Abstract
Phakopsora pachyrhizi is a biotrophic fungus provoking SBR disease. SBR poses a major threat to global soybean production. Though several R genes provided soybean immunity to certain P. pachyrhizi races, the pathogen swiftly overcame this resistance. Therefore, fungicides are the only current means to control SBR. However, insensitivity to fungicides is soaring in P. pachyrhizi and, therefore, alternative measures are needed for SBR control. In this article, we discuss the different approaches for fighting SBR and their potential, disadvantages, and advantages over other measures. These encompass conventional breeding for SBR resistance, transgenic approaches, exploitation of transcription factors, secondary metabolites, and antimicrobial peptides, RNAi/HIGS, and biocontrol strategies. It seems that an integrating approach exploiting different measures is likely to provide the best possible means for the effective control of SBR.
Keywords: Asian soybean rust, Phakopsora pachyrhizi, fungicide insensitivity, host resistance, non-host resistance, plant breeding, plant biotechnology
Introduction
SBR is currently the most severe soybean (Glycine max) disease. SBR is caused by Phakopsora pachyrhizi. The biotrophic basidiomycete threatens soybean production all over the globe, but the threat is most severe in the major soybean growing areas in South America. In Brazil SBR has caused crop losses of more than US$ 10 billion since its first endemic outbreak in 2001 (Yorinori et al., 2005; da Silva et al., 2014). Currently, three major strategies serve to manage SBR (Figure 1). First, applying chemical fungicides. Second, breeding or engineering of SBR-resistant soybean cultivars, and third, employing specific cultivation practices, such as planting early ripening varieties, monitoring fields, eliminating secondary hosts, and introducing soybean-free growth periods (60–90 days) in the threatened areas (Hartman et al., 2005; Godoy, 2011; Kendrick et al., 2011). Here, we elaborate on these strategies, and we also discuss the potential of AMPs, RNAi/HIGS, and biocontrol measures for controlling SBR. A detailed description of the life cycle, host range and distribution of P. pachyrhizi has been provided earlier (Goellner et al., 2010).
FIGURE 1.
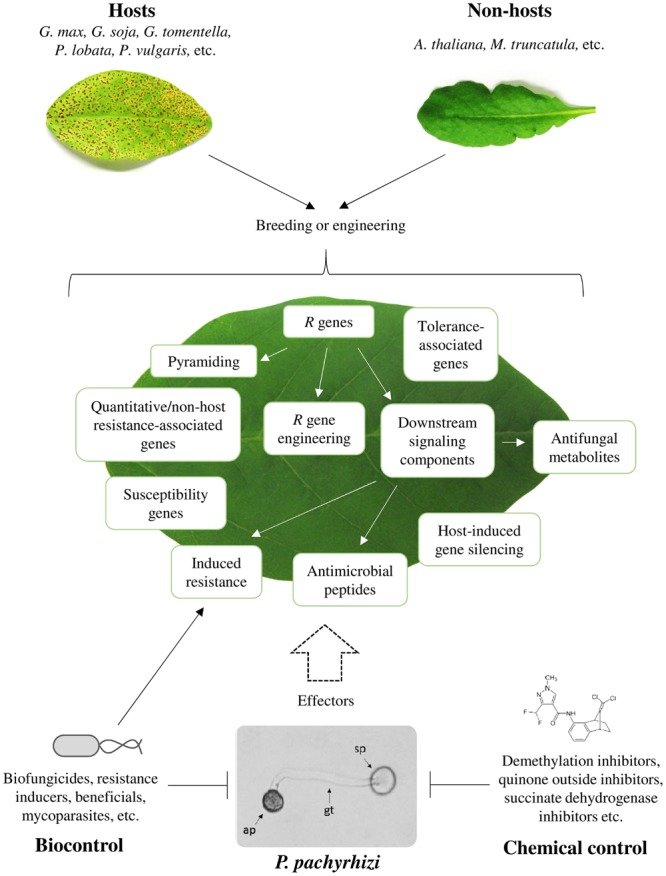
Strategies for controlling SBR. Exploitation of different genetic resources (host and non-host plants), biocontrol agents, and chemical fungicides to combat Phakopsora pachyrhizi. ap, appressorium; gt, germ tube; sp, uredospore.
Chemical Control of SBR
Fungicide use is the most effective means for controlling SBR these days. In Brazil, at least three fungicide applications are needed per season thus raising costs of ∼US$2 billion for soybean disease control annually (Godoy et al., 2015). In contrast to multisite fungicides (e.g., mancozeb) with comparatively low performance, the DMI and the QoI classes of fungicide are prime chemicals for fighting P. pachyrhizi. Since 2013, fungicides of the highly active SDHI class are available for SBR control (Guicherit et al., 2014). Because this new fungicide class performs extraordinarily well, the number of available SDHI fungicides and the intensity of their use is likely to steadily increase over the next couple of years (Godoy et al., 2015). However, the excessive use of fungicides increases the chance of fungal strains with evolved insensitivity to the fungicides in use. In the recent past, this was true for the azole-class fungicides to which P. pachyrhizi and other fungal pathogens have become insensitive (Godoy, 2012). The FRAC assigned rust fungi, including P. pachyrhizi, to the low-risk group of fungi (Brent and Holloman, 2007). However, P. pachyrhizi and other causes of polycyclic plant diseases are highly likely to evolve fungicide insensitivity because of the high number of spores they produce (Bradley, 2007).
The mechanism of fungal insensitivity to DMIs is highly complex and variable. After several years of fungicide use, a significant reduction in DMI efficacy to P. pachyrhizi was detected in Brazil (Scherm et al., 2009; Barbosa et al., 2013; Reis et al., 2015). The insensitivity is caused either by point mutations in the fungal cyp51 gene or by cyp51 overexpression (Schmitz et al., 2014). The major mechanism of QoI and SDHI insensitivity is by point mutations in the cyt b and sdh b/c/d genes, respectively. These mutations were reported for many plant-pathogenic fungi (Kim et al., 2003; Grasso et al., 2006a; Sierotzki et al., 2007; Sierotzki and Scalliet, 2013). The most common mutation for QoI insensitivity [substitution of glycine to alanine at position 143 of Cyt b] was not yet detected in rusts probably because of presence of a type-I intron after codon 143 (Grasso et al., 2006a,b,c; Oliver, 2014; Klosowski et al., 2015). Nucleotide substitutions in this codon would prevent intron splicing thus leading to a defective Cyt b protein (Grasso et al., 2006a). However, another cyt b mutation (F129L) was reported to confer QoI insensitivity in various fungi including P. pachyrhizi (Leiminger et al., 2014; Klosowski et al., 2015). For P. pachyrhizi SDHI insensitivity was not reported yet. However, the increased use of SDHIs is likely to further enhance the selection pressure for SDHI insensitivity in P. pachyrhizi (Godoy et al., 2015). MDR, as reported for Botrytis cinerea and other fungi (Kretschmer et al., 2009) also was not observed in P. pachyrhizi so far. To assess the risk and impact of fungicide-insensitive isolates, we recommend generating insensitive fungal mutants in the laboratory. Investigating such mutants is likely to disclose mechanisms underlying fungicide insensitivity, enable recommendations for avoiding selection of insensitive fungal populations, and developing novel mode-of-action fungicides. Applying fungicides preventively or as early as possible in the diseases cycle before or shortly after P. pachyrhizi infection is crucial for effective SBR control (Mueller et al., 2009; Godoy, 2012). Therefore, early SBR detection and precise forecasts are required for efficient SBR disease management.
Probably the best and most sustainable control of SBR is by providing soybean genotypes resisting P. pachyrhizi (see below). Growth of SBR resistant genotypes is likely to be associated with reduced fungicide use. This then might decrease soybean production costs, improve the CO2 footprint of soybean products, and minimize the potential risk of ecological and sanitary actions resulting from extensive use of fungicides (Maltby et al., 2009; Verweij et al., 2009; Wightwick et al., 2010).
Resources of SBR Resistance in Soybean
R Genes, R Gene Pyramids, and Engineered R Genes
Analysis of soybean genotypes disclosed six dominant R genes conferring immunity (no visible symptoms) or resistance (reddish brown lesions and reduced sporulation) to specific P. pachyrhizi isolates. Those loci were referred to as Rpp 1–6 genes (Bromfield and Hartwig, 1980; McLean and Byth, 1980; Bromfield and Melching, 1982; Hartwig and Bromfield, 1983; Hartwig, 1986; Garcia et al., 2008; Li et al., 2012). However, Rpp genes provide resistance exclusively to individual P. pachyrhizi isolates (race-specific disease resistance). Therefore, no currently available soybean genotype would ward off all P. pachyrhizi isolates (Monteros et al., 2007). In addition, Rpp gene-mediated resistance was swiftly overcome in the field (Yorinori et al., 2005; Garcia et al., 2008). Employing recessive R genes might represent another approach for providing stable SBR resistance (Calvo et al., 2008). In fact, three recessive R genes to P. pachyrhizi have been identified in the soybean genotypes PI 200456, PI 224270, and BR01-18437 (Calvo et al., 2008; Pierozzi et al., 2008). These genes are now awaiting exploitation in breeding and genetic engineering for SBR resistance.
Developing elite lines and varieties requires breeders to combine traits from multiple parents, a process called gene pyramiding or stacking (Francis et al., 2012). Pyramiding R genes into a single genetic background is another proposed strategy for conferring soybean resistance to multiple P. pachyrhizi isolates (Hartman et al., 2005; Garcia et al., 2008; Lemos et al., 2011; Maphosa et al., 2012; Yamanaka et al., 2013, 2015; Bhor et al., 2014). The SBR resistant Japanese soybean cultivar Hyuuga represents a natural example of R gene pyramiding (Kendrick et al., 2011). In line with this finding, soybean genotypes harboring two pyramided Rpp genes exhibited higher SBR resistance than their ancestors containing only single R genes (Maphosa et al., 2012; Bhor et al., 2015). Synergistic effects were also observed when three R genes were bred into a single soybean genotype (Lemos et al., 2011; Yamanaka et al., 2013, 2015). Remarkably, a combination of multiple R genes conferred resistance to different P. pachyrhizi isolates from various origin (including two highly virulent strains from Brazil; Yamanaka et al., 2015). Although molecular markers facilitate breeding approaches, traditional breeding is still time consuming, and introducing unwanted traits (Salomon and Sessa, 2012). Furthermore, SBR resistance based on static R gene pyramids will likely be overcome upon longer use in the field (McDonald, 2014) as has been reported for other crops like wheat or barley (McDonald and Linde, 2002). Therefore, transforming expression cassettes with alternative R gene combinations into elite soybean lines and dynamic turnover of such lines in the field might represent a promising strategy for providing sustainable and effective SBR resistance (McDonald and Linde, 2002). However, for cloning and utilization of such multi R gene expression cassettes the identity of Rpp genes needs to be revealed. Although SBR resistance loci have been mapped to different linkage groups on various chromosomes (reviewed by Bhor et al., 2014), the identity of Rpp genes has remained largely unknown. One exception is represented by the NB-LRR encoding gene Rpp4C4 that is likely responsible for Rpp4-mediated SBR resistance (Meyer et al., 2009).
