Abstract
In this study, the complete mitochondrial (mt) genomes of Chrysoporthe austroafricana (190,834 bp), C. cubensis (89,084 bp) and C. deuterocubensis (124,412 bp) were determined. Additionally, the mitochondrial genome of another member of the Cryphonectriaceae, namely Cryphonectria parasitica (158,902 bp), was retrieved and annotated for comparative purposes. These genomes showed high levels of synteny, especially in regions including genes involved in oxidative phosphorylation and electron transfer, unique open reading frames (uORFs), ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs), as well as intron positions. Comparative analyses revealed signatures of duplication events, intron number and length variation, and varying intronic ORFs which highlighted the genetic diversity of mt genomes among the Cryphonectriaceae. These mt genomes showed remarkable size polymorphism. The size polymorphism in the mt genomes of these closely related Chrysoporthe species was attributed to the varying number and length of introns, coding sequences and to a lesser extent, intergenic sequences. Compared to publicly available fungal mt genomes, the C. austroafricana mt genome is the second largest in the Ascomycetes thus far.
Introduction
Chrysoporthe species are economically important pathogens of Eucalyptus species and other Myrtales [1, 2]. Species in this genus include Chrysoporthe austroafricana [3], Chrysoporthe cubensis [4, 5], Chrysoporthe inopina [6], Chrysoporthe hodgesiana [1], Chrysoporthe doradensis [7], Chrysoporthe deuterocubensis [8], Chrysoporthe zambiensis and Chrysoporthe syzigiicola [9]. These species form part of the family Cryphonectriaceae, which also includes Cryphonectria [1, 2, 8, 10]. Although the molecular data used in these studies have helped to clarify the species boundaries in Chrysoporthe, the relationship among the members of this genus remains largely unresolved [8, 9].
Mitochondrial (mt) sequences have been used to determine and/or confirm phylogenetic relationships, especially in cases where nuclear genes have not accumulated sufficient phylogenetic signal, to clarify conflicting phylogenies [11, 12]. This is because mt genomes contain marker genes that are particularly suited to studies in evolutionary biology and systematics [13–15]. Additionally, due to the relatively small sizes of mt genomes, whole genome analysis can provide a large set of homologous genes that can be directly compared [16]. The advantages of using mt genes for phylogenetic inference are numerous–their evolution is largely independent from nuclear genes, they are uniparentally inherited, they display a uniform genetic background, and they have limited rates of recombination [17]. Furthermore, due to an increase in whole genome sequencing projects, there are a large number of complete mt genomes in the public domain (http://www.ncbi.nlm.nih.gov/genome/browse/?report=5). The availability of these genomic resources has enhanced their usage in evolutionary biology [12, 18–22].
Fungal mt genomes are circular A+T rich sequences that usually contain a set of 14 conserved genes that encode proteins involved in oxidative phosphorylation (OXPHOS) and electron transport [16, 23]. These include three ATP synthase subunits (atp6, atp8 and atp9), three subunits of cytochrome oxidase (cox1, cox2 and cox3), apocytochrome b (cob) and seven subunits of the nicotinamide adenine dinucleotide ubiquinone oxidoreductase subunits (nad1, nad2, nad3, nad4, nad4L, nad5 and nad6). Untranslated genes include the small subunit ribosomal RNA (rns) and the large subunit ribosomal RNA (rnl), as well as a varying number of transfer-RNA (tRNA) genes [16, 17]. Additionally, mitochondrial genomes are known to harbour intergenic ORFs, some of which are unique to each genome (uORFs), while others are mobile ORFs encoding endonucleases [24–27]. Linear or circular plasmids, plasmid-like and virus-like elements have also been reported from mitochondria and mt genomes [28–31].
The diversity of mt genomes is evident among Ascomycetes, where the smallest mt genome is 18,512 bp for Hanseniaspora uvarum [32] and the largest is 203,051 bp for Sclerotinia borealis [33]. These size differences appear to be directly correlated to the numbers and sizes of introns, the presence of dispersed repetitive regions, and new genes introduced through horizontal transfer [25, 34–36]. The divergence of these mt genomes is also associated with rearrangements in gene order [15, 22] and the presence or absence of auto-replicating plasmids [30].
Fungal mt introns encode various homing endonuclease genes (HEGs) [31, 37–39]. These intron-enclosed genes have homing endonuclease (HE) activity, thus are able to move or self-integrate (homing) themselves into specific locations in the genome [31, 38, 39]. The introns occurring in mt genomes are divided into two groups, namely groups I and II, both of which encode HEGs with signature LAGLIDADG or GIY-YIG domain motifs [40, 41]. These introns generally seem to affect the mt genome structure by influencing variations in genome size, gene order, gene duplications, and the introduction of exogenous genes via horizontal gene transfer [15, 33, 35, 36].
Despite the increasing number, and potential applications, of mtDNA in systems biology and evolutionary studies [14, 42, 43], the mt genome sequences of Chrysoporthe species have not yet been determined. The aim of this study was therefore, to provide insights into the structure, organization and source of variability in the mt genomes of these closely related species. Additionally, availability of these genomes also provides invaluable resources for evolutionary studies. For this reason, the complete mt genomes of C. austroafricana, C. cubensis and C. deuterocubensis were assembled and annotated. Also, for comparative purposes, the mt genome for C. parasitica was fully annotated. Comparisons were focused on genome organization, gene order and gene content of these four mt genomes.
Materials and Methods
Genome sequencing and assembly
The mt genome of C. austroafricana was sequenced as part of a whole genome sequencing project [44, 45]. Additionally, C. cubensis and C. deuterocubensis mt genome sequences were obtained from subsequent whole genome sequencing projects (Wingfield et al. 2015, submitted). The C. parasitica mt genome sequence was obtained from the Joint Genome Institute (JGI) database (http://genome.jgi.doe.gov/Crypa2/Crypa2.download.html). After assembly of the C. austroafricana, C. cubensis and C. deuterocubensis nuclear genomes, the mt genome of C. parasitica was used to identify conserved mt gene regions by reference assembly in CLC Genomics Workbench v 7.0.1 (CLC Bio, Arhus, Denmark). Identified mt sequences were further confirmed by BLASTn [46] searches against non-redundant nucleotide sequences in the NCBI GenBank database. The confirmed sequences were then used as seeds for iterative contig extension using MITObim v 1.7 beta [47] and PRICE v 1.2 [48] software. Extended contigs were aligned using the multiple sequence alignment program MAFFT v 7.182 [49] where contigs with more than 500 bp of overlapping sequences were merged into larger contigs. After confirming the presence of all the conserved mt genes in the assembled contigs, contigs that displayed overlaps at both ends were used to circularize the mt genomes. To verify the assemblies, raw paired-end sequence reads were mapped to the circularized genomes using CLC Genomics Workbench.
Mitochondrial genome annotation
The mt genomes of C. austroafricana, C. cubensis and C. deuterocubensis were annotated using the MFANNOT tool (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) with default settings. The C. parasitica mt genome was annotated via BLASTn searches using previously published C. parasitica mt genes deposited in GenBank, and completed using the MFANNOT tool. Additionally, open reading frames (ORFs) were predicted in CLC Genomics Workbench v 7.0.1 and compared to those predicted by MFANNOT. ORFs coding for hypothetical proteins were identified by BLASTP against the NCBI with a cut-off of 50% sequence identity. Introns and tRNA genes were characterized using the online program RNAweasel (http://megasun.bch.umontreal/ca/RNAweasel). ORFs encoding HEGs were identified based on their conserved domains through similarity searches in the Pfam (http://pfam.xfam.org/) and InterPro (http://www.ebi.ac.uk/interpro/) protein domain databases, as well as the NCBI (BLASTP) [46].
Sequence and phylogenetic analysis
Gene order among the four mt genomes was determined by comparing pairwise BLASTN results using genoPlotR [50]. Further, similarity information was obtained from pairwise sequence alignments using MAFFT. Codon usage was analysed using the web-based Sequence Manipulation Suite (http://www.bioinformatics.org/sms2/codon_usage.html) with the fungal mt genetic code 4. To identify the presence of direct sequence repeats and inverted sequence repeats in intergenic regions of the mt genomes, REPFIND (http://zlab.bu.edu/repfind/) and EMBOSS [51] were used respectively.
All phylogenetic analyses in this study were performed using maximum likelihood (ML) method implemented in RAxML [52]. Sequence alignments were generated from MAFFT were trimmed of poorly aligned regions using trimAL [53]. The best models of evolution were selected using jModelTest v 2.1.5 [54] for nucleotide sequences and ProtTest version 3.4 [55] for amino acid sequences. To determine the confidence of the recovered nodes, bootstrap analysis was performed with 1000 resampling steps.
