doi: 10.1073/pnas.252644599. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Roth JP, Wincek R, Nodet G, Edmondson DE, McIntire WS, Klinman JP. J. Am. Chem. Soc. 2004;126:15120–15131. doi: 10.1021/ja047050e. [DOI] [PubMed] [Google Scholar]; c Brinkley DW, Roth JP. J. Am. Chem. Soc. 2005;127:15720–15721. doi: 10.1021/ja056025l. [DOI] [PubMed] [Google Scholar]
- 98.Gas phase thermochemistry. http://webbook.nist.gov/chemistry/
- 99.a Costentin C, Robert M, Savéant J-M. Chem. Soc. Rev. 2013;42:2423–2436. doi: 10.1039/c2cs35360a. [DOI] [PubMed] [Google Scholar]; b Grice KA, Kubiak CP. Adv. Inorg. Chem. 2014;66:163–188. [Google Scholar]
- 100.a Schrock RR. Acc. Chem. Res. 2005;38:955–962. doi: 10.1021/ar0501121. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Anderson JS, Rittle J, Peters JC. Nature. 2013;501:84–87. doi: 10.1038/nature12435. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Hoffman BM, Lukoyanov D, Dean DR, Seefeldt LC. Acc. Chem. Res. 2013;46:587–595. doi: 10.1021/ar300267m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.a Collman JP, Boulatov R, Sunderland CJ, Fu L. Chem. Rev. 2004;104:561–588. doi: 10.1021/cr0206059. [DOI] [PubMed] [Google Scholar]; b Vasudevan P, Santosh, Mann N, Tyagi S. Transition Met. Chem. 1990;15:81–90. [Google Scholar]; c Dogutan DK, Stoian SA, McGuire R, Schwalbe M, Teets TS, Nocera DG. J. Am. Chem. Soc. 2011;133:131–140. doi: 10.1021/ja108904s. [DOI] [PubMed] [Google Scholar]; d Carver CT, Matson BD, Mayer JM. J. Am. Chem. Soc. 2012;134:5444–5447. doi: 10.1021/ja211987f. [DOI] [PubMed] [Google Scholar]; e Matson BD, Carver CT, Von Ruden A, Yang JY, Raugei S, Mayer JM. Chem. Commun. 2012;48:11100–11102. doi: 10.1039/c2cc35576k. [DOI] [PubMed] [Google Scholar]
- 102.Groves JT, Haushalter RC, Nakamura M, Nemo TE, Evans BJ. J. Am. Chem. Soc. 1981;103:2884–2886. [Google Scholar]
- 103.Groves JT. J. Inorg. Biochem. 2006;100:434–447. doi: 10.1016/j.jinorgbio.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 104.a Rittle J, Green MT. Science. 2010;330:933–937. doi: 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]; b Yosca TH, Rittle J, Krest CM, Onderko EL, Silakov A, Calixto JC, Behan RK, Green MT. Science. 2013;342:825–829. doi: 10.1126/science.1244373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.a Guengerich FP. Chem. Res. Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]; b Sono M, Roach MP, Coulter ED, Dawson JH. Chem. Rev. 1996;96:2841–2887. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 106.a Kirby JP, van D, Niels A, Chang CK, Nocera DG. Tet. Lett. 1995;36:3477–3480. [Google Scholar]; b Young ER, Rosenthal J, Hodgkiss JM, Nocera DG. J. Am. Chem. Soc. 2009;131:7678–7684. doi: 10.1021/ja809777j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Warren JJ, Mayer JM. J. Am. Chem. Soc. 2008;130:2774–2776. doi: 10.1021/ja711057t. [DOI] [PubMed] [Google Scholar]
- 108.Warren JJ. Ph.D. Dissertation. University of Washington; 2010. [Google Scholar]
- 109.Reid LS, Mauk MR, Mauk AG. J. Am. Chem. Soc. 1984;106:2182–2185. [Google Scholar]
- 110.Costentin C, Drouet S, Robert M, Savéant J-M. Science. 2012;338:90–94. doi: 10.1126/science.1224581. [DOI] [PubMed] [Google Scholar]
