Abstract
Membrane proteins are of outstanding importance in biology, drug discovery and vaccination. A common limiting factor in research and applications involving membrane proteins is the ability to solubilize and stabilize membrane proteins. Although detergents represent the major means for solubilizing membrane proteins, they are often associated with protein instability and poor applicability in structural and biophysical studies.
Here, we present a novel lipoprotein nanoparticle system that allows for the reconstitution of membrane proteins into a lipid environment that is stabilized by a scaffold of Saposin proteins. We showcase the applicability of the method on two purified membrane protein complexes as well as the direct solubilization and nanoparticle-incorporation of a viral membrane protein complex from the virus membrane. We also demonstrate that this lipid nanoparticle methodology facilitates high-resolution structural studies of membrane proteins in a lipid environment by single-particle electron cryo-microscopy (cryo-EM) and allows for the stabilization of the HIV-envelope glycoprotein in a functional state.
Introduction
Membrane proteins are encoded by approx. 30% of all open reading frames1. They are an important class of drug targets, as more than >60% of drugs in clinical use target this class of proteins2. Membrane proteins play essential roles in many cellular processes, such as signal transduction, cell-to-cell communication, membrane transport as well as lipid and energy metabolism. However, membrane proteins are difficult to study due to their instability and tendency to aggregate when extracted from their natural lipid bilayer environment. In order to maintain the integrity of membrane proteins, an artificial hydrophobic environment is needed. Detergent micelles are the most common approach to solubilize membrane proteins, but often have adverse effects on protein activity, stability and solubility or interfere with the experimental set-up3-5.
A major challenge is maintaining membrane proteins in a lipid-like environment while keeping them stable and monodisperse in solution, so that they become accessible for biochemical, biophysical and structural studies. Methods to reconstitute membrane proteins into lipid nanoparticles provide a potential solution for this challenge. Current nanoparticle technologies that address this problem involve liposomes and high-density lipoprotein (rHDL) particles6,7 based on Apolipoproteins, also termed Nanodiscs. Both approaches have been used routinely for biochemical and biophysical studies of membrane proteins and were also applied for structure determination by single particle cryo-EM8-10. However, both technologies are relatively laborious to optimize for individual membrane proteins.
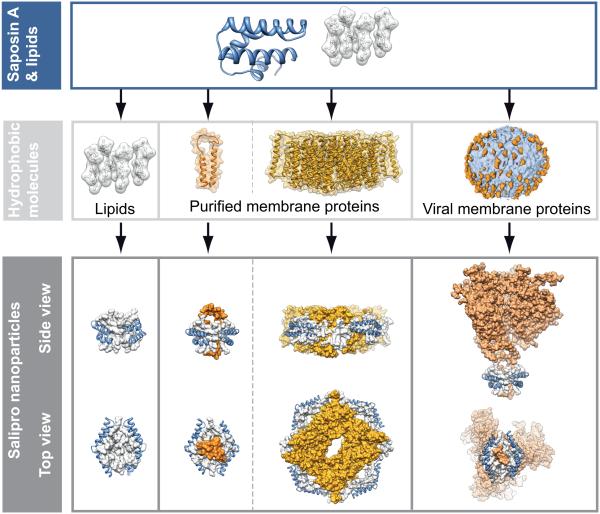
Here, we present a novel lipid nanoparticle system that is based on the Saposin protein family. Saposins are known to be modulators of lipid membranes11,12, mostly at an acidic pH within lysosomes. Given their lipid binding properties, we hypothesized that they could be used to generate a Saposin-based nanoparticle system for the incorporation of membrane proteins (Fig. 1). In this study, we developed a methodology to reconstitute membrane proteins into Saposin-lipoprotein (Salipro) nanoparticles, allowing to stabilize fragile membrane protein complexes in detergent-free buffer systems for functional and structural studies such as high-resolution structure determination by single particle cryo-EM.
Figure 1. Saposin-lipoprotein nanoparticles for the incorporation of lipids, membrane proteins and viral antigens.
By incubating Saposin A with lipids and membrane proteins it is possible to generate soluble and stable nanoparticles that adjust to the size of the incorporated molecule. Models of Saposin-Lipid-complexes were adapted from pdbs: 4ddj, 4aps, 4nco, 3din, 2dob; virus envelope structure modified49
Results
Generation of Saposin-lipid nanoparticles
Members of the Saposin protein family have membrane binding and lipid transport properties11,12. Several crystal structures of Saposin proteins have been reported, both in the absence and presence of lipids or detergents13-16. It was recently demonstrated that Saposin A forms lipid complexes at a lysosomal acidic pH, when incubated with unilamellar liposomes16.
Since Saposin proteins have the capability of forming lipid complexes, we rationalized that these could be used as scaffolding proteins for a lipid nanoparticle system to reconstitute membrane proteins into a lipid environment. Our aim was therefore to establish a more versatile system for incorporating membrane proteins into a lipid nanoparticle system based on Saposin proteins, without using liposomes and an acidic pH.
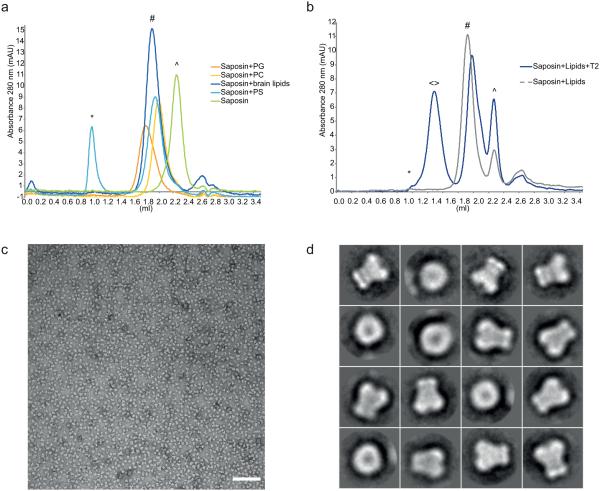
At first, we tested whether it would be possible to incorporate lipids into Saposin A complexes, in the absence of an acidic pH and in the absence of liposomes We incubated Saposin A (see Methods) with detergent-solubilised phospholipids at a physiological pH 7.4, followed by detergent removal via an initial dilution step and subsequent size-exclusion chromatography (SEC) without detergent in the buffer. We could observe a significant shift in the retention time of the Saposin A SEC peak after incubation with lipids such as phosphatidylcholine (PC), phosphatidylglycerol (PG), phosphatidylserine (PS) or brain lipid extract, supporting the incorporation of lipids to form soluble Saposin A based lipoprotein particles (Fig. 2a). In contrast, no shift was observed after incubation with POPE and an Escherichia coli (E. coli) total lipid extract (Supplementary Fig. 1a), indicating a certain degree of lipid preference for Saposin A at physiological pH11. Negative stain electron microscopy of Saposin-lipid complexes also revealed monodisperse particles (Supplementary Fig. 1b). Increasing the lipid-to-Saposin ratio leads to a slight increase in particle size (Supplementary Fig. 1c). In general, the gel-filtration analyses of the Saposin-lipid complexes displayed symmetric peaks, corresponding to the formation of homogenous nanoparticle populations. Salipro nanoparticles remain stable after concentration using standard centrifugal filter units. Moreover, particles that had been concentrated and frozen maintained their assembled state as judged by analytical SEC when thawed and incubated for 10 minutes at temperatures from 0-95 °C, indicating a high degree of thermostability (Supplementary Fig. 1d).
Figure 2. Salipro particles with lipids and the T2 channel.
(a) Gel filtration analysis of Saposin A after incubation with the indicated detergent solubilised lipid solutions. The void volume is marked with an asterisk, the peaks for monomeric Saposin with (^) and Saposin-lipid complexes with (#). (b) Gel filtration analysis of the incorporation of the pentameric T2 channel into Salipro nanoparticles. The void volume is marked with an asterisk, the peaks for monomeric Saposin with (^), Saposin-lipid complexes with (#) and Salipro-T2 with (◇). (c) A representative micrograph of T2 in Salipro discs stained with uranyl acetate. Individually side and top views of the reconstituted channel can be seen. Scale bar is 100 nm. (d) Selected 2D class averages of Salipro-T2, the boxsize is 243 Å.