Results
Mitochondrial genome structure and organization
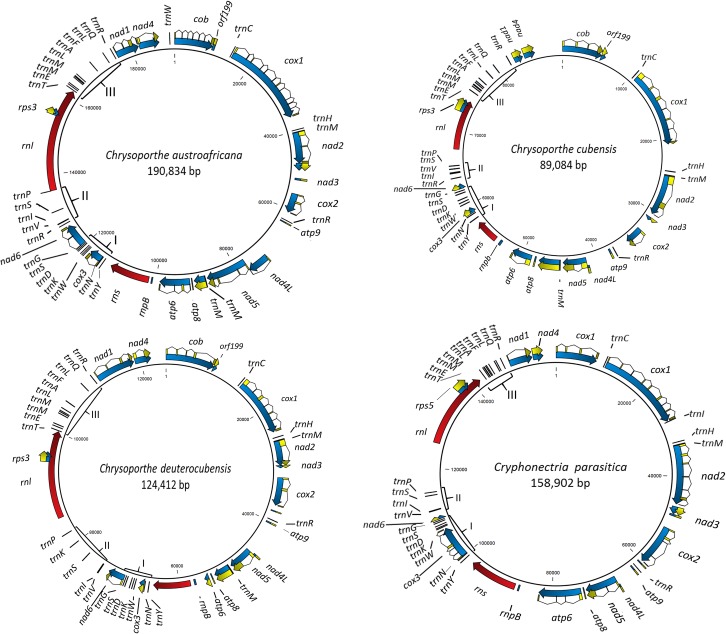
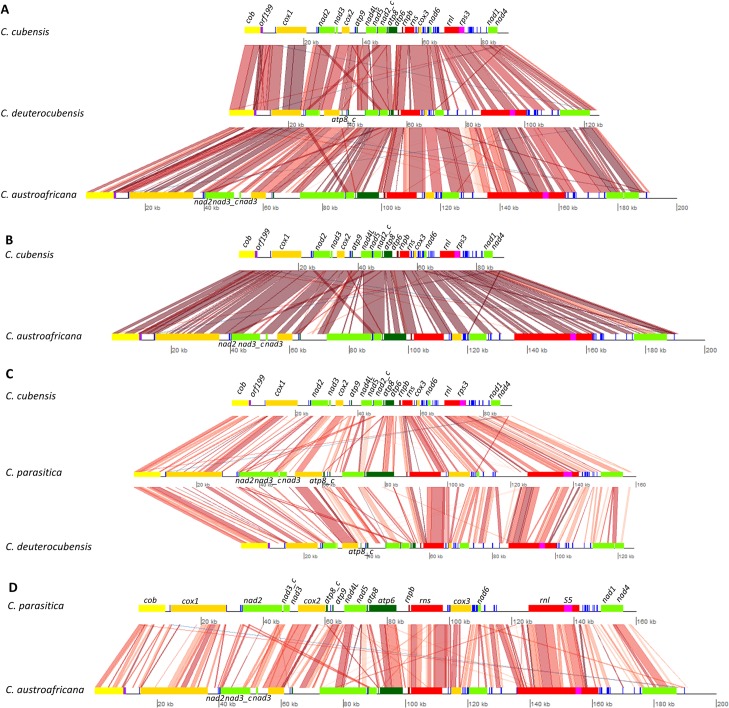
The mt genomes of C. austroafricana (GenBank accession no. KT380883), C. cubensis (GenBank accession no. KT380885), C. deuterocubensis (GenBank accession no. KT380884) and C. parasitica (Genbank accession no. KT428651) are circular mtDNA molecules of 190,834 bp, 89,084 bp, 124,412 bp, and 158,902 bp in size, respectively (Fig 1). The C. parasitica mt genome had the highest G+C content of 32.04%, followed by C. austroafricana with 30.25%, 29.98% for C. deuterocubensis, and 27.91% for C. cubensis. Pairwise whole mt genome sequence alignments revealed that C. austroafricana shared 41%, 46% and 55% sequence identity with C. parasitica, C. cubensis and C. deuterocubensis, respectively. The C. cubensis mt genome respectively shared 56% and 32% sequence identity with C. deuterocubensis and C. parasitica, while C. deuterocubensis and C. parasitica shared 35% sequence identity. Pairwise BLAST comparisons of these mt genomes identified conserved and syntenic blocks of sequences especially between OXPHOS genes (Fig 2). No sequence repeats of notable sizes were identified in any of the four mt genomes.
Fig 1. Physical maps of the mitochondrial (mt) genomes of Chrysoporthe austroafricana, C. cubensis, C. deuterocubensis and C. parasitica mitochondrial (mt) genomes.
The 14 genes involved in oxidative phosphorylation and electron transport are shown. Blue arrowed lines indicate full length genes and direction of transcription. Yellow blocks connected by lines indicate the coding sequences (CS), red arrowed lines show the large and small subunit ribosomal RNAs, and black lines indicate transfer RNA (tRNA) genes. Curly brackets labelled I, II and III indicate the major tRNA clusters.
Fig 2. Mitochondrial genome synteny maps.
A. Synteny comparisons between C. cubensis, C. deuterocubensis and C. austroafricana. B. Synteny comparison between C. cubensis and C. austroafricana. C. Comparisons between C. cubensis, C. parasitica and C. deuterocubensis. D. Comparison between C. parasitica and C. austroafricana. The Blast2Seq feature in NCBI was used to identify syntenic blocks using BLASTN. Alignments of length of greater than 100 bp and e-value > = 0.00005 are shown. Yellow boxes represent cox genes, gold; cob, lawn green; nad genes, green; atp synthase genes, blue; tRNAs, red; ribosomal RNAs and purple; rps3. Synteny graphs were generated using GenoPlotR.
All fourteen expected mt protein coding genes (atp6, atp8, atp9, cox1, cox2, cox3, cob, nad1, nad2, nad3, nad4, nad4L, nad5 and nad6) could be identified. Additionally, the small subunit RNA (rns), large subunit RNA (rnl) and RNA subunit of mt RNase P (rnpB) genes were identified in all four species (Fig 1). BLAST searches for other mitochondrial rnpb genes in GenBank database did not yield many hits however, six rnpb genes were identified by manually searching for annotated rnpb genes in ascomycete mt genomes deposited in GenBank. These genes were located in similar positions in the mt genomes where present (S1 Fig). ORFs homologous to rps3 gene, orf540, orf551 and orf551 were identified in mt genomes of C. austroafricana, C. cubensis and C. deuterocubensis, respectively. These ORFs encoded a protein that shared 88% amino acid sequence identity with the putative S5 ribosomal protein/maturase fusion protein identified in the C. parasitica mt genome [56]. However, from the conserved domain and BLAST analysis, these ORFs harbour a HEG, a feature that was observed in rps3 genes of other fungi (S2 Fig). Phylogenetic analysis grouped orf540, orf551 and orf551 with rps3/HEG fusion proteins (S2 Fig and S1 Table). Expression analysis based on available RNA-Seq data from C. austroafricana revealed that all these genes were actively expressed in minimal and complete media growth conditions (S3 Fig and Mangwanda et al., submitted).
All of the conserved mt genes were encoded on a single strand and were in the same direction and order (Fig 1). Although gene order was conserved among these four mt genomes, variations could be observed when compared to mt genomes of other filamentous fungi. However, there was conservation in the order of particular genes that are known to occur adjacent to each other, such as nad2-nad3, atp6-atp8, cob-tRNA-Cys-cox1, nad4L-nad5 and nad1-nad4 among Sordariomycetes [15, 22]. One notable difference was observed in the C. austroafricana mt genome, where the nad4L gene had an intron. This intron contained two ORFs, both encoding for HEGs with LAGLIDADG domains. Overall, the genes encoded in these mt genomes showed high levels of synteny (Fig 2).
A total of 28, 27, 27 and 26 tRNA genes (Table 1) coding for all amino acids were predicted in the mt genomes of C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica, respectively. All Chrysoporthe species had four tRNA-Met genes compared to three in C. parasitica. The tRNA genes in the four mt genomes were clustered, as is the case in most mt genomes of Pezizomycotina [15]. Three major clusters of tRNA genes were identified. The first cluster was between the rns and nad6 genes and contained genes encoding tRNA-Try, tRNA-Asn, tRNA-Trp, tRNA-Lys, tRNA-Asp, tRNA-Ser and tRNA-Gly (Fig 1). Cluster 2, which included genes for tRNA-Arg, tRNA-Val, tRNA-Ile, tRNA-Ser, and tRNA-Pro, was located between the nad6 and rnl genes (Fig 1). This cluster in the C. deuterocubensis mt genome did not have the tRNA-Arg gene, but instead, a tRNA-Lys gene was inserted between the tRNA-Ser and tRNA-Pro genes. Likewise, the tRNA-Arg gene was absent in the second tRNA cluster of four tRNA genes of the C. parasitica mt genome. The third cluster was the largest of the three, and had 10 tRNA genes located between the rnl and nad1 (Fig 1). These were the tRNA-Thr, tRNA-Glu, tRNA-Met, tRNA-Met, tRNA-Leu, tRNA-Ala, tRNA-Phe, tRNA-Leu, tRNA-Gln and tRNA-Arg genes. Only one tRNA-Ile gene was present in all three Chrysoporthe species, while two non-adjacent copies of this gene were observed in the C. parasitica mt genome (Fig 1 and S2 Table). Two tRNA-Gly genes were predicted in C. austroafricana, while only one was predicted in the other mt genomes. Likewise, two tRNA-Lys genes were predicted in the C. deuterocubensis mt genome, compared to one in the other mt genomes (Fig 1 and S2 Table).
