Summary
Escherichia coli (E. coli) mazEF is a stress-induced toxin-antitoxin (TA) module. The toxin MazF is an endoribonuclease that cleaves single-stranded mRNAs at ACA sequences. Here, we show that MazF cleaves at ACA sites at or closely upstream of the AUG start codon of some specific mRNAs and thereby generates leaderless mRNAs. Moreover, we provide evidence that MazF also targets 16S rRNA within 30S ribosomal subunits at the decoding center, thereby removing 43 nucleotides from the 3′ terminus. As this region comprises the anti-Shine-Dalgarno (aSD) sequence that is required for translation initiation on canonical mRNAs, a subpopulation of ribosomes is formed that selectively translates the described leaderless mRNAs both in vivo and in vitro. Thus, we have discovered a modified translation machinery that is generated in response to MazF induction and that probably serves for stress adaptation in Escherichia coli.
Introduction
Toxin-antitoxin modules are present in the chromosomes of many bacteria, including pathogens (Engelberg-Kulka and Glaser, 1999; Mittenhuber, 1999; Hayes, 2003; Pandey and Gerdes, 2005; Engelberg-Kulka et al., 2006; Agarwal et al., 2007; Ramage et al., 2009). Each of these modules consists of a pair of genes, usually cotranscribed as operons, in which, generally, the downstream gene encodes for a stable toxin and the upstream gene encodes for a labile antitoxin. In E. coli, seven toxin-antitoxin systems have been described (Metzger et al., 1988; Masuda et al., 1993; Aizenman et al., 1996; Mittenhuber, 1999; Christensen et al., 2001; Hayes, 2003; Pandey and Gerdes, 2005; Schmidt et al., 2007). Among these, one of the most studied is the chromosomal toxin-antitoxin system mazEF, which was the first to be described as regulatable and responsible for bacterial programmed cell death (Aizenman et al., 1996; Engelberg-Kulka et al., 2006). E. coli mazEF encodes the labile antitoxin MazE and the stable toxin MazF. Both mazE and mazF are coexpressed and negatively autoregulated at transcriptional level (Marianovsky et al., 2001). E. coli mazEF is triggered by various stressful conditions, as treatment with antibiotics affecting transcription or translation (Hazan et al., 2004), or by an increase of ppGpp upon severe amino acid starvation (Aizenman et al., 1996). Such stressful conditions prevent mazEF expression; thereby, the short-lived antitoxin MazE is degraded by the ATP-dependent ClpAP serine protease (Aizenman et al., 1996), permitting the stable MazF to exert its toxic effect (Engelberg-Kulka et al., 2006). mazEF-mediated cell death was also reported as a population phenomenon requiring a quorum-sensing factor called extracellular death factor (EDF) (Kolodkin-Gal et al., 2007).
MazF is a sequence-specific endoribonuclease that preferentially cleaves single-stranded mRNAs at ACA sequences (Zhang et al., 2003, 2004). As previously reported (Christensen et al., 2003; Zhang et al., 2003), MazF induction causes inhibition of protein synthesis. However, we have recently shown that, surprisingly, this inhibition was not complete; though MazF led to inhibition of synthesis of most proteins (about 90%), it selectively enabled specific synthesis of about 10% of proteins (Amitai et al., 2009). Some of those proteins were required for death of most cells within a population. However, we also found that MazF enabled the synthesis of proteins that permitted survival of a small subpopulation under stressful conditions that cause mazEF-mediated cell death for the majority of the population. Among the proteins involved in cell death were: (1) YfiD, a glycine radical protein known to be able to replace the oxidatively damaged pyruvate formate-lyase subunit (Wagner et al., 2001) and (2) YfbU, a protein of unknown function (Amitai et al., 2009). Among the proteins involved in cell survival were: (1) DeoC, a deoxy ribose-phosphate aldolase known to participate in the catabolism of deoxyribonucleosides (Hammer-Jespersen et al., 1971), and RsuA, a protein known to catalyze the pseudouridylation at position 516 in the 16S RNA (Wrzesinski et al., 1995). Because several ACA sites—the potential MazF cleavage sites—are located in the corresponding mRNAs, we were intrigued by the mechanism that is responsible for selective synthesis of these proteins.
Here, we have elucidated the underlying molecular mechanism leading to selective translation of a particular set of mRNAs upon MazF induction in E. coli. We found that, due to its endoribonucleolytic activity, MazF cleaves at ACA sites at or closely upstream of the AUG start codon of specific mRNAs. Thereby, short-leadered and leaderless mRNAs (lmRNAs) are generated, respectively. Surprisingly, the 16S rRNA of the 30S ribosomal subunit is another target of MazF endoribonuclease. Specifically, the toxin cleaves at an ACA triplet in the 16S rRNA located 5′ of helix 45. Thus, this cleavage leads to loss of 43 nucleotides (nts) at the 3′ terminus of 16S rRNA, including helix 45 and the anti-Shine-Dalgarno (aSD) sequence. As the SD-aSD interaction is required for translation initiation on canonical ribosome-binding sites, truncation of 16S rRNA yields a specialized protein synthesis machinery designated as “stress-ribosome,” which selectively translates lmRNAs generated by MazF. Because MazF is triggered under stressful conditions, our results uncovered a hitherto uncharacterized stress adaptation mechanism in E. coli, which is based on generation of a heterogeneous ribosome population that provides a means for selective synthesis of a subclass of proteins.
Results
MazF Cleaves yfiD and rpsU mRNAs at Specific Sites Directly Upstream of the AUG Start Codon In Vivo
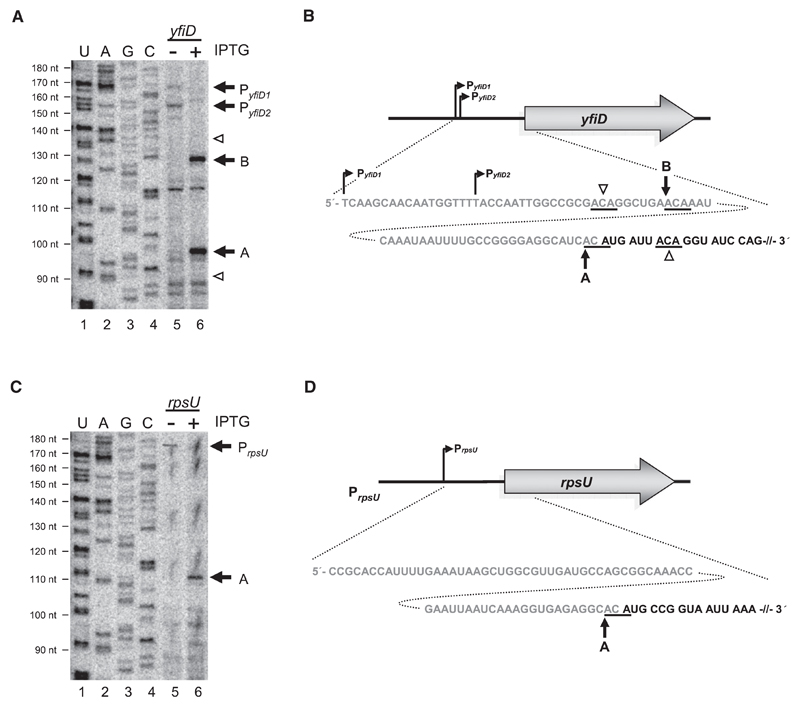
