Abstract
Chronic skin wounds are characterized by poor re-epithelialization, angiogenesis and granulation. Previous work has demonstrated that topical stromal cell-derived growth factor-1 (SDF1) promotes neovascularization, resulting in faster re-epithelialization of skin wounds in diabetic mice. However, the clinical usefulness of such bioactive peptides is limited because they are rapidly degraded in the wound environment due to high levels of proteases. Here, we describe the development of a recombinant fusion protein comprised of SDF1 and an elastin-like peptide that confers the ability to self-assemble into nanoparticles. The fusion protein and recombinant human SDF1 showed similar binding characteristics, as indicated by the measured equilibrium dissociation constant (Kd) for the binding of free SDF1 or the fusion protein to the CXCR4 receptor. The biological activity of SDF1-ELP, as measured by intracellular calcium release in HL60 cells was dose dependent, and also very similar to that of free SDF1. In contrast, the biological activity of SDF1-ELP in vivo was significantly superior to that of free SDF1. When applied to full thickness skin wounds in diabetic mice, wounds treated with SDF1-ELP nanoparticles were 95% closed by day 21, and fully closed by day 28, while wounds treated with free SDF1, ELP alone, or vehicle were only 80% closed by day 21, and took 42 days to fully close. In addition, the SDF1-ELP nanoparticles significantly increased the epidermal and dermal layer of the healed wound, as compared to the other groups. These results indicate SDF1-ELP fusion protein nanoparticles are promising agents for the treatment of chronic skin wounds.
Keywords: Stromal cell-derived factor-1 (SDF1), elastin like peptides (ELP), nanoparticles, wound healing, skin
Graphical Abstract
1. Introduction
In the United States, approximately $25 billion are spent annually on the treatment of chronic skin wounds [1], which are characterized by prolonged and excessive inflammation [2], and are prone to recurrent infections [3]. High levels of proteolytic activity degrade endogenous growth factors [4], resulting in poor angiogenesis, granulation and re-epithelialization [5]. Although this could be remediated by the application of exogenous growth factors at the wound site, potential therapeutic peptides are inactivated by the same proteolytic mechanisms.
Stromal cell-derived factor1 (SDF1), a key mediator of the wound healing response, has been reported by several literature sources to recruit endothelial progenitor cells that proliferate and differentiate into mature vascular endothelium [6, 7], which contributes to the revascularization which is needed to support re-epithelialization [8-10]. Others have suggested that are potentially many different cellular targets for SDF1 [11], making its role in the wound healing process more complicated.
Repeated high doses of topical SDF1, although costly and impractical, can achieve therapeutic efficacy [12]. Methodologies to increase SDF1 stability in vivo include mutating its protease cleavage sites [13-15], engineering derivatives of the molecule with a better stability profile [16, 17], [18] and incorporating it into biomaterials [19-21] or liposomes [22]. Nonetheless, all of these methods require expensive procedures for producing and purifying recombinant proteins.
Elastin-like peptides (ELPs) are nonimmunogenic, non-pyrogenic and biologically compatible [23] derivatives of tropoelastin with pentapeptide repeats of Valine-Proline-Glycine-(Xaa)-Glycine, where Xaa can be any natural amino acid except Proline. ELPs reversibly aggregate into nanoparticles and become insoluble above a transition temperature. ELPs can be also be expressed as fusion proteins together with a wide range of bioactive peptides, in which case they can be used as an inexpensive way to purify the protein [24],[25],[26]. ELP-based fusion proteins, by forming nanoparticles, have been shown to protect biomolecules from proteolysis [27], and can act as “drug depots” that supply the biomolecules over an extended period of time [23]. Here, we describe the development and characterization of an SDF1-ELP fusion protein that exhibits in vitro activity similar to that of free SDF, but is much more effective in vivo.
2. Materials and Methods
2.1. Cloning of SDF1-ELP
SDF1-ELP is a fusion protein which consists of the human growth factor SDF1 and an elastin-like peptide (ELP). A pET25B+ plasmid with 50 pentapeptide repeats of ELP as described by Koria et al [28] was used for cloning. The plasmid was obtained from the Center Engineering in Medicine, Massachusetts General Hospital. Sequencing of the plasmid by Genewiz Inc. indicated the presence of XbaI and NdeI restriction sites at the N-terminus of the ELP in the plasmid. SDF1 was fused to ELP via these 2 restriction sites. The NdeI and XbaI restriction enzymes and DNA ligation kit used for cloning were obtained from Thermo Fisher Scientific. The SDF1 gene string used was designed and ordered from Life Technologies as follows: GGCACCTCGATTAGTTCTCGTCTAGAATGAATGCGAAAGTCGTTGTCGTGCTGGTGTTGGTCTTAACTGCACTGTGTTTGTCTGATGGTAAACCGGTGAGTCTTTCGTACCGTTGCCCGTGCCGTTTCTTCGAATCACATGTTGCTCGCGCGAACGTGAAACACCTGAAAATTTTGAATACGCCGAATTGCGCACTGCAGATTGTGGCGCGTCTGAAAAACAATAACCGCCAGGTATGCATCGACCCTAAACTGAAGTGGATTCAAGAATATCTTGAAAAAGCACTTAACAAAGGTGGGGGTGGCTCTGGGGGCGGTGGTTCCGGAGGTGGTGGATCACATATGGAGTTCATGCGCTTCAAGGT
The SDF1 Gene string™ was amplified using Pfu Ultra II (Agilent Technologies) and Kapa Hifi (Kapa Biosystems) polymerases. Sequencing of the final cloned product was performed by Genewiz Inc. Subsequent to the successful cloning, site directed mutagenesis was performed by GenScript on the pET25B+ vector with SDF1-ELP, to bring an out of frame His tag on the plasmid in frame. The final cloned product, SDF1-ELP in pET25B+ vector, with an “in-frame” His tag was confirmed by Genewiz Inc.
2.2. Expression of SDF1-ELP Fusion Protein
The pET25B+ vector with SDF1-ELP was retransformed in E. coli (BL21 Star DE3), which was obtained from Invitrogen by Life Technologies. One bacteria colony was picked for an overnight culture in 5 ml LB medium containing 25μg/mL carbenicillin. The overnight culture was used to inoculate 500 mL of terrific broth supplemented with 100 mM of L-proline (Fisher Scientific) and with 25μg/mL carbenicillin. The culture was monitored until it reached an optical density at 600 nm of about 0.6, after which 0.5 mM of IPTG (Sigma) was added to induce the protein. The culture was left overnight. The next day, the culture was centrifuged at 3000 x g, and the pellet diluted with 40 ml of PBS and the suspension sonicated twice on ice for 9 min in cycles of 10 s on and 20 s off. Poly(ethyleneimine) solution (Sigma Aldrich) was added to a final concentration of 0.5% w/v to remove residual DNA, and after centrifuging, SDF1-ELP protein transition to nanoparticles was induced with the addition of 1M NaCl and warming to about 40°C.
2.3. Purification of SDF1-ELP Fusion Protein
2.3.1. Using ELP Inverse Transition Temperature Cycling
The inversion temperature of SDF1-ELP protein was obtained by warming up a sample of protein from 20°C to 50 °C while observing the change in optical density in a spectrophotometer (Spectramax, Molecular Devices), and was determined to be ~35°C. SDF1-ELP protein was purified by warming the protein to 40°C, thus inducing aggregation, centrifuging at the same temperature, and then resuspending the pellet in PBS at 4°C, thus disaggregating the particles. Two rounds of temperature cycling were used and the final SDF1-ELP nanoparticles were obtained by warming up the purified protein above its inverse temperature to ~40°C. For control studies, the ELP protein alone, and another fusion protein, KGF-ELP [28] were expressed and purified similarly.