Table 1. Mitochondrial genome features.
Number of genes, open reading frames (ORFs), rRNA, tRNAs and introns of the mitochondrial genomes of Chrysoporthe austroafricana, C. cubensis, C. deuterocubensis and Cryphonectria parasitica. Intergenic ORFs were considered uORFs if no HEG domains or significant BLAST hits could be identified.
| Species | Intergenic ORFs | Intronic ORFs | uORFs | rRNAs | tRNAs | GrpI/II Introns |
|---|---|---|---|---|---|---|
| C. austroafricana | 21 | 42 | 8 | 2 | 28 | 32/3 |
| C. cubensis | 7 | 11 | 5 | 2 | 27 | 14/0 |
| C. deuterocubensis | 17 | 20 | 10 | 2 | 27 | 19/0 |
| C. parasitica | 14 | 34 | 6 | 2 | 26 | 27/5 |
Codon usage
The codon usage frequencies for all the conserved protein coding mitochondrial genes were calculated from the respective CS (S2 Table). The start codon ATG was favoured by most CS across all four species. GTG was favoured as the start codon for nad4 in all four species, while cox1 favoured TTG. In the nad6 genes, C. parasitica had TTG as the start codon, in contrast to ATG in the Chrysoporthe species. The most frequently used stop codon was TAA, apart from cox1 and nad1 in all species, which had TAG as the stop codon. The TAG stop codon was also observed in atp9, nad5 and nad4 CS of C. austroafricana, C. deuterocubensis and C. parasitica, respectively. Among the intronic ORFs encoding proteins the preferred start codon was ATG, while TAA was the most frequent stop codon. Similarly, ATG was the start codon for all the intergenic ORFs encoding proteins in all species while TAA was the stop codon. Nonetheless, a few ORFs (4, 3, 2 and 7) had TAG as the stop codon C. austroafricana, C. deuterocubensis and C. parasitica, respectively. The most frequently used codons in the OXPHOS genes were those that code for hydrophobic amino acids, including Leu (TTA), Ile (ATA) and Phe (TTT), accounting for approximately 10%, 6% and 5% of all codons in all four species (S2 Table). The codon usage of intergenic ORFs, intronic ORFs, and duplicated genes did not differ from that of the conserved mt genes (data not shown).
The C. austroafricana and C. cubensis mt genomes shared three tRNA-Arg genes with similar anti-codons (TCT, TCT and ACG), unlike in C. deuterocubensis and C. parasitica where only two tRNA-Arg genes, with anticodons TCT and ACG, were observed. Most anticodons in the tRNA genes had a T or A nucleotide at the third position. Some amino acid codons in the mt genomes lacked corresponding tRNA genes (S2 Table).
Gene duplication
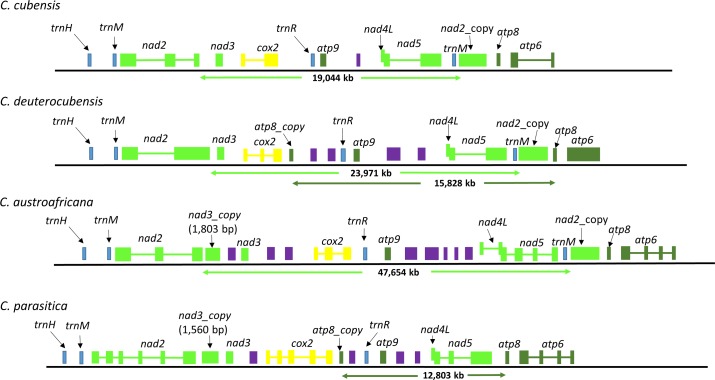
The mt genomes of the three Chrysoporthe species contained some genomic regions that were duplicated (Fig 3). One of these duplicated regions resulted in an ORF that appears to be partially, homologous to the nad2 gene. This ORF, located between nad5 and atp8, was 2,718 bp in size in all three mt genomes and shared between 97–99% pairwise DNA sequence identities between the three species. Both nad2 and the ORF with partially duplicated nad2 sequences were flanked by a stretch of homologous sequences including a tRNA-M gene in the upstream direction (Fig 3). This stretch was 154 bp, 153 bp, 150 bp and 151 bp, 153 bp, and 163 bp in C. austroafricana, C. cubensis and C. deuterocubensis respectively. The region between nad2 and this ORF was 19,044 bp, 23,971 bp and 47,654 bp in C. cubensis, C. deuterocubensis and C. cubensis respectively (Fig 3). Sequence alignment of nad2 gene and the duplicated region revealed up to 96% sequence identity over a region spanning 814 bp, 816 bp and 816 bp covering part exon1 of nad2 gene in C. austroafricana, C. cubensis and C. deuterocubensis, respectively.
Fig 3. Organization of genes around the nad2, nad3 and atp8 genes showing partially duplicated sequences.
Dark green, light green, olive green, red, yellow, light blue and purple boxes represent the nad2, nad3 and nad5, cox2, atp9, tRNAs genes and various open reading frames (ORFs), respectively. The duplicate genes are denoted as nad2_copy, atp8_copy and nad3_copy.
Two ORFs of were annotated as atp8 genes by MFANNOT in the C. deuterocubensis and C. parasitica mt genomes but only one copy in C. cubensis and C austroafricana. In the mt genomes of ascomycetes, atp8 gene is usually adjacent to the atp6 gene[15]. The additional ORFs located between the cox2 and atp9 genes of length 159 bp and 219 in C. deuterocubensis and C. parasitica respectively (Fig 3 and S4 Fig). The endogenous atp8 genes which were 165 bp and 168 bp long shared 93% and 100% sequence identity over a stretch of 130 bp and 147 bp respectively in C. deuterocubensis and C. parasitica. BLASTN comparison of these duplicate ORFs revealed 83% sequence identity over a 146 bp alignment. Analysis of sequences flanking both copies of the duplicated genes did not identify homologous sequences. The distance between the duplicate ORFs and atp8 gene was 15,828 bp in C. deuterocubensis and 12,803 bp C. parasitica.
ORFs of 1,803 bp and 1,560 bp with sequences partially similar to the nad3 genes C. austroafricana and C. parasitica were identified (Fig 3 and S4 Fig). Comparative nucleotide sequence analysis of these ORFs to the respective nad3 genes revealed a homologous region of 368 bp and 160 bp in the 5’ region both sharing 82% sequence identity in C. austroafricana and C. parasitica respectively. The 3’ region of both ORFs had sequences coding for GIY-YIG endonuclease domain. Pairwise comparison of the amino acid sequence product of these ORFs that were homologous to nad3 in C. austroafricana and C. parasitica, showed 48% sequence identity. 2,061 bp separated the ORFs with partially duplicated nad3 sequences and the endogenous nad3 genes was and 98 bp in C. austroafricana and C. parasitica respectively.
Introns and intron-encoded ORFs
Both group I and II introns [37] were identified in C. austroafricana and C. parasitica mt genomes, while only group I introns were identified in C. cubensis and C. deuterocubensis mt genomes (Tables 1 and 2). BLASTN analysis of introns from the four mt genomes revealed introns with sequence identities over 70% and alignment coverage over 50% (data not shown). However, only a few orthologous introns were identified. For example, intron 1 of cob in C. austroafricana and C. cubensis shared 100% sequence identity and had the same insertion point, but the same intron in the cob of C. deuterocubensis only showed 95% sequence identity with the C. austroafricana and C. cubensis introns (Fig 4). Intron 1 of the cob gene in C. parasitica shared up to 80% sequence identity with that of C. austroafricana, C. cubensis and C. deuterocubensis but had a different insertion point in (Fig 4) thus was not considered orthologous. Intron 5 of C. austroafricana cob had the same insertion point in C. cubensis (intron 3), C. deuterocubensis (intron 3) and C. parasitica (intron 2). Overall, only 17 introns could be considered orthologous based on sequence identity and insertion point in the exonic sequence. Only three introns located in the cox1, cob and cox2 genes were orthologous in all the four mt genomes analysed. Orthologous introns were mostly identified in the mt genomes of the closely related Chrysoporthe species compared to distantly related C. parasitica. BLAST analysis of the identified introns against NCBI GenBank database revealed sequence identities with sequences from other fungal mt genomes (S3 Table).
Table 2. Diversity of mitochondrial introns and intron encoded HEGs in the mt genomes of C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica.
