Abstract
Dengue viruses (DENVs) and their vectors are widely distributed throughout the tropical and subtropical regions of the world. An autochthonous case of DENV was reported in Tokyo, Japan, in 2014, for the first time in 70 years. A comprehensive database of DENV sequences containing both serotype and genotype data and epidemiological data is crucial to trace DENV outbreak isolates and promptly respond to outbreaks. We constructed a DENV database containing the serotype, genotype, year and country/region of collection by collecting all publically available DENV sequence information from the National Center for Biotechnology Information (NCBI) and assigning genotype information. We also implemented the web service Dengue Genographic Viewer (DGV), which shows the geographical distribution of each DENV genotype in a user-specified time span. DGV also assigns the serotype and genotype to a user-specified sequence by performing a homology search against the curated DENV database, and shows its homologous sequences with the geographical position and year of collection. DGV also shows the distribution of DENV-infected entrants to Japan by plotting epidemiological data from the Infectious Agents Surveillance Report (IASR), Japan. This overview of the DENV genotype distribution may aid in planning for the control of DENV infections. DGV is freely available online at: (https://gph.niid.go.jp/geograph/dengue/content/genomemap).
Keywords: NGS, dengue virus, genotyping, genograph, web service
Introduction
Dengue viruses (DENVs) are members of the genus Flavivirus in the family Flaviviridae and consist of four serotypes (DENV-1 to -4) (Lanciotti et al., 1992; Kuhn et al., 2002). Each serotype can be divided into five to six genotypes. However, there is no standard genotyping classification [DENV-1, (Goncalvez et al., 2002); DENV-2, (Anez et al., 2011; Khan et al., 2013); DENV-3, (Lanciotti et al., 1994; Wittke et al., 2002; Klungthong et al., 2008); DENV-4, (Abubakar et al., 2002)]. DENV has a positive-sense, single-stranded RNA genome that is ~11 kb in length and encodes a capsid protein (C), premembrane protein (prM), and envelope glycoprotein (E) in addition to seven non-structural proteins (NSs, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). DENV infection causes dengue illness, which may range from dengue fever (a mild illness) to dengue hemorrhagic fever and dengue shock syndrome (the severe forms of the illness) in addition to asymptomatic cases (Centers for Disease Control and Prevention; http://www.cdc.gov/dengue/clinicalLab/clinical.html). Infection results in lifetime immunity against the same serotype, but successive exposure to different DENVs increases the likelihood of contracting a severe form of dengue illness, such as dengue hemorrhagic fever or dengue shock syndrome (Chiappelli et al., 2014).
DENV and its vectors have become widely distributed throughout the tropical and subtropical regions of the world (Murray et al., 2013). An autochthonous case of DENV infection was reported in Tokyo, Japan, in 2014 for the first time in 70 years (Kutsuna et al., 2015). To ensure prompt action in response to a DENV outbreak, a comprehensive DENV database based on the genotypes would be essential for tracing the outbreak source. To date, only two DENV databases provide genotype information. The web service ViPR1 (Pickett et al., 2012) supports genetic analysis based on the viral genome for a tested input sequence, including DENV sequences. The second database is the Dengue virus genotyping database2 (Yamashita et al., 2013), which provides a summary table containing the DENV serotype/genotype, year and country of collection and accession number. There are many other DENV databases; however, no other sites provide summarized genotype information. The Dengue Virus Resource3 facilitates the retrieval of DENV sequences deposited in GenBank according to serotype, disease symptom, host, region/country, genome region, and collection and/or release data (Resch et al., 2009). DENVirDB4 provides sequence information and computationally curated information of dengue viral proteins (Asnet et al., 2014). DENVDB focuses on the dengue virus sequence database for keyword searches (no publication: http://proline.bic.nus.edu.sg/denvdb/). Finally, the Dengue Virus Portal is a sequence collection with metadata (no publication: https://www.broadinstitute.org/annotation/viral/Dengue/Home.html).
