Abstract
Mycoplasma genitalium is a cause of non-gonoccocal urethritis (NGU) in men and cervicitis and pelvic inflammatory disease in women. Recent international data also indicated that the first line treatment, 1 gram stat azithromycin therapy, for M. genitalium is becoming less effective, with the corresponding emergence of macrolide resistant strains. Increasing failure rates of azithromycin for M. genitalium has significant implications for the presumptive treatment of NGU and international clinical treatment guidelines. Assays able to predict macrolide resistance along with detection of M. genitalium will be useful to enable appropriate selection of antimicrobials to which the organism is susceptible and facilitate high levels of rapid cure. One such assay recently developed is the MG 23S assay, which employs novel PlexZyme™ and PlexPrime™ technology. It is a multiplex assay for detection of M. genitalium and 5 mutations associated with macrolide resistance. The assay was evaluated in 400 samples from 254 (186 males and 68 females) consecutively infected participants, undergoing tests of cure. Using the MG 23S assay, 83% (331/440) of samples were positive, with 56% of positives carrying a macrolide resistance mutation. Comparison of the MG 23S assay to a reference qPCR method for M. genitalium detection and high resolution melt analysis (HRMA) and sequencing for detection of macrolide resistance mutations, resulted in a sensitivity and specificity for M. genitalium detection and for macrolide resistance of 99.1/98.5% and 97.4/100%, respectively. The MG 23S assay provides a considerable advantage in clinical settings through combined diagnosis and detection of macrolide resistance.
Introduction
Mycoplasma genitalium was first isolated in 1980 from men with non-gonococcal urethritis (NGU) [1]. It has since been established as a common sexually transmitted infection responsible for 10–35% of non-chlamydial NGU in men [2] and also associated with cervicitis, endometritis, pelvic inflammatory disease (PID) and tubal factor infertility in women [3–7]. Studies also suggest M. genitalium plays an important role in HIV acquisition and transmission [8, 9].
Current recommended treatment for uncomplicated M. genitalium infection is a single 1 g oral dose of the macrolide antibiotic, azithromycin [10, 11]. In most developed countries, this is also the first line treatment for NGU. However cure rates following this treatment have declined to about 70% for genital M. genitalium infections, as macrolide resistance has emerged [12–14] [15].
Macrolide resistance occurs through mutations in nucleotide positions 2058 and 2059 (E.coli numbering) in region V of the 23S rRNA gene, inhibiting binding of the macrolide to M. genitalium [14, 16]. An important limitation in current clinical practice is the ability to effectively treat this organism in a significant proportion of individuals, when prevalence of resistance to first line therapies is so high. To address this, a high resolution melt analysis (HRMA) methodology was recently described to rapidly detect 23S rRNA gene mutations associated with macrolide resistance in M. genitalium positive specimens [17]. This HRMA assay allowed for screening of 23S rRNA mutations; however it needs to be performed as a separate assay after M. genitalium detection, delaying results, second line treatment and potentiating further transmission.
Development of an assay for the simultaneous, rapid and sensitive detection of M. genitalium and mutations associated with macrolide resistance would offer an enormous clinical advantage, facilitating rapid delivery of the most appropriate therapy. This study evaluates the MG 23S assay, which employs novel PlexZyme™ and PlexPrime™ technology (SpeeDx), is a single well, multiplex qPCR assay, which simultaneously detects M. genitalium and five 23S rRNA mutations associated with macrolide resistance.
Methods
Patient population
The assay was evaluated using clinical samples previously screened for both M. genitalium detection by 16S rRNA qPCR and 23S rRNA mutation status by HRMA and Sanger sequencing. Between July 2012-June 2013, 400 specimens (including baseline, day 14 and/or day 28 specimens) from 254 consecutive M. genitalium infected participants (186 males (300 urine/urethral and 12 anal samples) and 68 females (41 urine, 42 cervical/vaginal and 5 anal samples)) were collected at Melbourne Sexual Health Centre, Victoria, Australia. This clinic conducts routine testing for M. genitalium in patients with NGU, cervicitis, and/or PID, as well as sexual contacts of M. genitalium infected patients. Ethical approval for this study was granted by the Alfred Hospital Research Ethics Committee (number 150/12).