2.3.2. Using Nickel NTA Chromatography
We also purified SDF1-ELP using traditional nickel NTA chromatography using the protocol described in the Qiagen® Ni-NTA Spin Kit Handbook. The imidazole used to prepare the different buffers needed for purification was from Thermo Fisher Scientific, while the benzonase endonuclease was obtained from Merck KGaA. Lysosyme was obtained from Thermo Fisher.
2.4. Physical Characterization
2.4.1. SDS-PAGE
An 8–16% Mini-PROTEAN® TGX™ 10 well, 50 μl Gel from Bio-Rad Laboratories, Inc. was used in a Bio-Rad Mini Protean Tetracell. All relevant reagents for the assay were obtained from Bio-Rad. SDF1-ELP Protein in 1X PBS buffer was diluted with loading buffer and run under native conditions on the gel.
2.4.2. Western Blot
SDS-PAGE gels were transferred onto a nitrocellulose membrane (Bio-Rad Laboratories), blocked with blotting-grade blocker (Bio-Rad Laboratories), treated with anti-human SDF1 (Peprotech) and incubated overnight at 4°C. After thorough washing with TBST, goat anti-rabbit IgG HRP (Abcam) was added. The blots were rinsed, exposed to ECL substrate and exposed to film to detect the positive SDF1 bands.
2.4.3. Circular Dichroism
A Circular dichroism spectrometer (Model 420SF) was used to obtain secondary structure information on SDF1-ELP. The equipment was run at 4°C, and a CD signal obtained for wavelengths between 190 and 260 nm. In separate experiments, CD signals were also obtained for ELP and for SDF1. The raw CD signal was corrected for concentration of the individual proteins (SDF1-ELP: 15μM; SDF: 25μM; ELP: 4μM) and path length of the cuvette.
2.4.4. Particle Size and Charge
SDF1-ELP in PBS (~50 μM) was used to measure particle size in a Zetasizer Nano series (Malvern, Piscataway, NJ) set to 37°C. Gold nanoparticles (100 nm; Sigma Aldrich) was used for calibration. Particles were put on a 200 mesh Lacey Carbon Copper TEM Grid (SPI Supplies/Structure Probe Inc.) and transmission electron micrographs (TEM) images were obtained on a Topcon (Piscataway, NJ) microscope. SDF1-ELP in PBS (~20 μM) was used to measure nanoparticle charge in a Zetasizer Nano-ZS (Malvern).
2.5. Binding Activity
A Biacore™ T200 was used to measure binding affinity of SDF1-ELP to CXCR4. SDF1-ELP, free SDF1 (Peprotech) and ELP were mobilized on different channels on a Series S Sensor Chip CM4 (General Electric). The first channel on the sensor chip was left blank and used as a reference. The chip temperature was set to 37°C. Kinetic experiments were done with 5 different concentrations of recombinant human CXCR4 (Creative Biomart) diluted in PBS (0.74nM to 60nM). Sensograms obtained for SDF1-ELP and SDF1 were subtracted from the reference channel signal and the curves were fitted to a one-site interaction model using the Biacore T200 software.
2.6. Biological Activity - Calcium Flux Assay
The bottom of 12-well plates was coated with 125μL of fibronectin solution (200μg/mL in PBS and 1000X Pluronic® F-68; Sigma) each. HL-60 cells were washed in Hanks buffered saline solution supplemented with calcium and magnesium (HBSS+; Life Technologies). The cells were then suspended to 106 cells/mL in HBSS+, incubated with 4μM fluo-4 acetoxymethylester (AM) for 45 min at 37°C, washed again in HBSS+, and plated at a density of 5×105 cells/well on the fibronectin-coated wells. Cell were allowed to attach to the plates for 15 min at 37°C, unattached cells were aspirated, and 250 μL HBSS+ added. Background images were taken using an Olympus IX81® microscope, the HBSS+ was removed, and replaced with 250 μL of SDF1-ELP, free SDF1, ELP alone, or KGF-ELP. Images were taken for the next 3.5 min, the test solutions removed and replaced with 1μg/mL ionomycin (EMD Millipore), and imaged again for 3.5 min. Fluorescence intensity was quantified on the digital images by ImageJ software (NIH) after background was subtracted.
2.7. Nanoparticle versus Monomeric Activity of SDF1-ELP
To investigate whether biological activity resides in the nanoparticle vs. the monomeric form of SDF1-ELP, SDF1-ELP at a concentration of 8μM in 500 μL PBS was warmed up to 40°C to initiate nanoparticle formation and pipetted into a 1.5mL Nanosep® and Nanosep MF centrifuge tube with a 10nm nominal pore size (Pall Corporation). The tube was centrifuged at 5000 x g for 5 min at 40°C to separate monomers (which end up in the filtrate) from nanoparticles (which remain on top of the membrane). 600 μL of the filtered SDF1-ELP monomer, or SDF1-ELP nanoparticles, made to a concentration of 100nM in HBSS+ were used as the test solutions, with a control group using unfiltered SDF1-ELP (100nM).
2.8. Stability studies in Elastase
SDF1-ELP and SDF1 (both at 10 μM) were incubated with ~1 μM of elastase (197 units / mg protein; Sigma) at 37°C for 12 days. Samples were taken on day 0, 4, 8 and 12 and subjected to western blot analysis as explained in Section 2.4.2
2.9. Animal Studies
2.9.1. Diabetic mice wound assay
Animal studies were conducted in accordance with a protocol approved by the Rutgers University Institutional Animal Care and Use Committee (IACUC). Genetically modified diabetic mice (BKS.Cg-Dock7m +/+ Leprdb/J) were ordered from Jackson Laboratory and were used at the age of 10 weeks. On the day before surgery, the back of mice was shaved and depilated using clippers and Nair™ cream, followed by thorough rinsing with water. On the day of surgery (the next day), the mice were put under isoflurane anesthesia and betadine scrub (Purdue Products) and 70% ethanol were applied alternatively to prepare the dorsal skin area for surgery. Wounds were created by excising a 1 cm × 1 cm square of full thickness skin on the back the mice, using a pre-made template. Test solutions (SDF1-ELP, SDF, ELP and plain PBS) were prepared in fibrin gels to prevent them from leaking away when pipetted on the wound area. Fibrin gels were prepared as previously described [28]. Briefly, SDF1-ELP, SDF1, ELP, and plain PBS were mixed with 6.25 mg/mL of fibrinogen (Sigma Aldrich). The SDF1-ELP and ELP were incubated at 37°C for 1 hour to initiate particle formation. Prior to application to the wound, 120 μL of the individual fibrinogen with treatment solution was mixed with 30 μl of thrombin (12.5 U/mL, Sigma Aldrich). The mixture was immediately applied to the wound and allowed to gel for up to 2 min, after which the wounds were covered with Tegarderm™ (3M) and secured using sutures (Henry Schein). The wound was monitored over a period of 42 days. Digital photographs were captured weekly, and compared to the initial photographs using Image J (NIH). The wound closure percentage was calculated as .
2.9.2. Wound tissue histology
On post-wounding day 42, all the animals were sacrificed and the wound area excised. The tissues were placed in a surgical casket and fixed in 10% formalin (VWR) for 24 hours after which they were transferred to a jar with 70% ethanol and stored at 4°C. For histology, tissues were embedded in paraffin and thin sections were stained with picosirius red to visualize collagen deposition as well as morphological features of the skin. Image J was used to determine the epidermal and dermal thickness. Values shown are averages of two different tissue sections per group, with three 4x magnification fields evaluated per section.
2.10. Statistical Analysis
Statistical comparisons were performed using KaleidaGraph software. The Fisher Least Significant Difference was used to analyze the data from two independent groups, after performing a one way ANOVA. A p-value <0.05 is considered statistically significant. A p-value of <0.05 is represented by a star (*) on the graphs while a p-value of < 0.01 is represented by two stars (**) or by two plusses (++) on the graphs; both are considered statistically significant.
