Abstract
In all organisms the universal process of protein synthesis is performed by the ribosome, a complex multi-component assembly composed of RNA and protein elements. Although ribosome heterogeneity was observed already more than 40 years ago, the ribosome is still traditionally viewed as an unchangeable entity that has to be equipped with all ribosomal components and translation factors in order to precisely accomplish all steps in protein synthesis. In the recent years this concept was challenged by several studies highlighting a broad variation in the composition of the translational machinery in response to environmental signals, which leads to its adaptation and functional specialization. Here, we summarize recent reports on the variability of the protein synthesis apparatus in diverse organisms and discuss the multiple mechanisms and possibilities that can lead to functional ribosome heterogeneity. Collectively, these results indicate that all cells are equipped with a remarkable toolbox to fine tune gene expression at the level of translation and emphasize the physiological importance of ribosome heterogeneity for the immediate implementation of environmental information.
Keywords: Protein synthesis, Ribosome heterogeneity, Gene expression regulation, Translation regulation, Stress response
1. Introduction
According to the central dogma of molecular biology, in all living cells the genetic information is stored at the level of DNA and transcribed into messenger RNA (mRNA), which subsequently serves as a template for the translation of the encoded information into the amino acid sequence of proteins. This latter step is performed by the ribosome, a sophisticated cellular machinery composed of RNA and protein elements. In order to allow all organisms to respond and adapt protein synthesis to environmental cues, the coordinated regulation of gene expression is crucial to ensure the establishment of a productive metabolic network. Hitherto, the main scientific focus in the regulation of gene expression has been directed on the alteration of the transcriptional program. Thus, the role of adjustment at the translational level has been underestimated. However, in the past decades, this perception changed dramatically and it became widely accepted that mRNA levels do not necessarily correspond to the amount of proteins being made. Together, these observations implicate a profound regulation at the post-transcriptional level, which has mostly been attributed to features intrinsic to the mRNA or a variety of non-ribosomal protein and RNA factors that modulate diverse steps in protein synthesis like small RNAs, mRNA binding proteins, and cis- or trans-acting regulators [1,2]. However, still it was generally believed that the assembly of all ribosomal components is required and mandatory for the process of protein synthesis.
In striking contrast to this perception, in the recent years several lines of evidence strongly underpin the notion that the ribosome as well as genuine translation factors are likewise key players in post-transcriptional regulation. In this review we will focus on the intrinsic alteration of the translational program by the formation of distinct ribosomal subpopulations that differ in their protein or RNA complement, or which are equipped with differentially modified translation factors. Collectively, these results strongly underline the great potential of the translational machinery to serve as a hub for signal integration at the post-transcriptional level in a variety of different organisms.
2. Translation in pro- and eukaryotes
The ribosome is a highly conserved molecular machinery. In all organisms it is composed of two unequal subunits, which consist of a distinct set of ribosomal RNA (rRNA) and ribosomal protein (RP) components and perform a specific function during translation, as outlined below. The ribosome harbors three different tRNA binding sites: The A-site, where decoding occurs and the correct aminoacyl-tRNA (aa-tRNA) is selected on the basis of the mRNA codon displayed, the P-site, which carries the peptidyl-tRNA, and the E-site, which binds exclusively deacetylated tRNAs that are exiting the ribosome [3]. Thus, during translation the tRNA moves from the A-site through the P- and E-site, where it leaves the ribosome [4]. Conceptionally, the complexity of the ribosome structure is reflected in the process of protein synthesis, which can be intersected into three major steps: initiation, elongation and termination/recycling.
In prokaryotic translation initiation, the small ribosomal (30S) subunit binds the mRNA via direct interaction between the anti-Shine-Dalgarno (aSD) sequence located at the 3’-terminus of the 16S rRNA and the Shine-Dalgarno (SD) sequence in the 5’-untranslated region (5’-UTR) closely upstream of the start codon of the mRNA. The initiator tRNA fMet-tRNAfMet (tRNAi) is recruited by initiation factor 2 (IF2) to the ribosomal P-site where it interacts with the start codon, thus forming the 30S pre-initiation complex (PIC). The accuracy of the codon–anticodon recognition is controlled by IF3, while IF1 stimulates the activity of IF2. Subsequently, the large (50S) ribosomal subunit joins the PIC to result in the 70S initiation complex (IC) [5]. Interestingly, the exact chronological order of the PIC assembly is still a matter of debate and seems not to be strictly determined.
Contrary, eukaryotic translation initiation is mediated primarily via protein–protein interactions. First, the tRNAi is recruited to the small ribosomal subunit (40S) to form a ternary complex (TC) with the GTP-bound eukaryotic initiation factor 2 (eIF2) [6]. Formation of this 43S pre-initiation complex (PIC) is strongly enhanced by additional factors, such as eIF3 [7]. In contrast to the bacterial translation initiation complex, which is assembled directly at internal ribosome binding sites, the 43S PIC generally binds to the capped 5’-end of a eukaryotic mRNA and scans along the transcript in 5’–3’-direction until it encounters a start codon [8]. After recognition of the start codon, the large ribosomal subunit (60S) assembles to form the 80S initiation complex, which is ready for elongation. The diverse phases are assisted by 12 eIFs and additional auxiliary factors [6,9]. Alternatively, under distinct conditions or on certain transcripts internal initiation can occur in a cap-independent manner at so called internal ribosome entry sites (IRES) [10].
The step of translation elongation is well conserved in pro- and eukaryotes [11]. The aa-tRNA is recruited to the ribosome by elongation factor Tu (EF-Tu) in prokaryotes, or the ortholog eEF1A in eukaryotes. Subsequently, the growing peptide chain is transferred to the newly bound aa-tRNA in the peptidyl-transferase center (PTC) that is exclusively formed by rRNA of the large subunit. Then the ribosome translocates one codon downstream on the mRNA assisted by the orthologs EF-G and eEF2 in prokaryotes and eukaryotes, respectively [12,13]. When the elongating ribosome encounters a stop codon in its A-site, termination and recycling are initiated. In bacteria, either release factor 1 (RF1) or RF2 recognizes the stop codon and triggers hydrolysis of the ester-bond in the peptidyl-tRNA situated in the P-site resulting in the release of the synthesized polypeptide chain from the ribosome. Next, RF3 stimulates the rapid dissociation of RF1 or RF2 from the ribosome [14]. In eukaryotes the structurally different proteins eRF1 and eRF3 trigger these reactions [15]. The two subunits and the mRNA of the post-termination complex are then disassembled with the help of the ribosome recycling factor (RRF) together with IF3 in prokaryotes. In eukaryotes, where no such factor exists, the recycling process is more complex and involves the ATPase ABCE1 [16].
3. Heterogeneity of the translation machinery
As translation initiation is the rate-limiting step in protein synthesis, it is therefore the predominant target for regulation. As mentioned above, hitherto translational regulation was attributed to cis- and trans-acting non-ribosomal RNA and protein factors, which modulate the accessibility of the SD-sequence for the SD-aSD interaction in prokaryotes [17]. In eukaryotes, protein- or micro RNA (miRNA)-mediated translation regulation is more commonly involving the 3′-UTRs of mRNAs [9]. Additionally, mRNA structures like the 5′-cap, the 3′-poly(A)-tail, IRESs and upstream open reading frames (uORFs) play crucial roles in eukaryotic translation regulation [18].
In contrast to these regulatory mechanisms mediated by extraribosomal factors, in this review we will summarize and discuss the current scientific understanding concerning the functional heterogeneity of the translational apparatus, a concept that is considered to represent an accessory layer of gene expression regulation. We aim to emphasize that the translation machinery, built of the ribosome itself but also involving the essential factors that assist in the translation process, is not an inevitably unchangeable entity. In the recent years, a paradigm shift has taken place with increasing evidence that the ribosome is not one determined complex, but that its components can be altered resulting in translational fine-tuning to follow changing cellular needs.