Another possibility to enhance the resistance of soybean to SBR is to identify and exploit R genes conferring resistance to multiple pathogens. Several examples of such broadly active R genes exist in nature (Nombela et al., 2003; Narusaka et al., 2009; Atamian et al., 2012; Lozano-Torres et al., 2012). A complementary approach for broadened pathogen effector recognition uses random mutagenesis or rational design of synthetic NB-LRR immune receptors. Editing the potato NB-LRR receptor R3a at a single amino acid significantly expanded its response to Phytophthora infestans-derived effectors (Segretin et al., 2014). Effectively mutating the R3a orthologue I2 in tomato enhanced the response to the P. infestans AVR3a effector, conferred partial immunity to potato blight, and expanded the response spectrum to Fusarium oxysporum f. sp. lycopersici effectors compared to tomato plants expressing the wild-type I2 gene (Giannakopoulou et al., 2015). R gene engineering might also succeed in exploiting multiple Rpp genes for conferring an expanded response to multiple P. pachyrhizi isolates. Rpp4C4 (Meyer et al., 2009) may serve for engineering such R gene variants by untargeted protein evolution. Furthermore, genome editing may be used for the targeted evolution of NB-LRRs. In fact, genome-wide sequence analysis predicted nearly all soybean NB-LRR-encoding genes an be targeted specifically by CRISPR/Cas9 (Xie et al., 2014).
Signaling Components of R Gene-Mediated SBR Resistance
Several studies reported differential defense responses to SBR attack in susceptible and resistant soybean genotypes. The studies included analysis of transcriptional dynamics, proteome changes, or metabolic alterations to identify loci, genes, proteins, and metabolites associated with ETI to P. pachyrhizi in soybean.
Signaling network hubs and phytohormones
Transcriptome analysis disclosed different components of Rpp2-mediated resistance to SBR in soybean (van de Mortel et al., 2007; Pandey et al., 2011). Of 140 candidates tested by VIGS, eleven genes clearly contributed to Rpp2-mediated SBR resistance. The genes encompassed GmEDS1, GmPAD4, and GmNPR1.
NPR1 is a master regulator of SAR in Arabidopsis thaliana and some other plants (reviewed by Fu and Dong, 2013). When overexpressed in Arabidopsis, rice, tobacco, or apple, NPR1 enhances resistance to infectious oomycetes, bacteria, and fungi (including obligate biotrophic fungi such as powdery mildew; Cao et al., 1998; Chern et al., 2005; Chen et al., 2012). Because of possible side effects of NPR1 overexpression (Chern et al., 2005), such as yield reduction, the potential of this gene for generating SBR-resistant soybean varieties awaits assessment.
EDS1 and PAD4 are key regulators of several types of plant disease resistance (basal, R gene-mediated, and NHR). The two proteins are required for accumulation of SA, and they control various SA-dependent defense pathways (Falk et al., 1999; Jirage et al., 1999; Nawrath et al., 2002; Lipka et al., 2005; Wiermer et al., 2005; Langenbach et al., 2013; Wang et al., 2014). Because silencing of GmEDS1 or GmPAD4 lead to susceptibility of otherwise resistant soybean lines carrying Rpp2, EDS1 and PAD4 seem to control also Rpp2-mediated SBR resistance in soybean (Pandey et al., 2011). SA accumulation is thus likely to limit the growth and reproduction of P. pachyrhizi in soybean. Because PAD4 is also required for Arabidopsis postinvasion NHR to P. pachyrhizi (Langenbach et al., 2013), SA-associated defense responses seem to be highly effective in antagonizing SBR disease. However, overexpression of SA biosynthesis genes is likely not to provide a realistic agronomical solution for SBR control because constitutive SA accumulation often causes dwarfism (Bowling et al., 1994; Li et al., 2001).
In Arabidopsis and soybean, P. pachyrhizi activates expression of JA-responsive genes at early stages of infection (Loehrer et al., 2008; Alves et al., 2015) and before actual penetration [likely by secreted P. pachyrhizi effectors (Campe et al., 2014)]. Since JA is considered eliciting immune responses against necrotrophic pathogens (Pieterse et al., 2012) P. pachyrhizi pretends being a necrotroph at initial stages of colonization. By doing so, it may circumvent effective SA-dependent defense signaling which is known to be crucial to ward off biotrophic pathogens. Thus, engineering soybean plants for the fast and robust accumulation of SA, or exploiting SA-activated downstream signaling components for resistance might be a suited strategy for providing soybean varieties resisting SBR at low risks for energetic tradeoffs.
Transcription factors
The importance of TFs in conferring SBR resistance became obvious when van de Mortel et al. (2007) and Schneider et al. (2011) found that TF genes are being overrepresented among genes whose expression is activated in the biphasic transcriptional response in SBR-resistant soybean genotypes harboring Rpp2 or Rpp3. Amongst others, genes encoding WRKY, bHLH, and MYB TFs were activated in incompatible, but not compatible, soybean-P. pachyrhizi interactions. When GmWRKY36, GmWRKY40, GmWRKY45, and GmMYB84 were individually silenced using VIGS, Rpp2-mediated SBR resistance was gone (Pandey et al., 2011). Several other studies also revealed differential expression of TFs in incompatible or compatible soybean-P. pachyrhizi interactions (Panthee et al., 2009; Morales et al., 2013; Aoyagi et al., 2014). In fact, there seems to be considerable overlap of TF activity in Rpp2, Rpp3, and Rpp4-mediated soybean disease resistance (Morales et al., 2013). Therefore, these TFs seem to be excellent candidates for engineering SBR resistance. However, manipulation of TF balance may affect agronomic traits because TFs regulate a diverse array of loci.
In another approach, Cooper et al. (2011) compared nuclear proteome changes in a resistant vs. susceptible genotype at 24 h after inoculation with P. pachyrhizi. Their analysis disclosed more than 200 proteins that specifically accumulated in the nucleus of SBR-resistant soybean plants harboring Rpp1 (Cooper et al., 2011). Silencing two predicted soybean TFs (Glyma14g11400, PHD superfamily and Glyma12g30600, zinc finger TF) via VIGS partially compromised Rpp1-conferred SBR resistance (Cooper et al., 2013). Similarly, Bencke-Malato et al. (2014) demonstrated that accumulation of mRNA transcripts for several WRKY TFs was faster and more robust in a resistant than susceptible soybean accession. Consistently, the simultaneous silencing of four identified WRKY genes rendered soybean plants more susceptible to SBR disease. Because the authors did not succeed in producing WRKY-overexpressing soybean lines (Bencke-Malato et al., 2014), the potential of WRKY overexpression for providing SBR resistance to susceptible soybean genotypes remained unclear.
Secondary metabolism
Plants can halt or slow down infection by constitutive or inducible accumulation of antimicrobial and/or cell wall-fortifying secondary metabolites (Chiang and Norris, 1983; Hahlbrock and Scheel, 1989; Chang et al., 1995; Dixon et al., 2002; Boerjan et al., 2003; La Camera et al., 2004; Vogt, 2010). Secondary metabolites also contribute to the outcome of the soybean–P. pachyrhizi interaction. Daidzein, genistein, and glyceollin are isoflavonoids that accumulate in both resistant and susceptible soybean genotypes upon P. pachyrhizi infection (Lygin et al., 2009). Glyceollin efficiently reduces P. pachyrhizi uredospore germination in vitro (Lygin et al., 2009). Further evidence for a role of phytoalexins in SBR resistance was provided by Bilgin et al. (2009). The authors disclosed that SBR resistance in a Glycine tomentella accession correlated with the presence of a flavonoid that also inhibited P. pachyrhizi spore germination (Chung and Singh, 2008). The high potential of phytoalexins in defeating SBR is further supported by medicarpin accumulating in P. pachyrhizi-infected Medicago truncatula, a non-host of P. pachyrhizi. Consistently, medicarpin inhibits P. pachyrhizi spore germination (Ishiga et al., 2015). Providing such comparative large-scale metabolic profiles from resistant vs. susceptible soybean varieties, or other SBR-resistant species would likely identify more secondary metabolites inhibiting SBR. Genes in their biosynthesis pathways could be used to engineer SBR resistance in transgenic soybean. Alternatively, the compound(s) themselves could serve as natural fungicides in spray application, especially if they can be produced at low costs and in sufficient quantities for use in agriculture. In a variety of studies, genes in the phenylpropanoid and flavonoid metabolism were overrepresented when analyzing the transcriptional response of infected soybean genotypes with SBR resistance (van de Mortel et al., 2007; Choi et al., 2008; Panthee et al., 2009; Schneider et al., 2011). Overall, activation of these genes was faster and stronger in SBR-resistant accessions than in susceptible ones (van de Mortel et al., 2007; Schneider et al., 2011). Functional evidence for the importance of phenylpropanoid pathway genes in soybean’s SBR resistance was provided by Pandey et al. (2011). The authors demonstrated that silencing of soybean phenylalanine ammonia-lyase (GmPAL) or O-methyl transferase1 (GmOMT1) compromised Rpp2-mediated SBR resistance. OMT1 silencing also partially impaired Rpp1-mediated SBR resistance (Cooper et al., 2013) and significantly decreased lignin content (Pandey et al., 2011). The latter result points to an important role of lignification in rejecting SBR.
Susceptibility Genes and Effector Targets
Different from dominant R genes conferring effective, but exclusively race-specific and non-durable resistance (Yorinori et al., 2005; Garcia et al., 2008), the loss of functional S genes can eventually provide durable disease resistance (Pavan et al., 2010; Gawehns et al., 2013). For example, in barley absence of the S gene Mlo results in an incompatible interaction with Blumeria graminis f. sp. hordei that resembles NHR (Humphry et al., 2006). S genes function either as susceptibility factors or suppressors of plant defense. Thus they are potential targets of fungal effectors. Consistent with this assumption knocking out S genes leads to recessive resistance with effectivity to multiple races of a given pathogen (Pavan et al., 2010). This type of resistance is very stable. The resistance of plants harboring recessive alleles of Mlo (barley) or eIF4E (pepper) has not been overcome in the field for 30–50 years (Lyngkjær et al., 2000; Kang et al., 2005). Breeding for S gene variants insensitive to manipulation by pathogen effectors therefore has huge potential for durable, broad-spectrum disease resistance; although loss-of-function mutations in S genes may be associated with pleiotropic detrimental effects (Büschges et al., 1997).
Soybean S genes to SBR have not been identified so far. However, several approaches might identify potential S gene alleles for SBR resistance in soybean. Because most S genes of agricultural value were identified in screens for recessive resistance in wild species of plant (Bai et al., 2005), searching for such a resistance in wild Glycine might similarly provide genetic resources for breeding or engineering SBR resistance in G. max.