Incorporation of membrane proteins into Salipro particles
Studying integral membrane proteins in a lipid environment still represents one of the biggest challenges for structural biology, biochemical studies and drug development. Given the Salipro nanoparticles’ capacity of integrating detergent-solubilized lipids at a physiological pH, we focused on the potential of reconstituting membrane proteins. We incubated detergent purified membrane proteins with lipids and Saposin A, followed by detergent removal steps mentioned above. The model proteins used were an archaeal mechanosensitve channel17 (T2, 4 predicted transmembrane helices per monomer, 32.9 kDa, putative homopentamer) and a bacterial peptide transporter18,19 (POT1, 14 transmembrane helices per monomer, 56 kDa, homotetramer).
It should be noted that in the absence of detergent in the SEC buffer, membrane proteins are not soluble, but will aggregate and not elute from the column. In contrast, after incubation with Saposin A and lipids, the gel-filtration profile of the archaeal T2 channel displays three symmetric peaks (Fig. 2b), with the first peak corresponding to T2 channel incorporated into Salipro nanoparticles, while the second and third peak originate from lipid-only Salipro nanoparticles and monomeric Saposin A, respectively. Accordingly, Saposin A, lipids and the T2 channel associate in such way that they form water-soluble particles with incorporated membrane protein.
Peak fractions corresponding to Salipro-T2 were pooled, concentrated and flash-frozen prior to further analysis via electron microscopy (EM). Salipro-T2 particles that had been concentrated, frozen, thawed and diluted maintained their assembled state as judged by analytical SEC (Supplementary Fig. 2a). Negative stain EM revealed a homogenous population of single particles that adopt various orientations on the grid (Fig. 2c, d). The particles look significantly more distinct than the T2 channel embedded in micelles of the detergent n-Dodecyl β-D-maltoside (DDM)17. In side-views, both individual particles as well as two dimensional (2D) class averages of Salipro-T2 particles display two moieties of different sizes, representing a more compact domain and a bilobed domain (Fig. 2d).
Cryo-electron microscopy of Salipro-POT
Proton-coupled oligopeptide transporters (POTs) regulate the uptake of di- and tripeptides and are conserved among all kingdoms of life. We used the bacterial peptide transporter PeptTSo2, labeled POT, as model protein for our studies. For the integration of this POT into Salipro nanoparticles, we incubated the purified membrane protein with Saposin A and lipids. The gel-filtration profile of the bacterial peptide transporter exhibits three symmetric peaks (Supplementary Fig. 2b), with the first peak corresponding to the membrane protein incorporated into Salipro nanoparticles. Generally, the formation of Salipro particles with incorporated membrane proteins such as POT is feasible within a physiological pH range (Supplementary Fig. 2c). Prior to further analysis, peak fractions of Salipro-POT were pooled, concentrated and flash-frozen. Thermal unfolding analysis using dye-free differential scanning fluorimetry20 revealed that the peptide transporter is significantly more stable when embedded in Salipro nanoparticles (Tm 72°C) compared to detergent Nonyl-β-D-Maltopyranoside (Tm 43°C) (Supplementary Fig. 2d).
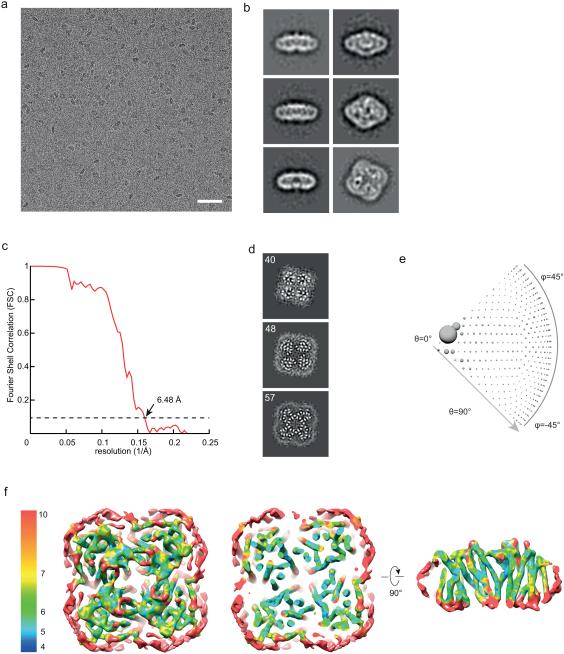
We used single-particle cryo electron microscopy (cryoEM) to determine the structure of the bacterial peptide transporter embedded within Salipro nanoparticles. From a cryoEM perspective, the POT transporter is a challenging target as it is mostly composed of transmembrane helices and contains only short loop regions that are not embedded in the membrane. Thus, there are no significant features outside of the membrane to align images of the molecules, which make it an extremely difficult target for single-particle cryo-EM. Cryo-EM grids of the Salipro-POT complex were prepared by the standard plunge-freezing method21 at a moderate concentration (0.7 mg/ml). In contrast, for single-particle cryoEM of membrane proteins in detergent, usually thick ice and high protein concentrations are required to obtain reasonable data22. Cryo-EM images revealed that frozen hydrated complexes of Salipro-POT are homogenous and monodisperse (Fig. 3a).
Figure 3. Single particle cryo-EM of Salipro-POT.
(a) Raw micrograph of PepTSO2 bacterial transporter collected using K2 Gatan direct electron camera. Scale bar is 50 nm. (b) Representative 2D class averages generated from the particles, the boxsize is 235 Å. (c) Gold-standard FSC curves between two independently refined half maps. (d) Selected slice views of the unsharpened 3D density map. The views are oriented in parallel with the membrane plane. The numbers of slices are marked. (e) Euler angle distribution of all particles included in calculating the final 3D reconstruction. The size of the ball is proportional to the number of particles in this specific orientation. (f) Final 3D reconstruction colored with local resolution, as seen from top (right), sliced parallel to the plane of the membrane (middle) and side view cut perpendicularly to the plane of the membrane (right).
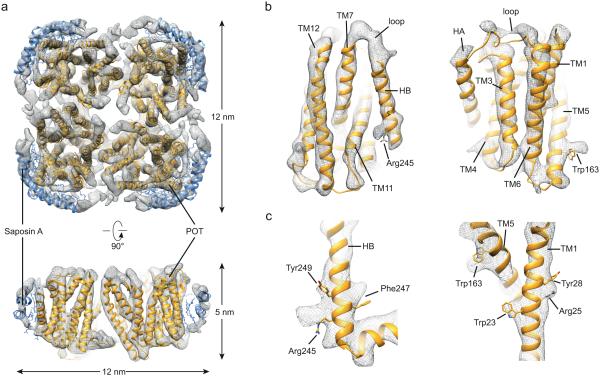
A cryo-EM data set was collected following established data acquisition and motion correction procedures23. Two-dimensional (2D) class averages display the features of secondary and tertiary structure, as well as the tetrameric organization of the peptide transporter (Fig. 3b). Using a total of 9,913 particles, we determined a 3D reconstruction with a 4-fold symmetry to a resolution of 6.5 Å (estimated with the gold-standard FSC = 0.143 criterion24 (Fig. 3c). All secondary structural features are clearly resolved in the final 3D density map (Fig. 3d). The Euler angle distribution shows that particles are evenly distributed in different orientations in vitreous ice (Fig. 3e). The local resolution in the core region of the nanoparticle, representing the transmembrane helices of the peptide transporter, is significantly higher than the regions that can be attributed to the more flexible Saposin-lipid scaffold (Fig. 3f, Supplementary Fig. 3). The cryo-EM density reveals a square-shaped disc with an overall dimension of 12 nm × 12 nm × 5 nm with well-resolved transmembrane helices (Fig. 4a, Supplemental Movie). Given the size (Fig. 4a) and the apparent stoichiometry (Supplementary Fig. 2e) of Salipro-POT1, each particle is composed of four POT molecules and four Saposin A molecules (Fig. 4a, Supplemental Movie).