Recent studies revealed that several mRNAs are translated upon induction of mazF in vivo (Amitai et al., 2009). However, the presence of potential MazF cleavage sites within these mRNAs would imply their immediate degradation following MazF cleavage. As several lines of evidence indicate that MazF cleaves only at ACA sequences located within unstructured regions and that stable secondary structures can shield the sites of cleavage (Zhang et al., 2004; Zhu et al., 2008), we first examined whether some candidate mRNAs are protected from mRNA cleavage. Therefore, we performed primer extension analysis on total RNA prepared upon mazF overexpression in vivo using the same conditions that determined the synthesis of the respective proteins (Amitai et al., 2009). We studied (1) mRNAs encoding proteins YfiD and YfbU, which are selectively translated in the presence of increased levels of MazF (Amitai et al., 2009), and (2) rpsU mRNA encoding ribosomal protein S21, a coding region that does not contain an ACA cleavage site. In brief, mazF expression was induced in E. coli strain MC4100relA+ harboring plasmid pSA1 that bears an IPTG-inducible mazF gene. Fifteen minutes thereafter, total RNA was purified and primer extension analysis was performed employing specific primers for yfiD (R48) (Table S1 available online and Figure 1A, lanes 5 and 6), rpsU (Y50) (Table S1 and Figure 1C, lanes 5 and 6), and yfbU (B6) (Table S1 and Figures S1C and S1D) mRNAs. In absence of mazF overexpression, primer extension reactions for yfiD mRNA (Figures 1A, lane 5, and 1B) and rpsU mRNA (Figures 1C, lane 5, and 1D) generate signals that correspond to the 5′ termini of the transcripts synthesized from the annotated promoters (PyfiD1 and PrpsU) (Green et al., 1998; Lupski et al., 1984). In addition, we determined a second transcriptional start point for yfiD mRNA, giving rise to an mRNA harboring a 57 nt long 5′ untranslated region (5′UTR) (PyfiD2) (Figures 1A, lane 5, and 1B). Surprisingly, upon induction of mazF expression, primer extension resulted in generation of cDNAs of 128 and 97 nts in length, which correspond to cleavage of yfiD mRNA at two ACA triplets 30 nts and directly upstream of the AUG start codon (Figures 1A, lane 6, and 1B). In contrast, we did not observe cleavage at two additional ACA sequences located 39 nts upstream and 3 nts downstream of the start codon (Figures 1A, lane 6, and 1B). We obtained similar results when primer extension was performed with the rpsU-specific primer (Figure 1C, lane 6). Again, MazF activity resulted in cleavage of rpsU mRNA at the ACA located directly upstream of the start codon (Figures 1C, lane 6, and 1D). Corresponding to absence of ACAs in the coding sequence of rpsU mRNA, we did not observe MazF cleavage within this region (Figures 1C, lane 6). To verify MazF cleavage at the sites determined in vivo, in addition, in vitro cleavage assays have been performed on yfiD and rpsU mRNAs. In vitro-transcribed mRNAs were incubated with purified MazF protein at 37°C. Subsequent primer extension analyses revealed MazF cleavage at the same positions as observed in vivo (Figures S1A and S1B, lane 7).
Figure 1. Determination of MazF Cleavage Sites on yfiD and rpsU mRNAs In Vivo and Schematic Depiction of Genomic Organization, 5′UTRs, and Proximal Coding Regions of the Respective mRNAs.
(A and C) Primer extensions employing primers (Table S1) specific for yfiD mRNA (A) and rpsU mRNA (C) performed on total RNA purified from E. coli strain MC4100relA+ comprising plasmid pSA1 without (lane 5) or with (lane 6) mazF overexpression. Extension signals corresponding to transcriptional start points of yfiD mRNA (PyfiD1 and PyfiD2) and rpsU mRNA (PrpsU) are indicated by black arrows (lane 5). Signals corresponding to MazF cleavage are indicated by black arrows and labeled analogous to Figures 1B and 1D, where “A” designates cleavage directly upstream of the AUG start codon, resulting in formation of a lmRNA, and “B” indicates cleavage further upstream (lane 6). White arrowheads indicate ACA triplets not cleaved by MazF (lane 6). (Lanes 1–4) Sequencing reactions of 16S rRNA employing primer V43 (Table S1) to determine length of primer extension (indicated to the left).
(B and D) Schematic depictions of promoter positions and sequence of 5′UTRs (in gray) and proximal coding regions (in black) of yfiD mRNA (B) and rpsU mRNA (D). MazF cleavage sites are underlined and indicted by arrows. The cleavage site directly upstream of the AUG start codon is marked with “A.” The cleavage site further upstream is indicated by “B.” Potential ACA triplets where MazF cleavage does not occur are indicated by white arrowheads.
See also Figure S1.
Surprisingly, primer extension using total RNA purified from cells with and without mazF overexpression employing a yfbU specific primer (B6) (Table S1) yielded a stop signal that corresponds to the A of the AUG start codon in both cases (Figure S1C, lanes 5 and 6). These results indicate that, in vivo, the yfbU mRNA might be transcribed as lmRNA. To entirely exclude MazF cleavage at the ACA present in the yfbU mRNA just upstream of the start codon (Figure S1D), primer extension analysis was repeated on total RNA purified from strain MC4100relA+ΔmazEF (Figure S1C, lane 7). Likewise, the extension signal indicates the presence of a leaderless yfbU mRNA (Figure S1C, lane 7). This result was verified by primer extension analysis at 55°C employing the heat stable reverse transcriptase Superscript III (Invitrogen) using a second yfbU specific primer (I6) (Table S1). As shown in Figure S1C, primer extension reactions generated cDNAs of 88 nts in length, again corresponding to transcription starting directly at the AUG codon (Figure S1C, lanes 8 and 9).
Taken together, these results tempted us to speculate that mazF induction results in selective translation of lmRNAs, which are either generally present, like yfbU mRNA, or generated by MazF, as shown for yfiD and rpsU mRNAs.
Selective Translation of an lmRNA upon mazF Overexpression
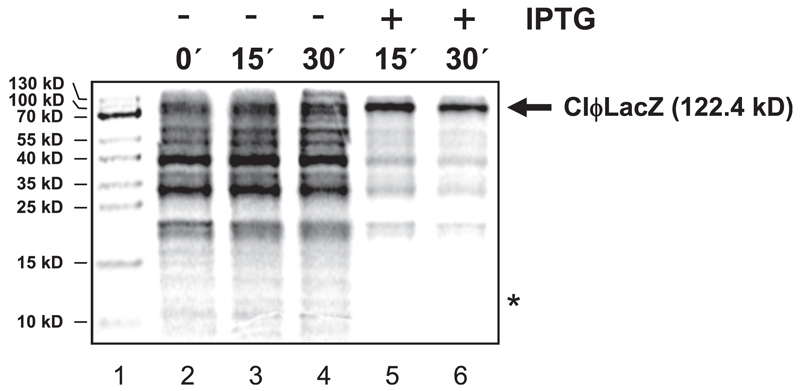
lmRNAs are selectively translated in the absence of ribosomal proteins S1 and/or S2 (Moll et al., 2002). Moreover, we have recently shown that lmRNAs are selectively translated by protein-depleted ribosomes, which are formed upon treatment of E. coli cells with the aminoglycoside antibiotic kasugamycin (Ksg) in vivo (Kaberdina et al., 2009). In light of the fact that the mazEF module can be triggered by antibiotics targeting the ribosome (Sat et al., 2001) and that MazF activity results in formation of lmRNAs, we hypothesized that MazF could likewise affect the protein synthesis machinery, thus rendering it selective for lmRNAs. To verify this notion, pulse labeling was performed employing E. coli strain MC4100relA+ harboring plasmid pSA1, which encodes the inducible mazF gene, and plasmid pKTplaccI. Plasmid pKTplaccI encodes the leaderless cI-lacZ fusion gene, giving rise to the 122.4 kD CI-LacZ fusion protein (Grill et al., 2000). The strain was grown in M9 minimal medium as specified in the Experimental Procedures. At OD600 of 0.5, the culture was divided and IPTG was added to one half to induce mazF expression. Before and 15 and 30 min after induction, pulse labeling was performed. Upon precipitation and separation of labeled proteins by SDS-PAGE, the autoradiography shown in Figure 2 reveals selective translation of the leaderless cI-lacZ mRNA upon overexpression of mazF. In contrast, translation of bulk mRNA is severely inhibited (Figure 2, lanes 5 and 6). It has to be noted that, despite induction of mazF overexpression, the protein cannot be detected (Figure 2, lanes 5 and 6) (Amitai et al., 2009).