Another option for identifying soybean S genes to SBR is via sequence homology search to known S genes. Functional analysis can be done using, for example, soybean insertion mutants (Mathieu et al., 2009), performing VIGS (Zhang and Ghabrial, 2006; Zhang et al., 2010, 2013; Pandey et al., 2011), TILLING (Cooper et al., 2008), or applying targeted genome editing techniques such as CRISPR/Cas9 (Jacobs et al., 2015). However, currently only one gene [the Cys(2)His(2) zinc finger TF palmate-like pentafoliata1, PALM1] that would classify as an SBR S gene is known from M. truncatula (Uppalapati et al., 2012). Alternatively, fungal effectors might serve as guides to identify novel S genes in soybean and other plants since several effectors of bacteria, fungi, or oomycetes were shown to target plant S genes (reviewed by Gawehns et al., 2013). Although various analyses identified stage-specific rust proteins that might have bona-fide effector function (Loehrer and Schaffrath, 2011; Stone et al., 2012; Link et al., 2014), their role as virulence factors awaits functional confirmation. Identification of effector proteins and corresponding S gene targets was likely hampered by missing P. pachyrhizi genome information (Loehrer et al., 2014). Transformation protocols enabling generation of P. pachyrhizi knockout mutants are also missing.
Another approach for identifying S gene alleles conferring SBR resistance is via screening of mutagenized soybean populations for loss-of-susceptibility mutants. The tetraploid nature of the soybean crop and the potential existence of multiple S gene copies might hamper this approach. Because 12 duplicated copies of a given DNA region might be present in the soybean genome (Cannon and Shoemaker, 2012), mutagenesis-induced phenotypic variation might be buffered by gene redundancy (Bolon et al., 2014). However, fast neuron irradiation recently provided more than 27,000 unique soybean mutants with significant phenotypic variation (Bolon et al., 2011, 2014). The mutants may facilitate genetic screens for loss of SBR susceptibility mutants with interesting resistance phenotypes similar to the M. truncatula irg1 mutant (Ishiga et al., 2015). Identified S gene alleles for SBR resistance in soybean might be engineered in elite soybean lines via genome editing (Jacobs et al., 2015).
Genes Providing Quantitative SBR Resistance or Tolerance
Forward genetic screens using activation-tagged soybean plants (Mathieu et al., 2009) could identify genes and loci that quantitatively contribute to SBR resistance. Genes and loci for SBR resistance can potentially also be found exploiting fungal effectors targeting proteins with a role in apoplastic immunity [e.g., the Ustilago maydis effector Pit2 targets maize apoplastic cysteine proteases (Mueller et al., 2013)].
PDR to SBR is found in ‘slow rusting’ soybean accessions such as SRE-Z-11A, SRE-Z-11B, and SRE-Z-15A (Tukamuhabwa and Maphosa, 2010). These genotypes can potentially provide useful genes and loci for quantitative SBR resistance. PDR is characterized by low infection frequency, long-lasting latency, small lesions, and reduced spore production per uredinium. Thus, PDR reduces SBR epidemics (Tukamuhabwa and Maphosa, 2010). Since PDR is polygenic and effective to multiple pathogen races (Long et al., 2006), identification, and transfer of genes from partially resistant to susceptible soybean varieties might provide only partial but durable resistance to diverse P. pachyrhizi isolates. Because of PDR’s polygenic nature and the time-consuming process for selecting partially resistant progeny, such soybean varieties have not attracted much attention as sources for SBR resistance in the past (Hartman et al., 2005).
Besides soybean genotypes with partial resistance, SBR-tolerant accessions also have not been a subject of molecular research. Although susceptible to SBR, these genotypes do better tolerate the presence of P. pachyrhizi and produce reasonably high yield even when severely infected. Yield may increase by 30–60% using SBR-tolerant varieties in the presence of P. pachyrhizi (Tukamuhabwa and Maphosa, 2010). Furthermore, planting tolerant varieties does not pose selection pressure on P. pachyrhizi, thus minimizing the risk of selecting adapted pathogen races (Arias et al., 2008). However, SBR disease tolerance of a given soybean accession is assessed with respect to its yield capacity. This requires multi-site field trials and hinders evaluation of a genotype’s tolerance and commercial value at small scale laboratory conditions (Tukamuhabwa and Maphosa, 2010). Nonetheless, identification of genes for SBR tolerance using, e.g., comparative transcriptome or proteome analysis, may enable provision of soybean varieties with capacity for enhanced yield at high SBR pressure.
Antimicrobial Peptides
AMPs can provide disease resistance to plants (Rahnamaeian, 2011). However, AMPs did not serve to fight SBR so far. Brand et al. (2012) introduced a method for the identification and employment of putative AMPs encrypted in soybean protein sequences. This approach was meant to provide an alternative to transgenic approaches that expressed AMPs from other organisms. Using in situ assays, Brand et al. (2012) found that IAPs conferred SBR resistance in a manner similar to AMPs from Phyllomedusa ssp. (dermaseptin SI) or Drosophila melanogaster (penetratin) when co-incubated with fungal uredospores on susceptible soybean leaves. In addition, soybean plants expressing a putative antimicrobial fragment of the G. max D-myo-inositol 3-phosphate synthase [IAP gb|ABM17058.1| (213–231)] showed enhanced resistance to P. pachyrhizi (Brand et al., 2012). These findings illustrate the feasibility of trans- or cisgenic AMP expression for SBR resistance.
Alternative Sources of SBR Resistance
Wild Glycine Species and Other Alternative Hosts
Wild perennial Glycine species might serve as valuable resources of germplasm for SBR resistance. This is because Glycine clandestina, Glycine canescens, Glycine tabacina, Glycine tomentella, and Glycine argyrea all display pathotype-specific resistance to P. pachyrhizi (Burdon and Speer, 1984; Burdon, 1987, 1988; Jarosz and Burdon, 1990). In G. clandestina, G. canescens, and G. argyrea differential SBR resistance phenotypes are linked to presence or absence of single or multiple (pyramided) R genes (Burdon and Speer, 1984; Burdon, 1987, 1988; Jarosz and Burdon, 1990). The resistance of G. tomentella accession PI 441001 to P. pachyrhizi, however, was associated with accumulation of an antifungal flavonoid inhibiting P. pachyrhizi spore germination (Chung and Singh, 2008). Because Singh and Nelson (2015) obtained fertile SBR-resistant plants from crosses of G. max and G. tomentella, transfer of R genes from wild perennial species to commercial soybean varieties via intersubgenic hybridization seems to be a powerful strategy for SBR resistance. The novel hybrid plant is still to be analyzed for its yield and resistance to multiple P. pachyrhizi isolates which will disclose the commercial value of the hybrid.
Other SBR resistance traits are present in G. soja. The species is closer related to G. max than its above mentioned wild perennial relatives (Bromfield, 1984). However, because of presence of undesired traits, generating hybrids for commercialization using G. soja or the wild, perennial Glycine species will likely require elaborate backcrossing and selection. Identifying the genetic basis of SBR resistance in wild species followed by engineered transfer of genes and/or traits to elite varieties might represent an alternative, more promising strategy for SBR resistance. The approach circumvents the drawbacks associated with hybridization strategies. However, only few attempts (e.g., Soria-Guerra et al., 2010) identified gene candidates to condition SBR resistance in wild Glycine species.
Kudzu (Pueraria lobata) is a leguminous weed that hosts P. pachyrhizi and could provide traits for SBR resistance. Genetic variation is high among different kudzu populations but low within a same population (Sun et al., 2005). As a consequence, individual kudzu plants are resistant/immune or susceptible to diverse P. pachyrhizi isolates (Bonde et al., 2009). In a kudzu genotype with immunity to SBR the early abrogation of P. pachyrhizi infection correlated with cell wall appositions and cell death in the leaf epidermis (Jordan et al., 2010). This finding suggests presence of early, effective defense responses in immune kudzu genotypes. Big differences in the response to P. pachyrhizi infection were also seen in several other legume species (Slaminko et al., 2008). Vigna adenantha PI 312898, for instance, is immune to SBR as are individual bean (Phaseolus vulgaris) cultivars (Miles et al., 2007; Souza et al., 2014). However, lack of genomic information and low genetic accessibility of alternative P. pachyrhizi hosts impede candidate gene identification and gene transfer.
Non-host Plants
Over the past decade, employing non-host plants has become a promising approach for identifying resistance traits. Due to the pervasive nature of NHR, the strategy explores a vast genetic resource. NHR is a multi-layered, complex type of plant disease resistance that shares signaling and defense mechanisms with host resistance (Schulze-Lefert and Panstruga, 2011). Classification of a given plant species as a host or non-host can be difficult because there seems to be a gradual continuum from host to non-host with many intermediate resistances (Bettgenhaeuser et al., 2014). Exploring the molecular basis of this variety of resistances and pyramiding underlying genes and loci in the soybean crop may represent a powerful approach for SBR resistance and provide an alternative to chemical fungicides and traditional breeding.
Arabidopsis and M. truncatula are the best described plants in terms of NHR to P. pachyrhizi. Since P. pachyrhizi does not produce macroscopic symptoms on any of 28 wild-type accessions tested, Arabidopsis can be considered a true non-host for P. pachyrhizi (Loehrer et al., 2008). Although initial stages of P. pachyrhizi development are identical on Arabidopsis and soybean, proliferation of P. pachyrhizi hyphae into the leaf mesophyll is rare in Arabidopsis (Loehrer et al., 2008). To determine the molecular basis of the preinvasion resistance to P. pachyrhizi in this plant, Loehrer et al. (2008) used Arabidopsis mutants with known compromised resistance to other non-adapted fungal pathogens. Colonization of the mesophyll occurred in Arabidopsis penetration mutant pen1, pen2, and pen3. However, despite hyphal growth and rarely observed haustoria in the mesophyll of pen mutants, the fungus failed to successfully colonize the plant. It also did not complete its life cycle, indicative of functional postinvasion resistance to P. pachyrhizi in these mutants. The postinvasion resistance was compromised in the Arabidopsis triple mutant pen2 pad4 sag101 in which P. pachyrhizi frequently developed haustoria (Langenbach et al., 2013). However, extensive mesophyll colonization and sporulation did not occur in any Arabidopsis mutants tested.
To identify components of Arabidopsis postinvasion resistance to P. pachyrhizi, Langenbach et al. (2013, 2016) performed comparative transcriptional profiling of genes specifically activated upon P. pachyrhizi infection in pen2 (a mutant with intact postinvasion resistance) but not pen2 pad4 sag101 (with compromised postinvasion resistance). The screen identified BRIGHT TRICHOMES 1 (BRT1), an UDP-glycosyltransferase in the phenylpropanoid metabolism. Postinvasion resistance to P. pachyrhizi was impaired in the pen2 brt1 double mutant. In this genotype the fungus developed more haustoria than in pen2. Since brt1 mutants were not affected in preinvasion SBR resistance (Langenbach et al., 2013), BRT1 seems to specifically contribute to postinvasion NHR to the disease.