Figure 4. Cryo-EM structure of Salipro-POT.
(a) 3D density map of the tetrameric bacterial transporter, filtered to 6.5 Å with each of the subunits and tentative placement of Saposin (orange and blue, respectively) in the density. Top view and side view cut perpendicularly to the plane of the membrane, dimensions are indicated. (b) Representative views of the cryo-EM density and rigidly fitted X-ray crystallography model (PDBID: 4TPH). The density shows a good fit of the model. The EM map reveals loop densities that could not be resolved in the X-ray structure. (c) Selected side chains densities of bulky residues such as Arg, Trp, Phe and Tyr.
The crystal structure of the peptide transporter19 could be directly docked into the cryo-EM density as a rigid body without modification (Fig. 4). While the transmembrane helices were well resolved, the Salipro scaffold of Saposin and lipids appears to be more flexible (Fig. 3f, Supplementary Fig. 3). Fitting of Saposin A into the scaffold density was therefore done with a map displayed at a lower isosurface level (Fig. 4a, Supplementary Video 1). The general dimensions of the Salipro scaffold are in agreement with a recently published structure of Saposin A16. At the current low resolution for the Saposin-lipid belt, it cannot be excluded that Saposin might also directly interact with the transmembrane helices of the bacterial transporter. However, we hypothesize that if Saposin would indeed bind directly to a transmembrane helix, it should be less flexible and thus, in turn, be possible to also resolve the α-helices of Saposin. However, this is not the case, the flexibility and hence lower resolution of the Saposin-lipid scaffold does not allow for resolving the α-helices of Saposin (Fig. 3f, Supplementary Fig. 3).
The cryo-EM density allows for the clear identification and positioning of the POT transmembrane helices (Fig. 4b). Moreover, the EM density reveals loop and helix densities that could not be modeled in the crystal structure of this peptide transporter19. The map also exhibits bulky side-chain densities from amino acids, positions of which are in good agreement to the rigid body fitting of the crystal structure of the peptide transporter (Fig. 4c).
Note that the Salipro-POT complex has almost no soluble domains exposed outside of the membrane. Generally, it was believed that membrane proteins ought to contain a soluble domain of considerable size exposed outside of the membrane to facilitate image alignment. While the four-fold symmetry of the tetrameric POT transporter facilitates cryo-EM 3-D reconstruction, it is still to our surprise that Salipro-POT complex can be reconstructed to subnanometer resolution from only 9,913 particles.
Stabilization of the functional HIV-1 spike protein
The membrane proteins of enveloped viruses often represent potent antigens that can be used for vaccination25. The trimeric HIV-1 envelope glycoprotein (HIV-1 spike), which mediates the viral entry into host target cells, may serve as a prominent example, as it is the only viral protein exposed to the immune system by intact HIV-1 particles. The HIV-1 spike is highly unstable in detergent26-28 (Supplementary Fig. 4).
Given the potential of the Saposin-based nanoparticle system to incorporate purified membrane proteins, we also investigated the prospect of incorporating membrane proteins from the original viral membrane. Here, we used the fragile HIV-1 spike as a model system (Fig. 5a). We established a rapid reconstitution protocol where we extracted the HIV-1 spike from the membrane of virus like particles (VLPs) and reconstituted the membrane protein into Salipro nanoparticles. To do so, we incubated VLPs containing HIV-1 spikes with detergent and Saposin A, followed by a fast removal of detergent-micelles using spin SEC columns, thereby forcing the hydrophobic moieties of the initial mixture to self-assemble into Salipro nanoparticles (Salipro-HIV-spikes) (Fig. 5b, Supplementary Fig. 5).
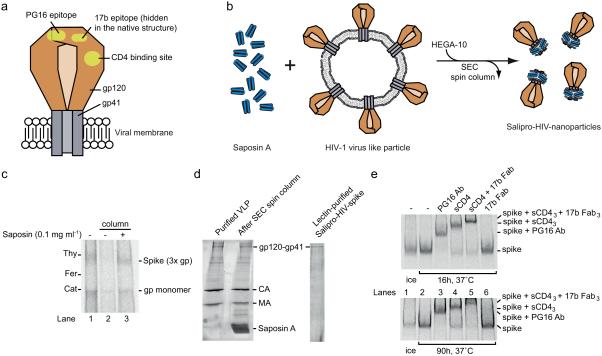
Figure 5. Stabilization of soluble and functional HIV-1 spike proteins within Salipro nanoparticles.
(a) Schematic illustration of the HIV-1 spike protein in the viral membrane. The HIV-1 Envelope glycoprotein, i.e. the spike, consists of two subunits, the peripheral gp120 subunit (brown) and the transmembrane gp41 subunit (grey), forming a heterotrimer. The binding site for the CD4 receptor and epitopes for the antibodies PG16 and 17b are shown (yellow). (b) Schematic illustration of the Salipro-HIV-spike reconstitution. Purified VLPs containing HIV-1 spikes (brown) were mixed with Saposin A (blue) and solubilized with HEGA-10 followed by detergent removal, leading to the formation of Salipro-HIV-spikes. (c) Reconstitution of the HIV-1 spike into Salipro nanoparticles. BN-PAGE analysis reveals that the solubility of the HIV-1 spike (3x gp) can be maintained by Saposin A upon detergent removal. (d) Lectin purification of Salipro-HIV-spikes. Particle purity was assayed using SDS-PAGE and Sypro Ruby protein staining. The gels show efficient removal of contaminating viral internal proteins. (e) The HIV-1 spike has a native fold and preserves its function in Salipro nanoparticles. The Salipro-HIV-spike particles were incubated at 37°C for 16 and 90 h followed by 2 h incubation at 37°C with 10 μg/ml of the HIV-1 spike ligands, PG16 antibody (150 kDa) (lane 3), sCD4 (50 kDa) (lane 4), sCD4 and 17b Fab (50 kDa) together (lane 5), 17b Fab alone (lane 6) or without a ligand (lane 2) and analyzed by BN-PAGE. A control sample without any ligand was kept on ice (lane 1). Binding of the ligands can be followed by the reduced migration of the Salipro-HIV-spike particles.
Initially six detergents with high critical micelle concentration (CMC) were screened. HEGA-10, MEGA-10 and C-HEGA-11 preserved the trimeric HIV-1 spike protein during solubilisation on ice for 10 min, as judged by BN-PAGE gel shift assay (Supplementary Fig. 4a). It should be noted that the viral spikes in detergent dissociated completely into monomers within 30 min at 37°C (Supplementary Fig. 4b). In order to stabilize the trimeric association, we reconstituted the HIV-1 spike protein into Salipro nanoparticles. Purified VLPs were solubilized with 9 mM HEGA-10 in the presence of 0.1 mg/ml Saposin A (for optimization see Supplementary Fig. 6), followed by removal of HEGA-10 using spin SEC columns (Supplementary Fig. 5). BN-PAGE analysis revealed that HIV-spikes eluted as soluble trimers only in the presence of Saposin A, indicating reconstitution into Salipro-HIV-spike particles (Fig. 5c). If Saposin A was omitted, HIV-spikes could not be resolved due to protein aggregation (Fig. 5c).
A second purification step using lectin affinity chromatography successfully removed viral matrix (MA) and capsid (CA) proteins as well as the excess of unbound Saposin A, leading to a pure Salipro-HIV-spike sample as determined by SDS-PAGE (Fig. 5d, Supplementary Fig. 5).
We investigated the stability and functionality of radiolabelled lectin-purified Salipro-HIV-spikes by BN-PAGE. While detergent solubilized HIV-spikes dissociated completely within 30 min at 37°C (Supplementary Fig. 4), Salipro-HIV-spikes remained intact for 16h and 90h at 37 °C as judged by BN-PAGE (Fig. 5e). Subsequent to the 16h and 90h incubation at 37 °C, we assessed the functionality and native features of the Salipro-HIV-spikes by binding to several protein ligands for additional 2h at 37°C. Binding of these ligands to the Salipro-HIV-spike increases its apparent molecular weight and therefore reduces the mobility in BN-PAGE correspondingly as shown before with detergent-HIV-spike complexes28.