3. Results
3.1. Cloning of SDF1-ELP
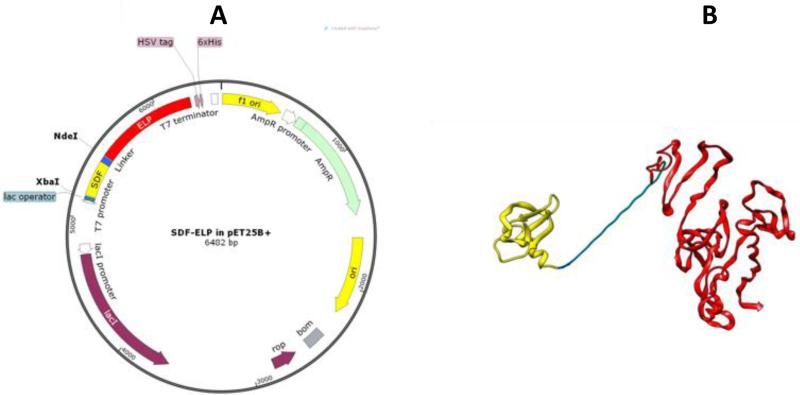
ELP was fused to the C-terminus of SDF1 via a linker sequence motif comprising of three repeats of four glycines and 1 serine (G4S)3 as shown in Fig. 1A. This relatively long linker (total of 15 amino acids) allows for a wide separation between the ELP chain and the binding region of SDF1, which is located on residues 1-9 at the N-terminus [29]. The pET25B+ expression plasmid used for the SDF1-ELP cloning is shown in Fig. 1B. After cloning, the plasmid and SDF1-ELP were mutated to bring a 6X Histidine tag in frame, to allow it to be used for epitope detection and as a purification tag.
Fig. 1.
Design and cloning of SDF1-ELP. (A) Cloning of SDF-ELP was done using a peT25B+ expression vector. SDF1 was fused to ELP using the XbaI and NdeI restriction sites. The plasmid with SDF1-ELP was mutated to put a 6X Histidine tag in frame with the protein sequence. The plasmid diagram was obtained using SnapGene® software (from GSL Biotech; available at snapgene.com). (B) Pymol rendition of SDF1-ELP. SDF1 (in yellow) is separated from ELP (red) by a linker (in blue) comprising 3 repeats of 4 glycines and 1 serine. SDF1 monomer sequence was extracted from Ryu et al.[30] (RSCB Protein Data Bank ID: 2J7Z). The ELP portion was modeled using I-TASSER software [31],[32],[33].
3.2. Purification and Characterization of SDF1-ELP
3.2.1. SDS-PAGE and Western Blot
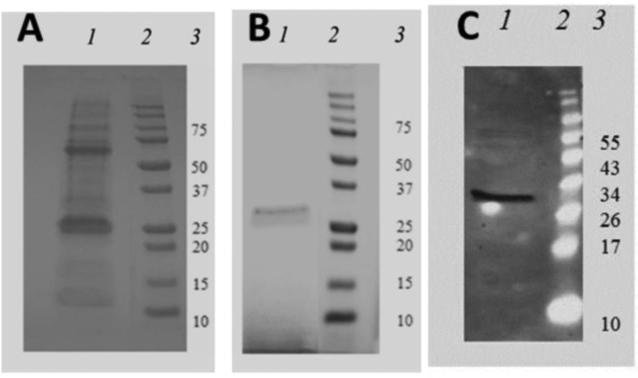
The bacterial lysate containing the SDF1-ELP product was initially separated using the histidinetag (6-His) on a nickel-NTA column (Fig.2A). The final product revealed multiple bands. We also used the ELP-dependent aggregation property; after two rounds of temperature cycling above and below the inverse temperature of 35°C, a single band was observed (Fig.2B). The purified protein by inverse temperature cycling was stained with a monoclonal anti-SDF1 antibody, showing a clear band at ~31 kDa (Fig.2C), consistent with the predicted molecular mass of SDF1-ELP.
Fig. 2.
Comparative purity assessment of SDF1-ELP by SDS-PAGE. (A) Representative gel image of protein purified using Nickel NTA Column. (B) Representative gel image of protein purified using inverse temperature cycling. Lane 1 is the profile of the purified protein; Lane 2 is the molecular weight (MW) ladder and lane 3 is the corresponding identification of the MW marker. (C) Representative gel image shown when the purified SDF1-ELP protein using the inverse temperature cycling is analyzed by Western blot. Lane 1 is the SDF1-ELP band at 31 kDA. Lane 2 is the molecular weight ladder, and Lane 3 is the corresponding identification of the MW marker.
3.2.2. Circular Dichroism (CD)
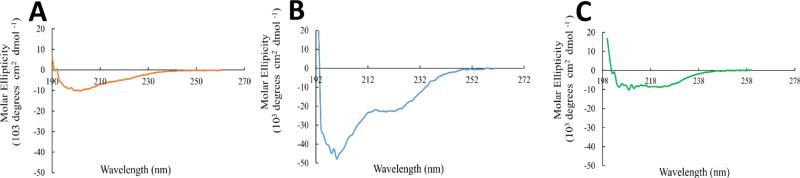
CD spectra were obtained for SDF1, ELP and SDF1-ELP to ascertain if secondary structure of SDF1 was retained in the fusion protein. Fig. 3 shows representative CD spectra for the 3 molecules. The result with SDF1 is consistent with a mixture of α helices, β sheets and random coils as depicted by Ryu et al [30]. In the case of ELP, a highly disordered structure is observed based on the very negative dip in the spectrum at around 205nm. For the SDF1-ELP, the spectrum suggests the presence of α helices and appears to have less random coils, thus at least some aspects of the secondary structure of SDF1 is preserved when fused to ELP.
Fig. 3.
CD spectra of (A) SDF1, (B) ELP and (C) SDF1-ELP. The raw CD spectra of the molecules were subtracted from their respective buffers, and normalized to the path length of the cuvette and their respective concentrations (SDF1-ELP: 15μM; SDF: 25μM; ELP: 4μM). CD signals with CD Dynodes above 700 were not included.
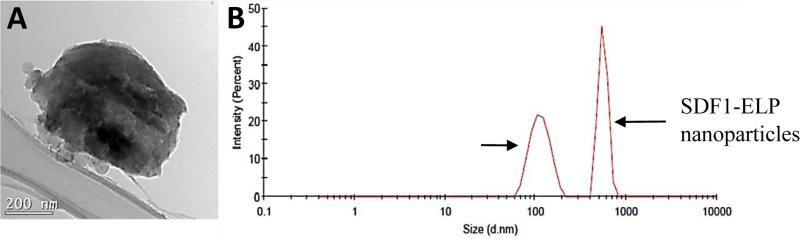
3.2.3. Particle Size and Charge
TEM images of the nanoparticles show a size of approximately 600 nm, which is corroborated with particle sizing data of 560 ± 28 nm obtained from the Zetasizer. The net charge on the protein surface was measured as approximately +3 mV.
3.3 Binding and Biological Activity
3.3.1 CXCR4 Receptor Binding Studies using Surface Plasmon Resonance
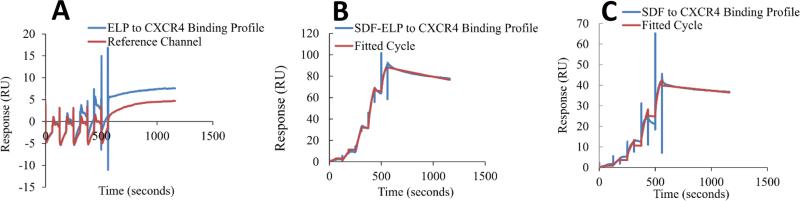
Binding affinity of the fusion protein to CXCR4 was compared to that of free SDF1, as well as ELP alone by surface plasmon resonance. ELP alone exhibited very little to no binding to CXCR4 as compared to the blank reference channel, as would be expected (Fig. 5A). SDF1-ELP bound to CXCR4 with a dissociation constant (KD) estimated to 1.14 nM (Fig. 5B). The binding of free SDF1 to CXCR4 yielded a KD of 0.3 nM (Fig. 5C). By way of comparison, values reported in the literature for SDF1 range from 1.32 to 6nM [34-36].