Assay description
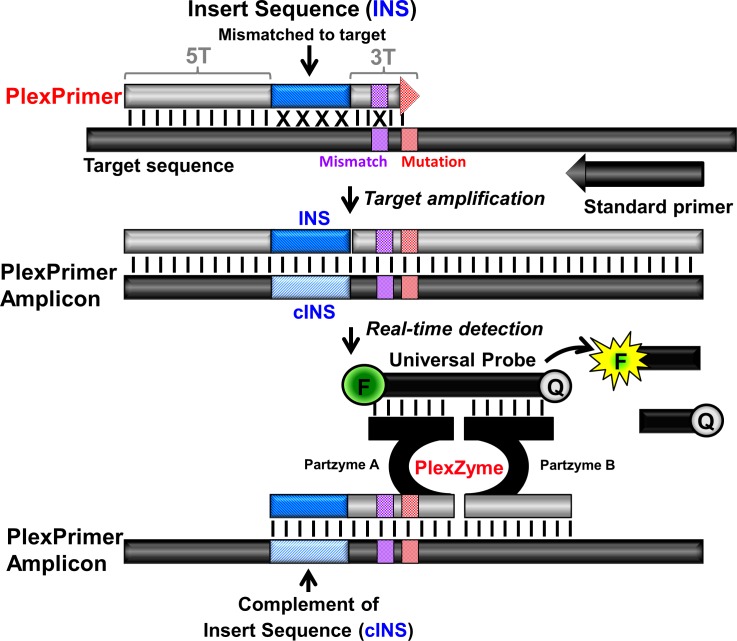
The MG 23S assay (SpeeDx Pty Ltd, Sydney, Australia) is a multiplex qPCR assay detecting across three fluorophore channels: 1) FAM (495–516) for detection of M. genitalium through detection of the MgPa gene, 2) Hex (535–556) for detection of 5 mutations that have been described in clinical samples (A2058G, A2059G, A2058C, A2059C and A2058T) in the 23S rRNA gene and 3) Cy5 (650–670) for detection of a heterologous extrinsic control to monitor extraction efficiency and qPCR inhibition. This assay employs novel PlexZyme (formerly known as MNAzyme) technology [18–20] and PlexPrimers which selectively amplify target sequences, coupled to enable high level multiplexing of mutant detection (Fig 1).
Fig 1. PlexPrimer and PlexZyme detection technology.
The Plexprimer contains three functional regions; a 5’ target recognition region (5T), a short 3’ target-specific sequence (3T) and an intervening Insert Sequence (INS) region, which is mismatched with respect to the target. The PlexPrimer binds to the mutation at the 3’ terminus (red box) and also contains a mismatched base (purple box). During amplification the INS and its complement (cINS) are incorporated into the PlexPrime amplicons and these can be detected in real-time using PlexZymes. PlexZymes are nucleic acid enzymes which only form, from their component partzymes A and B, when target amplicon are present. Each of the partzymes contain a probe binding arm, a partial catalytic core and a target binding arm, orientated such that partzyme A binds to the amplicon in the region containing the cINS whilst the Partzyme B binds adjacently downstream. Catalytically active PlexZymes bind and cleave universal reporter probes between fluorophore (F) and quencher (Q) moieties resulting in signal generation.