Figure 2. Overexpression of mazF Results in Stimulated and Selective Translation of the Leaderless cI-lacZ mRNA In Vivo.
Pulse labeling performed with E. coli strain MC4100relA+ harboring plasmids pKTplaccI, encoding the leaderless cI-lacZ fusion gene (Grill et al., 2000), and pSA1, encoding the mazF gene under control of the lac-operator. At time point 0 (lane 2), the culture was divided and one half remained untreated (lanes 3 and 4), whereas in the other half, mazF expression was induced with IPTG (lanes 5 and 6). At time points indicated, pulse labeling was performed. The position of the CI-LacZ fusion protein (122.4 kD) is indicated by an arrow to the right of the autoradiograph. The tentative position of MazF (12 kD) is marked by an asterisk. (Lane 1) Protein marker.
MazF Cleaves 16S rRNA of 30S Subunits and 70S Ribosomes
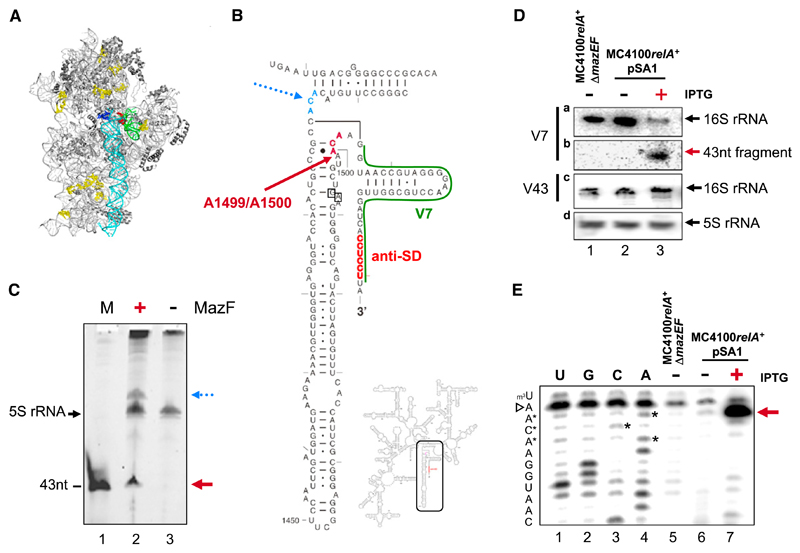
Taken together, MazF-mediated generation and selective translation of lmRNAs suggest that the endoribonuclease might directly affect ribosomes, as rRNA could represent a potential target. As indicated in the structure of the 30S ribosomal subunit (Figure 3A), several ACA triplets are present in the 16S rRNA, most of which are located within structured regions or are protected by ribosomal proteins. However, as shown in the secondary structure of the 3′-terminal 16S rRNA, two accessible ACA sites are located at positions 1500–1502 and 1396–1398 (Figure 3B). Cleavage at the latter site would be detrimental for ribosome activity, as it would result in loss of helix 44, which provides most intersubunit bridges (Gabashvili et al., 2000). However, given that this region is enclosed by tRNA and mRNA (Woodcock et al., 1991; Yusupova et al., 2006), we hypothesized that translationally active ribosomes might be protected at this position. In contrast, cleavage at position 1500–1502 in close proximity to the site for colicin E3 cleavage (between nts A1493 and G1494) (Figure 3B; Senior and Holland, 1971) would result in loss of the 3′ terminus of 16S rRNA containing helix 45 and the aSD sequence. The SD-aSD interaction is important for translation initiation complex formation on canonical mRNAs comprising structured 5′UTRs. Particularly in vivo, when several mRNAs have to compete for 30S subunits (Hui and de Boer, 1987; Calogero et al., 1988), the small subunit captures mRNAs via SD-aSD interaction (Kaminishi et al., 2007; Yusupova et al., 2001). Therefore, it is feasible that MazF activity could result in formation of ribosomes selective for translation of lmRNAs.
Figure 3. The 16S rRNA of Assembled 70S Ribosomes Represents a Target for MazF Activity.
(A) The structure of the 30S ribosomal subunit was modeled employing Polyview 3D (Porollo and Meller, 2007) and PyMOL molecular system software (DeLano, 2002) and PDB file 2AVY (Schuwirth et al., 2005). 16S rRNA (light gray), proteins (dark gray), helix 44 (cyan), and helix 45 (green) are shown. ACA sequences present in 16S rRNA, which are protected by proteins or structural features of rRNA, are indicated in yellow. Two potential MazF cleavage sites at positions 1396–1398 and 1500–1502 are indicated in blue and red, respectively.
(B) Secondary structure of 16S rRNA. Decoding region and helices 44 and 45 are enlarged. Potential MazF cleavage sites are indicated in blue and red, as in (A). Site of Colicin E3 cleavage (AG 1493/1494) is boxed, and aSD sequence is shown in red. Primer V7 (indicated in green) complementary to positions 1511–1535 of 16S rRNA was used for northern blot and primer extension analyses.
(C) Treatment of 70S ribosomes with MazF in vitro results in cleavage of 16S rRNA at positions indicated in (B). The 3′ end of rRNA was labeled with pC-Cy3. rRNA fragments obtained upon incubation of 70S ribosomes with (lane 2) or without (lane 3) MazF were separated by denaturing PAGE. (Lane 1) In vitro-synthesized Cy3-labeled RNA fragment of 43 nts in length (kindly provided by Dr. U. Bläsi) was used as a size marker. Red and blue arrows indicate fragments corresponding to MazF cleavage at positions shown in (B). The position of 5S rRNA is indicated by a black arrow.
(D) To verify MazF-mediated formation of the 43 nt fragment in vivo, total RNA prepared from untreated MC4100relA+ cells harboring plasmid pSA1 (lane 2) and upon induction of mazF expression with IPTG (lane 3) were separated by denaturing PAGE, blotted, and probed with oligonucleotide V7 (Figure 3B and Table S1) to determine the amount of 3′ terminus present in full-length 16S rRNA (a) and the cleaved 3′-terminal fragment (b). To determine the amount of total 16S rRNA, oligonucleotide V43 (Table S1; c) was used. Total RNA prepared from strain MC4100relA+ΔmazEF(lane 1) was included as control. Northern blot analysis of 5S rRNA employing primer R25 (Table S1; d) served as a loading control.
(E) The same RNA (D) was used for primer extension analysis employing primer V7 (Figure 3B). In contrast to strains MC4100relA+ΔmazEF (lane 5) and untreated MC4100relA+pSA1 (lane 6), a signal was obtained employing RNA purified upon mazF overexpression (lane 7) that indicates unambiguously the site of MazF cleavage 5′ of ACA between nts A1499 and A1500, thereby removing 43 nts at the 3′ terminus. (Lanes 1–4) Sequencing reactions. The sequence is given to the left; the ACA triplet and the signal corresponding to stop of reverse transcription due to the m3U1498 modification are indicated by asterisks and an open arrowhead, respectively.
See also Figure S2.
To pursue this idea, we treated 30S subunits (data not shown) and 70S ribosomes with purified MazF in vitro (Figure 3C). As specified in the Experimental Procedures, rRNAs within ribosomes were labeled 3′ terminally with pC-Cy3. Upon incubation with purified MazF protein, rRNA was extracted and separated on a denaturing gel. As shown in Figure 3C, upon MazF treatment, we observed formation of a fragment of about 44 nts in length (Figure 3C, lane 2). As we employed purified ribosomes, which do not harbor tRNAs or mRNAs protecting 16S rRNA at the decoding site (Woodcock et al., 1991; Yusupova et al., 2006), in addition, a minor fragment was generated that corresponds to cleavage at position 1396 of 16S rRNA (Figure 3C, lane 2). Additional experiments employing 16S as well as 23S rRNA radioactively labeled at the 5′ (data not shown) and 3′ end (Figure S2) revealed no further MazF cleavage.
