Abstract
Mallory-Denk bodies (MDBs) are aggresomes composed of undigested ubiqutinated short lived proteins which have accumulated because of a decrease in the rate of their degradation by the 26s proteasome. The decrease in the activity of the proteasome is due to a shift in the activity of the 26s proteasome to the immunoproteasome triggered by an increase in expression of the catalytic subunits of the immunoproteasome which replaces the catalytic subunits of the 26s proteasome. This switch in the type of proteasome in liver cells is triggered by the binding of IFNγ to the IFNγ sequence response element (ISRE) located on the FAT10 promoter. To determine if either FAT10 or IFNγ are essential for the formation of MDBs we fed both IFNγ and FAT10 knock out (KO) mice DDC added to the control diet for 10 weeks in order to induce MDBs. Mice fed the control diet and Wild type mice fed the DDC or control diet were compared. MDBs were located by immunofluorescent double stains using antibodies to ubiquitin to stain MDBs and FAT10 to localize the increased expression of FAT10 in MDB forming hepatocytes. We found that MDB formation occurred in the IFNγ KO mice but not in the FAT10 KO mice. Western blots showed an increase in the ubiquitin smears and decreases β 5 (chymotrypsin-like 26S proteasome subunit) in the Wild type mice fed DDC but not in the FAT10 KO mice fed DDC. To conclude, we have demonstrated that FAT10 is essential to the induction of MDB formation in the DDC fed mice.
Keywords: FAT10, Interferon gamma, Mallory-Denk Bodies
INTRODUCTION
This paper is written to honor Dr. Julius Cruse who helped many investigators publish their research results in Experimental Molecular Pathology.
The present report focuses on the question “what is the role of FAT10 in the mechanism of Mallory Denk body (MDB) formation which develops in chronic liver disease?” So far, we have determined that the following factors are essential for MDB formation in vivo or in vitro using the DDC fed mouse model: 1) the deacetylase inhibitor trichostatin (TSA) prevented MDB formation in vitro, whereas AZA, the DNA methylation inhibitor did not, indicating demethylation of DNA and histones is essential for MDB formation (Oliva et al., 2008). 2) NFκB inhibitor prevents MDB formation in vitro (Nan et al., 2005) and NFκB is activated when MDBs are formed in vivo and in vitro (Nan et al., 2006; Yuan, et al., 2006). NFκB activation is essential for MDB formation in vitro (Nan et al., 2005, Nan et al., 2006). FAT10 mediates NFκB activation in response to IFNγ binding the Interferon Sequence Response Element (ISRE) on the FAT10 promoter. TNFα-induced NFκB activation also results from the TNFα recdptor type 1 at the plasma membrane through IKBα phosphorylation, which leads to IKBα degradation and liberation of NFκB (Gong et al., 2010). 3) Inhibitors of phosphorylation of ERK and p38 prevent MDB formation in vitro (Nan et al., 2005; Nan et al., 2006, Wu et al., 2005). IFNγ binding to ISRE on the FAT10 promoter activates JNK and p38 (Oliva et al., 2010). 4) Epigenetic changes, primarily demethylated DNA and histones, also are essential for MDB formation (Bardag-Gorce et al., 2008; Li et al., 2008). For instances, S-adenosylmethionine (SAMe) prevents MDB formation in vitro (Li et al., 2008) presumably because SAMe, a major methyl donor which silences the molecular responses by methylating DNA and histones prevents MDB formation (Bardag-Gorce et al., 2008).
There are several other signaling pathways that are involved by IFNγ during MDB formation including TLR4 and TLR2 (Bardag-Gorce et al., 2010b). TLR4 and TLR2 knockout mice, however, form MDBs despite the absence of TLR4 and 2 (French et al., 2011).
In the present study we obtained IFNγ and FAT10 knockout mice and fed them DDC for 10 weeks to determine if either IFNγ or FAT10 were essential for MDB formation. FAT10 is likely to be essential for MDB formation because the promoter region signals the up regulation of the 3 catalytic subunits LMP2, LMP4 and MECL-1 of the immunoproteasome (Olive et al., 2010) that replace the 26s proteasome catalytic subunits. This reduces the activity of the 26S proteasome which leads to MDB formation (French et al., 2011).
METHODS
Animals
Two groups of knockout (KO) mice were fed 0.1% diethyl 1,4,-dihydro-2,4,6,- trimethyl-3,5-pyridine dicarboxylate (DDC, Aldrich, St Louis, MO) in a semi synthetic protein rich complete diet (Teklad, Madison, WI) (Yuan et al., 1996) for 10 weeks to induce Mallory-Denk body (MDB) formation in vivo. Controls were fed the same diet without DDC added. One group of 4 week old male mice was IFNγ KO mice supplied by the Jackson laboratory. They were fed the DDC diet or the control diet for 10 weeks. The other group was 4 week old female FAT10 KO C3H mice supplied by Dr. Canaan from Yale University (Canaan et el., 2006). They were fed the DDC diet or the control diet for 10 weeks. Wild type 4 week old C3H female mice were fed the DDC diet or the control diet for 10 weeks as strain controls. All mice were treated in a humane manner as approved by the Animal Care Committee at Harbor-UCLA LA Biomedical Research Institute according to the Guidelines of the National Academy of Science.