To identify more genes that function in Arabidopsis NHR to SBR, Langenbach et al. (2016) searched for genes co-regulated with BRT1. Upon confirming the genes’ importance in Arabidopsis postinvasion resistance, the authors expressed these genes in soybean. Four so-called postinvasion-induced NHR genes (PINGs) indeed reduced SBR disease severity. The supposed function of individual PING proteins is quite diverse and includes an EARLI4-like phospholipase (PING4), a group I receptor-like kinase (PING5), a GDSL-like lipase (PING7), and a germin-like protein (PING9). The exact mode of action of PINGs in conferring resistance to P. pachyrhizi has remained elusive (Langenbach et al., 2016). However, the study discloses that interspecies gene transfer is a promising strategy for conferring SBR resistance to soybean. Gene donor and receiver plant obviously do not need to be closely related, although it is likely that the successful transfer of a protein’s function from one species to another implies conservation or convergence of its physiological environment (e.g., signaling networks). Thus, employing phylogenetically related non-hosts might further enhance the success of interspecies NHR gene transfer as a means for SBR resistance. Because P. pachyrhizi infects many plants, non-hosts to the fungus are rare, especially in the legume family of plants. M. truncatula is the only reported leguminous non-host as sporulation of P. pachyrhizi has not been observed on this plant (Uppalapati et al., 2012; Ishiga et al., 2015). The former authors did a forward genetic screen to identify M. truncatula mutants with altered resistance to P. pachyrhizi. Because of its diploid genome, M. truncatula is better suited for forward genetic screening than the allopolyploid soybean crop (Gill et al., 2009). Furthermore, there is highly conserved microsynteny between soybean and M. truncatula (Yan et al., 2003). The screen by Uppalapati et al. (2012) identified an inhibitor of rust germ tube differentiation (irg)1 mutant on which P. pachyrhizi failed to promote preinfection structures. It turned out that the loss of abaxial epicuticular wax crystals and the reduced surface hydrophobicity inhibited fungal development on irg1 (Uppalapati et al., 2012). The mutation was mapped to PALM1 encoding a Cys(2)His(2) zinc finger TF controlling the expression of genes involved in long-chain fatty acid biosynthesis and transport (Uppalapati et al., 2012).
To further investigate the role of surface hydrophobicity or epicuticular waxes on P. pachyrhizi development, Ishiga et al. (2013) recorded the fungal transcriptome during germination on a hydrophobic surface (glass slides coated with epicuticular wax from wild-type plants and irg1/palm1 mutants) and on the leaf surface of M. truncatula wild-type plants and the irg1/palm1 mutant. They found expression of kinase family genes was activated on the hydrophobic surface and on the M. truncatula wild type but not on irg1/palm1. This result suggested that leaf hydrophobicity or epicuticular waxes may trigger expression of P. pachyrhizi genes involved in pre-penetration structure formation (Ishiga et al., 2013). Importance of cutin or cuticular waxes to both, germination and appressoria formation has also been reported for other fungal pathogens of plants (Mendoza-Mendoza et al., 2009; Hansjakob et al., 2011; Weidenbach et al., 2014). Further characterization of the irg1/palm1 mutant may help better understand asymmetric epicuticular wax loading on leaf surfaces and its importance to plant-pathogen interactions. Additionally, identifying IRG1/PALM1 orthologues and/or modifying epicuticular wax composition in soybean might be useful to conferring resistance to P. pachyrhizi.
Transcriptome analysis of the M. truncatula–P. pachyrhizi interaction revealed induction of many genes in the phenylpropanoid, flavonoid, and isoflavonoid pathways (Ishiga et al., 2015). Accompanying metabolome studies disclosed accumulation of the isoflavonoid derivative medicarpin and its intermediates in P. pachyrhizi-inoculated plants. Because medicarpin inhibited the germination and differentiation of P. pachyrhizi uredospores in vitro (Ishiga et al., 2015), the phytoalexin might contribute to NHR to P. pachyrhizi in M. truncatula. Various studies with P. pachyrhizi hosts also pointed to a role of phytoalexins in the interaction of plants with the fungus (Chung and Singh, 2008; Lygin et al., 2009). As the expression of genes in the secondary metabolism is strongly affected upon P. pachyrhizi infection in soybean (van de Mortel et al., 2007; Choi et al., 2008; Panthee et al., 2009; Schneider et al., 2011), secondary metabolites seem to be crucial to both host resistance and NHR to SBR.
RNA Interference and Host-Induced Gene Silencing
Another option for controlling SBR is by using RNAi to specifically silence essential P. pachyrhizi genes. HIGS, a specific RNAi technique, provided protection from sucking insects, nematodes, fungi, oomycetes, bacteria, and viruses (Koch and Kogel, 2014). To our knowledge there is not a single report on the application of HIGS in soybean for fighting SBR or other fungal diseases. However, knockdown of nematode genes by siRNAs expressed in soybean was demonstrated (Steeves et al., 2006; Li et al., 2010; Niu et al., 2012; Youssef et al., 2013). Moreover, the successful silencing of fungal genes, including those of the rust fungi Puccinia striiformis, P. triticina, and P. graminis in other crops (Yin et al., 2010; Panwar et al., 2013) is testament to the huge potential of this approach for fighting SBR. Various stage-specifically expressed fungal genes that may represent potential HIGS targets (e.g., genes encoding putative effectors like HESPs, kinase family proteins, cell wall degrading enzymes, metabolism-linked genes, succinate dehydrogenase, etc.) have already been identified in P. pachyrhizi (Posada-Buitrago and Frederick, 2005; Stone et al., 2012; Tremblay et al., 2012, 2013; Ishiga et al., 2013; Link et al., 2014). Since external application of dsRNAs has proven effective for the control of insect pests (Hunter et al., 2012), this approach might present a non-transgenic alternative to HIGS-mediated SBR control.
Biocontrol
In vitro studies and greenhouse and field trials reported protection by beneficial microbes with antagonistic properties to P. pachyrhizi. The fungus Simplicillium lanosoniveum preferentially colonizes P. pachyrhizi uredinia on infected soybean leaves and thereby significantly reduces SBR development in the field (Ward et al., 2012). Similarly, Kumar and Jha (2002) observed hypertrophy and shrinkage of P. pachyrhizi uredospores when colonized with Trichothecium rosae. Moreover, several strains of Bacillus spp. reduce SBR severity (Dorighello et al., 2015). One Bacillus strain that is the active ingredient in the organically approved commercial fungicide Ballad® provides SBR control. Besides antagonistic organisms, plant volatiles, such as farnesyl-acetate, can be used for biocontrol of SBR (Mendgen et al., 2006). Same is true for coffee oil and essential oils from Hyptis marrubioides, Aloysia gratissima, and Cordia verbenacea which suppressed spore germination in vitro and reduced SBR severity under greenhouse and/or field conditions (da Silva et al., 2014; Dorighello et al., 2015). Moreover, acibenzolar-S-methyl treatment or soil application of silicon reduced SBR severity on soybean leaves (da Cruz et al., 2013). Silicon most likely acts in two ways. First, it establishes a physical penetration barrier when deposited in the subcuticular layer and second, it primes plants for enhanced defense (Ma and Yamaji, 2006; da Cruz et al., 2013). Furthermore, soil application of saccharin and shale water were reported to induce SBR resistance in soybean (Srivastava et al., 2011; Mehta et al., 2015). These examples illustrate the potential of SBR biocontrol. However, the cost-benefit ratio and feasibility of field scale biocontrol needs to be determined to estimate the actual agronomic value of such approaches.
Conclusion
Phakopsora pachyrhizi is the causal agent of SBR and thus a major threat to global soybean production. Novel compounds in the SDHI class of fungicides hold promise for successful SBR control in the upcoming years, but P. pachyrhizi is likely to become increasingly insensitive to SDHI action as it has been observed for DMI and QoI fungicides. Similarly, the SBR resistance conferred by individual R genes was swiftly overcome in the field, but the pyramiding (stacking) of known and yet to be identified R genes might overcome traditional R gene inefficacy. Exploiting pathway components for the major plant defense hormones, SA and JA, seems not to be a realistic option for SBR control because component overexpression often impairs plant growth and yield. By contrast, transcription coactivator utilization could have huge potential but their efficacy for effective SBR control is still awaiting assessment in both the lab and field. Synthetic biology approaches to engineer R genes and phytoalexin biosynthesis pathways are promising, especially because several phytoalexins antagonize P. pachyrhizi both in vitro and in situ. Loss or elimination of S genes also is promising for SBR control but this approach has rarely been followed up. Same is true for the exploitation of soybean accessions with tolerance or PDR to SBR. Though promising, their potential for SBR control is currently unclear. Wild Glycine species, alternative P. pachyrhizi hosts, and especially non-host plants are promising sources of germplasm for SBR resistance while AMPs, RNAi/HIGS, and biocontrol approaches hold promise for sustainable soybean production in the future. It seems that an integrated approach exploiting different measures is likely to provide the best possible means for the effective control of SBR.
Author Contributions
CL, RC, SB, and AM contributed various chapters to the article. CL composed the review. CL, AM, and UC thoroughly reviewed the consecutive manuscript drafts.
Conflict of Interest Statement
Author RC is an employee of BASF Plant Science Company GmbH.
Acknowledgments
We thank Holger Schultheiss for valuable comments on the manuscript. We also appreciate provision of information by Cláudia Vierira Godoy and Francismar Corrêa Marcelino-Guimarães.
Abbreviations
- AMP
Antimicrobial peptide
- CRISPR
Clustered regularly interspersed short palindromic repeats
- DMI
Demethylation inhibitor
- dsRNA
Double stranded RNA
- ETI
Effector-triggered immunity
- FRAC
Fungicide Resistance Action Committee
- HESP
Haustoria-expressed secreted protein
- HIGS
Host-induced gene silencing
- IAP
Intragenic antimicrobial peptide
- JA
Jasmonic acid
- MDR
Muldidrug resistance
- NB-LRR
Nucleotide-binding leucine-rich repeat
- NHR
non-host resistance
- PDR
Partial disease resistance
- QoI
Quinone outside inhibitor
- R gene
Resistance gene
- RNAi
RNA interference
- Rpp
Resistance to P. pachyrhizi
- S gene
Susceptibility gene
- SA
Salicylic acid
- SAR
Systemic acquired resistance
- SBR
Asian soybean rust
- SDHI
Succinate dehydrogenase inhibitor
- siRNA
Small interfering RNA
- TF
Transcription factor
- TILLING
Targeting induced local lesions in genomes
- VIGS
Virus-induced gene silencing
Footnotes
Funding. Work on Phakopsora pachyrhizi-plant interactions in the Conrath lab is supported by BASF Plant Science Company GmbH, German Research Foundation (DFG, CO186/10-1), and the Bioeconomy Science Center (BioSC).