Here, we constructed the website Dengue Genographic Viewer (DGV), which presents DENV information based on the genotype and epidemiological data by using the geographic tool Google Maps© to update the recent dissemination of DENV genotypes from a global perspective.
Materials and methods
Construction of the DENV database
The DENV genotype database was constructed as follows: (1) all accessible DENV nucleotide sequences were collected; (2) the complete sequences of each protein region (C, prM, E, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) were extracted from the sequences; (3) a blastn homology search was performed against the genotype database and the genotype of the most homologous sequence was assigned; (4) and the genotype data were stored in a database using SQLite (https://www.sqlite.org/).
The DENV nucleotide sequences were downloaded from the NCBI database using key words (“Dengue virus”[porgn:__txid12637]).
A blastx homology search was performed to detect nucleotide regions that corresponded to each mature DENV protein; nucleotide regions that exhibited more than 85% sequence coverage to the protein were used for the subsequent analysis.
To reduce the time required to obtain the most homologous sequences in the genotype database, we reduced the number of sequences used in the blast search by clustering highly homologous sequences. We performed a uclust search (Edgar, 2010) against the nucleotide sequences of each protein region and selected one representative sequence for each homologous sequence group with a clustering threshold of 99% identity; then, the representative sequences were subjected to a homology search against the genotype database. The original genotype database was constructed according to the method proposed by previous report (Yamashita et al., 2013). Briefly, the representative sequences were aligned by using the mafft (Katoh and Standley, 2014) program and Neighbor joining (NJ) phylogenetic trees were constructed using the MEGA5 program (Tamura et al., 2011). The genotype of each gene was assigned manually according to the previous genotype database (Yamashita et al., 2013).
Sequence ID, country/region and year of collection were extracted from the deposited GenBank data and integrated into the SQL database by using an in house Perl script.
The above processes except for the original database construction are performed automatically every night to update the recent DENV database.
Interactive viewer using web interface
We implemented a set of viewer applications on DGV by using Google Maps©, which shows the data in a temporal and spatial manner. One application presents the geographical distribution of each DENV genotype on the map in a user-specified time span. Another option is a homology search program that searches for the most homologous DENV sequence in the DGV database and show the geographical positions of closely related sequences on the map. The other interface shows the sources of imported dengue cases on the map, according to the Infectious Agents Surveillance Report (IASR), Japan (http://www.nih.go.jp/niid/en/iasr-e.html). This set of applications is available at the DGV web site (https://gph.niid.go.jp/geograph/dengue/content/genomemap).
Results and discussion
Distribution of DENV genotypes
On March 4, 2016, DGV included a total of 10,514 DENV sequences, which consisted of 3872, 3414, 2309, and 919 DENV serotype-1, -2, -3, and -4 sequences, respectively (Table 1). Some genotypes have been abundantly sequenced and deposited in the public database, whereas other genotypes have rarely been sequenced (i.e., DENV-1 genotype II was reported in only seven records from 1960 to 2012, DENV-4 genotype III was also reported in only seven records from 1997 to 2001, and DENV-3 genotype IV has not been reported since 1977). These rare genotypes may have become minor populations or may be undergoing a silent transmission cycle (Lanciotti et al., 1994; Chen and Vasilakis, 2011; Santiago et al., 2012).
Table 1.
Number of DENV records (March 4, 2016).