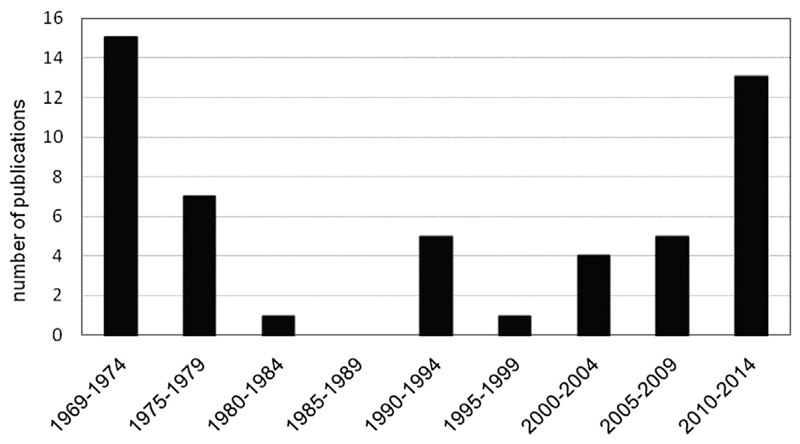
Ribosome heterogeneity has been observed already in the 1970s, where ribosomes purified from bacteria grown under different conditions were shown to lack certain RPs without losing their translational functions [19–21]. However, these observations have been neglected for a long time and the term ‘ribosome heterogeneity’ has not been established in the scientific community (Fig. 1). Over the years, many studies performed in eukaryotes presented evidence that ribosomes can vary in their protein and rRNA complement between different cell types and developmental states. These observations culminated in the postulation of the ‘ribosome filter hypothesis’ by Mauro and Edelman in the year 2002 [22]. The authors propose that the ribosome composition functions as translation determination factor. Depending on the RPs and rRNA sequences represented in the respective ribosome, the complex acts like a filter that selects for specific mRNAs and hence modulates translation [22,23]. A few years later, the Silver lab reported that different RP paralogs are functionally distinct and contribute to translational selectivity in Saccharomyces cerevisiae [24], which led to the proposal of the ‘ribosome code’, analogous to the ‘histone code’ that affects transcription. Subsequently, this theory that goes in line with the ribosome filter hypothesis was extended to the formation of specialized ribosomes via the incorporation of different forms or modifications of rRNA, or by post-translational modifications of RPs that allow regulated translation of specific mRNAs.
Fig. 1.
Scientific publications addressing ‘ribosome heterogeneity’ since 1969. The graph depicts that despite being already observed more than 40 years ago, ribosome heterogeneity as a means to regulate translation was not a scientific subject till the dawn of the new millennium.
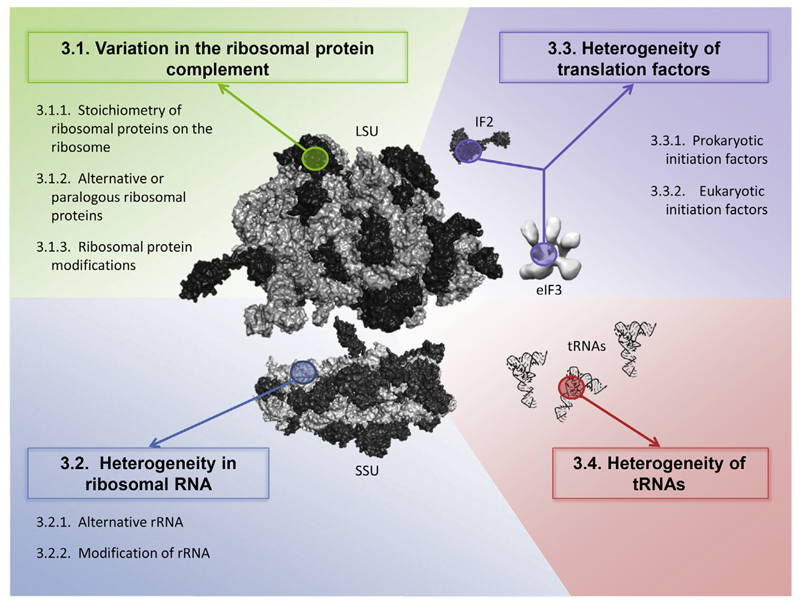
In the following chapters we aim to give a brief update on the recent progress in understanding functional heterogeneity of the translational machinery. Due to its complex nature there are different routes for potential modifications: on the ribosome itself, affecting either the rRNA or the RPs; via alteration of translation factors; or via modification of tRNAs (Fig. 2). Moreover, all components of the translation machinery can additionally have ancillary functions in non-translational processes.
Fig. 2.
Components of the translation machinery that have the potential to contribute to functional heterogeneity. Structures were visualized with Polyview 3D software [125] using the following maps: E. coli large ribosomal subunit (LSU; pdb accession code: 3d5a [126], rRNA in light gray, proteins in dark gray); small ribosomal subunit (SSU; pdb accession code: 3d5b [126]); E. coli IF2 (pdb accession code: 1g7r [127]); human eIF3 (EMD-2166 [128]); yeast tRNAPhe (pdb accession code: 6tna [129]).
3.1. Variation in the ribosomal protein complement
3.1.1. Stoichiometry of ribosomal proteins on the ribosome
The simplest way to modify a multi-subunit assembly is to alter the relative abundance of its individual components. Since the rRNA molecules are functionally indispensable for the active translation machinery, the RPs pose the sole ribosomal building blocks whose abundance on the complex can possibly be adapted. One mechanism to alter the RP complement is the differential expression of the respective genes. Previously, it was accepted that ribosomal proteins are produced in a coordinated manner to ensure that each ribosome contains a full complement of all components, which was considered to be mandatory for correct protein synthesis. However, this assumption was challenged by multiple observations that under distinct conditions as well as in diverse cell types some RPs are present in substoichiometric amounts on the ribosomes [21,25,26], indicative for an alteration of the translational activity and specificity by a heterogeneous RP complement. This notion is supported by the observed replacement of damaged RPs on Escherichia coli ribosomes that results in the repair of the multicomponent complex [27]. This phenomenon points towards the possibility to specifically re-equip the heterogeneous protein-depleted ribosomes.
In prokaryotes, the ribosomal protein complement was shown to differ with respect to the encountered environmental conditions or the growth phase [19–21,26,28]. Furthermore, antibiotic treatment can be added to the list of factors modifying the RP complement. In E. coli, the ribosome targeting antibiotic Kasugamycin was shown to mediate the formation of a specific ribosomal subclass, the so-called 61S ribosomes that lack the RPs S1, S2, S6, S12, S18, and S21 [29]. Due to the lack of protein S1 that is essential for translation initiation on canonical mRNAs [30] but is dispensable for translation of leaderless mRNAs (lmRNAs) harboring a 5’-terminal start codon [31], the 61S ribosomes are likewise selective for lmRNAs [29]. These findings tempted us to formulate a hypothesis that the presence or absence of a specific RP can modulate the translatome. In this context, the specific translation initiation on lmRNAs by a ribosomal subpopulation lacking protein S1 might pose a significant contribution to the post-transcriptional regulation in bacteria. This concept is further supported by findings that suggest the presence of ribosomes lacking S1 even under relaxed growth conditions [32]. Moreover, the relatively small boundary between protein S1 and the ribosome, which was recently determined at atomic resolution [33], opens the potential to modulate the affinity of protein S1 for the ribosome by post-translational protein modification.
A heterogeneous RP content as a means to remodel the translatome is not limited to prokaryotes. Already in the 1980s, work performed in Dictyostelium discoideum suggested that different sets of RPs assembled to the ribosome result in an alteration of the translational specificity [25]. This study indicates that the protein complement of ribosomes from vegetative amoeboid cells substantially differs when compared to the RPs present on ribosomes derived from spores. Besides the developmental stage, cell type dependent variations in the synthesis of several RPs have been reported in six different human tissues and several regions of the mouse embryo [34,35], which might likewise affect the composition of the ribosome. However, analyses of a selection of RNA sequencing data obtained from various organisms with a major focus on mammalian tissues and cell lines showed that the molar ratios of 80%–90% of RP transcripts vary less than threefold with little tissue specificity [36]. These results suggest that a post-transcriptional mechanism is required for the regulation of the expression of RP-coding genes either affecting mRNA translation or stability.
