Abstract
Higher alcohols and acetate esters belong to the most important yeast secondary metabolites that significantly contribute to the overall flavor and aroma profile of fermented products. In Saccharomyces cerevisiae, esterification of higher alcohols is catalyzed mainly by the alcohol acetyltransferases encoded by genes ATF1 and ATF2. Previous investigation has shown other Saccharomyces species, e.g., S. kudriavzevii and S. uvarum, to vary in aroma-active higher alcohols and acetate esters formation when compared to S. cerevisiae. Here, we aimed to analyze the enzymes encoded by the ATF1 and ATF2 genes from S. kudriavzevii (SkATF1, SkATF2) and S. uvarum (SuATF1, SuATF2). The heterologous expression of the individual ATF1 and ATF2 genes in a host S. cerevisiae resulted in the enhanced production of several higher alcohols and acetate esters. Particularly, an increase of 2-phenylethyl acetate production by the strains that harbored ATF1 and ATF2 genes from S. kudriavzevii and S. uvarum was observed. When grown with individual amino acids as the nitrogen source, the strain that harbored SkATF1 showed particularly high 2-phenylethyl acetate production and the strains with introduced SkATF2 or SuATF2 revealed increased production of isobutyl acetate, isoamyl acetate, and 2-phenylethyl acetate compared to the reference strains with endogenous ATF genes. The alcohol acetyltransferase activities of the individual Atf1 and Atf2 enzymes measured in the cell extracts of the S. cerevisiae atf1 atf2 iah1 triple-null strain were detected for all the measured substrates. This indicated that S. kudriavzevii and S. uvarum Atf enzymes had broad range substrate specificity as S. cerevisiae Atf enzymes. Individual Atf1 enzymes exhibited markedly different kinetic properties since SkAtf1p showed c. twofold higher and SuAtf1p c. threefold higher Km for isoamyl alcohol than ScAtf1p. Together these results indicated that the differences found among the three Saccharomyces species during the aroma-active acetate ester formation may be due, to some extent, to the distinct properties of Atf enzymes.
Keywords: yeasts, Saccharomyces, ATF1, ATF2, higher alcohols, acetate esters, synthetic wine must
Introduction
Amino acid metabolism, which occurs during yeast fermentation processes, plays an important role in fragrance, flavor and food industries. Its importance arises from the fact that amino acid catabolism leads to the production of higher alcohols, which are precursors of acetate esters (Dickinson et al., 1997, 1998, 2000, 2003; Hazelwood et al., 2008). Higher alcohols and acetate esters belong to the principal aroma-active substances that affect the complex flavor of fermented products (Welsh et al., 1989; Verstrepen et al., 2003a). The most significant acetate esters include isobutyl acetate (fruity-like aroma), isoamyl acetate (banana), and 2-phenylethyl acetate (flowery, rose-like), which result from the esterification of the corresponding higher alcohols: i.e., isobutanol (derived from valine), isoamyl alcohol (derived from leucine), and 2-phenylethanol (derived from phenylalanine), respectively (Nykanen, 1986; Lambrechts and Pretorius, 2000; Styger et al., 2011). The reaction of higher alcohols with acetyl-CoA, which results in acetyl esters, is catalyzed in S. cerevisiae by alcohol acetyltransferases I and II (AATase I and II, EC 2.3.1.84), codified by the ATF1 and ATF2 genes, respectively (Yoshioka and Hashimoto, 1981; Malcorps et al., 1991; Minetoki et al., 1993; Fujii et al., 1994; Nagasawa et al., 1998). It has been reported that Atf1p and Atf2p catalyze the acetylation of a wide variety of higher alcohols. Mainly isoamyl alcohol, but also other alcohols, such as propanol, isobutanol, hexanol, and 2-phenylethanol, are esterified by Atf1p and Atf2p (Verstrepen et al., 2003b). Nevertheless, the contributions of each enzyme seem to differ. The deletion and overexpression of ATF2 resulted only in minor changes in the formation of acetate esters, which indicated that the ATF2-encoded enzyme plays only a minor role compared with the ATF1-encoded enzyme (Verstrepen et al., 2003b; Lilly et al., 2006).
The most common yeast species employed in industrial applications, including food, flavor, and fragrance production, is S. cerevisiae. Our previous studies reported differences in the production of the higher alcohols and acetate esters directly derived from amino acids by S. kudriavzevii and S. uvarum compared to S. cerevisiae (Stribny et al., 2015). The analysis revealed, for instance, that S. kudriavzevii produced larger amounts of higher alcohols than S. cerevisiae, whereas S. uvarum excelled in acetate esters production. Similarly, differences in aroma compounds production by the three Saccharomyces species were observed during wine must fermentations (Gamero et al., 2013). To gain a better insight into the molecular aspects of the aforementioned differences, our recent study (Stribny et al., 2016) aimed to explore the nucleotide divergences in 23 ortholog genes (and, consequently, in the corresponding enzymes) from S. kudriavzevii/S. uvarum vs. S. cerevisiae involved in amino acid catabolism, which lead to aroma-active higher alcohols and esters production. An in silico analysis using Grantham scoring, which quantitatively evaluates (dis)similarity in amino acids substitutions on the basis of physiochemical properties (composition, polarity, and molecular volume; Grantham, 1974), detected the ATF1- and ATF2-encoded proteins among the hits with the highest Grantham scores.
In the present work, we cloned ATF1 and ATF2 genes from S. kudriavzevii and S. uvarum into S. cerevisiae to examine and compare their effect on higher alcohols and acetate esters production. Kinetic parameters together with substrate specificities of the encoded enzymes were also analyzed and compared with S. cerevisiae.
Materials and Methods
Yeast Strains
The yeast strains used in this study are listed in Table 1. S. cerevisiae Ta, a haploid strain derived from commercial wine strain T73, was previously constructed in the laboratory of A. Querol. Standard complex media (0.5% peptone, 2% glucose, and 0.5% yeast extract) or SC-Ura medium [0.67% YNB, 2% glucose, 1.92 g/L Drop-out –Ura (Formedium, Norfolk, UK)] were used to grow stock cultures. DNA isolation, restriction, and gel electrophoresis were carried out by standard genetic methods. Strains were transformed by the lithium acetate procedure (Gietz and Woods, 2002).
Table 1.
List of the yeast strains used in this study.