The broadly neutralizing antibody PG16 binds to the variable loops V1-V2 and V3 at the top of preferentially intact and functional trimeric HIV-1 spikes28-30.
We found that PG16 antibody bound efficiently to the Salipro-HIV-spikes even after 16h and 90h incubation at 37 °C (Fig. 5e). In addition, a soluble version of the primary HIV-1 receptor CD4 (sCD4) bound stoichiometrically to the Salipro-HIV-spikes (Fig. 5e). Binding of CD4 induces a conformational change in the spike, which is essential for the binding of the co-receptor site-directed monoclonal antibody 17b Fab31,32. In order to examine this functionality, we analysed the binding of the 17b Fab to Salipro-HIV-spike-sCD4 complexes via BN-PAGE. The analysis revealed efficient binding of 17b Fab to the Salipro-HIV-spike-sCD4 complex (Fig. 5e). Importantly, 17b Fab did not bind to the Salipro-HIV-1-spikes in the absence of sCD4 (Fig. 5e). This confirmed that sCD4 binding induced proper conformational changes to the reconstituted Salipro-HIV-spikes. Altogether, our data demonstrates that the HIV-1 spike can be properly reconstituted into Salipro nanoparticles, preserving the fragile trimeric HIV-1 spike structure in a native, stable and functional state for at least 90h at 37 °C.
Discussion
Integral membrane proteins are difficult to study in a lipid environment. Detergents may keep membrane proteins in a soluble state once extracted from the membrane, however they are associated with a range of problems with respect to membrane protein stability and applicability in biophysical or structural studies3-5.
Here, we demonstrate that it is possible to incorporate membrane proteins into Saposin-based lipid nanoparticles that allow for structural and functional studies of membrane proteins in detergent-free buffer systems. The formation of Salipro nanoparticles is comparably straightforward by mixing lipids with membrane proteins and Saposin A, which leads to a rapid self-assembly reconstitution process. Moreover, it is also possible to incorporate membrane proteins from the viral membrane by mixing Saposin A with virus particles, leading to a fully functional viral membrane protein embedded in Salipro nanoparticles.
The nanoparticles are composed of lipids and Saposin A, a member of the saposin-like protein family11-16. This class of proteins is composed of amphipathic helices and six invariable cysteine residues that form disulphide bridges, leading to an extraordinary stability of the domains11. In turn, we assume that the intrinsic stability of the Saposin proteins, which surround the lipid core of the nanoparticle, also renders the nanoscale protein-lipid complexes remarkably stable and uniform in a wide temperature range (Supplementary Fig. 1c).
Various methodologies have been developed to stabilize membrane proteins in an aqueous environment7,33-40 (Supplementary Table 1). To achieve high-resolution, both amphipoles and maltose-neopentyl glycol-3 analogs have been shown to facilitate structural studies of membrane proteins41,42, indicating their stabilizing effect on membrane proteins and their tight binding to the membrane protein. However, these systems do not provide a lipid environment, which is known to be of great importance for membrane protein structure and function.
In turn, discoidal HDL particles, also termed Nanodiscs7,43, provide a lipid environment, albeit do not bind tightly to the TM-helices of the membrane protein. Additional Apolipoprotein-based nanoparticle systems have been developed that may vary in diameter with respect to the classical Nanodisc33,36,40. Generally, the size of a nanodisc is dictated by the length of the scaffolding Apolipoprotein belt at an optimum lipid content43. The classical Nanodisc technology has been established more than 15 years ago43, but to date the application of Nanodiscs for single particle cryo-EM studies has been limited to relatively large targets at subnanometer resolution9,10.
In turn, the Salipro nanoparticle technology combines the advantages of adapting to the size of the incorporated membrane protein (like detergents or amphipols), while at the same time providing a lipid environment. Moreover, the methodology is comparably straightforward to adjust for individual membrane proteins. Using the Salipro system, in this study we determined a subnanometer resolution structure of a membrane protein without soluble domains and embedded in a lipid bilayer environment. The POT complex is a particularly challenging target for structural studies by single particle cryo-EM since it contains only short loop regions and almost no structural features that stick out of the plane of the membrane region. Therefore, image alignment was entirely driven by the structural features of the transmembrane domain. This is possibly facilitated by the relatively small dimension of the Saposin-lipid scaffold that does not significantly affect the alignment process, while at the same time this protein-lipid scaffold surrounding the membrane protein also enhances the contrast in cryo-conditions (Fig. 3 & 4, Supplementary Fig. 3).
Previously it had been shown that two Saposin A molecules encapsulate a lipid core16. Here, in the presence of a large membrane protein complex, we observe that several Saposin A proteins may encircle a larger hydrophobic lipid/protein assembly in the case of the homotetrameric POT transporter (Fig. 4a, Supplemental Video 1). This suggests a significant flexibility of Saposin A to assemble into homogenous and stable lipoprotein-complexes, by adapting to the size of the incorporated molecules. This intrinsic flexibility of Saposin A-lipid complexes is likely to facilitate the formation of nanoparticles that can accommodate membrane protein of different sizes and, at the same time, assists in the formation of homogenous lipid nanoparticles (Fig. 2-4).
Moreover, we also present a method to extract and stabilize fragile membrane proteins from the virus membrane. The HIV-1 spike is the only viral protein that is exposed to the immune system by intact virus particles and therefore an interesting target for HIV vaccine development25,44,45. Antigen candidates that could preserve or sufficiently well mimic the native conformation of the HIV-1 spike including the important membrane domain have long been sought-after44,45. It has been shown that those regions proximal to the membrane as well as the membrane domain of the HIV-1 spike are important for the generation of broadly neutralizing antibodies against this virus46-48.
To our knowledge, the HIV spike protein preparation presented here using the Salipro system represents the first approach that allows the stabilization of the HIV-1 spike, including the important membrane domains, in a soluble and functional state. Thus, the strategy applied here, may represent a novel approach for the generation of immunogen/vaccine candidates that might induce broadly neutralizing antibodies (BNAbs) against HIV-1. We envisage that this approach is applicable to other viral envelope proteins such as Influenza virus HA, Ebola virus G protein or Hepatitis C virus E protein.
In summary, Salipro particles provide a novel versatile nanoparticle platform for membrane proteins and may offer a wide range of potential applications, ranging from structural biology to the discovery of new pharmacological agents as well as the therapeutic delivery of protein-based therapeutics and vaccines.
Online Methods
Materials and reagents
All detergents were purchased from Affymetrix. Luria-Bertani, Miller (LB) and Terrific broth (TB) from Formedium. Kanamycin was obtained from Duchefa Biochemie and Isopropyl β-D-1-thiogalactopyranoside (IPTG) from Saveen Werner. Soluble human CD4 (4d-CD4) was obtained from Invitrogen. The PG16 and 17b antibody32 were obtained from NIH AIDS Research and Reference Reagent Program (ARRRP). We generated Fab fragments for ligand binding studies by digesting 18 μg of monoclonal antibody in 25 μl of phosphate-buffered saline (PBS) containing 10 mM EDTA, 5 mM Cys, and 0.5 μg of papain for 4 h at 37°C. The digestion was terminated by addition of iodoacetamide to 5 mM. Samples were diluted with 175 μl of HNC buffer (50 mM HEPES, 100 mM NaCl, 1.8 mM CaCl2 [pH 7.4]) and uncleaved antibodies and Fc fragments were removed by binding to protein A-Sepharose (GE Healthcare, Uppsala, Sweden) overnight at 4°C. All other chemicals were from Sigma-Aldrich, unless otherwise stated.