| Serotype | Genotype | Records on public database | Total sequences of each serotype |
|---|---|---|---|
| DENV-1 | |||
| I | 2508 | ||
| Ib | 13 | ||
| II | 7 | ||
| III | 6 | ||
| IV | 407 | ||
| V | 879 | ||
| Recombinant | 49 | ||
| Lab strain | 3 | 3872 | |
| DENV-2 | |||
| American | 101 | ||
| Asian-American | 1245 | ||
| Asian I | 858 | ||
| Asian II | 105 | ||
| Cosmopolitan | 1042 | ||
| Sylvatic | 26 | ||
| Recombinant | 35 | ||
| Unknown | 2 | 3414 | |
| DENV-3 | |||
| I | 326 | ||
| II | 742 | ||
| III | 1192 | ||
| IV | 5 | ||
| V | 34 | ||
| Recombinant | 9 | ||
| Unknown | 1 | 2309 | |
| DENV-4 | |||
| I | 295 | ||
| IIA | 223 | ||
| IIB | 385 | ||
| III | 7 | ||
| Sylvatic | 6 | ||
| Recombinant | 3 | 919 | |
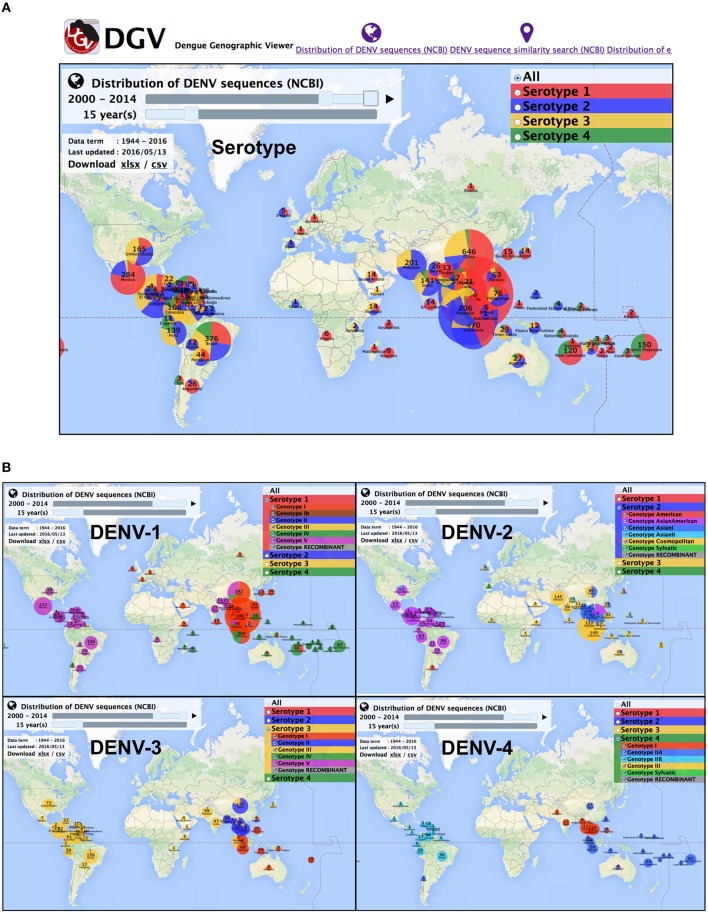
Fifteen years' worth of data from 2000 to 2014 for all serotypes showed that DENV sequences were primarily reported from South to Southeast Asia, Central to South America, and the countries of Oceania (Figure 1A). Some biases in DENV serotype compositions were observed in several countries. For instance, the dominant serotypes were DENV-1 and -2 in Mexico, DENV-1 and -4 in Polynesian countries with the exception of Fiji, and DENV-2 and -3 in Pakistan. In contrast, all serotypes were sampled in Brazil and Thailand.
Figure 1.
Overview of DGV for serotype distribution for all DENV (A) and genotype distribution for each serotype (B) in 2000–2014.
