Abstract
Orai channels are required for store-operated Ca2+ entry (SOCE) in multiple cell types. Septins are a class of GTP-binding proteins that function as diffusion barriers in cells. Here we show that Septin 7 acts as a ‘molecular brake’ on activation of Orai channels in Drosophila neurons. Lowering Septin 7 levels results in dOrai-mediated Ca2+ entry and higher cytosolic Ca2+ in resting neurons. This Ca2+ entry is independent of depletion of endoplasmic reticulum Ca2+ stores and Ca2+ release through the inositol-1,4,5-trisphosphate receptor. Importantly, store-independent Ca2+ entry through Orai compensates for reduced SOCE in the Drosophila flight circuit. Moreover, overexpression of Septin 7 reduces both SOCE and flight duration, supporting its role as a negative regulator of Orai channel function in vivo. Septin 7 levels in neurons can, therefore, alter neural circuit function by modulating Orai function and Ca2+ homeostasis.
 Orai channels are well known to mediate store-operated calcium entry. Here authors show that in neurons of the Drosophila flight circuit, Septin 7 acts as a negative regulator of Orai channels, surprisingly, by modulating store-independent calcium entry through Orai.
Orai channels are well known to mediate store-operated calcium entry. Here authors show that in neurons of the Drosophila flight circuit, Septin 7 acts as a negative regulator of Orai channels, surprisingly, by modulating store-independent calcium entry through Orai.
Physiological signals such as neurotransmitters, hormones and growth factors frequently regulate cellular function by the release of intracellular calcium from endoplasmic reticulum (ER) stores, followed by store-operated calcium entry (SOCE)1. This calcium signalling mechanism can be initiated by surface receptors that are coupled with phospholipase C, which generates inositol-1,4,5-trisphosphate (IP3) from plasma membrane (PM)-localized phosphatidyl inositol-4,5 bisphosphate or PIP2 (ref. 2). IP3 binds to and opens a ligand-gated intracellular Ca2+ channel, the IP3 receptor (IP3R) present on the membranes of ER stores, thus leading to the release of intracellular Ca2+2. The consequent drop in intracellular store Ca2+ is sensed by ER membrane-localized stromal interaction molecule (STIM) proteins, which subsequently oligomerize and translocate to the ER closely apposed to the PM3,4,5,6, where they bind to and open the Orai channel, leading to SOCE7,8,9,10,11,12.
Whereas STIM and Orai constitute integral components of SOCE, additional proteins regulate the strength and duration of SOCE signals1,13. Molecular screens for SOCE regulators have identified both positive13,14,15,16,17,18 and negative19,20 regulators of SOCE, some of which like SOCE associated regulatory factor (SARAF)19 are specific for vertebrates. Genetic studies in Drosophila21,22,23 have shown that IP3R mutants as well as flies with pan-neuronal knockdown of IP3R, dSTIM or dOrai exhibit reduced flight bouts21,22. Importantly, flight deficits of IP3R mutants can be rescued by overexpression of dSTIM and dOrai in neurons, indicating that IP3R-mediated Ca2+ release is coupled with SOCE through dSTIM/dOrai in neurons of the Drosophila flight circuit. Drosophila mutants for the IP3R (the gene is referred to as itpr) thus offer a genetic background for identifying additional regulators of SOCE via dSTIM/dOrai whose effect on IP3-mediated Ca2+ release and SOCE can be tested in cultured primary neurons derived from the central nervous system. Moreover, their physiological relevance can be validated in vivo by quantification of IP3R mutant flight deficits24 on introduction of mutants or overexpression constructs of the putative interacting partners.
A recent screen in HeLa cells identified mammalian septins, SEPT2, SEPT4 and SEPT5, as positive regulators of SOCE25. Septins are a class of evolutionarily conserved filament forming GTPases that primarily function as molecular adaptors or diffusion barriers in cells26. Five septin-encoding genes are reported in Drosophila27,28,29 among which the ‘pnut’ gene27 encodes the Drosophila homologue of mammalian Septin 7 (ref. 29), henceforth referred to as dSEPT7 (the gene is referred to as dSEPT7). Both in Drosophila and mammals, the SEPT7 family is represented by a single gene26,29 (Table 1). To investigate whether regulation of SOCE by septins is conserved in Drosophila neurons, a deficiency allele of the dSEPT7 gene, pnutxp (ref. 27), was tested for genetic interaction with itpr mutants. Systemic and cellular studies demonstrate a novel role for dSEPT7 in regulation of dOrai-mediated Ca2+ entry in neurons.