Expression and Purification of Saposin A
Protein expression was carried out using a vector with the coding region for Saposin A inserted into a pNIC-Bsa4 plasmid. The cloning vector pNIC28-Bsa4 adds an N-terminal hexahistidine tag with an integrated TEV protease cleavage site. The cleaved target protein represents the Saposin A polypeptide with three additional N-terminal Ser-Met-Gly residues as a result of the cloning strategy. The protein was expressed using E. coli Rosetta gami-2 (DE3) (Novagen). Cells were grown at 37 °C in TB medium supplemented with Tetracycline, Chloramphenicol and Kanamycin and induced with 0.7 mM IPTG. Three hours after induction the cells were collected by centrifugation at 12.000×g for 15 min and the supernatant discarded. The cell pellet was resuspended in lysis buffer (20 mM Hepes pH 7.5, 150 mM NaCl, 20 mM Imidazol) and disrupted by sonication. Lysates were subjected to centrifugation at 26.000×g for 30 min, the supernatant heated to 85 °C for 10 min, followed by an additional centrifugation step at 26.000×g for 30 min. Preparative IMAC purification was performed by batch-adsorption of the supernatant by end-over-end rotation with Ni SepharoseTM 6 Fast Flow medium for 60 min. After binding of Saposin A to the IMAC resin, the chromatography medium was packed in a 10-mm-(i.d.) open gravity flow column and unbound proteins were removed by washing with 15 bed volumes of lysis buffer. The resin was washed with 15 bed volumes of wash buffer WB2 (20 mM Hepes pH 7.5, 150 mM NaCl, 40 mM Imidazol). Saposin was eluted by addition of five bed volumes of elution buffer EB (20 mM Hepes pH 7.5, 150 mM NaCl, 400 mM Imidazol). The eluate was dialyzed overnight against gel filtration buffer GF (20 mM Hepes pH 7.5, 150 mM NaCl) supplemented with recombinant TEV protease. TEV protease containing an un-cleavable His-tag was removed from the eluate by passing it over 2 ml IMAC resin. Cleaved target protein was concentrated to a volume of 5 ml using centrifugal filter units and loaded onto a HiLoad SuperdexTM 200 16/60 GL column using an ÄKTAexplorer 10 chromatography system. Peak fractions were pooled and concentrated to 1.2 mg/ml protein. Typically, a 1-liter bacterial culture yields 5-10 mg of purified Saposin. The protein sample was flash frozen in liquid nitrogen and stored at − 80 °C.
Expression and Purification of POT1 and T2
POT1 was expressed and purified following a described recently18,19. T2 was purified as described recently17.
Generation of Salipro lipid particles
Detergent-solubilized lipids were prepared by dissolving lipid-powder (various lipids from Sigma-Aldrich) in buffer D (50 mM Hepes pH 7.5, 150 mM NaCl, 1 % DDM) to obtain a stock solution that contained 20 mg/ml lipids and incubated for 1h at 37 °C. If not otherwise stated, a brain lipid extract from bovine brain, Type I (Sigma-Aldrich) was used for nanoparticle generation. Prior to Salipro particle reconstitution, the lipid stock solutions were diluted to 5 mg/ml using buffer E (50 mM Hepes pH 7.5, 150 mM NaCl, 0.03 % DDM). For the reconstitution of Salipro particles, 10 μl of purified Saposin A and 5 μl of the respective detergent-solubilized lipids were incubated for 10 min at 37 °C. Subsequently, 50 μl of GF-buffer was added to the mixture and incubated for 10 min at room temperature. The reconstitution mixture was subjected to a gel-filtration analysis on Superdex S200 5/150 analytical gel filtration column (GE Healthcare) and eluted with GF-buffer, giving rise to a monodisperse peak of Salipro particles. For a medium scale preparation, 60 μl of a brain-lipid solution (5 mg/ml brain lipids, Sigma-Aldrich; 50 mM Hepes pH 7.5, 150 mM NaCl, 0.28% DDM) were incubated with 100 μl of purified Saposin A (1.2 mg/ml, 20 mM Hepes pH 7.5, 150 mM NaCl), incubated for 5 min at 37°C and 160 μl gel filtration buffer GF added, followed by incubation of the mixture for 10 min at RT. Then, 230 μl of GF-buffer were added and the sample was subjected to a gel-filtration step on a Superdex200 10/300 GL column using an ÄKTAexplorer 10 chromatography system equilibrated with GF-buffer. Fractions containing Salipro particles were pooled and concentrated to 1 mg/ml using Amicon Centricon filter devices at 3000 × g.
In one experiment, Salipro particles were generated after incubation with an increasing excess of lipids. Here, varying amounts ("Lipids 5": 5 μg, "Lipids 12.5": 12.5 μg, "Lipids 25": 25 μg, "Lipids 50": 50 μg, "Lipids 100": 100 μg) from a brain-lipid solution (5 mg/ml brain lipids, Sigma-Aldrich; 50 mM Hepes pH 7.5, 150 mM NaCl, 0.28% DDM) were incubated for 10 min at 37 °C. 10 μl of purified Saposin A (1.2 mg/ml, 20 mM Hepes pH 7.5, 150 mM NaCl) were added, incubated for 1 min at 37 °C and GF-buffer was added to a final volume of 41 μl. After 10 min incubation at RT, 24 μl of GF-buffer pH 7.5 were added and the samples were subjected to a gel-filtration step on a SuperdexTM 200 5/150 GL analytical gel filtration column using a ÄKTAmicroTM chromatography system (both GE Healthcare) equipped with the Autosampler A-905, which automatically injected 25 μl of protein containing sample. Analytical gel filtration runs were performed at 4 °C at a flow rate of 0.2 ml/min in GF-buffer.
Incorporation of membrane proteins into Salipro Particles
To reconstitute the archaeal mechanosensitve channel T2 into Salipro nanoparticles, 5 μl of a brain-lipid solution (5 mg/ml brain lipids, Sigma-Aldrich; 50 mM Hepes pH 7.5, 150 mM NaCl, 0.28% DDM) were incubated for 5 min at 37 °C, 2 μl of purified membrane protein T2 (10 mg/ml, 20 mM TrisHCl pH 7.5, 150 mM NaCl, 5 % glycerol, 0.03% DDM) were added and subjected to a second incubation step at 37°C. Subsequently, 20 μl of purified Saposin A (1.2 mg/ml, 20 mM Hepes pH 7.5, 150 mM NaCl) were added and incubated for 5 min at 37 °C. Then, 24 μl of 1xPBS pH 7.4 were added and the mixture was incubated for 5 min at RT, followed by adding 39 μl of 1xPBS pH 7.4 to the mixture. The sample was subjected to a gel-filtration step on a Superdex S200 5/150 analytical gel filtration column, equilibrated and eluted with 1x PBS pH 7.4.
For a medium scale preparation of Salipro-T2, 40 μl of a brain-lipid solution (5 mg/ml brain lipids, Sigma-Aldrich; 50 mM Hepes pH 7.5, 150 mM NaCl, 0.28% DDM) and 5 μl of protease inhibitor stock solution (1 cOmplete EDTA-free tablet in 1 ml H2O) were incubated for 5 min at 37 °C, 20 μl of purified membrane protein T2 (10 mg/ml, 20 mM TrisHCl pH 7.5, 150 mM NaCl, 5 % glycerol, 0.03% DDM) were added and subjected to a second incubation step at 37°C. Subsequently, 140 μl of purified Saposin A (1.2 mg/ml, 20 mM Hepes pH 7.5, 150 mM NaCl) were added and incubated for 5 min at 37 °C. Then, 145 μl of 1xPBS pH 7.4 were added and incubated for 5 min at RT, followed by adding 150 μl 1x PBS. The sample was subjected to a gel-filtration step on a Superdex200 10/300 GL column using an ÄKTAexplorer 10 chromatography system equilibrated with 1xPBS pH 7.4. Fractions containing Salipro-T2 were pooled and concentrated to 1 mg/ml using Amicon Centricon filter devices at 3000 × g. The protein sample was flash frozen in liquid nitrogen and stored at − 80 °C.
