In Gram-negative bacteria, IS26 recruits antibiotic resistance genes into the mobile gene pool by forming transposons carrying many different resistance genes. In addition to replicative transposition, IS26 was recently shown to use a novel conservative movement mechanism in which an incoming IS26 targets a preexisting one. Here, we have demonstrated how IS26-bounded class I transposons can be produced from translocatable units (TUs) containing only an IS26 and a resistance gene via the conservative reaction. TUs were incorporated next to an existing IS26, creating a class I transposon, and if the targeted IS26 is in a transposon, the product resembles two transposons sharing a central IS26, a configuration observed in some resistance regions and when a transposon is tandemly duplicated. Though homologous recombination could also incorporate a TU, Tnp26 is far more efficient. This provides insight into how IS26 builds transposons and brings additional transposons into resistance regions.
KEYWORDS: IS26, translocatable unit, transposition, transposons
ABSTRACT
The IS26 transposase, Tnp26, catalyzes IS26 movement to a new site and deletion or inversion of adjacent DNA via a replicative route. The intramolecular deletion reaction produces a circular molecule consisting of a DNA segment and a single IS26, which we call a translocatable unit or TU. Recently, Tnp26 was shown to catalyze an additional intermolecular, conservative reaction between two preexisting copies of IS26 in different plasmids. Here, we have investigated the relative contributions of homologous recombination and Tnp26-catalyzed reactions to the generation of a transposon from a TU. Circular TUs containing the aphA1a kanamycin and neomycin resistance gene or the tet(D) tetracycline resistance determinant were generated in vitro and transformed into Escherichia coli recA cells carrying R388::IS26. The TU incorporated next to the IS26 in R388::IS26 forms a transposon with the insertion sequence (IS) in direct orientation. Introduction of a second TU produced regions containing both the aphA1a gene and the tet(D) determinant in either order but with only three copies of IS26. The integration reaction, which required a preexisting IS26, was precise and conservative and was 50-fold more efficient when both IS26 copies could produce an active Tnp26. When both ISs were inactivated by a frameshift in tnp26, TU incorporation was not detected in E. coli recA cells, but it did occur in E. coli recA+ cells. However, the Tnp-catalyzed reaction was 100-fold more efficient than RecA-dependent homologous recombination. The ability of Tnp26 to function in either a replicative or conservative mode is likely to explain the prominence of IS26-bounded transposons in the resistance regions found in Gram-negative bacteria.
IMPORTANCE In Gram-negative bacteria, IS26 recruits antibiotic resistance genes into the mobile gene pool by forming transposons carrying many different resistance genes. In addition to replicative transposition, IS26 was recently shown to use a novel conservative movement mechanism in which an incoming IS26 targets a preexisting one. Here, we have demonstrated how IS26-bounded class I transposons can be produced from translocatable units (TUs) containing only an IS26 and a resistance gene via the conservative reaction. TUs were incorporated next to an existing IS26, creating a class I transposon, and if the targeted IS26 is in a transposon, the product resembles two transposons sharing a central IS26, a configuration observed in some resistance regions and when a transposon is tandemly duplicated. Though homologous recombination could also incorporate a TU, Tnp26 is far more efficient. This provides insight into how IS26 builds transposons and brings additional transposons into resistance regions.
INTRODUCTION
Most class I transposons in Gram-negative bacteria are bounded by two copies of IS26. In recent years, the precise origins of several antibiotic resistance genes have been uncovered. In two cases, the blaSHV and oqxAB genes have been picked up from the chromosome of a Klebsiella pneumoniae strain by IS26 and formed into transposons, which became part of the mobile gene pool (1, 2) (Fig. 1A). IS26 is known to move via a replicative mechanism and can use this mechanism (3, 4) to cause the deletion of sequence immediately adjacent to one of its ends (Fig. 1B) or invert adjacent DNA. The deletion reaction would create a circular product that includes a single copy of IS26 together with the deleted DNA segment. As this product is unable to replicate, it would be lost unless it was reincorporated. However, we have recently invoked this form, which we named a translocatable unit (TU), as the unit of movement for IS26-flanked transposons (5). To create a transposon, the TU would either be incorporated at a new location via replicative transposition or incorporated next to a preexisting IS26 using homologous recombination or a Tnp26-catalyzed conservative reaction as shown in Fig. 1B. We recently demonstrated that the IS26-encoded transposase, Tnp26, can catalyze a reaction involving two copies of IS26 that is conservative, i.e., neither the IS26 nor the target site is duplicated (5). It has been shown that replicative transposition occurs at least 50-fold less frequently than the IS26-targeted conservative reaction (5). However, the efficiency of the conservative reaction relative to RecA-dependent homologous recombination is not known.