Lower case a group stands for group 1A introns, b group 1B, c1 group 1C1, c2 group 1C2, d group 1D, dr group 1-derived introns. Lower case i stands for intron and the numbers indicate intron positions in the respective mitochondrial gene. Superscript letters l, g and rt stand for intron encoded LAGLIDADG, GIY-YIG type HEGs and reverse transcriptase encoding ORFs respectively.
| Intron type | site | C. austroafricana | C. cubensis | C. deuterocubensis | C. parasitica |
|---|---|---|---|---|---|
| Group1 | atp6 | i1c2ll, i2c2g, i3gg | i1g | i2c2g, i3c2gg | |
| cox1 | i1bll,i2bl,i3c2ll,i4bl,i5dgl,i6bl,i7bg,i8drll,i9bl,i10bg,i11bg | i1c2l,i2dl,i3bg, i4drl,i5bg,i6g | i1c2l,i2bl,i3bg,i4drl,i5bg,i6bg | i2bl,i4bg,i5gl,i6bg,i7drl,i8bg | |
| cox2 | i1bg, i2c1g | i1bg | i1g, i2c1g | i1bl, i2c1, i3l, i4c1g | |
| cox3 | i1bll | i1, i2al, i3c2,i4c1 g | |||
| cob | i1drl, i4dg, i5al, 6bl | i1drl, i2dg, i3al | i1drl, i2g, i3al, i4bgg | i1drl, i2al, i3bgg | |
| nad1 | i1bg, i2l, i3 | i1gb, i2ll, i3b, | i1c1 | ||
| nad2 | i1c2, i2c2g | i1c2, i2c2 | i1c2 | i1c2l, i2l, i3c2l, i5c2l, i6 | |
| nad4 | i1c2l | i1c2l | |||
| nad4L | i1c2ll | ||||
| nad5 | i1bl, i2lg,i3l | i1bl | i1dl | i1l, i2c2ll | |
| nad6 | i2l | i1l | |||
| Group 2 | cox1 | i1rt, i3rt | |||
| cob | i2, i3l | ||||
| nad2 | i4rt | ||||
| nad5 | i2 | ||||
| nad6 | i1rt | ||||
| atp6 | i1rt |
Fig 4. Conserved intron insertion position and intron type identified within the atp6, cox1, cob, nad1, nad2, cox2, nad6, and nad5 of C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica.
Blue lines with arrow heads represent genes compared. White vertical breaks within the blue lines represent intron position with the insertion position is shown below it. Intron type is indicated by letters in gold boxes. Amino acid residues bordering the exonic sequences are shown in red single letter amino acid notation. A;group 1A introns, b; group 1B, c1; group 1C1, c2; group 1C2, d; group 1D, dr; group I-derived.
Group I introns in the mt genomes of C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica contained 27, 6, 10 and 15 ORFs that encoded HEGs of the LAGLIDADG family, while 13, 6, 9 and 11 introns contained ORFs encoding for GIY-YIG type HEGs, respectively. Based on expression analysis of C. austroafricana RNA-Seq data (Mangwanda et al. submitted), these ORFs seem to be expressed (data not shown). 15, 6, 9 and 17 of these intron encoded ORFs in C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica respectively were in frame and did not have typical start codons. In the mt genomes of C. austroafricana, C. deuterocubensis and C. parasitica, 8, 2 and 4 introns respectively had two ORFs that encoded either LAGLIDADG or GIY-YIG type HEGs, or both ORFs encoded the same type of HEG (Table 2). Comparative analysis of these intron encoded HEGs in each species revealed sequence identities below 45% among themselves (data not shown). However, results from comparative analysis of these HEGs between the four species revealed homologous sequences with identities of up to 80% (S4 Table). Comparative BLAST analysis of the amino acid sequences of these HEGs against sequences deposited in the NCBI GenBank database revealed sequence identities of up to over 70% and alignment coverage of at least 50% (S5 Table).
Group II introns were not identified in the mt genomes of C. cubensis and C. deuterocubensis, however, three and five group II introns were identified in the mt genomes of C. austroafricana and C. parasitica (Tables 1 and 2). Of these group II introns only one intron in C. austroafricana mt genome had the typical reverse transcriptase encoding ORF commonly identified in this group of introns [37, 57]. In contrast four out of five group II introns in C. parasitica harboured an ORF encoding a reverse transcriptase protein (Table 2).
Intergenic and unique ORFs
Varying numbers of intergenic ORFs were predicted in the four fungal mt genomes: 21, 7, 17 and 14 free standing ORFs were detected from C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica, respectively. In the C. austroafricana mt genome, 13 of the 21 ORFs were identified as HEGs by means of their conserved GIY-YIG and LAGLIDADG domains. Two out of the seven intergenic ORFs predicted in C. cubensis were GIY-YIG and LAGLIDADG HEGs. Of the 17 and 14 intergenic ORFs in C. deuterocubensis and C. parasitica, respectively, three ORFs were GIY-YIG type HEGs in both mt genomes, while ORFs encoding LAGLIDADG type HEGs were four and five, respectively. BLAST analysis revealed that most of these ORFs could have been acquired independently due to lack of identities between the mt genomes. However, some intergenic ORFs encoding HEGs were shared. For example, orf368 of C. austroafricana encoding a LAGLIDADG type HEG shared 99% and 94% sequence identity with orf383 of C. deuterocubensis and orf427 of C. parasitica, respectively (S4 Table). In each mt genome, intergenic HEG sequences were compared with intron encoded HEGs to determine if there was any signature of recent movement within the genome. Results from the BLAST analysis revealed sequence identities below 50%. However, BLAST analysis of predicted proteins sequences from HEGs against mt genomes deposited in GenBank revealed hits of up to over 90% sequence identities (S5 Table).
In the mt genomes of C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica, 8, 4, 12 and 12 unique ORFs were identified, respectively. These ORFs had translation start sites similar to other mt genes and translated to amino acid sequences longer than 100 residues. Comparative amino acid sequence analysis of these ORFs identified one ORF that was shared across all Chrysoporthe species, but was not found in the C. parasitica mt genome. This ORF (orf199) was located immediately after the cob gene in all three mt genomes and was 600 bp in size. Sequence alignment of these three ORFs revealed sequence identities of up to 99%. BLASTP searches of this unique ORF in the NCBI protein database did not reveal sequence alignments with high sequence coverage or identity. Searches for functional domains in Pfam and InterPro protein databases were also unsuccessful for this ORFs.
Comparative analysis of mt genome size variation
Size polymorphism is a prominent feature of fungal mt genomes. Several factors have been associated with size variations including number and size invading introns and expanding intergenic regions. The mt genomes analysed in this study exhibited great size variation with C. austroafricana being the largest at 190,834 bp and C. cubensis the smallest at 89,084 bp. The mt genomes with more introns for example 35 and 32 in C. austroafricana and C. parasitica respectively had larger mt genomes compared to C. cubensis and C. deuterocubensis which had far fewer 14 and 19 (Table 1). In C. austroafricana, eleven of these were located in the cox1 gene, contributing to its large size (21,506 bp). Similarly, the C. cubensis, C. deuterocubensis and C. parasitica cox1 gene had 6, 6 and 8 introns, respectively, and this was the largest gene in all of these mt genomes (Table 3). The effect of introns on the gene sizes is evident from the mt genomes analysed (Table 1 and Table 3). Among the Chrysoporthe species and C. parasitica mt genomes, the longest intron (3,477 bp) was located in the C. austroafricana mt genome (intron 2 of the nad2 gene), while the shortest intron was 492 bp located in the cox3 gene of C. parasitica mt genome.
Table 3. Comparison of gene size and number of introns.
Comparisons were performed for the 14 mitochondrial genes of Chrysoporthe austroafricana, C. cubensis, C. deuterocubensis and Cryphonectria parasitica involved in oxidative phosphorylation and electron transport. Refer to Fig 1 and S4 Fig for physical maps of these genomes.
| C. austroafricana | C. cubensis | C. deuterocubensis | C. parasitica | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Size• | Introns | CDS | Size• | Introns | CDS | Size• | Introns | CDS | Size• | Introns | CDS |
| atp6 | 7,299 | 3 | 795 | 2,657 | 1 | 795 | 795 | - | 795 | 8602 | 3 | 795 |
| atp8 | 165 | - | 165 | 165 | - | 165 | 165 | - | 165 | 168 | - | 168 |
| atp9 | 225 | - | 225 | 225 | - | 225 | 225 | - | 225 | 252 | - | 252* |
| cox1 | 21,506 | 11 | 1,695 | 9,950 | 6 | 1,695 | 9,946 | 6 | 1,695 | 17,582 | 8 | 1,695 |
| cox2 | 4,881 | 3 | 753 | 2,319 | 1 | 753 | 4,867 | 2 | 753 | 8,574 | 4 | 753 |
| cox3 | 3,113 | 1 | 810 | 810 | - | 810 | 810 | - | 810 | 6,723 | 5 | 810 |
| cytb | 9,310 | 6 | 1,176 | 5,271 | 3 | 1,176 | 8,412 | 4 | 1,176 | 8,326 | 3 | 1,176 |
| nad1 | 5,656 | 3 | 1,182 | 1,182 | - | 1,182 | 6,815 | 3 | 1,182 | 4,738 | 1 | 1,161* |
| nad2 | 7,743 | 2 | 1,728 | 5,401 | 2 | 1,728 | 4,228 | 1 | 1,728 | 12,488 | 6 | 1,728 |
| nad3 | 429 | - | 429 | 429 | - | 429 | 429 | - | 429 | 348 | - | 348* |
| nad4 | 4,948 | 1 | 1,479 | 1,479 | - | 1,479 | 2,814 | 1 | 1,479 | 2,049 | - | 2,049† |
| nad4L | 4,945 | - | 270 | 270 | - | 270 | 270 | - | 270 | 270 | - | 270 |
| nad5 | 9,984 | 3 | 2,034* | 3,107 | 1 | 2,103* | 4,092 | 1 | 2,091* | 6,462 | 2 | 1,971* |
| nad6 | 5,684 | 2 | 681 | 681 | - | 681 | 2,867 | 1 | 699 | 666 | - | 666 |
• Gene sizes include intron sequences.