Next, we tested for cleavage of 16S rRNA at A1500 in vivo. Therefore, total RNA was prepared from strain MC4100relA+ ΔmazEF (Figure 3D, lane 1) or strain MC4100relA+ harboring plasmid pSA1 without (Figure 3D, lane 2) or upon induction of MazF synthesis (Figure 3D, lane 3) and subjected to northern blot analysis employing primers V7, specific for the 3′ end of 16S rRNA (Figure 3B and Table S1), and V43, binding to positions 939–955 in 16S rRNA (Table S1). As shown in Figure 3D, in contrast to total RNA prepared from untreated cells (Figure 3Db, lane 2), induction of MazF synthesis yielded the same fragment (Figure 3Db, lane 3) that we observed upon treatment of 70S ribosomes with MazF protein in vitro (Figure 3C, lane 2). Correspondingly, the signal for 16S rRNA obtained with primer V7 decreased upon MazF cleavage (Figure 3Da, lanes 2 and 3). However, the amount of total 16S rRNA, as verified with primer V43, remained constant (Figure 3Dc).
To unequivocally determine the site of cleavage, primer extension analysis employing primer V7 was performed on total RNA used for northern blotting. The result reveals that, upon overexpression of mazF, cleavage occurs upstream of A1500 (Figure 3E, lane 7), thus removing 43 nts at the 3′ end of 16S rRNA.
MazF Activity Results in Formation of Stress Ribosomes In Vivo
Next, we aimed to confirm that MazF-mediated removal of the 3′ end of 16S rRNA comprising the aSD sequence and helix 45 results in formation of a distinct ribosome population that is functionally specific for translation of lmRNAs, here referred to as “stress ribosomes” (70SΔ43). E. coli strain MC4100relA+ harboring plasmid pSA1 was grown in LB medium at 37°C. At OD600 of 0.5, the culture was divided and IPTG was added to one half to induce mazF expression, whereas the other half remained untreated. Intriguingly, the ribosome profile did not change upon overexpression of mazF except for reduction of polysome peaks, indicating inhibition of bulk mRNA translation (Figure S3). This result indicates that ribosomes are not degraded and that, in contrast to treatment with the antibiotic Ksg (Kaberdina et al., 2009), no protein-depleted ribosomes are formed upon mazF overexpression.
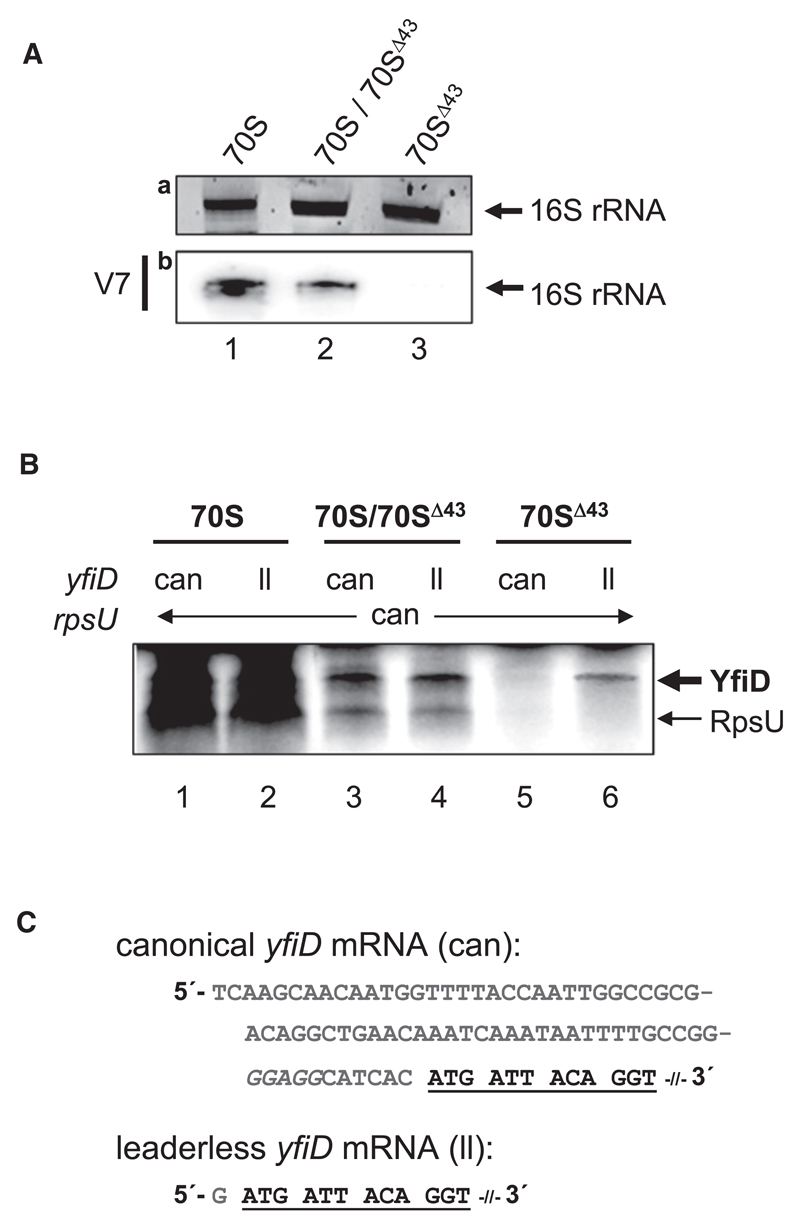
Concomitantly, ribosomes were prepared and their 16S rRNA was analyzed. First, the overall amount of 16S rRNA was determined upon denaturing gel electrophoresis by ethidium bromide staining (Figure 4Aa). The same samples were subsequently probed by northern blotting employing primer V7 for presence or absence of the 3′-terminal fragment (Figure 4Ab). In contrast to rRNA derived from 70S ribosomes purified without mazF expression (Figure 4Ab, lane 1), northern blot analysis employing rRNA from ribosomes purified upon mazF overexpression revealed a reduced signal for probe V7 (Figure 4Ab, lane 2; 70S/70SΔ43). This result supports our hypothesis that mazF overexpression yields a heterogeneous ribosome population still containing a substantial fraction of canonical 70S. Thus, we included a second purification step to remove uncleaved 70S ribosomes with the help of a biotinylated SD-oligonucleotide (V5) (Table S1) immobilized on magnetic beads. This additional step allowed clear separation of stress ribosomes from uncleaved 70S, as verified by the lack of a signal employing primer V7 in northern blot analysis (Figure 4Ab, lane 3).
Figure 4. Stress Ribosomes Formed upon Overexpression of mazF In Vivo Selectively Translate Leaderless yfiD mRNA.
(A) rRNA prepared from 10 pmoles of ribosomes purified from untreated cells (70S; lane 1) upon overexpression of mazF (70S/70SΔ43; lane 2) and upon further removal of uncleaved ribosomes employing a biotinylated SD-oligonucleotide (70SΔ43; lane 3), which were used for in vitro translation shown in (B), was separated by denaturing PAGE and stained with ethidium bromide (a) to determine amount and integrity of 16S rRNA. The same rRNA was probed using labeled oligonucleotide V7 (b) to verify presence and absence of the 3′-terminal 43 nt fragment in the individual ribosome preparations.
(B) In vitro translation of canonical (can; lanes 1, 3, and 5) and leaderless (ll; lanes 2, 4, and 6) variants of yfiD mRNA employing 70S ribosomes purified from untreated E. coli MC4100relA+ cells harboring plasmid pSA1 (70S; lanes 1 and 2), purified upon mazF overexpression (70S/70SΔ43; lanes 3 and 4) and upon removal of uncleaved ribosomes employing immobilized biotinylated SD oligonucleotides (70SΔ43; lanes 5 and 6). In all reactions, equimolar amounts of canonical rpsU mRNA were added as internal control. Positions of YfiD (14.3 kD) and RpsU (8.5 kD) proteins in the autoradiograph are indicated to the right.