Nevertheless, several studies underline the presence of this regulatory pathway in eukaryotes, exemplified by the observation that overexpression of the parkin gene in a human cell line leads to a specific decrease in abundance of protein RPSA [37]. The observed down-regulation of RPSA in response to parkin gene expression may explain Parkin’s proposed tumor suppressor activity, since RPSA was implicated in being enriched in cancer tissues and is considered to promote tumor progression via an unknown mechanism [38,39]. Along the same lines, the expression of the oncogene v-erbA in chicken erythrocytic progenitor cells was shown to affect the transcription of specific RP encoding genes and hence results in the formation of heterogeneous ribosomes that lack RPL11 [40]. Interestingly, proteomic analysis revealed that consequently the abundance of nine proteins was altered, suggesting that the modulation of the ribosome composition results in an alteration of the translatome. Moreover, it has been shown that in contrast to bulk mRNA translation the vesicular stomatitis virus specific cap-dependent translation requires RpL40 [41]. This study again exemplifies that the absence of certain RPs can have a very specific impact on translational properties. Collectively, the functional specialization of ribosomes by the alteration of their protein complement can be found in all domains of life. However, it is important to note that some results require a careful interpretation as they are based on the analysis of whole cell protein content, which does not necessarily reflect the actual ribosome composition and may neglect the fraction of free RPs. This is particularly relevant since described phenotypes can be likewise attributed to extra-ribosomal functions of RPs, which were reviewed in detail by Warner and McIntosh [42].
3.1.2. Alternative or paralogous ribosomal proteins
Several studies describing ribosome heterogeneity discuss the model of employing alternative RPs. This hypothesis is strengthened by the fact that many different prokaryotes and eukaryotes harbor multiple paralogs of RP coding genes [24,43,44], which are synthesized simultaneously or in response to certain environmental conditions [44]. In the Gram-positive soil bacterium Bacillus subtilis for example, the genes encoding ribosomal proteins L31 and S14 are duplicated, and the respective RP paralogs differ in the presence of a zinc-binding motif [45]. The authors hypothesize that incorporation of the protein variants into the ribosome is zinc-dependent. During zinc limitation, the zinc-bound variant is functionally replaced by the zinc-independent paralog. However, since hitherto no alteration of the translational specificity was described upon protein exchange, this mechanism could solely contribute to zinc storage and its mobilization under zinc-limiting conditions.
In yeast approximately 70% of all duplicated RP genes are asymmetrically expressed [44], potentially implying that the paralogous proteins are not merely functionally equivalent substitutes. Komili and co-workers underscored this hypothesis by reporting that the translation of localized mRNAs in S. cerevisiae is affected in a paralog-specific manner [24]. Besides, the analysis of cells lacking specific RP paralogs indicated functional differences between the paralogs that extend beyond mRNA localization [24]. Alongside, diploid yeast strains deficient in one or the other copy of RP genes, harbored distinct populations of specialized ribosomes with individual translational properties [46]. Interestingly, in S. cerevisiae, the ribosomal stalk protein paralogs P1a, P1b, P2a and P2b, form two heterodimers that preferentially bind to sites A and B of the P0 protein, respectively [47]. Each of the four possible P1/P2 combinations results in a specific phenotype indicating that they perform non-identical physiological roles. Moreover, the absence of one heterodimer reduced cell growth and hindered the synthesis of the 60S ribosomal subunit [47].
An exceptional example for the presence of RP paralogs are plants. In Arabidopsis thaliana, each ribosomal protein is encoded by two to seven genes [48], and recently 429 genes coding for potential RPs have been identified [49]. Many paralogs display sequence variations and are differentially expressed during development [50,51]. In addition, nearly half of the RPs identified in a proteomic approach are represented by two or more distinct spots in 2D gel analyses indicating different protein isoforms that are post-translationally modified [52]. In the same model organism, Zsögön and co-workers report that the levels of the two RPL27a paralogs with redundant function, RPL27aC and RPL27aB, influence the ovule development [53]. The reduction of the expression levels of one or the other has a distinct effect on fertility revealing a function of RPL27a in the coordination of ovule development.
Most genes coding for ribosomal proteins in mammals are represented in a single copy, with a few exceptions like RPS4 [54] or RPL39l [55], which was found to be highly expressed in differentiating mouse embryonic stem cells. Interestingly, the enrichment of RPL39l has been observed likewise in human hepatocellular carcinoma tumor (HCC) cells and cancer cell lines with high tumor grading and alpha-fetoprotein levels [55]. Moreover, the expression pattern of all RPs and their paralogs in mouse was tested across 22 different tissues indicating a high level of tissue-specific expression [55]. This finding goes in line with a proteomic survey of ribosomes in Mus musculus, revealing the testis-specific synthesis of the proteins RPL10-like and RPL39-like [56], which are paralogs of the X-linked ribosomal proteins RPL10 and RPL39, respectively. The same laboratory recently identified a new paralog of X-linked RPS4 [57]. They show that the autosomal, intronless gene was expressed predominantly in testis similar to RPL10-like and RPL39-like. However, in contrast to the paralog RPL10-like, the RPS4 paralog shows a partially different expression pattern in spermatogenic cells [57].
3.1.3. Ribosomal protein modifications
An alternative mechanism to alter the properties of the translational machinery is the modification of ribosomal components incorporated in the mature and active ribosome. Considering the energy demanding processes of de novo synthesis and assembly of alternative RPs to ribosomes, this mechanism might constitute a direct and energy effective option of adaptation.
In prokaryotes, comparative proteome analysis revealed the differential acetylation and phosphorylation of several RPs in the exponential or stationary growth phase [58,59]. Interestingly, the interaction of proteins L7/L12 with the ribosome is likewise affected by a growth phase dependent acetylation [60]. During stationary phase or growth in minimal medium the N-terminal acetylated protein L7 is the predominant form on the ribosome, resulting in stabilization of the ribosomal stalk complex during adverse conditions. Together, these findings strongly underscore the idea that protein modification might provide a powerful mechanism to fine tune protein synthesis. Nevertheless, no direct functional specialization has been attributed to RP modification in bacteria so far.
Correspondingly, methylation, acetylation, and hydroxylation of RPs were observed in yeast [61]. Some of these substitutions were found to appear in a growth phase dependent manner, for example the proteins RPS1B and RPS2 [62]. Interestingly, the dimethylation of protein RPS2 plays an additional role in processing and nuclear export of rRNA [63], and the hydroxylation of protein RPS23 was shown to affect translational accuracy in a stop codon context-dependent manner and determines the viability as a consequence of nonsense codon suppression under certain conditions [64]. Another example for ribosome heterogeneity based on RP modification was described by Ramagopal already in 1991, when differential ribosomal protein phosphorylation and methylation patterns were identified in ribosomes from different phases of the D. discoideum lifecycle [65]. This modulation is considered to facilitate the unique translational needs of the cell during the respective life phase.
Again, an exceptional high number of modifications in RPs was determined in plants. In A. thaliana, 23 of the 80 RP families were shown to contain at least one covalent modification that represent potential differential modification sites [66]. Moreover, UV-B exposure and the associated ribosome damage was described to lead to an increase of de novo synthesis of some RPs and phosphorylation of RPS6 and an S6 kinase in Zea mays leaves [67]. A similar response to UV-B light exposure was described for mammalian cells [68], making it tempting to envisage that ribosome damage induces a feedback-loop, which favors RP mRNA translation for de novo ribosome biogenesis in order to replace damaged ribosomes. This hypothesis is strengthened by recent observations that the expression of genes encoding mitoribosomal proteins in A. thaliana respond to the silencing of the mitochondrial gene encoding protein RPL10 [69]. This specific gene silencing leads to the formation of unstable small ribosomal subunits and consequently to a misbalance between the ribosomal subunits. Notably, these misregulated ribosomes display a translational selectivity as they preferentially translate mRNAs coding for mitochondrial RPs.
