Abstract
The components of the nervous system are assembled in development by the process of cell migration. Although the principles of cell migration are conserved throughout the brain, different subsystems may predominantly utilize specific migratory mechanisms, or may display unusual features during migration. Examining these subsystems offers not only the potential for insights into the development of the system, but may also help in understanding disorders arising from aberrant cell migration. The olfactory system is an ancient sensory circuit that is essential for the survival and reproduction of a species. The organization of this circuit displays many evolutionarily conserved features in vertebrates, including molecular mechanisms and complex migratory pathways. In this review, we describe the elaborate migrations that populate each component of the olfactory system in rodents and compare them with those described in the well-studied neocortex. Understanding how the components of the olfactory system are assembled will not only shed light on the etiology of olfactory and sexual disorders, but will also offer insights into how conserved migratory mechanisms may have shaped the evolution of the brain.
Keywords: Olfactory, Vomeronasal, Migration, Domains, Evolution, Neocortex
Introduction: cell migration in the developing forebrain
In the developing nervous system, neurons are born at specialized sites where progenitors reside. The central nervous system arises from the neural tube, and proliferating progenitors line the ventricle forming the ventricular zone (VZ). Neurons and glia, the postmitotic progeny of these cells, must often migrate to distant destinations to form mature brain structures. Cell migration is critical for proper circuit formation and functioning of the brain. Aberrant neuronal migration has been implicated in disorders such as epilepsy [1, 2], schizophrenia [3, 4], autism [5, 6] and in severe learning disabilities [7, 8]. Studying cell migration is therefore imperative for our understanding of brain development and the etiology of neurodevelopmental disorders.
The principles of neuronal migration are largely similar throughout the brain to the extent that they are currently understood. That said, neurons of the telencephalon and diencephalon, which together form the cerebral hemispheres, display extremely complex trajectories and elaborate migratory movements to reach their final destinations. As a result of these migrations, the telencephalon produces an array of diverse structures subserving distinct functions: the olfactory bulbs (OBs), the cerebral cortex, the basal ganglia, and the amygdaloid complex. The diencephalon forms the thalamus and the hypothalamus.
Cell migration in the brain may be broadly categorized based on the orientation of the migration trajectory with respect to the ventricular surface [9, 10]. Neurons may migrate either radially outward from this surface, or tangentially, in a direction orthogonal to the radial axis. Projection neurons, which are typically excitatory, primarily exhibit radial migration (Fig. 1) [10–12]. Interneurons, which are typically inhibitory, undertake tangential migration for much of their journey (Fig. 1) [9, 13–15].
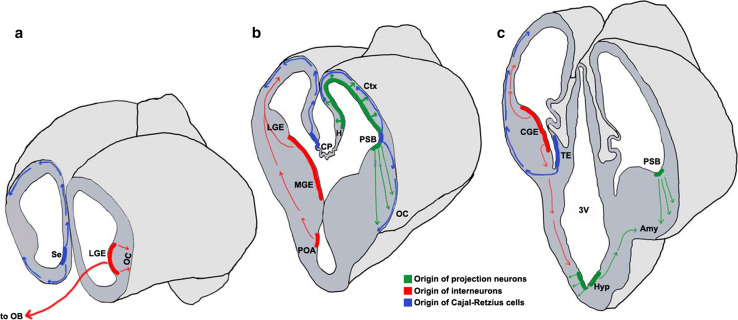
Fig. 1.
Neuronal migrations in the embryonic forebrain [13, 22, 23, 25–27, 29, 36, 67, 303–305]. Schematics representing the mouse brain at embryonic day (E)12.5 sectioned in the coronal plane at the rostral (a), mid (b), and caudal (c) levels. Domains of origin and migration routes for projection neurons (green), interneurons (red), and Cajal–Retzius cells (blue) are illustrated. Colored bands represent the ventricular zone and progenitors residing therein; arrows represent the route and direction of migration. 3V third ventricle, Amy amygdala, CGE caudal ganglionic eminence, CP choroid plexus, Ctx cortex, H hippocampus, Hyp hypothalamus, LGE lateral ganglionic eminence, MGE medial ganglionic eminence, OB olfactory bulb, OC olfactory cortex, POA pre-optic area, PSB pallial–subpallial boundary, Se septum, TE thalamic eminence
These two broad categories of migration are regulated by a spectrum of complex mechanisms that are well worth understanding, since it is cell migration that literally builds and shapes brain structures. Here, we review the migrations that contribute to the different components of the olfactory system in rodents. We compare and contrast the mechanisms underlying these migrations with those utilized in the well-studied neocortex and highlight features unique to the olfactory system. We conclude with developmental, disease, and evolutionary perspectives on cell migration in this system.
The main and accessory olfactory systems
The sense of smell is essential for a variety of behaviors such as mating, feeding, fear, and aggression. In rodents, the olfactory system has two distinct components: the main olfactory system, which is responsible for the sense of smell, and the vomeronasal system (VNS; also called the accessory olfactory system), which is essential for pheromone-based communication [16, 17]. These systems are tuned to discriminate between a variety of distinct odors and can do so at very low concentrations [18, 19]. Such efficient information processing requires the precise arrangement of a highly ordered circuit. In the sections below, we will examine the main and the accessory olfactory systems in terms of the cell migrations that create the mature circuits.
The olfactory system is unique among the sensory systems in how information enters the cortex. Whereas visual, auditory, and somatosensory input reaches the respective primary cortical areas via the thalamus, the olfactory cortex (OC) gets inputs directly via the OB. The OB is therefore the primary integration center of olfactory input in the brain.
Domains of origin
Throughout the central nervous system, neuronal cell fate is specified based on the domain of origin of the postmitotic cells in the VZ. In the telencephalon, the dorsal (pallial) VZ produces excitatory neurons from molecularly distinct domains called the medial, dorsal, lateral, and ventral pallia (MP, DP, LP, and VP, respectively) [20]. The ventral (subpallial) telencephalon is divided into the lateral, medial, and caudal ganglionic eminences (LGE, MGE, CGE, respectively) and the VZ of these domains produces distinct categories of interneurons based on an intricate transcription factor-based code [21–25]. At the rostral end of the telencephalon, the VZ of the septum has pallial as well as subpallial domains [26, 27]. Just dorsal to the septum is the rostromedial telencephalic wall (RMTW), which, together with the neuroepithelium of the septum, constitutes a rostral source of neurons for the forebrain [28, 29].
Broadly, excitatory projection neurons are pallial, and inhibitory interneurons are subpallial in origin. The DP produces excitatory neurons of the neocortical sensory areas (visual, auditory, somatosensory), the motor cortex, and higher cortical areas. In contrast, the OC, which processes the sense of smell, is populated by excitatory neurons from the LP and VP [26, 30–32]. The boundary between the pallium and subpallium, called the pallial–subpallial boundary (PSB), gives rise to the lateral cortical stream (LCS), which contributes both excitatory and inhibitory neurons to the OC [28, 32–35]. Neurons in the LCS migrate along a radial glial palisade that extends from the VZ of the PSB to the pial surface [35, 36]. This migration has similarities with mechanisms known to operate in neocortical projection neurons. Migrating LCS cells require doublecortin (Dcx), Lis1 [37], and filamin A [38] to maintain a bipolar morphology. Electroporation of shRNA in rat embryos to knockdown Dcx or Lis1 in the LCS leads to the aberrant accumulation of cells [37], similar to the effects of Dcx knockdown in the rat neocortex [39]. The LCS is not a unitary migration, however. It contains cells arising from multiple domains that lie on either side of the PSB, namely the LP, VP, and dorsal lateral ganglionic eminence (dLGE). A complex molecular code distinguishes the contributions of each domain of origin: cells arising in the LP express Tbr1, Emx1, and Pax6; the VP, Tbr1 and Pax6; and the dLGE, Pax6 and Dlx2 [26, 33, 34]. The persistent expression of Pax6 is a feature of migrating LCS cells that is not seen in the neocortex, wherein cells express Pax6 only when they are proliferating and turn off expression as they become postmitotic and commence migration [40–42].
The VZ of the LGE, MGE, and CGE contains discrete domains that generate inhibitory neurons that populate the entire telencephalon and also some diencephalic structures [23].
The domains of origin described above reside within the telencephalon. Other important sources of olfactory system neurons lie within the diencephalon or at structures located at the diencephalic–telencephalic boundary (DTB). These domains typically contribute to amygdaloid and hypothalamic nuclei that process olfactory information and will be discussed in later sections.
The main olfactory system
The main olfactory bulb (MOB) receives sensory input from olfactory sensory neurons (OSNs) in the olfactory epithelium (OE) via the olfactory nerve. Mitral/tufted (M/T) cells, the projection neurons of the MOB, receive OSN synapses and in turn project via the lateral olfactory tract (LOT) to the multiple components of the OC. The OC comprises five different regions, namely, the anterior olfactory nucleus (AON), the olfactory tubercle (OT), olfactory amygdala, piriform cortex (PC), and entorhinal cortex (Fig. 2) [43–46].
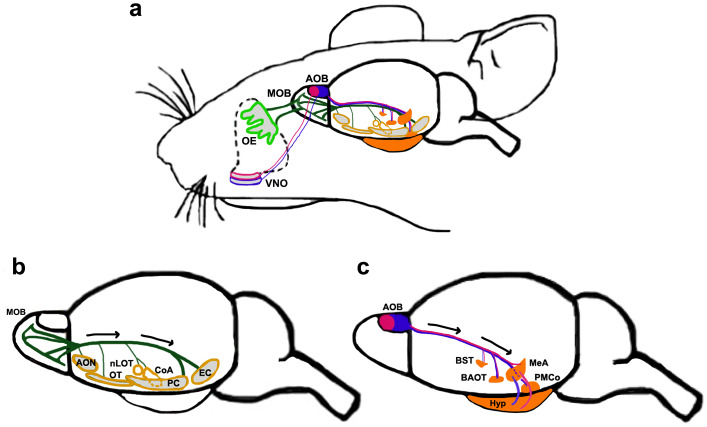
Fig. 2.
The main and accessory olfactory system [16, 43–46, 151, 189–192, 194, 205]. a Schematic depicting both systems of the mouse olfactory circuit. OSNs located in the OE (light green) project to the MOB, whereas VSNs in the VNO project to the AOB. Apical (pink outline) and basal (blue outline) VNO neurons project to the aAOB (solid pink) and pAOB (solid blue), respectively. b The main olfactory system. MOB M/T cells send their axons along the LOT in the direction of the black arrows to multiple targets (yellow outlined structures). These include different components of the olfactory cortex: the AON, OT, PC, EC, and the olfactory amygdaloid nuclei, CoA and nLOT. c The accessory olfactory system. Both the aAOB (solid pink) and the pAOB (solid blue) projection neurons send axons along the LOT (in the direction of the black arrows) to different parts of the vomeronasal amygdala, including the MeA, PMCo, and components of the extended amygdala, BST, BAOT (solid orange structures). AOB accessory olfactory bulb, aAOB anterior AOB, pAOB posterior AOB, AON anterior olfactory nucleus, BAOT bed nucleus of accessory olfactory tract, BST bed nucleus of stria terminalis, CoA cortical amygdaloid nucleus, EC entorhinal cortex, Hyp hypothalamus, LOT lateral olfactory tract, MeA medial amygdaloid nucleus, MOB main olfactory bulb, nLOT nucleus of the lateral olfactory tract, OE olfactory epithelium, OT olfactory tubercle, PC piriform cortex, PMCo posteromedial cortical nucleus, VNO vomeronasal organ
The main olfactory bulb
MOB morphogenesis is carried out in two steps. The projection neurons are born first, in the VZ at the rostral tip of the telencephalon, from where they migrate outward to create a small protrusion, the MOB anlage, by embryonic day (E)12–13 in the mouse. This is closely followed by the entry of interneurons (granule cells and periglomerular cells) which begins from E14 [47–49]. Cells of the MOB display a laminar arrangement such that the M/T projection neurons, granule cells and periglomerular cells all occupy distinct layers. A defect in the development and organization of any of these populations can lead to a lack of MOB protrusion [50] and severe functional consequences that we discuss further in the “Disease perspectives” section of this review.
MOB projection neurons
The M/T cells share interesting features with neocortical projection neurons. They are derived from pallial (dorsal telencephalic) progenitors at the local MOB VZ [51], arising from Pax6-positive radial glia [52], and their migration follows an inside-out pattern. Although distinct laminae that are characteristic of the neocortex have not been described within the M/T layer of the MOB, neurons residing deepest in this layer are born first, and later born neurons migrate past these and settle at more superficial locations [48, 50].
Transcription factors important for neocortical development, Pax6 and Lhx2, are required to regulate M/T cell migration in the OB. Loss of either of these results in no OB, but instead a misplaced, lateral olfactory bulb-like structure (OBLS) [53–56] that includes MOB and accessory OB (AOB) components. In addition to these shared genetic mechanisms required for the development of both the MOB and the neocortex, MOB-specific mechanisms also exist: transcription factors AP2-epsilon [55], Arx [56], and FezF1 [57] are necessary for proper orientation of M/T cells and organization of MCL, but are not known to be required for the development of neocortical projection neurons. Finally, there are some intriguing differences in the cellular and molecular mechanisms that mediate neocortical and M/T cell migration. Unlike in the neocortex, where radial glia have a uniform parallel arrangement, MOB radial glia display complex branched and intertwined morphologies, the function of which is not entirely clear (Fig. 3) [58]. Furthermore, newborn M/T neurons exhibit radial as well as tangential migration. In particular, later born cohorts migrate tangentially using the axons of earlier born cells to reach their proper location in the MCL (Fig. 3) [51]. Additionally, postmitotic M/T neurons express both Tbr2 and Tbr1 [52], unlike neocortical neurons, which switch off Tbr2 upon becoming postmitotic and express only Tbr1 [42]. Tbr2 expression in postmitotic neurons in the MOB is necessary for the proper migration of M/T cells and their organization in the MCL [59].
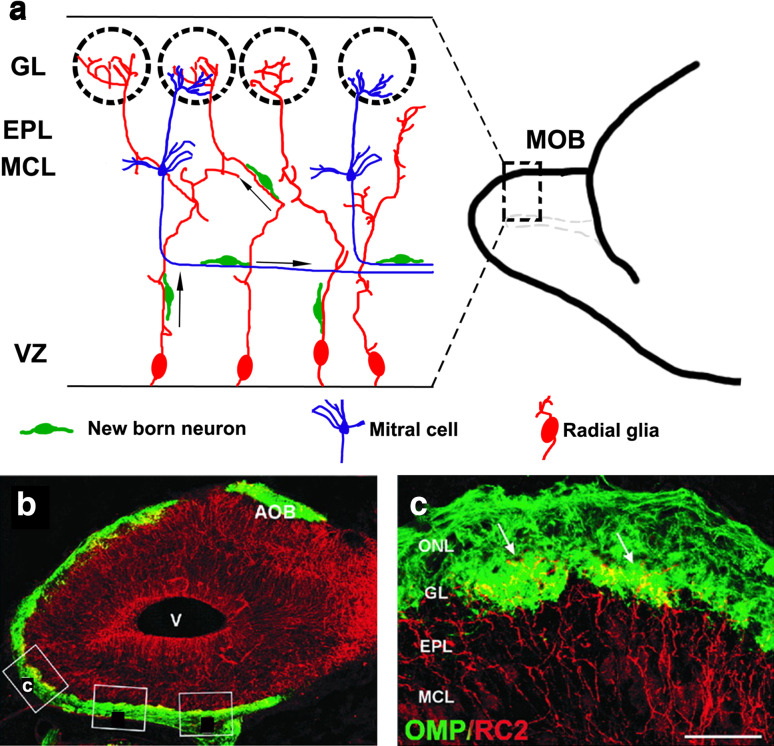
Fig. 3.
Migration of new neurons within the MOB [51, 52, 58]. a Schematic showing a sagittal section of an embryonic day (E)18.5 MOB. The radial glia (red) are convoluted and intertwined, with their endfeet merging in the glomeruli or in the EPL. The cell bodies of M/T cells (blue) are in the MCL, and their axons extend parallel to the ventricular zone. Newborn neurons (green) migrate radially to their destined laminar positions using the radial glia, or tangentially using M/T cells axons as scaffolds. b Confocal image of an E18.5 mouse MOB sagittal section immunostained for a radial glial marker, RC2 (red), and an OSN marker, the olfactory marker protein (OMP, green). c Magnified view of boxed area in b showing radial glial endfeet (arrows) penetrating glomeruli formed by OSN axons. Scale bar in c is 400 μm. Additional boxes in b are from the original artwork in [58]. AOB accessory olfactory bulb, EPL external plexiform layer, GL glomerular layer, MCL mitral cell layer, MOB main olfactory bulb, ONL olfactory nerve layer, OSN olfactory sensory neuron, V ventricle, VZ ventricular zone. The images in b, c are from Fig. 1 of [58], copyright 2001 Wiley-Liss, Inc. Reprinted with permission
MOB interneurons
MOB interneurons, like cortical interneurons, are born in the subpallium and undertake a tangential migration route to reach their destination [22, 60, 61]. Whereas cortical interneurons arise from the MGE and CGE [21–25], MOB interneurons arise from the dorsal segment of the lateral ganglionic eminence (dLGE), the LGE [62], and the septum [63]. Interneuron precursors born in these domains commence a rostral, tangential migration into the MOB. One portion of this migration, from the dLGE, continues into adulthood, constituting what is known as the rostral migratory stream (RMS) [64].