(C) 5′UTR and proximal coding region (underlined) of canonical and leaderless yfiD mRNAs used for in vitro translation. The SD sequence of the canonical mRNA is indicated in italics.
See also Figure S3.
Next, the purified ribosomes were tested in vitro for translational specificity using canonical and leaderless yfiD mRNA variants (Figure 4C, can and ll). Concomitantly, canonical rpsU mRNA containing a 135 nts 5′UTR (Figure S1B) was included in all reactions as internal control. As shown in Figure 4B, 70S ribosomes purified from untreated cells (lanes 1 and 2), as well as the heterogeneous ribosome population containing 70SΔ43 and 70S ribosomes roughly in a 1:1 ratio (O.V., unpublished data) purified upon mazF overexpression, were proficient in translating both canonical (can) and leaderless (ll) yfiD mRNA variants as well as the canonical rpsU mRNA (Figure 4B, lanes 3 and 4). Nevertheless, translational efficiency of heterogeneous 70S/70SΔ43 ribosomes was reduced for the canonical mRNA and lmRNA variant, respectively, when compared to canonical 70S ribosomes. As expected and consistent with lack of the aSD sequence, purified 70SΔ43 ribosomes did not translate canonical yfiD mRNA (Figure 4B, lane 5) as well as canonical rpsU mRNA (Figure 4B, lanes 5 and 6). In contrast, they were proficient in translation of the leaderless yfiD mRNA variant (Figure 4B, lane 6). These results clearly substantiate our notion that MazF activity generates a subpopulation of ribosomes that lack 43 nts of the 3′ end of 16S rRNA and thus selectively translate lmRNAs.
Induction of MazF Activity by Stress Conditions Leads to Formation of 70SΔ43 Ribosomes and lmRNAs
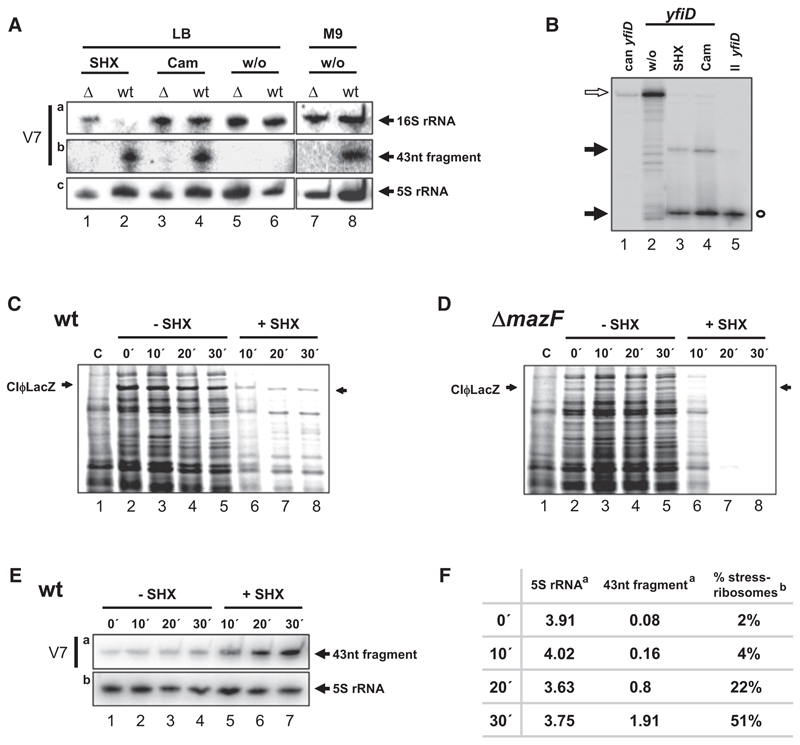
The results shown above were obtained upon artificial overexpression of mazF. Therefore, we next asked whether stress conditions that trigger the mazEF module (Kolodkin-Gal and Engelberg-Kulka, 2006) can likewise induce formation of stress ribosomes. Therefore, strains MC4100relA+ and MC4100relA+ ΔmazEF were treated with serine hydroxamate (SHX), which induces the stringent response, thus leading to ppGpp synthesis mediated by RelA (Wendrich et al., 2002) or chloramphenicol (Cam), an inhibitor of translation elongation (Moazed and Noller, 1987). Upon stress treatment, total RNA was purified and truncation of 16S rRNA was verified by northern blot analysis again employing probe V7 (Figure 5A). The result clearly shows that the 3′-terminal fragment is cleaved upon stress treatment in the wild-type strain, which correlates with 80% and 30% reduction of the signal obtained for 16S rRNA with the same probe upon SHX (lane 2) and Cam (lane 4) treatment, respectively (Figures 5Aa and 5Ab). In contrast, we did not detect the fragment in the mazEF deletion strain after addition of SHX or Cam (Figure 5Ab, lanes 1 and 3), and as expected, without stress treatment when both strains were grown in LB medium (Figure 5Ab, lanes 5 and 6). Because several lines of evidence indicate induction of MazF activity upon growth in minimal medium (Amitai et al., 2009), total RNA was isolated under this condition and likewise probed with primer V7. The result reveals that, in absence of stress treatment, growth in M9 induces MazF-mediated cleavage in 16S rRNA (Figure 5A, lane 8), which was not observed in the mazF-deletion mutant (Figure 5A, lane 7). A probe specific for 5S rRNA was used as loading control (Figure 5Ac), as only one out of eight 5S rRNAs contains an ACA site located at the very 3′ terminus in a double-stranded region (Baik et al., 2009).
Figure 5. Adverse Conditions Induce the MazF-Dependent Stress Response.
(A) Northern blot analyses of total RNA prepared from strains MC4100relA+ (lanes 2, 4, 6, and 8) and MC4100relA+ΔmazEF (lanes 1, 3, 5, and 7) grown in LB medium (lanes 1–6) treated with SHX (lanes 1 and 2), Cam (lanes 3 and 4), untreated (w/o; lanes 5 and 6), or grown in M9 minimal medium without treatment (w/o; lanes 7 and 8). Removal of the 3′ end of 16S rRNA (a) and generation of the 43 nt fragment (b) by MazF was determined using oligonucleotide V7. Northern blot analysis of 5S rRNA served as a loading control (c).
(B) Primer extension analysis on total RNA purified from untreated strain MC4100relA+ (lanes 2) upon treatment with SHX (lane 3) or Cam (lane 4) used for northern blotting shown in (A) with a yfiD mRNA-specific primer (Table S1). Extension signals indicate MazF cleavage upstream of the start codon upon stress treatment like shown in Figure 1A (black arrows; lanes 3 and 4). Primer extension of in vitro-transcribed canonical (open arrow; lane 1) and leaderless (open circle; lane 5) yfiD mRNAs serve as controls.
(C and D) Pulse labeling of strains MC4100relA+ (C) and MC4100relA+ΔmazEF (D) harboring plasmid pRB381cI encoding the leaderless cI-lacZ fusion gene. Strains were grown in M9 minimal medium and pulsed either in absence (lanes 2–5) or at time points indicated upon addition of SHX (lanes 6–8). (Lane 1) Pulse labeling of strain MC4100relA+ harboring the plasmid pRB381 without cI-lacZ fusion gene to determine the position of the CIΦLacZ fusion protein (indicated by arrows).
(E) At time points that pulse labeling was performed in (C), total RNA was isolated and subjected to northern blot analysis using primer V7 (a) and primer R25 (specific for 5S rRNA, b) to determine the amount of 43 nt fragment upon addition of SHX.