3.2. Heterogeneity in the rRNA
3.2.1. Alternative rRNA
The concept, that incorporation of alternative rRNA molecules into ribosomes might change the specificity of the translational machinery in response to external signals, was strengthened by the presence of multiple rRNA gene (rrn) copies in the genomes of a variety of organisms in all domains of life. The Streptomyces coelicolor genome harbors for example six copies of divergent large subunit (LSU) rRNA genes that constitute five LSU rRNA species in a cell, which are differentially transcribed during the morphological development [70]. Similarly, B. subtilis harbors ten rrn operons and their reduction to one copy increased the doubling time as well as the sporulation frequency and the motility [71]. Notably, mutants that carried different combinations of two rrn operons revealed a wide range of growth rates and sporulation frequencies, indicating distinct functional roles for all rrn operons.
An alternative mechanism for the adaptation of the translational machinery to environmental conditions has been shown for the halophilic archaeon Haloarcula marismortui, which harbors three rRNA operons [72]. Here, the rrnB operon, which is GC-rich in contrast to the rrnA and rrnC operons, is selectively transcribed at high temperatures. By this means, heat-stable ribosomes are generated during temperature stress. However, no specific functional differences can be attributed to the presence of the specific rRNA variants.
The parasite Plasmodium berghei has two structurally distinct genes that code for cytoplasmatic small subunit (SSU) rRNAs [73]. The expression of one rRNA gene was almost exclusively found when living in the mosquito host, while the alternative transcript arises when the parasite infects the mammalian host, becoming the predominant SSU rRNA species. Interestingly, no structural differences between the ribosomes containing one or the other rRNA were detected and parasites lacking the mosquito specific rRNA gene were able to complete their development in both hosts, declining the hypothesis of functionally distinct ribosome subspecies and rather indicating a dose dependent role for the prevalence of two distinct rRNA genes [74]. Similarly, the sea urchin Paracentrotus lividus harbors three different 5S rRNA clusters, the transcription of which leads to several 5S rRNA variants that are incorporated in functional ribosomes resulting in a high heterogeneity in animal ribosomes [75].
3.2.2. Modification of rRNA
The rRNA is heavily modified in particular at functionally relevant positions, mainly by 2-hydroxyl methylation and the conversion of uridine to pseudouridine [76]. In general, these modifications occur during rRNA maturation in the process of riboneogenesis, and are considered as check points during ribosome assembly. In eukaryotes, the modifications are facilitated by snoRNAs and their tissue specific expression might be a source for ribosome specialization [77]. Recently, a new method was established to determine pseudouridinylated positions in RNA using next generation sequencing that allows to test whether specific sites in rRNA are differently modified in response to environmental cues representing translational adaptation [78,79]. Nevertheless, Yoon and co-workers already showed that altering the pseudouridylation-state of rRNA affects translation and moreover, that reduced rRNA pseudouridylation led to a pathological syndrome and deficiencies in IRES-dependent translation [80]. However, the next generation sequencing based methodology allows to globally identify functional heterogeneity mediated by alternative pseudouridylation.
In contrast to the above mentioned modifications that occur during assembly, an intriguing mechanism of rRNA processing occurs on already translationally active ribosomes in E. coli [81]. When E. coli cells encounter stress conditions, the endoribonuclease MazF, the toxin component of the toxin–antitoxin module mazEF, becomes activated [82]. Subsequently, MazF targets the 16S rRNA within 30S ribosomal subunits at the decoding center, thereby removing the 3′-terminal 43 nucleotides [81]. As this region comprises the aSD sequence that is required for translation initiation on canonical mRNAs, a subpopulation of ribosomes is engendered that selectively translates lmRNAs both in vivo and in vitro. Concomitantly, MazF generates lmRNAs by removing the 5′-UTRs of distinct transcripts. Collectively, the translational program is remodeled in response to stressful conditions [81].
Considering that rRNA interacts with a variety of different proteins, it is obvious that rRNA modifications have the potential to modulate the respective affinities and consequently alter ribosomal properties. The erythromycin resistance methyltransferases (Erm), for example, dimethylate nucleotide A2058 in the LSU rRNA [83,84]. Despite conferring resistance to macrolide antibiotics, this modification reduces cell fitness, as it mediates abnormal interactions of the nascent peptide with the modified rRNA in the peptide exit tunnel. These findings depict the two-edged nature of this ribosome modification and explain the necessity why erm genes have evolved to be inducible [83].
In S. cerevisiae, a variation in ribose methylation was described within the 18S rRNA. 32% of molecules incorporated in ribosomes are not methylated at 2′-O-ribose of the A100 residue, however, both 40S ribosomal subunits, with and without Am100, participate in translation [85]. A different study showed a more severe impact of the lack of an rRNA modification. Ribosomes isolated from a yeast strain, which harbors a catalytically impaired rRNA pseudouridine synthase, display decreased affinities for tRNAs as well as for the cricket paralysis virus IRES [86]. In general, these ribosomes reveal a decreased translational fidelity and IRES-dependent translational initiation [86].
3.3. Heterogeneity of translation factors
As protein synthesis is assisted by several genuine translation factors, it is obvious to envision regulatory mechanisms by virtue of modified translation factors. In this chapter we want to summarize and discuss heterogeneity within translation factors and their possible impacts on translation regulation.
3.3.1. Prokaryotic initiation factors
In bacteria, there is ample evidence that the ratio between the individual IFs and the ribosomes play crucial roles in selective translation of certain mRNAs. This mechanism was described in E. coli by Giuliodori and co-workers [87]. The authors observed the preferential translation of distinct mRNAs that encode proteins important for cold-shock and cold-tolerance under cold-shock conditions. Their results further indicate that this selectivity can be attributed to a stoichiometric imbalance of the IF to ribosome ratio, contradicting the dogma of equimolar ratios between ribosome and IFs by Howe and Hershey [88]. Interestingly, increased amounts of IF3, together with IF1, preferentially stimulate translation of cold-shock mRNAs [87]. Alternatively, IF2, which recruits the tRNAi to the PIC, has been shown to selectively stimulate translation initiation on lmRNAs [89]. This result clearly suggests that recognition of the 5′-terminally AUG start codon via codon–anticodon interaction is crucial for translation initiation on lmRNAs. In contrast, IF3 appears to antagonize start codon selection on lmRNAs by destabilizing the translation initiation complex assembled at the 5′-end of the transcript [90]. These results were confirmed in a subsequent study indicating that the ratio between IF2 and IF3 is crucial for efficient translation initiation on 5′-terminal AUG start codons of lmRNAs in E. coli [91]. Together, these studies exemplify that alterations of the IF stoichiometry have the potential to affect the selectivity of the translational machinery in bacteria resulting in the preferred translation of a subset of mRNAs.
3.3.2. Eukaryotic initiation factors
The alteration of the translational specificity by means of variation or modification of translation factors is a well-established mechanism in eukaryotes. For example the stress-induced phosphorylation of the α-subunit of eIF2, which recruits the tRNAi to the 40S ribosome, leads to a general inhibition of translation [92]. However, in hepatitis C virus (HCV) infections, an alternative tRNAi-binding protein, eIF2A, has been reported to guide translation initiation to HCV IRES sites [93]. By this means the viral mRNA is selectively translated and overcomes the general translation inhibition by eIF2α phosphorylation during stress induced by the virus infection itself. Interestingly, eIF2A seems likewise to be involved in translation initiation with elongator tRNAs as shown for Leu-tRNA initiation on CUG-start codons [94]. This mechanism has been exemplified by Liang and co-workers, who have shown the synthesis of an N-terminal elongated variant of the human tumor suppressor PTEN, by translation initiation at an upstream and inframe CUG-start codon [95]. Another alternative eIF2 initiation factor, namely eIF2D, acts in a GTP-independent manner and guides translation initiation to unconventional mRNAs like HCV IRES or lmRNAs [96]. Employing an alternative initiation mechanism, eIF2D recruits tRNAi to the 40S ribosome after the AUG start codon has been positioned at the P-site. Collectively, eIF2 and its alternatives appear to add another regulatory level to translation initiation, as the choice of the one or the other variant guides initiation towards different classes of mRNAs. This mechanism appears to be conceptionally similar to the regulation of transcription, where the use of alternative sigma factors guides the RNA polymerase to another set of promoters upon encountering external stimuli.