There is a large diversity of MOB interneuron subtypes, in terms of morphology and neurochemical content [65, 66] as is the case for cortical interneurons [67, 68]. There is also a surprising temporal and spatial control of MOB interneuron diversity, which includes up to seven distinct subtypes based on neurochemical and neuropeptide markers [65]. Different interneuron subtypes are generated depending on the age of the animal, with particular subtypes being born at specific embryonic or postnatal stages [65, 69]. Spatially distinct progenitors with unique molecular signatures produce the diversity of MOB interneurons. The LGE produces interneurons from Gsh2-positive progenitors which are also Pax6 positive (from the dLGE) [32], or Dlx2 positive (from the rest of the LGE) [70]. Conditional removal of Pax6 alters the postnatal production of dLGE-derived interneurons [71]. There is also a locally generated pool of Pax6 expressing progenitors in the OB VZ, which produces both GABAergic granule cells and dopaminergic periglomerular interneurons [70, 72]. An unusual pool of MOB interneurons arises from the pallial Emx1 lineage. These progenitors arise from E15 and integrate with the Dlx2 expressing LGE progenitors within the striatal germinal zone. In this new subpallial location, these cells begin to express Dlx2 and then contribute to the MOB interneuron pool through adulthood [72, 73]. The MOB therefore displays an unexpected complexity and temporal dynamics in the molecular identity of its interneuron population.
Embryonic MOB interneurons utilize similar molecular mechanisms to those employed by cortical interneurons to regulate their migration. These include transcription factors Dlx1, Dlx2, and Mash1 and also the Robo–Slit and neuregulin signaling systems [74–76].
From postnatal to adult stages, the progenitors of the RMS migration reside in the anterior subventricular zone (aSVZ) which is derived from the embryonic dLGE [22]. Despite this developmental continuity of the domain of origin, distinct mechanisms are utilized by embryonic versus adult cells for migration. In the postnatal and mature RMS, interneuron precursors, or neuroblasts, migrate in a closely associated neurophilic or chain migration pattern, along blood vessels with the aid of astrocytes which form a glial tunnel ensheathing the migrating cells [77–82]. These astrocytes are detected only by early postnatal ages and are not seen embryonically [83]. The polysialylated form of neural cell adhesion molecule (PSA-NCAM) is necessary for chain migration and is therefore expressed robustly by the neuroblasts from perinatal stages [84–86]. In addition, adhesion molecules such as integrins are differentially expressed during the migration of embryonic versus adult neuroblasts; α1 and β1 subunits are expressed in the embryo, whereas αv, β3 and β6 subunits are expressed in the adult [87, 88]. An unusual mechanism is employed in the last phase of migration when the cells reach the MOB and must migrate radially outward into their destined layers to differentiate into mature interneurons [89]. For this stage of migration, adult neuroblasts are guided along blood vessels, in contrast to embryonic cells which use radial glial-guided migration [90].
OB projection neurons, interneurons/neuroblasts therefore demonstrate the use of cellular substrates other than radial glia for their migration, i.e., the axons of M/T neurons, other neuroblasts in chain migration, and blood vessels. This contrasts with the neocortex in which radial glia are the only reported cellular substrates utilized by projection neurons and interneurons [9, 91–94]. About 50 % of MGE-derived interneurons can utilize axons for their migration in vitro [95, 96], but there is no direct evidence of axon-mediated migration of cortical interneurons in vivo. Another major point of difference with cortical interneurons is that from perinatal stages, RMS migration comprises neuroblasts-specified precursors that will produce interneurons, but which retain proliferative capability and indeed do proliferate during their chain migration. This unusual feature is seen in only a few sites in the entire central nervous system—dentate granule cells, cells from the olfactory placode (OP), and cells migrating along the LCS are the only other populations that exhibit simultaneous proliferation and migration of neuroblasts [35, 97–100]. The latter two populations are part of the olfactory migrations described in this review.
Olfactory placode: a cell source for the OB
The OE, a derivative of the OP in the snout, is one of the few regions outside the neural tube that generates neurons. The OE produces OSNs which relay sensory information to the OB throughout the life of an organism [101–105]. The OP also produces multiple cell types that populate the OB. These cells, termed the “migratory mass” (MM) [106], migrate together with the OSN axons as they extend from the OE toward the OB bundled in the olfactory nerve [106–109]. The MM is well characterized in rodents and chick, and known to contain cells with molecularly distinct identities. Differentiated cells within the mesenchyme of mouse embryos are observed as early as E10–10.5 [110]. Cells of the MM express different combinations of markers such as Doublecortin, Notch1 and its effector Hes5, Delta/Notch-like EGFR receptor (DNER), OMP, Lhx2, and GnRH [110, 111]. The MM includes putative guidepost neurons for OSNs; olfactory ensheathing cells (OECs) and their precursors; neurons expressing the olfactory marker protein (OMP); and several other distinct cell types expressing Dlx5, Six1, NCAM, GAP43, or vGlut2 whose fate and function are not well understood [100, 106, 110, 112–123].
The OECs ensheath the OSN axons throughout their growth, during their extension through the cribriform plate, and into the olfactory nerve layer and glomerular layer of the OB [100, 124–126]. OECs express BLBP and S100β, and have a range of functions in the development and immunity of the olfactory system. OECs envelope the OSN axons along their entire length forming a complex extracellular matrix containing laminin and fibronectin, express cell adhesion molecules such as PSA-NCAM and N-cadherin, produce neurotrophic factors including the p75 neurotrophin receptor and nerve growth factor, and express guidance cues such as ephrin B2 and semaphorin 3A. All these molecules promote axon growth and fasciculation [100, 127–135]. OECs also release soluble factors such as fibroblast growth factor, FGF2, which are thought to regulate the proliferation and differentiation of OSN progenitors [136–138]. They also participate in innate immunity and thereby protect the peripheral olfactory system from pathogens. They release neuropeptide Y, show inflammatory signaling cascades in response to bacterial trigger, and can lyse bacteria following endocytosis [139–144]. A subpopulation of OECs are precursors that express Sox2 and nestin, and continue to proliferate during migration.
An intriguing guidance role for OECs is suggested by reports of OR expression in this population. Each OSN axon expresses a specific individual odorant receptor (OR), which serves not only as a receptor for odorant molecules, but also guides the OSN axon to a specific glomerulus within the OB where it synapses with an M/T cell [145–148]. Interestingly, OMP expressing cells among the OECs contain a subpopulation that also expresses individual ORs. A curious feature of these cells is that they associate with OSN fibers that express the same OR. This suggests the speculative possibility that these cells may be involved in the regulation of OSN guidance via as yet unidentified mechanisms that may include a guidepost-like role [120, 149]. Though OMP expression is not found in birds, OECs that express individual ORs and associate with the growing OSN fibers have been reported in chick [118, 119], suggesting that this population may be conserved in evolution.
OECs may have additional roles into adulthood. OECs can attract RMS interneuron progenitors over short length scales in vitro, comparable to the distance interneurons travel during their radial migration phase after they enter the OB. It is possible that OECs may provide such an activity in vivo as well [100, 150].
A special class of differentiated neurons within the MM is the gonadotropin-releasing hormone (GnRH) neurons, which migrate past the OB to the hypothalamus [108, 109]. These GnRH neurons will be discussed further in “Hypothalamic nuclei”.
In summary, the MM is an interesting mix of dividing, postmitotic, and fully differentiated cells. The identities and functional implications of the diversity within this population are yet to be completely understood, but it reveals the broad range of regulatory and functional contributions of placode-derived neurons and non-neuronal cells to the development of the olfactory system.
Migration of “lot cells”
Axons from the MOB and AOB project to their targets via the LOT [151]. These are restricted to a tight corridor created by a group of guidepost neurons known as the “lot cells” (Fig. 4) [152]. The lot cells are an intriguing population, sharing molecular features with Cajal–Retzius cells and posterior AOB (pAOB) M/T cells, yet serving a highly specialized function in olfactory development. Lot cells are believed to arise in the dorsal neocortical VZ; from here, they migrate ventrally and tangentially along the telencephalic surface and finally align themselves at the PSB along the entire rostrocaudal extent [153]. A recent study (Ruiz-Reig et al., under revision, cerebral cortex) offers evidence that lot cells may arise from a different source, the thalamic eminence (TE). This transient structure located at the DTB is the source of several distinct migratory populations described later in this review. Intriguingly, a subpopulation of the TE-derived lot cells may later differentiate into pAOBM/T cells (Ruiz-Reig et al., under revision, cerebral cortex), which highlights a new evolutionary interpretation of the pAOB, discussed at the end of this review. Proneural transcription factors neurogenin 1 and 2, necessary for patterning and cortical neuron specification [154, 155], are required for the differentiation of lot cells [156].
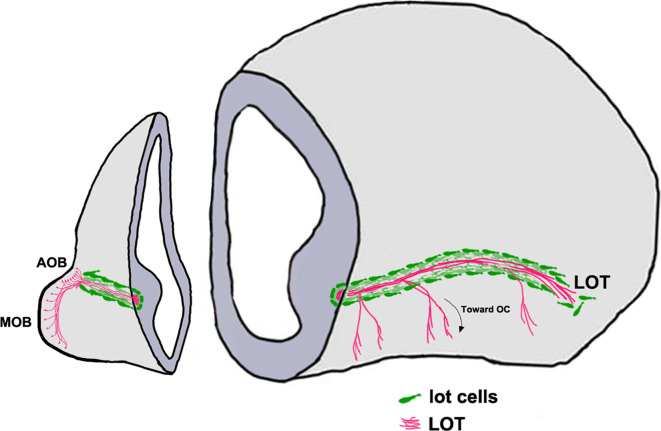
Fig. 4.
“Lot cell” array and formation of the LOT [152, 153]. Schematic representing one hemisphere of an embryonic day (E)14.5 mouse brain. The projection neurons of the MOB and the AOB extend their axons along the LOT and innervate different olfactory cortical and vomeronasal structures. The “lot cells” (green) form a “permissive corridor” along the lateral face of the telencephalon through which the LOT axons (pink) grow. AOB accessory olfactory bulb, LOT lateral olfactory tract, MOB main olfactory bulb
The lot cells assemble at the prospective LOT position prior to the incoming OB M/T cell axons forming the LOT. Proper alignment of the lot cells is essential for guiding the LOT. Ablating these cells using 6-hydroxydopamine disrupts the formation of a proper LOT [152].
It is therefore not surprising that complex cell-autonomous and cell non-autonomous mechanisms guide the positioning of the lot cell array. Netrin1/DCC guidance is necessary for lot cell alignment at the PSB [157]. Semaphorin 3F, which is expressed in the mantle of the lateral telencephalon, restricts the lot cells to the telencephalic surface [158]. The lot cell array is profoundly disrupted when transcription factors Gli3 or Lhx2 are lost [54, 153, 157]. These transcription factors may directly control lot cell migration via cell-autonomous mechanisms, or indirectly via regulation of signaling at the PSB. Transcription factor Gli3 is required for dorsoventral patterning, such that the subpallial component of the PSB expands dorsally in the Gli3 mutant [159, 160]. Disruption of lot cell array in this mutant is cell non-autonomous [153, 157] and may be due to perturbed signaling cues at the PSB. Loss of transcription factor Lhx2 does not affect the position of the PSB [161], but causes upregulation of Semaphorin 6A expression in the lateral telencephalic region where the lot cells accumulate, which may underlie the profoundly disrupted lot cell array in this mutant [54].
Olfactory cortex: many structures and multiple migrations
The OC extends along the entire rostrocaudal length of the ventral telencephalon and consists of five structures—the AON, OT, PC, entorhinal cortex, and the olfactory amygdala [44]. OB projection neurons make connections with different rostrocaudal portions of the OC depending on their location in the OB and birth order. Mitral cells residing in the ventral OB project to the OT, whereas those residing in the dorsal OB preferentially project to the PC [162, 163]. The birth order of these neurons plays a role in determining the strength of projections to particular OC areas, such that later born cells project more axons to the OT than those born earlier [51]. An additional complexity is that within the class of projection neurons, mitral cells connect with more posterior and tufted cells with more anterior OC regions [163, 164].
The components of the OC display either a nuclear or a trilaminar cortical organization. Of the latter type, the trilaminar PC is best studied in terms of its cytoarchitectonics and connectivity. Layer 1 is a cell-sparse zone and contains dendrites from the underlying cellular layer 2, long distance axons from the OB bundled in the LOT, and intracortical feedback connections. Layer 2, the principal cellular layer, is densely packed with pyramidal neurons and granule cells. Layer 3 has sparse pyramidal and polymorphic cells with no apical dendrites. It is primarily involved in intracortical communication rostrocaudally within the OC [165].
The olfactory amygdala, which receives input from the MOB comprises two amygdaloid nuclei: the cortical amygdala (CoA) and nucleus of the lateral olfactory tract (nLOT) [166]. Both these nuclei are considered to be “cortical” since they appear laminated and have radially oriented pyramidal neurons [167].
OC receives cells from multiple regions of the forebrain, some of which originate at E10.5 [28, 29], the same time as the Cajal–Retzius cells and subplate cells which are the earliest born cells of the neocortex. OC neurogenesis in the rodent embryo continues until late gestation [168]. The deeper neurons of OC (layer 3) are born earlier than the superficial neurons (layer 2), particularly in the PC [168–170]. Therefore, the OC displays a rudimentary inside-out pattern of neurogenesis, similar to the neocortex [171].
Migrations to the components of the OC
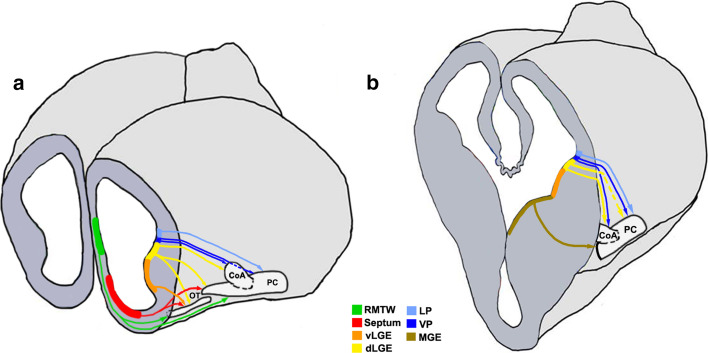
Lineage tracing using vital dyes, genetic approaches, or in utero electroporation in the mouse reveals an array of distinct VZ domains in the forebrain that contribute to the OC (Fig. 5; Table 1). These include the LP, VP, and dorsal and ventral segments of the LGE (dLGE and vLGE, respectively), which migrate together in the LCS, MGE, septum, RMTW, dorsal telencephalon, caudal telencephalon, and the diencephalon–telencephalon boundary (DTB). We will now discuss the different components of the OC formed as a result of migrations from these domains of origin.
Fig. 5.
Cell migrations to the olfactory cortex [26, 29, 30, 34, 35, 37, 172]. Schematics representing an embryonic day (E)12.5 brain sectioned at rostral (a) and mid (b) levels in the coronal plane to reveal neuroepithelial domains and cell migrations (arrows) that populate different olfactory cortical structures. CoA cortical amygdaloid nucleus, LP lateral pallium, dLGE dorsal lateral ganglionic eminence, vLGE ventral lateral ganglionic eminence, MGE medial ganglionic eminence, OT olfactory tubercle, PC piriform cortex, RMTW rostromedial telencephalic wall, VP ventral pallium
Table 1.
Domains of origin for the components of the OC
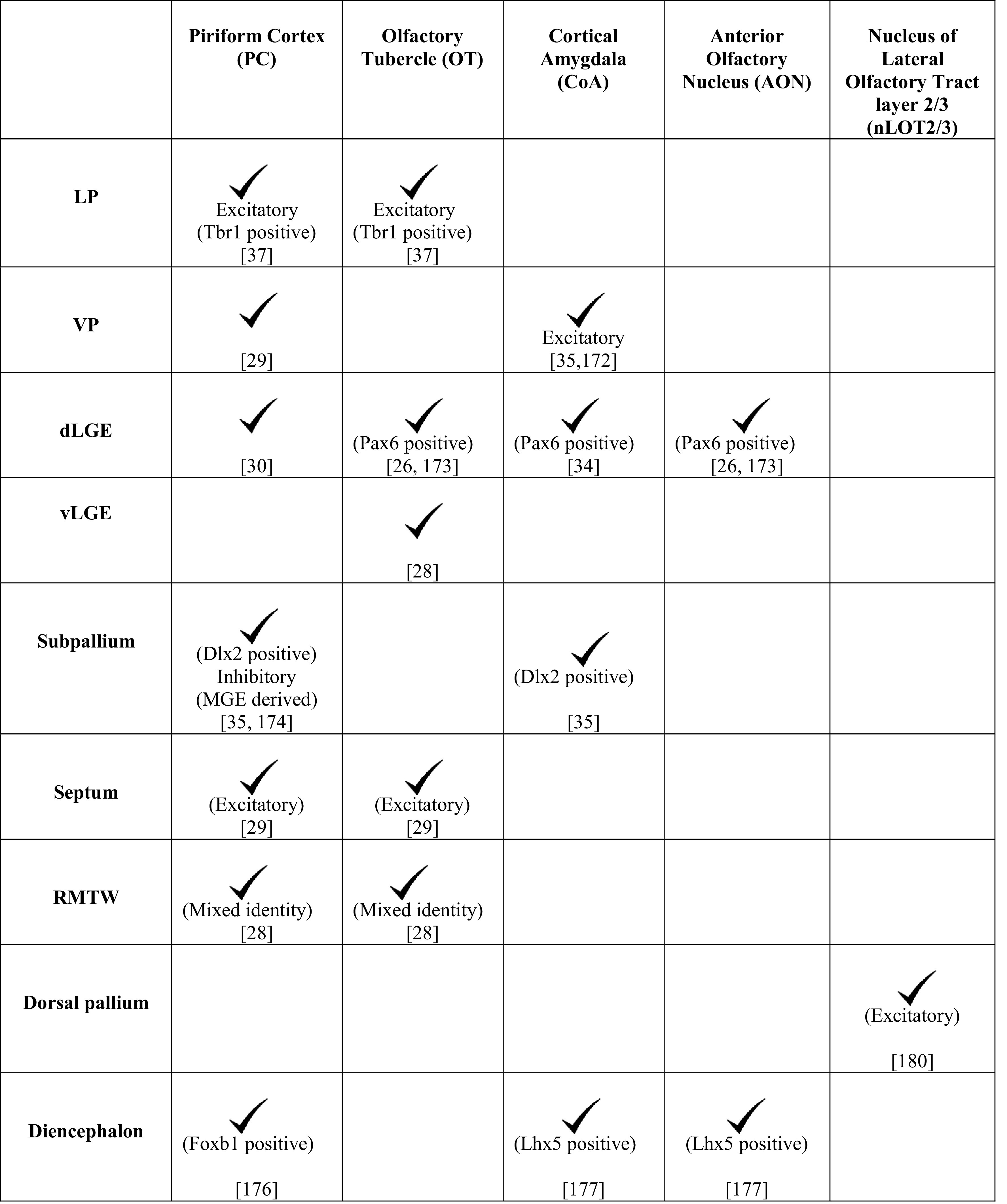
The different pallial and subpallial components of the LCS contribute excitatory and inhibitory neurons, respectively, to distinct structures of the OC (Table 1). A Tbr1-positive population arising from the LP contributes excitatory neurons to the OT and PC [37] whereas the VP produces excitatory projection neurons for the CoA [35, 172]. Pax6-positive interneurons from the dLGE are suggested to contribute to the AON, OT [26, 173], and the anterior CoA, which is reduced in the Pax6 sey/sey mutant [34]. Cells migrating from the dLGE [30] and VP [29] also contribute to the PC, whereas neurons born from the vLGE contribute only to the OT (Fig. 5) [28]. The interneurons of the LCS display different modes of migration. A subset of the Dlx2-expressing population follows chain migration and proliferates en route to the CoA and PC [35], similar to the cells in the RMS. Interneurons migrating to the PC also originate from the MGE [174], a well-characterized source of cortical interneurons [175].