Table 1. Nomenclature of septin subunits in mammals and Drosophila.
| Septin subgroups | SEPT2 | SEPT6 | SEPT7 | SEPT3 |
|---|---|---|---|---|
| Homo sapiens | SEPT1, SEPT2, SEPT4, SEPT5 | SEPT6, SEPT8, SEPT10, SEPT11 | SEPT7 | SEPT3, SEPT9, SEPT12 |
| Drosophila melanogaster | dSEPT1, dSEPT4 | dSEPT2, dSEPT5 | dSEPT7 | No representatives |
Results
dSEPT7 reduction can compensate for reduced IP3R function
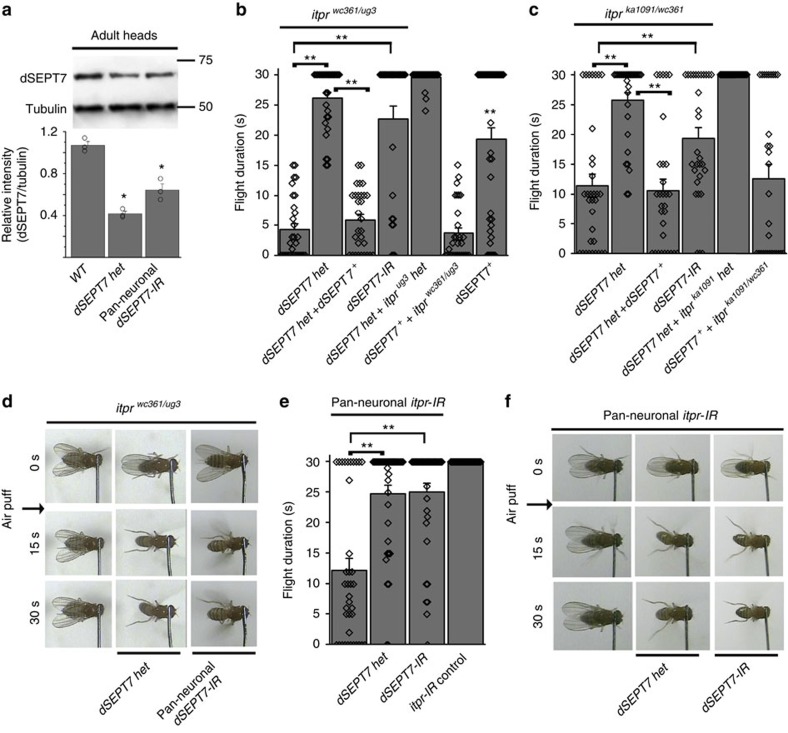
Animals homozygous for the dSEPT7 deficiency allele (pnutxp/xp, here after referred to as ‘dSEPT7 null’) are lethal during development and fail to survive till adulthood27. In animals heterozygous for this deficiency allele (pnutxp/+, hereafter referred to as ‘dSEPT7 het’), dSEPT7 protein levels are reduced to ∼50% of wild-type (WT) levels in adult heads and the larval central nervous system (CNS) (Fig. 1a; Supplementary Fig. 1c). The role of dSEPT7 in intracellular Ca2+ signalling was investigated by introducing a single copy of the deficiency allele for dSEPT7 (ref. 27; dSEPT7 het) in two viable IP3R mutant combinations, itprwc361/ug3 and itprka1091/wc361 (refs 24, 30). The wc361 allele has a deletion of 15 residues at the C terminus due to the mutation of an arginine residue at position 2,814 to a stop codon. The ug3 allele converts a serine to a phenylalanine at position 224 in the ligand-binding domain of the Drosophila IP3R, whereas in the ka1091 mutant allele, a glycine residue at position 1,891 in the modulatory domain of the protein is mutated to a serine30. As with other viable combinations of the IP3R, the heteroallelic combinations tested here exhibit defective wing postures and significantly reduced flight durations (Fig. 1b–d; Supplementary Fig. 1a), but appear normal as heterozygotes24 (Supplementary Fig. 1b). Interestingly, introduction of a single copy of the dSEPT7 deletion allele rescued flight (Fig. 1b–d) and wing-posture defects (Supplementary Fig. 1a) to a significant extent in both itpr mutants. Control genotypes consisting of dSEPT7 het in combination with either WT or heterozygotes of itpr gene mutants exhibit normal flight durations (Fig. 1b; Supplementary Fig. 1b). The focus of this genetic interaction is the nervous system because targeted reduction of dSEPT7 in neurons with an RNA interference (RNAi; dSEPT7-IR) also rescued flight deficits of the tested itpr mutants (Fig. 1b–d). Moreover, flight deficits of animals where IP3R was reduced in the CNS by pan-neuronal knockdown with an RNAi against itpr (itpr-IR)22,31 could be rescued by knockdown of dSEPT7 or partial genetic depletion of dSEPT7 (Fig. 1e,f; Supplementary Fig. 1a). dSEPT7-IR significantly reduced dSEPT7 protein in lysates from both adult fly heads and larval CNS (Fig. 1a; Supplementary Fig. 1c). Together, these data suggest that partial loss of dSEPT7 compensates for the reduced IP3R function in neurons of the Drosophila flight circuit. Conversely, restoring dSEPT7 levels by pan-neuronal overexpression of a dSEPT7+ transgene in itpr mutant animals