| Strain | Species | Description | Reference |
|---|---|---|---|
| T73 | S. cerevisiae | Wine strain, Alicante, Spain | Querol et al., 1992 |
| IFO1802 | S. kudriavzevii | Type strain, NCBI | Kaneko and Banno, 1991 |
| CECT12600 | S. uvarum | Wine strain, Alicante, Spain | Spanish culture collection (CECT) |
| Ta | S. cerevisiae | T73hoΔ::loxP | A. Querol |
| JET02 | S. cerevisiae | Ta atf1Δ::NAT1 | This study |
| JET02Sk | S. cerevisiae | Ta atf1Δ::SkATF1-kX | This study |
| JET02Su | S. cerevisiae | Ta atf1Δ::SuATF1-kX | This study |
| JET02Sc | S. cerevisiae | Ta atf1Δ::ScATF1-kX | This study |
| JET03 | S. cerevisiae | Ta atf2Δ::NAT1 | This study |
| JET03Sk | S. cerevisiae | Ta atf2Δ::SkATF2-kX | This study |
| JET03Su | S. cerevisiae | Ta atf2Δ::SuATF2-kX | This study |
| JET03Sc | S. cerevisiae | Ta atf2Δ::SuATF2-kX | This study |
| BY4741atf1atf2iah1 | S. cerevisiae | MATa leu2-Δ0 his3-Δ1 met15-Δ0 ura3-Δ0 atf1Δ1::loxP atf2Δ1::loxP iah1Δ1::loxP | Uber-Garcia, 2005 |
| CLpSkATF1 | S. cerevisiae | BY4741atf1atf2iah1 pG-SkATF1-TDH3p | This study |
| CLpSuATF1 | S. cerevisiae | BY4741atf1atf2iah1 pG-SuATF1-TDH3p | This study |
| CLpScATF1 | S. cerevisiae | BY4741atf1atf2iah1 pG-ScATF1-TDH3p | This study |
| CLpSkATF2 | S. cerevisiae | BY4741atf1atf2iah1 pG-SkATF2-TDH3p | This study |
| CLpSuATF2 | S. cerevisiae | BY4741atf1atf2iah1 pG-SuATF2-TDH3p | This study |
| CLpScATF2 | S. cerevisiae | BY4741atf1atf2iah1 pG-ScATF2-TDH3p | This study |
Plasmid and Strain Construction
The S. kudriavzevii ATF1 and ATF2 genes (SkATF1 and SkATF2) were amplified from the genomic DNA of S. kudriavzevii IFO1802. The S. uvarum ATF1 and ATF2 genes (SuATF1 and SuATF2) were amplified from the genomic DNA of S. uvarum CECT12600. The S. cerevisiae ATF1 and ATF2 genes (ScATF1 and ScATF2) were amplified by the PCR from the genomic DNA of S. cerevisiae T73. The primers that we used are listed in Table 2. PCR fragments were then independently cloned into the pGREG526 vector (Jansen et al., 2005), previously cut with NotI/SalI. Before the start codon of the individual ATF genes within the pGREG526 vectors, the TDH3 promoter (amplified by PCR from genomic DNA from the S288C strain) was inserted by homologous recombination. The resulting plasmids are listed in Table 3. The constructed plasmids that contained ATF genes behind the TDH3 promoter were then introduced into the BY4741atf1atf2iah1 strain (Table 1) used in the enzyme assays.
Table 2.
The primers used in this study.
| Primer | Sequence 5′–3′ |
|---|---|
| Cloning into pGREG526 | |
| SkATF1-F | CCTAGTACGGATTAGAAGCCGCCGAGCGGGTGACATCTTCAAAAGCCTCCTCATAC |
| SkATF1-R | GCGTGACATAACTAATTACATGACTCGAGGTCGACGCCTAAGGAATGACGATACC |
| SuATF1-F | CCTAGTACGGATTAGAAGCCGCCGAGCGGGTGACAACAAAACCATAACCGAATACG |
| SuATF1-R | GCGTGACATAACTAATTACATGACTCGAGGTCGACCGGCTAAAAGAACGATACAA |
| ScATF1-F | CCTAGTACGGATTAGAAGCCGCCGAGCGGGTGACAGGTACTCATCGTAAAAGATTGC |
| ScATF1-R | GCGTGACATAACTAATTACATGACTCGAGGTCGACTAACCAACCAAAGCCGAG |
| SkATF2-F | CCTAGTACGGATTAGAAGCCGCCGAGCGGGTGACATTATCACCAGACGGCTCAC |
| SkATF2-R | GCGTGACATAACTAATTACATGACTCGAGGTCGACGCTCTGTCCGATACACCG |
| SuATF2-F | CCTAGTACGGATTAGAAGCCGCCGAGCGGGTGACAATCACCAAAGTAACCACCAT |
| SuATF2-R | GCGTGACATAACTAATTACATGACTCGAGGTCGACATACCGCTTCCTTGCTGT |
| ScATF2-F | CCTAGTACGGATTAGAAGCCGCCGAGCGGGTGACAAGGAAGCACGTCAGAAAAAG |
| ScATF2-R | GCGTGACATAACTAATTACATGACTCGAGGTCGACGCTCTGTCCGATACACTGC |
| ATF1/ATF2 deletion cassettes | |
| TaATF1-NAT1-F | ATGAATGAAATCGATGAGAAAAATCAGGCGCCCGTGCAACGGTGTTTAGGTCGATGCCATC |
| TaATF1-NAT1-R | CTAAGGGCCTAAAAGGAGAGCTTTATAAATGGAGCAAAGCGGATGGCGGCGTTAGTATCG |
| TaATF2-NAT1-F | ATGGAAGATATAGAAGGATACGAACCACATATCACTCAAGGGTGTTTAGGTCGATGCCATC |
| TaATF2-NAT1-R | TTAAAGCGACGCAAATTCGCCGATGGTTTGGTAGAAGAGCGGATGGCGGCGTTAGTATCG |
| Integration fragments | |
| pGSkATF1f | CTTCATCAGTATCACAAATACCATCAATTTATCAGCTCTCATGACTAAAATCAGCGAAGAG |
| pGSuATF1f | CTTCATCAGTATCACAAATACCATCAATTTATCAGCTCTCATGAATACCTATAGTGAAAA |
| pGScATF1f | CTTCATCAGTATCACAAATAC |
| pG-ATF1-R | TCATATTGTCGAATAATATCAGTCAAGCATCATGTGAGATCTCACTATAGGGCGAATTGG |
| pGSkATF2f | CGAATAATAACTTCAGCAATAAAAATTGTCCAGGTTAATTATGGATGATATAGAGGAATAC |
| pGSuATF2f | CGAATAATAACTTCAGCAATAAAAATTGTCCAGGTTAATTATGGATAGTTTAGAGGAATAC |
| pGScATF2f | CGAATAATAACTTCAGCAAT |
| pG-ATF2-R | TCGGCCGAGCTATACGAAGGCCCGCTACGGCAGTATCGCACTCACTATAGGGCGAATTGG |
| Integration of TDH3 promoter | |
| pG526-TDH3p-Fa | GAGCTGATACCGCTCGCCGCAGCCGAACGACCGAGCGCAGCAGTTCGAGTTTATCATTATC |
| pG-TDH3p-CF1-R | CTGATAAATTGATGGTATTTGTGATACTGATGAAGCGAAACTAAGTTCTTGGTGTT |
| pG-TDH3p-KF1-R | GGTAGATTGATGATATTTGTTATACTGATGAGTGGCGAAACTAAGTTCTTGGTGTT |
| pG-TDH3p-UF1-R | GTTGGATTGATGGTTTTGTTAAACTGCTAAATTGGCGAAACTAAGTTCTTGGTGTT |
| pG-TDH3p-CF2-R | CCTGGACAATTTTTATTGCTGAAGTTATTATTCGTCGAAACTAAGTTCTTGGTGTT |
| pG-TDH3p-KF2-R | CGCAATTTTTTGTTCGTTTGAAGATCTAGTAGAGCCGAAACTAAGTTCTTGGTGTT |
| pG-TDH3p-UF2-R | GCAGTTTTTTGTTCTGAAACCTATTGTTCGTTTGTCGAAACTAAGTTCTTGGTGTT |
| Diagnostic | |
| T73ATF1-UF | GGTACTCATCGTAAAAGATTGC |
| T73ATF2-UF | AGGAAGCACGTCAGAAAAAG |
| K2 | GGGACAATTCAACGCGTCTG |
| K3 | CCTCGACATCATCTGCCC |
| SkATF1-R1 | GCCTAAGGAATGACGATACC |
| SuATF1-R1 | CGGCTAAAAGAACGATACAA |
| ScATF1-R1 | TTTGTGATACTGATGAAGTGCCG |
| SkATF2-R1 | GCTCTGTCCGATACACCG |
| SuATF2-R1 | ATACCGCTTCCTTGCTGT |
| ScATF2-R1 | GCTCTGTCCGATACACTGC |
Table 3.