The PC and OT have a contribution from a rostral origin consisting of excitatory neurons arising from the septum [29] and neurons of mixed identity from a pallial domain immediately dorsal to it, called the RMTW (Fig. 5; Table 1) [28]. The PC also receives cells from the dorsal telencephalon [169] and a diencephalic population of the Foxb1-lineage [176]. An unusual population of cells exhibiting a tangential, surface migration arises caudally, possibly at the DTB, and migrates along the lateral aspect of the telencephalon to populate the rostral OC. This population expresses Lhx5 like the DTB and displays similarities with Cajal–Retzius cells in its surface migration and Reelin expression [177].
Cell migrations to the nLOT
The nLOT is a trilaminar component of the olfactory amygdala and is bidirectionally connected to the OB and PC. It is implicated in non-pheromonal olfactory behaviors, especially feeding or ingestive behavior [178, 179]. Layers 2 and 3 of the nLOT (nLOT2/3) are the major output layers of this excitatory nucleus [179]. Whereas most of the amygdala develops either from the PSB or the subpallium, in utero electroporation of the caudal telencephalic neuroepithelium showed that the nLOT2/3 develops from the DP and therefore shares its origin and mechanisms of development with the neocortex [180]. Consistent with this interpretation, transcription factors required for proper development of the neocortex, such as Tbr1, Lhx2, and Pax6, are also required for the specification of the nLOT2/3 [34, 180, 181]. The neurons of the nLOT2/3 migrate along the caudal amygdaloid stream (CAS) and follow two modes of migration sequentially: a radial glia-independent phase that is parallel to the ventricular surface, followed by migration along the radial glia to their destination. Interestingly, this second phase requires Reelin and Cdk5 [180] similar to neurons of the neocortex [182, 183].
During development, the nLOT2 and 3 are indistinguishable [181], but on maturity, the nLOT3 appears as an ovoid structure surrounded by the crescent-like nLOT2 (Fig. 6). It would be useful to elucidate whether the origins of the nLOT2 and nLOT3 are indeed distinct from each other, since no fate-mapping study, either using genetic drivers or electroporation, distinguishes these two sub-nuclei. Layer 1 of the nLOT (nLOT1) is born at E10.5, a day earlier than the nLOT2/3 [180] and its origin and migration route are not well understood. The nLOT1 and nLOT2/3 express mutually exclusive markers and utilize distinct developmental mechanisms [33, 34, 179–181]. Therefore, the nLOT1 and nLOT2/3 may in fact be distinct nuclei that happen to assemble in close proximity gaining the collective name “the nucleus of the LOT.”
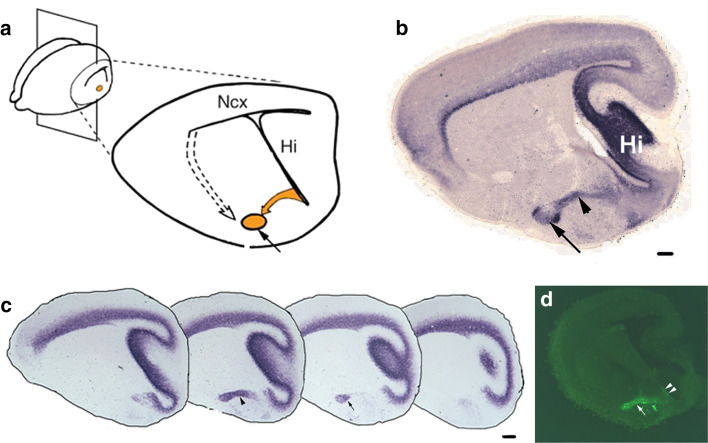
Fig. 6.
The caudal amygdaloid stream and migration to the nLOT2/3 [180]. a A sagittal section of an E17.5 mouse brain shows the caudal amygdaloid stream (CAS; yellow arrow) arising in the caudal telencephalic VZ and terminating in the globular nLOT2/3 (yellow circle, black arrow). Dashed arrow depicts the migration route from the VP to other amygdaloid nuclei. b Both the CAS (arrowhead) and the nLOT2/3 (arrow) are identified by NeuroD expression, c NeuroD expression in a lateral-to-medial series of sagittal sections at E15.5, d in utero electroporation of an EGFP-expressing construct in the caudal telencephalic neuroepithelium at E11.5, and examination of the brain at E15.5, reveals GFP-positive cells migrating along the CAS (white arrow). Note the residual GFP-positive neuroepithelium at the site of electroporation indicating the origin of the nLOT2/3 cells (white arrowheads). Scale bars are 200 μm. nLOT nucleus of lateral olfactory tract, Hi hippocampus, Ncx neocortex. All images in this figure are from [180], copyright 2007 Nature Publishing Group. Reprinted with permission
The accessory olfactory system
Though the MOB has been traditionally thought to process common odors and the AOB to process pheromonal odors, there is increasing evidence of cross-talk between these two arms of the olfactory system at multiple stages, including the OB, amygdala and OC [184–188]. However, here we will treat the accessory system as a distinct entity, since this is more appropriate from a developmental perspective. Pheromonal odors are detected by vomeronasal sensory neurons (VSNs) in the vomeronasal organ (VNO). VSNs project via the olfactory nerve to the M/T cells, which are the projection neurons of the AOB. The M/T cells in turn project along the LOT together with the MOB axons. Targets of the AOB are nuclei of the vomeronasal amygdala, which in turn project to specific regions of the hypothalamus (Fig. 2) [189, 190]. We will describe cell migrations to each structure in the accessory olfactory circuit in the sections below.
The accessory olfactory bulb
In rodents, the AOB is located on the dorsal aspect of the MOB and is the first recipient of vomeronasal innervation in the brain. VSNs project to the AOB in an ordered fashion such that the apical neurons project to the anterior AOB (aAOB) and basal neurons to the pAOB [191, 192]. The aAOB and pAOB also express a battery of mutually exclusive molecular markers and display an apparent functional segregation, such that the aAOB mediates mating behavior whereas the pAOB processes defensive/aggressive cues [16]. A developmental rationale for this functional dichotomy was provided by the discovery that the projection neurons of the aAOB and pAOB are produced in independent and widely separated domains of origin. Projection neurons of the aAOB originate from the local OB neuroepithelium, similar to those of the MOB. In contrast, projection neurons of the pAOB arise in a distant location at the DTB, migrating the entire rostrocaudal extent of the telencephalic surface to reach their destination [193].
The AOB was presumed to share developmental mechanisms with the MOB, in part due to the close juxtaposition and similar cellular composition of these two structures [194, 195]. However, the identification of disparate origins of aAOB and pAOB projection neurons implies that they may utilize distinct mechanisms for their development. Indeed, the aAOB requires mechanisms of specification that are similar to the MOB, whereas the pAOB depends on a different set of regulatory genes, e.g., Tbr1 is required for the specification of the aAOB and MOB, but not the pAOB. The opposite is true for Lhx5, which is required for the specification of the pAOB, but not the aAOB or MOB [47, 193]. One unusual feature of pAOB neurons is that they perform tangential migration, a feature characteristic to interneurons that migrate over large distances [196] and not usually seen in other populations of projection neurons which migrate radially from the local VZ. Intriguingly, Cdk5, a molecule required for cell shape changes and necessary for radial glia-dependent migration [180, 183, 197] is also necessary for the pAOB neurons to migrate tangentially along the telencephalic surface, in the absence of which they remain accumulated at the caudal telencephalic pial surface [193].
The dichotomy between the domains of origin of the aAOB and pAOB is seen only for projection neurons. Interneurons of both the aAOB and pAOB are derived from the rostral LGE and from the anterior SVZ, similar to those populating the MOB [198, 199].
The vomeronasal amygdala
Gene expression and fate-mapping studies have identified an assortment of nuclei of the amygdala and the extended amygdala [200, 201] to be part of the vomeronasal amygdala [189, 202–204]. These nuclei are all part of the vomeronasal circuit. The axons of AOB M/T cells project along the LOT to distinct nuclei of the vomeronasal amygdala. The MeA is the primary target of the AOB. Other targets include the posteromedial cortical nucleus (PMCo; Fig. 2) and components of the extended amygdala such as the bed nucleus of stria terminalis (BST) and the bed nucleus of the accessory olfactory tract (BAOT), all of which comprise the vomeronasal amygdala [190, 205].
Migrations to the MeA and PMCo
The MeA is anatomically divided into anterior, posterodorsal, and posteroventral divisions (MeAA, MeAPD, and MeAPV, respectively), which send projections to functionally distinct hypothalamic nuclei. The MeAPD projects to the reproductive hypothalamic nuclei and the MeAA and the MeAPV project mainly to the nuclei processing defense and aggression [189, 206, 207].
In utero electroporation, lineage tracing using vital dyes, and genetic mapping studies show that each of these subnuclei is populated by cells from multiple pallial and subpallial neuroepithelial domains (Table 2). The Lhx9 expressing VP populates both the MeA and PMCo [33, 202]. The rostral portion of the VP contributes to the MeAA and a more caudal portion of the VP contributes to both the MeAPD and the MeAPV [29, 202, 204]. The MeAA receives neurons of the Nkx2.1 lineage [211] from the subpallial anterior entopeduncular area (AEP) and preoptic area (POA). In contrast, the MeAPD is populated by cells of the Nkx2.1 lineage from the AEP, and the MeAPV receives Dbx1-positive neurons of the sonic hedgehog (Shh) lineage from the POA (Fig. 7) [173, 202, 204, 209]. A population of glutamatergic neurons of MeA and PMCo is derived from the third ventricle. These cells express a diencephalic transcription factor, orthopedia (Otp), which is necessary for their migration across the DTB [203]. This migration takes place along radial glia which extend from the neuroepithelium underlying the paraventricular hypothalamic nucleus (PVH) to the MeA [36, 202–204].
Table 2.
Domains of origin for the components of the vomeronasal amygdala
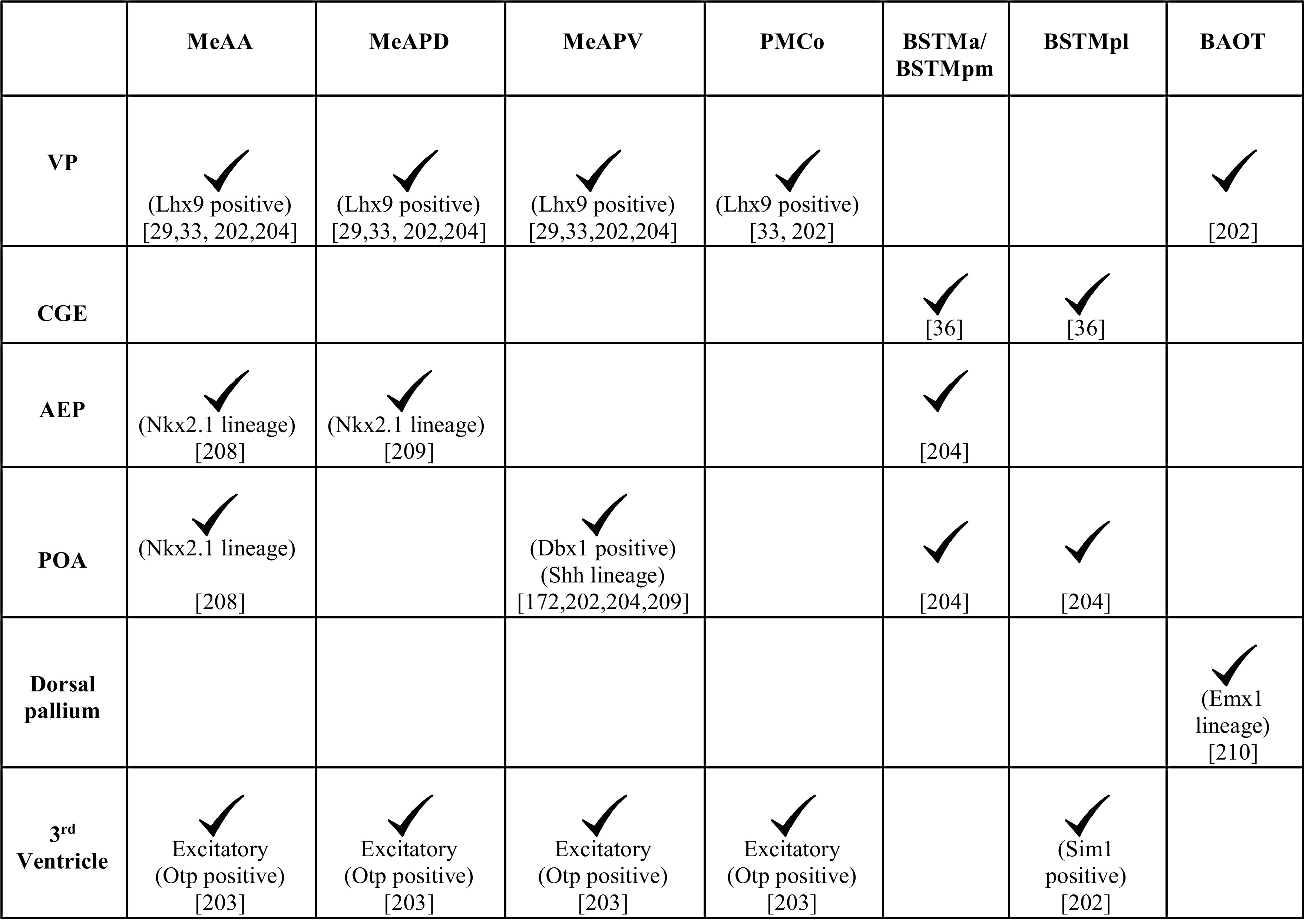
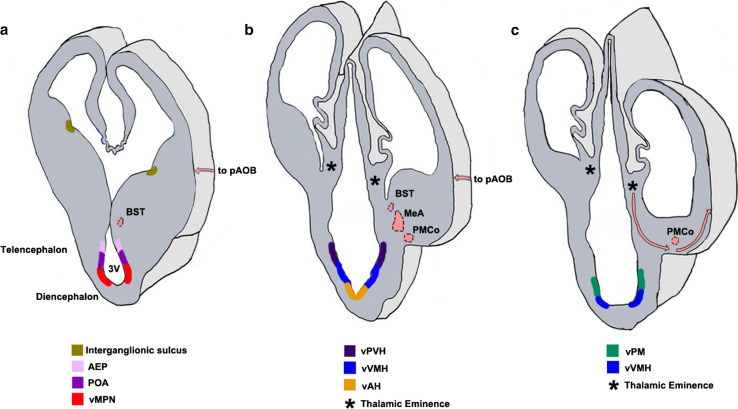
Fig. 7.