Fig. 5. Surface plasmon resonance analysis of CXCR4-SDF1 binding.
SDF1-ELP, SDF1 and ELP were captured on different channels of a CM4 chip and the binding of different concentrations of CXCR4 (0.74 nM to 60 nM) to the individual proteins was measured in a single kinetic experiment. The first channel was left blank and used as reference channel. The reference channel subtracted binding curves were fitted to a one site interaction model. Each step on the sensogram represents binding to a specific concentration of CXCR4, starting from the smallest concentration (0.74 nM) and ending at the highest concentration (60 nM). (A) Sensogram for free ELP and the reference channel, where no binding is expected to occur. N=3. (B) SDF1-ELP binding to CXCR4 fitted sensogram. N=3. (C) Representative binding of free SDF1 to CXCR4 fitted sensogram.
3.3.2 Calcium Flux Study
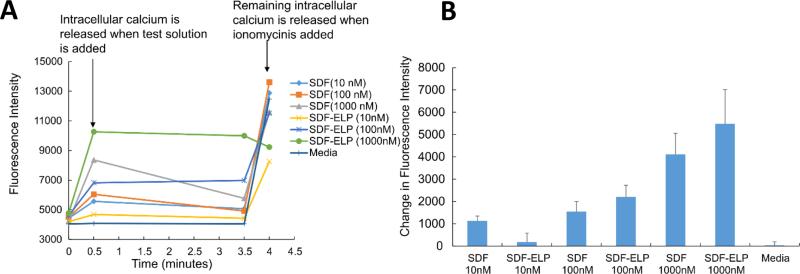
To characterize the biological activity of SDF1-ELP, we measured its effect on intracellular calcium release in HL60 cells. A dose response of SDF1-ELP and free SDF1 in the range of 100 to 1000 nM for each was performed on HL60 cells preloaded with the cytosolic calcium ion sensitive dye Fluo 4. We noted that SDF1-ELP at 1000nM caused the highest intracellular calcium release (Fig. 6). Furthermore, SDF1-ELP at 100nM and 1000nM exhibited slightly higher responses compared to free SDF1 at the same concentrations.
Fig. 6. Dose response using SDF1-ELP (and SDF1) on intracellular calcium release as measured by Fluo-4 in HL60 cells.
(A) Time course of cell manipulations and Fluo-4 fluorescence intensity. (B) Fluo-4 fluorescence measured 30 s into the assay minus fluorescence measured at the starting point. N=6.
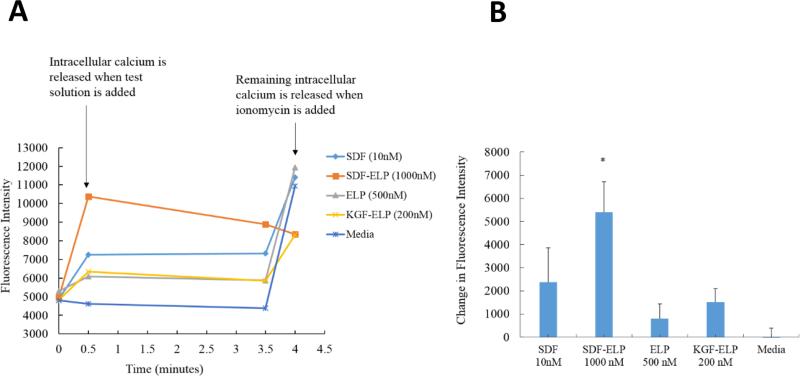
We then directly compared the effect of SDF1-ELP at two representative doses (the highest response of SDF1-ELP - 1000nM; a typical dose of SDF1 used in the literature [37] – 10 nM) to several negative controls, namely ELP alone, KGF-ELP, and plain medium. As shown in Fig. 7, ELP and KGF-ELP triggered a small rise in calcium levels and plain medium had no effect at all. In addition, 1000nM SDF1-ELP caused a significantly higher intracellular calcium rise as compared to 10 nM SDF1 as well as any of the negative controls.
Fig. 7. Effect of SDF1-ELP, SDF1 and plain medium on intracellular calcium release as measured by Fluo-4 in HL60 cells.
(A) Time course of cell manipulations and Fluo-4 fluorescence intensity. (B) Fluo-4 fluorescence measured 30 s into the assay minus fluorescence measured at the starting point. N=6. (*: p < 0.05, one way ANOVA, Fisher's LSD post-test).
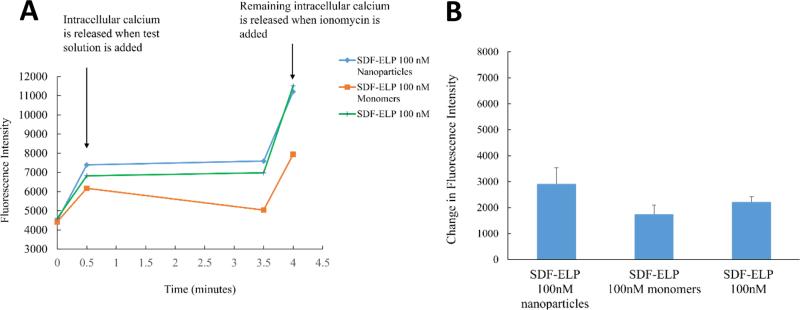
SDF1-ELP preparations are expected to contain primarily SDF1-ELP nanoparticles that are at equilibrium with monomers. To attempt to determine whether biological activity was primarily in the monomeric vs. nanoparticle fractions, we separated freshly aggregated SDF1-ELP by filtration through a 10nm pore membrane, and compared the calcium rise triggered by the filtrate (assumed to mainly consist of SDF1-ELP monomers) vs. the material on top of the membrane (assumed to mainly consist of SDF1-ELP nanoparticles). A higher calcium release was obtained from the top fraction containing the SDF-ELP nanoparticles, as compared to the bottom fraction containing the SDF1-ELP monomers although the difference was not statistically significant. (Fig. 8).
Fig. 8. Comparison of the effect of SDF1-ELP nanoparticles versus SDF1-ELP monomers on intracellular calcium release as measured by Fluo-4 in HL60 cells.
(A) Time course of calcium concentration as measured by Fluo 4AM fluorescence. (B) Fluo-4 fluorescence measured 30 s into the assay minus fluorescence measured at the starting point. N=6.