- 111.a McDonald AR, Que J, Lawrence Coord. Chem. Rev. 2013;257:414–428. [Google Scholar]; b Collins TJ. Acc. Chem. Res. 2002;35:782–790. doi: 10.1021/ar010079s. [DOI] [PubMed] [Google Scholar]; c Decker A, Clay MD, Solomon EI. J. Inorg. Biochem. 2006;100:697–706. doi: 10.1016/j.jinorgbio.2006.01.013. [DOI] [PubMed] [Google Scholar]; d Nam W. Acc. Chem. Res. 2007;40:522–531. doi: 10.1021/ar700027f. [DOI] [PubMed] [Google Scholar]
- 112.Jonas RT, Stack TDP. J. Am. Chem. Soc. 1997;119:8566–8567. [Google Scholar]
- 113.a Roth JP, Mayer JM. Inorg. Chem. 1999;38:2760–2761. doi: 10.1021/ic990251c. [DOI] [PubMed] [Google Scholar]; b Roth JP, Lovell S, Mayer JM. J. Am. Chem. Soc. 2000;122:5486–5498. [Google Scholar]; c Yoder JC, Roth JP, Gussenhoven EM, Larsen AS, Mayer JM. J. Am. Chem. Soc. 2003;125:2629–2640. doi: 10.1021/ja0273905. [DOI] [PubMed] [Google Scholar]
- 114.Manner VW, Markle TF, Freudenthal JH, Roth JP, Mayer JM. Chem. Commun. 2008:256–258. doi: 10.1039/b712872j. [DOI] [PubMed] [Google Scholar]
- 115.a Roth JP, Yoder JC, Won T-J, Mayer JM. Science. 2001;294:2524–2526. doi: 10.1126/science.1066130. [DOI] [PubMed] [Google Scholar]; b Mader EA, Larsen AS, Mayer JM. J. Am. Chem. Soc. 2004;126:8066–8067. doi: 10.1021/ja049246k. [DOI] [PubMed] [Google Scholar]
- 116.Warren JJ, Mayer JM. Proton-Coupled Electron Transfer: A Carrefour of Chemical Reactivity Traditions. 2012:1–31. [Google Scholar]
- 117.a Mader EA, Davidson ER, Mayer JM. J. Am. Chem. Soc. 2007;129:5153–5166. doi: 10.1021/ja0686918. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Mader EA, Manner VW, Markle TF, Wu A, Franz JA, Mayer JM. J. Am. Chem. Soc. 2009;131:4335–4345. doi: 10.1021/ja8081846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Abu-Omar MM, Loaiza A, Hontzeas N. Chem. Rev. 2005;105:2227–2252. doi: 10.1021/cr040653o. [DOI] [PubMed] [Google Scholar]
- 119.e.g., in bleomycin. Burger RM. Chem. Rev. 1998;98:1153–1170. doi: 10.1021/cr960438a.
- 120.Solomon EI, Decker A, Lehnert N. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3589–3594. doi: 10.1073/pnas.0336792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Costas M, Mehn MP, Jensen MP, Que L., Jr. Chem. Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 122.de Oliveira FT, Chanda A, Banerjee D, Shan X, Mondal S, Que L, Bominaar EL, Münck E, Collins TJ. Science. 2007;315:835–838. doi: 10.1126/science.1133417. [DOI] [PubMed] [Google Scholar]
- 123.Park J, Morimoto Y, Lee Y-M, Nam W, Fukuzumi S. Inorg. Chem. 2014;53:3618–3628. doi: 10.1021/ic403124u. [DOI] [PubMed] [Google Scholar]
- 124.a Wang D, Ray K, Collins MJ, Farquhar ER, Frisch JR, Gomez L, Jackson TA, Kerscher M, Waleska A, Comba P, Costas M, Que L. Chem. Sci. 2013;4:282–291. doi: 10.1039/C2SC21318D. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Que L. Acc. Chem. Res. 2007;40:493–500. doi: 10.1021/ar700024g. [DOI] [PubMed] [Google Scholar]
- 125.Venkateswara R, P., Holm RH. Chem. Rev. 2003;104:527–560. [Google Scholar]
- 126.For recent reviews, see: Gloaguen F, Rauchfuss TB. Chem. Soc. Rev. 2009;38:100–108. doi: 10.1039/b801796b.DuBois DL, Bullock RM. European Journal of Inorganic Chemistry Eur. J. Inorg. Chem. 2011;2011:1017–1027.