(F) Quantification of the 43 nt fragment and 5S rRNA present at time points of pulse labeling upon addition of SHX indicated in (E) (Figure S4) to estimate the amount of cleaved and total ribosomes, respectively, given in pmoles (a). The percentage of cleaved ribosomes is given (b).
Next, we tested whether these conditions concomitantly result in formation of lmRNAs. Primer extension analyses with primers specific for yfiD (Figure 5B, lanes 1–5) and rpsU mRNAs (data not shown) show that mazF-mediated cleavage of both mRNAs occurs directly upstream of the start codon upon treatment with SHX (Figure 5B, lane 3) or Cam (Figure 5B, lane 4). In contrast, without stress treatment, we were not able to detect the signal corresponding to the lmRNA (Figure 5B, lane 2).
Pulse-labeling experiments performed in the presence of SHX (Figures 5C and 5D) and Cam (data not shown) likewise support the selective translation of lmRNAs caused by MazF under adverse conditions. Strains MC4100relA+ (Figure 5C) and MC4100relA+ΔmazEF (Figure 5D) harboring plasmid pRB381cI encoding the leaderless cI-lacZ fusion gene (Moll et al., 2004) were grown in M9 minimal medium. At OD600 of 0.25, the cultures were divided and one half was treated with SHX. Before and at time points indicated in Figures 5C and 5D, pulse labeling was carried out. Intriguingly, even before SHX treatment, translation of the leaderless cI-lacZ mRNA was more efficient in the WT strain (Figure 5C, lanes 2–5) when compared to the mazEF deletion strain (Figure 5D, lanes 2–5), consistent with the observed induction of MazF activity upon growth in minimal media (Figure 5A, lanes 7 and 8). Moreover, upon SHX treatment, expression of the leaderless reporter gene as well as other particular genes continued, whereas bulk mRNA translation was reduced in strain MC4100relA+ (Figure 5C, lanes 6–8). In contrast, employing the mazF deletion strain, translation was reduced 10 min after addition of SHX (Figure 5D, lane 6). Surprisingly, upon prolonged SHX treatment, translation ceased completely and no specific translation was detectable (Figure 5D, lanes 7 and 8).
Next, total RNA was isolated at the same time points that pulse labeling was performed in strain MC4100relA+ pRB381cI (Figure 5C) and was subjected to Northern blot analysis using primer V7 (specific for the 43 nt fragment) (Figure 5Ea) and R25 (specific for 5S rRNA) (Figure 5Eb). Corresponding to the generation of the 43 nt fragment upon growth in minimal medium, we observed a faint signal using primer V7 before SHX treatment (Figure 5Ea, lanes 1–4). However, upon stress treatment, the amount of the 43 nt fragment increased (Figure 5Ea, lanes 5–7), and further quantification revealed that, upon 20 min treatment, when translation is specific for a distinct pool of mRNAs (Figure 5C, lane 7), 22% of ribosomes are cleaved by MazF (Figure 5F and Figure S4). The fact that only a minor population of ribosomes has to be cleaved in order to result in selective protein synthesis might be explained by inhibition of canonical ribosomes by 5′UTRs containing SD sequences that were cleaved off by MazF (Figure 1). As these RNA fragments were determined to be rather stable (O.V. and I.M., unpublished data), it is conceivable that these fragments bind to “uncleaved” 30S subunits via SD-aSD interaction, thus blocking translation of canonical mRNAs. This idea is supported by work of Mawn et al. (2002), who showed that overexpression of RNA fragments containing SD-like sequences is detrimental for cell viability, as it leads to depletion of free 30S ribosomal subunits. Experiments scrutinizing our hypothesis are currently ongoing.
To unambiguously verify that cleavage at position A1500 in the 16S rRNA is pivotal for the posttranscriptional stress response pathway presented here, we introduced mutations at positions A1500 and A1502 in the rrsB gene encoding the 16S rRNA in plasmid pKK3535 containing the rrnB ribosomal RNA operon (Brosius et al., 1981). However, in contrast to results obtained by Vila-Sanjurjo and Dahlberg (2001), strain SQZ10Δ7 (Cochella and Green, 2004) lacking all rRNA operons was not viable when the only sources for 16S rRNA were plasmids containing mutant rrsB genes, supporting our notion that the presence of the ACA site is crucial for cell viability.
Discussion
mazEF as a Master Regulatory Element that Leads to Alteration of the Translation Program under Stress Conditions in E. coli
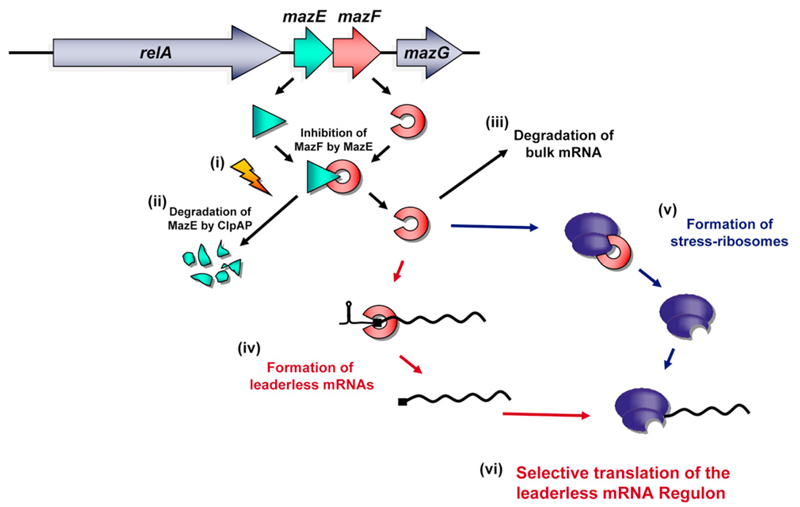
We have previously shown that, though induction of the endoribonuclease MazF leads to translation inhibition of the majority of mRNAs, the synthesis of an exclusive group of about 50 proteins is still permitted (Amitai et al., 2009). Here, we discovered a molecular mechanism leading to selective translation of specific mRNAs upon MazF induction. We show for the first time that MazF activity leads to an alteration of the translation program by generating a modified translational apparatus composed of functionally specialized ribosomes on one hand and lmRNAs on the other, as illustrated in Figure 6. The E. coli mazEF module is located downstream of the relA gene (Metzger et al., 1988), whose product is responsible for ppGpp synthesis upon amino acid starvation (Wendrich et al., 2002). Stressful conditions that inhibit mazEF expression, such as antibiotics inhibiting transcription and/or translation or increased ppGpp concentration upon severe amino acid starvation (Figure 6, i, indicated by an arrow) (Engelberg-Kulka et al., 2006; Hazan et al., 2004; Christensen et al., 2003) prevent de novo synthesis of both MazE and MazF. Subsequently, the labile MazE is degraded by the ClpAP protease (Figure 6, ii), thereby permitting MazF to act freely as endoribonuclease, which results in degradation of the majority of mRNAs (Figure 6, iii). In the work presented here, we show that, in addition, MazF removes the 5′UTR of specific mRNAs, thus rendering them leaderless (Figure 6, iv). Moreover, we were able to demonstrate that another target of the endoribonuclease is the ribosome: MazF specifically removes the 3′-terminal 43 nts of 16S rRNA containing helix 45 as well as the aSD sequence (Figure 6, v), which is essential for formation of a translation initiation complex on canonical ribosome-binding sites (Hui and de Boer, 1987; Calogero et al., 1988). Consequently, (Figure 6, vi) MazF activity leads to selective translation of a “leaderless mRNA regulon” by a subpopulation of specialized ribosomes.
Figure 6. A Model for the Generation of Leaderless mRNAs and Stress Ribosomes by MazF.