References
- Alves M. S., Soares Z. G., Vidigal P. M. P., Barros E. G., Poddanosqui A. M. P., Aoyagi L. N., et al. (2015). Differential expression of four soybean bZIP genes during Phakopsora pachyrhizi infection. Funct. Integr. Genomics 15 685–696. 10.1007/s10142-015-0445-0 [DOI] [PubMed] [Google Scholar]
- Aoyagi L. N., Lopes-Caitar V. S., de Carvalho M. C. C. G., Darben L. M., Polizel-Podanosqui A., Kuwahara M. K., et al. (2014). Genomic and transcriptomic characterization of the transcription factor family R2R3-MYB in soybean and its involvement in the resistance responses to Phakopsora pachyrhizi. Plant Sci. 229 32–42. 10.1016/j.plantsci.2014.08.005 [DOI] [PubMed] [Google Scholar]
- Arias C. A. A., Toledo J. F. F., Almeida L. A., Pipolo G. E. S., Carneiro R. V., Abdelnoor R. V., et al. (2008). “Asian rust in Brazil: varietal resistance,” in Facing the Challenge of Soybean Rust in South America eds Kudo H., Suenaga K., Soares R. M. S., Toledo A. (Tsukuba: JIRCAS; ) 29–30. [Google Scholar]
- Atamian H. S., Eulgem T., Kaloshian I. (2012). SlWRKY70 is required for Mi-1-mediated resistance to aphids and nematodes in tomato. Planta 235 299–309. 10.1007/s00425-011-1509-6 [DOI] [PubMed] [Google Scholar]
- Bai Y., van der Hulst R., Bonnema G., Marcel T. C., Meijer-Dekens F., Niks R. E., et al. (2005). Tomato defense to Oidium neolycopersici: dominant Ol genes confer isolate-dependent resistance via a different mechanism than recessive ol-2. Mol. Plant Microbe Interact. 18 354–362. 10.1094/MPMI-18-0354 [DOI] [PubMed] [Google Scholar]
- Barbosa G. F., da Cruz Centurion M. A. P., Marin B. T., Barbosa G. F. (2013). Effect of reduced fungicide doses on control of soybean Asian rust and bean yield. Interciencia 38 347–352. [Google Scholar]
- Bencke-Malato M., Cabreira C., Wiebke-Strohm B., Bücker-Neto L., Mancini E., Osorio M. B., et al. (2014). Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol. 14:236 10.1186/s12870-014-0236-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettgenhaeuser J., Gilbert B., Ayliffe M., Moscou M. J. (2014). Nonhost resistance to rust pathogens – a continuation of continua. Front. Plant Sci. 5:664 10.3389/fpls.2014.00664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhor T. J., Chimote V. P., Deshmukh M. P. (2014). Inheritance of rust (Phakopsora pachyrhizi) resistance in soybean. J. Food Legum 27 177–185. 10.4238/2014.July.25.18 [DOI] [Google Scholar]
- Bhor T. J., Chimote V. P., Deshmukh M. P. (2015). Molecular tagging of Asiatic soybean rust resistance in exotic genotype EC 241780 reveals complementation of two genes. Plant Breed. 134 70–77. 10.1111/pbr.12240 [DOI] [Google Scholar]
- Bilgin D., de Lucia E. H., Zangerl A. R., Singh R. J. (2009). Plant-Derived Biofungicide Against Soybean Rust Disease. U.S. Provisional Patent Application No 12/370,373. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54 519–546. 10.1146/annurev.arplant.54.031902.134938 [DOI] [PubMed] [Google Scholar]
- Bolon Y., Stec A. O., Michno J., Roessler J., Bhaskar P. B., Ries L., et al. (2014). Genome resilience and prevalence of segmental duplications following fast neutron irradiation of soybean. Genetics 198 967–981. 10.1534/genetics.114.170340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolon Y.-T., Haun W. J., Xu W. W., Grant D., Stacey M. G., Nelson R. T., et al. (2011). Phenotypic and genomic analyses of a fast neutron mutant population resource in soybean. Plant Physiol. 156 240–253. 10.1104/pp.110.170811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonde M. R., Nester S. E., Moore W. F., Allen T. W. (2009). Comparative susceptibility of kudzu accessions from the southeastern United States to infection by Phakopsora pachyrhizi. Plant Dis. 93 593–598. 10.1094/PDIS-93-6-0593 [DOI] [PubMed] [Google Scholar]
- Bowling S. A., Guo A., Cao H., Gordon A. S., Klessig D. F., Dong X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6 1845–1857. 10.1105/tpc.6.12.1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley C. A. (2007). “Fungicide resistance management in soybean,” in Using Foliar Fungicides to Manage Soybean Rust eds Dorrance A. E., Draper M. A., Hershman D. E. (Columbus, OH: Land-Grant Universities Cooperating NCERA-208 and OMAF; ) 57–60. [Google Scholar]
- Brand G. D., Magalhães M. T. Q., Tinoco M. L. P., Aragão F. J. L., Nicoli J., Kelly S. M., et al. (2012). Probing protein sequences as sources for encrypted antimicrobial peptides. PLoS ONE 7:e45848 10.1371/journal.pone.0045848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent K. J., Holloman D. W. (2007). Fungicide Resistance in Crop Pathogens: How Can It Be Managed? Brussels: CropLife International. [Google Scholar]
- Bromfield K. R. (1984). Soybean Rust. St. Paul, MN: American Phytopathological Society. [Google Scholar]
- Bromfield K. R., Hartwig E. E. (1980). Resistance to soybean rust (Phakopsora pachyrhizi) and mode of inheritance. Crop Sci. 20 254–255. 10.1071/AR9800951 [DOI] [Google Scholar]
- Bromfield K. R., Melching J. S. (1982). Sources of specific resistance to soybean rust. Phytopathology 72:706. [Google Scholar]
- Burdon J. J. (1987). Phenotypic and genetic patterns of resistance to the pathogen Phakopsora pachyrhizi in populations of Glycine canescens. Oecologia 73 257–267. 10.1007/BF00377516 [DOI] [PubMed] [Google Scholar]
- Burdon J. J. (1988). Major gene resistance to Phakopsora pachyrhizi in Glycine canescens, a wild relative of soybean. Theor. Appl. Genet. 75 923–928. 10.1007/BF00258055 [DOI] [Google Scholar]
- Burdon J. J., Speer S. S. (1984). A set of differential Glycine hosts for the identification of races of Phakopsora pachyrhizi Syd. Euphytica 33 891–896. 10.1007/BF00021917 [DOI] [Google Scholar]
- Büschges R., Hollricher K., Panstruga R., Simons G., Wolter M., Frijters A., et al. (1997). The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88 695–705. 10.1016/S0092-8674(00)81912-1 [DOI] [PubMed] [Google Scholar]
- Calvo ÉS., Kiihl R. A. S., Garcia A., Harada A., Hiromoto D. M. (2008). Two major recessive soybean genes conferring soybean rust resistance. Crop Sci. 48 1350–1354. 10.2135/cropsci2007.10.0589 [DOI] [Google Scholar]
- Campe R., Loehrer M., Conrath U., Goellner K. (2014). Phakopsora pachyrhizi induces defense marker genes to necrotrophs in Arabidopsis thaliana. Physiol. Mol. Plant Pathol. 87 1–8. 10.1016/j.pmpp.2014.04.005 [DOI] [Google Scholar]
- Cannon S. B., Shoemaker R. C. (2012). Evolutionary and comparative analyses of the soybean genome. Breed. Sci. 61 437–444. 10.1270/jsbbs.61.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Li X., Dong X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. U.S.A. 95 6531–6536. 10.1073/pnas.95.11.6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Nair M. G., Nitiss J. L. (1995). Metabolites of daidzein and genistein and their biological activities. J. Nat. Prod. 58 1901–1905. 10.1021/np50126a016 [DOI] [PubMed] [Google Scholar]
- Chen X.-K., Zhang J.-Y., Zhang Z., Du X.-L., Du B.-B., Qu S.-C. (2012). Overexpressing MhNPR1 in transgenic Fuji apples enhances resistance to apple powdery mildew. Mol. Biol. Rep. 39 8083–8089. 10.1007/s11033-012-1655-3 [DOI] [PubMed] [Google Scholar]
- Chern M., Fitzgerald H. A., Canlas P. E., Navarre D. A., Ronald P. C. (2005). Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol. Plant Microbe. Interact. 18 511–520. 10.1094/MPMI-18-0511 [DOI] [PubMed] [Google Scholar]
- Chiang H. S., Norris D. M. (1983). Phenolic and tannin contents as related to anatomical parameters of soybean resistance to agromyzid bean flies. J. Agric. Food Chem. 31 726–730. 10.1021/jf00118a012 [DOI] [Google Scholar]
- Choi J. J., Alkharouf N. W., Schneider K. T., Matthews B. F., Frederick R. D. (2008). Expression patterns in soybean resistant to Phakopsora pachyrhizi reveal the importance of peroxidases and lipoxygenases. Funct. Integr. Genomics 8 341–359. 10.1007/s10142-008-0080-0 [DOI] [PubMed] [Google Scholar]
- Chung G., Singh R. J. (2008). Broadening the genetic base of soybean: a multidisciplinary approach. Crit. Rev. Plant Sci. 27 295–341. 10.1080/07352680802333904 [DOI] [Google Scholar]
- Cooper B., Campbell K. B., Feng J., Garrett W. M., Frederick R. (2011). Nuclear proteomic changes linked to soybean rust resistance. Mol. Biosyst. 7 773–783. 10.1039/C0MB00171F [DOI] [PubMed] [Google Scholar]
- Cooper B., Campbell K. B., McMahon M. B., Luster D. G. (2013). Disruption of Rpp1-mediated soybean rust immunity by virus-induced gene silencing. Plant Signal. Behav. 8:e27543 10.4161/psb.27543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. L., Till B. J., Laport R. G., Darlow M. C., Kleffner J. M., Jamai A., et al. (2008). TILLING to detect induced mutations in soybean. BMC Plant Biol. 8:9 10.1186/1471-2229-8-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cruz M. F. A., Rodrigues F. Á, Polanco L. R., da Silva Curvêlo C. R., Nascimento K. J. T., Moreira M. A., et al. (2013). Inducers of resistance and silicon on the activity of defense enzymes in the soybean-Phakopsora pachyrhizi interaction. Bragantia 72 162–172. 10.1590/S0006-87052013005000025 [DOI] [Google Scholar]
- da Silva A. C., de Souza P. E., Amaral D. C., Zeviani W. M., Pinto J. E. B. P. (2014). Essential oils from Hyptis marrubioides, Aloysia gratissima and Cordia verbenacea reduce the progress of Asian soybean rust. Acta Sci. Agron. 36 159–166. 10.4025/actasciagron.v36i2.17441 [DOI] [Google Scholar]
- Dixon R. A., Achnine L., Kota P., Liu C.-J., Reddy M. S. S., Wang L. (2002). The phenylpropanoid pathway and plant defence-a genomics perspective. Mol. Plant Pathol. 3 371–390. 10.1046/j.1364-3703.2002.00131.x [DOI] [PubMed] [Google Scholar]
- Dorighello D. V., Bettiol W., Maia N. B., de Campos Leite R. M. V. B. (2015). Controlling Asian soybean rust (Phakopsora pachyrhizi) with Bacillus spp. and coffee oil. Crop Prot. 67 59–65. 10.1016/j.cropro.2014.09.017 [DOI] [Google Scholar]