Liver homogenates
Mouse liver homogenates were prepared by homogenizing 100 mg of liquid nitrogen frozen liver in 2 ml of 20 mM Tris-HCl pH 7.5; glycerol 10% EGTA1 mM; DTT 1 mM; sodium-fluoride 50 mM; protease and phosphatases inhibitor cocktail (Sigma, St Louis, MO). The livers were homogenized using the Ultra-Turrax T25 homogenizer. Protein concentrations were quantitated using the Bradford method (Bradford, 1976).
Western blot analysis
Proteins (50 µg) from liquid nitrogen frozen stored livers and nuclear and histone extracts were separated by SDS-PAGE gels and transferred to a PVDF membrane (Bio-Rad, Hercules, CA) for 1 h in 25 mM Tris-HC1 (pH 8.3), 192 mM Glycine and 20% methanol. The membranes were stained using primary antibodies to antigens. The antibody used was: ubiquitin (DAKO, Carpentaria, CA).
Appropriate species polyclonal and monoclonal HRP-conjugated antibodies were used as the secondary antibodies. The membranes were subjected to chemiluminescence detection using luminal, according to the manufacturer’s instructions (Amersham Pharmacia Biotech, Piscataway, NJ).
Immunohistochemistry
Liver biopsy sections were double stained with the primary antibodies rabbit anti UbD (FAT10) (BioMol, Plymouth Meeting, PA) and mouse anti ubiquitin (Chemicon, Temecula, CA). Secondary antibodies conjugated with FITC or Texas Red (Jackson, West Grove, PA) were used to detect binding of the primary antibodies. DAPI was used as the nuclear stain. The slides were examined using a Nikon-400 fluorescent microscope with a triple color band cube to detect simultaneously FITC, Texas Red and DAPI staining. UbD positive hepatocytes were detected.