The mazEF module can be triggered by stressful conditions (i, indicated by an arrow) (Engelberg-Kulka et al., 2006; Christensen et al., 2003), which results in (ii) degradation of the antidote MazE by the ClpAP protease (Aizenman et al., 1996). The activity of released MazF leads to degradation of the majority of transcripts (iii). In addition, it removes the 5′UTR of specific mRNAs, thus rendering them leaderless (iv), and moreover, specifically removes the 3′-terminal 43 nts of 16S rRNA comprising helix 45 as well as the aSD sequence (v), which is essential for the formation of a translation initiation complex on canonical ribosome-binding sites. Consequently, (vi) MazF activity leads to selective translation of a “leaderless mRNA regulon.”
Stress Adaptation via the Generation of Functionally Specialized Ribosomes and the “Leaderless Stress Regulon”
Bacteria must cope with environments that undergo perpetual alterations in temperature, osmolarity, pH, availability of nutrients, and antibiotics, as well as a variety of other adverse agents and conditions. A strategy that bacteria had developed in order to cope with such environmental changes is called stress response. It involves activation of specific sets of genes, mainly at the level of transcription (Storz and Hengge-Aronis, 2000). This report provides a paradigm for posttranscriptional stress response in bacteria based on ribosome specialization: the heterogeneity of the translational machinery caused by MazF results in selective synthesis of proteins encoded by the “leaderless stress regulon.” Thus, the destructive endonuclease MazF turned out to be an instructive element by its ability to generate a subpopulation of distinct ribosomes. Therefore, it should be emphasized that MazF does not cause a complete change but a crucial modulation of the translation program, thereby coupling protein synthesis to the physiological state of the cell.
As expected for every newly discovered mechanism, there still remain critical questions, some of which are under our current investigation and two of which are here described. First, our results shows that, under stressful conditions that may be less drastic than the one obtained by overproduction of MazF, the ribosomal population is heterogeneous, including canonical ribosomes and the described specialized ribosomes. The question of whether the bifurcation of the ribosomal population occurs inside of individual cells or is distributed among subpopulations of cells still remains to be elucidated. In addition, what is the cellular fate of specialized ribosomes lacking the 3′ end of 16S rRNA? One possibility is that accumulation of such ribosomes is part of the mazEF-mediated death program, whereby we found “a point of no return” in MazF lethality, particularly in minimal medium (Amitai et al., 2004; Kolodkin-Gal and Engelberg-Kulka, 2006). However, there is an initial stage in which the effect of MazF can still be reversed by the antitoxin MazE (Amitai et al., 2004; Kolodkin-Gal and Engelberg-Kulka, 2006). Therefore, a “ribosome repair system” might exist that enables recovering from stressful conditions. Further studies on the leaderless stress regulon will shed light on bacterial pathways in which this regulon is involved and their relation to general physiological phenomena in E. coli. These include cell death (Engelberg-Kulka et al., 2006; Kolodkin-Gal, et al., 2007; Amitai et al., 2009), growth arrest (Gerdes et al., 2005), biofilm formation (Kolodkin-Gal et al., 2009), and persistence (Keren et al., 2004), which were previously described as being related to the mazEF module.
Experimental Procedures
Bacterial Strains and Plasmids Used in This Study
E. coli strains MC4100relA+ and MC4100relA+ΔmazEF (both Engelberg-Kulka et al., 1998), BL21 (DE3) (Invitrogen), TG1 (Gibson, 1984), and MG1655 (Blattner et al., 1997) have been described. Unless otherwise indicated, bacterial cultures were grown in LB broth at 37°C supplemented with 100 μg/ml ampicillin or 15 μg/ml tetracycline, as appropriate for plasmid maintenance. Growth of liquid cultures was monitored photometrically by measuring the optical density at 600 nm.
Plasmids pKTplaccI (Grill et al., 2000) and pRB381cI (Moll et al., 2004) harbor the first 189 nts of the λ cI gene fused to the eighth codon of the lacZ gene under control of a constitutive lac promoter. Plasmid pSA1 is a derivative of pQE30 (QIAGEN) harboring the mazF gene under control of the T5 promoter and the lac operator (Amitai et al., 2009).
Plasmid pET28a-mazEF(His)6 was constructed from pET28a (Novagen) to coexpress MazE and MazF(His)6 under the control of the T7 promoter, using the SD sequence of the mazEF operon.
Purification of Ribosomes upon In Vivo mazF Expression and Verification of MazF Cleavage
To verify MazF cleavage in 16S rRNA in ribosomes in vivo, E. coli strain MC4100relA+pSA1 was grown in LB. At OD600 of 0.5, the culture was divided, and in one half, mazF expression was induced by addition of 500 μM IPTG. Thirty minutes later, cells were harvested, resuspended in Tico buffer (20 mM HEPES-KOH [pH 7.4], 6 mM magnesium acetate, 30 mM ammonium acetate, and 4 mM β-mercapto-ethanol), and lysed by the lysozyme freeze-thaw method. Upon separation of the S30 extract through a 10% sucrose cushion made up in Tico buffer, the pellet containing crude ribosomes was resuspended in Tico buffer. 70S ribosomes still containing the aSD sequence were removed employing biotinylated SD oligonucleotides (Table S1) that were immobilized on streptavidin-coated magnetic beads.
The absence of the 3′-terminal 16S rRNA fragment form ribosomes purified upon MazF treatment was determined by northern blot analysis employing oligonucleotides V43 and V7, which bind to nts 939–955 in the central part of 16S rRNA and to nts 1511–1535 within the 3′-terminal fragment, respectively (Table S1). In brief, rRNA was prepared from ribosomes used for in vitro translation analysis (Figure 4C) by phenol-chloroform extraction. Upon ethanol-precipitation, the rRNA (0.5 μg each) was fractionated on a 4% denaturing polyacrylamide gel, transferred to Hybond-membrane (Amersham) using the Trans-Blot Semi-Dry Transfer Cell (Bio-Rad), and then hybridized to [32P]-labeled oligos V7 and V43. The signals obtained with the labeled probes were visualized by a PhosphorImager (Molecular Dynamics) and quantified employing ImageQuant software. Likewise, total RNA prepared under the same conditions employed for ribosome purification was used to verify the presence of the 3′-terminal fragment upon induction of mazF expression. Northern blot analysis was performed exactly as described before to optimize for the short RNA fragment (Pall and Hamilton, 2008).
Supplemental Information
Supplemental Information includes Extended Experimental Procedures, four figures, and one table and is available with this article online at doi:10.1016/j.cell.2011.07.047.
Acknowledgments
This work was supported by grants P20112-B03 and P22249-B20 from the Austrian Science Fund to I.M., by grant number 66/10 from the Israel Science Foundation (ISF) administrated by the Israel Academy of Science and Humanities, by the USA Army grant W911NF0910212, and by NIH grant GM069509 to H.E.-K.