The plasmids used in this study.
| Plasmid name | Description | Reference |
|---|---|---|
| pGREG526 | URA3, kanMX, AmpR | Jansen et al., 2005 |
| pGREG526 containing ATF orthologs | ||
| pG-SkATF1-kX | Containing S. kudriavzevii ATF1 | This study |
| pG-SuATF1-kX | Containing S. uvarum ATF1 | This study |
| pG-ScATF1-kX | Containing S. cerevisiae ATF1 | This study |
| pG-SkATF2-kX | Containing S. kudriavzevii ATF2 | This study |
| pG-SuATF2-kX | Containing S. uvarum ATF2 | This study |
| pG-ScATF2-kX | Containing S. cerevisiae ATF2 | This study |
| pGREG526 harboring ATF orthologs with the TDH3 promoter | ||
| pG-SkATF1-TDH3p | Containing S. kudriavzevii ATF1 | This study |
| pG-SuATF1-TDH3p | Containing S. uvarum ATF1 | This study |
| pG-ScATF1-TDH3p | Containing S. cerevisiae ATF1 | This study |
| pG-SkATF2-TDH3p | Containing S. kudriavzevii ATF2 | This study |
| pG-SuATF2-TDH3p | Containing S. uvarum ATF2 | This study |
| pG-ScATF2-TDH3p | Containing S. cerevisiae ATF2 | This study |
The strategy of replacing endogenous ATF1 and ATF2 in the Ta genome with the corresponding ATF genes from S. kudriavzevii or S. uvarum involved two steps (i) deletion of either ATF1 or ATF2 gene; and (ii) integration of the S. kudriavzevii or S. uvarum orthologs into the locus. The ATF genes deletions in the Ta genome were performed by integrating a nourseothricin resistance cassette by homologous recombination. Deletion cassettes were amplified using pAG25 (Goldstein and McCusker, 1999) as a template and specific primers (Table 2). The resulting strains were named JET02 (atf1Δ) and JET03 (atf2Δ). The integration of SkATF1 into JET02 is described to illustrate the second step. The integration of the other orthologs was performed in the same way. The integration cassette was amplified from plasmid pG-SkATF1-kX with primers pGSkATF1f and pG-ATF1-R. The resulting PCR fragment included the SkATF1 gene, followed by kanamycin resistance marker, which was used for selection in the subsequent JET02 strain transformation. The final Ta mutant that held SkATF1 gene was named JET02Sk. The same procedure was carried out with ScATF1 (and ScATF2), which resulted in the restoration of the endogenous ATF1 (and ATF2) gene by the undergone process. This strain, named JET02Sc, was used as a reference in the subsequent assays.
Cultivation to Study the Production of the Higher Alcohols and Acetate Esters That Derived from the Corresponding Amino Acids
Cultivations were performed in triplicate with a synthetic medium that contained 0.17% YNB w/o AAs and (NH4)2SO4 (BD DIFCOTM, Madrid, Spain) and 2% glucose as the carbon source, as previously described (Stribny et al., 2015) with minimal modifications. Media were supplemented by individual amino acids leucine, phenylalanine, and valine as the nitrogen source. Concentrations were proportional to 5 g/L (NH4)2SO4 to obtain the same nitrogen content, as follows: 10 g/L leucine, 12.5 g/L phenylalanine, 8.9 g/L valine (Bolat et al., 2013).
Starter cultures were prepared by pregrowing yeast in 15-mL tubes that contained 4 mL of standard complex media. Before inoculating the experimental culture, the grown precultures were washed with water and resuspended in the same synthetic medium (with a certain nitrogen source), as used in the assay. Cells were resuspended in a volume that allowed an OD600 of 1.7 to be achieved. These precultures (100 μL) were used to inoculate 1.6 mL of synthetic media, when the initial OD600 was 0.1. Cultivation was performed in 96-well plates with 2-mL-deep wells. Wells were covered by microplate sealer (Greiner bio-one, Frickenhausen, Germany) to allow yeast growth and avoid evaporation and loss of volatile flavor compounds. Cultures were incubated for 5 days at 25°C. The individual 1.7-mL cultures were later transferred to 2-mL tubes and were stored at -20°C for the analysis.
Yeast Growth Analysis
Yeast cells growth was followed using a 96-well plate. Synthetic media were supplemented with amino acids as described above. Then 100 μL of media were inoculated in a well with 2 μL of cell suspension with OD600 = 1. Growth was monitored in a Spectrostar Nano absorbance reader (BMG Labtech, Ortenberg, Germany).
Synthetic Wine Must Fermentation
A synthetic wine must was prepared as described previously (Riou et al., 1997), but with 200 g/L of reducing sugars (100 g/L glucose + 100 g/L fructose) and with no anaerobic factors (Beltran et al., 2004). Nitrogen source 300 mg N/L was composed of amino acids (180 mg/L) and NH4Cl (120 mg/L). The composition of the amino acids mixture was that described by Beltran et al. (2004). The following mineral salts were used: KH2PO4 750 mg/L, K2SO4 500 mg/L, MgSO4 250 mg/L, CaCl2 155 mg/L, NaCl 200 mg/L, MnSO4 4 mg/L, ZnSO4 4 mg/L, CuSO4 1 mg/L, KI 1 mg/L, CoCl2 0.4 mg/L, H3BO3 1 mg/L, (NH4)6Mo7O24 1 mg/L. The following organic acids were used: malic acid 5 g/L, citric acid 0.5 g/L, and tartaric acid 3 g/L. The following vitamins were used: myo-inositol 20 mg/L, calcium pantothenate 1.5 mg/L, nicotinic acid 2 mg/L, chlorohydrate thiamine 0.25 mg/L, chlorohydrate pyridoxine 0.25 mg/L, and biotin 0.003 mg/L. The final pH was adjusted to 3.3 with NaOH.
Fermentations were performed in 250-mL glass bottles that contained 200 mL of synthetic must. Fermentations were done in triplicate at 25°C with continuous orbital shaking (150 rpm). Flasks were closed with Müller valves and monitored by weight loss until a constant weight was obtained. Immediately after fermentation ended, yeast cells were removed by centrifugation, and the contents of higher alcohols and esters in the supernatants were analyzed by gas chromatography.
Higher Alcohols and Esters Determination
The samples stored in the 2-mL tubes were centrifuged (13,000 rpm, 2 min) and 1.5 mL of the supernatant was transferred to the 15-mL vials with 0.35 g of NaCl. The 20-μl volume of 2-heptanone (0.005%) was added as an internal standard. Higher alcohols and esters were analyzed by the headspace solid phase microextraction (HS-SPME) technique using a 100-μm poly-dimethylsiloxane (PDMS) fiber (Supelco, Sigma–Aldrich, Madrid, Spain). Solutions were maintained for 2 h at 25°C to establish the headspace-liquid equilibrium. The fiber was inserted through a vial septum into the headspace and was held for 7 min. The fiber was then inserted into the gas chromatograph inlet port for 4 min at 220°C with helium flow (1 mL/min) to desorb analytes. A Thermo Science TRACE GC Ultra gas chromatograph with a flame ionization detector (FID) was used, equipped with an HP INNOWax 30 m × 0.25 m capillary column coated with a 0.25-m layer of cross-linked polyethylene glycol (Agilent Technologies, Valencia, Spain). The oven temperature program was: 5 min at 35°C, 2°C/min to 150°C, 20°C/min to 250°C and 2 min at 250°C. The detector temperature was kept constant at 300°C. A chromatograph was recorded by the ChromQuest program. Volatile compounds were identified by the retention time for reference compounds. Quantification of the volatile compounds was determined using the calibration graphs of the corresponding standard volatile compounds.