- Falk A., Feys B. J., Frost L. N., Jones J. D. G., Daniels M. J., Parker J. E. (1999). EDS1 an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. U.S.A. 96 3292–3297. 10.1073/pnas.96.6.3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D. M., Merk H. L., Namuth-Covert D. (2012). Gene pyramiding using molecular markers. Plant Breed. Genomics. Available at: http://articles.extension.org/pages/32465/gene-pyramiding-using-molecular-markers [Google Scholar]
- Fu Z. Q., Dong X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64 839–863. 10.1146/annurev-arplant-042811-105606 [DOI] [PubMed] [Google Scholar]
- Garcia A., Calvo E. S., de Souza Kiihl R. A., Harada A., Hiromoto D. M., Vieira L. G. (2008). Molecular mapping of soybean rust (Phakopsora pachyrhizi) resistance genes: discovery of a novel locus and alleles. Theor. Appl. Genet. 117 545–553. 10.1007/s00122-008-0798-z [DOI] [PubMed] [Google Scholar]
- Gawehns F., Cornelissen B. J. C., Takken F. L. W. (2013). The potential of effector-target genes in breeding for plant innate immunity. Microb. Biotechnol. 6 223–229. 10.1111/1751-7915.12023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakopoulou A., Steele J. F. C., Segretin M. E., Bozkurt T., Zhou J., Robatzek S., et al. (2015). Tomato I2 immune receptor can be engineered to confer partial resistance to the oomycete Phytophthora infestans in addition to the fungus Fusarium oxysporum. Mol. Plant Microbe Interact. 27 624–637. 10.1094/MPMI-07-15-0147-R [DOI] [PubMed] [Google Scholar]
- Gill N., Findley S., Walling J. G., Hans C., Ma J., Doyle J., et al. (2009). Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol. 151 1167–1174. 10.1104/pp.109.137935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy C. V. (2011). “Phakopsora pachyrhizi: the performance of soybean rust fungicides over years and regions in Brazil,” in Modern Fungicides and Antifungal Compounds VI eds Dehne H. W., Deising H. B., Gisi U., Kuck K. H., Russell P. E., Lyr H. (Braunschweig: Deutsche Phytomedizinische Gesellschaft e.V. Selbstverlag; ) 203–209. [Google Scholar]
- Godoy C. V. (2012). “Risk and management of fungicide resistance in the Asian soybean rust fungus Phakopsora pachyrhizi,” in Fungicide Resistance in Crop Protection: Risk and Management ed. Thind T. S. (Wallingford: CABI; ) 87–95. 10.1079/9781845939052.0000 [DOI] [Google Scholar]
- Godoy C. V., Bueno A. F., Gazziero D. L. P. (2015). Brazilian soybean pest management and threats to its sustainability. Outlooks Pest Manag. 26 113–117. 10.1564/v26_jun_06 [DOI] [Google Scholar]
- Goellner K., Loehrer M., Langenbach C., Conrath U., Koch E., Schaffrath U. (2010). Phakopsora pachyrhizi, the causal agent of Asian soybean rust. Mol. Plant Pathol. 11 169–177. 10.1111/j.1364-3703.2009.00589.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso V., Palermo S., Sierotzki H., Garibaldi A., Gisi U. (2006a). Cytochrome b gene structure and consequences for resistance to Qo inhibitor fungicides in plant pathogens. Pest Manag. Sci. 62 465–472. 10.1002/ps.1236 [DOI] [PubMed] [Google Scholar]
- Grasso V., Sierotzki H., Garibaldi A., Gisi U. (2006b). Characterization of the cytochrome b gene fragment of Puccinia species responsible for the binding site of QoI fungicides. Pestic. Biochem. Physiol. 84 72–82. 10.1016/j.pestbp.2005.05.005 [DOI] [Google Scholar]
- Grasso V., Sierotzki H., Garibaldi A., Gisi U. (2006c). Relatedness among agronomically important rusts based on mitochondrial cytochrome b gene and ribosomal ITS sequences. J. Phytopathol. 154 110–118. 10.1111/j.1439-0434.2006.01070.x [DOI] [Google Scholar]
- Guicherit E., Bartlett D., Dale S. M., Haas H. U., Scalliet G., Walter H., et al. (2014). “Solatenol-the second generation benzonorbornene SDHI carboxamide with outstanding performance against key crop diseases,” in Modern Fungicides and Antifungal Compounds VII eds Dehne H. W., Deising H. B., Fraaije B., Gisi U., Hermann D., Mehl A., et al. (Braunschweig: Deutsche Phytomedizinische Gesellschaft e.V. Selbstverlag; ) 67–72. [Google Scholar]
- Hahlbrock K., Scheel D. (1989). Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40 347–369. 10.1146/annurev.pp.40.060189.002023 [DOI] [Google Scholar]
- Hansjakob A., Riederer M., Hildebrandt U. (2011). Wax matters: absence of very-long-chain aldehydes from the leaf cuticular wax of the glossy11 mutant of maize compromises the prepenetration processes of Blumeria graminis. Plant Pathol. 60 1151–1161. 10.1111/j.1365-3059.2011.02467.x [DOI] [Google Scholar]
- Hartman G. L., Frederick R. D., Miles M. R. (2005). Breeding for resistance to soybean rust. Plant Dis. 89 664–666. 10.1094/PD-89-0664 [DOI] [PubMed] [Google Scholar]
- Hartwig E., Bromfield K. (1983). Relationships among three genes conferring specific resistance to rust in soybeans. Crop Sci. 23 237–239. 10.2135/cropsci1983.0011183X002300020012x [DOI] [Google Scholar]
- Hartwig E. E. (1986). Identification of a fourth major gene conferring resistance to soybean rust. Crop Sci. 26 1135–1136. 10.2135/cropsci1986.0011183X002600060010x [DOI] [Google Scholar]
- Humphry M., Consonni C., Panstruga R. (2006). mlo-based powdery mildew immunity: silver bullet or simply non-host resistance? Mol. Plant Pathol. 7 605–610. 10.1111/j.1364-3703.2006.00362.x [DOI] [PubMed] [Google Scholar]
- Hunter W. B., Glick E., Paldi N., Bextine B. R. (2012). Advances in RNA interference: dsRNA treatment in trees and grapevines for insect pest suppression. Southwest. Entomol. 37 85–87. 10.3958/059.037.0110 [DOI] [Google Scholar]
- Ishiga Y., Rao Uppalapati S., Gill U. S., Huhman D., Tang Y., Mysore K. S. (2015). Transcriptomic and metabolomic analyses identify a role for chlorophyll catabolism and phytoalexin during Medicago nonhost resistance against Asian soybean rust. Sci. Rep. 5:13061 10.1038/srep13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga Y., Uppalapati S. R., Mysore K. S. (2013). Expression analysis reveals a role for hydrophobic or epicuticular wax signals in pre-penetration structure formation of Phakopsora pachyrhizi. Plant Signal. Behav. 8:e26959 10.4161/psb.26959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs T. B., LaFayette P. R., Schmitz R. J., Parrott W. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15:16 10.1186/s12896-015-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz A. M., Burdon J. J. (1990). Predominance of a single major gene for resistance to Phakopsora pachyrhizi in a population of Glycine argyrea. Heredity 64 347–353. 10.1038/hdy.1990.43 [DOI] [Google Scholar]
- Jirage D., Tootle T. L., Reuber T. L., Frost L. N., Feys B. J., Parker J. E., et al. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 96 13583–13588. 10.1073/pnas.96.23.13583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S. A., Mailhot D. J., Gevens A. J., Marois J. J., Wright D. L., Harmon C. L., et al. (2010). Characterization of kudzu (Pueraria spp.) resistance to Phakopsora pachyrhizi, the causal agent of soybean rust. Phytopathology 100 941–948. 10.1094/PHYTO-100-9-0941 [DOI] [PubMed] [Google Scholar]
- Kang B.-C., Yeam I., Frantz J. D., Murphy J. F., Jahn M. M. (2005). The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 42 392–405. 10.1111/j.1365-313X.2005.02381.x [DOI] [PubMed] [Google Scholar]
- Kendrick M. D., Harris D. K., Ha B.-K., Hyten D. L., Cregan P. B., Frederick R. D., et al. (2011). Identification of a second Asian soybean rust resistance gene in Hyuuga soybean. Phytopathology 101 535–543. 10.1094/PHYTO-09-10-0257 [DOI] [PubMed] [Google Scholar]
- Kim Y.-S., Dixon E. W., Vincelli P., Farman M. L. (2003). Field resistance to strobilurin (QoI) fungicides in Pyricularia grisea caused by mutations in the mitochondrial cytochrome b gene. Phytopathology 93 891–900. 10.1094/PHYTO.2003.93.7.891 [DOI] [PubMed] [Google Scholar]
- Klosowski A. C., May De Mio L. L., Miessner S., Rodrigues R., Stammler G. (2015). Detection of the F129L mutation in the cytochrome b gene in Phakopsora pachyrhizi. Pest Manag. Sci. 72 1211–1215. 10.1002/ps.4099 [DOI] [PubMed] [Google Scholar]
- Koch A., Kogel K. (2014). New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol. J. 12 821–831. 10.1111/pbi.12226 [DOI] [PubMed] [Google Scholar]
- Kretschmer M., Leroch M., Mosbach A., Walker A.-S., Fillinger S., Mernke D., et al. (2009). Fungicide-driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea. PLoS Pathog. 5:e1000696 10.1371/journal.ppat.1000696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Jha D. K. (2002). Trichothecium roseum: a potential agent for the biological control of soybean rust. Indian Phytopathol. 55 232–234. [Google Scholar]
- La Camera S., Gouzerh G., Dhondt S., Hoffmann L., Fritig B., Legrand M., et al. (2004). Metabolic reprogramming in plant innate immunity: the contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 198 267–284. 10.1111/j.0105-2896.2004.0129.x [DOI] [PubMed] [Google Scholar]
- Langenbach C., Campe R., Schaffrath U., Goellner K., Conrath U. (2013). UDP-glucosyltransferase UGT84A2/BRT1 is required for Arabidopsis nonhost resistance to the Asian soybean rust pathogen Phakopsora pachyrhizi. New Phytol. 198 536–545. 10.1111/nph.12155 [DOI] [PubMed] [Google Scholar]
- Langenbach C., Schultheiss H., Rosendahl M., Tresch N., Conrath U., Goellner K. (2016). Interspecies gene transfer provides soybean resistance to a fungal pathogen. Plant Biotechnol. J. 14 699–708. 10.1111/pbi.12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiminger J. H., Adolf B., Hausladen H. (2014). Occurrence of the F129L mutation in Alternaria solani populations in Germany in response to QoI application, and its effect on sensitivity. Plant Pathol. 63 640–650. 10.1111/ppa.12120 [DOI] [Google Scholar]
- Lemos N. G., de Lucca e Braccini A., Abdelnoor R. V., de Oliveira M. C. N., Suenaga K., Yamanaka N. (2011). Characterization of genes Rpp2 Rpp4 and Rpp5 for resistance to soybean rust. Euphytica 182 53–64. 10.1007/s10681-011-0465-3 [DOI] [Google Scholar]