FIG 1 .
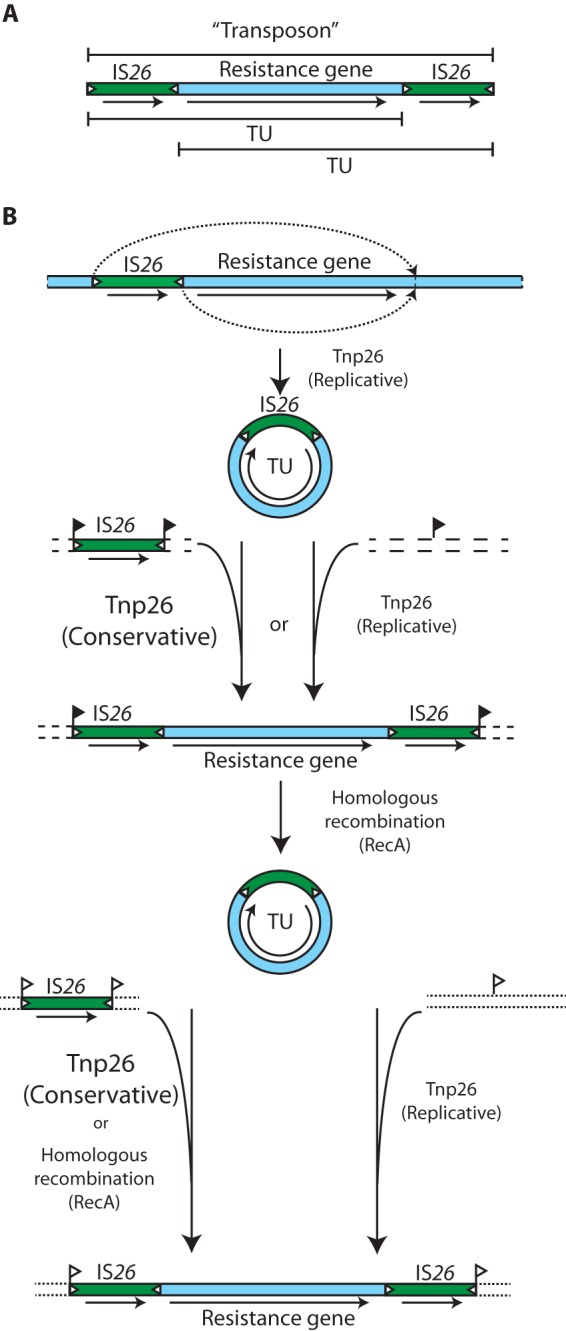
Building IS26-bounded transposons. (A) Schematic of a typical IS26-bounded class I transposon. IS26 (green box) and the 14-bp inverted repeats of IS26 (open triangles) are indicated. An arrow indicates the position and orientation of tnp26. The extent of the compound transposon and two alternate translocatable units (TUs) are shown above and below the schematic representation. (B) Two pathways to IS26-mediated formation of compound transposons. The relative frequency of the Tnp26-mediated and RecA-dependent reactions is indicated by the size of the label. Eight-base-pair direct repeats of the target sequence are indicated by a flag.