To reconstitute the bacterial peptide transporter POT1 into Salipro nanoparticles, 5 μl of a brain-lipid solution (5 mg/ml brain lipids, Sigma-Aldrich; 50 mM Hepes pH 7.5, 150 mM NaCl, 0.28% DDM) were incubated for 5 min at 37 °C, 2 μl of purified membrane protein POT1 (10 mg/ml, 20 mM TrisHCl pH 7.5, 150 mM NaCl, 5 % glycerol, 0.4% NM) were added and subjected to a second incubation step at 37°C. Subsequently, 15 μl of purified Saposin A (1.2 mg/ml, 20 mM Hepes pH 7.5, 150 mM NaCl) were added and incubated for 5 min at 37 °C. Then, 19 μl of 1xPBS pH 7.4 were added and the mixture was incubated for 5 min at RT, followed by adding 39 μl of 1xPBS pH 7.4 to the mixture. The sample was subjected to a gel-filtration step on a Superdex S200 5/150 analytical gel filtration column, equilibrated and eluted with 1x PBS pH 7.4.
For a medium scale preparation of Salipro-POT1, 40 μl of a brain-lipid solution (5 mg/ml brain lipids, Sigma-Aldrich; 50 mM Hepes pH 7.5, 150 mM NaCl, 0.28% DDM) were incubated for 5 min at 37 °C, 20 μl of purified membrane protein POT1 (10 mg/ml, 20 mM TrisHCl pH 7.5, 150 mM NaCl, 5 % glycerol, 0.4% NM) were added and subjected to a second incubation step at 37°C. Subsequently, 100 μl of purified Saposin A (1.2 mg/ml, 20 mM Hepes pH 7.5, 150 mM NaCl) were added and incubated for 5 min at 37 °C. Then, 150 μl of 1xPBS pH 7.4 were added and incubated for 5 min at RT, followed by adding 220 μl 1x PBS. The sample was subjected to a gel-filtration step on a Superdex200 10/300 GL column using an ÄKTAexplorer 10 chromatography system equilibrated with 1xPBS pH 7.4, 5% glycerol. Fractions containing Salipro-POT1 were pooled and concentrated to 0.7 mg/ml using Amicon Centricon filter devices at 3000 × g. The protein sample was flash frozen in liquid nitrogen and stored at − 80 °C.
In one experiment, the effect of pH on POT1 reconstitution into Salipro particles was tested. Here, to reconstitute the bacterial peptide transporter POT1 into Salipro nanoparticles, 18 μl of a brain-lipid solution (5 mg/ml brain lipids, Sigma-Aldrich; 50 mM Hepes pH 7.5, 150 mM NaCl, 0.28% DDM) were incubated for 5 min at 37 °C, 5 μl of purified membrane protein POT1 (10 mg/ml, 20 mM TrisHCl pH 7.5, 150 mM NaCl, 5 % glycerol, 0.03% DDM) were added and subjected to a second incubation step at 37°C. Subsequently, 80 μl of purified Saposin A (1.2 mg/ml, 20 mM Hepes pH 7.5, 150 mM NaCl) were added and incubated for 5 min at 37 °C. From this mixture, 23 ul were transferred into a single tube each and 50 μl of buffers with various pH added (pH 4.5: 20 mM Trisodium citrate, 100 mM NaCl; pH 6: 20 mM Trisodium citrate, 100 mM NaCl; pH 7.5: 20 mM TrisHCl, 100 mM NaCl; pH 9: 20 mM TrisHCl, 100 mM NaCl). After 5 min incubation, the samples were subjected to a gel-filtration step on a Superdex S200 5/150 analytical gel filtration column, equilibrated and eluted with buffers at the corresponding pH (4.5-9).
Negative stain electron microscopy
Salipro lipid-only discs: Carbon coated copper grids (400 mesh) were glow discharged in low pressure air to render them hydrophilic for 20 sec before application of 4 μl sample for 30 sec. Washing was done with three drops of water before staining with 4 μl of uranyl formate (1%, Polysciences, USA) for 30 sec. The grids were air dried after removal of excess stain solution by torn filter paper. Negative stain image data were collected with a JEOL2100F field emission gun transmission electron microscope at an accelerating voltage of 200kV and a final magnification of 69500 on a CCD camera (4K × 4K, Tiez Video and Image Processing System, GmbH Gauting, Germany) with 15 mm pixel size (corresponding to 2.16 Å on the specimen level).
Salipro-T2 samples were stained with uranyl acetate and imaged on an FEI TECNAI Polara TEM at 100 kV at a magnification of 59kx and a pixel size of 1.91 A on a UltraScan 4000 CCD (Gatan, Inc.). Individual particles were windowed semi-automatically with the swarm option of e2boxer (EMAN2 package50). After normalization, 33000 particles were subjected to reference-free 2D classification and class-averaging using the EMAN2 routines (e2refine2d).
Thermal unfolding analysis
Thermal unfolding analysis was performed using dye-free differential scanning fluorimetry20 using two samples each for POT in detergent (1x PBS, pH 7.4, 0.4% NM) and Salipro-POT (1x PBS pH 7.4) with a sample volume of 10 μl per capillary at concentrations of 0.2 mg/ml. The temperature was increased by 1°C/min from 20°C to 95°C using a Prometheus NT.48 instrument (Nanotemper).
Cryo-EM
The PepT data was collected and processed as described51. 2.5 μl of purified sample at 0.7 mg/ml concentration was applied to a glow discharged Quantifoil holey carbon grid (400 mesh, 1.2 um hole size) and blotted at 100% humidity using FEI Vitrobot Mark I for 5.5 s blotting time. Images of the vitrified sample were subsequently collected on FEI TF30 Polara electron microscope operated at 300 kV, equipped with a Gatan K2 Summit direct electron detector camera. The images were recorded with a defocus in a range from 1.5 to 2.5 mm at a nominal magnification of 31,000, corresponding to a calibrated physical pixel size of 1.2156 Å/pixel on the specimen at a dose rate on the camera set to 8.2 counts per physical pixel per second. The sample was exposed for a total time of 6s, with each image fractionated into 30 subframes, each with an accumulation time of 0.2 s per frame, with the total accumulated dose of 41e-/Å2 on the specimen.
Image processing
Dose-fractionated super-resolution image frames were drift-corrected using whole-frame motion correction23; a sum of all subframes in each image stack was used for further processing. The particles embedded in vitreous ice were monodisperse and exhibited a good contrast (Fig. 3a), even at low defocii.
For all 2D image display and particle picking, SamViewer, an interactive image analysis program written in Python was used. Defocus was determined using CTFFIND452. Particle extraction and class averaging was performed in RELION53. Using 12200 particles, we performed 2D classification (Fig. 3b) and obtained classes with a large number of inner-membrane details. Selected classes were extracted and probabilistic initial 3D model generation procedure (PRIME) implemented in SIMPLE54 package was used for an ab initio 3D reconstruction generation; This served as a reference model for the maximum likelihood-based 3D classification and auto-refinement procedure implemented in RELION. For the final 3D reconstruction (Fig. 4a,b) 9,913 particles were used and C4 symmetry was imposed; resolutions were estimated using the FSC 0.143 criterion24 on corrected FSC curves to obtain the map at 6.48 Å (Fig. 3e). ResMap was used to estimate local resolution from unbinned and unsharpened raw density map55. Figures were prepared using UCSF Chimera56.
HIV virus like particle production
VLPs carrying HIV-1 JR-FL spikes with a deleted cytoplasmic tail (truncated at residue 708), disulfide linked gp120 and gp41 (Cys501-Cys605), and E168K and N189A mutations (facilitates binding of the PG16 antibody to the JR-FL spike) were produced in 293T cells by calcium phosphate precipitation-mediated cotransfection. For production of [35S]Cys labelled VLPs one 150 cm2 culture flask semi confluent 293T cells was transfected using 10 μg of pCAGGS JRFL gp160SOS,△CT,E168K,N189A and 10 μg of pNL4-3.Luc.R-E DNA57,58. [35S]Cys was incorporated into the VLPs through metabolic labeling in Cys-free Dulbecco′s modified Eagle medium (National Veterinary Institute, Uppsala, Sweden) supplemented with 10 μM unlabeled Cys and 100 μCi of [35S]Cys/ml (Fisher Scientific, Gothenburg, Sweden) from 24 to 48 h after the transfection. Cell debris was removed from the virus-contain media using low speed centrifugation, 1000 rpm for 10 min. The cleared supernatant was layered on top of a step gradient composed of 1 ml of 60% and 4.5 ml of 20% sucrose (wt/wt) in HN buffer (50 mM HEPES, 100 mM NaCl, pH 7.4,) and centrifuged at 4°C for 2 h at 93,000 × g (22,000 rpm) in a Beckman SW28.1 rotor. Radiolabeled virus was collected from the 20/60% sucrose interphase. For unlabelled VLP production 15 culture flasks (150 cm2) with 293T cells were transfected and the virus collected and concentrated as above. To increase VLP concentration the virus was diluted to 15-20% sucrose in HN buffer and added on top of a 1 ml 60% (wt/wt) sucrose cushion followed by a second ultra centrifugation at 4°C for 6 h at 22,000 rpm using the SW28.1 rotor. The virus was collected by fractionation from the bottom and fractions were analysed using SDS-PAGE and Sypro Ruby protein stain to visualize the VLP peak for collection.