heterozygous for dSEPT7 (dSEPT7 het+itprmutant) reverted the rescue obtained on reducing dSEPT7 and resulted in flies with flight and wing-posture defects similar to itpr mutants (Fig. 1b–d; Supplementary Fig. 1a). dSEPT7 overexpression in neurons of WT flies also resulted in significant flight deficits, suggesting a negative regulatory function of dSEPT7 in flight circuit neurons (Fig. 1b; Supplementary Fig. 1d). The genetic interactions observed between dSEPT7 and IP3R are not due to the changes in expression of the interacting genes, because itpr knockdown did not alter dSEPT7 levels in adult fly heads (Supplementary Fig. 1d) and reducing dSEPT7 levels in flies with pan-neuronal knockdown of IP3R did not alter the extent of IP3R knockdown (Supplementary Fig. 1e). The cellular basis of the genetic interaction between dSEPT7 and IP3R was investigated next.
Figure 1. Flight deficits in IP3R mutants and knockdown flies can be modulated by dSEPT7 expression.
(a) A representative western blot and quantification of three such blots measuring dSEPT7 protein levels in extracts from heads of adult flies of the indicated genotypes. dSEPT7 levels are significantly reduced in ‘dSEPT7 het’ flies and in flies with pan-neuronal knockdown of dSEPT7 when compared with the wild type (Canton-S); *P<0.05, Student’s t-test, N=3. (b,c) Flight duration in two itpr mutant strains tested, itprwc361/ug3 (b) and itprka1091/wc361 (c), improved significantly on reducing dSEPT7 with either a single copy of the deficiency allele, ‘dSEPT7 het’, or pan-neuronal knockdown of dSEPT7 (dSEPT7-IR) in neurons. Overexpression of dSEPT7 (dSEPT7+) in neurons restored flight deficits in itprwc361/ug3 or itprka1091/wc361 flies with reduced dSEPT7 (dSEPT7 het+itprwc361/ug3 or dSEPT7 het+itprka1091/wc361). The pan-neuronal elavC155GAL4 driver was used for expressing dSEPT7-IR or dSEPT7+ in all neurons of the fly. Flight times of 30 or more flies, monitored for 30 s are shown as mean±s.e.m. The diamonds within each bar represent the flight duration of a single fly. **P<0.01, Mann–Whitney U-test with Bonferroni correction. Brackets indicate the genotypes being compared. Flight times in dSEPT7+ animals are compared with wild-type animals (shown in Supplementary Fig. 1b). (d) Snapshots from video recordings of air-puff-induced flight of the indicated genotypes at 0 s (start), 15 and 30 s after delivery of the air-puff. (e) Reduced flight times observed by pan-neuronal knockdown of itpr with an RNAi (itpr-IR) were rescued by reduction of dSEPT7 with either a single copy of the deficiency allele, ‘dSEPT7 het’ or pan-neuronal knockdown of dSEPT7. itpr-IR control refers to animals with the itpr-IR transgene, but no GAL4 driver, resulting in no expression of the RNAi and no reduction in IP3R levels (Supplementary Fig. 1e). **P<0.01, Mann–Whitney U-test with Bonferroni correction. (f) Snapshots from video recordings of air-puff-induced flight of the indicated genotypes at 0 s (start), 15 and 30 s after delivery of the air-puff.