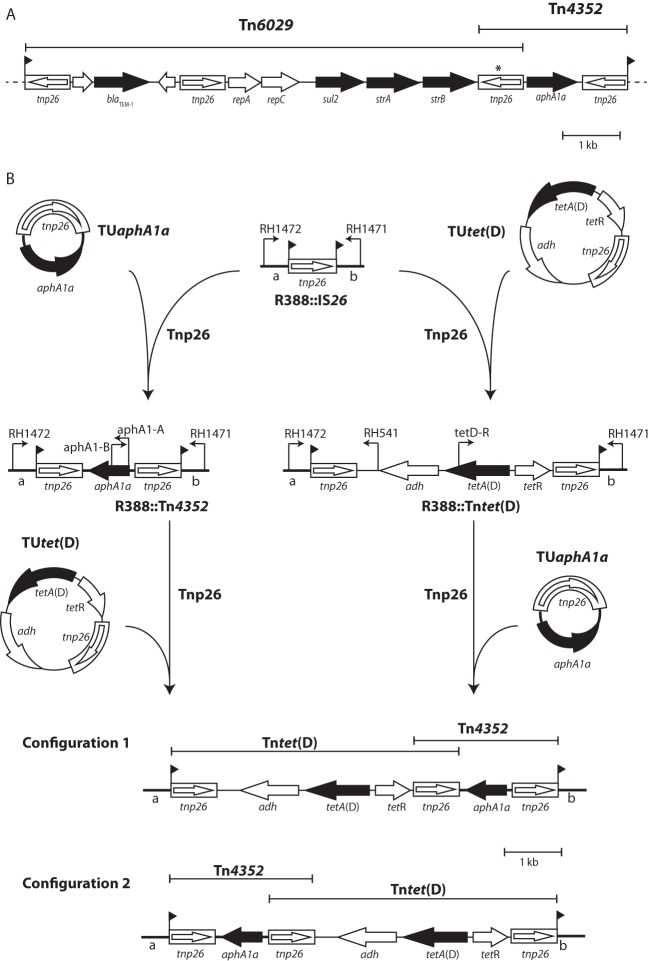
Once a transposon has been generated, it can then move to a new location via a TU intermediate (Fig. 1B, bottom). Though in rare cases Tnp26 can excise a TU, in other cases this does not occur (6), and generation of a TU from a preexisting transposon would necessarily occur via homologous recombination. Again, the TU can be incorporated next to an existing IS26 using either homologous recombination or the conservative Tnp26-catalyzed mechanism. Repetition of the incorporation process with a second TU should lead to the formation of overlapping transposons such as Tn4352 and Tn6029 as shown in Fig. 2A. This structure, found in an IncHI2 plasmid from Salmonella enterica, is flanked by a target site duplication of 8 bp (Fig. 2A) (7).
FIG 2 .
Formation of complex antibiotic resistance regions. (A) Structure of Tn6026. (B) Building transposons using TUs derived from Tn4352 and Tntet(D). The origin of each IS26-bounded structure is shown above. Genes and open reading frames are shown as arrows indicating the direction of transcription. Genes conferring antibiotic resistance are black. IS26 elements are shown as open boxes with an arrow indicating the position and orientation of tnp26.
In this study, we investigated the role of the conservative Tnp26 reaction in the generation of transposons from TUs. TUs were generated in vitro containing the aphA1a kanamycin and neomycin resistance determinant or the tet(D) tetracycline resistance determinant and used to generate transposons, which were then built into a multiple resistance region containing overlapping IS26-bounded transposons. The efficiency of the Tnp26-catalyzed reaction was compared to that of homologous recombination.
RESULTS
IS26-mediated incorporation of a TU.
To demonstrate targeted incorporation of a TU adjacent to an IS26, the circular form of TUaphA1a containing only an IS26 and a 1,040-bp fragment that includes aphA1a (Kmr) was generated in vitro from Tn4352 as described in Materials and Methods. To ensure that any reaction was catalyzed by Tnp26, and not the result of RecA-dependent homologous recombination, the TU was introduced into the RecA-deficient Escherichia coli strain UB1637 (Smr) containing R388::IS26 (contains the dfrB2 gene conferring resistance to trimethoprim [Tpr]). Transformation of cells with 1 µg of the circular TU yielded an average of 156 Kmr Smr Tpr transformants per µg of TU in four independent experiments (Table 1). UB1637 cells containing R388 devoid of IS26 were also transformed with 1 µg of TUaphA1a to determine the frequency of TU incorporation when IS26 was not present in the recipient. No kanamycin-resistant transformants were recovered from three independent experiments (Table 1), indicating that IS26 is required as a target.