Template and Targets
Quantified synthetic DNA templates from the MG 23S Positive Control (beta version) (SpeeDx) were utilized for the limit of detection testing of M. genitalium (MgPa gene) and mutations from the 23S rRNA gene; A2058G, A2059G, A2058C, A2059C and A2058T. Testing of cross-reactivity of the MG 23S assay was performed with 104 copies of Amplirun® DNA (Vircell, Granada, Spain) for Mycoplasma hominis (Cat# MBC084), Trichomonas vaginalis (Cat# MBC079), Chlamydia trachomatis (Cat# MBC012), Neisseria gonorrhoeae (Cat# MBC075) and Mycoplasma pneumoniae (Cat# MBC035), and 105 copies of extracted DNA from clinical isolates of Ureaplasma parvum and Ureaplasma urealyticum. We have further tested extracted DNA from clinical isolates of further 24 unrelated species including Salmonella typhimurium, Klebsiella pneumoniae, Haemophilus influenzae, Enterococcus faecalis, Proteus mirabilis, Staphylococcus aureus, Staphylococcus epidermidis, Candida albicans, Streptococcus pyogenes, Pseudomonas aeruginosa, Acinetobacter calcoaceticus, Neisseria gonorrhoeae, Bacteroides fragilis, E. coli, Viridans streptococci, Streptococcus Group A and B, Campylobacter jejuni, Neisseria meningitidis, Neisseria cinerea, Neisseria subflava, Neisseria lactamica, Chlamydophila pneumoniae and Chlamydia trachomatis.
Analytical Sensitivity
The sensitivity of each of the targets of the MG 23S assay was determined as the lowest number of synthetic DNA template copies tested with ≥95% detection.
Processing and PCR
Samples were processed and extracted as described previously [12]. The reference method for detection of M. genitalium was an established qPCR assay targeting the 16S rRNA gene [21]. The reference standard for determining the presence of a mutation was HRMA and if any were unable to be determined using this method, the samples underwent sequencing of the region V of 23S rRNA as described previously [16, 17]. The MG 23S assay was performed as described by the manufacturer. Briefly, a 5 μl aliquot of the extracted DNA was mixed with Plex Mastermix in 20 μl final volume. All testing was performed in 96 well plates on the LightCycler® 480 Instrument II (Roche Diagnostic, Indianapolis, USA) using parameters of 95°C for 2 min, followed by 10 cycles of 95°C for 5 s, 61°C for 30 s (-0.5°C per cycle), and 40 cycles of 95°C for 5 s, 52°C for 40 s. Data analysis was performed using an analysis algorithm provided with the assay, resulting in reporting of the presence or absence of M. genitalium, any of the five mutations in the 23S rRNA gene, and the internal control. Controls for the assay were comprised of synthetic double stranded DNA targets for each 23S rRNA gene mutation type and M. genitalium G37 genomic DNA (ATCC 33530D) serving as the wild type and M. genitalium control.
Results
The limit of detection for the M. genitalium MgPa target was 10 copies. The limit of detection of 23S rRNA gene targets was 10 copies for A2058C, A2058T and A2059G, 12 copies for A2058G, and 15 copies for A2059C (Table 1). Cross-reactivity of primer and probe sequences for both MgPa and 23S rRNA were first analyzed in silico using BLAST to ensure specificity. Subsequent testing of the assay using DNA from related organisms and other clinically relevant organisms showed no cross-reacting agents for the MgPa assay or the 23S rRNA mutant assay.
Table 1. Analytical sensitivity of the MG 23S assay as determined by detection of replicate of different concentration of each template.
| Copy number Detected/replicate tested | Copy number (detected/ replicates tested) | ||||||
|---|---|---|---|---|---|---|---|
| Template | 105 | 104 | 103 | 102 | 10 | 1 | |
| A2058G | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 2/3 | 12 (20/20) |
| A2058C | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 10 (20/20) |
| A2058T | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 1/3 | 10 (19/20) |
| A2059G | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 1/3 | 12 (20/20) |
| A2059C | 3/3 | 3/3 | 3/3 | 3/3 | 2/3 | 0/3 | 15 (20/20) |
| MgPa | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 2/3 | 10 (20/20) |
Detection of M. genitalium by the MG 23S assay was highly concordant (99.0%) with the reference method (qPCR targeting the 16S rRNA gene) with a Kappa value of 0.965 (95% confidence interval (CI) 0.930 to 0.999) and sensitivity/specificity of 99.1% and 98.5% respectively (Table 2). Only 4 (1.0%) of the specimens were discordant with one positive only by the MG 23S assay and 3 positive only by the reference assay. All of the discordant samples had low copy number of M. genitalium. Among the 330 specimens positive for M. genitalium, 325 (98.5%) were concordant for detection of a macrolide mutation, with a Kappa value of 0.969 (95% CI 0.942–0.996) and sensitivity/specificity of 97.4% and 100% respectively (Table 3). Only 5 cases with macrolide mutations detected by the reference method were classified as wild type by the MG 23S assay, and there were no cases with macrolide resistance detected by the MG 23S assay, where the reference method had shown a wild type status.