* Differences in CDS are due to the presence or absence of additional codons at the 3’ borders of genes.
† The C. parasitica nad4 gene seems to be fused to a homing endonuclease gene (HEG). Only one ORF could be predicted which included both genes.
The total size of introns, intergenic regions and coding sequences were calculated. Introns constitute 72,468 bp (37.97%), 20,458 bp (22.96%), 33,238 bp (26.72%) and 63,373 bp (39.88%) in C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica respectively (Table 4). The percentage of intergenic sequences (including all OXPHOS genes, intronic ORFs and intergenic ORFs) was 48,854 bp (25.60%), 38,350 bp (43.05%), 43,340 bp (34.84%) and 41,930 bp (26.39%) in C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica respectively. Coding sequences constituted 49.17% (93,848 bp), 38.39% (34,202), 45.45% (56,550) and 48.30%) (76,752). The length of sequences in the region covering the ribosomal RNAs was largest in C. austroafricana and smallest in C. cubensis (Table 4). To determine factors majorly contributing to size polymorphisms in the four mt genomes, the proportion of coding sequences, intergenic sequences and introns was compared using the method described in [58]. From this analysis, introns and coding sequences were the primary source of genome size variation (Table 5). This result was consistent with BLAST comparisons in Fig 2 which showed more variation in intron regions. The comparison between C. austroafricana and C. parasitica revealed that intergenic sequences contributed more compared to comparisons between C. austroafricana and C. cubensis or C. austroafricana and C. deuterocubensis. In the Chrysoporthe mt genomes, the size variation of intergenic sequences ranged between 8% and 14%.
Table 4. Total sizes of genes, coding sequences, and intergenic regions of C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica mt genomes.
The size of the intergenic region was calculated by subtracting the sum total of the 14 OXPHOS genes, tRNAs, rRNAs and rnpb from the mt genome size.
| Species | mt genome size (bp) | OXPHOS genes (including introns) | CS | Introns | Intergenic region | rRNA genes (without introns)+rnpb | tRNA genes |
|---|---|---|---|---|---|---|---|
| C. austroafricana | 190,834 | 85,888 (44.34%) | 93,848 (49.17%) | 72,468 (37.97%) | 48,854 (25.60%) | 36,228 (18.98%) | 2,083 (1.09%) |
| C. cubensis | 89,084 | 33,946 (38.11%) | 34,202 (38.39%) | 20,458 (22.96%) | 38,350 (43.05%) | 9,990 (11.21%) | 2,010 (2.26%) |
| C. deuterocubensis | 124,412 | 46,735 (37.56%) | 56,550 (45.45%) | 33,238 (26.72%) | 43,340 (34.84%) | 21,957 (17.65%) | 2,018 (1.62%) |
| C. parasitica | 158,902 | 77,248 (48.61%) | 76,752 (48.30%) | 63,373 (39.88%) | 41,930 (26.39%) | 26,345 (16.58%) | 1,937 (1.22%) |
Table 5. Comparison of factors contributing to genome size polymorphism.
Pairwise comparisons were calculated for introns, intergenic regions (IR) and coding sequences (CS).
| Species | C. austroafricana (CA) | C. cubensis (CC) | C. deuterocubensis (CD) | C. parasitica (CP) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Introns | IR | CS | Introns | IR | CS | Introns | IR | CS | Introns | IR | CS | |
| CA | 51% | 10% | 58% | 59% | 8% | 56% | 24% | 22% | 53% | |||
| CC | 51% | 10% | 58% | 36% | 14% | 63% | 61% | 5% | 60% | |||
| CD | 59% | 8% | 56% | 36% | 14% | 63% | 87% | 4% | 58% | |||
| CP | 24% | 22% | 53% | 61% | 5% | 60% | 87% | 4% | 58% | |||
Discussion
In this study the mt genome sequences of C. austroafricana, C. cubensis and C. deuterocubensis were determined. This is the first report of complete mt genome sequences in the genus Chrysoporthe. The sizes of these genomes were within the range of other fungal mt genomes [32, 33]. The diversity of mt genome size was evident in the mt genome sequences of C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica, which were 190,834 bp, 89 084 bp, 124 412 bp, and 158 902 bp in size, respectively. Notably, compared to all mt genomes in the NCBI organelle database, the C. austroafricana mt genome is the second largest fungal mt genome to date after that of Sclerotinia borealis [33]. The mt genomes of C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica seem plastic, containing only 40%, 34%, 35% and 42% protein CS, respectively. Large numbers of introns contributed up to 28% of the total genome size, while the remaining portion of noncoding sequence was comprised of highly variable intergenic regions.
Chrysoporthe austroafricana, C. cubensis and C. deuterocubensis are closely related species [1, 8, 9]. However, their mt genomes showed extensive size differences. Fungal mt genome size differences are largely attributed to the number and size of introns and the lengths of intergenic sequences, which consequently lead to variations associated with mt genome diversity [37, 59, 60]. In the current study, variations in mt genome size was consistent with other studies that attributed expansions of fungal mt genomes to intron sequences [25, 33, 35, 42, 61, 62]. The extent to which the intron sizes and numbers influence mt genome size variation is evident in the cox1 gene of C. austroafricana, which was the largest reservoir of 11 introns in all four mt genomes. Comparing the cox1 gene of C. austroafricana to those of C. cubensis, C. deuterocubensis and C. parasitica, the C. austroafricana cox1 gene was more than double in size due to additional introns. This observation was not limited to cox1 genes, but to all other mt genes harbouring these introns. Long stretches of intergenic sequences also seemed to influence the mt genome sizes of these species. In other fungi, these regions harbour ORFs that have been associated with genome size variations [33].
The mt genome sequences of C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica have highly similar gene content. These genomes encode fourteen genes associated with oxidative phosphorylation and electron transport in fungi. Also, genes encoding the rns, rnl, and tRNA genes, were contained in these mt genome sequences. This was consistent with previously described mt genome sequences [20, 25]. The mt genomes investigated in the current study all had an rnpB gene that was located in a similar location, immediately before the small subunit ribosomal RNA. BLAST searches for this gene in GenBank did not yield many hits which could be either because it was not identified in the fungal mt genomes. ORFs identified in the Chrysoporthe species mt genomes analysed contained a putative rps5/maturase fusion protein similar to that of previously identified in C. parasitica. Phylogenetic analysis however showed that these rps5/maturase like ORFs grouped together with rps3/HEG fusion proteins. The rps3 gene is commonly identified in other Sordariomycetes [22, 24, 42].
Gene order in the four mt genomes investigated in the current study was highly conserved, despite the considerable size differences. This was in agreement with previous observations that mt genome size variations do not affect gene order [15]. When compared to mt genomes of other Sordariomycetes, a comparable level of synteny was observed, especially with regard to genes known to occur in pairs, such as nad4L-nad5, cox3-nad6, nad1-nad4, nad2-nad3, atp8-atp6 and cob-cox1 [15, 22]. A high level of gene order conservation was also observed for tRNA genes which were clustered the same as in other Sordariomycetes [12, 15, 42]. The organisation of tRNA genes was highly conserved, with differences emerging only due to intervening ORFs. The higher number of tRNAs in C. austroafricana mt genome was due to duplicated tRNAs that were absent in the mt genomes of C. cubensis, C. deuterocubensis and C. parasitica.
Results of this study revealed duplications of some mt genome regions. One such duplication was a 2,718 bp ORF with sequences homologous to the nad2 gene in the mt genomes of C. austroafricana, C. cubensis and C. deuterocubensis. This duplication could not be identified in the C. parasitica mt genome, suggesting that it could have occured after the divergence of Chrysoporthe and Cryphonectria. The high sequence identity (between 97–99%) of this duplication in all three genomes of C. austroafricana, C. cubensis and C. deuterocubensis, combined with the high level of synteny around the duplicated gene region, suggests a single duplication event in the most recent common ancestor, which was then transmitted vertically when these species diverged. Notably, the sequences flanking the 5’ region of this ORF included a duplicate tRNA-M gene, which also flanks the 5’ region of the nad2 gene. The occurrence of these duplications in regions around tRNAs is in agreement with studies that have associated tRNA genes with editing, excision and integration capabilities in mt genomes and are also linked to horizontal gene transfer events [63].