- Li J., Todd T. C., Oakley T. R., Lee J., Trick H. N. (2010). Host-derived suppression of nematode reproductive and fitness genes decreases fecundity of Heterodera glycines Ichinohe. Planta 232 775–785. 10.1007/s00425-010-1209-7 [DOI] [PubMed] [Google Scholar]
- Li S., Smith J. R., Ray J. D., Frederick R. D. (2012). Identification of a new soybean rust resistance gene in PI 567102B. Theor. Appl. Genet. 125 133–142. 10.1007/s00122-012-1821-y [DOI] [PubMed] [Google Scholar]
- Li X., Clarke J. D., Zhang Y., Dong X. (2001). Activation of an EDS1-mediated R -gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant Microbe Interact. 14 1131–1139. 10.1094/MPMI.2001.14.10.1131 [DOI] [PubMed] [Google Scholar]
- Link T. I., Lang P., Scheffler B. E., Duke M. V., Graham M. A., Cooper B., et al. (2014). The haustorial transcriptomes of Uromyces appendiculatus and Phakopsora pachyrhizi and their candidate effector families. Mol. Plant. Pathol. 15 379–393. 10.1111/mpp.12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka V., Dittgen J., Bednarek P., Bhat R., Wiermer M., Stein M., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310 1180–1183. 10.1126/science.1119409 [DOI] [PubMed] [Google Scholar]
- Loehrer M., Langenbach C., Goellner K., Conrath U., Schaffrath U. (2008). Characterization of nonhost resistance of Arabidopsis to the Asian soybean rust. Mol. Plant Microbe Interact. 21 1421–1430. 10.1094/MPMI-21-11-1421 [DOI] [PubMed] [Google Scholar]
- Loehrer M., Schaffrath U. (2011). “Asian soybean rust – meet a prominent challenge in soybean cultivation,” in Soybean - Biochemistry, Chemistry and Physiology ed. Ng T. -B. (Rijeka: InTech; ) 83–100. 10.5772/15651 [DOI] [Google Scholar]
- Loehrer M., Vogel A., Huettel B., Reinhardt R., Benes V., Duplessis S., et al. (2014). On the current status of Phakopsora pachyrhizi genome sequencing. Front. Plant Sci. 5:377 10.3389/fpls.2014.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Holland J. B., Munkvold G. P., Jannink J.-L. (2006). Responses to selection for partial resistance to crown rust in oat. Crop Sci. 46 1260–1265. 10.2135/cropsci2005.06-0169 [DOI] [Google Scholar]
- Lozano-Torres J. L., Wilbers R. H. P., Gawronski P., Boshoven J. C., Finkers-Tomczak A., Cordewener J. H. G., et al. (2012). Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. U.S.A. 109 10119–10124. 10.1073/pnas.1202867109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygin A. V., Li S., Vittal R., Widholm J. M., Hartman G. L., Lozovaya V. V. (2009). The importance of phenolic metabolism to limit the growth of Phakopsora pachyrhizi. Phytopathology 99 1412–1420. 10.1094/PHYTO-99-12-1412 [DOI] [PubMed] [Google Scholar]
- Lyngkjær M. F., Newton A. C., Atzema J. L., Baker S. J. (2000). The barley mlo-gene: an important powdery mildew resistance source. Agronomy 20 745–756. 10.1051/agro:2000173 [DOI] [Google Scholar]
- Ma J. F., Yamaji N. (2006). Silicon uptake and accumulation in higher plants. Trends Plant Sci. 11 392–397. 10.1016/j.tplants.2006.06.007 [DOI] [PubMed] [Google Scholar]
- Maltby L., Brock T. C. M., van den Brink P. J. (2009). Fungicide risk assessment for aquatic ecosystems: importance of interspecific variation, toxic mode of action, and exposure regime. Environ. Sci. Technol. 43 7556–7563. 10.1021/es901461c [DOI] [PubMed] [Google Scholar]
- Maphosa M., Talwana H., Tukamuhabwa P. (2012). Enhancing soybean rust resistance through Rpp2 Rpp3 and Rpp4 pair wise gene pyramiding. African J. Agric. Res. 7 4271–4277. 10.5897/AJAR12.1123 [DOI] [Google Scholar]
- Mathieu M., Winters E. K., Kong F., Wan J., Wang S., Eckert H., et al. (2009). Establishment of a soybean (Glycine max Merr. L) transposon-based mutagenesis repository. Planta 229 279–289. 10.1007/s00425-008-0827-9 [DOI] [PubMed] [Google Scholar]
- McDonald B. A. (2014). Using dynamic diversity to achieve durable disease resistance in agricultural ecosystems. Trop. Plant Pathol. 39 191–196. 10.1590/S1982-56762014000300001 [DOI] [Google Scholar]
- McDonald B. A., Linde C. (2002). Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40 349–379. 10.1146/annurev.phyto.40.120501.101443 [DOI] [PubMed] [Google Scholar]
- McLean R. J., Byth D. E. (1980). Inheritance of resistance to rust (Phakopsora pachyrhizi) in soybeans. Aust. J. Agric. Res. 31 951–956. 10.1071/AR9800951 [DOI] [Google Scholar]
- Mehta Y. R., Marangoni M. S., Matos J. N., Mandarino J. M. G., Galbieri R. (2015). Systemic acquired resistance of soybean to soybean rust induced by shale water. Am. J. Plant Sci. 6 2249–2256. 10.4236/ajps.2015.614227 [DOI] [Google Scholar]
- Mendgen K., Wirsel S. G. R., Jux A., Hoffmann J., Boland W. (2006). Volatiles modulate the development of plant pathogenic rust fungi. Planta 224 1353–1361. 10.1007/s00425-006-0320-2 [DOI] [PubMed] [Google Scholar]
- Mendoza-Mendoza A., Berndt P., Djamei A., Weise C., Linne U., Marahiel M., et al. (2009). Physical-chemical plant-derived signals induce differentiation in Ustilago maydis. Mol. Microbiol. 71 895–911. 10.1111/j.1365-2958.2008.06567.x [DOI] [PubMed] [Google Scholar]
- Meyer J. D. F., Silva D. C. G., Yang C., Pedley K. F., Zhang C., van de Mortel M., et al. (2009). Identification and analyses of candidate genes for Rpp4-mediated resistance to Asian soybean rust in soybean. Plant Physiol. 150 295–307. 10.1104/pp.108.134551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles M. R., Pastor-Corrales M. A., Hartman G. L., Frederick R. D. (2007). Differential response of common bean cultivars to Phakopsora pachyrhizi. Plant Dis. 91 698–704. 10.1094/PDIS-91-6-0698 [DOI] [PubMed] [Google Scholar]
- Monteros M. J., Missaoui A. M., Phillips D. V., Walker D. R., Boerma H. R. (2007). Mapping and confirmation of the “Hyuuga” red-brown lesion resistance gene for Asian soybean rust. Crop Sci. 47 829–834. 10.2135/cropsci06.07.0462 [DOI] [Google Scholar]
- Morales A. M. A. P., O’Rourke J. A., van de Mortel M., Scheider K. T., Bancroft T. J., Borém A., et al. (2013). Transcriptome analyses and virus induced gene silencing identify genes in the Rpp4-mediated Asian soybean rust resistance pathway. Funct. Plant Biol. 40 1029–1047. 10.1071/FP12296 [DOI] [PubMed] [Google Scholar]
- Mueller A. N., Ziemann S., Treitschke S., Aßmann D., Doehlemann G. (2013). Compatibility in the Ustilago maydis–maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog. 9:e1003177 10.1371/journal.ppat.1003177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller T. A., Miles M. R., Morel W., Marois J. J., Wright D. L., Kemerait R. C., et al. (2009). Effect of fungicide and timing of application on soybean rust severity and yield. Plant Dis. 93 243–248. 10.1094/PDIS-93-3-0243 [DOI] [PubMed] [Google Scholar]
- Narusaka M., Shirasu K., Noutoshi Y., Kubo Y., Shiraishi T., Iwabuchi M., et al. (2009). RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J. 60 218–226. 10.1111/j.1365-313X.2009.03949.x [DOI] [PubMed] [Google Scholar]
- Nawrath C., Heck S., Parinthawong N., Metraux J. P. (2002). EDS5 an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14 275–286. 10.1105/tpc.010376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J., Jian H., Xu J., Chen C., Guo Q., Liu Q., et al. (2012). RNAi silencing of the Meloidogyne incognita Rpn7 gene reduces nematode parasitic success. Eur. J. Plant Pathol. 134 131–144. 10.10007/s10658-012-9971-y [DOI] [Google Scholar]
- Nombela G., Williamson V. M., Muñiz M. (2003). The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol. Plant Microbe Interact. 16 645–649. 10.1094/MPMI.2003.16.7.645 [DOI] [PubMed] [Google Scholar]
- Oliver R. P. (2014). A reassessment of the risk of rust fungi developing resistance to fungicides. Pest Manag. Sci. 70 1641–1645. 10.1002/ps.3767 [DOI] [PubMed] [Google Scholar]
- Pandey A. K., Yang C., Zhang C., Graham M. A., Horstman H. D., Lee Y., et al. (2011). Functional analysis of the Asian soybean rust resistance pathway mediated by Rpp2. Mol. Plant Microbe Interact. 24 194–206. 10.1094/MPMI-08-10-0187 [DOI] [PubMed] [Google Scholar]
- Panthee D. R., Marois J. J., Wright D. L., Narváez D., Yuan J. S., Stewart C. N. (2009). Differential expression of genes in soybean in response to the causal agent of Asian soybean rust (Phakopsora pachyrhizi Sydow) is soybean growth stage-specific. Theor. Appl. Genet. 118 359–370. 10.1007/s00122-008-0905-1 [DOI] [PubMed] [Google Scholar]
- Panwar V., McCallum B., Bakkeren G. (2013). Endogenous silencing of Puccinia triticina pathogenicity genes through in planta-expressed sequences leads to the suppression of rust diseases on wheat. Plant J. 73 521–532. 10.1111/tpj.12047 [DOI] [PubMed] [Google Scholar]
- Pavan S., Jacobsen E., Visser R. G. F., Bai Y. (2010). Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 25 1–12. 10.1007/s11032-009-9323-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierozzi P. H. B., Ribeiro A. S., Moreira J. U. V., Laperuta L. D. C., Rachid B. F., Lima W. F., et al. (2008). New soybean (Glycine max Fabales, Fabaceae) sources of qualitative genetic resistance to Asian soybean rust caused by Phakopsora pachyrhizi (Uredinales, Phakopsoraceae). Genet. Mol. Biol. 31 505–511. 10.1590/S1415-47572008000300018 [DOI] [Google Scholar]
- Pieterse C. M. J., van der Does D., Zamioudis C., Leon-Reyes A., van Wees S. C. M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28 489–521. 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- Posada-Buitrago M. L., Frederick R. D. (2005). Expressed sequence tag analysis of the soybean rust pathogen Phakopsora pachyrhizi. Fungal Genet. Biol. 42 949–962. 10.1016/j.fgb.2005.06.004 [DOI] [PubMed] [Google Scholar]
- Rahnamaeian M. (2011). Antimicrobial peptides: modes of mechanism, modulation of defense responses. Plant Signal. Behav. 6 1325–1332. 10.4161/psb.6.9.16319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis E. M., Deuner E., Zanatta M., Reis E. M., Deuner E., Zanatta M. (2015). In vivo sensitivity of Phakopsora pachyrhizi to DMI and QoI fungicides. Summa Phytopathol. 41 21–24. 10.1590/0100-5405/1975 [DOI] [Google Scholar]