Reduced dSEPT7 increases the uptake of extracellular Ca2+
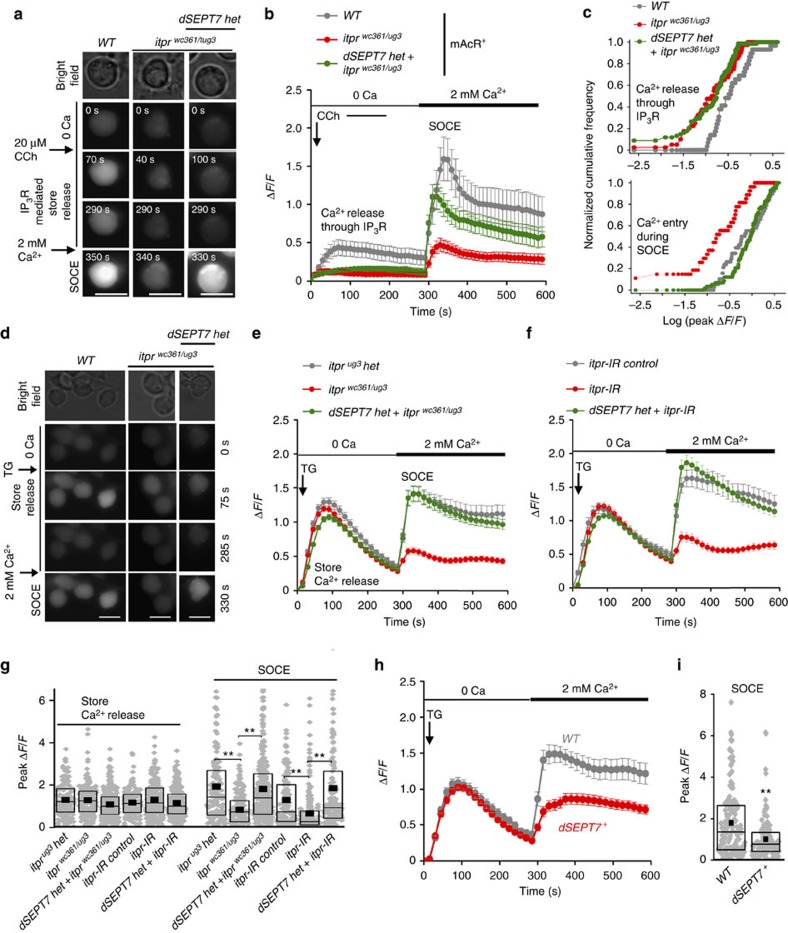
Ca2+ release through the IP3R was measured by agonist stimulation of the muscarinic acetylcholine receptor with carbachol (CCh) in primary cultured neurons (Fig. 2a–c)21. An initial peak of intracellular Ca2+ release through the IP3R was observed on CCh stimulation in WT neurons. This peak was significantly reduced in itprwc361/ug3 mutant neurons (Fig. 2a–c). Reduction of dSEPT7 did not affect Ca2+ release through the IP3R in WT or itprwc361/ug3 neurons (Fig. 2a–c; Supplementary Fig. 2a,b). SOCE was observed on Ca2+ add-back in the extracellular medium as a second Ca2+ peak in WT neurons (Fig. 2a–c; Supplementary Fig. 2b). SOCE induced by IP3R-mediated ER store depletion was significantly lower in itpr mutant neurons, when compared with WT (Fig. 2a–c; Supplementary Fig. 2b). Interestingly, dSEPT7 reduction resulted in significant uptake of extracellular Ca2+ in itpr mutant neurons, although the Ca2+ release through the mutant IP3R remained low and unaltered (Fig. 2b,c). Thus, reducing levels of dSEPT7 restores extracellular Ca2+ entry in itpr mutant neurons despite attenuated depletion of store Ca2+ (Fig. 2a–c; Supplementary Fig. 2b).
Figure 2. dSEPT7 reduction improves uptake of extracellular Ca2+ in neurons with reduced IP3R function.
(a) Representative images of primary Drosophila larval neurons of the indicated genotypes. Images were obtained before and after carbachol (CCh, 20 μM)-induced ER store Ca2+ release and entry of extracellular Ca2+ by SOCE at the indicated time points. Scale bar, 5 μm. (b) SOCE in itprwc361/ug3 neurons with and without dSEPT7 reduction. Traces represent the mean (±s.e.m.) ΔF/F values obtained at each time point during CCh-induced ER store Ca2+ release and SOCE for the indicated genotypes (N≥70 cells). (c) Kolmogorov–Smirnov (K–S) plots comparing the distribution of peak ΔF/F values obtained during IP3R-mediated Ca2+ release (top) and Ca2+ entry during SOCE (bottom) in the indicated genotypes. P<0.0001, K–S test for itprwc361/ug3 and ‘dSEPT7 het+itprwc361/ug3’ as compared with WT, (top) and itprwc361/ug3compared with WT and ‘dSEPT7 het+itprwc361/ug3’ (bottom). (d) Representative images of primary Drosophila larval neurons of the indicated genotypes with changes in cytosolic Ca2+ levels obtained after ER store depletion by thapsigargin (TG, 10 μM) and entry of extracellular Ca2+ during SOCE. Scale bar, 5 μm. (e,f) Reduced SOCE observed in itprwc361/ug3 neurons (e) and neurons with itpr knockdown (itpr-IR) (f) was restored to control levels on reduction of dSEPT7. itprug3 het (or itprug3/+) refers to animals heterozygous for the ug3 mutation in the itpr gene and is a genotypic control for the itprwc361/ug3 mutant. Pan-neuronal elavC155GAL4 was used to drive expression of the itprRNAi (itpr-IR). itpr-IR control refers to neurons with the RNAi construct, but no GAL4 driver, resulting in the absence of RNAi expression. Traces represent the mean (±s.e.m.) ΔF/F values obtained at each time point after TG-induced ER store Ca2+ depletion and SOCE for the indicated genotypes (N≥150 cells). (g) Quantification of peak ΔF/F values obtained by ER store Ca2+ depletion (0–285 s) and SOCE (300–600 s) for the indicated genotypes; **P<0.001, Mann–Whitney U-test with Bonferroni correction. (h) Reduced SOCE in primary neurons with overexpression of dSEPT7+; N≥150 cells. dSEPT7+ was expressed using the pan-neuronal elavC155 GAL4. (i) Quantification of peak ΔF/F values obtained during SOCE in the indicated genotypes; **P<0.01, Mann–Whitney U-test. Diamonds represent peak ΔF/F values of individual cells.