TABLE 1 .
Frequency of TU incorporation in a recA background
| TU | Plasmid in UB1637 | No. of transformants/µg of TUa |
|
|---|---|---|---|
| Values for individual experimentsb | Avg value | ||
| TUaphA1a | R388::IS26 | 166, 153, 142, 162 | 156 |
| R388 | 0, 0, 0 | 0 | |
| TUtet(D) | R388::IS26 | 7, 9, 5 | 7 |
| R388 | 0, 0, 0 | 0 | |
| R388::Tn4352 | 5, 11, 12 | 9 | |
| TUaphA1a | R388::Tntet(D) | 148, 176, 161 | 162 |
Kmr Tpr transformants for TU4352 or TU4352B transformed into cells containing R388::IS26, Tcr Tpr transformants for TUtet(D) transformed into cells containing R388::IS26, Kmr Tcr Tpr for TU4352 or TU4352B transformed into cells containing R388:TUtet(D) and for TUtet(D) transformed into cells containing R388::Tn4352.
Four experiments were performed for TUaphA1a and UB1637-R388::IS26, and three experiments were performed for all other combinations of TU and plasmid.
Plasmid DNA from 15 transformants (5 from each of 3 replicates) was amplified with the R388 backbone primers RH1471 and RH1472 combined with outward-facing primers aphA1-A and aphA1-B in the aphA1a fragment (Fig. 2B). This screening confirmed that, in all cases, R388::Tn4352 had been reformed by incorporation of the TU adjacent to the existing IS26 in R388::IS26 (Fig. 2B, top). Sequencing of the PCR amplicons confirmed that the IS26 elements were always in direct orientation and that this was a precise conservative reaction such that the 8-bp direct repeats flanking the IS26 in R388::IS26 now flanked Tn4352. Primers RH1471 and RH1472 yielded an amplicon of 3.4 kb in each transformant tested, demonstrating that only a single copy of the TU had inserted adjacent to the IS26.
A second 4.3-kb circular TU derived from Tntet(D) (8) (Tcr) was incorporated into R388::IS26. Transformation with 1 µg of the TUtet(D) yielded between five and nine Tcr Tpr transformants in three independent experiments (Table 1). This frequency is lower than that observed for the incorporation of TUaphA1a, and this may reflect a lower transformation efficiency due to the larger size of TUtet(D). Again, when R388 replaced R388::IS26, no tetracycline-resistant transformants were detected in three independent experiments (Table 1). PCR screening (primers RH541 with RH1472 and primers tetD-R with RH1471 [Fig. 2B]) and sequencing confirmed that, in all cases, R388::Tntet(D) had been reformed by incorporation of the TU adjacent to the existing IS26 via a precise conservative reaction.
IS26-mediated accumulation of resistance genes.
To demonstrate IS26-mediated accumulation of resistance genes, TUtet(D) was incorporated into R388::Tn4352. Transformation of UB1637 cells containing R388::Tn4352 with 1 µg of the TUtet(D) yielded between 5 and 12 Kmr Tcr Tpr transformants in three independent experiments (Table 1). Plasmid DNA from five transformants from each of the independent experiments was amplified with the R388 backbone primers RH1471 and RH1472 in combination with outward-facing primers located within Tn4352 (aphA1-A and aphA1-B) and Tntet(D) (RH541 and tetD-R) to determine the location of the incorporated TU. In nine cases, the tet(D) TU had inserted adjacent to the left-hand IS26 of Tn4352 (Fig. 2B, configuration 1), and in six cases, it had inserted adjacent to the right-hand IS26 (Fig. 2B, configuration 2). Sequencing of the PCR amplicons confirmed that the IS26 elements were all in direct orientation and the 8-bp direct repeats flanking the IS26 in R388::Tn4352 now flanked the whole Tn4352-Tntet(D) structure.