Incorporation of the trimeric HIV-spike into the Salipro nanoparticles
VLPs were lysed for 10 min on ice in HN buffer supplemented with 9 mM HEGA-10 in the presence of Saposin A, and in some cases brain lipids. HEGA-10 was then removed by two successive spin SEC steps (Zeba Spin Desalting Columns, 7K MWCO). The columns were washed and centrifuged according to manufacturer's instructions (Pierce). The column removes the HEGA-10 detergent but not saposin A, which with viral lipids replaces the detergent in the spike detergent complexes. Salipro-HIV-spike particles eluted from the column void volume together with the other soluble virus proteins. The sample was then incubated for 15 min at 37°C before lectin purification of the Salipro-HIV-spike particles.
Purification of the Salipro-HIV-spike particles
Salipro-HIV-spikes were purified using Galanthis Nivalis lectin (Sigma-Aldrich) affinity chromatography. The lectin binds to high mannose sugar moieties on the spike protein. The column was washed with 50 column volumes HN buffer followed by elution of the Salipro-HIV-spike particles using 0.3M methyl-αD-mannopyranoside. The methyl-αD-mannopyranoside was removed from the sample using a spin SEC column (Zeba Spin Desalting Columns, 7K MWCO) and changed to HN buffer. The Salipro-HIV-spikes were concentrated by ultra filtration using an Amicon Ultra-4 50K MWCO centrifugation filter (Millipore) and sample purity was assayed with SDS-PAGE using Sypro Ruby protein staining (Lonza group Ltd) and BN-PAGE using silver staining (Thermo Scientific).
Functional assays of the Salipro-HIV-spikes
Salipro-HIV-spike particles were incubated with 10 μg/ml of PG16 antibody, sCD4 and 17b Fab, respectively, in HN buffer for 2 h at 37°C. The formation of ligand spike complexes was followed by BN-PAGE and phosphorimaging. One volume of two times-concentrated BN-PAGE sample buffer was added, and the samples were incubated for 10 min at room temperature and analyzed by gradient BN-PAGE containing 3 to 8% total acrylamide as described previously59. The dried gels were exposed to phosphorimage screens (BAS MS2025; Fujifilm, Science Imaging, Nacka, Sweden) and the radiolabeled bands visualized using a Molecular Imager FX and the Quantity One software (Bio-Rad, Hercules, CA).
Supplementary Material
Acknowledgements
J.F was supported by a European Molecular Biology Organization (EMBO) long-term fellowship. This research was further supported by the Swedish Research Council (Grant 2014-5583 and 2013-3922 to PN, Grant 2010-4483 to C.J.), by the Knut and Alice Wallenberg foundation (Grant 2014.0112), the Swedish Cancer Society (Grant 13 0401), the Swedish Childhood Cancer Foundation (Grant PR2014-0156) and a Start-up grant from Nanyang Technological University to P.N. We also acknowledge the Protein Science Facility at the Karolinska Institutet for cloning assistance and protein purification. Furthermore, the Swedish Science Foundation (Grant 2778), Swedish Cancer Foundation (Grant 0525), EU FP7-People-ITN-2008 Marie Curie actions project Virus Entry 235649 to H.G. and Stiftelsen Läkare mot AIDS Forskningsfond to R.L. supported this work. Cryo-EM work was supported by Howard Hughes Medical Institute and a grant from National Institute of Health (R01GM098672 and 1S10OD020054) to Y.C. Funding support to C.J. by the Karolinska Institutet Center for Biosciences and to L.Z. by a fellowship by the China Scholarship Council are gratefully acknowledged. A.S. was supported by the Molecular Medicine Partnership Unit (MMPU) of the University Clinic Heidelberg and the European Molecular Biology Laboratory. Y.C. is an investigator of Howard Hughes Medical Institute.
Footnotes
Accession Codes
The 3D cryo-EM density map of Salipro-POT has been deposited in the Electron Microscopy Data Bank under the accession number EMD-3302.
Author Contributions
J.F. developed the concept, performed experiments, analyzed data and wrote the manuscript. R.L. developed, performed and analyzed HIV-related experiments, J.-P.A. carried out all cryo-EM experiments, including data acquisition, processing and data interpretation. F.G., P.M., A.F., and C.L. purified membrane proteins, A.S. prepared grids and collected negative-stain data on Salipro-T2. C.J., L.Z. prepared grids and collected negative-stain data on lipid-only Salipro nanoparticles. J.A.G.B., H.G., Y.C. and P.N. contributed project feedback and comments on the manuscript. All authors contributed to interpreting the data and to preparing the manuscript.
Competing Financial Interests
J.F., R.L. and H.G. have filed patent applications related to this work. J.F. is the founder of Salipro Biotech AB.
References
- 1.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein science : a publication of the Protein Society. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Overington J, Al-Lazikani B, Hopkins A. How many drug targets are there? Nature Reviews Drug Discovery. 2006;5:993–996. doi: 10.1038/nrd2199. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 3.Tate CG. Practical considerations of membrane protein instability during purification and crystallisation. Methods in molecular biology. 2010;601:187–203. doi: 10.1007/978-1-60761-344-2_12. doi:10.1007/978-1-60761-344-2_12. [DOI] [PubMed] [Google Scholar]
- 4.Breyton C, Pucci B, Popot JL. Amphipols and fluorinated surfactants: Two alternatives to detergents for studying membrane proteins in vitro. Methods in molecular biology. 2010;601:219–245. doi: 10.1007/978-1-60761-344-2_14. doi:10.1007/978-1-60761-344-2_14. [DOI] [PubMed] [Google Scholar]
- 5.Raschle T, Hiller S, Etzkorn M, Wagner G. Nonmicellar systems for solution NMR spectroscopy of membrane proteins. Current opinion in structural biology. 2010;20:471–479. doi: 10.1016/j.sbi.2010.05.006. doi:10.1016/j.sbi.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rigaud JL, Levy D. Reconstitution of membrane proteins into liposomes. Methods in enzymology. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. doi:10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- 7.Bayburt TH, Carlson JW, Sligar SG. Reconstitution and imaging of a membrane protein in a nanometer-size phospholipid bilayer. Journal of structural biology. 1998;123:37–44. doi: 10.1006/jsbi.1998.4007. doi:10.1006/jsbi.1998.4007. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Sigworth FJ. Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy. Nature. 2009;461:292–295. doi: 10.1038/nature08291. doi:10.1038/nature08291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frauenfeld J, et al. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nature structural & molecular biology. 2011;18:614–621. doi: 10.1038/nsmb.2026. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efremov RG, Leitner A, Aebersold R, Raunser S. Architecture and conformational switch mechanism of the ryanodine receptor. Nature. 2015;517:39–43. doi: 10.1038/nature13916. doi:10.1038/nature13916. [DOI] [PubMed] [Google Scholar]
- 11.Bruhn H. A short guided tour through functional and structural features of saposin-like proteins. The Biochemical journal. 2005;389:249–257. doi: 10.1042/BJ20050051. doi: 10.1042/bj20050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olmeda B, Garcia-Alvarez B, Perez-Gil J. Structure-function correlations of pulmonary surfactant protein SP-B and the saposin-like family of proteins. Eur Biophys J. 2013;42:209–222. doi: 10.1007/s00249-012-0858-9. doi:10.1007/s00249-012-0858-9. [DOI] [PubMed] [Google Scholar]
- 13.Ahn VE, Faull KF, Whitelegge JP, Fluharty AL, Prive GG. Crystal structure of saposin B reveals a dimeric shell for lipid binding. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:38–43. doi: 10.1073/pnas.0136947100. doi:10.1073/pnas.0136947100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawkins CA, de Alba E, Tjandra N. Solution structure of human saposin C in a detergent environment. Journal of molecular biology. 2005;346:1381–1392. doi: 10.1016/j.jmb.2004.12.045. doi:10.1016/j.jmb.2004.12.045. [DOI] [PubMed] [Google Scholar]