Table 2. Evaluation of the MG 23S assay for the detection of M. genitalium.
| Reference Assaya | |||||
|---|---|---|---|---|---|
| Pos | Neg | Total N (%) | Sensitivity % (95% CI) / Specificity % (95% CI) | ||
| PlexPCR | Pos | 330 | 1 | 331 (83) | 99.1 (97.4–99.8) / 98.5 (92.0–100.0) |
| Neg | 3 | 66 | 69 (17) | ||
| Total | 333 | 67 | 400 | ||
a The reference assay for M. genitalium detection was an in house qPCR assay targeting the 16S rRNA gene [21].
Table 3. Evaluation of the MG 23S assay for the detection of 23S rRNA mutations.
| Reference Assaya | |||||
|---|---|---|---|---|---|
| MTb | WTb | Total N (%) | Sensitivity % (95% CI) / Specificity % (95% CI) | ||
| PlexPCR | MTb | 184 | 0 | 184 (56) | 97.4 (93.9–99.1) / 100.0 (97.4–100.0) |
| WTb | 5 | 141 | 146 (44) | ||
| Total | 189 | 141 | 330c | ||
b MT refers to 5 common mutation detection of adenine to another base in 2058 or 2059 position of 23S rRNA. WT refers to adenine in both 2058 and 2059 positions of 23S rRNA.
c Only includes M. genitalium positive samples determined by reference method and excludes three samples in which 23S sequence status could not be confirmed.
Discussion
Overall, the MG 23S assay showed excellent performance for the simultaneous detection of M. genitalium and 23S rRNA mutations by qPCR, with high sensitivity and specificity compared to established gold standards.
Macrolide resistance is highly prevalent in M. genitalium and exceeds 30% in many countries. As culture is difficult and rarely performed to allow performance of susceptibility testing, a molecular approach has been utilized to predict resistance to azithromycin. All azithromycin treatment failures to date have been shown to carry one of the five mutations in the 2058 and 2059 positions in the 23S rRNA gene [12], therefore detection of these mutations can be used to infer macrolide resistance. Molecular methodologies such as pyrosequencing, HRMA or FRET methods [17, 22] have been used to detect the presence of macrolide resistance mutations [23]. However these assays must be performed after the initial M. genitalium diagnostic test, delaying reporting of the mutation status. Other qPCR technologies are able to detect mutations such as mutation-specific primers and mutation-specific probes [24–26]. However, it is difficult to efficiently multiplex mutation-specific primers as they may only differ by a single base. Mutation-specific probes are also similarly limited in multiplexing due to competition. The novel design of the PlexPrimer overcomes this limitation by incorporating a unique sequence into the amplicon, to increase differentiation and reduce competition. PlexPrimers coupled with PlexZymes enables high level multiplexing, for specific mutation amplification and detection in qPCR.