- 127.Ginovska-Pangovska B, Dutta A, Reback ML, Linehan JC, Shaw WJ. Acc. Chem. Res. 2014;47:2621–2630. doi: 10.1021/ar5001742. [DOI] [PubMed] [Google Scholar]
- 128.Helm ML, Stewart MP, Bullock RM, DuBois MR, DuBois DL. Science. 2011;333:863–866. doi: 10.1126/science.1205864. [DOI] [PubMed] [Google Scholar]
- 129.Liu T, DuBois DL, Bullock RM. Nat Chem. 2013;5:228–233. doi: 10.1038/nchem.1571. [DOI] [PubMed] [Google Scholar]
- 130.a Chen S, Ho M-H, Bullock RM, DuBois DL, Dupuis M, Rousseau R, Raugei S. ACS Catal. 2014;4:229–242. [Google Scholar]; b Wiese S, Kilgore UJ, Ho M-H, Raugei SR, DuBois DL, Bullock RM, Helm ML. ACS Catal. 2013;3:2527–2535. [Google Scholar]; c Amélie Kochem A, Neese F, van Gastel M. J. Phys. Chem. C. 2014;118:2350–2360. [Google Scholar]
- 131.a Solomon EI, Augustine AJ, Yoon J. Dalton Trans. 2008:3921–3932. doi: 10.1039/b800799c. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Bento I, Silva C, Chen Z, Martins L, Lindley P, Soares C. BMC Structural Biology. 2010;10:28–42. doi: 10.1186/1472-6807-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.a Saouma CT, Kaminsky W, Mayer JM. J. Am. Chem. Soc. 2012;134:7293–7296. doi: 10.1021/ja3019324. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Saouma CT, Pinney MM, Mayer JM. Inorg. Chem. 2014;53:3153–3161. doi: 10.1021/ic403131p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.a Ballmann J, Albers A, Demeshko S, Dechert S, Bill E, Bothe E, Ryde U, Meyer F. Angew. Chem. Int. Ed. 2008;47:9537–9541. doi: 10.1002/anie.200803418. [DOI] [PubMed] [Google Scholar]; b Albers A, Bayer T, Demeshko S, Dechert S, Meyer F. Chem. Eur. J. 2013;19:10101–10106. doi: 10.1002/chem.201301760. [DOI] [PubMed] [Google Scholar]; c Albers A, Demeshko S, Dechert S, Saouma CT, Mayer JM, Meyer F. J. Am. Chem. Soc. 2014;136:3946–3954. doi: 10.1021/ja412449v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.a Zu Y, Fee JA, Hirst J. J. Am. Chem. Soc. 2001;123:9906–9907. doi: 10.1021/ja016532c. [DOI] [PubMed] [Google Scholar]; b Zu Y, Couture MM-J, Kolling DRJ, Crofts AR, Eltis LD, Fee JA, Hirst J. Biochemistry. 2003;42:12400–12408. doi: 10.1021/bi0350957. [DOI] [PubMed] [Google Scholar]
- 135.Umena Y, Kawakami K, Shen J-R, Kamiya N. Nature. 2011;473:55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- 136.Kern J, Alonso-Mori R, Tran R, Hattne J, Gildea RJ, Echols N, Gl√∂ckner C, Hellmich J, Laksmono H, Sierra RG, Lassalle-Kaiser B, Koroidov S, Lampe A, Han G, Gul S, DiFiore D, Milathianaki D, Fry AR, Miahnahri A, Schafer DW, Messerschmidt M, Seibert MM, Koglin JE, Sokaras D, Weng T-C, Sellberg J, Latimer MJ, Grosse-Kunstleve RW, Zwart PH, White WE, Glatzel P, Adams PD, Bogan MJ, Williams GJ, Boutet S, Messinger J, Zouni A, Sauter NK, Yachandra VK, Bergmann U, Yano J. Science. 2013;340:491–495. doi: 10.1126/science.1234273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.a Pokhrel R, Brudvig GW. Phys. Chem. Chem. Phys. 2014;16:11812–11821. doi: 10.1039/c4cp00493k. [DOI] [PubMed] [Google Scholar]; b Tsui EY, Tran R, Yano J, Agapie T. Nature Chem. 2013;5:293–299. doi: 10.1038/nchem.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.March J. Advanced Organic Chemistry. 3rd ed. Wiley-Interscience; New York: 1985. [Google Scholar]
- 139.Gardner KA, Kuehnert LL, Mayer JM. Inorg. Chem. 1997;36:2069–2078. doi: 10.1021/ic961297y. [DOI] [PubMed] [Google Scholar]
- 140.Yocum CF, Blankenship RE, Ferguson-Miller S. Dedication/Perspective in Photosystem II: The light-driven water:plastoquinone oxidoreductase Adv. Photosynth. Resp. 2005;22:1–10. [Google Scholar]
- 141.Batinic-Haberle I, Reboucas JS, Spasojevic I. Antioxid. Redox. Signal. 2010;13:877–918. doi: 10.1089/ars.2009.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. Oxford University Press; Oxford: 2007. [Google Scholar]
- 143.Riley DP, Lennon PJ, Neumann WL, Weiss RH. J. Am. Chem. Soc. 1997;119:6522–6528. [Google Scholar]