eIF3 is the largest initiation factor, consisting of 13 different subunits in mammals, of which only 6 build the functional core complex [97]. It generally plays a regulatory role in translation initiation as it stimulates the formation of the TC and the 43S PIC. eIF3 is also involved in cap-independent translation initiation as on IRES-mediated initiation of protein synthesis. The diverse roles of eIF3 might in part be explained by its heterogeneous composition. Already in the 1970s, researchers have described heterogeneity in association of eIF3 to the ribosomes in rat liver homogenates [98]. The results suggested that eIF3 was preferentially bound to newly synthesized 40S ribosomes, but not to recycled ribosomes. However, eIF3 is not only involved in the association of the TC eIF2-GTP-tRNAi to the 40S ribosome, but acts likewise as a ribosome-dissociation factor after termination and hinders re-association of the free 40S and 60S subunits [99,100]. Kolupaeva and co-workers have proposed that the ability of eIF3 to bind the 40S subunit and to prevent its re-association with 60S subunits is dependent on the presence of the subunit eIF3j in mammals [101]. As observed more recently in a mammalian cell line, the subunit eIF3a shows oscillating levels within the cell cycle and appears to be a translational regulator for the S-phase entrance [102]. The subunits eIF3e and eIF3g have been reported to associate with translational regulators like p56 or poly(A)-binding proteins, respectively, thereby stimulating or inhibiting translation initiation [103,104]. For other eIF3 subunits a role in cancer cells was suggested. The mRNA levels encoding the subunits eIF3a, -b, -c, -h, and -I are increased in a wide variety of human tumors and moreover, their overexpression was shown to induce oncogenic malignancy [105]. In contrast, eIF3f seems to have tumor suppressive activity by affecting cap-dependent and cap-independent translation initiation [106]. In addition, the factor is involved in translation inhibition during apoptosis ([107] and references therein). Intriguingly, eIF3 also is remarkably heterogeneous in terms of localization: nuclear eIF3 has a different subunit composition than cytosolic eIF3, which is lacking the subunits eIF3a and -f [108]. The authors suggest that the cytosolic eIF3f is phosphorylated during apoptosis, which results in a stronger association of eIF3f to the eIF3 core complex leading to translation inhibition. The pro-apoptotic function of eIF3f seems furthermore to be regulated by direct interaction with the anti-apoptotic factor Mss4 [109]. Thus, eIF3 is an intriguing example for regulation of protein synthesis mediated by modified translation factors, as induced heterogeneity of a single IF can adapt the translational program to a vast variety of conditions.
3.4. Heterogeneity of tRNAs
During translation, tRNAs act as the adaptor molecules in decoding of the base triplets on the mRNA into the corresponding amino acids. But their role is not merely mechanistical and – although highly conserved – they show remarkable structural diversity amongst species [110]. In the recent years, it has been shown that tRNAs do not only exist in their aminoacylated or uncharged full length form, but also short fragments of tRNAs or tRNA halves have been observed (reviewed in Ref. [111]). The stress-induced 5′-tRNA halves inhibit translation in mammalian cells by interfering with initiation factors eIF4E/G/A [112], and tRNA fragments can bind to the ribosome and inhibit translation as it was shown for the archaeon Haloferax volcanii [113].
However, in correspondence to genes encoding RPs, there are also several copies for tRNA genes. The E. coli genome encodes four copies for the tRNAi. Samhita and co-workers found that one of these genes (metY) appeared to be dispensable [114]. However, in nutrient-rich conditions the presence of all four genes is advantageous over only three. On the other hand, the lack of one tRNAi-gene has been shown to be beneficial during nutrient-deprived conditions and during long-term growth [114]. These findings strongly imply that different variants of tRNAi are used for translation initiation under different environmental conditions. It is important to note that ribosomes derived from an E. coli strain lacking three tRNAi genes (metZVW) are slightly depleted of ribosomal protein S1 [115], which is required for translation initiation on canonical mRNAs [30,116]. Also higher eukaryotes harbor multiple tRNA gene copies coding for so-called isodecoders, different tRNA-bodies with the same anti-codon. The function of these isodecoders appears not to be redundant as loss of one particular tRNA (tRNAArgUCU) cannot be compensated by its isodecoders in mice [117].
Several lines of evidence indicate that tRNAs can also be targeted by bacterial stress-dependent toxins. VapC is an endoribonuclease of the toxin–antitoxin system vapBC. Upon activation it cleaves the tRNAi at the anticodon stem-loop thus resulting in the inhibition of translation [118]. Besides, VapC activation stimulates translation initiation on elongation codons of the dksA mRNA, however, the underling mechanism still remains elusive. Taken together, these findings indicate an alternative mechanism of stress-induced adaptation of the translational program by tRNA intrinsic variations.
3.5. Extra-translational functions of ribosomal proteins and translation factors
As mentioned above, heterogeneous expression of RPs can result in the formation of specialized ribosomes, and has been observed in prokaryotes and eukaryotes in dependence of growth conditions and cell type [34,119]. Considering the extraribosomal functions of several RPs (summarized in Ref. [42]) and translation factors, one is tempted to speculate that these extra-ribosomal functions could likewise represent a means to reduce the amount of RPs or factors available for the translational machinery. For example, the largest RP present in Gram-negative bacteria, namely protein S1, is, besides its important mRNA-binding function in translation initiation, a subunit of the RNA-directed RNA polymerase of the bacteriophage Qβ [120]. In this context, S1 plays a role in the termination of the polymerase reaction. Moreover, the mammalian RPS3 acts additionally as an endonuclease and can become part of the nuclear NFκB complex, thereby interfering with transcription regulation (reviewed in Ref. [42]).
Likewise, translation factors have been implicated with non-translational functions, like eIF3f, which has been proposed to create a link between translation and RNA degradation during stress [106]. During apoptosis, eIF3f might interact with heterogeneous nuclear ribonucleoprotein K blocking its RNA protective function. Additionally, eIF3f possesses a de-ubiquitinase activity by itself, which links it to Notch signaling pathways [121] and moreover, it seems to be a regulatory factor in the balance between muscle atrophy and hypertrophy [122]. Another factor involved in several extra-ribosomal functions is eEF1A, which besides the delivery of aa-tRNAs to the ribosomal A-site is responsible for non-canonical processes like quality control of newly synthesized proteins, apoptosis, and viral propagation [123]. Moreover, it was shown recently that a methylated version of eEF1A is required for nodavirus RNA replication in yeast [124]. Taken together, these results add significant weight to the audacious hypothesis that extra-translational functions could withdraw RPs or translation factors from the translational machinery and thus result in an adjustment of translation.
4. The multifaceted translational machinery: a perspective
In light of the increasing evidence, ribosome heterogeneity, though still far from being entirely understood, proves to be an integral mechanism to modulate and fine tune protein synthesis in response to environmental signals in all organisms. Considering the time and energy consuming steps of the transcriptional stress response mechanisms, which involve the synthesis of alternative sigma factors followed by the selective transcription and translation of regulatory factors, it is conceivable that the strategies summarized here provide a novel ‘fast-track reaction’ for the cell to cope with the immediate changes in external conditions. Thus, the alteration of the transcriptional program would constitute the second level of stress response required for the mid- and long-term adjustment of the metabolic network.
Despite the multitude of examples for ribosome heterogeneity available, there is still ample need for further studies to gain comprehensive insights into the functional reprogramming of the translational machinery. Nevertheless, the above mentioned studies dramatically object the long-cherished tenet that the ribosome is the unchangeable operating unit of protein synthesis and strongly favor the conception that multifaceted ribosomes represent a central hub for signal integration in cellular physiology.