Neuroepithelial domains contributing to components of the vomeronasal system [36, 193, 203, 204, 212–214, 225, 226]. a–c Schematics of sections of the forebrain at three rostrocaudal levels in the coronal plane showing the different neuroepithelial domains that contribute to the hypothalamic nuclei of the VNS. Newborn neurons migrate radially from the designated neuroepithelial ventricular zones (vMPN, vAH, vVMH, vPM) to populate the MPN, AH, VMH, and PM, respectively. a The AEP and POA provide neurons to the BST, MeA, and PMCo. The interganglionic sulcus, between the LGE and MGE, generates interneurons destined for the AH/POA. b The vPVH produces neurons for the vomeronasal amygdala (MeA, PMCo and BST). c the TE (asterisks, b, c) generates M/T neurons destined for the pAOB, which migrate to the telencephalic surface at caudal levels (pink arrows). These neurons undertake a tangential migration along the telencephalic surface (pink arrows, a, b) to the rostrally located pAOB which is not seen in the schematic. The pAOB, MeA, PMCo, BST, and the hypothalamic nuclei are all generated from the VZ of the third ventricle and are all part of the VNS. 3V third ventricle, AEP anterior entopeduncular area, BST bed nucleus of stria terminalis, MeA medial amygdaloid nucleus, pAOB accessory olfactory bulb, posterior division, PMCo posteromedial cortical nucleus, POA preoptic area, TE thalamic eminence, vAH ventricular zone for anterior hypothalamic nucleus, vMPN ventricular zone for medial preoptic nucleus, vPM ventricular zone for pre-mammillary nucleus, vPVH ventricular zone for paraventricular hypothalamic nucleus, vVMH ventricular zone for ventromedial hypothalamic nucleus
Migrations to the BST and BAOT
The BST is an important relay structure and a target of the MeA [187, 189]. It is divided into lateral (BSTL) and medial components (BSTM) which are involved in mediating autonomic and vomeronasal responses, respectively. The BST has heterogeneous origins that have been revealed by lineage tracing using vital dyes, in utero electroporation, and genetic mapping studies (Table 2) [36, 202–204].
Neurons of the BSTM are derived from the AEP, commissural POA, CGE and the neuroepithelium of the third ventricle underlying the PVH (Fig. 7) [36, 203, 204]. The third ventricle-derived population is part of the Otp-dependent excitatory neuron migration that also populates the MeA and the PMCo, described earlier [203]. The BSTM is parcellated functionally into anterior (BSTMa), posterolateral (BSTMpl), and posteromedial (BSTMpm) components. Studies based on gene expression patterns suggest that BSTMa and BSTMpm receive cells in large part from the AEP. The diencephalic population is thought to contribute largely to the BSTMpl based on the expression of Pax6 and Tbr1 in this subregion, and their coexpression with Sim1 at the hypothalamic neuroepithelium [202].
BAOT, a lesser studied component of the vomeronasal amygdala, displays a distinct developmental profile from the other three components. Based on gene expression studies, cells for the BAOT appear to be derived from the VP [202] and the Emx1-lineage [210], indicating a pallial origin for this nucleus.
Hypothalamic nuclei
The hypothalamus is a rostral diencephalic structure that receives input from a variety of systems including the olfactory system. In particular, the MeA projects densely to the medial preoptic nucleus (MPN), anterior hypothalamic nucleus (AH), ventromedial hypothalamic nucleus (VMH), and premammillary nucleus (PM). Of these, the MPN, ventrolateral VMH, and ventral PM process reproductive cues, whereas the AH, dorsomedial VMH, and dorsal PM process defensive/aggressive cues [187, 189, 211]. With the exception of the MPN, described below, the entire hypothalamus is largely born from the neuroepithelium of the third ventricle between E10 and E16 in mice (Fig. 7) [212, 213]. Newborn neurons migrate along radial glia to their respective positions in an outside-in pattern, such that the earliest born neurons migrate the farthest from VZ [214, 215].
Migrations to the hypothalamic nuclei
The MPN is a sexually dimorphic nucleus located in the POA and is considered to be more of a boundary structure at the DTB than a diencephalic region. In contrast to the migration pattern seen in the rest of the POA or other hypothalamic nuclei, the MPN develops using an inside-out pattern of radial migration similar to that seen in the dorsal telencephalon [214, 216]. The MPN also hosts neurons arising from an unusual migration from an unexpected source: GnRH neurons that control reproductive behavior, arising from the OP. These neurons undergo what may possibly be the longest tangential migration in the forebrain, arising at E10.5 in the OP. They undertake an axonophilic migration from E11.5 to E13.5, along a transient branch of the vomeronasal nerve from the VNO, penetrating the cribriform plate as part of the “migratory mass” described in an earlier section. They eventually settle in the MPN and lamina terminalis of the hypothalamus by E16.5 [108]. Not surprisingly, proper migration of GnRH neurons requires multiple guidance cues en route: adhesion molecules such as PSA-NCAM [217]; anosmin-1, an extracellular glycoprotein [218, 219]; semaphorin 4D–plexin B1 coupling [220]; ephrin receptor EphA5 [221]; and fibroblast growth factor 8 [222, 223]. The mechanisms of this complex migration have been reviewed elsewhere [224]. Once they settle in the MPN, they extend their axons into the median eminence thereby controlling the release of anterior pituitary hormones responsible for reproductive behavior.
The AH is located caudal to the POA with which it shares several developmental characteristics. The embryonic AH and the POA display a complementary expression of Foxd1 and Foxg1, respectively. There are, however, some Foxd1 expressing cells sprinkled in the AH, suggesting a possible migration of cells from the POA [225]. Though the closest ventricular domain to the AH and POA is the third ventricle, both structures receive radially migrating as well as tangentially migrating cells of telencephalic origin. Migrating cells are reported along the radial glia extending from telencephalic lateral ventricles adjacent to the septum and terminating at the pial surface of AH/POA [215]. Interneurons for the AH/POA migrate tangentially from the telencephalic inter-ganglionic sulcus between the LGE and the MGE, guided by nuclear receptors COUP-TF I and II [226]. The POA and the ventral midline of the AH both express Shh embryonically. The AH appears to be critically dependent on this factor, since the entire anterior hypothalamic area is severely reduced in the hypothalamus-specific knockout of Shh [227].
The VMH is a midline structure of the tuberal hypothalamic region and is derived from Nkx2.1 expressing neuroepithelium [225] from E10 to E15 in mice [212]. Neurons that populate this region are from the Shh-lineage, arising from the neuroepithelium lining the third ventricle, and migrating along the radial glia to the mantle [228]. Though the VMH neurons all appear to arise from this single domain, the mature VMH contains molecularly distinct populations of cells in its different sub-regions. Neurons of the ventrolateral VMH express the estrogen receptor, ERα, and also the GABAA receptor subunits. Neurons of the central and dorsomedial VMH express GABAB receptor subunits. Not surprisingly, GABA plays an important role in regulating migration and positioning of the neurons, a function that is specific to VMH neurons and not those of adjacent hypothalamic nuclei [213, 229–231]. Central and dorsomedial VMH neurons also release brain-derived neurotrophic factor (BDNF) and selectively express an orphan nuclear receptor, steroidogenic factor SF-1/Nr5a1 which is required for the positioning and coalescence of both SF-1 expressing and non-expressing VMH neurons [230, 232].
The PM is located in the mammillary region and is the most caudal of all the vomeronasal hypothalamic targets. Neurons of the PM are produced in the Nkx2.1 positive neuroepithelium of the third ventricle. Lef1, a mediator and a target of Wnt signaling, is detected specifically in the PM neuroepithelium at the time of hypothalamic neurogenesis as well as in differentiated PM neurons later [225]. Wnt signaling may therefore play an important role in the formation of the PM. Shh is necessary for the differentiation of the PM, though its role for the AH and VMH development may be more prominent, since these latter nuclei are more severely affected in the absence of Shh [225, 227, 233, 234].
Insights from olfactory system migrations: a developmental perspective
Creative use of boundaries
A common developmental feature across all systems in which developmental patterning is examined is that cell lineage restriction boundaries or “compartment” boundaries play critical roles in defining the identities of adjacent regions. Boundaries prevent the intermixing of cells with other compartments, thereby spatially restricting cells destined to form particular structures and providing signaling cues to surrounding cell populations. When certain cell populations cross such boundaries, they add a layer of complexity to the system and deserve special attention. The olfactory system is rich in intriguing examples of this. Three boundaries in the forebrain have been well studied: the PSB in the telencephalon; the DTB at the telencephalic–diencephalic boundary; and the zona limitans intrathalamica (ZLI) in the diencephalon that demarcates the thalamus from the prethalamus. Each of these are known or proposed to be signaling centers that emanate cues to adjacent domains [26, 235–240].
The DTB has recently been proposed to be part of a “forebrain hem system” [27] and is witness to many migrations in each direction. Cells from the TE that cross the DTB and enter the telencephalon include cells destined for the pAOB, MeA, and OC. Lot cells that form a corridor in the lateral telencephalon to guide the axons of the LOT may also arise from the TE. Migrations in the opposite direction include cells arising from telencephalic domains that migrate across the DTB to the diencephalic AH and POA. The role of the boundary itself in these migrations is worth close examination in future studies.
The PSB is suggested to be a telencephalic signaling center [241, 242]. The domain abutting the PSB on the pallial side, the VP, expresses a number of morphogens and guidance cues such as the secreted form of Wnt antagonist, soluble Frizzled-related protein, sfrp2 [236], chemokine SDF1 [243], and members of epidermal growth factor (EGF) family [241, 244]. Multiple migrations to components of olfactory system, the pAOB projection neurons [193], lot cells [153] (Ruiz-Reig et al., under revision, cerebral cortex), Lhx5-positive cells migrating to the rostral OC [177], and the axons of the LOT themselves follow a trajectory that traces the PSB on the lateral aspect of the telencephalon and may utilize cues secreted from this structure for their migration.
The olfactory system therefore illustrates that “boundaries” can play a wide range of roles in the development not necessarily limited to the canonical definition of segregating compartments.
A hypothesis for programming connectivity at the domain of origin
Is connectivity between the individual components of a circuit encoded in their progenitors? Sokolowski and Corbin [195] proposed an elegant mechanism for the formation of complex circuits. They propose that if progenitors from a single domain expressing a specific set of transcription factors give rise to multiple structures of a circuit, the shared molecular code of the neurons may provide a mechanism to mediate the establishment of connectivity between those structures. There are some circuits in which such a mechanism could be examined. One example is the reproductive arm of the accessory olfactory system, in which several nuclei express Lhx6 [189]. These include the MeAPD division of the MeA as well as its target, the BSTMpm division of the BST [187, 189], both of which arise from a common domain of origin, the AEP [202, 204], and connect to each other as part of a functional circuit.
Another example is the AOB–MeA circuit. In mouse and Xenopus, cells originating in the TE encounter the MeA in their migration route that ends in the AOB [193] and may contribute to it. The TE, migrating neurons, MeA, and AOB all express Lhx5. It may not be mere coincidence that the MeA is a major target of the AOB. Rather, their connectivity may be linked to a common domain of origin, the TE, which itself is known to control olfactory processing in amphibians [245].
Similarly, cell migrations from closely juxtaposed domains at the DTB populate many interconnected components of the VNS. The TE, MPN and POA are all located at the DTB in rodents (Fig. 7), in close proximity with the hypothalamic ventricle from which all of the hypothalamic targets of the VNS arise, indicating that much of the vomeronasal circuit originates from the diencephalic ventricle/DTB.
The olfactory system therefore offers an appropriately complex set of circuits to examine the hypothesis that the connectivity of a circuit may be linked to the domain of origin of its components. An exciting parallel has been demonstrated at a clonal level in the neocortex, wherein neurons born from the same progenitor preferentially connect to each other [246]. This process can have implications on function, e.g., sibling neurons in the primary visual cortex respond to similar visual stimuli for both orientation and direction [247]. Future studies could aim at finding downstream targets which direct connectivity between individual structures in the olfactory system.
Insights from olfactory system migrations: a disease perspective
Understanding the nature and the mechanisms of cell migration in the olfactory system is critical for an insight into the etiology of disorders such as Kallmann syndrome, which is characterized by hypogonadotropic hypogonadism and anosmia. This disorder is caused by a failure of olfactory nerve formation, which results in a migration defect in GnRH neurons. There is concomitant aplasia of the OB itself, since pioneer axons from the olfactory nerve are known to stimulate OB evagination [30, 248, 249], causing defects in olfaction [218, 219, 250]. Mutations in KAL1, which encodes an NCAM anosmin-1, result in the X-linked form of the disease. Mutations in KAL2, which encodes the fibroblast growth factor receptor FGFR1, leads to the autosomal dominant form of Kallmann syndrome [219, 250–252].
Other developmental pleiotropic diseases such as the CHARGE syndrome, trisomy 13 or Patau syndrome, and trisomy 18 or Edward syndrome in which the OB is hypoplasic or aplasic also show migration defects in GnRH neurons [253], which leads to a decrease in the levels of circulating sex hormones causing hypogonadism in these patients. In contrast, when there is hyperplasia of the OB, it results in a different set of disorders such as fetal immobility/Pena–Shokeir syndrome. The enlarged OBs display lamination defects and the absence of glomeruli [254, 255]. In addition to developmental disorders, a number of neurodegenerative disorders are accompanied by deficits in proliferation or migration of neuroblasts from the SVZ to the OB. These deficits may be used for predicting the onset of disorders such as Alzheimer’s disease [256]. The hypothalamus is the seat of neuroendocrine control for the body and, therefore, disruptions of migration to the hypothalamic components of the circuit may underlie eating disorders such as anorexia nervosa, bulimia nervosa, neurohypophyseal diabetes insipidus, disorders of sexual behavior, and mood disorders [257, 258]. Some patients with eating disorders show inability to detect and/or identify odors [259], which may be due to abnormal proliferation or migration of the olfactory components of the system, underscoring the olfactory–hypothalamic relationship in development. Understanding cell migrations in the olfactory system can therefore provide insights into disease etiology and treatment.
OECs, which envelop the OSN axons, are being evaluated as potential donor cells for transplantation therapy in peripheral nerve and spinal cord injuries [260–262]. OECs derived from the adult rodent OB express myelin-associated proteins and have been shown to myelinate axons of co-cultured dorsal root ganglion cells in vitro [263, 264]. Furthermore, in vivo OEC transplants in rodents appear to promote spinal cord regeneration and recovery of behaviors affected due to spinal cord injuries [265–267]. One study in humans performed intraspinal grafting of autologous OECs and fibroblasts isolated from the olfactory mucosa in paralyzed patients with complete spinal cord injury. Both control and transplant recipients received intense neurorehabilitation, but only the latter displayed some recovery of neurological function [262]. Therefore, migratory cells of the olfactory system may have properties that are not utilized in normal life, but may be harnessed in translational paradigms for therapeutic applications.
Insights from olfactory system migrations: an evolutionary perspective
The olfactory system is the most ancient sensory system and is evolutionarily conserved in terms of function, connectivity, and development across vertebrates [268, 269]. The conservation of origins for different structures within the olfactory system across disparate vertebrate classes is remarkable. This is particularly evident in OB development. OB M/T cells are pallial, and OB interneurons are subpallial in origin in rodents, chicks, Xenopus, and fish [26, 270–276]. Several vertebrate species including fish [277–279], reptiles [280, 281], rodents [77, 78], and non-human primates [282, 283] demonstrate postnatal and adult olfactory neurogenesis and migration of neuroblasts along the RMS. Adult humans also have a proliferative SVZ [284, 285] and there is evidence of migrating olfactory neuroblasts along the RMS to the OB [286].
Some differences in the nature of RMS migration are intriguing and may hint at how this phenomenon evolved: neuronal precursors migrate along the RMS from the telencephalic VZ to the OB in the adult zebrafish brain [279, 287], but this migration is along radial glia and not surrounded by glial tubes [279, 288, 289], in contrast to the rodent RMS [78].
In rodents, the AOB is a distinct substructure from the MOB. This distinction is not observed in fish except lungfish [290, 291], nor is it seen in some reptiles such as crocodiles and turtles, and birds [292]. While amphibians do have an AOB, it does not appear to be divided into the aAOB and pAOB [273, 276] except in the common Japanese toad, Bufo japonicus [293]. However, projection neurons of Xenopus AOB originate in the TE and migrate from the caudal end of the telencephalon rostrally to the AOB, similar to the migration of mouse pAOB projection neurons [193]. This leads to the tantalizing speculation that the amphibian AOB may correspond to the pAOB in rodents, and that the rodent aAOB may be an added-on specialization derived from the MOB, with which it shares its domain of origin and migratory mechanisms [193].
Multiple amygdaloid nuclei are implicated in olfactory and vomeronasal behaviors in rodents [16, 167]. The MeA in particular has been studied across several species of tetrapods and anurans. In anurans, the MeA is the only nucleus that receives input from the olfactory system [294–296]. Consistent with its subpallial identity, the developing MeA expresses Nkx2.1 in Xenopus and rodents [208, 209, 268, 274, 276]. However, the MeA also receives migratory cells from disparate sources, making it a highly mixed structure. In rodents and Xenopus, pallial origin cells from the VP marked by Lhx9 expression populate the MeA [268, 274, 297]. In addition, cells from the diencephalic VZ also migrate to the MeA in rodents [36], and this migration requires Otp [203]. A similar migration is suggested by Otp expression in Rana perezi and axolotl [298].
The apparent diencephalic-origin, Otp expression, subpallium-derived Nkx2.1/Lhx6 expression, and the VP-derived Lhx9 expression are also seen in the MeA of reptiles [298–300] and chicks [26, 298, 301, 302], suggesting that multiple origins for the MeA are conserved in different vertebrate species. These observations bring the MeA to the center stage for studies of behavior arising from circuitry that may also be similarly conserved.
Concluding remarks
The olfactory system mediates a variety of social, motivational, and emotional behaviors including innate behaviors that are important for the survival of an organism and for the propagation of its species. This fundamental purpose may explain why the organization of the olfactory circuit is similar across most vertebrates. This is also consistent with the considerable degree of conservation of developmental origins and cell migrations to diverse structures within the system [193, 298, 299]. The olfactory system is composed of both nuclear and laminated structures. The mechanisms that shape the assembly of this ancient sensory system may have laid the foundation of developmental mechanisms for evolutionarily more recent structures such as the neocortex.