References
- Agarwal S, Agarwal S, Bhatnagar R. Identification and characterization of a novel toxin-antitoxin module from Bacillus anthracis. FEBS Lett. 2007;581:1727–1734. doi: 10.1016/j.febslet.2007.03.051. [DOI] [PubMed] [Google Scholar]
- Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai S, Yassin Y, Engelberg-Kulka H. MazF-mediated cell death in Escherichia coli: a point of no return. J Bacteriol. 2004;186:8295–8300. doi: 10.1128/JB.186.24.8295-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, Engelberg-Kulka H. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genet. 2009;5:e1000390. doi: 10.1371/journal.pgen.1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik S, Inoue K, Ouyang M, Inouye M. Significant bias against the ACA triplet in the tmRNA sequence of Escherichia coli K-12. J Bacteriol. 2009;191:6157–6166. doi: 10.1128/JB.00699-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Brosius J, Ullrich A, Raker MA, Gray A, Dull TJ, Gutell RR, Noller HF. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- Calogero RA, Pon CL, Canonaco MA, Gualerzi CO. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc Natl Acad Sci USA. 1988;85:6427–6431. doi: 10.1073/pnas.85.17.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci USA. 2001;98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Pedersen K, Hansen FG, Gerdes K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol. 2003;332:809–819. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- Cochella L, Green R. Isolation of antibiotic resistance mutations in the rRNA by using an in vitro selection system. Proc Natl Acad Sci USA. 2004;101:3786–3791. doi: 10.1073/pnas.0307596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- Engelberg-Kulka H, Glaser G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu Rev Microbiol. 1999;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Reches M, Narasimhan S, Schoulaker-Schwarz R, Klemes Y, Aizenman E, Glaser G. rexB of bacteriophage lambda is an anti-cell death gene. Proc Natl Acad Sci USA. 1998;95:15481–15486. doi: 10.1073/pnas.95.26.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Gabashvili IS, Agrawal RK, Spahn CM, Grassucci RA, Svergun DI, Frank J, Penczek P. Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- Gibson TJ. Studies on the Epstein-Barr virus genome. PhD thesis; University of Cambridge, UK: 1984. [Google Scholar]
- Green J, Baldwin ML, Richardson J. Downregulation of Escherichia coli yfiD expression by FNR occupying a site at -93.5 involves the AR1-containing face of FNR. Mol Microbiol. 1998;29:1113–1123. doi: 10.1046/j.1365-2958.1998.01002.x. [DOI] [PubMed] [Google Scholar]
- Grill S, Gualerzi CO, Londei P, Bläsi U. Selective stimulation of translation of leaderless mRNA by initiation factor 2: evolutionary implications for translation. EMBO J. 2000;19:4101–4110. doi: 10.1093/emboj/19.15.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer-Jespersen K, Munch-Petersen A, Schwartz M, Nygaard P. Induction of enzymes involed in the catabolism of deoxyribonucleosides and ribonucleosides in Escherichia coli K 12. Eur J Biochem. 1971;19:533–538. doi: 10.1111/j.1432-1033.1971.tb01345.x. [DOI] [PubMed] [Google Scholar]
- Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- Hazan R, Sat B, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J Bacteriol. 2004;186:3663–3669. doi: 10.1128/JB.186.11.3663-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui A, de Boer HA. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:4762–4766. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaberdina AC, Szaflarski W, Nierhaus KH, Moll I. An unexpected type of ribosomes induced by kasugamycin: a look into ancestral times of protein synthesis? Mol Cell. 2009;33:227–236. doi: 10.1016/j.molcel.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminishi T, Wilson DN, Takemoto C, Harms JM, Kawazoe M, Schluenzen F, Hanawa-Suetsugu K, Shirouzu M, Fucini P, Yokoyama S. A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine-Dalgarno interaction. Structure. 2007;15:289–297. doi: 10.1016/j.str.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Engelberg-Kulka H. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J Bacteriol. 2006;188:3420–3423. doi: 10.1128/JB.188.9.3420-3423.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Hazan R, Gaathon A, Carmeli S, Engelberg-Kulka H. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science. 2007;318:652–655. doi: 10.1126/science.1147248. [DOI] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Verdiger R, Shlosberg-Fedida A, Engelberg-Kulka H. A differential effect of E. coli toxin-antitoxin systems on cell death in liquid media and biofilm formation. PLoS ONE. 2009;4:e6785. doi: 10.1371/journal.pone.0006785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Ruiz AA, Godson GN. Promotion, termination, and anti-termination in the rpsU-dnaG-rpoD macromolecular synthesis operon of E. coli K-12. Mol Gen Genet. 1984;195:391–401. doi: 10.1007/BF00341439. [DOI] [PubMed] [Google Scholar]
- Marianovsky I, Aizenman E, Engelberg-Kulka H, Glaser G. The regulation of the Escherichia coli mazEF promoter involves an unusual alternating palindrome. J Biol Chem. 2001;276:5975–5984. doi: 10.1074/jbc.M008832200. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawn MV, Fournier MJ, Tirrell DA, Mason TL. Depletion of free 30S ribosomal subunits in Escherichia coli by expression of RNA containing Shine-Dalgarno-like sequences. J Bacteriol. 2002;184:494–502. doi: 10.1128/JB.184.2.494-502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger S, Dror IB, Aizenman E, Schreiber G, Toone M, Friesen JD, Cashel M, Glaser G. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J Biol Chem. 1988;263:15699–15704. [PubMed] [Google Scholar]
- Mittenhuber G. Occurrence of mazEF-like antitoxin/toxin systems in bacteria. J Mol Microbiol Biotechnol. 1999;1:295–302. [PubMed] [Google Scholar]
- Moazed D, Noller HF. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- Moll I, Grill S, Gründling A, Bläsi U. Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol Microbiol. 2002;44:1387–1396. doi: 10.1046/j.1365-2958.2002.02971.x. [DOI] [PubMed] [Google Scholar]
- Moll I, Hirokawa G, Kiel MC, Kaji A, Bläsi U. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res. 2004;32:3354–3363. doi: 10.1093/nar/gkh663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porollo A, Meller J. Versatile annotation and publication quality visualization of protein complexes using POLYVIEW-3D. BMC Bioinformatics. 2007;8:316. doi: 10.1186/1471-2105-8-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sat B, Hazan R, Fisher T, Khaner H, Glaser G, Engelberg-Kulka H. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J Bacteriol. 2001;183:2041–2045. doi: 10.1128/JB.183.6.2041-2045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Schuenemann VJ, Hand NJ, Silhavy TJ, Martin J, Lupas AN, Djuranovic S. prlF and yhaV encode a new toxin-antitoxin system in Escherichia coli. J Mol Biol. 2007;372:894–905. doi: 10.1016/j.jmb.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Senior BW, Holland IB. Effect of colicin E3 upon the 30S ribosomal subunit of Escherichia coli. Proc Natl Acad Sci USA. 1971;68:959–963. doi: 10.1073/pnas.68.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Hengge-Aronis R. Bacterial Stress Responses. Washington, DC: ASM Press; 2000. [Google Scholar]
- Vila-Sanjurjo A, Dahlberg AE. Mutational analysis of the conserved bases C1402 and A1500 in the center of the decoding domain of Escherichia coli 16S rRNA reveals an important tertiary interaction. J Mol Biol. 2001;308:457–463. doi: 10.1006/jmbi.2001.4576. [DOI] [PubMed] [Google Scholar]
- Wagner AF, Schultz S, Bomke J, Pils T, Lehmann WD, Knappe J. YfiD of Escherichia coli and Y06I of bacteriophage T4 as autonomous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase. Biochem Biophys Res Commun. 2001;285:456–462. doi: 10.1006/bbrc.2001.5186. [DOI] [PubMed] [Google Scholar]
- Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Dissection of the mechanism for the stringent factor RelA. Mol Cell. 2002;10:779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- Woodcock J, Moazed D, Cannon M, Davies J, Noller HF. Interaction of antibiotics with A- and P-site-specific bases in 16S ribosomal RNA. EMBO J. 1991;10:3099–3103. doi: 10.1002/j.1460-2075.1991.tb07863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzesinski J, Bakin A, Nurse K, Lane BG, Ofengand J. Purification, cloning, and properties of the 16S RNA pseudouridine 516 synthase from Escherichia coli. Biochemistry. 1995;34:8904–8913. doi: 10.1021/bi00027a043. [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- Zhang JJ, Zhang YL, Inouye M. Characterization of the interactions within the mazEF addiction module of Escherichia coli. J Biol Chem. 2003;278:32300–32306. doi: 10.1074/jbc.M304767200. [DOI] [PubMed] [Google Scholar]
- Zhang JJ, Zhang YL, Zhu L, Suzuki M, Inouye M. Interference of mRNA function by sequence-specific endoribonuclease PemK. J Biol Chem. 2004;279:20678–20684. doi: 10.1074/jbc.M314284200. [DOI] [PubMed] [Google Scholar]
- Zhu L, Phadtare S, Nariya H, Ouyang M, Husson RN, Inouye M. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol Microbiol. 2008;69:559–569. doi: 10.1111/j.1365-2958.2008.06284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.