Neurons cultured from an itpr mutant, itprka1091/ug3, exhibit reduced SOCE after passive depletion of ER Ca2+ stores by thapsigargin (TG), an inhibitor of the sarco-endoplasmic reticulum Ca2+ATPase pump21,23.This finding contrasts with previous reports from non-excitable DT-40 cells32. TG-induced SOCE was reduced in neurons cultured from itprwc361/ug3 larvae and in neurons with knockdown of the IP3R (Fig. 2d–g). dSEPT7 reduction, in either itpr mutant or knockdown neurons, significantly improved extracellular Ca2+ entry after TG-mediated store depletion, without affecting passive ER Ca2+ release (Fig. 2e–g). The cellular specificity of this rescue was tested next. dSEPT7 knockdown with dSEPT7 RNAi significantly improved extracellular Ca2+ entry in itprwc361/ug3 neurons (Supplementary Fig. 2c,d). Second, overexpression of dSEPT7 reverted rescue of Ca2+ entry in itprwc361/ug3 neurons with reduced dSEPT7 back to the reduced levels observed in itprwc361/ug3 neurons (Supplementary Fig. 2c,d). Initial rate of Ca2+ entry on Ca2+ add-back was significantly reduced in both itpr mutant and knockdown neurons, and was rescued significantly in neurons with dSEPT7 reduction (Supplementary Fig. 2e). This increase in the rate of Ca2+ entry is possibly a consequence of faster dOrai channel activation in dSEPT7 reduced neurons when compared with IP3R mutant or knockdown neurons. Together, these data demonstrate that dSEPT7 reduction facilitates extracellular Ca2+ entry after either passive or IP3R-mediated ER store Ca2+ release. The effect of increasing dSEPT7 levels on neuronal SOCE was investigated next.
Overexpression of dSEPT7 reduces SOCE in neurons
SOCE obtained after TG-mediated passive store depletion in dSEPT7 het, dSEPT7 null or dSEPT7-IR neurons was not significantly different from that of WT neurons (Supplementary Fig. 2f,g). However, overexpression of dSEPT7 resulted in significant reduction of SOCE (Fig. 2h,i). These data, together with the earlier observation (Fig. 2b,d,e), suggest that dSEPT7 could function as a negative regulator of SOCE in neurons. We have earlier demonstrated that upregulation of the SOCE components dSTIM and dOrai in neurons improve SOCE and restore flight in a flightless itpr mutant22. In agreement with this, expression of dSTIM+ significantly improved SOCE after passive store depletion in neurons with itpr knockdown (Supplementary Fig. 2h,i). It is possible that dSEPT7 affects Ca2+ entry by regulating levels of SOCE components. dSTIM and dOrai levels were, however, unchanged on either reduction or overexpression of dSEPT7 (Supplementary Fig. 2j). These data support a direct effect of dSEPT7 on the function of either one or both of these SOCE proteins.