The reciprocal experiment, in which TUaphA1a was incorporated into R388::Tntet(D), was also performed. An average of 162 Kmr Tcr Tpr transformants/µg of TU was recovered from three independent experiments. PCR screening of 15 Kmr Tcr Tpr transformants (five from each of three independent experiments) again revealed transformants with configuration 1 (eight) and configuration 2 (seven) as shown in Fig. 2B. Again, the number of transformants was approximately 20-fold higher with TUaphA1a than with TUtet(D).
Tnp26 is required for targeted TU incorporation.
The requirement for Tnp26 was confirmed by utilizing a TU and target plasmid that cannot produce active Tnp26. E. coli UB1637 (recA) cells containing R388::IS26 were first transformed with 1 µg of TUaphA1a from pRMH761 to generate a baseline frequency for the incorporation of the Tn4352B-derived TU. This yielded an average of 175 Kmr Smr Tpr transformants per µg of TU in three independent experiments (Table 2).
TABLE 2 .
Transformation of recA+ and recA mutant strains carrying R388::IS26
| TU (transposase) |
Plasmid in UB1637 | Expt no. or parameter |
No. of Kmr Tpr transformants/µg of TU for strain: |
|
|---|---|---|---|---|
| UB1637 (recA) | E294 (recA+) | |||
| TUaphA1a (Tnp26) |
R388::IS26 | 1 | 100 | 195 |
| 2 | 272 | 126 | ||
| 3 | 153 | 181 | ||
| Avg | 175 | 167.3 | ||
| TUaphA1a (Tnp26-FS-La) |
R388::IS26 | 1 | 1 | 2 |
| 2 | 2 | 0 | ||
| 3 | 0 | 0 | ||
| 4 | 1 | 2 | ||
| Avg | 1 | 1 | ||
| TUaphA1a
(Tnp26) |
R388::IS26-FS-Rb | 1 | 2 | 1 |
| 2 | 2 | 2 | ||
| 3 | 1 | 0 | ||
| 4 | 1 | 2 | ||
| Avg | 1.5 | 1.3 | ||
| TUaphA1a
(Tnp26-FS-L) |
R388::IS26-FS-R | 1 | 0 | 1 |
| 2 | 0 | 2 | ||
| 3 | 0 | 1 | ||
| 4 | 0 | 2 | ||
| 5 | 0 | 0 | ||
| Avg | 0 | 1.5 | ||
IS26-FS-L, frameshift mutation in tnp26 in the TU, producing a truncated 30-aa protein.
IS26-FS-R, frameshift mutation in R388::IS26 tnp26.
UB1637 (recA) cells containing R388::IS26-FS-R with a frameshift truncating Tnp26 were transformed with 1 µg of TUaphA1a derived from pRMH990 (6), which contains a variant of Tn4352B with a frameshift in tnp26 that introduces a premature stop codon allowing only 30 amino acids (aa) to be translated. TU incorporation was below the limit of detection, and no Kmr Tpr transformants were recovered from five independent transformations (Table 2). Hence, a complete Tnp26 is required for TU incorporation to occur.
To determine whether targeted TU incorporation is possible with only one active Tnp26, the UB1637 strain containing R388::IS26 was transformed with 1 µg of TUaphA1a that cannot produce Tnp26 due to FS-L (FS stands for frameshift mutation). This yielded a total of only four Kmr Tpr transformants from four independent experiments (Table 2). In the reciprocal experiment, recipient cells carrying R388::IS26-FS-R, which cannot produce Tnp26, were transformed with TUaphA1a from pRMH761, which can produce full-length Tnp26. Only a total of six Kmr Tpr transformants were recovered from four independent experiments (Table 2). These frequencies are >100-fold lower than when both molecules participating in the reaction can produce Tnp26. Screening of plasmid DNA from the 10 Kmr Tpr transformants recovered in these experiments showed that the TU had incorporated adjacent to the existing IS26 in all instances. This shows that while targeted TU incorporation is possible with only one active Tnp26, the frequency of incorporation is dramatically decreased compared to when both participating molecules can produce Tnp26.