STATISTICS
Statistical analyses were performed using ANOVA+Test and Bonferoni for multiple group comparison (Sigma Stat Software). Significance was defined as P<0.05.
RESULTS
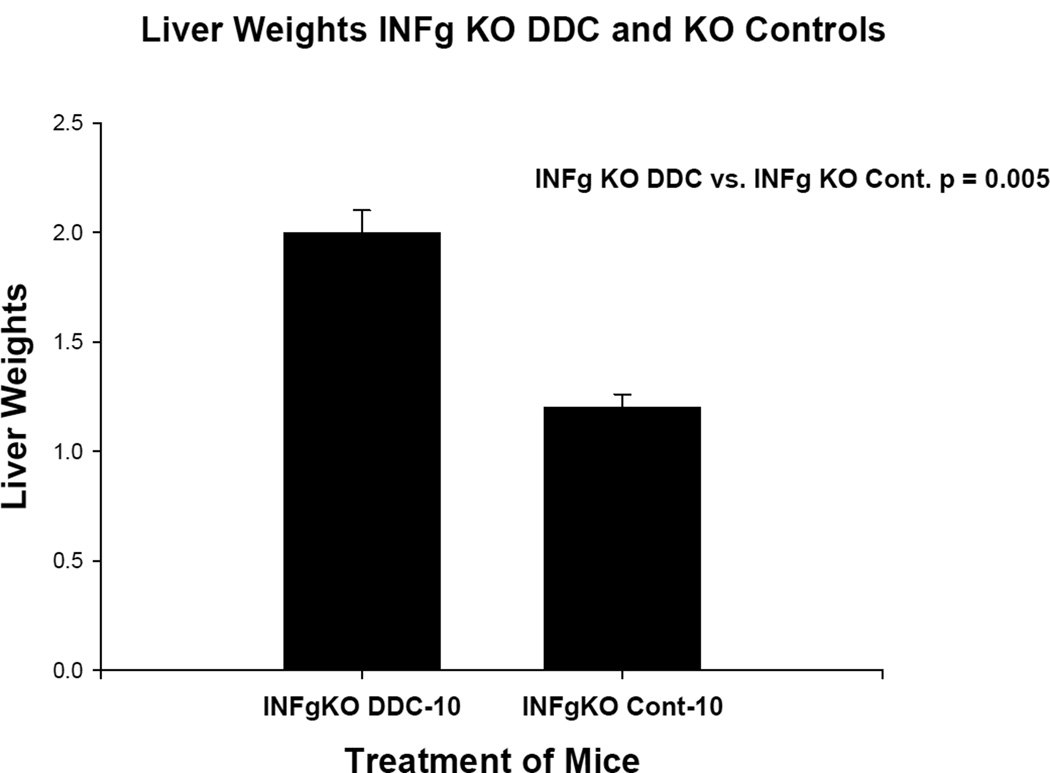
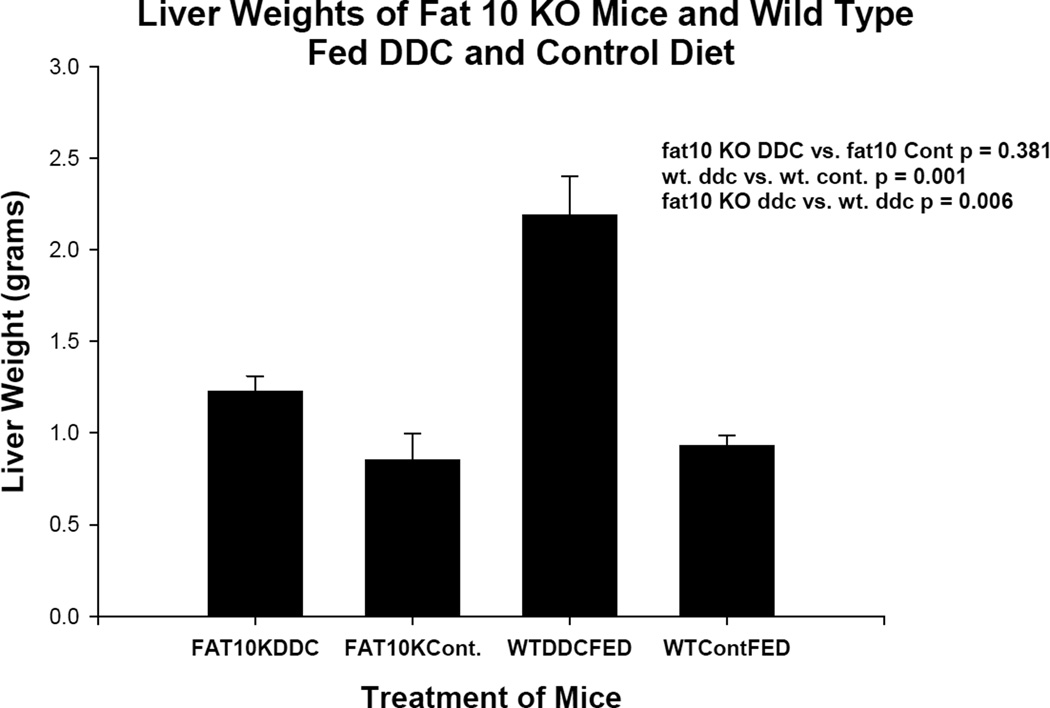
The liver weights for the two experiments are shown in Figures 1 and 2. In the case of IFNγ KO mice (group 1) there was no difference in these parameters. In the case of the FAT10 KO mice (group 2) the KO mice fed DDC body weights were reduced. One mouse died during the 10 week feeding period.
Fig 1.
Liver weights are shown comparing Interferon γ KO mice fed DDC for 10 weeks with Interferon γ wild type mice fed the diet without DDC added. The difference is statistically significant.
Fig 2.
Liver weights are shown comparing four groups of mice. Group 1 was FAT10 KO mice fed DDC for 10 weeks, Group 2 was the FAT10 KO mice fed the diet without DDC added. Group 3 was wild type strain control mice fed DDC for 10 weeks. Group 4 was wild type strain control mice fed the diet without DDC added. Group 3 livers were significantly enlarged compared to the other 3 groups.
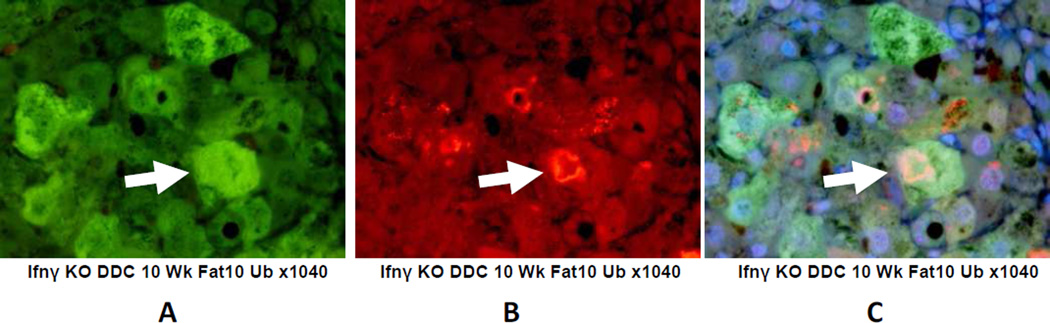
The livers of the IFNγ KO mice fed DDC and the KO mice fed the control diet (group 1) both showed numerous liver cells which over expressed FAT10 and had formed ubiquitin positive MDBs of various sizes and shapes (Fig 3). From this it can be concluded that IFNγ was not essential for MDB formation.
Fig 3.