- 15.Rossmann M, et al. Crystal structures of human saposins C andD: implications for lipid recognition and membrane interactions. Structure. 2008;16:809–817. doi: 10.1016/j.str.2008.02.016. doi:10.1016/j.str.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Popovic K, Holyoake J, Pomes R, Prive GG. Structure of saposin A lipoprotein discs. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2908–2912. doi: 10.1073/pnas.1115743109. doi:1115743109 [pii] 10.1073/pnas.1115743109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Low C, et al. Nanobody mediated crystallization of an archeal mechanosensitive channel. PloS one. 2013;8:e77984. doi: 10.1371/journal.pone.0077984. doi:10.1371/journal.pone.0077984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guettou F, et al. Structural insights into substrate recognition in proton-dependent oligopeptide transporters. EMBO reports. 2013;14:804–810. doi: 10.1038/embor.2013.107. doi:10.1038/embor.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guettou F, et al. Selectivity mechanism of a bacterial homolog of the human drug-peptide transporters PepT1 and PepT2. Nature structural & molecular biology. 2014;21:728–731. doi: 10.1038/nsmb.2860. doi:10.1038/nsmb.2860. [DOI] [PubMed] [Google Scholar]
- 20.Alexander CG, et al. Novel microscale approaches for easy, rapid determination of protein stability in academic and commercial settings. Biochimica et biophysica acta. 2014;1844:2241–2250. doi: 10.1016/j.bbapap.2014.09.016. doi:10.1016/j.bbapap.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cyrklaff M, Adrian M, Dubochet J. Evaporation during preparation of unsupported thin vitrified aqueous layers for cryo-electron microscopy. Journal of electron microscopy technique. 1990;16:351–355. doi: 10.1002/jemt.1060160407. doi:10.1002/jemt.1060160407. [DOI] [PubMed] [Google Scholar]
- 22.Rubinstein JL. Structural analysis of membrane protein complexes by single particle electron microscopy. Methods. 2007;41:409–416. doi: 10.1016/j.ymeth.2006.07.019. doi:10.1016/j.ymeth.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 23.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. doi:10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. doi:10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plotkin SA, Plotkin SL. The development of vaccines: how the past led to the future. Nature reviews. Microbiology. 2011;9:889–893. doi: 10.1038/nrmicro2668. doi:10.1038/nrmicro2668. [DOI] [PubMed] [Google Scholar]
- 26.Agrawal N, et al. Functional stability of unliganded envelope glycoprotein spikes among isolates of human immunodeficiency virus type 1 (HIV-1) PloS one. 2011;6:e21339. doi: 10.1371/journal.pone.0021339. doi:10.1371/journal.pone.0021339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leaman DP, Zwick MB. Increased functional stability and homogeneity of viral envelope spikes through directed evolution. PLoS pathogens. 2013;9:e1003184. doi: 10.1371/journal.ppat.1003184. doi:10.1371/journal.ppat.1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loving R, Sjoberg M, Wu SR, Binley JM, Garoff H. Inhibition of the HIV-1 spike by single-PG9/16-antibody binding suggests a coordinated-activation model for its three protomeric units. Journal of virology. 2013;87:7000–7007. doi: 10.1128/JVI.00530-13. doi:10.1128/JVI.00530-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julien JP, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4351–4356. doi: 10.1073/pnas.1217537110. doi:10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. doi:10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. doi:10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thali M, et al. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. Journal of virology. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagn F, Etzkorn M, Raschle T, Wagner G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. Journal of the American Chemical Society. 2013;135:1919–1925. doi: 10.1021/ja310901f. doi:10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knowles TJ, et al. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. Journal of the American Chemical Society. 2009;131:7484–7485. doi: 10.1021/ja810046q. doi:10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- 35.McGregor CL, et al. Lipopeptide detergents designed for the structural study of membrane proteins. Nature biotechnology. 2003;21:171–176. doi: 10.1038/nbt776. doi:10.1038/nbt776. [DOI] [PubMed] [Google Scholar]
- 36.Park SH, et al. Nanodiscs versus macrodiscs for NMR of membrane proteins. Biochemistry. 2011;50:8983–8985. doi: 10.1021/bi201289c. doi:10.1021/bi201289c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schafmeister CE, Miercke LJ, Stroud RM. Structure at 2.5 A of a designed peptide that maintains solubility of membrane proteins. Science. 1993;262:734–738. doi: 10.1126/science.8235592. [DOI] [PubMed] [Google Scholar]
- 38.Tao H, et al. Engineered nanostructured beta-sheet peptides protect membrane proteins. Nature methods. 2013;10:759–761. doi: 10.1038/nmeth.2533. doi:10.1038/nmeth.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tribet C, Audebert R, Popot JL. Amphipols: polymers that keep membrane proteins soluble in aqueous solutions. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Mu Z, Li Y, Bi Y, Wang Y. Smaller Nanodiscs are Suitable for Studying Protein Lipid Interactions by Solution NMR. The protein journal. 2015;34:205–211. doi: 10.1007/s10930-015-9613-2. doi:10.1007/s10930-015-9613-2. [DOI] [PubMed] [Google Scholar]
- 41.Paulsen CE, Armache JP, Gao Y, Cheng Y, Julius D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature. 2015;520:511–517. doi: 10.1038/nature14367. doi:10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rollauer SE, et al. Structure of the TatC core of the twin-arginine protein transport system. Nature. 2012;492:210–214. doi: 10.1038/nature11683. doi:10.1038/nature11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. doi:10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haynes BF, Verkoczy L. AIDS/HIV. Host controls of HIV neutralizing antibodies. Science. 2014;344:588–589. doi: 10.1126/science.1254990. doi:10.1126/science.1254990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwong PD, Mascola JR, Nabel GJ. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nature reviews. Immunology. 2013;13:693–701. doi: 10.1038/nri3516. doi:10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- 46.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. doi:10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muster T, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. Journal of virology. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rujas E, et al. Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Non-Neutralizing Forms of the Anti-Hiv-1 Antibody 4e10. Journal of virology. 2015 doi: 10.1128/JVI.01793-15. doi:10.1128/JVI.01793-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS pathogens. 2006;2:e83. doi: 10.1371/journal.ppat.0020083. doi:10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. Software tools for macromolecular microscopy. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. doi:10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. bioRxiv. 2015 doi: 10.1016/j.jsb.2015.08.008. doi:10.1101/020917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. doi:10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elmlund D, Elmlund H. SIMPLE: Software for ab initio reconstruction of heterogeneous single-particles. Journal of structural biology. 2012;180:420–427. doi: 10.1016/j.jsb.2012.07.010. doi:10.1016/j.jsb.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 55.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nature methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. doi:10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. doi:10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Moore PL, et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. Journal of virology. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. doi:10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu SR, et al. Single-particle cryoelectron microscopy analysis reveals the HIV-1 spike as a tripod structure. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18844–18849. doi: 10.1073/pnas.1007227107. doi:10.1073/pnas.1007227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loving R, Wu SR, Sjoberg M, Lindqvist B, Garoff H. Maturation cleavage of the murine leukemia virus Env precursor separates the transmembrane subunits to prime it for receptor triggering. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7735–7740. doi: 10.1073/pnas.1118125109. doi:10.1073/pnas.1118125109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.