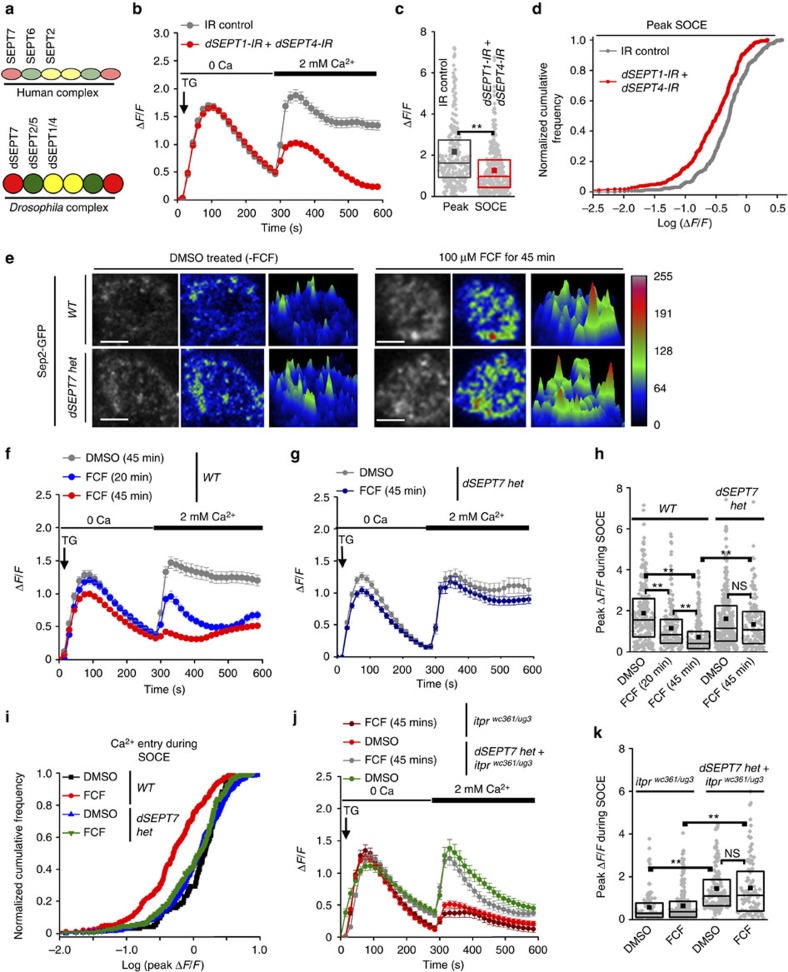
Mammalian septins, SEPT2, SEPT4 and SEPT5, belong to the SEPT2 class of septins26,29, and are positive regulators of SOCE in HeLa and T cells25. Because dSEPT7 is also a constituent of Drosophila septin complexes containing dSEPT1 (ref. 33), a Drosophila septin of the SEPT2 group (Table 1; Fig. 3a), we tested whether positive regulation of SOCE by septins of the SEPT2 subgroup is conserved between mammalian cells and Drosophila neurons.
Figure 3. Reducing dSEPT7 restores Ca2+ entry in neurons with pharmacological inhibition of septin dynamics.
(a) Positions occupied by septins of different subgroups (Table 1) in a septin complex formed by human septin subunits and their Drosophila homologues. (b) SOCE after TG-induced depletion of ER stores in neurons with knockdown of dSEPT1 and dSEPT4. IR control refers to neurons carrying RNAi constructs for both genes without the GAL4 driver. N>300 cells for each genotype. (c) Quantification of peak ΔF/F values obtained during SOCE. (d) K–S plots analysing peak ΔF/F values during SOCE in the indicated genotypes. The distribution for neurons with dSEPT1 and dSEPT4 knockdown is significantly shifted to the left compared with the IR control indicating a greater proportion of cells with lower ΔF/F values. P<0.001, K–S test. (e) Sep2-GFP localization in WT and neurons with reduced dSEPT7 treated with either DMSO (control) or 100 μM FCF for 45 min. Original grey scale images have been pseudo-coloured for visualizing the intensity of Sep2-GFP distribution where warmer colours represent higher intensities. A surface plot constructed with the pseudo-coloured image is shown for better resolution. The calibration bar represents the grey scale intensities corresponding to each colour. Scale bar, 3 μm. (f,g) Changes in ΔF/F (corresponding to changes in cytosolic [Ca2+]) during TG (10 μM)-induced passive ER Ca2+ release and SOCE in neurons of the indicated genotypes treated with 100 μM FCF for 20 or 45 min or DMSO for 45 min. N>100 cells. (h) Box plots quantifying peak ΔF/F values during SOCE in neurons of the indicated genotypes with or without FCF treatment; **P<0.001, Mann–Whitney U-test with Bonferroni correction. (i) K–S plots analysing peak ΔF/F values during SOCE in the indicated genotypes and treatment conditions. P<0.0001, K–S test. (j) Changes in cytosolic [Ca2+] indicated as ΔF/F during TG (10 μM)-induced passive ER Ca2+ release and SOCE in neurons of the indicated genotypes and treatment conditions. (k) Box plots quantifying peak ΔF/F values during SOCE in the indicated genotypes; **P<0.001, Mann–Whitney U-test; NS, not significant.