Immunohistochemical double stains (IHC) showing MDB formation in FAT10 positive hepatocytes. A) FAT10 antibody filter (FITC). B) Ubiquitin filter (Texas red) and C) tricolor filter showing MDB formed in FAT10 positive hepatocytes (arrows). X1040
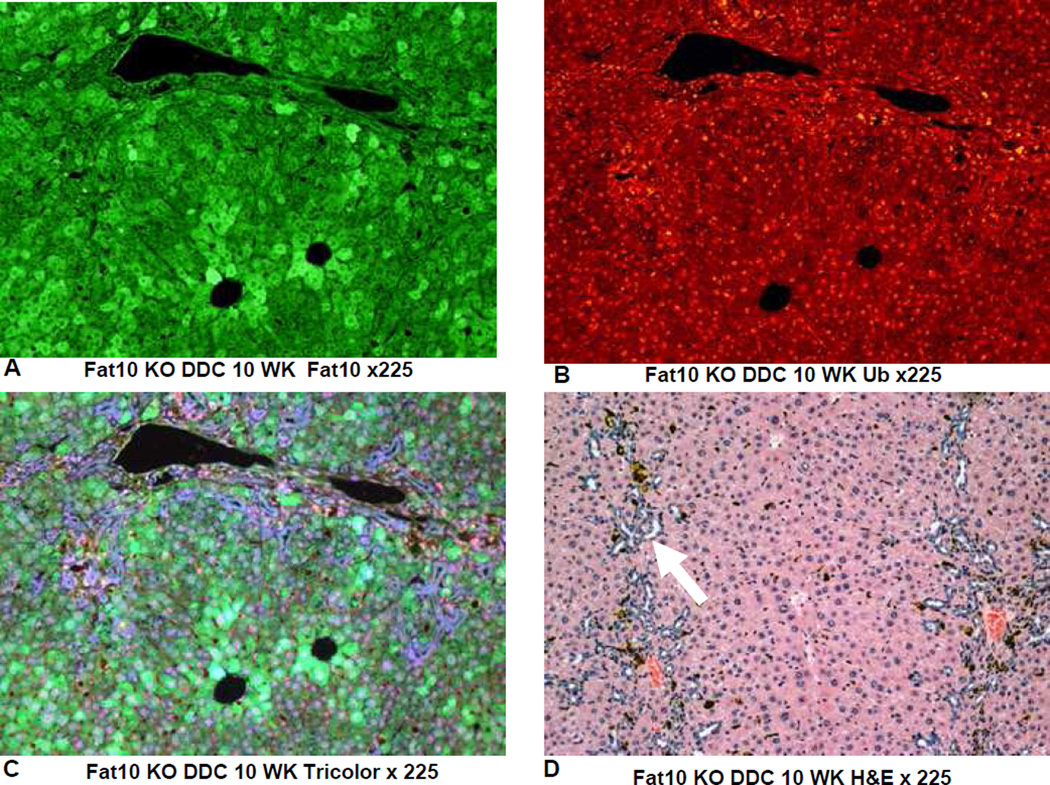
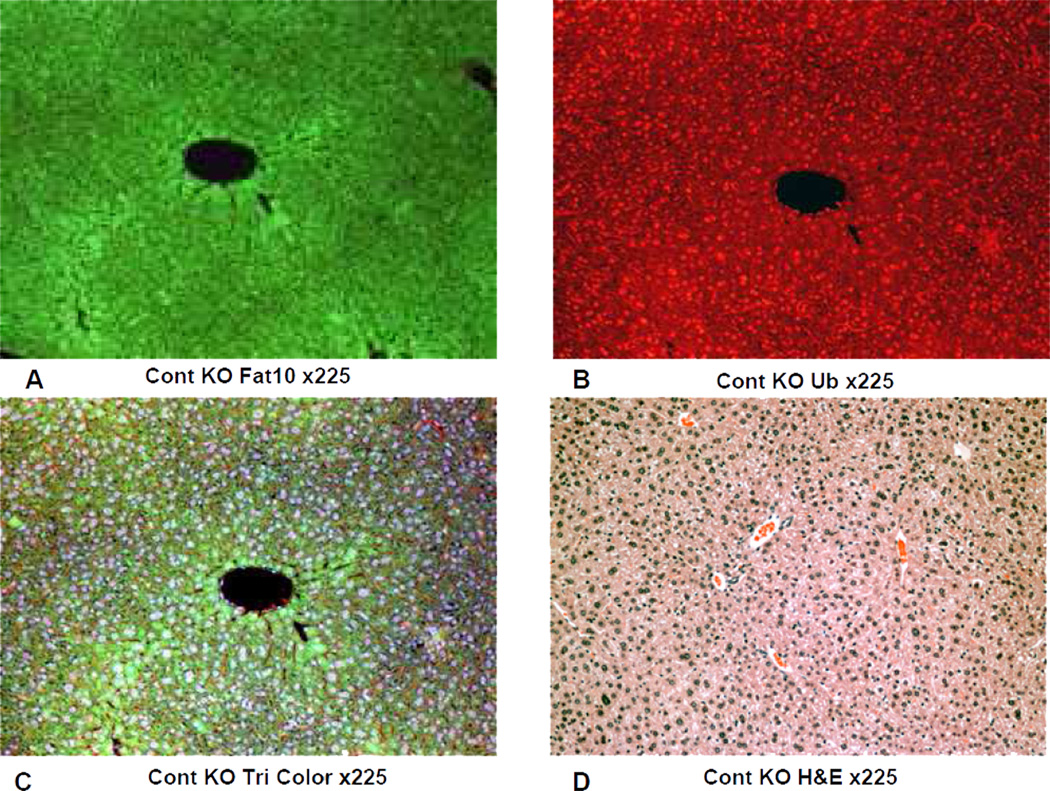
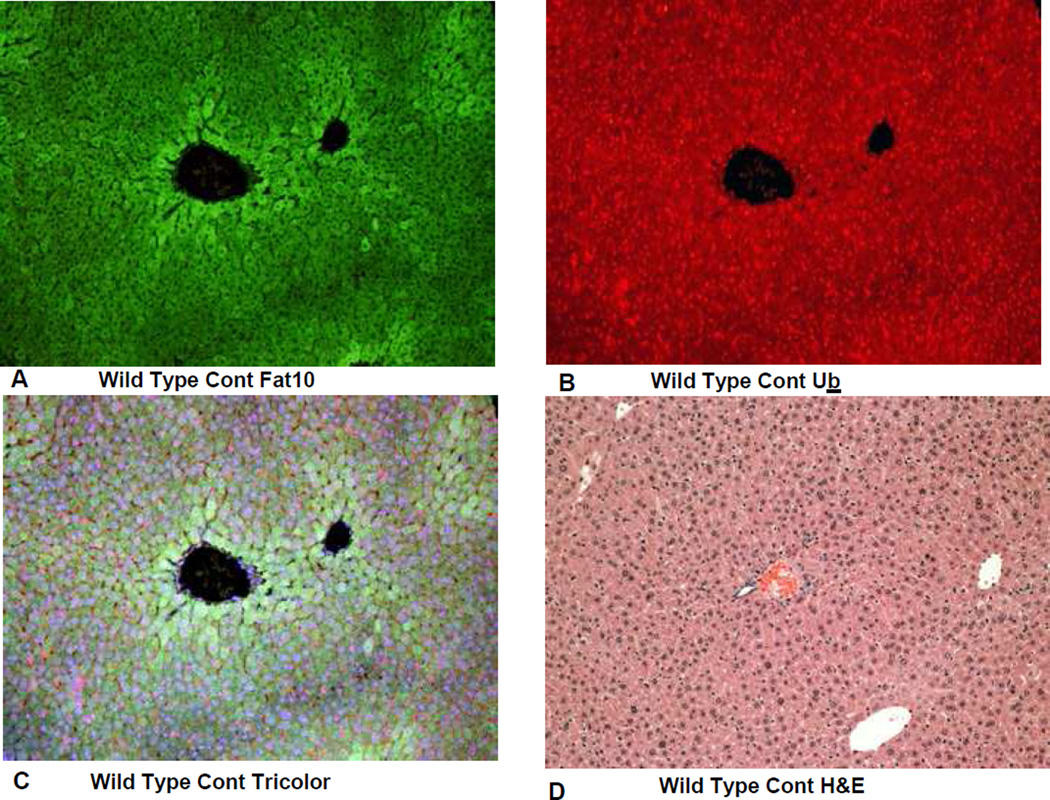
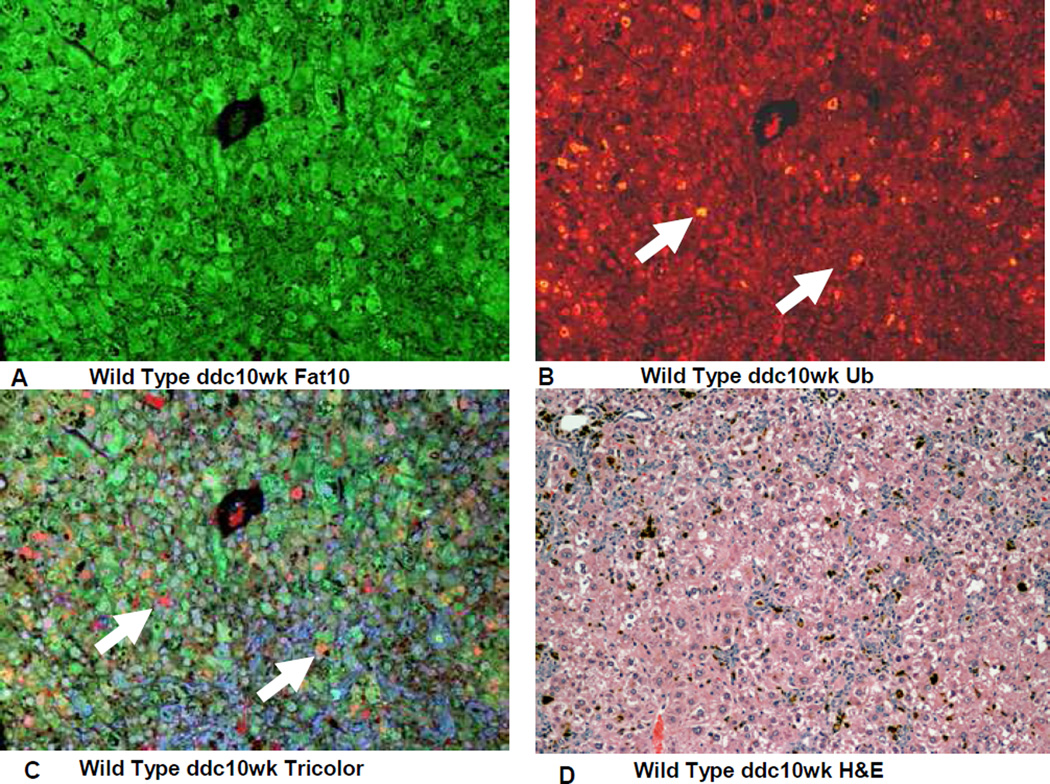
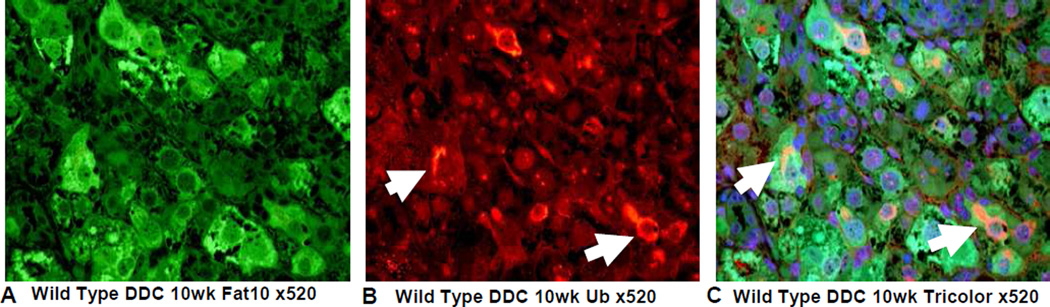
On the other hand the FAT10 KO (group 2) mice failed to form MDBs when fed DDC (Fig 4). This was also true for the FAT10 KOs fed the control diet (Fig 1, 2, 5) as well as the wild type strain control mice fed the control diet (Fig 6). The wild type strain control mice fed DDC formed numerous MDBs in the FAT10 over expressing liver cells (Fig 7, 8) indicating that the strain and gender differences did not modify the DDC induced FAT10/MDB forming response of the mice studied. These results indicate that MDB formation depends on the presence of FAT10.