Preparation of Yeast Cell Extracts
Cell extracts were prepared as described previously (Rojas et al., 2002) with minor modifications. Yeasts were grown in SC-Ura at 25°C up to the late log phase and by taking optical density at 600 nm as a reference. Yeast cells were collected by centrifugation, washed twice with cold 0.85% NaCl solution and resuspended in ice-chilled disruption buffer [10 mM potassium phosphate, pH 7.5, which contained 0.8 mM MgCl2, 10% (w/v) glycerol, 5 mM DTT]. Protease inhibitor cocktail tablet Complete Mini, EDTA-free (Roche Diagnostics, Mannheim, Germany) was also added to the disruption buffer (one tablet per 10 mL). Disruption was performed with glass beads in a MillMix 20 Bead Beater cell disrupter (Domel Tehtnica, Zelezniki, Slovenia) at a frequency of 30 s-1 in 1-min intervals over a 10-min period. The resulting homogenate was centrifuged at 15,000 g for 30 min at 4°C and the resulting supernatant was used to assay alcohol acetyltransferase activity.
The protein concentrations in the cell extracts were estimated by the Bradford method (Bradford, 1976) using bovine serum albumin as a standard.
Enzyme Assays
Enzyme assays were performed in a 25-ml glass syringe provided with a Luer lock cap, as described previously (Rojas et al., 2002) with minor modifications. Reaction mixtures consisted in glycerol buffer [50 mM potassium phosphate, pH 7.5, 10% (w/v) glycerol] that contained higher alcohol as a cosubstrate, glycerol buffer that contained acetyl-CoA as the other cosubstrate and a cell extract. The higher alcohol, acetyl-CoA and cell extract showed the following volume ratio 1:0.1:0.4. The final volume was determined by the number of samples (1.5 mL). Isoamyl alcohol (at a final concentration of 0.01–100 mM), isobutanol (60 mM) or 2-phenylethanol (30 mM) was used as the substrate together with acetyl-CoA (0.8 mM). Substrate concentrations were determined according to Rojas (2002). After adding all the components, entrapped air was removed by the plunger and the syringe was attached to an orbital shaker. After 30 min, the 1.5-mL samples were transferred to 15-mL vials with 0.35 g NaCl for ester quantification with gas chromatography. Enzyme activity was stopped by adding 60 μL of a saturated KSCN solution.
Statistical Analysis
The presented values are averages of biological triplicates with standard errors. The differences between the measured volatile compounds were determined by a one-way ANOVA, followed by Tukey’s HSD test (statistical level of significance was set at P ≤ 0.05). The analysis was performed using the STATISTICA 7.0 software (StatSoft, Inc., Tulsa, OK, USA).
Results
Impact of the ATF1 and ATF2 Homologous Genes on the Formation of Higher Alcohols and Esters
To verify the effect of the Atf1 and Atf2 enzymes from S. kudriavzevii and S. uvarum on the final content of higher alcohols and/or acetate esters, individual genes (SkATF1, SuATF1, SkATF2, and SuATF2) were cloned into the locus of ATF1 or ATF2 in a haploid strain of the wine S. cerevisiae T73 strain, maintaining in all strains the wild S. cerevisiae promoter. The constructed mutants were named JET02Sk, JET02Su, JET03Sk, and JET03Su, respectively (Table 1). The formation of the major aroma-active higher alcohols and acetate esters was compared to that of S. cerevisiae. To exclude any other mutations that may have occurred in the gene replacement step, the original ATF1 (or ATF2) gene was returned to its native position which resulted in the strain JET02Sc (or JET03Sc) that was then used as a reference in the aroma comparison.
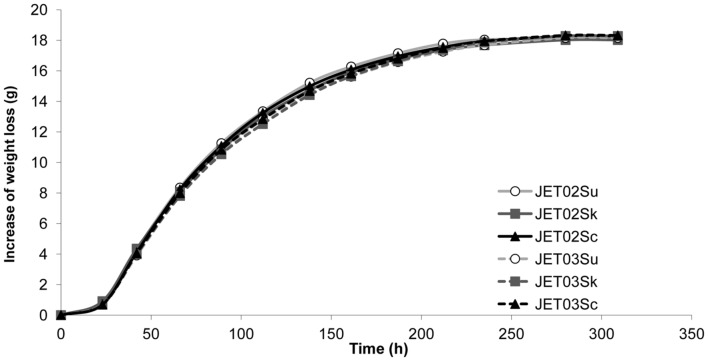
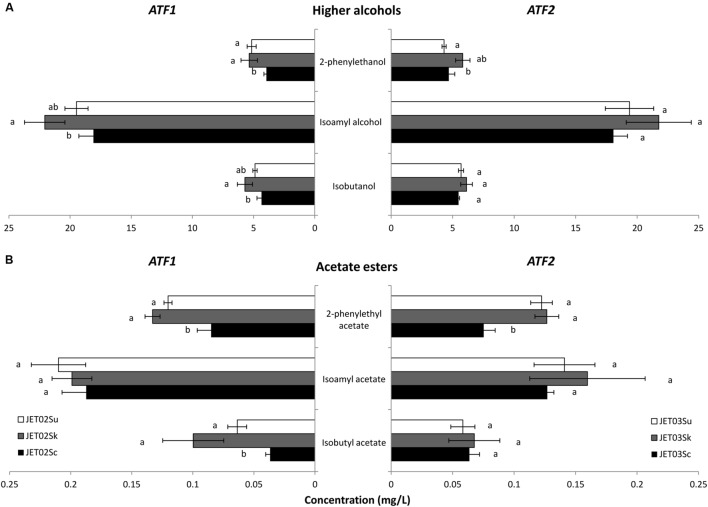
Synthetic wine must fermentations by the strains with the different ATF genes were performed, and the production of higher alcohols and acetate esters was analyzed. Weight loss monitoring showed all the strains to have similar fermentation rates with no significant differences (Figure 1). Regarding aroma compounds production, the most significant differences were observed during 2-phenylethyl acetate production (Figure 2). The strains that harbored ATF1 or ATF2 from S. kudriavzevii and S. uvarum produced almost twofold larger amounts of 2-phenylethyl acetate compared to the corresponding reference strains with ScATF1 or ScATF2. Strain JET02Sk also revealed c. threefold and JET02Su c. twofold larger amounts of isobutyl acetate than JET02Sc. During synthetic must fermentation several ethyl esters (ethyl hexanoate, ethyl decanoate, and ethyl octanoate) were also detected. Nevertheless, the comparison of the concentrations of the ethyl esters produced by the strains with the different ATF genes did not reveal any significant differences (data not shown).
FIGURE 1.
Progress of synthetic wine must fermentation. Fermentations were monitored by weight loss until a constant weight was achieved.
FIGURE 2.
Production of higher alcohols (A) and acetate esters (B) by the JET02-derived strains that harbored the individual ATF1 orthologs (left-oriented bars) and by the JET03-derived strains that harbored the individual ATF2 orthologs (right-oriented bars) during synthetic wine must fermentation. The statistically significant differences among the strains were determined independently for each compound and are indicated by labels beside the columns.