Is Tnp26-mediated TU incorporation more efficient than RecA-dependent recombination?
TU incorporation via homologous recombination between two copies of IS26 should be possible. To assess the contribution of RecA-dependent recombination to TU integration, TU incorporation was explored using recombination-proficient E. coli E294 cells carrying R388::IS26. When both insertion sequences (ISs) produced an active Tnp26, an average of 167 Kmr Tpr transformants per µg of TUaphA1a were recovered (Table 2), similar to the frequency observed in recA cells. The frequency of TU incorporation was >100-fold lower (an average of 1.5 Kmr Tpr transformants per µg of TU) when both the TU and the target plasmid contained a frameshift in the tnp26 gene, i.e., when the reaction relied upon RecA-dependent homologous recombination (Table 2). Hence, while RecA-mediated homologous recombination can, as expected, lead to TU incorporation, an active Tnp26 in each participating molecule is the major contributor to targeted TU incorporation.
DISCUSSION
The formation of transposons containing resistance genes is a major force in the development of multiply and extensively resistant strains of Gram-negative bacteria. This study has demonstrated that a TU containing only an IS26 and a resistance gene preferentially inserts adjacent to an existing IS26 in the same cell, creating an IS26-bounded transposon. TUs generated from Tn4352, Tn4352B, and Tntet(D) were able to incorporate adjacent to an IS26 in R388. Incorporation of a second TU generated a resistance array with overlapping IS26-bounded compound transposons, and the 8-bp direct repeat originally flanking the IS26 in R388 now flanked the whole structure.
In complex resistance regions, the IS26-bounded compound transposons Tn4352 and Tn6029 are often seen together (7). We found that in the nine sequenced examples in GenBank, Tn4352 is always incorporated adjacent to the strB end of Tn6029 (Fig. 2A), suggesting that there may be a preference for a TU to target a particular end of an existing IS26-bounded structure. However, we detected no such preference in this study using Tn4352 and Tntet(D). The TUs generated from Tn4352 and Tntet(D) both incorporated equally well on the left and right ends of existing IS26-bounded structures. Hence, it is possible that the Tn4352-Tn6029 structure observed today originated once and has then disseminated as a single unit into multiple strains and species. Regions containing IS26-bounded transposons are increasingly being reported in other chromosomes and plasmids, though homologous recombination, rather than the Tnp26-mediated conservative reaction, is often invoked as the mechanism responsible (9). The role of TUs in disseminating genetic material is becoming more evident, with recent studies reporting the detection of circular TUs containing multiple resistance genes (9, 10).
As in our previous study using two plasmids each carrying an IS26 (5), Tnp26 was essential for TU incorporation at the site of a preexisting IS26 in a recombination-deficient background. Targeted TU incorporation could also occur via RecA-dependent homologous recombination in RecA+ wild-type cells. However, this reaction makes only a minor contribution, as it occurs at a frequency at least 2 orders of magnitude lower than the same reaction catalyzed by Tnp26. Hence, the ability of IS26 to participate in both replicative transposition and self-targeted transposition can explain the abundance of IS26-bounded transposons in the mobile gene pool. Moreover, the findings of this study should be applicable to other members of the IS6 family, including IS257 and IS1216, that play a major role in the mobilization of antibiotic resistance genes in staphylococci and enterococci, respectively.
MATERIALS AND METHODS
Plasmids and plasmid construction.
The plasmids used in this study are listed in Table 3. pRMH761, pRMH976, pRMH990, R388::IS26, R388::IS26-FS-R, R388::Tn4352, and R388::Tn4352B have been described previously (5, 6, 11). pUC19::Tntet(D) was constructed by cloning a 6,038-bp Tntet(D)-containing SacI fragment (bases 134454 to 140495 in GenBank accession number KP276584) from the A/C2 plasmid p39R861-4 (8) into SacI-digested pUC19. Tcr transformants were screened via PCR and restriction digestion with PstI and NdeI to confirm the identity and orientation of the insert.