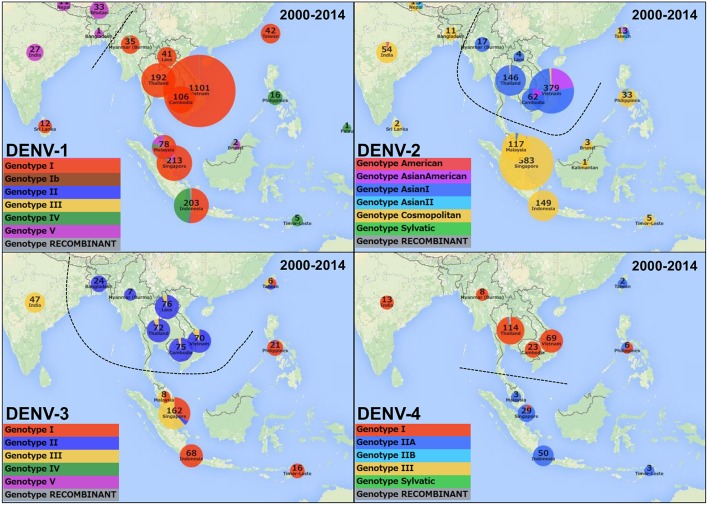
Intriguingly, when focusing on the genotype instead of the serotype, the data from 2000 to 2014 showed at least three potential geographical genotype distribution border lines in Asia (Figures 1B,2). The first border is between the American continents and other regions (Figure 1B), the second is located between Bangladesh and Myanmar for the genotype distributions of DENV-1 and -2 and India and Myanmar for DENV-3, and the third is located between Indochina and the Malay Peninsula (Figure 2). There seem to be differences in the DENV-1 and -3 distributions between Malaysia, Singapore and Indonesia; however, the border line is not clear because Malaysia and Indonesia consist of many islands and share Kalimantan Island and the deposited sequence data do not specify the original island isolation site. Although, some boundaries are not clear, these boundaries are roughly conserved among all serotypes except for the Bangladesh-Myanmar border line for DENV-4, suggesting potential barriers against the vector mosquitos' movements or human activities between the countries.
Figure 2.
Genotype distribution for each DENV serotype in Asia in 2000–2014. Possible borderline of genotype distribution in each serotype is shown as dash line.
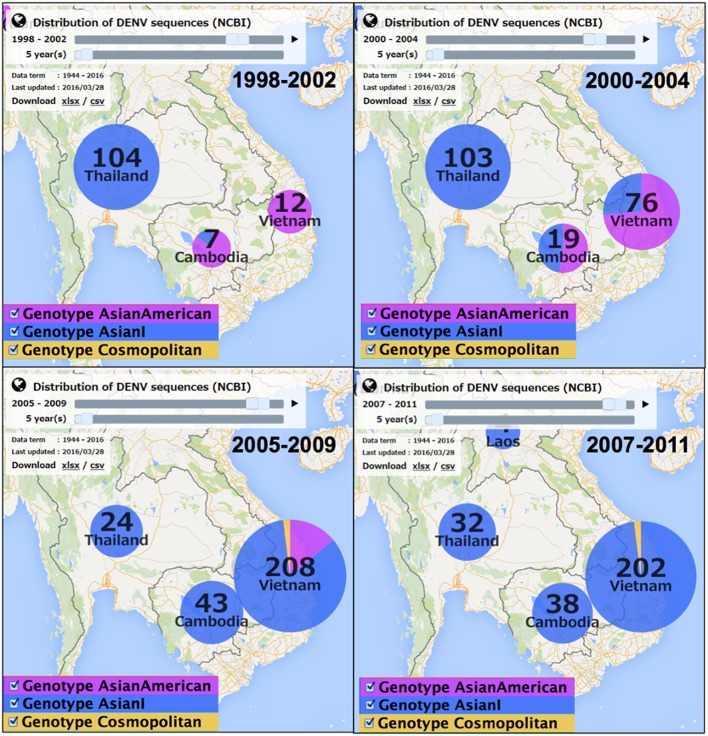
We also found a timeline change in the predominant genotypes. From 1998 to 2007, the dominant genotype in Asia was Cosmopolitan, although India-Pakistan-Sri Lanka and Southeast-Oceania belonged to different lineages (Khan et al., 2013). The major genotypes in the Indochina countries were different from those of the other Asian countries; genotype Asian I was predominant in Thailand, whereas genotype Asian American was predominant in Cambodia and Vietnam (Figure 3 and Movie S1). From 2001, Asian I increased in Cambodia and Vietnam until finally in 2007 Asian I became the predominant genotype in Indochina. The genotype Asian I viruses in Thailand seemed to be widely disseminated into Vietnam via Cambodia but did not reach Malaysia and Bangladesh (Figure 2). Thus, the Asian American genotype was replaced by Asian I in Cambodia and Vietnam between 1998 and 2011. This example also suggests the idea of genotype transition, which probably reflects the mosquito vector habitat and human activities in the Indochinese Peninsula.