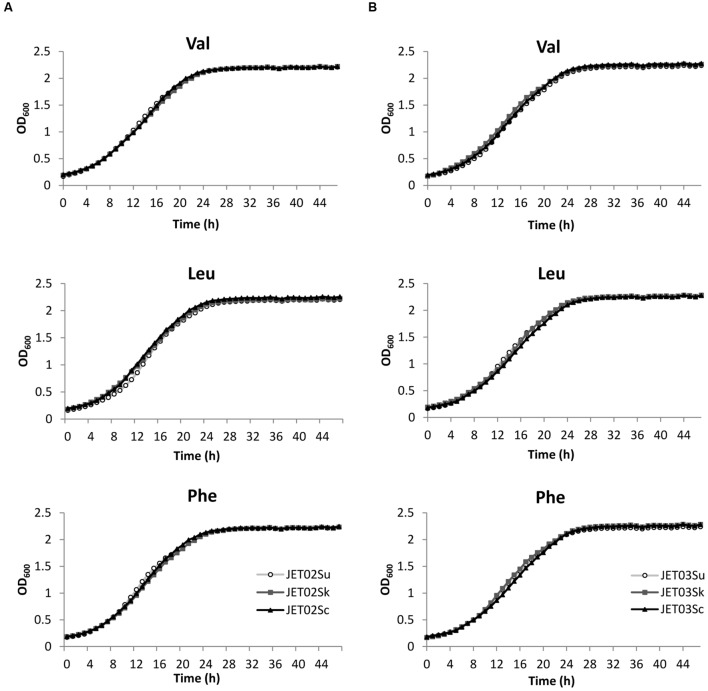
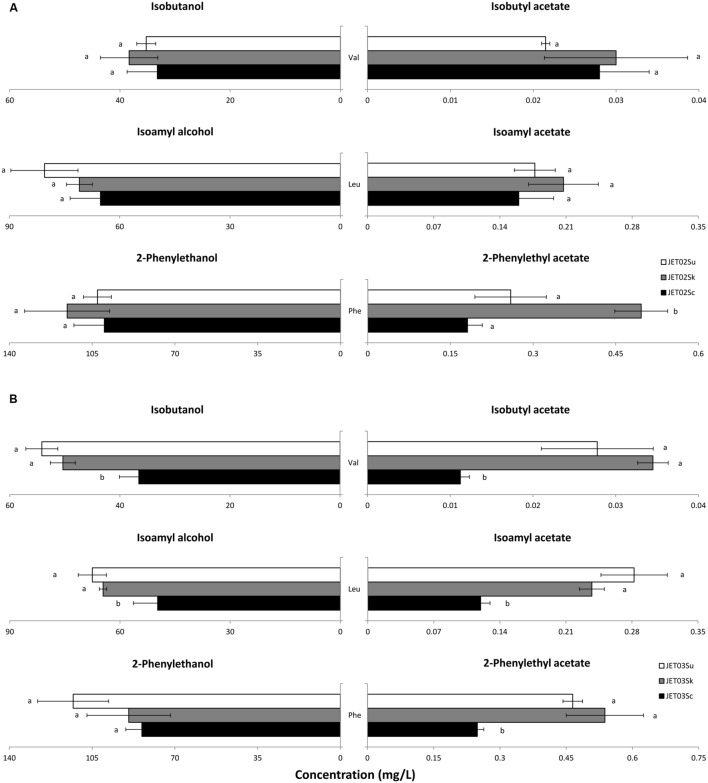
Since aroma active higher alcohols, and subsequently acetate esters, derive directly from their amino acidic precursors, our aim was to verify whether, and to what extent, the individual ATF genes played a role in the production of higher alcohols and acetate esters from their corresponding amino acidic precursors. Valine, leucine, or phenylalanine was therefore used individually as the sole nitrogen source in the cultivations, and subsequently production of the corresponding higher alcohols and their esters was analyzed. In the assay, all the strains presented a normal growth pattern under these conditions (Figure 3). This confirmed their capability to use these individual amino acids as the sole nitrogen source. The analysis of the higher alcohols and acetate esters produced by the JET02-derived strains (strains that harbored ATF1 genes from S. kudriavzevii or S. uvarum) showed significantly higher concentrations of 2-phenylethyl acetate in the phenylalanine-grown culture of the JET02Sk strain (Figure 4A). This strain, which harbored SkATF1, produced c. 2.7-fold higher concentrations than reference strain JET02Sc. With the other analyzed compounds only small and statistically insignificant differences were detected. In contrast, the JET03-derived strains (strains with introduced SkATF2 or SuATF2) grown with the individual amino acids exhibited more differences during higher alcohols and acetate esters production. When grown with valine as the sole nitrogen source, the strain that carried SkATF2 or SuATF2 genes produced both corresponding derivatives, isobutanol and isobutyl acetate, in larger amounts than the strain with ScATF2 (Figure 4B). Particularly, the isobutyl acetate concentrations were around threefold higher in both strains than that produced by JET03Sc. A similar upward trend in favor of JET03Sk and JET03Su was detected during isoamyl acetate production when their amino acidic precursor leucine was used as the nitrogen source. JET03Sk produced a twofold and JET03Su a 2.5-fold increase in the isoamyl acetate concentration. The phenylalanine-grown cultures of JET03Sk and JET03Su exhibited 2.2 and 1.9-fold larger amounts of 2-phenylethyl acetate than the reference JET03Sc.
FIGURE 3.
Growth of the JET02-derived strains that harbored the individual ATF1 orthologs (A) and of the JET03-derived strains that harbored the individual ATF2 orthologs (B) with the indicated amino acids as the nitrogen source.
FIGURE 4.
Production of the higher alcohols and esters derived from the corresponding amino acidic precursors used as the nitrogen source (indicated in the graphs). (A) Production of the higher alcohols and esters by the JET02-derived strains that harbored the individual ATF1 orthologs. (B) Production of the higher alcohols and esters by the JET03-derived strains that harbored the individual ATF2 orthologs. The statistically significant differences among the species were determined independently for each nitrogen source and are indicated by labels beside the columns.
Comparison of the AATase Substrate Specificities
To determine and compare the AATase activities of Atf1p and Atf2p from the three different Saccharomyces species, the individual ATF genes were cloned into the plasmid pGREG526 and expressed in a host S. cerevisiae strain with the deleted ATF1 and ATF2 genes. In order to obtain sufficient AATase activity, the ATF genes were expressed under the control of the constitutive TDH3 promoter. The host S. cerevisiae strain was also transformed with the empty pGREG526 vector as a negative control. In all assays with cell extracts of empty vector transformants, the concentrations of the measured acetate esters formed from the corresponding higher alcohols (isobutanol, isoamyl alcohol, and 2-phenylethanol) were below detection limit. This confirmed none or negligible alcohol acetyltransferase activity in these control strain cell extracts. The kinetic parameters (Km and Vmax) were assessed for isoamyl alcohol as a substrate since isoamyl alcohol acetylation is known to be catalyzed only by Atf1p and Atf2p. To avoid the isoamyl acetate breakdown, the measured catalytic reaction product, IAH1 gene that codifies ester hydrolase, was also deleted in the host strain (BY4741atf1atf2iah1). AATase activity was determined in cell extracts over a wide range of isoamyl alcohol concentrations (0.01–100 mM) and was measured by isoamyl acetate formation which was analyzed by head space gas chromatography. The activity measured in the cell extracts that expressed the ATF1 orthologs showed typical Michaelis–Menten saturation kinetics (see Supplementary File 1). In contrast, the activity of all the AATases encoded by ATF2 displayed an increasing tendency over the entire range of tested concentrations, without reaching saturation. Therefore, Km and Vmax were determined only for the enzymes encoded by the ATF1 orthologs. The Km and Vmax values calculated for isoamyl alcohol displayed considerable differences among the individual Atf1 enzymes (Table 4). SkAtf1p showed almost twofold higher and SuAtf1p c. threefold higher Km than ScAtf1p. Regarding Vmax, both SkAtf1p and SuAtf1p exhibited around twofold lower values than ScAtf1p.