The implementation of combined diagnostic and resistance testing to optimize rapid selection of more effective antimicrobials is considered a priority in the field. This would help to reduce recurrent clinical presentations to services, sequelae such as infertility, and secondary transmission of resistant strains. The MG 23S assay offers an enormous advantage in clinical settings, as it can be incorporated into clinical algorithms. Detection of 23S rRNA mutations would allow patients to be rapidly treated with an appropriate second line antibiotic such as the fluoroquinolone moxifloxacin, rather than waiting 3–4 weeks for a test of cure to show azithromycin treatment failure. Patients without evidence of macrolide resistance can currently be treated with azithromycin with high cure rates. There are however, up to 10% of patients that do not have detectable pre-treatment mutations, and appear to develop induced or selected resistance following azithromycin treatment. Treatment failure in these cases will not be avoided by the use of pre-treatment resistance assays. This will only be averted by the use of non-macrolide first line regimens; however identification of suitable alternatives has proven difficult. In addition to macrolides, M. genitalium is also developing resistance to moxifloxacin [12]. Development and inclusion of further markers for fluoroquinolone resistance would be greatly beneficial and is currently under development.
Acknowledgments
SpeeDx are the developer and manufacturer of the evaluated MG 23S assay and supplied the reagents utilized for this study.
Data Availability
All relevant data are within the paper.
Funding Statement
In regards to the funding statement the authors would like to confirm that the funder (SpeedX) provided support in the form of salaries for authors (LT, SW, EM, AT). The funder did not have any role in the clinical evaluation including data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Tully JG, Taylor-Robinson D, Cole RM, Rose DL. A newly discovered mycoplasma in the human urogenital tract. Lancet. 1981;1(8233):1288–91. . [DOI] [PubMed] [Google Scholar]
- 2.Horner PJ, Taylor-Robinson D. Association of Mycoplasma genitalium with balanoposthitis in men with non-gonococcal urethritis. Sex Transm Infect. 2011;87(1):38–40. 10.1136/sti.2010.044487 . [DOI] [PubMed] [Google Scholar]
- 3.Cohen CR, Manhart LE, Bukusi EA, Astete S, Brunham RC, Holmes KK, et al. Association between Mycoplasma genitalium and acute endometritis. Lancet. 2002;359(9308):765–6. 10.1016/S0140-6736(02)07848-0 . [DOI] [PubMed] [Google Scholar]
- 4.Deguchi T, Maeda S. Mycoplasma genitalium: another important pathogen of nongonococcal urethritis. J Urol. 2002;167(3):1210–7. . [DOI] [PubMed] [Google Scholar]
- 5.Jensen JS. Mycoplasma genitalium: the aetiological agent of urethritis and other sexually transmitted diseases. J Eur Acad Dermatol Venereol. 2004;18(1):1–11. . [DOI] [PubMed] [Google Scholar]
- 6.Taylor-Robinson D. Mycoplasma genitalium—an up-date. Int J STD AIDS. 2002;13(3):145–51. . [DOI] [PubMed] [Google Scholar]
- 7.Uuskula A, Kohl PK. Genital mycoplasmas, including Mycoplasma genitalium, as sexually transmitted agents. Int J STD AIDS. 2002;13(2):79–85. . [DOI] [PubMed] [Google Scholar]
- 8.Manhart LE, Mostad SB, Baeten JM, Astete SG, Mandaliya K, Totten PA. High Mycoplasma genitalium organism burden is associated with shedding of HIV-1 DNA from the cervix. J Infect Dis. 2008;197(5):733–6. 10.1086/526501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mavedzenge SN, Van Der Pol B, Weiss HA, Kwok C, Mambo F, Chipato T, et al. The association between Mycoplasma genitalium and HIV-1 acquisition in African women. AIDS. 2012;26(5):617–24. 10.1097/QAD.0b013e32834ff690 . [DOI] [PubMed] [Google Scholar]
- 10.Shahmanesh M, Moi H, Lassau F, Janier M, Iusti/Who. 2009 European guideline on the management of male non-gonococcal urethritis. Int J STD AIDS. 2009;20(7):458–64. 10.1258/ijsa.2009.009143 . [DOI] [PubMed] [Google Scholar]