Figure 3.
Genotype distribution of DENV-2 in Indochina from 1998 to 2011. The movie of the genotype distribution in 1986–2014 can be downloaded as Movie S1.
DGV currently does not support the prediction of Dengue epidemics, because number of deposited sequence data does not always reflect the actual number of events, in addition, it takes long time to be a public sequence through isolation, sequencing, and publication.
DENV sequence similarity search
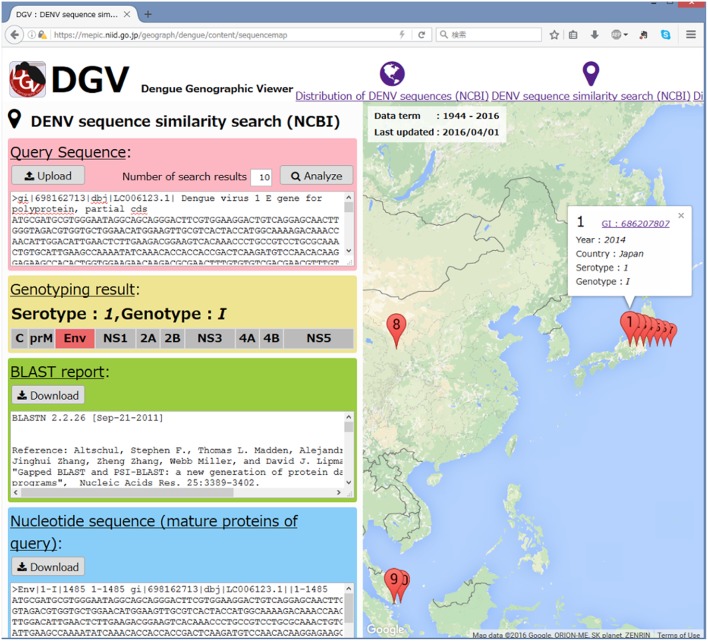
DGV provides a search engine for the assignment of the DENV serotype, genotype, and origin country according to the most homologous sequence on the basis of a blastn search against the DENV database. The search results are shown as text and are also plotted through Google Maps©. Subsequently, the query sequence is divided into mature protein regions and displayed with a serotype/genotype assignment. The homology search results and the divided nucleotide sequences in fasta format can be downloaded.
Here, we present an example similarity search for an Env sequence derived from an autochthonous case in Japan (LC006123 or gi: 698162713). DGV assigned the sequence as the Env region of the DENV-1 genotype I and identified homologous sequences from Japan, China, Singapore and Indonesia. These results are consistent with those from a previous study (Figure 4; Kutsuna et al., 2015).
Figure 4.
A screenshot of the DENV sequence similarity search. An Env sequence derived from an autochthonous case in Japan (LC006123 or gi: 698162713) was used as a sample query. The query was assigned as the Env region of DENV-1 genotype I.