Table 4.
The kinetic parameters of SkAtf1p, SuAtf1p, and ScAtf1p measured in the cell extracts of the S. cerevisiae strain with atf1 atf2 iah1 deletion.
| Cell extract | Km (mM) | Vmax [nmol.min-1.(mg protein)-1] |
|---|---|---|
| CLpSkATF1 | 57.4 ± 7.1 | 4.80 ± 0.26 |
| CLpSuATF1 | 92.9 ± 10.9 | 5.69 ± 0.35 |
| CLpScATF1 | 32.2 ± 2.2 | 9.99 ± 0.24 |
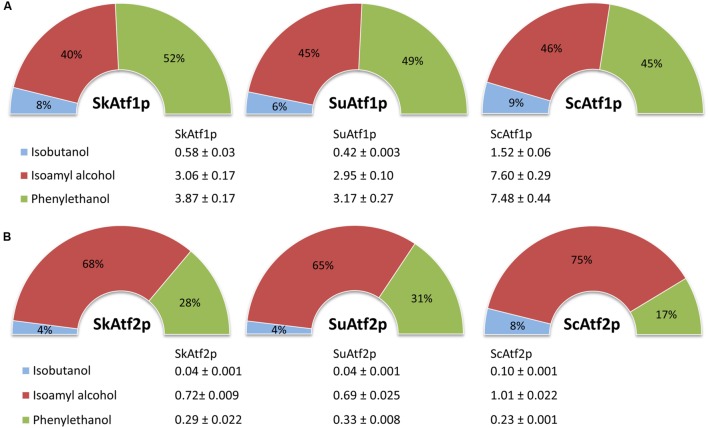
To further characterize enzymatic properties, substrate specificities were compared for three different amino-acid-derived higher alcohols as follows: isobutanol, isoamyl alcohol, and 2-phenylethanol. Specific activities were analyzed at fixed substrate concentrations of 60 mM for isobutanol, 100 mM for isoamyl alcohol, and 30 mM for 2-phenylethanol. The cell extracts of all the strains that expressed the individual ATF1 and ATF2 orthologs exhibited activities for all the measured substrates. Interesting differences were observed between enzymes Atf1 and enzymes Atf2. When individual enzymatic activities were expressed as a percentage distributed among the total enzymatic activity of the measured substrates, enzymes Atf1 showed approximately equal percentage portions of the catalytic activities for isoamyl alcohol and 2-phenylethanol (Figure 5A). In contrast, enzymes Atf2 showed considerably stronger activity toward isoamyl alcohol than the other substrates. Moreover, while all three Atf1 enzymes displayed an almost identical percentage distribution of individual enzymatic activities, the proportion of the enzyme activity of SkAtf2p and SuAtf2p to 2-phenylethanol was almost twofold greater, and twofold lower to isobutanol than for ScAtf2p (Figure 5B).
FIGURE 5.
The relative alcohol acetyltransferase activities of the Atf1 (A) and Atf2 (B) enzymes expressed as a percentage distributed among the total enzymatic activity of the measured substrates. The presented values [nmol⋅min-1(mg protein)-1] are averages and mean deviations of three replicates. The activities of the different Atf1 and Atf2 enzymes were measured in the cell extracts of the S. cerevisiae strain with atf1 atf2 iah1 deletion at a concentration of 60 mM for isobutanol, 100 mM for isoamyl alcohol and 30 mM for 2-phenylethanol.
Discussion
Our previous study focused on a computational comparative analysis of the DNA sequences of the ortholog genes involved in higher alcohols and acetate ester formation in S. cerevisiae, S. kudriavzevii, and S. uvarum (Stribny et al., 2016). The individual changes noted in the S. kudriavzevii and S. uvarum sequences (with the S. cerevisiae orthologs taken as references) were evaluated by Grantham’s score, which quantifies the biochemical divergence between two amino acids on the basis of their molecular volume, polarity and composition (Grantham, 1974). Equally for S. kudriavzevii and S. uvarum, the alcohol acetyltransferases encoded by ATF1 and ATF2 clearly yielded the largest number of amino acid substitutions, as well as the highest Grantham scores (together with the 2-keto acid decarboxylase encoded by ARO10) among the 23 assessed sequences.
Hence, this work aimed to investigate and compare the effects of ATF1 and ATF2 from S. kudriavzevii and S. uvarum on the production of the aroma-active higher alcohols and acetate esters that derived from the corresponding amino acids. The gas chromatography analysis revealed increasing amounts of acetate esters for the strains that expressed genes SkATF2 and SuATF2. One possible explanation is the amino acid substitutions observed during the aforementioned in silico analysis. Both sequences, SkAtf2p and SuAtf2p, revealed a large number of substitutions (20 and 25%, respectively) compared to ScAtf2p. Only four substitutions in SkAtf2p and five in SuAtf2p were classified as radical according to the Grantham scale. None of the substitutions were found in the two conserved regions, WRLICLP and HXXXD, hypothesized as parts of the active site (Nagasawa et al., 1998; D’Auria, 2006). Thus, the large number of total substitutions, or combinations of some, might play role in the differences observed during acetate ester production. While the Atf1p sequences also showed a large number of amino acid changes (SkAtf1p vs. ScAtf1p 17%, SuAtf1p vs. ScAtf1p 18%), the comparison of the strains that expressed the individual ATF1 orthologs gave significant differences only for 2-phenylethyl acetate during the production of higher alcohols and acetate esters. Further research by mutagenesis assays could clarify a possible correlation between amino acid substitutions and the different higher alcohol and acetate ester productions.
The JET03-derived strains with introduced SkATF2 and SuATF2 (JET03Sk and JET03Su, respectively) also exhibited larger amounts of higher alcohols than JET03Sc. One possible explanation could be the catalytic activity of esterases that catalyzed ester breakdown, such as Iah1p (Fukuda et al., 2000; Lilly et al., 2006). In hypothetical terms, the superior concentrations of the higher alcohols produced by JET03Sk and JET03Su could result from Iah1p, which degraded the large amounts of acetate esters previously produced as a result of SkAtf2p and SuAtf2p activity. As previously described (Fukuda et al., 1998), the balance between ester-synthesizing and ester-degrading enzymes leads to an optimal balance between higher alcohols and acetate esters.
It has been shown that Atf1p and Atf2p in S. cerevisiae are able to transfer an activated acetate group to a wide variety of substrates with an alcohol group, and thus display wide substrate specificity for alcohol cosubstrates (Verstrepen et al., 2003b). Similarly, our findings showed wide substrate specificity for Atf1p and Atf2p from S. kudriavzevii and S. uvarum. The activities observed in the extracts of the cells that expressed the individual ATF2 genes were an order of magnitude lower than the activities in the cell extracts that expressed ATF1 genes. It has been hypothesized that Atf2p might be important for different metabolic processes other than Atf1p. For instance, Cauet et al. (1999) suggested that Atf2p plays a key role in the detoxification of 3β-hydroxysteroids. Our results also indicate that the Atf2 proteins from S. kudriavzevii and S. uvarum play only a secondary role in acetate ester formation compared to Atf1p, similarly to that observed in S. cerevisiae (Verstrepen et al., 2003b; Lilly et al., 2006).