3.4 Stability Studies in Elastase
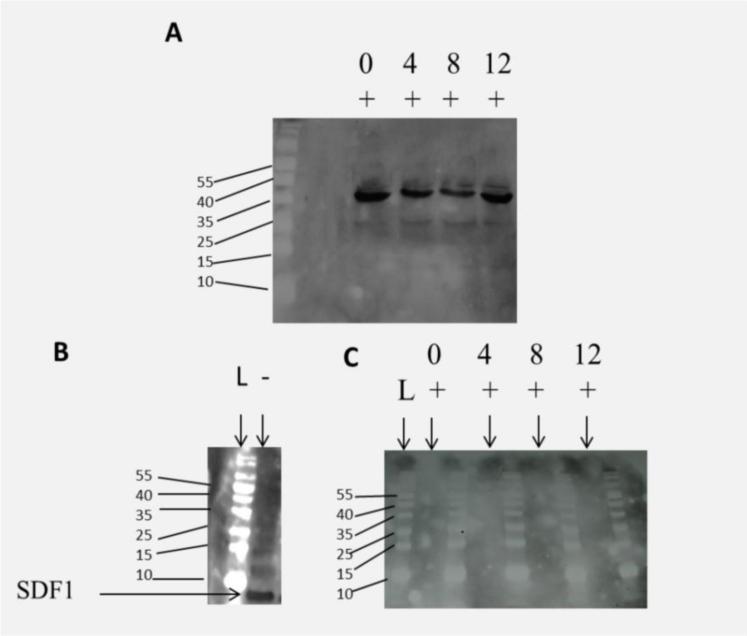
To investigate the stability of SDF1-ELP in elastase, one of the proteases that are known to degrade SDF1 in vivo [38], we incubated SDF1-ELP and SDF1 in elastase over a period of 12 days. Samples collected at 4 day intervals were subjected to a Western blot analysis. We noted that SDF1-ELP remained intact throughout the incubation period (Fig (9A), while no positive bands were seen for the SDF1 samples (Fig 9C).
Fig. 9.
Degradation of SDF-ELP or SDF by elastase. SDF1-ELP and SDF1 were incubated in elastase over a 12 day period. Samples were pulled at 4 day intervals and subjected to Western blot analysis. (A) Representative blot of SDF1-ELP samples after incubation in elastase. (B) Lane 1, labelled L is the molecular weight ladder. Lane 2, labelled (−) is SDF1 with no elastase. (C) Representative blot of SDF1 samples in elastase. No SDF positive bands are seen in any of the lanes.
3.5 In Vivo Activity
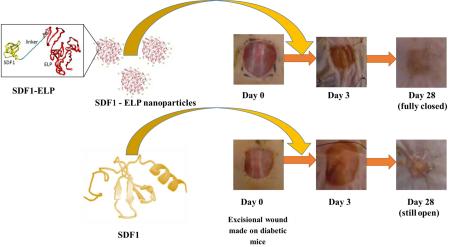
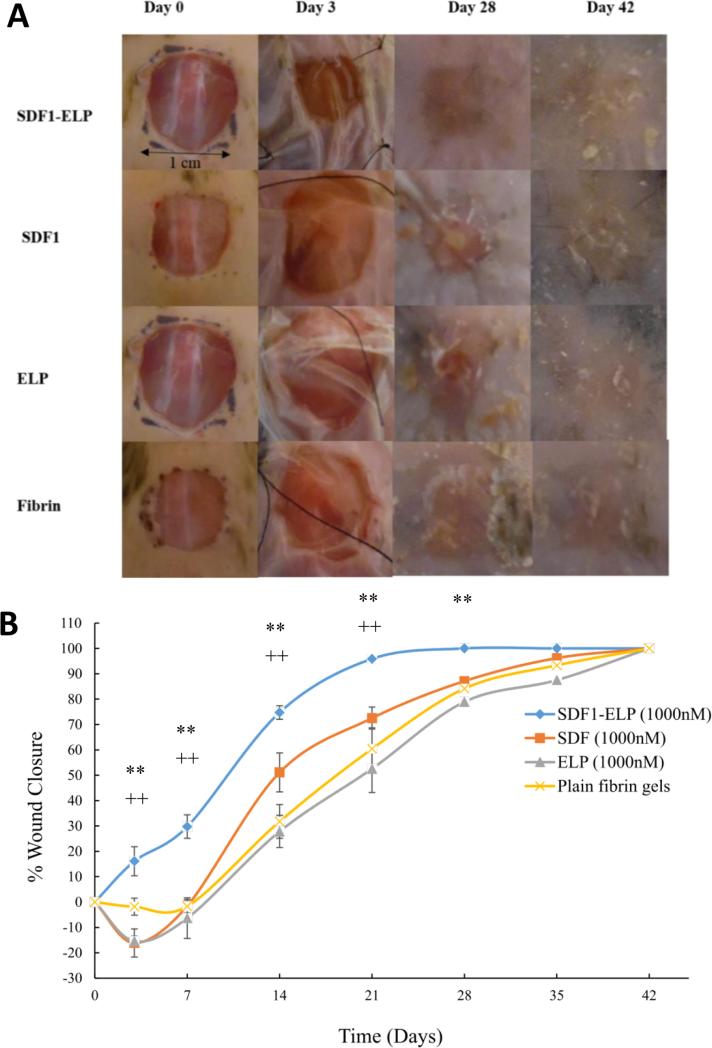
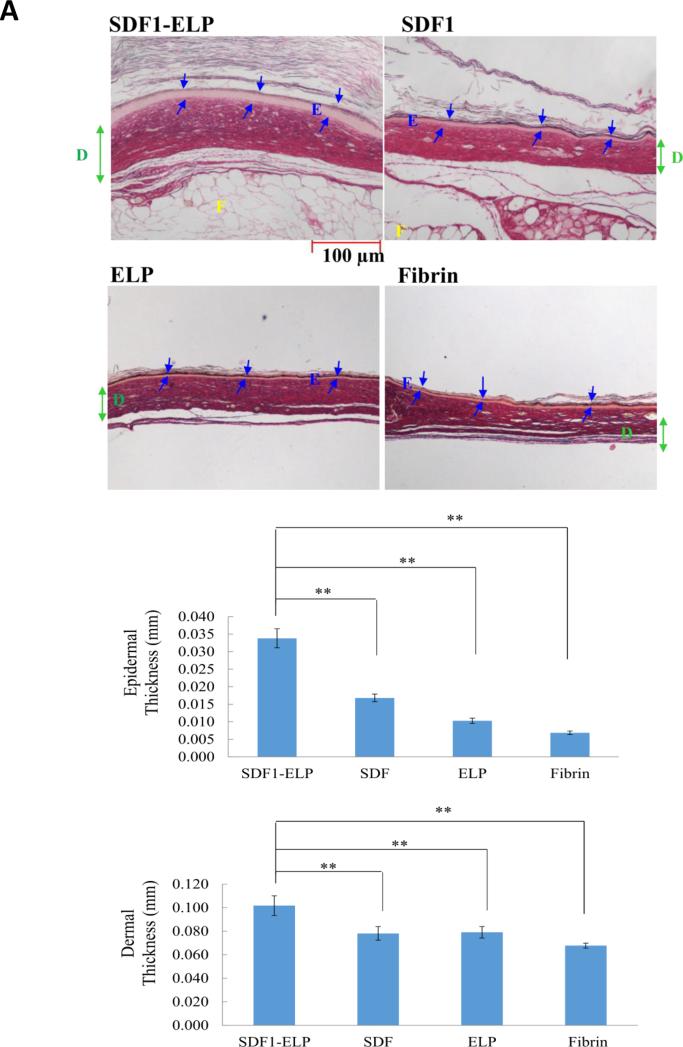
The bioactivity of SDF1-ELP was tested in vivo using a diabetic mouse model. Excisional wounds (1 × 1 cm2) were created on the back of diabetic mice and were treated with 1000nM SDF1-ELP nanoparticles in fibrin gels, 1000nM of free SDF1 in fibrin gels, 1000nM ELP nanoparticles in fibrin gels, or fibrin gels with plain medium (used as vehicle control). The closure of the wound was monitored over a period of 42 days. We noted that the wounds treated with SDF1-ELP were more closed than any other group at all time points of observation (Fig. 10). In fact, by postwounding day 21, the SDF1-ELP treated wounds were about 95% closed, and 100% closed on day 28, while the mice in the remaining groups did not fully close until day 42. Although the SDF1 group exhibited a trend towards faster closure at postwounding day 14, this group was essentially the same as the ELP and vehicle control at days 21 and beyond. The ELP and vehicle control groups followed closely each other during the entire study. Wound tissues harvested and stained on day 42 exhibited a continuous epidermis, confirming wound closure in all groups. However, both the epidermis and dermis were significantly thicker in the SDF-ELP group (Fig. 11).