Fig 4.
The IHC double stains showing the absence of MDB formation in a liver from a FAT10 KO mouse fed DDC for 10 weeks. A) FAT10 antibody (FITC). B) Ubiquitin antibody filter (Texas red). C) Tricolor filter. D) The same liver stained with Hematoxylin and eosin showing proliferation of oval cells (arrow) induced by DDC feeding. X225
Fig 5.
IHC double stains showing the FAT10 and ubiquitn of a liver from a FAT10 KO mouse fed the control diet without DDC added for 10 weeks. A) FAT10 antibody filter (FITC). B) Ubiquitin antibody filter (Texas red). C) Tricolor filter. D) The same liver stained with Hematoxylin and eosin showing normal liver histology. X225
Fig 6.
IHC double stains showing the FAT10 and ubiquitin of a liver from a FAT10 wild type strain control mouse fed the control diet without DDC added. A) FAT10 antibody filter (FITC). B) Ubiquitin antibody filter (Texas red). C) Tricolor filter. D) The same liver stained with Hematoxylin and eosin showing normal liver histology. X225
Fig 7.
IHC double stains showing the FAT10and ubiqutin of a liver from a FAT10 wild type strain control mouse fed the control diet with DDC added for 10 weeks. A) FAT10 antibody filter (FITC). B) Ubiquitin antibody filter (Texas red). C) Tricolor filter. Multiple ubiquitin positive MDBs are present (arrows) D) The same liver stained with Hematoxylin and eosin showing changed liver histology. X225
Fig 8.
IHC double stains for FAT10 and ubiquitin of a liver from a wild type mouse fed DDC for 10 weeks show numerous MDBs formed by FAT10 positive liver cells (arrows). A) FAT10 antibody (Fitc). B) Ubiquitin antibody (Texas red). C) Tricolor filter. X520
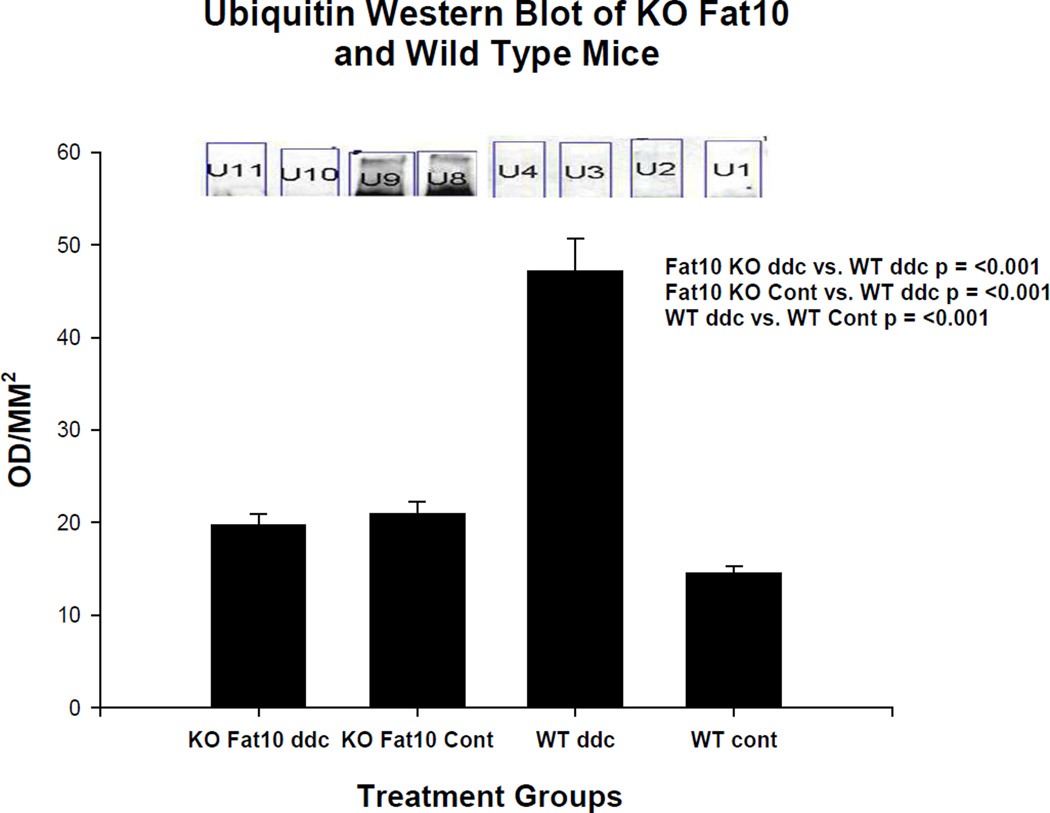
Western blots of the livers in group 2 FAT10 KO mice showed that the high molecular weight ubiquitinated protein smear was markedly increased in the livers of the strain control mice fed DDC but not in the FAT10 KO mice fed DDC or the mice fed the control diet (Fig 9). This indicates that the FAT10 KO mice fed DDC did not develop the decrease in the 26s proteasome whereas the wild type refed DDC did. Consequently, digestion of proteins caused by the shift to the immunoproteasome catalytic proteins and the accumulation of polyubiquinated proteins to form aggresomes (MDBs) in the FAT10 KO mice did not occur (Bardag-Gorce et al., 2010a).