An ORF with partially duplicated sequences of atp8 gene was predicted in the mt genome of C. deuterocubensis and C. parasitica between cox2 and atp9. The absence of this duplication in C. cubensis and C. austroafricana suggests independent evolutionary processes involving either gain or loss after the divergence of these species. The ORFs homologous to nad3 in C. austroafricana and C. parasitica but absent in C. cubensis and C. deuterocubensis could be due to subsequent loss after these species diverged. The presence of these duplications is consistent with what has been reported for Sclerotinia borealis [33] where these duplications have been associated with genome expansion and in Fusarium species [24] contributing to mt genome diversity. The presence of duplicated genes in these mt genomes is evidence of independent evolutionary events that may have happened after the divergence of these species. No functional or phenotypic adaptations have thus far been suggested for such duplications.
Fungal mt introns show extensive diversity even among species in the same genus [19, 24, 25]. Group I introns in the four Cryphonectriaceae mt genomes showed known diversity of mitochondrial introns [31, 37, 39, 62]. A high level of diversity was also observed in the few group II introns identified in the mt genomes of C. austroafricana and C. parasitica. Intron content showed remarkable diversity especially within the cox1, cob, nad1, nad2 and nad5 genes. Differences in intron content can occur in species and even subspecies groups [24, 25, 42, 61, 64, 65]. Thus, the diverse range of introns and intron content in the mt genomes of Cryphonectriaceae seems to be a common feature with potential influence in mt genome size variations.
In fungal mt genomes, intron acquisition can occur via both vertical and horizontal transmission, and appears to occur at homologous gene positions [39, 66, 67]. A high level of sequence similarity was observed between the group I introns of the three Chrysoporthe mt genomes, some of which encoded similar HEGs and had similar insertion points. Introns that lacked sequence similarities could be indicative of independent evolutionary histories involving multiple acquisitions [33, 36]. The few group II introns identified in these mt genomes did not share sequence similarities, but similar sequences could be identified from other fungal species. This suggests that group II introns were acquired by horizontal transfer. The few intron sequences from the Chrysoporthe species mt genomes homologous to those of the C. parasitica mt genome was consistent with the notion that introns are either fixed or lost after divergence events [24, 68].
Fungal mt introns contain ORFs that encode HEGs of the LAGLIDADG or GIY-YIG families [38, 39, 69]. In frame ORFs encoding HEGs, did not have start codons a feature that is common in fungal mt genomes. These ORFs are also found free standing in genomic regions other than those related to OXPHOS [33]. Consistent with other studies [24, 61], results of this study revealed widespread diversity of these genes in Cryphonectriaceae mt genomes. This, coupled with the low sequence identities (< 45%) of HEGs within each mt genome seems to rule out intragenomic proliferation, indicating that these genes could have diverse origins. Similarly, low sequence identities were observed between intron encoded and free standing HEGs within and between these mt genomes. Therefore, acquisition of these genes seems to have independent evolutionary origins which was then followed by the accumulation of mutations [67, 70].
In other fungal mt genomes, unique ORFs (uORFs) have been identified and are implicated in genome size expansion [24, 42]. One uORF was shared among the three mt genomes of Chrysoporthe spp. This uORF (orf199) was 600 bp long in all three mt genomes, and was located immediately after the cob gene. Comparative analysis using both amino acid and nucleotide sequences could not reveal significant BLAST matches, thus the origin and function of this uORF could not be identified. However, this uORF seems to have been transmitted vertically among the Chrysoporthe spp. from the most recent common ancestor and could potentially be used as a marker for this lineage.
Analysis of factors hugely contributing to size polymorphism in the mt genomes of C. austroafricana, C. cubensis, C. deuterocubensis and C. parasitica revealed introns and coding sequences as the primary source of genome size variation. Coding sequences included sequences from the 14 OXPHOS genes, intronic ORFs and free standing ORFs. Intergenic sequences seemed to contribute more in the size variation between C. austroafricana and C. parasitica. Also, introns did not seem to contribute much between these two mt genomes which could be attributed to the high number of introns whose size difference was the least compared to C. cubensis and C. deuterocubensis. Generally, the size of the mt genomes seemed to drastically increase with increasing number of introns and coding sequences.
Conclusions
In this study we were able to determine the mt genome sequences of C. austroafricana, C. cubensis, and C. deuterocubensis. The C. austroafricana genome was the second largest fungal mt genome to date, and the large size could be attributed to the numbers and sizes of invading introns varying size of coding sequences and intergenic regions. Despite the size polymorphism, results from comparative analyses of these genomes indicated a conserved gene order and direction of transcription. From all analysed introns, only a few introns were orthologous, while the rest seem to have been acquired independently. The general lack of high sequence identity between proteins sequences of HEGs and uORFs identified in this study but high sequence identity when compared to protein sequences in NCBI GenBank suggest possible acquisition via horizontal transfer. However, further evolutionary analysis is required for these genes. These genomes present invaluable resources for future studies focusing on the evolution and population biology of Chrysoporthe species.
Supporting Information
Rnpb genes were mannualy retrieved from annotated fungal mt genomes publicly available in GenBank. Phylogenetic analysis was performed using maximum likelihood method implemented in RAxML with HKY model of selection. Branch support was calculated using 1000 bootstrap replicates. Green boxes depict atp6 gene, red; small sub-unit of ribosomal RNA (rns) and dark red; small sub-unit of ribosomal RNA (rnl).
(PDF)
Phylogenetic analysis of orf540, orf551 and orf551 from C. austroafricana, C. cubensis and C. deuterocubensis which show sequence similarity to C. parasitica S5 ribosomal protein/maturase fusion protein. Sequences used in this phylogeny were retrieved from GenBank using BLAST. The LG+G model of substitution was used. Branch support for was calculated using the bootstrap method with 1000 replicates.
(PDF)
The graph shows the average expression for genes that are involved in oxidative phosphorylation and electron transport and the rnpb gene of Chrysoporthe austroafricana grown in complete and minimal media.
(PDF)
Intronic ORFs are depicted by yellow arrowed boxes on introns of genes where found and are labelled according to the gene and intron position. Intergenic ORFs are also depicted by yellow arrowed boxes and are labelled with “IN” prefix.
(PDF)
(PDF)
Comparison of codon usage and tRNAs for the 14 genes involved in oxidative phosphorylation and electron transport in the mitochondrial genomes of Chrysoporthe austroafricana, C. cubensis, C. deuterocubensis and Cryphonectria parasitica.
(PDF)
The best hits for each query (intron) is shown. Default BLAST parameters were used. The prefix CA, CC, CD and CP is added to the intron names for clarity.
(PDF)
Results from all possible comparisons were calculated. Only Blast hits with a threshold of over 50% alignment coverage, 50% sequence identity and E-value of 0.00005 are shown.
(PDF)
HEG type were annotated using the NCBI Conserved Domain Database (CDD).
(PDF)
Acknowledgments
This work was co-funded by the Genomics Research Institute (GRI) University of Pretoria, the University of Pretoria Research Development Programme, the DST/NRF Centre of Excellence in Tree Health Biotechnology (FABI, University of Pretoria), and the National Research Foundation (NRF) (Grant number 87332).
Data Availability
The mitochondrial genome sequences of Chrysoporthe austroafricana, C. cubensis, C. deuterocubensis and Cryphonectria parasitica are available at NCBI GenBank accession numbers KT380883, KT380885, KT380884, and KT428651, respectively.