- 144.a Yikilmaz E, Porta J, Grove LE, Vahedi-Faridi A, Bronshteyn Y, Brunold TC, Borgstahl GEO, Miller A-F. J. Am. Chem. Soc. 2007;129:9927–9940. doi: 10.1021/ja069224t. [DOI] [PubMed] [Google Scholar]; b Grove LE, Brunold TC. Comm. Inorg. Chem. 2008;29:134–168. [Google Scholar]
- 145.a Cramer CJ, Tolman WB. Acc. Chem. Res. 2007;40:601–608. doi: 10.1021/ar700008c. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kim E, Chufán EE, Kamaraj K, Karlin KD. Chem. Rev. 2004;104:1077–1134. doi: 10.1021/cr0206162. [DOI] [PubMed] [Google Scholar]; c Liang H–C, Dahan M, Karlin KD. Curr. Op. Chem. Biol. 1999;3:168–175. doi: 10.1016/S1367-5931(99)80029-5. [DOI] [PubMed] [Google Scholar]; d Lyons CT, Stack TDP. Coord. Chem. Rev. 2013;257:528–540. doi: 10.1016/j.ccr.2012.06.003. (e) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Waidmann CR, Zhou X, Tsai EA, Kaminsky W, Hrovat DA, Borden WT, Mayer JM. J. Am. Chem. Soc. 2009;131:4729–4743. doi: 10.1021/ja808698x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bakac A. J. Am. Chem. Soc. 2000;122:1092–1097. [Google Scholar]
- 148.Gray HB, Winkler JR. Q. Rev. Biophys. 2003;36:341–372. doi: 10.1017/s0033583503003913. [DOI] [PubMed] [Google Scholar]
- 149.a Kuss-Petermann M, Wolf H, Stalke D, Wenger OS. J. Am. Chem. Soc. 2012;134:12844–12854. doi: 10.1021/ja3053046. [DOI] [PubMed] [Google Scholar]; b Kuss-Petermann M, Wenger OS. J. Phys. Chem. A. 2013;117:5726–5733. doi: 10.1021/jp402567m. [DOI] [PubMed] [Google Scholar]; c Chen J, Kuss-Petermann M, Wenger OS. Chem. Eur. J. 2014;20:4098–4104. doi: 10.1002/chem.201304256. [DOI] [PubMed] [Google Scholar]
- 150.Wenger OS. Chem. Soc. Rev. 2011;40:3538–3550. doi: 10.1039/c1cs15044h. [DOI] [PubMed] [Google Scholar]
- 151.Warren JJ, Menzeleev AR, Kretchmer JS, Miller TF, Gray HB, Mayer JM. J. Phys. Chem. Lett. 2013;4:519–523. doi: 10.1021/jz400029w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.a Manner VW, Dipasquale AG, Mayer JM. J. Am. Chem. Soc. 2008;130:7210–7211. doi: 10.1021/ja801672w. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Manner VW, Mayer JM. J. Am. Chem. Soc. 2009;131:9874–9875. doi: 10.1021/ja902942g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.e.g., a References 45, 101, 110. DuBois DL, Bullock RM. Eur. J. Inorg. Chem. 2011;2011:1017–1027.Costentin C, Robert M, Savéant J-M, Tard C. Acc. Chem. Res. 2013;47:271–280. doi: 10.1021/ar4001444.
- 154.a Pagba CV, Barry BA. J. Phys. Chem. B. 2012;116:10590–10599. doi: 10.1021/jp303607b. [DOI] [PubMed] [Google Scholar]; b Sibert RS, Josowicz M, Barry BA. ACS Chem. Biol. 2010;5:1157–1168. doi: 10.1021/cb100138m. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Sibert R, Josowicz M, Porcelli F, Veglia G, Range K, Barry BA. J. Am. Chem. Soc. 2007;129:4393–4400. doi: 10.1021/ja068805f. [DOI] [PubMed] [Google Scholar]
- 155.a Berry BW, Martínez-Rivera MC, Tommos C. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9739–9743. doi: 10.1073/pnas.1112057109. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Martínez-Rivera MC, Berry BW, Valentine KG, Westerlund K, Hay S, Tommos C. J. Am. Chem. Soc. 2011;133:17786–17795. doi: 10.1021/ja206876h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ginovska-Pangovska B, Dutta A, Reback ML, Linehan JC, Shaw WJ. Acc. Chem. Res. 2014;47:2621–2630. doi: 10.1021/ar5001742. [DOI] [PubMed] [Google Scholar]
- 157.Green MT, Dawson JH, Gray HB. Science. 2004;304:1653–1656. doi: 10.1126/science.1096897. [DOI] [PubMed] [Google Scholar]
- 158.Esselborn J, Lambertz C, Adamska-Venkatesh A, Simmons T, Berggren G, Noth J, Siebel J, Hemschemeier A, Artero V, Reijerse E, Fontecave M, Lubitz W, Happe T. Nat. Chem. Biol. 2013;9:607–609. doi: 10.1038/nchembio.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Layfield JP, Hammes-Schiffer S. Chem. Rev. 2014;114:3466–3494. doi: 10.1021/cr400400p. [DOI] [PMC free article] [PubMed] [Google Scholar]