TABLE 3 .
Plasmids used in this study
| Plasmid | Description | Resistance phenotypea |
Reference |
|---|---|---|---|
| pRMH761 | 8.8-kb BamHI fragment of pRMH760 containing Tn4352B cloned into pUC19 |
Ap Km Nm | 5 |
| pRMH976 | pRMH761 derivative containing Tn4352
in tniAb |
Ap Km Nm | 6 |
| pRMH990 | pRMH761 derivative with frameshifts in both tnp26c |
Ap Km Nm | 6 |
| R388::IS26 | R388 with IS26d | Su Tp | 5 |
| R388::IS26-FS-R | R388::IS26 frameshift mutante | Su Tp | 5 |
| R388::Tn4352B | R388 containing Tn4352Bf | Km Nm Su Tp | 5 |
| R388::Tn4352 | R388 containing Tn4352f | Km Nm Su Tp | 6 |
| p39R861-4 | Type 2 A/C2 plasmid carrying Tntet(D) | Cm Fl Su Tc | 8 |
| pUC19::Tntet(D) | 6.0-kb SacI fragment of p39R861-4 containing Tntet(D) cloned into pUC19g |
Ap Tc | This study |
Ap, ampicillin; Cm, chloramphenicol; Fl, florfenicol; Km, kanamycin; Nm, neomycin; Su, sulfamethoxazole; Tc, tetracycline; Tp, trimethoprim.
1.8-kb central SwaI Tn4352B fragment replaced with Tn4352 from pDGO100.
Frameshift generated by end filling the BsiWI site and duplicating 116 to 119 bp from the left end of IS26 as shown in Fig. 5 of Harmer et al. (5).
IS26 8-bp duplication of bases 26745 to 26752 in R388 (GenBank accession no. BR000038).
Lacks 13 bp (bases 624 to 636 from the left end of IS26).
Tn4352B together with 8-bp duplication of bases 26745 to 26752 in R388 (GenBank accession number BR000038).
Bases 134454 to 140495 from p39R861-4 cloned into pUC19.
DNA manipulation.
Plasmid DNA was isolated by alkaline lysis, digested, and gel purified as previously described (5). PCR and routine sequencing of PCR products were performed as previously described using published primers RH1471, RH1472, aphA1-A, aphA1-B, RH541, and tetD-R (5, 8, 12). The positions and orientations of the primers are marked in Fig. 2B.
In vitro construction and transformation of a TU.
Escherichia coli UB1637 (recA Smr) and E. coli E294 (Rifr) (13) were used as recipients in transformation experiments. The presence of a unique SwaI restriction site in IS26 was exploited to generate a circular TU in vitro. A similar strategy has been used successfully to study IntI-dependent insertion of gene cassettes into integrons (14). A Tn4352B- or Tn4352-derived TUaphA1a was generated in vitro by SwaI digestion of pRMH761 (Tn4352B) or pRMH976 (Tn4352) plasmid DNA extracted from cells grown overnight with kanamycin selection. The 1.8-kb fragment containing aphA1a and two partial copies of IS26 was isolated via gel extraction. One microgram of the 1.8-kb fragment was religated using 40 U of T4 DNA ligase (New England Biolabs) to form a circular TU containing aphA1a and a complete copy of IS26. Similarly, a 4.3-kb fragment was recovered from pUC19::Tntet(D) plasmid DNA digested with SwaI and religated to form TUtet(D). pRMH990, a derivative of pRMH761 with a frameshift in both copies of IS26, was similarly used to generate TUaphA1a encoding an inactive truncated Tnp26.
One microgram of ligation mixture containing the appropriate TU was electroporated into E. coli UB1637 (RecA-deficient) or E294 (RecA-proficient) recipient cells that had been rendered electrocompetent as described previously (15). Electroporation was performed using a BioRad MicroPulser electroporator according to the manufacturer’s instructions. After recovery in 1-ml Luria broth for 90 min at 37°C, transformed cells were plated onto Mueller-Hinton agar supplemented with trimethoprim (10 µg ml−1) and either kanamycin (50 µg ml−1) to select for transformants containing TUaphA1a and/or tetracycline (10 µg ml−1) to select for TUtet(D).