Funding Statement
This work was co-funded by the Genomics Research Institute (GRI) University of Pretoria, the University of Pretoria Research Development Programme, the DST/NRF Centre of Excellence in Tree Health Biotechnology (FABI, University of Pretoria), and the National Research Foundation (NRF) (grant number 87332). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gryzenhout M, Myburg H, van der Merwe NA, Wingfield BD, Wingfield MJ. Chrysoporthe, a new genus to accommodate Cryphonectria cubensis. Stud. Mycol. 2004;50:119–42. [Google Scholar]
- 2.Gryzenhout M, Myburg H, Wingfield BD, Wingfield MJ. Cryphonectriaceae (Diaporthales), a new family including Cryphonectria, Chrysoporthe, Endothia and allied genera. Mycologia. 2006;98:239–49. [DOI] [PubMed] [Google Scholar]
- 3.Wingfield MJ, Swart WJ, Abear BJ. First record of Cryphonectria canker of Eucalyptus in South Africa. Phytophylactica. 1989;20:311–3. [Google Scholar]
- 4.Hodges C, Reis M, Ferreira F, Henfling J. O cancro do Eucalipto causado por Diaporthe cubensis [Eucalyptus spp.; Fungo; Brasil]. Fitopatologia Brasileira. 1976;1:129–70. [Google Scholar]
- 5.Hodges CS, Geary TF, Cordell CE. The occurrence of Diaporthe cubensis on Eucalyptus in Florida, Hawaii, and Puerto Rico. Plant Dise. Rep. 1979;63:216–20 [Google Scholar]
- 6.Gryzenhout M, Rodas CA, Mena P, Julio, Clegg P, Wingfield BD, Wingfield MJ. Novel hosts of the Eucalyptus canker pathogen Chrysoporthe cubensis and a new Chrysoporthe species from Colombia. Mycol Res. 2006;110:833–45. [DOI] [PubMed] [Google Scholar]
- 7.Gryzenhout M, Myburg H, Wingfield BD, Montenegro F, and Wingfield MJ. Chrysoporthe doradensis sp. Nov. Pathogenic to Eucalyptus in ecuador. Fungal Diversity. 2005;20:39–57. [Google Scholar]
- 8.van der Merwe NA, Gryzenhout M, Steenkamp ET, Wingfield BD, Wingfield MJ. Multigene phylogenetic and population differentiation data confirm the existence of a cryptic species within Chrysoporthe cubensis. Fungal Biology. 2010;114:966–79. 10.1016/j.funbio.2010.09.007 [DOI] [PubMed] [Google Scholar]
- 9.Chungu D, Gryzenhout M, Muimba-Kankolongo A, Wingfield MJ, Roux J. Taxonomy and pathogenicity of two novel Chrysoporthe species from Eucalyptus grandis and Syzygium guineense in Zambia. Mycol Progress. 2010;9:379–93. [Google Scholar]
- 10.Myburg H, Gryzenhout M, Wingfield BD, Wingfield MJ. B-tubulin and histone h3 gene sequences distinguish Cryphonectria cubensis from South Africa, Asia, and South America. Can J. Bot. 2002;80:590–6. [Google Scholar]
- 11.Stone CL, Buitrago MLP, Boore JL, Frederick RD. Analysis of the complete mitochondrial genome sequences of the soybean rust pathogens Phakopsora pachyrhizi and P. Meibomiae. Mycologia. 2010;102:887–97. [DOI] [PubMed] [Google Scholar]
- 12.van de Sande WW. Phylogenetic analysis of the complete mitochondrial genome of Madurella mycetomatis confirms its taxonomic position within the order sordariales. PLOS ONE. 2012;7:e38654 10.1371/journal.pone.0038654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Steenkamp E, Brinkmann H, Forget L, Philippe H, Lang BF. Phylogenomic analyses predict sistergroup relationship of nucleariids and fungi and paraphyly of zygomycetes with significant support. BMC Evol Biol. 2009;9:272 10.1186/1471-2148-9-272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pantou M, Kouvelis V, Typas M. The complete mitochondrial genome of the vascular wilt fungus Verticillium dahliae: A novel gene order for Verticillium and a diagnostic tool for species identification. Curr Genet. 2006;50:125–36. [DOI] [PubMed] [Google Scholar]
- 15.Kouvelis VN, Ghikas DV, Typas MA. The analysis of the complete mitochondrial genome of Lecanicillium muscarium (synonym Verticillium lecanii) suggests a minimum common gene organization in mtdnas of Sordariomycetes: Phylogenetic implications. Fungal Genet Biol. 2004;41:930–40. [DOI] [PubMed] [Google Scholar]
- 16.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–81. [DOI] [PubMed] [Google Scholar]
- 17.Burger G, Gray MW, Franz Lang B. Mitochondrial genomes: Anything goes. Trends Genet. 2003;19:709–16. [DOI] [PubMed] [Google Scholar]
- 18.Kouvelis VN, Sialakouma A, Typas MA. Mitochondrial gene sequences alone or combined with its region sequences provide firm molecular criteria for the classification of Lecanicillium species. Mycol Res. 2008;112:829–44. 10.1016/j.mycres.2008.01.016 [DOI] [PubMed] [Google Scholar]
- 19.Wu Y, Yang J, Yang F, Liu T, Leng W, Chu Y, et al. Recent dermatophyte divergence revealed by comparative and phylogenetic analysis of mitochondrial genomes. BMC Genomics. 2009;10:238 10.1186/1471-2164-10-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantou M, Kouvelis V, Typas M. The complete mitochondrial genome of Fusarium oxysporum: Insights into fungal mitochondrial evolution. Gene. 2008;419:7–15. 10.1016/j.gene.2008.04.009 [DOI] [PubMed] [Google Scholar]
- 21.Youssar L, Grüning BA, Günther S, Hüttel W. Characterization and phylogenetic analysis of the mitochondrial genome of Glarea lozoyensis indicates high diversity within the order Helotiales. PLOS ONE. 2013;8:e74792 10.1371/journal.pone.0074792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguileta G, de Vienne DM, Ross ON, Hood ME, Giraud T, Petit E, et al. High variability of mitochondrial gene order among fungi. Genome Biology and Evolution. 2014;6:451–65. 10.1093/gbe/evu028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hausner G. Fungal mitochondrial genomes, plasmids and introns In: Dilip KA, George GK, editors. Applied mycology and biotechnology. Volume 3: Elsevier; 2003. p. 101–31. [Google Scholar]
- 24.Al-Reedy RM, Malireddy R, Dillman CB, Kennell JC. Comparative analysis of Fusarium mitochondrial genomes reveals a highly variable region that encodes an exceptionally large open reading frame. Fungal Genet Biol. 2012;49:2–14. 10.1016/j.fgb.2011.11.008 [DOI] [PubMed] [Google Scholar]
- 25.Joardar V, Abrams N, Hostetler J, Paukstelis P, Pakala S, Pakala S, et al. Sequencing of mitochondrial genomes of nine Aspergillus and Penicillium species identifies mobile introns and accessory genes as main sources of genome size variability. BMC Genomics. 2012;13:698 10.1186/1471-2164-13-698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barr CM, Neiman M, Taylor DR. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 2005;168:39–50. [DOI] [PubMed] [Google Scholar]
- 27.Basse CW. Mitochondrial inheritance in fungi. Curr Opin Microbiol. 2010;13:712–9. 10.1016/j.mib.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 28.Cole TE, Hong Y, Brasier CM, Buck KW. Detection of an rna-dependent rna polymerase in mitochondria from a mitovirus-infected isolate of the dutch elm disease fungus, Ophiostoma novo-ulmi. Virology. 2000;268:239–43. [DOI] [PubMed] [Google Scholar]
- 29.Hong Y, Dover SL, Cole TE, Brasier CM, Buck KW. Multiple mitochondrial viruses in an isolate of the dutch elm disease fungus Ophiostoma novo-ulmi. Virology. 1999;258:118–27. [DOI] [PubMed] [Google Scholar]
- 30.Monteiro-Vitorello CB, Hausner G, Searles DB, Gibb EA, Fulbright DW, Bertrand H. The Cryphonectria parasitica mitochondrial rns gene: Plasmid-like elements, introns and homing endonucleases. Fungal Genet Biol. 2009;46:837–48. 10.1016/j.fgb.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 31.Hausner G. Introns, mobile elements, and plasmids In: Bullerwell CE, editor. Organelle genetics: Springer; Berlin Heidelberg; 2012. p. 329–57. [Google Scholar]
- 32.Pramateftaki PV, Kouvelis VN, Lanaridis P, Typas MA. The mitochondrial genome of the wine yeast Hanseniaspora uvarum: A unique genome organization among yeast/fungal counterparts2006 2006-01-01 00:00:00. 77–90 p. [DOI] [PubMed]
- 33.Mardanov AV, Beletsky AV, Kadnikov VV, Ignatov AN, Ravin NV. The 203 kbp mitochondrial genome of the phytopathogenic fungus Sclerotinia borealis reveals multiple invasions of introns and genomic duplications. PLOS One. 2014;9:e107536 10.1371/journal.pone.0107536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Himmelstrand K, Olson A, Brandstrom Durling M, Karlsson M, Stenlid J. Intronic and plasmid-derived regions contribute to the large mitochondrial genome sizes of Agaricomycetes. Current Genetics. 2014;60:303–13. 10.1007/s00294-014-0436-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beaudet D, Nadimi M, Iffis B, Hijri M. Rapid mitochondrial genome evolution through invasion of mobile elements in two closely related species of arbuscular mycorrhizal fungi. PLOS ONE. 2013;8:e60768 10.1371/journal.pone.0060768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferandon C, Moukha S, Callac P, Benedetto J, Castroviejo M, Barroso G. The Agaricus bisporus cox1 gene: The longest mitochondrial gene and the largest reservoir of mitochondrial group I introns. PLOS ONE. 2010;5:e14048 10.1371/journal.pone.0014048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lang BF, Laforest M-J, Burger G. Mitochondrial introns: A critical view. Trends Genet. 2007;23:119–25. [DOI] [PubMed] [Google Scholar]