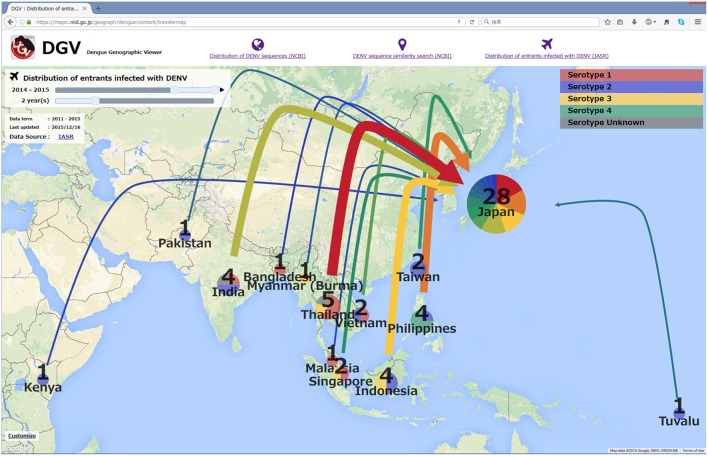
Distribution of entrants infected with DENV
To aid in visualizing the source countries of dengue infection cases imported to Japan, the number of annual imported cases was also mapped on Google Maps©. The serotype (but not genotype), year, and visiting country/area are also indicated based on the Infectious Agents Surveillance Report (IASR), which releases monthly data and information obtained from prefectural and municipal public health institutes and quarantine stations to the public (Figure 5).
Figure 5.
A screenshot of the distribution of entrants infected with DENV, which shows the cumulative number of infected entrants from 2014 to 2015 based on the IASR.
Author contributions
AY performed the experimental design, participated in the analysis and drafted the manuscript. TS1 implemented the application, performed the data collection, constructed the original genotype database, and participated in the analysis. TS2, KK, and TT reviewed the application and participated in the discussion. MK contributed to the experimental design, performed the analysis and drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by a grant for Research on Emerging and Re-emerging Infectious Diseases (H25 Shinko-Ippan-015/H26 Shinko-Gyosei-Shitei-002) from the Ministry of Health, Labor and Welfare, Japan, and was also supported by the Research Program on Emerging and Re-emerging Infectious Diseases (15fk0108011h0003 and 15fm0108022h0001) from the Japan Agency for Medical Research and Development, AMED. This work was also partially supported by JSPS KAKENHI Grant Number 15K08488.The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Ms. Inamine from NIID for drawing the DGV icon. We thank Prof. Ikuta and Prof. Yasunaga from BIKEN for allowing us to use the original dataset “dengue virus genotyping database.”
Glossary
Abbreviations
- DENV
Dengue virus
- C
capsid protein
- prM
premembrane protein
- E
envelope glycoprotein
- NS
non-structural protein
- NCBI
national center for biotechnology information
- IASR
Infectious Agents Surveillance Report.
Footnotes
1ViPR (Virus Pathogen Database and Analysis Resource): http://www.viprbrc.org/brc/home.spg?decorator=flavi_dengue
2Dengue virus genotyping database: http://www.gen-info.osaka-u.ac.jp/~uhmin/genotyping/index.html
3Dengue Virus Resource: https://www.ncbi.nlm.nih.gov/genome/viruses/variation/dengue/
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00875
Genotype transition of DENV-2 in Asia from 1986–1990 to 2010–2014.