Fig. 10. Effect of SDF1-ELP on skin wound closure in diabetic mice.
Full-thickness excisional wounds were treated with fibrin gel with SDF1-ELP particles, fibrin gel containing free SDF1, fibrin gel containing ELP particles or plain fibrin gel (vehicle control). (A) Representative images of the wounds on different days. On postwounding day 28, the wound treated with SDF1-ELP was fully closed, while in the other groups it was still open, only fully closing by day 42. (B) Quantified wound closure as a function of time. N = 5. (** and ++: p <0.01, one way ANOVA, Fisher's LSD post-test; (++) = SDF1-ELP compared to SDF1, (**) = SDF1-ELP compared to ELP or plain fibrin).
Fig. 11. Morphology of wounds excised on post-wounding day 42.
(A) Wounds were stained with the collagen stain picosirius red, which also makes it easy to identify the main skin layers. Structures are labeled as: E= epidermis; D= dermis; F = fat. Representative images are shown. Blue arrows represent epidermal layer thickness. Green arrow represents dermal layer thickness. (B, C) Thickness of the epidermis and dermis, respectively, as quantified by ImageJ. Values shown are averages of two (2) different tissue sections per group, with three (3) 4x magnification fields evaluated per section (N = 10). (**: p <0.01, one way ANOVA, Fisher's LSD post-test)
4. Discussion
In this study, we generated a fusion protein with SDF1 and ELP domains that forms nanoparticles of ~600 nm in size above its inverse transition temperature. We verified that SDF1-ELP binds the SDF1 receptor CXCR4 with similar affinity compared to free SDF1, and that the in vitro biological activity (intracellular calcium release) of SDF1-ELP is very similar to that of free SDF1 when using HL60 cells as responders, which express CXCR4 [39]. When applied to excisional wounds on the back of diabetic mice, the SDF1-ELP nanoparticles significantly accelerated wound closure as compared to free SDF1, ELP alone, or vehicle. Wounds treated with SDF1-ELP nanoparticles closed around 21 days post wounding, representing a 50% decrease compared to the other groups, which required up to 42 days to fully close. Furthermore, the SDF1-ELP treated wounds healed with a significantly thicker epidermal and dermal layer as compared to the other groups.
Previous work has demonstrated the ability of topically applied recombinant SDF1 to promote wound healing in experimental animals when used in high and repeated doses [12]. This is however impractical and very costly [40]. Our goal was to design an SDF1 derivative that would have similar activity compared to pure recombinant SDF1, but would also have a simpler purification process (thereby reducing manufacturing costs), and could be used as a substitute for SDF1 therapeutic applications. For this purpose, we used a fusion protein approach whereby ELP was chosen as the fusion partner because the ELP portion has a tendency to self-assemble above a transition temperature to form nanoparticles that can be separated by simple centrifugation [26], [28], [41],[42]. Thus, the SDF1-ELP fusion protein can be purified using a non-chromatographic, but thermally driven, method based on the phase transition property of ELP. We fused the ELP at the C-terminus of SDF1 with an intervening 15 amino acid residue linker to limit potential interference of the long ELP chain length on the activity of the SDF1 binding domain which is known to be at the N-terminus [29]. We chose a 50 pentapeptide repeat for the ELP sequence motif based on previous work which showed that this elastin cassette has an inverse transition temperature which is lower than physiological [28]; in fact we measured an inverse temperature of about 35°C, thus ensuring that the majority of the protein is in nanoparticle form in the wound.
The SDF1-ELP nanoparticles bound the CXCR4 receptor with high affinity, with a KD =1.14nM, which is close to the reported values for free SDF1 ranging from 1.32 to 6nM [34-36]. The biological activity of SDF1-ELP, as measured by intracellular calcium release in HL60 cells, was dose dependent and very similar to that of free SDF1. Furthermore, we noted that the system was not saturated and more calcium was released when the concentration of either SDF1 or SDF1-ELP was increased to 1000nM. Similar observations have been reported in calcium imaging studies of cardiomyocytes stimulated with SDF1, where a saturation of the response did not occur until SDF1 concentration reached 5000 nM [43]. Based on the low KD for the binding to CXCR4, we would however expect the CXCR4 receptors to be about 99% bound in the presence of 100 nM SDF1 or SDF1-ELP. It is therefore likely that other processes besides ligand-receptor binding, such as endocytosis and further intracellular processing of the SDF1-CXCR4 complexes play a quantitative role in mediating the cellular response [44].
While the binding and cellular effects of SDF1-ELP were similar to that of SDF1 in vitro, SDF1-ELP significantly outperformed SDF1 in vivo. The wounds treated with SDF1-ELP were about 95% closed by postwounding day 21, while those treated with SDF1 were only about 70% closed. By day 28, the wounds treated with SDF1-ELP were 100% closed, while those treated with free SDF1 were only about 80% closed. Wound treated with SDF1, ELP, or vehicle, took 42 days to fully close. It is noteworthy that wound cross-sections exhibited a significantly thicker epidermis and dermis compared to the other groups. This finding is similar to that previously reported by Koria et al [28], where KGF-ELP induced a higher proliferation of keratinocytes, which resulted significant increase in reepithelialization in full-thickness wounds made on diabetic mice as compared to the controls. While angiogenesis is the most commonly presumed mechanism of SDF1, a recent review suggests potentially many different cellular targets for SDF1 [11], making the role of SDF1-CXCR4 in the wound healing process more complicated. The mechanism of action of SDF1-ELP is therefore unclear.
Because SDF1-ELP had superior in vivo performance compared to SDF1, while both behaved almost identically in vitro, we hypothesized that SDF1-ELP nanoparticles are more stable in the diabetic wound environment compared to SDF1. Prior studies suggest that the ELP fusion proteins can serve as “drug depots” with a better stability profile and/or in vivo half-life than the free target protein [23]. For example, glucagon-like peptide-1 (GLP1; a potential type-2 diabetes drug) fused to ELP is more resistant to proteolysis by neutral endopeptidase, which is known to degrade GLP1 in vivo, as compared to free GLP1 [27]. The same study also shows that a single injection of the GLP1-ELP fusion protein was able to reduce blood glucose levels in mice for 5 days, which is about 120 times longer than what has been observed for the free GLP1. Similarly, we observed that SDF1-ELP was very stable in elastase which is known to degrade free SDF1 in vivo [38].
We used our in vitro intracellular calcium release assay to quantify the activity of SDF1-ELP monomers as compared to the nanoparticles. We noted that more activity appeared to be in the nanoparticle versus the monomeric fractions, although the difference was not statistically significant. In fact, it seemed that bioactivity was contributed by both nanoparticle and monomeric forms. Based on this we cannot conclude that release from the nanoparticle was necessary for bioactivity.
In conclusion, we have developed an SDF1-ELP fusion protein that has comparable biological and binding activities to recombinant human SDF1, and also has the ability to self-assemble into nanoparticles below physiological temperatures. Furthermore, SDF1-ELP was stable in the presence of elastase, making it a potential drug depot for use in chronic wound treatment. Although in this work we observed that the release of the monomers from the nanoparticles was not necessary for bioactivity, it may potentially be needed for other cellular responses which contribute to wound healing such as cell migration and/or proliferation, which will have to be further investigated. While our research focus is on skin wounds, our SDF1-ELP fusion protein nanoparticles may be useful for other wound healing applications, such as in myocardial infarction, where SDF1 has been reported to recruit stem cells to promote local tissue regeneration [14, 45].
Fig. 4. Size of SDF1-ELP Nanoparticles.