Acknowledgments
The authors would like to thank all colleagues and past and present members of the Moll group who contributed to our studies on ribosome heterogeneity. The work was supported by grants W1207-B09, P20112-B03, P22249-B20, F4316-B09, P26946-B20, and P27043-B20 from the Austrian Science Fund to I.M.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuersten S, Radek A, Vogel C, Penalva LO. Translation regulation gets its ‘omics’ moment. Wiley Interdiscip Rev RNA. 2013;4:617–630. doi: 10.1002/wrna.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkhardt N, Junemann R, Spahn CM, Nierhaus KH. Ribosomal tRNA binding sites: three-site models of translation. Crit Rev Biochem Mol Biol. 1998;33:95–149. doi: 10.1080/10409239891204189. [DOI] [PubMed] [Google Scholar]
- 4.Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simonetti A, Marzi S, Jenner L, Myasnikov A, Romby P, et al. A structural view of translation initiation in bacteria. Cell Mol Life Sci. 2009;66:423–436. doi: 10.1007/s00018-008-8416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lomakin IB, Steitz TA. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature. 2013;500:307–311. doi: 10.1038/nature12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang SK. Internal initiation: IRES elements of picornaviruses and hepatitis c virus. Virus Res. 2006;119:2–15. doi: 10.1016/j.virusres.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Rodnina MV, Wintermeyer W. Recent mechanistic insights into eukaryotic ribosomes. Curr Opin Cell Biol. 2009;21:435–443. doi: 10.1016/j.ceb.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 12.Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agirrezabala X, Frank J. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q Rev Biophys. 2009;42:159–200. doi: 10.1017/S0033583509990060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zavialov AV, Buckingham RH, Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 15.Korostelev AA. Structural aspects of translation termination on the ribosome. RNA. 2011;17:1409–1421. doi: 10.1261/rna.2733411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nurenberg E, Tampe R. Tying up loose ends: ribosome recycling in eukaryotes and archaea. Trends Biochem Sci. 2013;38:64–74. doi: 10.1016/j.tibs.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Romby P, Springer M. Bacterial translational control at atomic resolution. Trends Genet TIG. 2003;19:155–161. doi: 10.1016/S0168-9525(03)00020-9. [DOI] [PubMed] [Google Scholar]
- 18.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Duin J, Kurland CG. Functional heterogeneity of the 30S ribosomal subunit of E. coli. Mol Gen Genet. 1970;109:169–176. doi: 10.1007/BF00269653. [DOI] [PubMed] [Google Scholar]
- 20.Kurland CG, Voynow P, Hardy SJ, Randall L, Lutter L. Physical and functional heterogeneity of E. coli ribosomes. Cold Spring Harb Symp Quant Biol. 1969;34:17–24. doi: 10.1101/sqb.1969.034.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Deusser E. Heterogeneity of ribosomal populations in Escherichia coli cells grown in different media. Mol Gen Genet. 1972;119:249–258. doi: 10.1007/BF00333862. [DOI] [PubMed] [Google Scholar]
- 22.Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc Natl Acad Sci U S A. 2002;99:12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mauro VP, Edelman GM. The ribosome filter redux. Cell Cycle. 2007;6:2246–2251. doi: 10.4161/cc.6.18.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramagopal S, Ennis HL. Regulation of synthesis of cell-specific ribosomal proteins during differentiation of Dictyostelium discoideum. Proc Natl Acad Sci U S A. 1981;78:3083–3087. doi: 10.1073/pnas.78.5.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deusser E, Wittmann HG. Ribosomal proteins: variation of the protein composition in Escherichia coli ribosomes as function of growth rate. Nature. 1972;238:269–270. doi: 10.1038/238269a0. [DOI] [PubMed] [Google Scholar]
- 27.Pulk A, Liiv A, Peil L, Maivali U, Nierhaus K, et al. Ribosome reactivation by replacement of damaged proteins. Mol Microbiol. 2010;75:801–814. doi: 10.1111/j.1365-2958.2009.07002.x. [DOI] [PubMed] [Google Scholar]
- 28.Bickle TA, Howard GA, Traut RR. Ribosome heterogenecity. The nonuniform distribution of specific ribosomal proteins among different functional classes of ribosomes. J Biol Chem. 1973;248:4862–4864. [PubMed] [Google Scholar]
- 29.Kaberdina AC, Szaflarski W, Nierhaus KH, Moll I. An unexpected type of ribosomes induced by kasugamycin: a look into ancestral times of protein synthesis? Mol Cell. 2009;33:227–236. doi: 10.1016/j.molcel.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorensen MA, Fricke J, Pedersen S. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J Mol Biol. 1998;280:561–569. doi: 10.1006/jmbi.1998.1909. [DOI] [PubMed] [Google Scholar]
- 31.Moll I, Resch A, Blasi U. Discrimination of 5’-terminal start codons by translation initiation factor 3 is mediated by ribosomal protein S1. FEBS Lett. 1998;436:213–217. doi: 10.1016/s0014-5793(98)01131-4. [DOI] [PubMed] [Google Scholar]
- 32.Delvillani F, Papiani G, Deho G, Briani F. S1 ribosomal protein and the interplay between translation and mRNA decay. Nucleic Acids Res. 2011;39:7702–7715. doi: 10.1093/nar/gkr417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Byrgazov K, Grishkovskaya I, Arenz S, Coudevylle N, Temmel H, Wilson DN, Djinovic-Carugo K, Moll I. Structural basis for the flexible interaction of protein S1 with the Escherichia coli ribosome. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bortoluzzi S, d’Alessi F, Romualdi C, Danieli GA. Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics. 2001;17:1152–1157. doi: 10.1093/bioinformatics/17.12.1152. [DOI] [PubMed] [Google Scholar]
- 35.Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta V, Warner JR. Ribosome-omics of the human ribosome. RNA. 2014;20:1004–1013. doi: 10.1261/rna.043653.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song DG, Kim YS, Jung BC, Rhee KJ, Pan CH. Parkin induces upregulation of 40S ribosomal protein SA and posttranslational modification of cytokeratins 8 and 18 in human cervical cancer cells. Appl Biochem Biotechnol. 2013;171:1630–1638. doi: 10.1007/s12010-013-0443-4. [DOI] [PubMed] [Google Scholar]
- 38.Nelson J, McFerran NV, Pivato G, Chambers E, Doherty C, et al. The 67 kDa laminin receptor: structure, function and role in disease. Biosci Rep. 2008;28:33–48. doi: 10.1042/BSR20070004. [DOI] [PubMed] [Google Scholar]
- 39.Li D, Chen J, Gao Z, Li X, Yan X, et al. 67-kDa laminin receptor in human bile duct carcinoma. Eur Surg Res. 2009;42:168–173. doi: 10.1159/000198234. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen-Lefebvre AT, Leprun G, Morin V, Vinuelas J, Coute Y, et al. V-erbA generates ribosomes devoid of RPL11 and regulates translational activity in avian erythroid progenitors. Oncogene. 2014;33:1581–1589. doi: 10.1038/onc.2013.93. [DOI] [PubMed] [Google Scholar]
- 41.Lee AS, Burdeinick-Kerr R, Whelan SP. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci U S A. 2013;110:324–329. doi: 10.1073/pnas.1216454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warner JR, Mcintosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 44.Parenteau J, Durand M, Morin G, Gagnon J, Lucier JF, et al. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell. 2011;147:320–331. doi: 10.1016/j.cell.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 45.Natori Y, Nanamiya H, Akanuma G, Kosono S, Kudo T, et al. A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol Microbiol. 2007;63:294–307. doi: 10.1111/j.1365-2958.2006.05513.x. [DOI] [PubMed] [Google Scholar]
- 46.Bauer JW, Brandl C, Haubenreisser O, Wimmer B, Weber M, et al. Specialized yeast ribosomes: a customized tool for selective mRNA translation. PLoS One. 2013;8:e67609. doi: 10.1371/journal.pone.0067609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cardenas D, Revuelta-Cervantes J, Jimenez-Diaz A, Camargo H, Remacha M, et al. P1 and P2 protein heterodimer binding to the P0 protein of Saccharomyces cerevisiae is relatively non-specific and a source of ribosomal heterogeneity. Nucleic Acids Res. 2012;40:4520–4529. doi: 10.1093/nar/gks036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, et al. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 2001;127:398–415. [PMC free article] [PubMed] [Google Scholar]