SEPT2 subgroup and dSEPT7 functions appear distinct
Simultaneous knockdown of dSEPT1 (Sep1 (refs 33, 34)) and dSEPT4 (Sep4 (refs 28, 35)), homologues of mammalian SEPT1 and SEPT4 in flies29 (Table 1), significantly attenuated SOCE in neurons (Fig. 3b–d) with resulting flight deficits in adult flies (Supplementary Fig. 3a). Thus, septins of the SEPT2 subgroup, which nucleate the septin complex and filament formation36 (Fig. 3a), function as positive regulators of SOCE in both vertebrates and Drosophila. This is in contrast to dSEPT7, which functions as a negative regulator of SOCE in Drosophila neurons (Fig. 2).
Conservation of septin function in SOCE was further tested in Drosophila neurons by application of forchlorfenuron (FCF), a pharmacological agent that perturbs septin filament organization and dynamics37,38, and leads to the loss of SOCE in mammalian cells25. Aggregation of Sep2-GFP39 was used as a cellular read-out for the efficacy of FCF treatment on inhibition of septin filament dynamics. Treatment of WT Drosophila neurons with 100 μM FCF for 45 min lead to Sep2-GFP aggregation (Fig. 3e) and a concurrent loss of SOCE (Fig. 3f,h,i), with no significant change in passive ER store release (Supplementary Fig. 3b), whereas FCF treatment for a shorter period of 20 min resulted in partial reduction of SOCE (Fig. 3f,h). These data support a requirement for septin filament dynamics during SOCE in Drosophila neurons, in agreement with their observed requirement for SOCE in non-excitable mammalian cells25. Interestingly, neurons with reduced dSEPT7 exhibit similar extent of Sep2-GFP aggregation as WT neurons (Fig. 3e), but normal SOCE after FCF treatment (Fig. 3g–i). In agreement with these data, SOCE in itpr mutant neurons with reduced dSEPT7 also remained unaffected on FCF treatment (Fig. 3j,k). Together, these data suggest that, although septin filament dynamics are required for activation of SOCE in Drosophila neurons, reduction of dSEPT7 overrides this requirement. The mechanism of dSEPT7 regulated Ca2+ uptake was investigated next.
Ca2+ uptake in neurons with reduced dSEPT7 requires dOrai
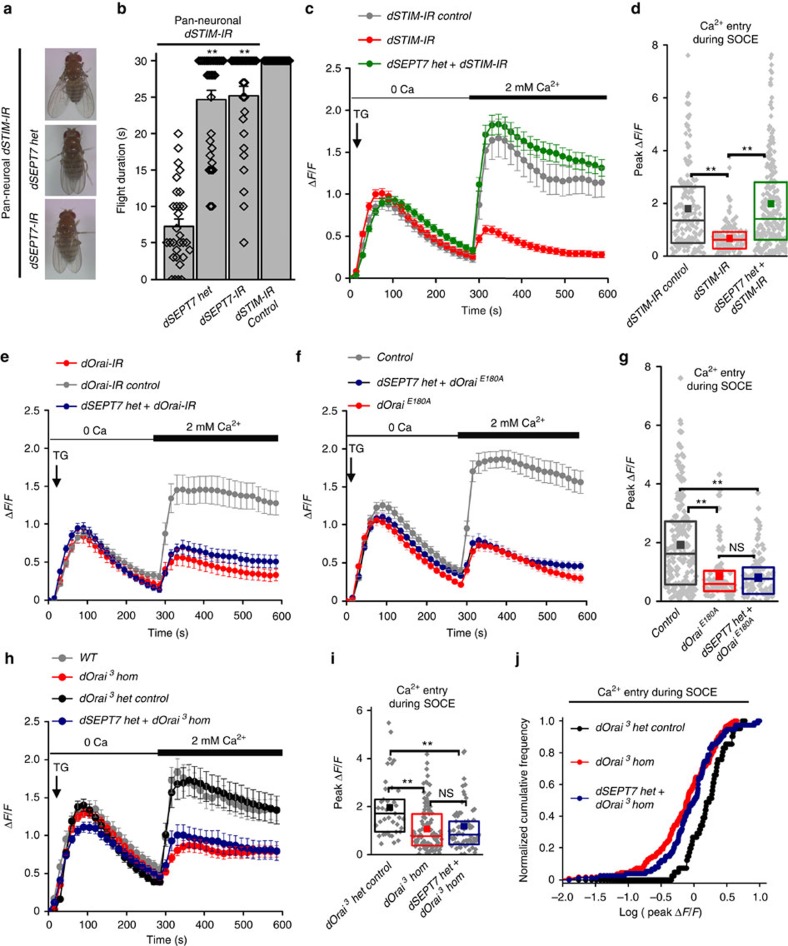
Flies with pan-neuronal knockdown of dSTIM exhibit wing-posture and flight deficits21, similar to adult phenotypes of itpr mutants (Fig. 4a,b). dSEPT7 reduction either by genetic deletion or RNAi-mediated knockdown rescued both phenotypes in dSTIM knockdown flies (Fig. 4a,b). dSEPT7 reduction also restored robust extracellular Ca2+ uptake in neurons with dSTIM knockdown after store depletion (Fig. 4c,d; Supplementary Fig. 4a). This rescue was not accompanied by a concomitant increase of dSTIM protein in the CNS (Supplementary Fig. 4b), indicating that the rescue was not by upregulation of dSTIM levels. Reduction of dSEPT7, however, failed to improve Ca2+ entry in neurons with either reduced levels of dOrai or compromised dOrai function. Neurons cultured from animals with RNAi-mediated knockdown of dOrai (Fig. 4e; Supplementary Fig. 4c) and from a