(A) TEM image of a single SDF1-ELP nanoparticle. Bar = 200 nm; (B) Chromatogram of SDF1-ELP nanoparticles (~600nm) and 100nm diameter gold nanoparticles run on the Zetasizer.
Key.
Forward primer sequence added for PCR: GGCACCTCGATTAGTTCTCG
Xba1 site: TCTAGA
SDF1 Gene string™: Underlined sequence
Linker (G4,S)3: GGTGGGGGTGGCTCTGGGGGCGGTGGTTCCGGAAGGTGGTGGATCA
NdeI site: CATAGA
Reverse primer sequence added for PCR: GAGTTCATGCGCTTCAAGGT
Acknowledgements
This work was partially supported by the Shriners Hospitals for Children and the New Jersey Commission for Spinal Cord Research. Agnes Yeboah was supported by a National Institutes of Health–funded biotechnology training fellowship, and a Schlumberger Faculty for the Future fellowship. We would like to thank Dr. Jafar Al-Sharab for helping with the TEM measurements, and the School of Environmental and Biological Sciences for allowing us to use their Biacore equipment, and also for helping with the experiments. Our sincere thanks also go to the School of Marine and Coastal Sciences, Rutgers University, for the use of their spectrophotometer, the Center for Advanced Biotechnology and Medicine, Rutgers University, for the use of their circular dichroism equipment, the Material Science Engineering Department and the Ernest Mario School of Pharmacy, Rutgers University for the use of their Zetasizer equipment, and the Digital Imaging and Histology Core at the Rutgers-New Jersey Medical School Cancer Center for their histological staining services.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sen CK, Gordillo GM, Roy S, Kirsner R, Lambert L, Hunt TK, Gottrup F, Gurtner GC, Longaker MT. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. 2009;17:763–771. doi: 10.1111/j.1524-475X.2009.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tellechea A, Leal E, Veves A, Carvalho E. Inflammatory and Angiogenic Abnormalities in Diabetic Wound Healing: Role of Neuropeptides and Therapeutic Perspectives. TOCVJ. 2010;3:43–55. [Google Scholar]
- 3.Edwards R, Harding KG. Bacteria and wound healing. Curr Opin Infect Dis. 2004;17:91–96. doi: 10.1097/00001432-200404000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Eming SA, Krieg T, Davidson JM. Inflammation in wound repair: molecular and cellular mechanisms. J Invest Dermatol. 2007;127:514–525. doi: 10.1038/sj.jid.5700701. [DOI] [PubMed] [Google Scholar]
- 5.Brem H, Tomic-Canic M. Cellular and molecular basis of wound healing in diabetes. J Clin Invest. 2007;117:1219–1222. doi: 10.1172/JCI32169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X, Zhu F, Zhang M, Zeng D, Luo D, Liu G, Cui W, Wang S, Guo W, Xing W, Liang H, Li L, Fu X, Jiang J, Huang H. Stromal cell-derived factor-1 enhances wound healing through recruiting bone marrow-derived mesenchymal stem cells to the wound area and promoting neovascularization. Cells Tissues Organs. 2013;197:103–113. doi: 10.1159/000342921. [DOI] [PubMed] [Google Scholar]
- 7.George AL, Bangalore-Prakash P, Rajoria S, Suriano R, Shanmugam A, Mittelman A, Tiwari RK. Endothelial progenitor cell biology in disease and tissue regeneration. J Hematol Oncol. 2011;4:24. doi: 10.1186/1756-8722-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalka C, Masuda H, Takahashi T, Kalka-Moll WM, Silver M, Kearney M, Li T, Isner JM, Asahara T. Transplantation of ex vivo expanded endothelial progenitor cells for therapeutic neovascularization. Proc Natl Acad Sci U S A. 2000;97:3422–3427. doi: 10.1073/pnas.070046397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majka SM, Jackson KA, Kienstra KA, Majesky MW, Goodell MA, Hirschi KK. Distinct progenitor populations in skeletal muscle are bone marrow derived and exhibit different cell fates during vascular regeneration. J Clin Invest. 2003;111:71–79. doi: 10.1172/JCI16157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi T, Kalka C, Masuda H, Chen D, Silver M, Kearney M, Magner M, Isner JM, Asahara T. Ischemia- and cytokine-induced mobilization of bone marrow-derived endothelial progenitor cells for neovascularization. Nat Med. 1999;5:434–438. doi: 10.1038/7434. [DOI] [PubMed] [Google Scholar]
- 11.Bollag WB, Hill WD. CXCR4 in Epidermal Keratinocytes: Crosstalk within the Skin. Journal of Investigative Dermatology. 2013;133:2505–2508. doi: 10.1038/jid.2013.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar A, Tatlidede S, Scherer SS, Orgill DP, Berthiaume F. Combination of stromal cell-derived factor-1 and collagen-glycosaminoglycan scaffold delays contraction and accelerates reepithelialization of dermal wounds in wild-type mice. Wound Repair Regen. 2011;19:71–79. doi: 10.1111/j.1524-475X.2010.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziegler M, Elvers M, Baumer Y, Leder C, Ochmann C, Schonberger T, Jurgens T, Geisler T, Schlosshauer B, Lunov O, Engelhardt S, Simmet T, Gawaz M. The bispecific SDF1-GPVI fusion protein preserves myocardial function after transient ischemia in mice. Circulation. 2012;125:685–696. doi: 10.1161/CIRCULATIONAHA.111.070508. [DOI] [PubMed] [Google Scholar]
- 14.Segers VF, Tokunou T, Higgins LJ, MacGillivray C, Gannon J, Lee RT. Local delivery of protease-resistant stromal cell derived factor-1 for stem cell recruitment after myocardial infarction. Circulation. 2007;116:1683–1692. doi: 10.1161/CIRCULATIONAHA.107.718718. [DOI] [PubMed] [Google Scholar]
- 15.Yang OO, Swanberg SL, Lu Z, Dziejman M, McCoy J, Luster AD, Walker BD, Herrmann SH. Enhanced inhibition of human immunodeficiency virus type 1 by Met-stromal-derived factor 1beta correlates with down-modulation of CXCR4. J Virol. 1999;73:4582–4589. doi: 10.1128/jvi.73.6.4582-4589.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiesinger W, Perez-Aguilar JM, Atluri P, Marotta NA, Frederick JR, Fitzpatrick JR, 3rd, McCormick RC, Muenzer JR, Yang EC, Levit RD, Yuan LJ, Macarthur JW, Saven JG, Woo YJ. Computational protein design to reengineer stromal cell-derived factor-1alpha generates an effective and translatable angiogenic polypeptide analog. Circulation. 2011;124:S18–26. doi: 10.1161/CIRCULATIONAHA.110.009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiesinger W, Goldstone AB, Woo YJ. Re-engineered stromal cell-derived factor-1alpha and the future of translatable angiogenic polypeptide design. Trends Cardiovasc Med. 2012;22:139–144. doi: 10.1016/j.tcm.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumann L, Prokoph S, Gabriel C, Freudenberg U, Werner C, Beck-Sickinger AG. A novel, biased-like SDF-1 derivative acts synergistically with starPEG-based heparin hydrogels and improves eEPC migration in vitro. J Control Release. 2012;162:68–75. doi: 10.1016/j.jconrel.2012.04.049. [DOI] [PubMed] [Google Scholar]
- 19.Rabbany SY, Pastore J, Yamamoto M, Miller T, Rafii S, Aras R, Penn M. Continuous delivery of stromal cell-derived factor-1 from alginate scaffolds accelerates wound healing. Cell Transplant. 2010;19:399–408. doi: 10.3727/096368909X481782. [DOI] [PubMed] [Google Scholar]