References
- Abubakar S., Wong P. F., Chan Y. F. (2002). Emergence of dengue virus type 4 genotype IIA in Malaysia. J. Gen. Virol. 83, 2437–2442. 10.1099/0022-1317-83-10-2437 [DOI] [PubMed] [Google Scholar]
- Anez G., Morales-Betoulle M. E., Rios M. (2011). Circulation of different lineages of dengue virus type 2 in Central America, their evolutionary time-scale and selection pressure analysis. PLoS ONE 6:e27459. 10.1371/journal.pone.0027459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnet M. J., Rubia A. G., Ramya G., Nagalakshmi R. N., Shenbagarathai R. (2014). DENVirDB: a web portal of dengue virus sequence information on Asian isolates. J. Vector Borne Dis. 51, 82–85. [PubMed] [Google Scholar]
- Chen R., Vasilakis N. (2011). Dengue–quo tu et quo vadis? Viruses 3, 1562–1608. 10.3390/v3091562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappelli F., Santos S. M., Caldeira Brant X. M., Bakhordarian A., Thames A. D., Maida C. A., et al. (2014). Viral immune evasion in dengue: toward evidence-based revisions of clinical practice guidelines. Bioinformation 10, 726–733. 10.6026/97320630010726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Goncalvez A. P., Escalante A. A., Pujol F. H., Ludert J. E., Tovar D., Salas R. A., et al. (2002). Diversity and evolution of the envelope gene of dengue virus type 1. Virology 303, 110–119. 10.1006/viro.2002.1686 [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2014). MAFFT: iterative refinement and additional methods. Methods Mol. Biol. 1079, 131–146. 10.1007/978-1-62703-646-7_8 [DOI] [PubMed] [Google Scholar]
- Khan M. A., Ellis E. M., Tissera H. A., Alvi M. Y., Rahman F. F., Masud F., et al. (2013). Emergence and diversification of dengue 2 cosmopolitan genotype in Pakistan, 2011. PLoS ONE 8:e56391. 10.1371/journal.pone.0056391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klungthong C., Putnak R., Mammen M. P., Li T., Zhang C. (2008). Molecular genotyping of dengue viruses by phylogenetic analysis of the sequences of individual genes. J. Virol. Methods 154, 175–181. 10.1016/j.jviromet.2008.07.021 [DOI] [PubMed] [Google Scholar]
- Kuhn R. J., Zhang W., Rossmann M. G., Pletnev S. V., Corver J., Lenches E., et al. (2002). Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108, 717–725. 10.1016/S0092-8674(02)00660-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsuna S., Kato Y., Moi M. L., Kotaki A., Ota M., Shinohara K., et al. (2015). Autochthonous dengue fever, Tokyo, Japan, 2014. Emerg. Infect. Dis. 21, 517–520. 10.3201/eid2103.141662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti R. S., Calisher C. H., Gubler D. J., Chang G. J., Vorndam A. V. (1992). Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J. Clin. Microbiol. 30, 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti R. S., Lewis J. G., Gubler D. J., Trent D. W. (1994). Molecular evolution and epidemiology of dengue-3 viruses. J. Gen. Virol. 75(Pt 1), 65–75. 10.1099/0022-1317-75-1-65 [DOI] [PubMed] [Google Scholar]
- Murray N. E., Quam M. B., Wilder-Smith A. (2013). Epidemiology of dengue: past, present and future prospects. Clin. Epidemiol. 5, 299–309. 10.2147/CLEP.S34440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett B. E., Greer D. S., Zhang Y., Stewart L., Zhou L., Sun G., et al. (2012). Virus pathogen database and analysis resource (ViPR): a comprehensive bioinformatics database and analysis resource for the coronavirus research community. Viruses 4, 3209–3226. 10.3390/v4113209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch W., Zaslavsky L., Kiryutin B., Rozanov M., Bao Y., Tatusova T. A. (2009). Virus variation resources at the National Center for Biotechnology Information: dengue virus. BMC Microbiol 9:65. 10.1186/1471-2180-9-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago G. A., Mcelroy-Horne K., Lennon N. J., Santiago L. M., Birren B. W., Henn M. R., et al. (2012). Reemergence and decline of dengue virus serotype 3 in Puerto Rico. J. Infect. Dis. 206, 893–901. 10.1093/infdis/jis426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittke V., Robb T. E., Thu H. M., Nisalak A., Nimmannitya S., Kalayanrooj S., et al. (2002). Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology 301, 148–156. 10.1006/viro.2002.1549 [DOI] [PubMed] [Google Scholar]
- Yamashita A., Sasaki T., Kurosu T., Yasunaga T., Ikuta K. (2013). Origin and distribution of divergent dengue virus: novel database construction and phylogenetic analyses. Future Virol. 8, 1061–1083. 10.2217/fvl.13.99 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotype transition of DENV-2 in Asia from 1986–1990 to 2010–2014.