REFERENCES
- 1.Norman A, Hansen LH, She Q, Sørensen SJ. 2008. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60:59–74. doi: 10.1016/j.plasmid.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Ford PJ, Avison MB. 2004. Evolutionary mapping of the SHV beta-lactamase and evidence for two separate IS26-dependent blaSHV mobilization events from the Klebsiella pneumoniae chromosome. J Antimicrob Chemother 54:69–75. doi: 10.1093/jac/dkh251. [DOI] [PubMed] [Google Scholar]
- 3.Blackwell GA, Nigro SJ, Hall RM. 2015. Evolution of AbGRI2-0, the progenitor of the AbGRI2 resistance island in global clone 2 of Acinetobacter baumannii. Antimicrob Agents Chemother 60:1421–1429. doi: 10.1128/AAC.02662-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He S, Hickman AB, Varani AM, Siguier P, Chandler M, Dekker JP, Dyda F. 2015. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6:e00762. doi: 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harmer CJ, Moran RA, Hall RM. 2014. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 5:e01801-14. doi: 10.1128/mBio.01801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harmer CJ, Hall RM. 2015. IS26-mediated precise excision of the IS26-aphA1a translocatable unit. mBio 6:e01866-15. doi: 10.1128/mBio.01866-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cain AK, Liu X, Djordjevic SP, Hall RM. 2010. Transposons related to Tn1696 in IncHI2 plasmids in multiply antibiotic resistant Salmonella enterica serovar Typhimurium from Australian animals. Microb Drug Resist 16:197–202. doi: 10.1089/mdr.2010.0042. [DOI] [PubMed] [Google Scholar]
- 8.Anantham S, Harmer CJ, Hall RM. 2015. p39R861-4, a type 2 A/C plasmid carrying a segment from the A/C1 plasmid RA1. Microb Drug Resist 21:571–576. doi: 10.1089/mdr.2015.0133. [DOI] [PubMed] [Google Scholar]
- 9.Karah N, Dwibedi CK, Sjöström K, Edquist P, Johansson A, Wai SN, Uhlin BE. 2016. Novel aminoglycoside resistance transposons and transposon-derived circular forms detected in carbapenem-resistant Acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother 60:1801–1818. doi: 10.1128/AAC.02143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh H, Doijad S, Bunk B, Falgenhauer L, Yao Y, Spröer C, Gentil K, Schmiedel J, Imirzalioglu C, Overmann J, Chakraborty T. 2016. Detection of translocatable units in a blaCTX-M-15 extended-spectrum β-lactamase-producing ST131 Escherichia coli isolate using a hybrid sequencing approach. Int J Antimicrob Agents 47:245–247. doi: 10.1016/j.ijantimicag.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Partridge SR, Hall RM. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob Agents Chemother 47:342–349. doi: 10.1128/AAC.47.1.342-349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng LK, Martin I, Alfa M, Mulvey M. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes 15:209–215. doi: 10.1006/mcpr.2001.0363. [DOI] [PubMed] [Google Scholar]
- 13.Walker MJ, Pemberton JM. 1987. Construction of a transposon containing a gene for polygalacturonate trans-eliminase from Klebsiella oxytoca. Arch Microbiol 146:390–395. doi: 10.1007/BF00410941. [DOI] [PubMed] [Google Scholar]
- 14.Collis CM, Grammaticopoulos G, Briton J, Stokes HW, Hall RM. 1993. Site-specific insertion of gene cassettes into integrons. Mol Microbiol 9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 15.Gonzales MF, Brooks T, Pukatzki SU, Provenzano D. 2013. Rapid protocol for preparation of electrocompetent Escherichia coli and Vibrio cholerae. J Vis Exp 2013:e50684. doi: 10.3791/50684. [DOI] [PMC free article] [PubMed] [Google Scholar]