- 49.Sormani R, Masclaux-Daubresse C, Daniel-Vedele F, Chardon F. Transcriptional regulation of ribosome components are determined by stress according to cellular compartments in Arabidopsis thaliana. PLoS One. 2011;6:e28070. doi: 10.1371/journal.pone.0028070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falcone Ferreyra ML, Pezza A, Biarc J, Burlingame AL, Casati P. Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress. Plant Physiol. 2010;153:1878–1894. doi: 10.1104/pp.110.157057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, et al. An Arabidopsis minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development. 2001;128:4289–4299. doi: 10.1242/dev.128.21.4289. [DOI] [PubMed] [Google Scholar]
- 52.Giavalisco P, Wilson D, Kreitler T, Lehrach H, Klose J, et al. High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol Biol. 2005;57:577–591. doi: 10.1007/s11103-005-0699-3. [DOI] [PubMed] [Google Scholar]
- 53.Zsogon A, Szakonyi D, Shi X, Byrne ME. Ribosomal protein RPL27a promotes female gametophyte development in a dose-dependent manner. Plant Physiol. 2014;165:1133–1143. doi: 10.1104/pp.114.241778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopes AM, Miguel RN, Sargent CA, Ellis PJ, Amorim A, et al. The human RPS4 paralogue on Yq11.223 encodes a structurally conserved ribosomal protein and is preferentially expressed during spermatogenesis. BMC Mol Biol. 2010;11:33. doi: 10.1186/1471-2199-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong QW, Li J, Ng SR, Lim SG, Yang H, et al. RPL39L is an example of a recently evolved ribosomal protein paralog that shows highly specific tissue expression patterns and is upregulated in ESCs and HCC tumors. RNA Biol. 2014;11:33–41. doi: 10.4161/rna.27427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugihara Y, Honda H, Iida T, Morinaga T, Hino S, et al. Proteomic analysis of rodent ribosomes revealed heterogeneity including ribosomal proteins L10-like, L22-like 1, and L39-like. J Proteome Res. 2010;9:1351–1366. doi: 10.1021/pr9008964. [DOI] [PubMed] [Google Scholar]
- 57.Sugihara Y, Sadohara E, Yonezawa K, Kugo M, Oshima K, et al. Identification and expression of an autosomal paralogue of ribosomal protein S4, X-linked, in mice: potential involvement of testis-specific ribosomal proteins in translation and spermatogenesis. Gene. 2013;521:91–99. doi: 10.1016/j.gene.2013.02.040. [DOI] [PubMed] [Google Scholar]
- 58.Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, et al. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Yu BJ, Kim JA, Moon JH, Ryu SE, Pan JG. The diversity of lysine-acetylated proteins in Escherichia coli. J Microbiol Biotechnol. 2008;18:1529–1536. [PubMed] [Google Scholar]
- 60.Gordiyenko Y, Deroo S, Zhou M, Videler H, Robinson CV. Acetylation of L12 increases interactions in the Escherichia coli ribosomal stalk complex. J Mol Biol. 2008;380:404–414. doi: 10.1016/j.jmb.2008.04.067. [DOI] [PubMed] [Google Scholar]
- 61.Lee SW, Berger SJ, Martinovic S, Pasa-Tolic L, Anderson GA, et al. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc Natl Acad Sci U S A. 2002;99:5942–5947. doi: 10.1073/pnas.082119899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ladror DT, Frey BL, Scalf M, Levenstein ME, Artymiuk JM, et al. Methylation of yeast ribosomal protein S2 is elevated during stationary phase growth conditions. Biochem Biophys Res Commun. 2014;445:535–541. doi: 10.1016/j.bbrc.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lipson RS, Webb KJ, Clarke SG. Rmt1 catalyzes zinc-finger independent arginine methylation of ribosomal protein Rps2 in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2010;391:1658–1662. doi: 10.1016/j.bbrc.2009.12.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loenarz C, Sekirnik R, Thalhammer A, Ge W, Spivakovsky E, et al. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc Natl Acad Sci U S A. 2014;111:4019–4024. doi: 10.1073/pnas.1311750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramagopal S. Covalent modifications of ribosomal proteins in growing and aggregation-competent Dictyostelium discoideum: phosphorylation and methylation. Biochem Cell Biol. 1991;69:263–268. doi: 10.1139/o91-040. [DOI] [PubMed] [Google Scholar]
- 66.Carroll AJ, Heazlewood JL, Ito J, Millar AH. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol Cell Proteomics. 2008;7:347–369. doi: 10.1074/mcp.M700052-MCP200. [DOI] [PubMed] [Google Scholar]
- 67.Casati P, Walbot V. Crosslinking of ribosomal proteins to RNA in maize ribosomes by UV-B and its effects on translation. Plant Physiol. 2004;136:3319–3332. doi: 10.1104/pp.104.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brenneisen P, Wenk J, Wlaschek M, Krieg T, Scharffetter-Kochanek K. Activation of p70 ribosomal protein S6 kinase is an essential step in the DNA damage-dependent signaling pathway responsible for the ultraviolet B-mediated increase in interstitial collagenase (MMP-1) and stromelysin-1 (MMP-3) protein levels in human dermal fibroblasts. J Biol Chem. 2000;275:4336–4344. doi: 10.1074/jbc.275.6.4336. [DOI] [PubMed] [Google Scholar]
- 69.Kwasniak M, Majewski P, Skibior R, Adamowicz A, Czarna M, et al. Silencing of the nuclear RPS10 gene encoding mitochondrial ribosomal protein alters translation in Arabidopsis mitochondria. Plant Cell. 2013;25:1855–1867. doi: 10.1105/tpc.113.111294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim HL, Song WS, Kim K, Lee K. Characterization of heterogeneous LSU rRNA profiles in Streptomyces coelicolor under different growth stages and conditions. Curr Microbiol. 2008;57:537–541. doi: 10.1007/s00284-008-9238-1. [DOI] [PubMed] [Google Scholar]
- 71.Yano K, Wada T, Suzuki S, Tagami K, Matsumoto T, et al. Multiple rRNA operons are essential for efficient cell growth and sporulation as well as outgrowth in Bacillus subtilis. Microbiology. 2013;159:2225–2236. doi: 10.1099/mic.0.067025-0. [DOI] [PubMed] [Google Scholar]
- 72.Lopez-Lopez A, Benlloch S, Bonfa M, Rodriguez-Valera F, Mira A. Intra-genomic 16S rDNA divergence in Haloarcula marismortui is an adaptation to different temperatures. J Mol Evol. 2007;65:687–696. doi: 10.1007/s00239-007-9047-3. [DOI] [PubMed] [Google Scholar]
- 73.Gunderson JH, Sogin ML, Wollett G, Hollingdale M, de la Cruz VF, et al. Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science. 1987;238:933–937. doi: 10.1126/science.3672135. [DOI] [PubMed] [Google Scholar]
- 74.van Spaendonk RM, Ramesar J, van Wigcheren A, Eling W, Beetsma AL, et al. Functional equivalence of structurally distinct ribosomes in the malaria parasite, Plasmodium berghei. J Biol Chem. 2001;276:22638–22647. doi: 10.1074/jbc.M101234200. [DOI] [PubMed] [Google Scholar]
- 75.Dimarco E, Cascone E, Bellavia D, Caradonna F. Functional variants of 5S rRNA in the ribosomes of common sea urchin Paracentrotus lividus. Gene. 2012;508:21–25. doi: 10.1016/j.gene.2012.07.067. [DOI] [PubMed] [Google Scholar]
- 76.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 77.Castle JC, Armour CD, Lower M, Haynor D, Biery M, et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS One. 2010;5:e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515(7525):143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 81.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, et al. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011;147:147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3’,5’-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci U S A. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gupta P, Sothiselvam S, Vazquez-Laslop N, Mankin AS. Deregulation of translation due to post-transcriptional modification of rRNA explains why erm genes are inducible. Nat Commun. 2013;4:1984. doi: 10.1038/ncomms2984. [DOI] [PubMed] [Google Scholar]
- 84.Starosta AL, Karpenko VV, Shishkina AV, Mikolajka A, Sumbatyan NV, et al. Interplay between the ribosomal tunnel, nascent chain, and macrolides influences drug inhibition. Chem Biol. 2010;17:504–514. doi: 10.1016/j.chembiol.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 85.Buchhaupt M, Sharma S, Kellner S, Oswald S, Paetzold M, et al. Partial methylation at Am100 in 18S rRNA of baker’s yeast reveals ribosome heterogeneity on the level of eukaryotic rRNA modification. PLoS One. 2014;9:e89640. doi: 10.1371/journal.pone.0089640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell. 2011;44:660–666. doi: 10.1016/j.molcel.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giuliodori AM, Brandi A, Gualerzi CO, Pon CL. Preferential translation of cold-shock mRNAs during cold adaptation. RNA. 2004;10:265–276. doi: 10.1261/rna.5164904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Howe JG, Hershey JW. Initiation factor and ribosome levels are coordinately controlled in Escherichia coli growing at different rates. J Biol Chem. 1983;258:1954–1959. [PubMed] [Google Scholar]