The olfactory system presents a combination of ancient origins, complex migrations leading to intricate circuitry, evolutionarily conserved circuit components and regulatory mechanisms, and fundamental behaviors critical for the maintenance of a species. These features make the olfactory system an attractive model for understanding both, the developmental mechanisms of circuit assembly in the forebrain and the possible evolution of these strategies in more recent structures such as the neocortex. Insight into the mechanisms underlying disorders arising from aberrant olfactory system development may also inform our understanding of disorders arising from defective neocortical development.
Acknowledgments
We thank Zeba Khatri, Hillary Schiff, and Vidita Vaidya for critical comments and Leora D’Souza for assistance with the references. Work on olfactory migrations in the Tole lab was funded by a Wellcome Trust Senior Fellowship (056684/Z/99/Z) and a Grant from the Department of Biotechnology, Government of India (BT/PR/14266/Med/30/442/2010).
Abbreviations
- AEP
Anterior entopeduncular area
- AH
Anterior hypothalamic nucleus
- AOB
Accessory olfactory bulb
- aAOB
Anterior division, accessory olfactory bulb
- pAOB
Posterior division, accessory olfactory bulb
- AON
Anterior olfactory nucleus
- aSVZ
Anterior sub-ventricular zone
- BAOT
Bed nucleus of accessory olfactory tract
- BST
Bed nucleus of stria terminalis
- BSTL
Bed nucleus of stria terminalis, lateral division
- BSTM
Bed nucleus of stria terminalis, medial division
- BSTMa
Bed nucleus of stria terminalis, medial division, anterior portion
- BSTMpl
Bed nucleus of stria terminalis, medial division, posterolateral portion
- BSTMpm
Bed nucleus of stria terminalis, medial division, posteromedial portion
- CGE
Caudal ganglionic eminence
- CoA
Cortical amygdaloid nucleus
- DTB
Diencephalic–telencephalic boundary
- DP
Dorsal pallium
- EGF
Epidermal growth factor
- FGF
Fibroblast growth factor
- LCS
Lateral cortical stream
- LGE
Lateral ganglionic eminence
- dLGE
Dorsal lateral ganglionic eminence
- vLGE
Ventral lateral ganglionic eminence
- LOT
Lateral olfactory tract
- LP
Lateral pallium
- MCL
Mitral cell layer
- MeA
Medial amygdaloid nucleus
- MeAA
Medial amygdaloid nucleus, anterior division
- MeAPD
Medial amygdaloid nucleus, posterodorsal division
- MeAPV
Medial amygdaloid nucleus, posteroventral division
- MGE
Medial ganglionic eminence
- MOB
Main olfactory bulb
- MP
Medial pallium
- MPN
Medial preoptic nucleus
- nLOT
Nucleus of lateral olfactory tract
- OBLS
Olfactory bulb-like structure
- OC
Olfactory cortex
- OT
Olfactory tubercle
- PC
Piriform cortex
- PMCo
Posteromedial cortical nucleus
- PM
Pre-mammillary nucleus
- POA
Preoptic area
- PSB
Pallial–subpallial boundary
- PSA-NCAM
Polysialic acid–neural cell adhesion molecule
- PVH
Para-ventricular hypothalamic nucleus
- RMS
Rostral migratory stream
- RMTW
Rostromedial telencephalic wall
- Shh
Sonic hedgehog
- TE
Thalamic eminence
- VMH
Ventromedial hypothalamic nucleus
- VNO
Vomeronasal organ
- VNS
Vomeronasal system
- VP
Ventral pallium
- VZ
Ventricular zone
- ZL
Zona limitans intrathalamica
References
- 1.Haas CA, et al. Role for reelin in the development of granule cell dispersion in temporal lobe epilepsy. J Neurosci. 2002;22(14):5797–5802. doi: 10.1523/JNEUROSCI.22-14-05797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bozzi Y, Casarosa S, Caleo M. Epilepsy as a neurodevelopmental disorder. Front Psychiatry. 2012;3:19. doi: 10.3389/fpsyt.2012.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jamadar S, et al. Genetic influences of cortical gray matter in language-related regions in healthy controls and schizophrenia. Schizophr Res. 2011;129(2–3):141–148. doi: 10.1016/j.schres.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee FH, et al. Abnormal interneuron development in disrupted-in-schizophrenia-1 L100P mutant mice. Mol Brain. 2013;6:20. doi: 10.1186/1756-6606-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Penagarikano O, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147(1):235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fatemi SH, et al. Consensus paper: pathological role of the cerebellum in autism. Cerebellum. 2012;11(3):777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grove M, et al. ABI2-deficient mice exhibit defective cell migration, aberrant dendritic spine morphogenesis, and deficits in learning and memory. Mol Cell Biol. 2004;24(24):10905–10922. doi: 10.1128/MCB.24.24.10905-10922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitch RH, et al. Persistent spatial working memory deficits in rats with bilateral cortical microgyria. Behav Brain Funct. 2008;4:45. doi: 10.1186/1744-9081-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marin O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 10.Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27(7):392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Berry M, Rogers AW. The migration of neuroblasts in the developing cerebral cortex. J Anat. 1965;99(Pt 4):691–709. [PMC free article] [PubMed] [Google Scholar]
- 12.Rakic P. Principles of neural cell migration. Experientia. 1990;46(9):882–891. doi: 10.1007/BF01939380. [DOI] [PubMed] [Google Scholar]
- 13.Corbin JG, Nery S, Fishell G. Telencephalic cells take a tangent: non-radial migration in the mammalian forebrain. Nat Neurosci. 2001;4(Suppl):1177–1182. doi: 10.1038/nn749. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka DH, et al. Multidirectional and multizonal tangential migration of GABAergic interneurons in the developing cerebral cortex. Development. 2006;133(11):2167–2176. doi: 10.1242/dev.02382. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka DH, Nakajima K. GABAergic interneuron migration and the evolution of the neocortex. Dev Growth Differ. 2012;54(3):366–372. doi: 10.1111/j.1440-169X.2012.01351.x. [DOI] [PubMed] [Google Scholar]
- 16.Halpern M, Martinez-Marcos A. Structure and function of the vomeronasal system: an update. Prog Neurobiol. 2003;70(3):245–318. doi: 10.1016/S0301-0082(03)00103-5. [DOI] [PubMed] [Google Scholar]
- 17.Munger SD, Leinders-Zufall T, Zufall F. Subsystem organization of the mammalian sense of smell. Annu Rev Physiol. 2009;71:115–140. doi: 10.1146/annurev.physiol.70.113006.100608. [DOI] [PubMed] [Google Scholar]
- 18.Abraham NM, et al. Maintaining accuracy at the expense of speed: stimulus similarity defines odor discrimination time in mice. Neuron. 2004;44(5):865–876. doi: 10.1016/j.neuron.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 19.He J, et al. Distinct signals conveyed by pheromone concentrations to the mouse vomeronasal organ. J Neurosci. 2010;30(22):7473–7483. doi: 10.1523/JNEUROSCI.0825-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puelles L, et al. Comparison of the mammalian and avian telencephalon from the perspective of gene expression data. Eur J Morphol. 1999;37(2–3):139–150. doi: 10.1076/ejom.37.2-3.0139. [DOI] [PubMed] [Google Scholar]
- 21.Anderson SA, et al. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128(3):353–363. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- 22.Wichterle H, et al. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 2001;128(19):3759–3771. doi: 10.1242/dev.128.19.3759. [DOI] [PubMed] [Google Scholar]
- 23.Flames N, et al. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27(36):9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanatani S, et al. COUP-TFII is preferentially expressed in the caudal ganglionic eminence and is involved in the caudal migratory stream. J Neurosci. 2008;28(50):13582–13591. doi: 10.1523/JNEUROSCI.2132-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyoshi G, et al. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J Neurosci. 2010;30(5):1582–1594. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puelles L, et al. Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol. 2000;424(3):409–438. doi: 10.1002/1096-9861(20000828)424:3<409::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 27.Roy A, et al. Lhx2 regulates the development of the forebrain hem system. Cereb Cortex. 2014;24(5):1361–1372. doi: 10.1093/cercor/bhs421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Moreno F, Lopez-Mascaraque L, de Carlos JA. Early telencephalic migration topographically converging in the olfactory cortex. Cereb Cortex. 2008;18(6):1239–1252. doi: 10.1093/cercor/bhm154. [DOI] [PubMed] [Google Scholar]
- 29.Ceci ML, Pedraza M, de Carlos JA. The embryonic septum and ventral pallium, new sources of olfactory cortex cells. PLoS One. 2012;7(9):e44716. doi: 10.1371/journal.pone.0044716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Carlos JA, Lopez-Mascaraque L, Valverde F. Early olfactory fiber projections and cell migration into the rat telencephalon. Int J Dev Neurosci. 1996;14(7–8):853–866. doi: 10.1016/S0736-5748(96)00055-X. [DOI] [PubMed] [Google Scholar]
- 31.Rubenstein JL, et al. Genetic control of cortical regionalization and connectivity. Cereb Cortex. 1999;9(6):524–532. doi: 10.1093/cercor/9.6.524. [DOI] [PubMed] [Google Scholar]
- 32.Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128(2):193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
- 33.Medina L, et al. Expression of Dbx1, neurogenin 2, semaphorin 5A, cadherin 8, and Emx1 distinguish ventral and lateral pallial histogenetic divisions in the developing mouse claustroamygdaloid complex. J Comp Neurol. 2004;474(4):504–523. doi: 10.1002/cne.20141. [DOI] [PubMed] [Google Scholar]
- 34.Tole S, et al. Selective requirement of Pax6, but not Emx2, in the specification and development of several nuclei of the amygdaloid complex. J Neurosci. 2005;25(10):2753–2760. doi: 10.1523/JNEUROSCI.3014-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carney RS, et al. Cell migration along the lateral cortical stream to the developing basal telencephalic limbic system. J Neurosci. 2006;26(45):11562–11574. doi: 10.1523/JNEUROSCI.3092-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soma M, et al. Development of the mouse amygdala as revealed by enhanced green fluorescent protein gene transfer by means of in utero electroporation. J Comp Neurol. 2009;513(1):113–128. doi: 10.1002/cne.21945. [DOI] [PubMed] [Google Scholar]
- 37.Bai J, et al. The role of DCX and LIS1 in migration through the lateral cortical stream of developing forebrain. Dev Neurosci. 2008;30(1–3):144–156. doi: 10.1159/000109859. [DOI] [PubMed] [Google Scholar]
- 38.Nagano T, Morikubo S, Sato M. Filamin A and FILIP (filamin A-interacting protein) regulate cell polarity and motility in neocortical subventricular and intermediate zones during radial migration. J Neurosci. 2004;24(43):9648–9657. doi: 10.1523/JNEUROSCI.2363-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramos RL, Bai J, LoTurco JJ. Heterotopia formation in rat but not mouse neocortex after RNA interference knockdown of DCX. Cereb Cortex. 2006;16(9):1323–1331. doi: 10.1093/cercor/bhj074. [DOI] [PubMed] [Google Scholar]
- 40.Gotz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21(5):1031–1044. doi: 10.1016/S0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 41.Warren N, et al. The transcription factor, Pax6, is required for cell proliferation and differentiation in the developing cerebral cortex. Cereb Cortex. 1999;9(6):627–635. doi: 10.1093/cercor/9.6.627. [DOI] [PubMed] [Google Scholar]
- 42.Englund C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25(1):247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mombaerts P. Axonal wiring in the mouse olfactory system. Annu Rev Cell Dev Biol. 2006;22:713–737. doi: 10.1146/annurev.cellbio.21.012804.093915. [DOI] [PubMed] [Google Scholar]
- 44.Wilson RI, Mainen ZF. Early events in olfactory processing. Annu Rev Neurosci. 2006;29:163–201. doi: 10.1146/annurev.neuro.29.051605.112950. [DOI] [PubMed] [Google Scholar]
- 45.Mitsui S, et al. Genetic visualization of the secondary olfactory pathway in Tbx21 transgenic mice. Neural Syst Circuits. 2011;1(1):5. doi: 10.1186/2042-1001-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Igarashi KM, et al. Parallel mitral and tufted cell pathways route distinct odor information to different targets in the olfactory cortex. J Neurosci. 2012;32(23):7970–7985. doi: 10.1523/JNEUROSCI.0154-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bayer SA. 3H-thymidine-radiographic studies of neurogenesis in the rat olfactory bulb. Exp Brain Res. 1983;50(2–3):329–340. doi: 10.1007/BF00239197. [DOI] [PubMed] [Google Scholar]
- 48.Hinds JW. Autoradiographic study of histogenesis in the mouse olfactory bulb. II. Cell proliferation and migration. J Comp Neurol. 1968;134(3):305–322. doi: 10.1002/cne.901340305. [DOI] [PubMed] [Google Scholar]
- 49.Tucker ES, Polleux F, LaMantia AS. Position and time specify the migration of a pioneering population of olfactory bulb interneurons. Dev Biol. 2006;297(2):387–401. doi: 10.1016/j.ydbio.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 50.Bulfone A, et al. An olfactory sensory map develops in the absence of normal projection neurons or GABAergic interneurons. Neuron. 1998;21(6):1273–1282. doi: 10.1016/S0896-6273(00)80647-9. [DOI] [PubMed] [Google Scholar]
- 51.Imamura F, et al. Timing of neurogenesis is a determinant of olfactory circuitry. Nat Neurosci. 2011;14(3):331–337. doi: 10.1038/nn.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Imamura F, Greer CA. Pax6 regulates Tbr1 and Tbr2 expressions in olfactory bulb mitral cells. Mol Cell Neurosci. 2013;54:58–70. doi: 10.1016/j.mcn.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nomura T, Haba H, Osumi N. Role of a transcription factor Pax6 in the developing vertebrate olfactory system. Dev Growth Differ. 2007;49(9):683–690. doi: 10.1111/j.1440-169X.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 54.Saha B, et al. Dual role for LIM-homeodomain gene Lhx2 in the formation of the lateral olfactory tract. J Neurosci. 2007;27(9):2290–2297. doi: 10.1523/JNEUROSCI.5571-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng W, et al. Disorganized olfactory bulb lamination in mice deficient for transcription factor AP-2epsilon. Mol Cell Neurosci. 2009;42(3):161–171. doi: 10.1016/j.mcn.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshihara S, et al. Arx homeobox gene is essential for development of mouse olfactory system. Development. 2005;132(4):751–762. doi: 10.1242/dev.01619. [DOI] [PubMed] [Google Scholar]
- 57.Shimizu T, Hibi M. Formation and patterning of the forebrain and olfactory system by zinc-finger genes Fezf1 and Fezf2. Dev Growth Differ. 2009;51(3):221–231. doi: 10.1111/j.1440-169X.2009.01088.x. [DOI] [PubMed] [Google Scholar]
- 58.Puche AC, Shipley MT. Radial glia development in the mouse olfactory bulb. J Comp Neurol. 2001;434(1):1–12. doi: 10.1002/cne.1160. [DOI] [PubMed] [Google Scholar]
- 59.Arnold SJ, et al. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22(18):2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anderson SA, et al. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278(5337):474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 61.Nobrega-Pereira S, Marin O. Transcriptional control of neuronal migration in the developing mouse brain. Cereb Cortex. 2009;19(Suppl 1):i107–i113. doi: 10.1093/cercor/bhp044. [DOI] [PubMed] [Google Scholar]
- 62.Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci. 2003;23(1):167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Long JE, et al. DLX5 regulates development of peripheral and central components of the olfactory system. J Neurosci. 2003;23(2):568–578. doi: 10.1523/JNEUROSCI.23-02-00568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Batista-Brito R, et al. The distinct temporal origins of olfactory bulb interneuron subtypes. J Neurosci. 2008;28(15):3966–3975. doi: 10.1523/JNEUROSCI.5625-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krosnowski K, et al. Diverse populations of intrinsic cholinergic interneurons in the mouse olfactory bulb. Neuroscience. 2012;213:161–178. doi: 10.1016/j.neuroscience.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rudy B, et al. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011;71(1):45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taniguchi H, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71(6):995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Marchis S, et al. Generation of distinct types of periglomerular olfactory bulb interneurons during development and in adult mice: implication for intrinsic properties of the subventricular zone progenitor population. J Neurosci. 2007;27(3):657–664. doi: 10.1523/JNEUROSCI.2870-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vergano-Vera E, et al. Generation of GABAergic and dopaminergic interneurons from endogenous embryonic olfactory bulb precursor cells. Development. 2006;133(21):4367–4379. doi: 10.1242/dev.02601. [DOI] [PubMed] [Google Scholar]