Interesting differences were observed in the kinetic properties of the alcohol acetyltransferases among the Atf1p from S. cerevisiae, S. kudriavzevii, and S. uvarum toward isoamyl alcohol. Of the two cosubstrates, isoamyl alcohol was chosen for determining the kinetic properties because previous studies have shown alcohol to be a rate-limiting cosubstrate during alcohol acetyltransferases reactions (Minetoki et al., 1993). However, we did not observe any correlation between the different Km values and isoamyl acetate formation. The strains that expressed the individual ATF2 orthologs revealed marked differences in isoamyl acetate concentrations. However, unlike the kinetic data of the Atf1 enzymes, the individual Atf2p showed a linear increase of isoamyl acetate synthesis with isoamyl alcohol concentrations, and did not reach saturation, not even with the highest concentration (100 mM) used in the assay. Higher concentrations resulted in non-specific peaks, as detected by gas chromatography. As Malcorps et al. (1991) explained, one possible reason is that a high isoamyl alcohol concentration in the enzyme assay could modify the enzyme properties through conformational changes or denaturation, or could induce non-specific alcoholysis of the acetyl-enzyme intermediate. The higher isoamyl alcohol concentrations needed to determine the Atf2p kinetic properties indicated that the Km values of the Atf2 enzymes were higher than those of Atf1p, which is in accordance with the observations made by Nagasawa et al. (1998). Regarding the Atf1 enzymes, the Km (32.2 mM) for S. cerevisiae Atf1p calculated in our study was similar to that observed for S. cerevisiae sake strain Kyokai No. 7 (29.8 mM; Minetoki et al., 1993) and for S. cerevisiae beer strain NCYC 366 (25 mM; Malcorps and Dufour, 1992). These similar values indicate that the range of isoamyl alcohol concentrations used in our assays did not lead to the aforementioned modifications in the Atf1 enzymatic properties.
In a recent study we observed differences in the production of higher alcohols and acetate esters by S. kudriavzevii and S. uvarum compared to S. cerevisiae (Stribny et al., 2015). These differences could be explained by the different kinetic properties of the individual Atf1 enzymes and/or the activities of the Atf2 enzymes observed in the present work. Another reason could be variations in the expression levels of the ATF genes as the expression levels have been demonstrated to be an important factor for ester synthesis (Malcorps et al., 1991; Verstrepen et al., 2003b). For instance, Gamero et al. (2013) observed considerable differences in aroma production during wine fermentation by these three Saccharomyces species. The subsequent expression analysis revealed, besides others, the up-regulation of ATF1 in S. uvarum and the up-regulation of ATF2 in both S. uvarum and S. kudriavzevii compared to S. cerevisiae (Gamero et al., 2014). The existence of another as yet unknown enzyme with AATase activity in the yeast proteome has also been proposed (Malcorps and Dufour, 1992; Verstrepen et al., 2003b). Its putative effect on acetate ester production by the three different Saccharomyces species cannot be ruled out.
Conclusion
The amino acid variations noted in the orthologous Atf1p and Atf2p of S. kudriavzevii, S. uvarum, and S. cerevisiae indicated a possible impact on the distinct properties of the enzymes characterized herein. Together with differences in gene expression levels, these distinct enzymatic properties appear to play an important role in the differences among these three Saccharomyces species during acetate ester formation. The knowledge on the important enzymes involved in aroma development by closely related Saccharomyces yeasts is of scientific as well as of applied interest, and uncover new possibilities to enhance biotechnological flavor production.
Author Contributions
JS, RP-T, and AQ conceived and designed the experiments. JS performed the experiments. JS, AQ, and RP-T participated in the analysis and interpretation of the data. JS wrote the first manuscript version. AQ and RP-T participated in the final manuscript version. All authors read and approved the final manuscript version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Helen L. Warburton for checking the English, who specializes in scientific works.
Footnotes
Funding. This work has been supported by the European Commission FP7: Marie Curie Initial Network CORNUCOPIA no. 264717 and by CICYT grant (ref. AGL2015-67504-C3-1-R) from Ministerio de Economía y Competitividad.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00897
References
- Beltran G., Novo M., Rozes N., Mas A., Guillamon J. M. (2004). Nitrogen catabolite repression in Saccharomyces cerevisiae during wine fermentations. FEMS Yeast Res. 4 625–632. 10.1016/j.femsyr.2003.12.004 [DOI] [PubMed] [Google Scholar]
- Bolat I., Romagnoli G., Zhu F. B., Pronk J. T., Daran J. M. (2013). Functional analysis and transcriptional regulation of two orthologs of ARO10, encoding broad-substrate-specificity 2-oxo-acid decarboxylases, in the brewing yeast Saccharomyces pastorianus CBS1483. FEMS Yeast Res. 13 505–517. 10.1111/1567-1364.12051 [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Cauet G., Degryse E., Ledoux C., Spagnoli R., Achstetter T. (1999). Pregnenolone esterification in Saccharomyces cerevisiae – A potential detoxification mechanism. Eur. J. Biochem. 261 317–324. 10.1046/j.1432-1327.1999.00282.x [DOI] [PubMed] [Google Scholar]
- D’Auria J. C. (2006). Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant. Biol. 9 331–340. 10.1016/j.pbi.2006.03.016 [DOI] [PubMed] [Google Scholar]
- Dickinson J. R., Eshantha L., Salgado J., Hewlins M. J. E. (2003). The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J. Biol. Chem. 278 8028–8034. 10.1074/jbc.M211914200 [DOI] [PubMed] [Google Scholar]
- Dickinson J. R., Harrison S. J., Dickinson J. A., Hewlins M. J. E. (2000). An investigation of the metabolism of isoleucine to active amyl alcohol in Saccharomyces cerevisiae. J. Biol. Chem. 275 10937–10942. 10.1074/jbc.275.15.10937 [DOI] [PubMed] [Google Scholar]
- Dickinson J. R., Harrison S. J., Hewlins M. J. E. (1998). An investigation of the metabolism of valine to isobutyl alcohol in Saccharomyces cerevisiae. J. Biol. Chem. 273 25751–25756. 10.1074/jbc.273.40.25751 [DOI] [PubMed] [Google Scholar]
- Dickinson J. R., Lanterman M. M., Danner D. J., Pearson B. M., Sanz P., Harrison S. J., et al. (1997). A C-13 nuclear magnetic resonance investigation of the metabolism of leucine to isoamyl alcohol in Saccharomyces cerevisiae. J. Biol. Chem. 272 26871–26878. 10.1074/jbc.272.43.26871 [DOI] [PubMed] [Google Scholar]