- 89.Grill S, Gualerzi CO, Londei P, Blasi U. Selective stimulation of translation of leaderless mRNA by initiation factor 2: evolutionary implications for translation. EMBO J. 2000;19:4101–4110. doi: 10.1093/emboj/19.15.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tedin K, Moll I, Grill S, Resch A, Graschopf A, et al. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol Microbiol. 1999;31:67–77. doi: 10.1046/j.1365-2958.1999.01147.x. [DOI] [PubMed] [Google Scholar]
- 91.Grill S, Moll I, Hasenohrl D, Gualerzi CO, Blasi U. Modulation of ribosomal recruitment to 5′-terminal start codons by translation initiation factors IF2 and IF3. FEBS Lett. 2001;495:167–171. doi: 10.1016/s0014-5793(01)02378-x. [DOI] [PubMed] [Google Scholar]
- 92.Schmitt E, Naveau M, Mechulam Y. Eukaryotic and archaeal translation initiation factor 2: a heterotrimeric tRNA carrier. FEBS Lett. 2010;584:405–412. doi: 10.1016/j.febslet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 93.Kim JH, Park SM, Park JH, Keum SJ, Jang SK. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 2011;30:2454–2464. doi: 10.1038/emboj.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, et al. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science. 2012;336:1719–1723. doi: 10.1126/science.1220270. [DOI] [PubMed] [Google Scholar]
- 95.Liang H, He S, Yang J, Jia X, Wang P, et al. PTENalpha, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab. 2014;19:836–848. doi: 10.1016/j.cmet.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, et al. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Masutani M, Sonenberg N, Yokoyama S, Imataka H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26:3373–3383. doi: 10.1038/sj.emboj.7601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hinton RH, Mullock BM. Differences between newly formed and recycled free small ribosome subunits in liver cytoplasm. Biochem J. 1976;158:97–103. doi: 10.1042/bj1580097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Trachsel H, Staehelin T. Initiation of mammalian protein synthesis. The multiple functions of the initiation factor eIF-3. Biochim Biophys Acta. 1979;565:305–314. doi: 10.1016/0005-2787(79)90207-7. [DOI] [PubMed] [Google Scholar]
- 100.Thompson HA, Sadnik I, Scheinbuks J, Moldave K. Studies on native ribosomal subunits from rat liver. Purification and characterization of a ribosome dissociation factor. Biochemistry. 1977;16:2221–2230. doi: 10.1021/bi00629a028. [DOI] [PubMed] [Google Scholar]
- 101.Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dong Z, Liu Z, Cui P, Pincheira R, Yang Y, et al. Role of eIF3a in regulating cell cycle progression. Exp Cell Res. 2009;315:1889–1894. doi: 10.1016/j.yexcr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Martineau Y, Derry MC, Wang X, Yanagiya A, Berlanga JJ, et al. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol Cell Biol. 2008;28:6658–6667. doi: 10.1128/MCB.00738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guo J, Hui DJ, Merrick WC, Sen GC. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 2000;19:6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang L, Pan X, Hershey JW. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem. 2007;282:5790–5800. doi: 10.1074/jbc.M606284200. [DOI] [PubMed] [Google Scholar]
- 106.Wen F, Zhou R, Shen A, Choi A, Uribe D, et al. The tumor suppressive role of eIF3f and its function in translation inhibition and rRNA degradation. PLoS One. 2012;7:e34194. doi: 10.1371/journal.pone.0034194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Marchione R, Leibovitch SA, Lenormand JL. The translational factor eIF3f: the ambivalent eIF3 subunit. Cell Mol Life Sci. 2013;70:3603–3616. doi: 10.1007/s00018-013-1263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shi J, Hershey JW, Nelson MA. Phosphorylation of the eukaryotic initiation factor 3f by cyclin-dependent kinase 11 during apoptosis. FEBS Lett. 2009;583:971–977. doi: 10.1016/j.febslet.2009.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Walter BM, Nordhoff C, Varga G, Goncharenko G, Schneider SW, et al. Mss4 protein is a regulator of stress response and apoptosis. Cell Death Dis. 2012;3:e297. doi: 10.1038/cddis.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fujishima K, Kanai A. tRNA gene diversity in the three domains of life. Front Genet. 2014;5:142. doi: 10.3389/fgene.2014.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gebetsberger J, Zywicki M, Kunzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909. doi: 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Samhita L, Nanjundiah V, Varshney U. How many initiator tRNA genes does Escherichia coli need? J Bacteriol. 2014;196:2607–2615. doi: 10.1128/JB.01620-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Samhita L, Virumae K, Remme J, Varshney U. Initiation with elongator tRNAs. J Bacteriol. 2013;195:4202–4209. doi: 10.1128/JB.00637-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tedin K, Resch A, Blasi U. Requirements for ribosomal protein S1 for translation initiation of mRNAs with and without a 5′ leader sequence. Mol Microbiol. 1997;25:189–199. doi: 10.1046/j.1365-2958.1997.4421810.x. [DOI] [PubMed] [Google Scholar]
- 117.Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, et al. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Winther KS, Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A. 2011;108:7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Milne AN, Mak WW, Wong JT. Variation of ribosomal proteins with bacterial growth rate. J Bacteriol. 1975;122:89–92. doi: 10.1128/jb.122.1.89-92.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vasilyev NN, Kutlubaeva ZS, Ugarov VI, Chetverina HV, Chetverin AB. Ribosomal protein S1 functions as a termination factor in RNA synthesis by Qbeta phage replicase. Nat Commun. 2013;4:1781. doi: 10.1038/ncomms2807. [DOI] [PubMed] [Google Scholar]
- 121.Moretti J, Chastagner P, Gastaldello S, Heuss SF, Dirac AM, et al. The translation initiation factor 3f (eIF3f) exhibits a deubiquitinase activity regulating Notch activation. PLoS Biol. 2010;8:e1000545. doi: 10.1371/journal.pbio.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Csibi A, Leibovitch MP, Cornille K, Tintignac LA, Leibovitch SA. MAFbx/Atrogin-1 controls the activity of the initiation factor eIF3-f in skeletal muscle atrophy by targeting multiple C-terminal lysines. J Biol Chem. 2009;284:4413–4421. doi: 10.1074/jbc.M807641200. [DOI] [PubMed] [Google Scholar]
- 123.Sasikumar AN, Perez WB, Kinzy TG. The many roles of the eukaryotic elongation factor 1 complex. Wiley Interdiscip Rev RNA. 2012;3:543–555. doi: 10.1002/wrna.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li Z, Gonzalez PA, Sasvari Z, Kinzy TG, Nagy PD. Methylation of translation elongation factor 1A by the METTL10-like See1 methyltransferase facilitates tombusvirus replication in yeast and plants. Virology. 2014;448:43–54. doi: 10.1016/j.virol.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 125.Porollo A, Meller J. Versatile annotation and publication quality visualization of protein complexes using POLYVIEW-3D. BMC Bioinform. 2007;8:316. doi: 10.1186/1471-2105-8-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 127.Roll-Mecak A, Cao C, Dever TE, Burley SK. X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell. 2000;103:781–792. doi: 10.1016/s0092-8674(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 128.Querol-Audi J, Sun C, Vogan JM, Smith MD, Gu Y, et al. Architecture of human translation initiation factor 3. Structure. 2013;21:920–928. doi: 10.1016/j.str.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sussman JL, Holbrook SR, Warrant RW, Church GM, Kim SH. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J Mol Biol. 123:607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]