- 71.Cocas LA, et al. Pax6 is required at the telencephalic pallial–subpallial boundary for the generation of neuronal diversity in the postnatal limbic system. J Neurosci. 2011;31(14):5313–5324. doi: 10.1523/JNEUROSCI.3867-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kohwi M, et al. A subpopulation of olfactory bulb GABAergic interneurons is derived from Emx1- and Dlx5/6-expressing progenitors. J Neurosci. 2007;27(26):6878–6891. doi: 10.1523/JNEUROSCI.0254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Willaime-Morawek S, et al. Embryonic cortical neural stem cells migrate ventrally and persist as postnatal striatal stem cells. J Cell Biol. 2006;175(1):159–168. doi: 10.1083/jcb.200604123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andrews W, et al. Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development. 2006;133(11):2243–2252. doi: 10.1242/dev.02379. [DOI] [PubMed] [Google Scholar]
- 75.Li H, et al. Neuregulin repellent signaling via ErbB4 restricts GABAergic interneurons to migratory paths from ganglionic eminence to cortical destinations. Neural Dev. 2012;7:10. doi: 10.1186/1749-8104-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Long JE, et al. Dlx-dependent and -independent regulation of olfactory bulb interneuron differentiation. J Neurosci. 2007;27(12):3230–3243. doi: 10.1523/JNEUROSCI.5265-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luskin MB. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron. 1993;11(1):173–189. doi: 10.1016/0896-6273(93)90281-U. [DOI] [PubMed] [Google Scholar]
- 78.Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264(5162):1145–1148. doi: 10.1126/science.8178174. [DOI] [PubMed] [Google Scholar]
- 79.Lois C, Garcia-Verdugo JM, Alvarez-Buylla A. Chain migration of neuronal precursors. Science. 1996;271(5251):978–981. doi: 10.1126/science.271.5251.978. [DOI] [PubMed] [Google Scholar]
- 80.Snapyan M, et al. Vasculature guides migrating neuronal precursors in the adult mammalian forebrain via brain-derived neurotrophic factor signaling. J Neurosci. 2009;29(13):4172–4188. doi: 10.1523/JNEUROSCI.4956-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whitman MC, et al. Blood vessels form a migratory scaffold in the rostral migratory stream. J Comp Neurol. 2009;516(2):94–104. doi: 10.1002/cne.22093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaneko N, et al. New neurons clear the path of astrocytic processes for their rapid migration in the adult brain. Neuron. 2010;67(2):213–223. doi: 10.1016/j.neuron.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Law AK, et al. Neurogenesis and neuronal migration in the neonatal rat forebrain anterior subventricular zone do not require GFAP-positive astrocytes. Dev Biol. 1999;216(2):622–634. doi: 10.1006/dbio.1999.9498. [DOI] [PubMed] [Google Scholar]
- 84.Ono K, et al. N-CAM mutation inhibits tangential neuronal migration and is phenocopied by enzymatic removal of polysialic acid. Neuron. 1994;13(3):595–609. doi: 10.1016/0896-6273(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 85.Pencea V, Luskin MB. Prenatal development of the rodent rostral migratory stream. J Comp Neurol. 2003;463(4):402–418. doi: 10.1002/cne.10746. [DOI] [PubMed] [Google Scholar]
- 86.Battista D, Rutishauser U. Removal of polysialic acid triggers dispersion of subventricularly derived neuroblasts into surrounding CNS tissues. J Neurosci. 2010;30(11):3995–4003. doi: 10.1523/JNEUROSCI.4382-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jankovski A, Sotelo C. Subventricular zone-olfactory bulb migratory pathway in the adult mouse: cellular composition and specificity as determined by heterochronic and heterotopic transplantation. J Comp Neurol. 1996;371(3):376–396. doi: 10.1002/(SICI)1096-9861(19960729)371:3<376::AID-CNE3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 88.Murase S, Horwitz AF. Deleted in colorectal carcinoma and differentially expressed integrins mediate the directional migration of neural precursors in the rostral migratory stream. J Neurosci. 2002;22(9):3568–3579. doi: 10.1523/JNEUROSCI.22-09-03568.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Petreanu L, Alvarez-Buylla A. Maturation and death of adult-born olfactory bulb granule neurons: role of olfaction. J Neurosci. 2002;22(14):6106–6113. doi: 10.1523/JNEUROSCI.22-14-06106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bovetti S, et al. Blood vessels form a scaffold for neuroblast migration in the adult olfactory bulb. J Neurosci. 2007;27(22):5976–5980. doi: 10.1523/JNEUROSCI.0678-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 1972;145(1):61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- 92.Noctor SC, et al. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409(6821):714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 93.Poluch S, Juliano SL. A normal radial glial scaffold is necessary for migration of interneurons during neocortical development. Glia. 2007;55(8):822–830. doi: 10.1002/glia.20488. [DOI] [PubMed] [Google Scholar]
- 94.Yokota Y, et al. Radial glial dependent and independent dynamics of interneuronal migration in the developing cerebral cortex. PLoS One. 2007;2(8):e794. doi: 10.1371/journal.pone.0000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McManus MF, et al. Axon mediated interneuron migration. J Neuropathol Exp Neurol. 2004;63(9):932–941. doi: 10.1093/jnen/63.9.932. [DOI] [PubMed] [Google Scholar]
- 96.Luccardini C, et al. N-cadherin sustains motility and polarity of future cortical interneurons during tangential migration. J Neurosci. 2013;33(46):18149–18160. doi: 10.1523/JNEUROSCI.0593-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Altman J, Bayer SA. Mosaic organization of the hippocampal neuroepithelium and the multiple germinal sources of dentate granule cells. J Comp Neurol. 1990;301(3):325–342. doi: 10.1002/cne.903010302. [DOI] [PubMed] [Google Scholar]
- 98.Altman J, Bayer SA. Migration and distribution of two populations of hippocampal granule cell precursors during the perinatal and postnatal periods. J Comp Neurol. 1990;301(3):365–381. doi: 10.1002/cne.903010304. [DOI] [PubMed] [Google Scholar]
- 99.Li G, et al. Identification of a transient subpial neurogenic zone in the developing dentate gyrus and its regulation by Cxcl12 and reelin signaling. Development. 2009;136(2):327–335. doi: 10.1242/dev.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blanchart A, et al. Peripheral contributions to olfactory bulb cell populations (migrations towards the olfactory bulb) Glia. 2011;59(2):278–292. doi: 10.1002/glia.21100. [DOI] [PubMed] [Google Scholar]
- 101.Graziadei PP, Graziadei GA. Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J Neurocytol. 1979;8(1):1–18. doi: 10.1007/BF01206454. [DOI] [PubMed] [Google Scholar]
- 102.Calof AL, Chikaraishi DM. Analysis of neurogenesis in a mammalian neuroepithelium: proliferation and differentiation of an olfactory neuron precursor in vitro. Neuron. 1989;3(1):115–127. doi: 10.1016/0896-6273(89)90120-7. [DOI] [PubMed] [Google Scholar]
- 103.Calof AL, et al. Neurogenesis and cell death in olfactory epithelium. J Neurobiol. 1996;30(1):67–81. doi: 10.1002/(SICI)1097-4695(199605)30:1<67::AID-NEU7>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 104.Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci. 2007;10(6):720–726. doi: 10.1038/nn1882. [DOI] [PubMed] [Google Scholar]
- 105.Iwai N, et al. Horizontal basal cells are multipotent progenitors in normal and injured adult olfactory epithelium. Stem Cells. 2008;26(5):1298–1306. doi: 10.1634/stemcells.2007-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Valverde F, Santacana M, Heredia M. Formation of an olfactory glomerulus: morphological aspects of development and organization. Neuroscience. 1992;49(2):255–275. doi: 10.1016/0306-4522(92)90094-I. [DOI] [PubMed] [Google Scholar]
- 107.Marin-Padilla M, Amieva MR. Early neurogenesis of the mouse olfactory nerve: Golgi and electron microscopic studies. J Comp Neurol. 1989;288(2):339–352. doi: 10.1002/cne.902880211. [DOI] [PubMed] [Google Scholar]
- 108.Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone–releasing hormone neurons. Nature. 1989;338(6211):161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- 109.Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone–releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci USA. 1989;86(20):8132–8136. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miller AM, Treloar HB, Greer CA. Composition of the migratory mass during development of the olfactory nerve. J Comp Neurol. 2010;518(24):4825–4841. doi: 10.1002/cne.22497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schwarting GA, Gridley T, Henion TR. Notch1 expression and ligand interactions in progenitor cells of the mouse olfactory epithelium. J Mol Histol. 2007;38(6):543–553. doi: 10.1007/s10735-007-9110-9. [DOI] [PubMed] [Google Scholar]
- 112.Doucette JR. The glial cells in the nerve fiber layer of the rat olfactory bulb. Anat Rec. 1984;210(2):385–391. doi: 10.1002/ar.1092100214. [DOI] [PubMed] [Google Scholar]
- 113.Doucette R. Development of the nerve fiber layer in the olfactory bulb of mouse embryos. J Comp Neurol. 1989;285(4):514–527. doi: 10.1002/cne.902850407. [DOI] [PubMed] [Google Scholar]
- 114.Valverde F, Lopez-Mascaraque L. Neuroglial arrangements in the olfactory glomeruli of the hedgehog. J Comp Neurol. 1991;307(4):658–674. doi: 10.1002/cne.903070411. [DOI] [PubMed] [Google Scholar]
- 115.Schwanzel-Fukuda M, et al. Immunocytochemical demonstration of neural cell adhesion molecule (NCAM) along the migration route of luteinizing hormone–releasing hormone (LHRH) neurons in mice. J Comp Neurol. 1992;321(1):1–18. doi: 10.1002/cne.903210102. [DOI] [PubMed] [Google Scholar]
- 116.Valverde F, Heredia M, Santacana M. Characterization of neuronal cell varieties migrating from the olfactory epithelium during prenatal development in the rat. Immunocytochemical study using antibodies against olfactory marker protein (OMP) and luteinizing hormone–releasing hormone (LH–RH) Brain Res Dev Brain Res. 1993;71(2):209–220. doi: 10.1016/0165-3806(93)90173-8. [DOI] [PubMed] [Google Scholar]
- 117.Pellier V, Astic L. Histochemical and immunocytochemical study of the migration of neurons from the rat olfactory placode. Cell Tissue Res. 1994;275(3):587–598. doi: 10.1007/BF00318827. [DOI] [PubMed] [Google Scholar]
- 118.Leibovici M, et al. Avian olfactory receptors: differentiation of olfactory neurons under normal and experimental conditions. Dev Biol. 1996;175(1):118–131. doi: 10.1006/dbio.1996.0100. [DOI] [PubMed] [Google Scholar]
- 119.Nef S, et al. Olfaction in birds: differential embryonic expression of nine putative odorant receptor genes in the avian olfactory system. Mech Dev. 1996;55(1):65–77. doi: 10.1016/0925-4773(95)00491-2. [DOI] [PubMed] [Google Scholar]
- 120.Conzelmann S, et al. Extraepithelial cells expressing distinct olfactory receptors are associated with axons of sensory cells with the same receptor type. Cell Tissue Res. 2002;307(3):293–301. doi: 10.1007/s00441-001-0507-6. [DOI] [PubMed] [Google Scholar]
- 121.Honma S, et al. Expression and immunohistochemical localization of vesicular glutamate transporter 2 in the migratory pathway from the rat olfactory placode. Eur J Neurosci. 2004;20(4):923–936. doi: 10.1111/j.1460-9568.2004.03544.x. [DOI] [PubMed] [Google Scholar]
- 122.Ikeda K, et al. Six1 is essential for early neurogenesis in the development of olfactory epithelium. Dev Biol. 2007;311(1):53–68. doi: 10.1016/j.ydbio.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 123.Merlo GR, et al. The role of Dlx homeogenes in early development of the olfactory pathway. J Mol Histol. 2007;38(4):347–358. doi: 10.1007/s10735-007-9109-2. [DOI] [PubMed] [Google Scholar]
- 124.Raisman G. Olfactory ensheathing cells—another miracle cure for spinal cord injury? Nat Rev Neurosci. 2001;2(5):369–375. doi: 10.1038/35072576. [DOI] [PubMed] [Google Scholar]
- 125.Au WW, Treloar HB, Greer CA. Sublaminar organization of the mouse olfactory bulb nerve layer. J Comp Neurol. 2002;446(1):68–80. doi: 10.1002/cne.10182. [DOI] [PubMed] [Google Scholar]
- 126.Raisman G, Li Y. Repair of neural pathways by olfactory ensheathing cells. Nat Rev Neurosci. 2007;8(4):312–319. doi: 10.1038/nrn2099. [DOI] [PubMed] [Google Scholar]
- 127.Astic L, Pellier-Monnin V, Godinot F. Spatio-temporal patterns of ensheathing cell differentiation in the rat olfactory system during development. Neuroscience. 1998;84(1):295–307. doi: 10.1016/S0306-4522(97)00496-X. [DOI] [PubMed] [Google Scholar]
- 128.Franceschini IA, Barnett SC. Low-affinity NGF-receptor and E-N-CAM expression define two types of olfactory nerve ensheathing cells that share a common lineage. Dev Biol. 1996;173(1):327–343. doi: 10.1006/dbio.1996.0027. [DOI] [PubMed] [Google Scholar]
- 129.Gong Q, Shipley MT. Expression of extracellular matrix molecules and cell surface molecules in the olfactory nerve pathway during early development. J Comp Neurol. 1996;366(1):1–14. doi: 10.1002/(SICI)1096-9861(19960226)366:1<1::AID-CNE1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 130.Kafitz KW, Greer CA. Role of laminin in axonal extension from olfactory receptor cells. J Neurobiol. 1997;32(3):298–310. doi: 10.1002/(SICI)1097-4695(199703)32:3<298::AID-NEU4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 131.Key B, Akeson RA. Olfactory neurons express a unique glycosylated form of the neural cell adhesion molecule (N-CAM) J Cell Biol. 1990;110(5):1729–1743. doi: 10.1083/jcb.110.5.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Murdoch B, Roskams AJ. A novel embryonic nestin-expressing radial glia-like progenitor gives rise to zonally restricted olfactory and vomeronasal neurons. J Neurosci. 2008;28(16):4271–4282. doi: 10.1523/JNEUROSCI.5566-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rela L, Bordey A, Greer CA. Olfactory ensheathing cell membrane properties are shaped by connectivity. Glia. 2010;58(6):665–678. doi: 10.1002/glia.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schwarting GA, et al. Semaphorin 3A is required for guidance of olfactory axons in mice. J Neurosci. 2000;20(20):7691–7697. doi: 10.1523/JNEUROSCI.20-20-07691.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.St John JA, Key B. EphB2 and two of its ligands have dynamic protein expression patterns in the developing olfactory system. Brain Res Dev Brain Res. 2001;126(1):43–56. doi: 10.1016/S0165-3806(00)00136-X. [DOI] [PubMed] [Google Scholar]
- 136.Key B, et al. Expression and localization of FGF-1 in the developing rat olfactory system. J Comp Neurol. 1996;366(2):197–206. doi: 10.1002/(SICI)1096-9861(19960304)366:2<197::AID-CNE1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 137.MacDonald KP, et al. FGF2 promotes neuronal differentiation in explant cultures of adult and embryonic mouse olfactory epithelium. J Neurosci Res. 1996;44(1):27–39. doi: 10.1002/(SICI)1097-4547(19960401)44:1<27::AID-JNR4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 138.Mackay-Sima A, Chuahb MI. Neurotrophic factors in the primary olfactory pathway. Prog Neurobiol. 2000;62(5):527–559. doi: 10.1016/S0301-0082(00)00009-5. [DOI] [PubMed] [Google Scholar]
- 139.Ubink R, et al. Neuropeptide tyrosine is expressed in ensheathing cells around the olfactory nerves in the rat olfactory bulb. Neuroscience. 1994;60(3):709–726. doi: 10.1016/0306-4522(94)90499-5. [DOI] [PubMed] [Google Scholar]
- 140.Ubink R, Hokfelt T. Expression of neuropeptide Y in olfactory ensheathing cells during prenatal development. J Comp Neurol. 2000;423(1):13–25. doi: 10.1002/1096-9861(20000717)423:1<13::AID-CNE2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 141.Teare KA, et al. Alpha-MSH inhibits inflammatory signalling in olfactory ensheathing cells. NeuroReport. 2003;14(17):2171–2175. doi: 10.1097/00001756-200312020-00008. [DOI] [PubMed] [Google Scholar]
- 142.Vincent AJ, et al. Bacteria and PAMPs activate nuclear factor kappaB and Gro production in a subset of olfactory ensheathing cells and astrocytes but not in Schwann cells. Glia. 2007;55(9):905–916. doi: 10.1002/glia.20512. [DOI] [PubMed] [Google Scholar]
- 143.Leung JY, et al. Olfactory ensheathing cells are attracted to, and can endocytose, bacteria. Cell Mol Life Sci. 2008;65(17):2732–2739. doi: 10.1007/s00018-008-8184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Harris JA, West AK, Chuah MI. Olfactory ensheathing cells: nitric oxide production and innate immunity. Glia. 2009;57(16):1848–1857. doi: 10.1002/glia.20899. [DOI] [PubMed] [Google Scholar]
- 145.Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79(7):1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 146.Vassar R, et al. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79(6):981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 147.Feinstein P, et al. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell. 2004;117(6):833–846. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 148.Strotmann J, et al. Olfactory receptor proteins in axonal processes of chemosensory neurons. J Neurosci. 2004;24(35):7754–7761. doi: 10.1523/JNEUROSCI.2588-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schwarzenbacher K, et al. Expression of olfactory receptors in the cribriform mesenchyme during prenatal development. Gene Expr Patterns. 2004;4(5):543–552. doi: 10.1016/j.modgep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 150.Zhu Y, et al. Olfactory ensheathing cells: attractant of neural progenitor migration to olfactory bulb. Glia. 2010;58(6):716–729. doi: 10.1002/glia.20957. [DOI] [PubMed] [Google Scholar]