- Fujii T., Nagasawa N., Iwamatsu A., Bogaki T., Tamai W., Hamachi M. (1994). Molecular cloning, sequence analysis, and expression of the yeast alcohol acetyltransferase gene. Appl. Environ. Microbiol. 60 2786–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K., Kiyokawa Y., Yanagiuchi T., Wakai Y., Kitamoto K., Inoue Y., et al. (2000). Purification and characterization of isoamyl acetate-hydrolyzing esterase encoded by the IAH1 gene of Saccharomyces cerevisiae from a recombinant Escherichia coli. Appl. Microbiol. Biotechnol. 53 596–600. 10.1007/s002530051662 [DOI] [PubMed] [Google Scholar]
- Fukuda K., Yamamoto N., Kiyokawa Y., Yanagiuchi T., Wakai Y., Kitamoto K., et al. (1998). Balance of activities of alcohol acetyltransferase and esterase in Saccharomyces cerevisiae is important for production of isoamyl acetate. Appl. Environ. Microbiol. 64 4076–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamero A., Belloch C., Ibanez C., Querol A. (2014). Molecular analysis of the genes involved in aroma synthesis in the species S. cerevisiae, S. kudriavzevii and S. bayanus var. uvarum in winemaking conditions. Plos ONE 9:e97626 10.1371/journal.pone.0097626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamero A., Tronchoni J., Querol A., Belloch C. (2013). Production of aroma compounds by cryotolerant Saccharomyces species and hybrids at low and moderate fermentation temperatures. J. Appl. Microbiol. 114 1405–1414. 10.1111/jam.12126 [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Woods R. A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350 87–96. 10.1016/S0076-6879(02)50957-5 [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 1541–1553. [DOI] [PubMed] [Google Scholar]
- Grantham R. (1974). Amino acid difference formula to help explain protein evolution. Science 185 862–864. 10.1126/science.185.4154.862 [DOI] [PubMed] [Google Scholar]
- Hazelwood L. A., Daran J. M., van Maris A. J. A., Pronk J. T., Dickinson J. R. (2008). The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74 2259–2266. 10.1128/aem.02625-2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G., Wu C. L., Schade B., Thomas D. Y., Whiteway M. (2005). Drag&Drop cloning in yeast. Gene 344 43–51. 10.1016/j.gene.2004.10.016 [DOI] [PubMed] [Google Scholar]
- Kaneko Y., Banno I. (1991). Re-examination of Saccharomyces bayanus strains by DNA-DNA hybridization and electrophoretic karyotyping. IFO Res. Comm. 15 30–41. [Google Scholar]
- Lambrechts M. G., Pretorius I. S. (2000). Yeast and its importance to wine aroma – a review. S. Afr. J. Enol. Viticul. 21 97–125. [Google Scholar]
- Lilly M., Bauer F. F., Lambrechts M. G., Swiegers J. H., Cozzolino D., Pretorius I. S. (2006). The effect of increased yeast alcohol acetyltransferase and esterase activity on the flavour profiles of wine and distillates. Yeast 23 641–659. 10.1002/yea.1382 [DOI] [PubMed] [Google Scholar]
- Malcorps P., Cheval J. M., Jamil S., Dufour J. P. (1991). A new model for the regulation of ester synthesis by alcohol acetyltransferase in Saccharomyces cerevisiae during fermentation. J Am. Soc. Brew. Chem. 49 47–53. [Google Scholar]
- Malcorps P., Dufour J. P. (1992). Short-chain and medium -chain aliphatic-ester synthesis in Saccharomyces cerevisiae. Eur. J. Biochem. 210 1015–1022. 10.1111/j.1432-1033.1992.tb17507.x [DOI] [PubMed] [Google Scholar]
- Minetoki T., Bogaki T., Iwamatsu A., Fujii T., Hamachi M. (1993). The purification, properties and internal peptide sequences of alcohol acetytransferase isolated from Saccharomyces cerevisiae Kyokai No. 7. Biosci. Biotechnol. Biochem. 57 2094–2098. 10.1271/bbb.57.2094 [DOI] [PubMed] [Google Scholar]
- Nagasawa N., Bogaki T., Iwamatsu A., Hamachi M., Kumagai C. (1998). Cloning and nucleotide sequence of the alcohol acetyltransferase II gene (ATF2) from Saccharomyces cerevisiae Kyokai No. 7. Biosci. Biotechnol. Biochem. 62 1852–1857. 10.1271/bbb.62.1852 [DOI] [PubMed] [Google Scholar]
- Nykanen L. (1986). Formation and occurence of flavor compounds in wine and distilled alcoholic beverages. Am. J. Enol. Vitic. 37 84–96. [Google Scholar]
- Querol A., Huerta T., Barrio E., Ramon D. (1992). Dry yeast-strain for use in fermentation of Alicante wines – selection and DNA patterns. J. Food Sci. 57 183–185. 10.1111/j.1365-2621.1992.tb05451.x [DOI] [Google Scholar]
- Riou C., Nicaud J. M., Barre P., Gaillardin C. (1997). Stationary-phase gene expression in Saccharomyces cerevisiae during wine fermentation. Yeast 13 903–915. [DOI] [PubMed] [Google Scholar]
- Rojas V. (2002). Actividades Esterásicas en Levaduras Vínicas. Ph.D., thesis, University of Valencia; Spain. [Google Scholar]
- Rojas V., Gil J. V., Manzanares P., Gavara R., Pinaga F., Flors A. (2002). Measurement of alcohol acetyltransferase and ester hydrolase activities in yeast extracts. Enzyme Microb. Technol. 30 224–230. 10.1016/s0141-0229(01)00483-485 [DOI] [Google Scholar]
- Stribny J., Gamero A., Perez-Torrado R., Querol A. (2015). Saccharomyces kudriavzevii and Saccharomyces uvarum differ from Saccharomyces cerevisiae during the production of aroma-active higher alcohols and acetate esters using their amino acidic precursors. Int. J. Food Microbiol. 205 41–46. 10.1016/j.ijfoodmicro.2015.04.003 [DOI] [PubMed] [Google Scholar]
- Stribny J., Romagnoli G., Perez-Torrado R., Daran J. M., Querol A. (2016). Characterisation of the broad substrate specificity 2-keto acid decarboxylase Aro10p of Saccharomyces kudriavzevii and its implication in aroma development. Microb. Cell Fact. 15 51 10.1186/s12934-016-0449-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styger G., Prior B., Bauer F. F. (2011). Wine flavor and aroma. J. Ind. Microbiol. Biotechnol. 38 1145–1159. 10.1007/s10295-011-1018-1014 [DOI] [PubMed] [Google Scholar]
- Uber-Garcia G. (2005). Modificación Genética De Levaduras Vínicas Industriales Par Mejorar La Producción De Aroma Secundario. Ph.D., thesis, University of Valencia; Spain. [Google Scholar]
- Verstrepen K. J., Derdelinckx G., Dufour J. P., Winderickx J., Thevelein J. M., Pretorius I. S., et al. (2003a). Flavor-active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 96 110–118. 10.1016/s1389-1723(03)90112-90115 [DOI] [PubMed] [Google Scholar]
- Verstrepen K. J., Van Laere S. D. M., Vanderhaegen B. M. P., Derdelinckx G., Dufour J. P., Pretorius I. S., et al. (2003b). Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg-ATF1, and ATF2 control the formation of a broad range of volatile esters. Appl. Environ. Microbiol. 69 5228–5237. 10.1128/aem.69.9.5228-5237.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh F. W., Murray W. D., Williams R. E. (1989). Microbiological and enzymatic production of flavor and fragrance chemicals. Crit. Rev. Biotechnol. 9 105–169. 10.3109/07388558909040617 [DOI] [Google Scholar]
- Yoshioka K., Hashimoto N. (1981). Ester formation by alcohol acetyltransferase from brewers’ yeast. Agri. Biol. Chem. 45 2183–2190. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.