Fig 9.
Ubiquitin smear is present on a Western blot done on the wild type mouse fed DDC for 10 weeks (WT ddc) (U8 and U9). The other 3 groups of mice including the FAT10 KO mice fed DDC (KO FAT10 ddc) did not develop a ubiquitin smear indicating the activity of the 26s proteasome was not diminished in these groups of mice.
DISCUSSION
The FAT10 gene is proven to be essential for the formation of MDBs because the binding of cytokines to its promoter leads to activities of many genes which are required for MDBs to form i.e. NFκB, p38, and the immunoproteasome catalytic subunits LMP2, LMP7 and MECL-1 (Oliva et al., 2010; Bardag-Gorce et al., 2010a; Nan et al., 2005; Nan et al., 2006) (Canaan et al., 2006. The induction of MDBs by DDC is through a mechanism which depends on the replacement of the 26s proteasome catalytic subunits by the immunoproteasome subunits LMP2, LMP7 and MECL-1 (Bardag-Gorce et al., 2010a) and consequently the loss of the proteolytic activity of the 26s proteasome. This results in the accumulation of undigested keratins, as well as numerous other proteins that are normally turned over by the 26s proteasome. The aggregate of accumulating ubiquitinated proteins forms the MDB and also the characteristic polyubiquitin smear of undigested ubiquitinated proteins seen on the Western blots (Bardag-Gorce et al., 2010a).
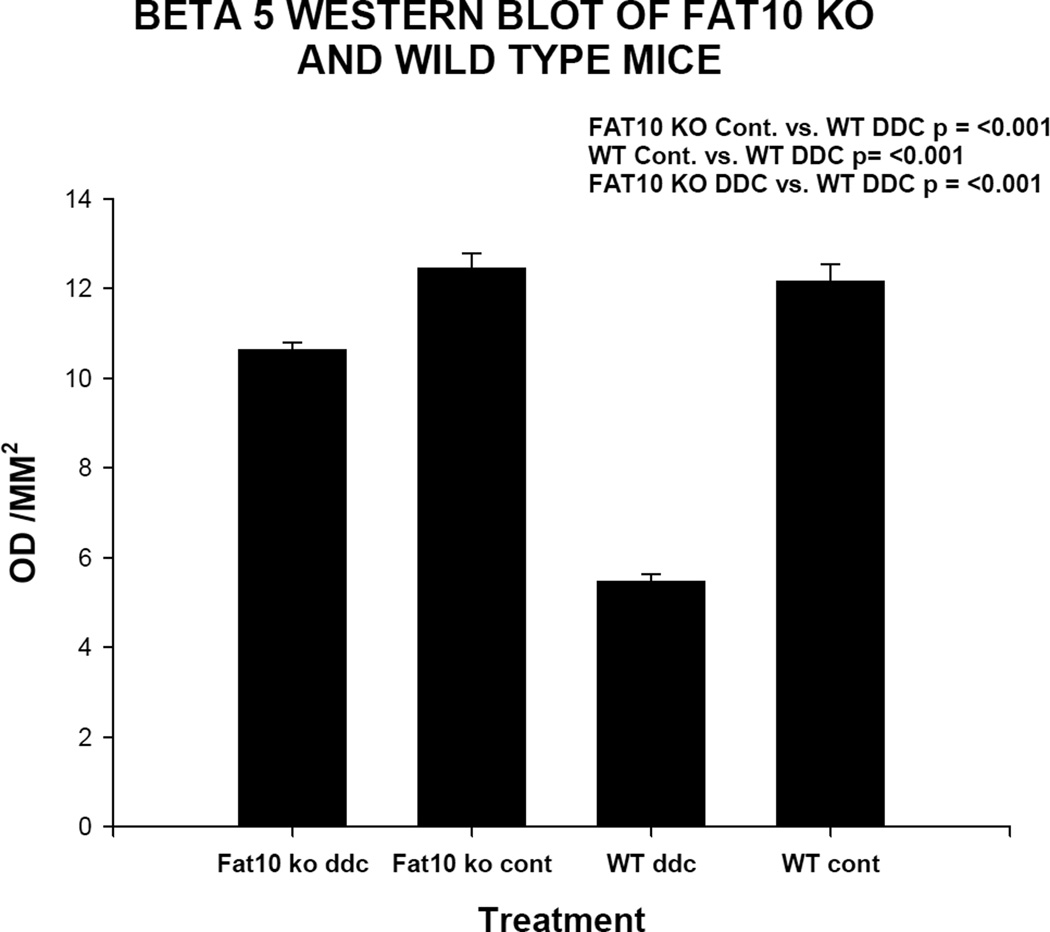
Fig 10.