- 151.Inaki K, et al. Laminar organization of the developing lateral olfactory tract revealed by differential expression of cell recognition molecules. J Comp Neurol. 2004;479(3):243–256. doi: 10.1002/cne.20270. [DOI] [PubMed] [Google Scholar]
- 152.Sato Y, et al. Requirement for early-generated neurons recognized by monoclonal antibody lot1 in the formation of lateral olfactory tract. J Neurosci. 1998;18(19):7800–7810. doi: 10.1523/JNEUROSCI.18-19-07800.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tomioka N, et al. Neocortical origin and tangential migration of guidepost neurons in the lateral olfactory tract. J Neurosci. 2000;20(15):5802–5812. doi: 10.1523/JNEUROSCI.20-15-05802.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fode C, et al. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14(1):67–80. [PMC free article] [PubMed] [Google Scholar]
- 155.Schuurmans C, et al. Sequential phases of cortical specification involve neurogenin-dependent and -independent pathways. EMBO J. 2004;23(14):2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dixit R, et al. Neurog1 and Neurog2 control two waves of neuronal differentiation in the piriform cortex. J Neurosci. 2014;34(2):539–553. doi: 10.1523/JNEUROSCI.0614-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kawasaki T, Ito K, Hirata T. Netrin 1 regulates ventral tangential migration of guidepost neurons in the lateral olfactory tract. Development. 2006;133(5):845–853. doi: 10.1242/dev.02257. [DOI] [PubMed] [Google Scholar]
- 158.Ito K, et al. Semaphorin 3F confines ventral tangential migration of lateral olfactory tract neurons onto the telencephalon surface. J Neurosci. 2008;28(17):4414–4422. doi: 10.1523/JNEUROSCI.0372-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rallu M, et al. Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and hedgehog signaling. Development. 2002;129(21):4963–4974. doi: 10.1242/dev.129.21.4963. [DOI] [PubMed] [Google Scholar]
- 160.Tole S, Ragsdale CW, Grove EA. Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes(J) Dev Biol. 2000;217(2):254–265. doi: 10.1006/dbio.1999.9509. [DOI] [PubMed] [Google Scholar]
- 161.Bulchand S, et al. LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech Dev. 2001;100(2):165–175. doi: 10.1016/S0925-4773(00)00515-3. [DOI] [PubMed] [Google Scholar]
- 162.Haberly LB, Price JL. The axonal projection patterns of the mitral and tufted cells of the olfactory bulb in the rat. Brain Res. 1977;129(1):152–157. doi: 10.1016/0006-8993(77)90978-7. [DOI] [PubMed] [Google Scholar]
- 163.Scott JW, McBride RL, Schneider SP. The organization of projections from the olfactory bulb to the piriform cortex and olfactory tubercle in the rat. J Comp Neurol. 1980;194(3):519–534. doi: 10.1002/cne.901940304. [DOI] [PubMed] [Google Scholar]
- 164.Scott JW. Electrophysiological identification of mitral and tufted cells and distributions of their axons in olfactory system of the rat. J Neurophysiol. 1981;46(5):918–931. doi: 10.1152/jn.1981.46.5.918. [DOI] [PubMed] [Google Scholar]
- 165.Schikorski T, Stevens CF. Quantitative fine-structural analysis of olfactory cortical synapses. Proc Natl Acad Sci USA. 1999;96(7):4107–4112. doi: 10.1073/pnas.96.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sanchez-Andrade G, Kendrick KM. The main olfactory system and social learning in mammals. Behav Brain Res. 2009;200(2):323–335. doi: 10.1016/j.bbr.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 167.Swanson LW, Petrovich GD. What is the amygdala? Trends Neurosci. 1998;21(8):323–331. doi: 10.1016/S0166-2236(98)01265-X. [DOI] [PubMed] [Google Scholar]
- 168.Bayer SA. Neurogenesis in the rat primary olfactory cortex. Int J Dev Neurosci. 1986;4(3):251–271. doi: 10.1016/0736-5748(86)90063-8. [DOI] [PubMed] [Google Scholar]
- 169.Nomura T, et al. Pax6-dependent boundary defines alignment of migrating olfactory cortex neurons via the repulsive activity of ephrin A5. Development. 2006;133(7):1335–1345. doi: 10.1242/dev.02290. [DOI] [PubMed] [Google Scholar]
- 170.Valverde F, Santacana M. Development and early postnatal maturation of the primary olfactory cortex. Brain Res Dev Brain Res. 1994;80(1–2):96–114. doi: 10.1016/0165-3806(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 171.Greig LC, et al. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14(11):755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hirata T, et al. Identification of distinct telencephalic progenitor pools for neuronal diversity in the amygdala. Nat Neurosci. 2009;12(2):141–149. doi: 10.1038/nn.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yun K, et al. Patterning of the lateral ganglionic eminence by the Gsh1 and Gsh2 homeobox genes regulates striatal and olfactory bulb histogenesis and the growth of axons through the basal ganglia. J Comp Neurol. 2003;461(2):151–165. doi: 10.1002/cne.10685. [DOI] [PubMed] [Google Scholar]
- 174.Legaz I, Garcia-Lopez M, Medina L. Subpallial origin of part of the calbindin-positive neurons of the claustral complex and piriform cortex. Brain Res Bull. 2005;66(4–6):470–474. doi: 10.1016/j.brainresbull.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 175.Batista-Brito R, Fishell G. The developmental integration of cortical interneurons into a functional network. Curr Top Dev Biol. 2009;87:81–118. doi: 10.1016/S0070-2153(09)01203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhao T, et al. Genetic mapping of Foxb1-cell lineage shows migration from caudal diencephalon to telencephalon and lateral hypothalamus. Eur J Neurosci. 2008;28(10):1941–1955. doi: 10.1111/j.1460-9568.2008.06503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Miquelajauregui A, et al. LIM-homeobox gene Lhx5 is required for normal development of Cajal–Retzius cells. J Neurosci. 2010;30(31):10551–10562. doi: 10.1523/JNEUROSCI.5563-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Martinez-Garcia F, Martinez-Marcos A, Lanuza E. The pallial amygdala of amniote vertebrates: evolution of the concept, evolution of the structure. Brain Res Bull. 2002;57(3–4):463–469. doi: 10.1016/S0361-9230(01)00665-7. [DOI] [PubMed] [Google Scholar]
- 179.Santiago AC, Shammah-Lagnado SJ. Efferent connections of the nucleus of the lateral olfactory tract in the rat. J Comp Neurol. 2004;471(3):314–332. doi: 10.1002/cne.20028. [DOI] [PubMed] [Google Scholar]
- 180.Remedios R, et al. A stream of cells migrating from the caudal telencephalon reveals a link between the amygdala and neocortex. Nat Neurosci. 2007;10(9):1141–1150. doi: 10.1038/nn1955. [DOI] [PubMed] [Google Scholar]
- 181.Remedios R, Subramanian L, Tole S. LIM genes parcellate the embryonic amygdala and regulate its development. J Neurosci. 2004;24(31):6986–6990. doi: 10.1523/JNEUROSCI.0001-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Caviness VS., Jr Neocortical histogenesis in normal and reeler mice: a developmental study based upon [3H]thymidine autoradiography. Brain Res. 1982;256(3):293–302. doi: 10.1016/0165-3806(82)90141-9. [DOI] [PubMed] [Google Scholar]
- 183.Ohshima T, et al. Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc Natl Acad Sci USA. 1996;93(20):11173–11178. doi: 10.1073/pnas.93.20.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lin DY, et al. Encoding social signals in the mouse main olfactory bulb. Nature. 2005;434(7032):470–477. doi: 10.1038/nature03414. [DOI] [PubMed] [Google Scholar]
- 185.Yoon H, Enquist LW, Dulac C. Olfactory inputs to hypothalamic neurons controlling reproduction and fertility. Cell. 2005;123(4):669–682. doi: 10.1016/j.cell.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 186.Kang N, Baum MJ, Cherry JA. A direct main olfactory bulb projection to the ‘vomeronasal’ amygdala in female mice selectively responds to volatile pheromones from males. Eur J Neurosci. 2009;29(3):624–634. doi: 10.1111/j.1460-9568.2009.06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pardo-Bellver C, et al. Differential efferent projections of the anterior, posteroventral, and posterodorsal subdivisions of the medial amygdala in mice. Front Neuroanat. 2012;6:33. doi: 10.3389/fnana.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Thompson JA, et al. Second-order input to the medial amygdala from olfactory sensory neurons expressing the transduction channel TRPM5. J Comp Neurol. 2012;520(8):1819–1830. doi: 10.1002/cne.23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Choi GB, et al. Lhx6 delineates a pathway mediating innate reproductive behaviors from the amygdala to the hypothalamus. Neuron. 2005;46(4):647–660. doi: 10.1016/j.neuron.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 190.von Campenhausen H, Mori K. Convergence of segregated pheromonal pathways from the accessory olfactory bulb to the cortex in the mouse. Eur J Neurosci. 2000;12(1):33–46. doi: 10.1046/j.1460-9568.2000.00879.x. [DOI] [PubMed] [Google Scholar]
- 191.Del Punta K, et al. Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes. Nature. 2002;419(6902):70–74. doi: 10.1038/nature00955. [DOI] [PubMed] [Google Scholar]
- 192.Wagner S, et al. A multireceptor genetic approach uncovers an ordered integration of VNO sensory inputs in the accessory olfactory bulb. Neuron. 2006;50(5):697–709. doi: 10.1016/j.neuron.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 193.Huilgol D, et al. Dual origins of the mammalian accessory olfactory bulb revealed by an evolutionarily conserved migratory stream. Nat Neurosci. 2013;16(2):157–165. doi: 10.1038/nn.3297. [DOI] [PubMed] [Google Scholar]
- 194.Meisami E, Bhatnagar KP. Structure and diversity in mammalian accessory olfactory bulb. Microsc Res Tech. 1998;43(6):476–499. doi: 10.1002/(SICI)1097-0029(19981215)43:6<476::AID-JEMT2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 195.Sokolowski K, Corbin JG. Wired for behaviors: from development to function of innate limbic system circuitry. Front Mol Neurosci. 2012;5:55. doi: 10.3389/fnmol.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Miyoshi G, Fishell G. GABAergic interneuron lineages selectively sort into specific cortical layers during early postnatal development. Cereb Cortex. 2011;21(4):845–852. doi: 10.1093/cercor/bhq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gilmore EC, et al. Cyclin-dependent kinase 5-deficient mice demonstrate novel developmental arrest in cerebral cortex. J Neurosci. 1998;18(16):6370–6377. doi: 10.1523/JNEUROSCI.18-16-06370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Peretto P, et al. Sexually dimorphic neurogenesis is topographically matched with the anterior accessory olfactory bulb of the adult rat. Cell Tissue Res. 2001;306(3):385–389. doi: 10.1007/s00441-001-0471-1. [DOI] [PubMed] [Google Scholar]
- 199.Oboti L, et al. Integration and sensory experience-dependent survival of newly-generated neurons in the accessory olfactory bulb of female mice. Eur J Neurosci. 2009;29(4):679–692. doi: 10.1111/j.1460-9568.2009.06614.x. [DOI] [PubMed] [Google Scholar]
- 200.Alheid GF, Heimer L. New perspectives in basal forebrain organization of special relevance for neuropsychiatric disorders: the striatopallidal, amygdaloid, and corticopetal components of substantia innominata. Neuroscience. 1988;27(1):1–39. doi: 10.1016/0306-4522(88)90217-5. [DOI] [PubMed] [Google Scholar]
- 201.Shammah-Lagnado SJ, et al. Supracapsular bed nucleus of the stria terminalis contains central and medial extended amygdala elements: evidence from anterograde and retrograde tracing experiments in the rat. J Comp Neurol. 2000;422(4):533–555. doi: 10.1002/1096-9861(20000710)422:4<533::AID-CNE5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 202.Garcia-Lopez M, et al. Histogenetic compartments of the mouse centromedial and extended amygdala based on gene expression patterns during development. J Comp Neurol. 2008;506(1):46–74. doi: 10.1002/cne.21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Garcia-Moreno F, et al. A neuronal migratory pathway crossing from diencephalon to telencephalon populates amygdala nuclei. Nat Neurosci. 2010;13(6):680–689. doi: 10.1038/nn.2556. [DOI] [PubMed] [Google Scholar]
- 204.Bupesh M, et al. Multiple telencephalic and extratelencephalic embryonic domains contribute neurons to the medial extended amygdala. J Comp Neurol. 2011;519(8):1505–1525. doi: 10.1002/cne.22581. [DOI] [PubMed] [Google Scholar]
- 205.Mohedano-Moriano A, et al. Segregated pathways to the vomeronasal amygdala: differential projections from the anterior and posterior divisions of the accessory olfactory bulb. Eur J Neurosci. 2007;25(7):2065–2080. doi: 10.1111/j.1460-9568.2007.05472.x. [DOI] [PubMed] [Google Scholar]
- 206.Swanson LW. Cerebral hemisphere regulation of motivated behavior. Brain Res. 2000;886(1–2):113–164. doi: 10.1016/S0006-8993(00)02905-X. [DOI] [PubMed] [Google Scholar]
- 207.Canteras NS. The medial hypothalamic defensive system: hodological organization and functional implications. Pharmacol Biochem Behav. 2002;71(3):481–491. doi: 10.1016/S0091-3057(01)00685-2. [DOI] [PubMed] [Google Scholar]
- 208.Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008;506(1):16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- 209.Carney RS, et al. Sonic hedgehog expressing and responding cells generate neuronal diversity in the medial amygdala. Neural Dev. 2010;5:14. doi: 10.1186/1749-8104-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gorski JA, et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22(15):6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Dong HW, et al. Basic organization of projections from the oval and fusiform nuclei of the bed nuclei of the stria terminalis in adult rat brain. J Comp Neurol. 2001;436(4):430–455. doi: 10.1002/cne.1079. [DOI] [PubMed] [Google Scholar]
- 212.Shimada M, Nakamura T. Time of neuron origin in mouse hypothalamic nuclei. Exp Neurol. 1973;41(1):163–173. doi: 10.1016/0014-4886(73)90187-8. [DOI] [PubMed] [Google Scholar]
- 213.McClellan KM, Calver AR, Tobet SA. GABAB receptors role in cell migration and positioning within the ventromedial nucleus of the hypothalamus. Neuroscience. 2008;151(4):1119–1131. doi: 10.1016/j.neuroscience.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Altman J, Bayer SA. The development of the rat hypothalamus. Adv Anat Embryol Cell Biol. 1986;100:1–178. doi: 10.1007/978-3-642-71301-9_1. [DOI] [PubMed] [Google Scholar]
- 215.Tobet SA, et al. Telencephalic and diencephalic origin of radial glial processes in the developing preoptic area/anterior hypothalamus. J Neurobiol. 1995;26(1):75–86. doi: 10.1002/neu.480260107. [DOI] [PubMed] [Google Scholar]
- 216.Bayer SA, Altman J. Development of the preoptic area: time and site of origin, migratory routes, and settling patterns of its neurons. J Comp Neurol. 1987;265(1):65–95. doi: 10.1002/cne.902650106. [DOI] [PubMed] [Google Scholar]
- 217.Yoshida K, et al. Polysialic acid facilitates migration of luteinizing hormone–releasing hormone neurons on vomeronasal axons. J Neurosci. 1999;19(2):794–801. doi: 10.1523/JNEUROSCI.19-02-00794.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Franco B, et al. A gene deleted in Kallmann’s syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353(6344):529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 219.Legouis R, et al. The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell. 1991;67(2):423–435. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- 220.Giacobini P, et al. Semaphorin 4D regulates gonadotropin hormone–releasing hormone-1 neuronal migration through plexinB1–Met complex. J Cell Biol. 2008;183(3):555–566. doi: 10.1083/jcb.200806160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Herbison AE, et al. Gonadotropin-releasing hormone neuron requirements for puberty, ovulation, and fertility. Endocrinology. 2008;149(2):597–604. doi: 10.1210/en.2007-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tsai PS, et al. Targeted expression of a dominant-negative fibroblast growth factor (FGF) receptor in gonadotropin-releasing hormone (GnRH) neurons reduces FGF responsiveness and the size of GnRH neuronal population. Mol Endocrinol. 2005;19(1):225–236. doi: 10.1210/me.2004-0330. [DOI] [PubMed] [Google Scholar]
- 223.Falardeau J, et al. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Investig. 2008;118(8):2822–2831. doi: 10.1172/JCI34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wierman ME, Kiseljak-Vassiliades K, Tobet S. Gonadotropin-releasing hormone (GnRH) neuron migration: initiation, maintenance and cessation as critical steps to ensure normal reproductive function. Front Neuroendocrinol. 2011;32(1):43–52. doi: 10.1016/j.yfrne.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Shimogori T, et al. A genomic atlas of mouse hypothalamic development. Nat Neurosci. 2010;13(6):767–775. doi: 10.1038/nn.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tripodi M, et al. The COUP-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Development. 2004;131(24):6119–6129. doi: 10.1242/dev.01530. [DOI] [PubMed] [Google Scholar]