- 20.Henderson PW, Singh SP, Krijgh DD, Yamamoto M, Rafii DC, Sung JJ, Rafii S, Rabbany SY, Spector JA. Stromal-derived factor-1 delivered via hydrogel drug-delivery vehicle accelerates wound healing in vivo. Wound Repair Regen. 2011;19:420–425. doi: 10.1111/j.1524-475X.2011.00687.x. [DOI] [PubMed] [Google Scholar]
- 21.Dalonneau F, Liu XQ, Sadir R, Almodovar J, Mertani HC, Bruckert F, Albiges-Rizo C, Weidenhaupt M, Lortat-Jacob H, Picart C. The effect of delivering the chemokine SDF-1alpha in a matrix-bound manner on myogenesis. Biomaterials. 2014;35:4525–4535. doi: 10.1016/j.biomaterials.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olekson MA, Faulknor R, Bandekar A, Sempkowski M, Hsia HC, Berthiaume F. SDF-1 Liposomes Promote Sustained Cell Proliferation in Mouse Diabetic Wounds. Wound Repair Regen. 2015 doi: 10.1111/wrr.12334. [DOI] [PubMed] [Google Scholar]
- 23.Shamji MF, Betre H, Kraus VB, Chen J, Chilkoti A, Pichika R, Masuda K, Setton LA. Development and characterization of a fusion protein between thermally responsive elastin-like polypeptide and interleukin-1 receptor antagonist: sustained release of a local antiinflammatory therapeutic. Arthritis Rheum. 2007;56:3650–3661. doi: 10.1002/art.22952. [DOI] [PubMed] [Google Scholar]
- 24.MacEwan SR, Hassouneh W, Chilkoti A. Non-chromatographic purification of recombinant elastin-like polypeptides and their fusions with peptides and proteins from Escherichia coli. J Vis Exp. 2014 doi: 10.3791/51583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassouneh W, Christensen T, Chilkoti A. Elastin-like polypeptides as a purification tag for recombinant proteins. Curr Protoc Protein Sci. 2010 doi: 10.1002/0471140864.ps0611s61. Chapter 6 Unit 6 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer DE, Chilkoti A. Purification of recombinant proteins by fusion with thermally-responsive polypeptides. Nat Biotechnol. 1999;17:1112–1115. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- 27.Amiram M, Luginbuhl KM, Li X, Feinglos MN, Chilkoti A. A depot-forming glucagon-like peptide-1 fusion protein reduces blood glucose for five days with a single injection. J Control Release. 2013;172:144–151. doi: 10.1016/j.jconrel.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koria P, Yagi H, Kitagawa Y, Megeed Z, Nahmias Y, Sheridan R, Yarmush ML. Self-assembling elastin-like peptides growth factor chimeric nanoparticles for the treatment of chronic wounds. Proc Natl Acad Sci U S A. 2011;108:1034–1039. doi: 10.1073/pnas.1009881108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loetscher P, Gong JH, Dewald B, Baggiolini M, Clark-Lewis I. N-terminal peptides of stromal cell-derived factor-1 with CXC chemokine receptor 4 agonist and antagonist activities. J Biol Chem. 1998;273:22279–22283. doi: 10.1074/jbc.273.35.22279. [DOI] [PubMed] [Google Scholar]
- 30.Ryu EK, Kim TG, Kwon TH, Jung ID, Ryu D, Park YM, Kim J, Ahn KH, Ban C. Crystal structure of recombinant human stromal cell-derived factor-1alpha. Proteins. 2007;67:1193–1197. doi: 10.1002/prot.21350. [DOI] [PubMed] [Google Scholar]
- 31.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nature Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salcedo R, Wasserman K, Young HA, Grimm MC, Howard OM, Anver MR, Kleinman HK, Murphy WJ, Oppenheim JJ. Vascular endothelial growth factor and basic fibroblast growth factor induce expression of CXCR4 on human endothelial cells: In vivo neovascularization induced by stromal-derived factor-1alpha. Am J Pathol. 1999;154:1125–1135. doi: 10.1016/s0002-9440(10)65365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crump MP, Gong JH, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier JL, Baggiolini M, Sykes BD, Clark-Lewis I. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McQuibban GA, Butler GS, Gong JH, Bendall L, Power C, Clark-Lewis I, Overall CM. Matrix metalloproteinase activity inactivates the CXC chemokine stromal cell-derived factor-1. J Biol Chem. 2001;276:43503–43508. doi: 10.1074/jbc.M107736200. [DOI] [PubMed] [Google Scholar]
- 37.Aiuti A. The Chemokine SDF-1 Is a Chemoattractant for Human CD34+ Hematopoietic Progenitor Cells and Provides a New Mechanism to Explain the Mobilization of CD34+ Progenitors to Peripheral Blood. J Exp Med. 1997;185:111–120. doi: 10.1084/jem.185.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valenzuela-Fernández A, Planchenault T, Baleux F, Staropoli I, Le-Barillec K, Leduc D, Delaunay T, Lazarini F, Virelizier J, Chignard M, Pidard D, Arenzana-Seisdedos F. Leukocyte elastase negatively regulates stromal cell-derived factor-1 (SDF-1)/CXCR4 binding and functions by amino-terminal processing of SDF-1 and CXCR4. J Biol Chem. 2002;277:15677–15689. doi: 10.1074/jbc.M111388200. [DOI] [PubMed] [Google Scholar]
- 39.Bogani C, Ponziani V, Guglielmelli P, Desterke C, Rosti V, Bosi A, Le Bousse-Kerdiles MC, Barosi G, Vannucchi AM, Myeloproliferative Disorders Research C. Hypermethylation of CXCR4 promoter in CD34+ cells from patients with primary myelofibrosis. Stem Cells. 2008;26:1920–1930. doi: 10.1634/stemcells.2008-0377. [DOI] [PubMed] [Google Scholar]
- 40.Hiesinger W, Frederick JR, Atluri P, McCormick RC, Marotta N, Muenzer JR, Woo YJ. Spliced stromal cell-derived factor-1alpha analog stimulates endothelial progenitor cell migration and improves cardiac function in a dose-dependent manner after myocardial infarction. J Thorac Cardiovasc Surg. 2010;140:1174–1180. doi: 10.1016/j.jtcvs.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang HJ, Kim JH, Chang WJ, Kim ES, Koo YM. Heterologous expression and optimized one-step separation of levansucrase via elastin-like polypeptides tagging system. J Microbiol Biotechnol. 2007;17:1751–1757. [PubMed] [Google Scholar]
- 42.Hu F, Ke T, Li X, Mao PH, Jin X, Hui FL, Ma XD, Ma LX. Expression and purification of an antimicrobial peptide by fusion with elastin-like polypeptides in Escherichia coli. Appl Biochem Biotechnol. 2010;160:2377–2387. doi: 10.1007/s12010-009-8850-2. [DOI] [PubMed] [Google Scholar]
- 43.Hadad I, Veithen A, Springael JY, Sotiropoulou PA, Mendes Da Costa A, Miot F, Naeije R, De Deken X, Entee KM. Stroma cell-derived factor-1alpha signaling enhances calcium transients and beating frequency in rat neonatal cardiomyocytes. PLoS ONE. 2013;8:e56007. doi: 10.1371/journal.pone.0056007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou L, Guo X, Ba J, Zhao L. CD44 is involved in CXCL-12 induced acute myeloid leukemia HL-60 cell polarity. Biocell. 2010;34:91–94. [PubMed] [Google Scholar]
- 45.Prokoph S, Chavakis E, Levental KR, Zieris A, Freudenberg U, Dimmeler S, Werner C. Sustained delivery of SDF-1alpha from heparin-based hydrogels to attract circulating pro-angiogenic cells. Biomaterials. 2012;33:4792–4800. doi: 10.1016/j.biomaterials.2012.03.039. [DOI] [PubMed] [Google Scholar]