- 38.Bonen L, Vogel J. The ins and outs of group II introns. Trends Genet. 2001;17:322–31. [DOI] [PubMed] [Google Scholar]
- 39.Haugen P, Simon DM, Bhattacharya D. The natural history of group I introns. Trends Genet. 2005;21:111–9. [DOI] [PubMed] [Google Scholar]
- 40.Sethuraman J, Majer A, Friedrich N, Edgell D, Hausner G. Genes within genes: Multiple laglidadg homing endonucleases target the ribosomal protein s3 gene encoded within an rnl group I intron of Ophiostoma and related taxa. Mol Biol Evol. 2009;26:2299–315. 10.1093/molbev/msp145 [DOI] [PubMed] [Google Scholar]
- 41.Shimko N, Liu L, Lang BF, Burger G. Gobase: The organelle genome database. Nucleic Acids Res. 2001;29:128–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fourie G, Merwe NAvd, Wingfield BD, Bogale M, Tudzynski B, Wingfield MJ, et al. Evidence for inter-specific recombination among the mitochondrial genomes of Fusarium species in the Gibberella fujikuroi complex. BMC Genomics. 2013;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salavirta H, Oksanen I, Kuuskeri J, Makela M, Laine P, Paulin L, et al. Mitochondrial genome of Phlebia radiata is the second largest (156 kbp) among fungi and features signs of genome flexibility and recent recombination events. PLOS ONE. 2014;9:e97141 10.1371/journal.pone.0097141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wingfield BD, Ades PK, Al-Naemi FA, Beirn LA, Bihon W, Crouch JA, et al. IMA genome-f 4. Draft genome sequences of Chrysoporthe austroafricana, Diplodia scrobiculata, Fusarium nygamai, Leptographium lundbergii, Limonomyces culmigenus, Stagonosporopsis tanaceti, and Thielaviopsis punctulata. IMA Fungus 2015;6:233–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wingfield BD, Barnes I, de Beer ZW, De Vos L, Duong TA, Kanzi AM, et al. IMA genome-f 5: Draft genome sequences of Ceratocystis eucalypticola, Chrysoporthe cubensis, C. deuterocubensis, Davidsoniella virescens, Fusarium temperatum, Graphilbum fragrans, Penicillium nordicum, and thielaviopsis Thielaviopsis musarum. IMA Fungus. 2015;6:493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. [DOI] [PubMed] [Google Scholar]
- 47.Hahn C, Bachmann L, Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 2013;41:e129–e. 10.1093/nar/gkt371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruby JG, Bellare P, Derisi JL. Price: Software for the targeted assembly of components of (meta) genomic sequence data. G3: Genes|Genomes|Genetics. 2013;3:865–80. 10.1534/g3.113.005967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katoh K, Misawa K, Kuma Ki, Miyata T. Mafft: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002;30:3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guy L, Roat Kultima J, Andersson SGE. Genoplotr: Comparative gene and genome visualization in r. Bioinformatics. 2010;26:2334–5. 10.1093/bioinformatics/btq413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice P, Longden I, Bleasby A. Emboss: The european molecular biology open software suite. Trends Genet. 2000;16:276–7. [DOI] [PubMed] [Google Scholar]
- 52.Stamatakis A. Raxml version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. Trimal: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darriba D, Taboada GL, Doallo R, Posada D. Jmodeltest 2: More models, new heuristics and parallel computing. Nat Meth. 2012;9:772–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abascal F, Zardoya R, Posada D. Prottest: Selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–5. [DOI] [PubMed] [Google Scholar]
- 56.Hausner G, Monteiro-Vitorello CB, Searles DB, Maland M, Fulbright DW, Bertrand H. A long open reading frame in the mitochondrial lsu rrna group-i intron of Cryphonectria parasitica encodes a putative s5 ribosomal protein fused to a maturase. Curr Genet. 1999;35:109–17. [DOI] [PubMed] [Google Scholar]
- 57.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. [DOI] [PubMed] [Google Scholar]
- 58.Lin R, Liu C, Shen B, Bai M, Ling J, Chen G, et al. Analysis of the complete mitochondrial genome of Pochonia chlamydosporia suggests a close relationship to the invertebrate-pathogenic fungi in hypocreales. BMC Microbiol. 2015;15:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bullerwell CE, Forget L, Lang BF. Evolution of monoblepharidalean fungi based on complete mitochondrial genome sequences. Nucleic Acids Res. 2003;31:1614–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bullerwell CE, Gray MW. Evolution of the mitochondrial genome: Protist connections to animals, fungi and plants. Curr Opin Microbiol. 2004;7:528–34. [DOI] [PubMed] [Google Scholar]
- 61.Torriani SF, Penselin D, Knogge W, Felder M, Taudien S, Platzer M, et al. Comparative analysis of mitochondrial genomes from closely related Rhynchosporium species reveals extensive intron invasion. Fungal Genet Biol. 2014;62:34–42. 10.1016/j.fgb.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 62.Férandon C, Xu J, Barroso G. The 135 kbp mitochondrial genome of agaricus bisporus is the largest known eukaryotic reservoir of group I introns and plasmid-related sequences. Fungal Genet Biol. 2013;55:85–91. 10.1016/j.fgb.2013.01.009 [DOI] [PubMed] [Google Scholar]
- 63.Tuller T, Girshovich Y, Sella Y, Kreimer A, Freilich S, Kupiec M, et al. Association between translation efficiency and horizontal gene transfer within microbial communities. Nucleic Acids Res. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gilmore SR, Gräfenhan T, Louis-Seize G, Seifert KA. Multiple copies of cytochrome oxidase 1 in species of the fungal genus Fusarium. Molecular Ecology Resources. 2009;9:90–8. 10.1111/j.1755-0998.2009.02636.x [DOI] [PubMed] [Google Scholar]
- 65.Cunnington J. The distribution of optional mitochondrial introns encoding putative homing endonuclease genes in the Fusarium oxysporum complex. Sydowia. 2009;61:1–9. [Google Scholar]
- 66.Paquin B, Laforest M, Forget L, Roewer I, Wang Z, Longcore J, et al. The fungal mitochondrial genome project: Evolution of fungal mitochondrial genomes and their gene expression. Curr Genet. 1997;31:380–95. [DOI] [PubMed] [Google Scholar]
- 67.Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13880–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yin L-F, Hu M-J, Wang F, Kuang H, Zhang Y, Schnabel G, et al. Frequent gain and loss of introns in fungal cytochrome b genes. PLOS ONE. 2012;7:e49096 10.1371/journal.pone.0049096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stoddard BL. Homing endonucleases: From microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. 10.1016/j.str.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burt A, Koufopanou V. Homing endonuclease genes: The rise and fall and rise again of a selfish element. Curr Opin Genet Dev. 2004;14:609–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rnpb genes were mannualy retrieved from annotated fungal mt genomes publicly available in GenBank. Phylogenetic analysis was performed using maximum likelihood method implemented in RAxML with HKY model of selection. Branch support was calculated using 1000 bootstrap replicates. Green boxes depict atp6 gene, red; small sub-unit of ribosomal RNA (rns) and dark red; small sub-unit of ribosomal RNA (rnl).
(PDF)
Phylogenetic analysis of orf540, orf551 and orf551 from C. austroafricana, C. cubensis and C. deuterocubensis which show sequence similarity to C. parasitica S5 ribosomal protein/maturase fusion protein. Sequences used in this phylogeny were retrieved from GenBank using BLAST. The LG+G model of substitution was used. Branch support for was calculated using the bootstrap method with 1000 replicates.
(PDF)
The graph shows the average expression for genes that are involved in oxidative phosphorylation and electron transport and the rnpb gene of Chrysoporthe austroafricana grown in complete and minimal media.
(PDF)
Intronic ORFs are depicted by yellow arrowed boxes on introns of genes where found and are labelled according to the gene and intron position. Intergenic ORFs are also depicted by yellow arrowed boxes and are labelled with “IN” prefix.
(PDF)
(PDF)
Comparison of codon usage and tRNAs for the 14 genes involved in oxidative phosphorylation and electron transport in the mitochondrial genomes of Chrysoporthe austroafricana, C. cubensis, C. deuterocubensis and Cryphonectria parasitica.
(PDF)
The best hits for each query (intron) is shown. Default BLAST parameters were used. The prefix CA, CC, CD and CP is added to the intron names for clarity.
(PDF)
Results from all possible comparisons were calculated. Only Blast hits with a threshold of over 50% alignment coverage, 50% sequence identity and E-value of 0.00005 are shown.
(PDF)
HEG type were annotated using the NCBI Conserved Domain Database (CDD).
(PDF)
Data Availability Statement
The mitochondrial genome sequences of Chrysoporthe austroafricana, C. cubensis, C. deuterocubensis and Cryphonectria parasitica are available at NCBI GenBank accession numbers KT380883, KT380885, KT380884, and KT428651, respectively.