- 227.Zhao L, et al. Disruption of SoxB1-dependent sonic hedgehog expression in the hypothalamus causes septo-optic dysplasia. Dev Cell. 2012;22(3):585–596. doi: 10.1016/j.devcel.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Alvarez-Bolado G, Paul FA, Blaess S. Sonic hedgehog lineage in the mouse hypothalamus: from progenitor domains to hypothalamic regions. Neural Dev. 2012;7:4. doi: 10.1186/1749-8104-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ikeda Y, et al. The nuclear receptor steroidogenic factor 1 is essential for the formation of the ventromedial hypothalamic nucleus. Mol Endocrinol. 1995;9(4):478–486. doi: 10.1210/mend.9.4.7659091. [DOI] [PubMed] [Google Scholar]
- 230.Dellovade TL, et al. Disruption of the gene encoding SF-1 alters the distribution of hypothalamic neuronal phenotypes. J Comp Neurol. 2000;423(4):579–589. doi: 10.1002/1096-9861(20000807)423:4<579::AID-CNE4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 231.McClellan KM, Parker KL, Tobet S. Development of the ventromedial nucleus of the hypothalamus. Front Neuroendocrinol. 2006;27(2):193–209. doi: 10.1016/j.yfrne.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 232.Davis AM, et al. Loss of steroidogenic factor 1 alters cellular topography in the mouse ventromedial nucleus of the hypothalamus. J Neurobiol. 2004;60(4):424–436. doi: 10.1002/neu.20030. [DOI] [PubMed] [Google Scholar]
- 233.Szabo NE, et al. Role of neuroepithelial Sonic hedgehog in hypothalamic patterning. J Neurosci. 2009;29(21):6989–7002. doi: 10.1523/JNEUROSCI.1089-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Blackshaw S, et al. Molecular pathways controlling development of thalamus and hypothalamus: from neural specification to circuit formation. J Neurosci. 2010;30(45):14925–14930. doi: 10.1523/JNEUROSCI.4499-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Figdor MC, Stern CD. Segmental organization of embryonic diencephalon. Nature. 1993;363(6430):630–634. doi: 10.1038/363630a0. [DOI] [PubMed] [Google Scholar]
- 236.Kim AD, et al. Pax-6 regulates expression of SFRP-2 and Wnt-7b in the developing CNS. J Neurosci. 2001;21(5):RC132. doi: 10.1523/JNEUROSCI.21-05-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Zeltser LM, Larsen CW, Lumsden A. A new developmental compartment in the forebrain regulated by lunatic fringe. Nat Neurosci. 2001;4(7):683–684. doi: 10.1038/89455. [DOI] [PubMed] [Google Scholar]
- 238.Kiecker C, Lumsden A. Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat Neurosci. 2004;7(11):1242–1249. doi: 10.1038/nn1338. [DOI] [PubMed] [Google Scholar]
- 239.Fotaki V, Price DJ, Mason JO. Newly identified patterns of Pax2 expression in the developing mouse forebrain. BMC Dev Biol. 2008;8:79. doi: 10.1186/1471-213X-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Roy A, et al. LHX2 is necessary for the maintenance of optic identity and for the progression of optic morphogenesis. J Neurosci. 2013;33(16):6877–6884. doi: 10.1523/JNEUROSCI.4216-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Assimacopoulos S, Grove EA, Ragsdale CW. Identification of a Pax6-dependent epidermal growth factor family signaling source at the lateral edge of the embryonic cerebral cortex. J Neurosci. 2003;23(16):6399–6403. doi: 10.1523/JNEUROSCI.23-16-06399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Subramanian L, et al. Signals from the edges: the cortical hem and antihem in telencephalic development. Semin Cell Dev Biol. 2009;20(6):712–718. doi: 10.1016/j.semcdb.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Daniel D, et al. Stromal cell-derived factor-1 (SDF-1) expression in embryonic mouse cerebral cortex starts in the intermediate zone close to the pallial–subpallial boundary and extends progressively towards the cortical hem. Gene Expr Patterns. 2005;5(3):317–322. doi: 10.1016/j.modgep.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 244.Kornblum HI, et al. Prenatal ontogeny of the epidermal growth factor receptor and its ligand, transforming growth factor alpha, in the rat brain. J Comp Neurol. 1997;380(2):243–261. doi: 10.1002/(SICI)1096-9861(19970407)380:2<243::AID-CNE7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 245.Krug L, Wicht H, Northcutt RG. Afferent and efferent connections of the thalamic eminence in the axolotl, Ambystoma mexicanum . Neurosci Lett. 1993;149(2):145–148. doi: 10.1016/0304-3940(93)90757-C. [DOI] [PubMed] [Google Scholar]
- 246.Yu YC, et al. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 2009;458(7237):501–504. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Li Y, et al. Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature. 2012;486(7401):118–121. doi: 10.1038/nature11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.De Carlos JA, Lopez-Mascaraque L, Valverde F. The telencephalic vesicles are innervated by olfactory placode-derived cells: a possible mechanism to induce neocortical development. Neuroscience. 1995;68(4):1167–1178. doi: 10.1016/0306-4522(95)00199-S. [DOI] [PubMed] [Google Scholar]
- 249.Gong Q, Shipley MT. Evidence that pioneer olfactory axons regulate telencephalon cell cycle kinetics to induce the formation of the olfactory bulb. Neuron. 1995;14(1):91–101. doi: 10.1016/0896-6273(95)90243-0. [DOI] [PubMed] [Google Scholar]
- 250.Schwanzel-Fukuda M, Bick D, Pfaff DW. Luteinizing hormone–releasing hormone (LHRH)-expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Brain Res Mol Brain Res. 1989;6(4):311–326. doi: 10.1016/0169-328X(89)90076-4. [DOI] [PubMed] [Google Scholar]
- 251.Dode C, et al. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2003;33(4):463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 252.Sato N, et al. Clinical assessment and mutation analysis of Kallmann syndrome 1 (KAL1) and fibroblast growth factor receptor 1 (FGFR1, or KAL2) in five families and 18 sporadic patients. J Clin Endocrinol Metab. 2004;89(3):1079–1088. doi: 10.1210/jc.2003-030476. [DOI] [PubMed] [Google Scholar]
- 253.Teixeira L, et al. Defective migration of neuroendocrine GnRH cells in human arrhinencephalic conditions. J Clin Investig. 2010;120(10):3668–3672. doi: 10.1172/JCI43699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Andrews PI, Hulette CM. An infant with macrocephaly, abnormal neuronal migration and persistent olfactory ventricles. Clin Neuropathol. 1993;12(1):13–18. [PubMed] [Google Scholar]
- 255.Yamanouchi H, et al. Olfactory bulb dysplasia: a novel subtype of neuronal migration disorder. Ann Neurol. 1999;46(5):783–786. doi: 10.1002/1531-8249(199911)46:5<783::AID-ANA16>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 256.Curtis MA, Faull RL, Eriksson PS. The effect of neurodegenerative diseases on the subventricular zone. Nat Rev Neurosci. 2007;8(9):712–723. doi: 10.1038/nrn2216. [DOI] [PubMed] [Google Scholar]
- 257.Michaud JL. The developmental program of the hypothalamus and its disorders. Clin Genet. 2001;60(4):255–263. doi: 10.1034/j.1399-0004.2001.600402.x. [DOI] [PubMed] [Google Scholar]
- 258.Tobet S, et al. Brain sex differences and hormone influences: a moving experience? J Neuroendocrinol. 2009;21(4):387–392. doi: 10.1111/j.1365-2826.2009.01834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Fedoroff IC, et al. Olfactory dysfunction in anorexia and bulimia nervosa. Int J Eat Disord. 1995;18(1):71–77. doi: 10.1002/1098-108X(199507)18:1<71::AID-EAT2260180108>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 260.Boyd JG, Doucette R, Kawaja MD. Defining the role of olfactory ensheathing cells in facilitating axon remyelination following damage to the spinal cord. FASEB J. 2005;19(7):694–703. doi: 10.1096/fj.04-2833rev. [DOI] [PubMed] [Google Scholar]
- 261.Radtke C, Kocsis JD, Vogt PM. Chapter 22: Transplantation of olfactory ensheathing cells for peripheral nerve regeneration. Int Rev Neurobiol. 2009;87:405–415. doi: 10.1016/S0074-7742(09)87022-0. [DOI] [PubMed] [Google Scholar]
- 262.Tabakow P, et al. Transplantation of autologous olfactory ensheathing cells in complete human spinal cord injury. Cell Transplant. 2013;22(9):1591–1612. doi: 10.3727/096368912X663532. [DOI] [PubMed] [Google Scholar]
- 263.Ramon-Cueto A, Nieto-Sampedro M. Glial cells from adult rat olfactory bulb: immunocytochemical properties of pure cultures of ensheathing cells. Neuroscience. 1992;47(1):213–220. doi: 10.1016/0306-4522(92)90134-N. [DOI] [PubMed] [Google Scholar]
- 264.Santos-Silva A, Cavalcante LA. Expression of the non-compact myelin protein 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNPase) in olfactory bulb ensheathing glia from explant cultures. Neurosci Res. 2001;40(2):189–193. doi: 10.1016/S0168-0102(01)00204-8. [DOI] [PubMed] [Google Scholar]
- 265.Li Y, Field PM, Raisman G. Repair of adult rat corticospinal tract by transplants of olfactory ensheathing cells. Science. 1997;277(5334):2000–2002. doi: 10.1126/science.277.5334.2000. [DOI] [PubMed] [Google Scholar]
- 266.Ramon-Cueto A. Olfactory ensheathing glia transplantation into the injured spinal cord. Prog Brain Res. 2000;128:265–272. doi: 10.1016/S0079-6123(00)28024-2. [DOI] [PubMed] [Google Scholar]
- 267.Mackay-Sim A. Olfactory ensheathing cells and spinal cord repair. Keio J Med. 2005;54(1):8–14. doi: 10.2302/kjm.54.8. [DOI] [PubMed] [Google Scholar]
- 268.Moreno N, Gonzalez A. Evolution of the amygdaloid complex in vertebrates, with special reference to the anamnio-amniotic transition. J Anat. 2007;211(2):151–163. doi: 10.1111/j.1469-7580.2007.00780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Suarez R, Garcia-Gonzalez D, de Castro F. Mutual influences between the main olfactory and vomeronasal systems in development and evolution. Front Neuroanat. 2012;6:50. doi: 10.3389/fnana.2012.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Mione M, et al. Overlapping expression of zebrafish T-brain-1 and eomesodermin during forebrain development. Mech Dev. 2001;100(1):93–97. doi: 10.1016/S0925-4773(00)00501-3. [DOI] [PubMed] [Google Scholar]
- 271.Boyd JD, Delaney KR. Tyrosine hydroxylase-immunoreactive interneurons in the olfactory bulb of the frogs Rana pipiens and Xenopus laevis . J Comp Neurol. 2002;454(1):42–57. doi: 10.1002/cne.10428. [DOI] [PubMed] [Google Scholar]
- 272.Brox A, et al. Expression of the genes GAD67 and Distal-less-4 in the forebrain of Xenopus laevis confirms a common pattern in tetrapods. J Comp Neurol. 2003;461(3):370–393. doi: 10.1002/cne.10688. [DOI] [PubMed] [Google Scholar]
- 273.Moreno N, et al. Pallial origin of mitral cells in the olfactory bulbs of Xenopus. NeuroReport. 2003;14(18):2355–2358. doi: 10.1097/00001756-200312190-00013. [DOI] [PubMed] [Google Scholar]
- 274.Brox A, et al. Expression of the genes Emx1, Tbr1, and Eomes (Tbr2) in the telencephalon of Xenopus laevis confirms the existence of a ventral pallial division in all tetrapods. J Comp Neurol. 2004;474(4):562–577. doi: 10.1002/cne.20152. [DOI] [PubMed] [Google Scholar]
- 275.Mione M, et al. How neuronal migration contributes to the morphogenesis of the CNS: insights from the zebrafish. Dev Neurosci. 2008;30(1–3):65–81. doi: 10.1159/000109853. [DOI] [PubMed] [Google Scholar]
- 276.Moreno N, et al. Anuran olfactory bulb organization: embryology, neurochemistry and hodology. Brain Res Bull. 2008;75(2–4):241–245. doi: 10.1016/j.brainresbull.2007.10.027. [DOI] [PubMed] [Google Scholar]
- 277.Byrd CA, Brunjes PC. Neurogenesis in the olfactory bulb of adult zebrafish. Neuroscience. 2001;105(4):793–801. doi: 10.1016/S0306-4522(01)00215-9. [DOI] [PubMed] [Google Scholar]
- 278.Adolf B, et al. Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon. Dev Biol. 2006;295(1):278–293. doi: 10.1016/j.ydbio.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 279.Grandel H, et al. Neural stem cells and neurogenesis in the adult zebrafish brain: origin, proliferation dynamics, migration and cell fate. Dev Biol. 2006;295(1):263–277. doi: 10.1016/j.ydbio.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 280.Penafiel A, et al. A tangential neuronal migration in the olfactory bulbs of adult lizards. NeuroReport. 1996;7(7):1257–1260. doi: 10.1097/00001756-199605170-00007. [DOI] [PubMed] [Google Scholar]
- 281.Font E, et al. Neurogenesis and neuronal regeneration in the adult reptilian brain. Brain Behav Evol. 2001;58(5):276–295. doi: 10.1159/000057570. [DOI] [PubMed] [Google Scholar]
- 282.Kornack DR, Rakic P. Cell proliferation without neurogenesis in adult primate neocortex. Science. 2001;294(5549):2127–2130. doi: 10.1126/science.1065467. [DOI] [PubMed] [Google Scholar]
- 283.Pencea V, et al. Neurogenesis in the subventricular zone and rostral migratory stream of the neonatal and adult primate forebrain. Exp Neurol. 2001;172(1):1–16. doi: 10.1006/exnr.2001.7768. [DOI] [PubMed] [Google Scholar]
- 284.Sanai N, et al. Unique astrocyte ribbon in adult human brain contains neural stem cells but lacks chain migration. Nature. 2004;427(6976):740–744. doi: 10.1038/nature02301. [DOI] [PubMed] [Google Scholar]
- 285.Quinones-Hinojosa A, et al. Cellular composition and cytoarchitecture of the adult human subventricular zone: a niche of neural stem cells. J Comp Neurol. 2006;494(3):415–434. doi: 10.1002/cne.20798. [DOI] [PubMed] [Google Scholar]
- 286.Curtis MA, et al. Human neuroblasts migrate to the olfactory bulb via a lateral ventricular extension. Science. 2007;315(5816):1243–1249. doi: 10.1126/science.1136281. [DOI] [PubMed] [Google Scholar]
- 287.Kishimoto N, et al. Migration of neuronal precursors from the telencephalic ventricular zone into the olfactory bulb in adult zebrafish. J Comp Neurol. 2011;519(17):3549–3565. doi: 10.1002/cne.22722. [DOI] [PubMed] [Google Scholar]
- 288.Pellegrini E, et al. Identification of aromatase-positive radial glial cells as progenitor cells in the ventricular layer of the forebrain in zebrafish. J Comp Neurol. 2007;501(1):150–167. doi: 10.1002/cne.21222. [DOI] [PubMed] [Google Scholar]
- 289.Marz M, et al. Heterogeneity in progenitor cell subtypes in the ventricular zone of the zebrafish adult telencephalon. Glia. 2010;58(7):870–888. doi: 10.1002/glia.20971. [DOI] [PubMed] [Google Scholar]
- 290.Gonzalez A et al (2010) Lungfishes, like tetrapods, possess a vomeronasal system. Front Neuroanat 4 [DOI] [PMC free article] [PubMed]
- 291.Nakamuta S, et al. Histological and ultrastructural characteristics of the primordial vomeronasal organ in lungfish. Anat Rec (Hoboken) 2012;295(3):481–491. doi: 10.1002/ar.22415. [DOI] [PubMed] [Google Scholar]
- 292.Eisthen HL. Evolution of vertebrate olfactory systems. Brain Behav Evol. 1997;50(4):222–233. doi: 10.1159/000113336. [DOI] [PubMed] [Google Scholar]
- 293.Saito S, Kobayashi N, Atoji Y. Subdivision of the accessory olfactory bulb in the Japanese common toad, Bufo japonicus, revealed by lectin histochemical analysis. Anat Embryol (Berl) 2006;211(5):395–402. doi: 10.1007/s00429-006-0088-y. [DOI] [PubMed] [Google Scholar]
- 294.Moreno N, Gonzalez A. Hodological characterization of the medial amygdala in anuran amphibians. J Comp Neurol. 2003;466(3):389–408. doi: 10.1002/cne.10887. [DOI] [PubMed] [Google Scholar]
- 295.Moreno N, Gonzalez A. Localization and connectivity of the lateral amygdala in anuran amphibians. J Comp Neurol. 2004;479(2):130–148. doi: 10.1002/cne.20298. [DOI] [PubMed] [Google Scholar]
- 296.Moreno N, et al. Lateral and medial amygdala of anuran amphibians and their relation to olfactory and vomeronasal information. Brain Res Bull. 2005;66(4–6):332–336. doi: 10.1016/j.brainresbull.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 297.Moreno N, Gonzalez A. The common organization of the amygdaloid complex in tetrapods: new concepts based on developmental, hodological and neurochemical data in anuran amphibians. Prog Neurobiol. 2006;78(2):61–90. doi: 10.1016/j.pneurobio.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 298.Bardet SM, et al. Conserved pattern of OTP-positive cells in the paraventricular nucleus and other hypothalamic sites of tetrapods. Brain Res Bull. 2008;75(2–4):231–235. doi: 10.1016/j.brainresbull.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 299.Abellan A, Desfilis E, Medina L. The olfactory amygdala in amniotes: an evo-devo approach. Anat Rec (Hoboken) 2013;296(9):1317–1332. doi: 10.1002/ar.22744. [DOI] [PubMed] [Google Scholar]
- 300.Moreno N, et al. Subdivisions of the turtle Pseudemys scripta subpallium based on the expression of regulatory genes and neuronal markers. J Comp Neurol. 2010;518(24):4877–4902. doi: 10.1002/cne.22493. [DOI] [PubMed] [Google Scholar]
- 301.Abellan A, et al. Olfactory and amygdalar structures of the chicken ventral pallium based on the combinatorial expression patterns of LIM and other developmental regulatory genes. J Comp Neurol. 2009;516(3):166–186. doi: 10.1002/cne.22102. [DOI] [PubMed] [Google Scholar]
- 302.Abellan A, et al. Differential expression of LIM-homeodomain factors in Cajal–Retzius cells of primates, rodents, and birds. Cereb Cortex. 2010;20(8):1788–1798. doi: 10.1093/cercor/bhp242. [DOI] [PubMed] [Google Scholar]
- 303.Angevine JB, Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 304.Bielle F, et al. Multiple origins of Cajal–Retzius cells at the borders of the developing pallium. Nat Neurosci. 2005;8(8):1002–1012. doi: 10.1038/nn1511. [DOI] [PubMed] [Google Scholar]
- 305.Yoshida M, et al. Massive loss of Cajal–Retzius cells does not disrupt neocortical layer order. Development. 2006;133(3):537–545. doi: 10.1242/dev.02209. [DOI] [PubMed] [Google Scholar]