- Salomon D., Sessa G. (2012). “Biotechnological strategies for engineering plants with durable resistance to fungal and bacterial pathogens,” in Plant Biotechnology and Agriculture -Prospects for the 21st Century- eds Altman A., Hasegawa P. M. (Cambridge, MS: Academic Press; ) 329–342. 10.1016/B978-0-12-381466-1.00021-3 [DOI] [Google Scholar]
- Scherm H., Christiano R. S. C., Esker P. D., Del Ponte E. M., Godoy C. V. (2009). Quantitative review of fungicide efficacy trials for managing soybean rust in Brazil. Crop Prot. 28 774–782. 10.1016/j.cropro.2009.05.006 [DOI] [Google Scholar]
- Schmitz H. K., Medeiros C.-A., Craig I. R., Stammler G. (2014). Sensitivity of Phakopsora pachyrhizi towards quinone-outside-inhibitors and demethylation-inhibitors, and corresponding resistance mechanisms. Pest Manag. Sci. 70 378–388. 10.1002/ps.3562 [DOI] [PubMed] [Google Scholar]
- Schneider K. T., van de Mortel M., Bancroft T. J., Braun E., Nettleton D., Nelson R. T., et al. (2011). Biphasic gene expression changes elicited by Phakopsora pachyrhizi in soybean correlates with fungal penetration and haustoria formation. Plant Physiol. 157 355–371. 10.1104/pp.111.181149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P., Panstruga R. (2011). A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 16 117–125. 10.1016/j.tplants.2011.01.001 [DOI] [PubMed] [Google Scholar]
- Segretin M. E., Pais M., Franceschetti M., Chaparro-garcia A., Bos J. I. B., Banfield M. J., et al. (2014). Single amino acid mutations in the potato immune receptor R3a expand response to Phytophthora effectors. Mol. Plant Microbe Interact. 27 624–637. 10.1094/MPMI-02-14-0040-R [DOI] [PubMed] [Google Scholar]
- Sierotzki H., Frey R., Wullschleger J., Palermo S., Karlin S., Godwin J., et al. (2007). Cytochrome b gene sequence and structure of Pyrenophora teres and P. tritici-repentis and implications for QoI resistance. Pest Manag. Sci. 63 225–233. 10.1002/ps.1330 [DOI] [PubMed] [Google Scholar]
- Sierotzki H., Scalliet G. (2013). A review of current knowledge of resistance aspects for the next-generation succinate dehydrogenase inhibitor fungicides. Phytopathology 103 880–887. 10.1094/PHYTO-01-13-0009-RVW [DOI] [PubMed] [Google Scholar]
- Singh R. J., Nelson R. L. (2015). Intersubgeneric hybridization between Glycine max and G. tomentella: production of F1 amphidiploid, BC1 BC2 BC3 and fertile soybean plants. Theor. Appl. Genet. 128 1117–1136. 10.1007/s00122-015-2494-0 [DOI] [PubMed] [Google Scholar]
- Slaminko T. L., Miles M. R., Frederick R. D., Bonde M. R., Hartman G. L. (2008). New legume hosts of Phakopsora pachyrhizi based on greenhouse evaluations. Plant Dis. 92 767–771. 10.1094/PDIS-92-5-0767 [DOI] [PubMed] [Google Scholar]
- Soria-Guerra R. E., Rosales-Mendoza S., Chang S., Haudenshield J. S., Padmanaban A., Rodriguez-Zas S., et al. (2010). Transcriptome analysis of resistant and susceptible genotypes of Glycine tomentella during Phakopsora pachyrhizi infection reveals novel rust resistance genes. Theor. Appl. Genet. 120 1315–1333. 10.1007/s00122-009-1258-0 [DOI] [PubMed] [Google Scholar]
- Souza T. L. P. O., Dessaune S. N., Moreira M. A., Barros E. G. (2014). Soybean rust resistance sources and inheritance in the common bean (Phaseolus vulgaris L.). Genet. Mol. Res. 13 5626–5636. 10.4238/2014.July.25.18 [DOI] [PubMed] [Google Scholar]
- Srivastava P., George S., Marois J. J., Wright D. L., Walker D. R. (2011). Saccharin-induced systemic acquired resistance against rust (Phakopsora pachyrhizi) infection in soybean: effects on growth and development. Crop Prot. 30 726–732. 10.1016/j.cropro.2011.02.023 [DOI] [Google Scholar]
- Steeves R. M., Todd T. C., Essig J. S., Trick H. N. (2006). Transgenic soybeans expressing siRNAs specific to a major sperm protein gene suppress Heterodera glycines reproduction. Funct. Plant Biol. 33 991–999. 10.1071/FP06130 [DOI] [PubMed] [Google Scholar]
- Stone C. L., McMahon M. B., Fortis L. L., Nuñez A., Smythers G. W., Luster D. G., et al. (2012). Gene expression and proteomic analysis of the formation of Phakopsora pachyrhizi appressoria. BMC Genomics 13:269 10.1186/1471-2164-13-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. H., Li Z., Jewett D. K., Britton K. O., Ye W. H., Ge X. (2005). Genetic diversity of Pueraria lobata (kudzu) and closely related taxa as revealed by inter-simple sequence repeat analysis. Weed Res. 45 255–260. 10.1111/j.1365-3180.2005.00462.x [DOI] [Google Scholar]
- Tremblay A., Hosseini P., Li S., Alkharouf N. W., Matthews B. F. (2012). Identification of genes expressed by Phakopsora pachyrhizi, the pathogen causing soybean rust, at a late stage of infection of susceptible soybean leaves. Plant Pathol. 61 773–786. 10.1111/j.1365-3059.2011.02550.x [DOI] [Google Scholar]
- Tremblay A., Hosseini P., Li S., Alkharouf N. W., Matthews B. F. (2013). Analysis of Phakopsora pachyrhizi transcript abundance in critical pathways at four time-points during infection of a susceptible soybean cultivar using deep sequencing. BMC Genomics 14:614 10.1186/1471-2164-14-614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukamuhabwa P., Maphosa M. (2010). State of Knowledge on Breeding for Durable Resistance to Soybean Rust Disease in the Developing World. Rome: FAO. [Google Scholar]
- Uppalapati S. R., Ishiga Y., Doraiswamy V., Bedair M., Mittal S., Chen J., et al. (2012). Loss of abaxial leaf epicuticular wax in Medicago truncatula irg1/palm1 mutants results in reduced spore differentiation of anthracnose and nonhost rust pathogens. Plant Cell 24 353–370. 10.1105/tpc.111.093104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Mortel M., Recknor J. C., Graham M. A., Nettleton D., Dittman J. D., Nelson R. T., et al. (2007). Distinct biphasic mRNA changes in response to Asian soybean rust infection. Mol. Plant Microbe Interact. 20 887–899. 10.1094/MPMI-20-8-0887 [DOI] [PubMed] [Google Scholar]
- Verweij P. E., Snelders E., Kema G. H. J., Mellado E., Melchers W. J. G. (2009). Azole resistance in Aspergillus fumigatus: a side-effect of environmental fungicide use? Lancet. Infect. Dis. 9 789–795. 10.1016/S1473-3099(09)70265-8 [DOI] [PubMed] [Google Scholar]
- Vogt T. (2010). Phenylpropanoid biosynthesis. Mol. Plant 3 2–20. 10.1093/mp/ssp106 [DOI] [PubMed] [Google Scholar]
- Wang J., Shine M. B., Gao Q. M., Navarre D., Jiang W., Liu C., et al. (2014). Enhanced disease susceptibility1 mediates pathogen resistance and virulence function of a bacterial effector in soybean. Plant Physiol. 165 1269–1284. 10.1104/pp.114.242495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward N. A., Robertson C. L., Chanda A. K., Schneider R. W. (2012). Effects of Simplicillium lanosoniveum on Phakopsora pachyrhizi, the soybean rust pathogen, and its use as a biological control agent. Phytopathology 102 749–760. 10.1094/PHYTO-01-11-0031 [DOI] [PubMed] [Google Scholar]
- Weidenbach D., Jansen M., Franke R. B., Hensel G., Weissgerber W., Ulferts S., et al. (2014). Evolutionary conserved function of barley and Arabidopsis 3-KETOACYL-CoA SYNTHASES in providing wax signals for germination of powdery mildew fungi. Plant Physiol. 166 1621–1633. 10.1104/pp.114.246348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M., Feys B. J., Parker J. E. (2005). Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8 383–389. 10.1016/j.pbi.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Wightwick A., Walters R., Allinson G., Reichman S., Menzies N. (2010). “Environmental risks of fungicides used in horticultural production systems,” in Fungicides ed. Carisse O. (Rijeka: InTech; ) 273–304. [Google Scholar]
- Xie K., Zhang J., Yang Y. (2014). Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol. Plant 7 923–926. 10.1093/mp/ssu009 [DOI] [PubMed] [Google Scholar]
- Yamanaka N., Lemos N. G., Uno M., Akamatsu H., Yamaoka Y., Abdelnoor R. V., et al. (2013). Resistance to Asian soybean rust in soybean lines with the pyramided three Rpp genes. Crop Breed. Appl. Biotechnol. 13 75–82. 10.1590/S1984-70332013000100009 [DOI] [Google Scholar]
- Yamanaka N., Morishita M., Mori T., Lemos N. G., Hossain M. M., Akamatsu H., et al. (2015). Multiple Rpp-gene pyramiding confers resistance to Asian soybean rust isolates that are virulent on each of the pyramided genes. Trop. Plant Pathol. 40 283–290. 10.1007/s40858-015-0038-4 [DOI] [Google Scholar]
- Yan H. H., Mudge J., Kim D.-J., Larsen D., Shoemaker R. C., Cook D. R., et al. (2003). Estimates of conserved microsynteny among the genomes of Glycine max, Medicago truncatula and Arabidopsis thaliana. Theor. Appl. Genet. 106 1256–1265. 10.1007/s00122-002-1183-y [DOI] [PubMed] [Google Scholar]
- Yin C., Jurgenson J. E., Hulbert S. H. (2010). Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 24 554–561. 10.1094/MPMI-10-10-0229 [DOI] [PubMed] [Google Scholar]
- Yorinori J. T., Paiva W. M., Frederick R. D., Costamilan L. M., Bertagnolli P. F., Hartman G. E. (2005). Epidemics of soybean rust (Phakopsora pachyrhizi) in Brazil and Paraguay from 2001 to 2003. Plant Dis. 89 675–677. 10.1094/PD-89-0675 [DOI] [PubMed] [Google Scholar]
- Youssef R. M., Kim K. H., Haroon S. A., Matthews B. F. (2013). Post-transcriptional gene silencing of the gene encoding aldolase from soybean cyst nematode by transformed soybean roots. Exp. Parasitol. 134 266–274. 10.1016/j.exppara.2013.03.009 [DOI] [PubMed] [Google Scholar]
- Zhang C., Bradshaw J. D., Whitham S. A., Hill J. H. (2010). The development of an efficient multipurpose Bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol. 153 52–65. 10.1104/pp.109.151639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Ghabrial S. A. (2006). Development of Bean pod mottle virus-based vectors for stable protein expression and sequence-specific virus-induced gene silencing in soybean. Virology 344 401–411. 10.1016/j.virol.2005.08.046 [DOI] [PubMed] [Google Scholar]
- Zhang C., Whitham S. A., Hill J. H. (2013). Virus-induced gene silencing in soybean and common bean. Methods Mol. Biol. 975 149–156. 10.1007/978-1-62703-278-0_11 [DOI] [PubMed] [Google Scholar]