- 11.Workowski KA, Bolan GA, Centers for Disease C, Prevention. Sexually transmitted diseases treatment guidelines, 2015. MMWR Recomm Rep. 2015;64(RR-03):1–137. . [PMC free article] [PubMed] [Google Scholar]
- 12.Bissessor M, Tabrizi SN, Twin J, Abdo H, Fairley CK, Chen MY, et al. Macrolide resistance and azithromycin failure in a Mycoplasma genitalium-infected cohort and response of azithromycin failures to alternative antibiotic regimens. Clin Infect Dis. 2015;60(8):1228–36. 10.1093/cid/ciu1162 . [DOI] [PubMed] [Google Scholar]
- 13.Bradshaw CS, Chen MY, Fairley CK. Persistence of Mycoplasma genitalium following azithromycin therapy. PLoS One. 2008;3(11):e3618 10.1371/journal.pone.0003618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradshaw CS, Jensen JS, Tabrizi SN, Read TR, Garland SM, Hopkins CA, et al. Azithromycin failure in Mycoplasma genitalium urethritis. Emerg Infect Dis. 2006;12(7):1149–52. 10.3201/eid1207.051558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manhart LE, Gillespie CW, Lowens MS, Khosropour CM, Colombara DV, Golden MR, et al. Standard treatment regimens for nongonococcal urethritis have similar but declining cure rates: a randomized controlled trial. Clin Infect Dis. 2013;56(7):934–42. 10.1093/cid/cis1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen JS, Bradshaw CS, Tabrizi SN, Fairley CK, Hamasuna R. Azithromycin treatment failure in Mycoplasma genitalium-positive patients with nongonococcal urethritis is associated with induced macrolide resistance. Clin Infect Dis. 2008;47(12):1546–53. 10.1086/593188 . [DOI] [PubMed] [Google Scholar]
- 17.Twin J, Jensen JS, Bradshaw CS, Garland SM, Fairley CK, Min LY, et al. Transmission and selection of macrolide resistant Mycoplasma genitalium infections detected by rapid high resolution melt analysis. PLoS One. 2012;7(4):e35593 10.1371/journal.pone.0035593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mokany E, Bone SM, Young PE, Doan TB, Todd AV. MNAzymes, a versatile new class of nucleic acid enzymes that can function as biosensors and molecular switches. J Am Chem Soc. 2010;132(3):1051–9. 10.1021/ja9076777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mokany E, Tan YL, Bone SM, Fuery CJ, Todd AV. MNAzyme qPCR with superior multiplexing capacity. Clin Chem. 2013;59(2):419–26. 10.1373/clinchem.2012.192930 . [DOI] [PubMed] [Google Scholar]
- 20.Mokany E, Todd AV. MNAzyme qPCR: a superior tool for multiplex qPCR. Methods Mol Biol. 2013;1039:31–49. 10.1007/978-1-62703-535-4_3 . [DOI] [PubMed] [Google Scholar]
- 21.Twin J, Taylor N, Garland SM, Hocking JS, Walker J, Bradshaw CS, et al. Comparison of two Mycoplasma genitalium real-time PCR detection methodologies. J Clin Microbiol. 2011;49(3):1140–2. 10.1128/JCM.02328-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen JS. Protocol for the detection of Mycoplasma genitalium by PCR from clinical specimens and subsequent detection of macrolide resistance-mediating mutations in region V of the 23S rRNA gene. Methods Mol Biol. 2012;903:129–39. 10.1007/978-1-61779-937-2_8 . [DOI] [PubMed] [Google Scholar]
- 23.Touati A, Peuchant O, Jensen JS, Bebear C, Pereyre S. Direct detection of macrolide resistance in Mycoplasma genitalium isolates from clinical specimens from France by use of real-time PCR and melting curve analysis. J Clin Microbiol. 2014;52(5):1549–55. 10.1128/JCM.03318-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ. Allelic discrimination using fluorogenic probes and the 5' nuclease assay. Genet Anal. 1999;14(5–6):143–9. . [DOI] [PubMed] [Google Scholar]
- 25.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 1989;17(7):2503–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16(1):49–53. 10.1038/nbt0198-49 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.