B5 (chymotrypsin) was diminished in the wild type mice fed DDC for 10 weeks (WT ddc) compared to the other 3 groups of mice by ½ including the FAT10 KO mice fed DDC (KO FAT ddc) indicating that the FAT10 KO mice fed DDC were prevented from developing a decrease in the 26s proteasome catalytic subunit because of the absence of FAT10 dependent decrease in the expression of Beta 5. The level of Beta 5 was slightly lower in the FAT10 KO DDC fed mice compared to the FAT10 KO controls and the WT controls.
Acknowledgments
The authors thank Adriana Flores for typing the manuscript. Supported by Grants NIAAA 8116 and P50-011999, Morphology Core.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bardag-Gorce F, Oliva J, French BA, Li J, Dedes J, French SW. SAMe prevents the induction of immunoproteasome and preserves the 26s proteasomes in the DDC induced MDB; Mouse Model. Exp Mol Pathol. 2010a;88:352–362. doi: 10.1016/j.yexmp.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardag-Gorce F, Oliva J, Lin A, Li J, French BA, French SW. SAMe prevents the up regulation of toll-like receptor signaling in Mallory-Denk body forming hepatocytes. Exp Mol Pathol. 2010b;88:376–379. doi: 10.1016/j.yexmp.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardag-Gorce F, Oliva J, Villegas J, Fraley S, Amidi F, Li J, Dedes J, French B, French SW. Epigenetic mechanisms regulate Mallory Denk body formation in the livers of drug-primed mice. Exp Mol Pathol. 2008;94:113–121. doi: 10.1016/j.yexmp.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford HM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Canaan A, Yu X, Booth CJ, Lian I, Lazar I, Gamfi SL, Castille K, Kohya N, Nakayama Y, Liu Y-C, Eynon E, Flavell R, Weissman SM. FAT10/diubiquitin-like protein-deficient mice exhibit minimal phenotypic differences. Mol Cell Biol. 2006;26:5180–5189. doi: 10.1128/MCB.00966-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SW, Bardag-Gorce F, French BA, Li J, Oliva J. The role of innate immunity in the pathogenesis of preneoplasia in drug-induced chronic hepatitis based on a mouse model. Exp Molec Pathol. 2011;91:653–659. doi: 10.1016/j.yexmp.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French BA, Oliva J, Bardag-Gorce F, Li J, Zhong J, Buslon V, French SW. Mallory-Denk bodies form when EZH2/H3k27me3 fails to methylate DNA in the nuclei of human and mice liver cells. Exp Mol Pathol. 2012;92:318–326. doi: 10.1016/j.yexmp.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P, Canaan KA, Wang B, Leventhal J, Snyder A, Nair V, Cohen CC, Kretziet M, Agati V, Weissman S, Ross MJ. The ubiquitin-like protein FAT10 mediates NFκB activation. J Am Soc Nephrol. 2010;21:316–326. doi: 10.1681/ASN.2009050479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Bardag-Gorce F, Dedes J, French BA, Oliva J, Amidi F, French SW. S-adenosylmethionine prevents Mallory Denk body formation in drug-primed mice by inhibiting epigenetic memory. Hepatology. 2008;47:613–624. doi: 10.1002/hep.22029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan L, Wu Y, Bardag-Gorce F, Li J, French BA, Wilson LT, French SW. The p105/50NF-κB pathway is essential for Mallory body formation. Exp Mol Pathol. 2005;78:198–206. doi: 10.1016/j.yexmp.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Nan L, Dedes J, French BA, Bardag-Gorce F, Li J, Wu Y, French SW. Mallory body (cytokeratin aggresomes) formation is prevented in vitro by p38 inhibitor. Exp Mol Pathol. 2006;80:228–240. doi: 10.1016/j.yexmp.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, French BA, Li J, McPhaul L, Amidi F, Dedes J, Habibi A, Nguyen S, French SW. FAT10 is an epigenetic marker for liver preneoplasia in a drug-primed mouse model of tumorigenesis. Exp Mol Pathol. 2008;94:101–112. doi: 10.1016/j.yexmp.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, Lin A, French BA, French SW. Role of cytokines in UBD promoter regulation and Mallory-Denk body-like aggresomes. Exp Mol Pathol. 2010;89:1–8. doi: 10.1016/j.yexmp.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan QX, Marceau N, French BA, Fu P, French SW. Mallory body induction in drug primed mouse liver. Hepatology. 1996;24:603–612. doi: 10.1002/hep.510240324. [DOI] [PubMed] [Google Scholar]
- Yuan QX, Nagao Y, French BA, Wan Y-JY, French SW. Dexamethasone enhances Mallory body formation in drug primed mouse liver. Exp Mol Path. 2000;69:202–210. doi: 10.1006/exmp.2000.2320. [DOI] [PubMed] [Google Scholar]