hypomorphic dOrai mutant (dOrai3/3 or dOrai3 hom; Fig. 4h) exhibit significantly reduced SOCE and this remained low on reducing dSEPT7 (Fig. 4e,h–j; Supplementary Fig. 4c). Similarly, reduction of dSEPT7 did not restore SOCE in neurons with overexpression of a mutant dOraiE180A, which functions as a dominant-negative in vivo40 (Fig. 4f,g). In agreement with this, dSEPT7 reduction also failed to improve flight in flies with pan-neuronal knockdown of dOrai (Supplementary Fig. 4d). Together, these data suggest that Ca2+ entry on reduction of dSEPT7 requires functional dOrai channels. Moreover, reduced dSEPT7 can compensate for reduced levels of the ER Ca2+ sensor dSTIM.
Figure 4. Extracellular Ca2+ entry obtained on dSEPT7 reduction requires functional dOrai channels.
(a,b) Wing-posture defects (a) and reduced flight duration (b) in flies with pan-neuronal knockdown of dSTIM (dSTIM-IR) was rescued by dSEPT7 reduction with either a single copy of the deficiency allele or RNAi-mediated targeted knockdown in neurons. N≥30; **P<0.01, Mann–Whitney U-test; flight duration of dSTIM knockdown flies with reduced dSEPT7 using either dSEPT7 het or dSEPT7-IR compared with that of dSTIM knockdown flies. (c) Reduced SOCE on dSTIM knockdown in primary neurons was restored on dSEPT7 reduction; N≥150 cells. Pan-neuronal elavC155GAL4 was used to drive expression of dSTIM-IR. (d) Quantification of peak ΔF/F values obtained during SOCE (300–600 s) for the indicated genotypes; **P<0.001, Mann–Whitney U-test with Bonferroni correction. (e) Reduced SOCE obtained by knockdown of dOrai was not rescued on dSEPT7 reduction. Traces represent the mean (±s.e.m.) ΔF/F values obtained at each time point during TG-induced ER store Ca2+ depletion and SOCE for the indicated genotypes (N≥150 cells). Pan-neuronal elavC155 GAL4 was used to drive expression of dOrai-IR. (f) Reduced SOCE obtained by expression of dOraiE180A was not rescued by dSEPT7 reduction; N≥150 cells. Pan-neuronal elavC155GAL4 was used to drive expression of dOraiE180A. Control refers to neurons with the dOraiE180A transgenic construct, but no GAL4 driver, resulting in no expression of the transgene. (g) Box plots quantifying the peak ΔF/F values for SOCE (300–600 s) computed from the Ca2+ traces shown in f **P<0.001, Mann–Whitney U-test with Bonferroni correction. (h) Reduced SOCE obtained in neurons homozygous for a hypomorphic dOrai mutant allele (dOrai3 hom or dOrai3/3) was not rescued on dSEPT7 reduction. Traces represent the mean (±s.e.m.) ΔF/F values obtained at each time point during TG-induced ER store Ca2+ depletion and SOCE for the indicated genotypes (N≥150 cells). dOrai3 het control refers to neurons heterozygous for the dOrai3 hypomorphic allele (recessive). (i) Box plot quantifying the peak ΔF/F obtained during SOCE in the indicated genotypes. **P<0.001, Mann–Whitney U-test. (j) K–S plot analysing the peak ΔF/F obtained during SOCE in the indicated genotypes. P<0.001, K–S test.
dSEPT7 reduction leads to reorganization of dSTIM and dOrai
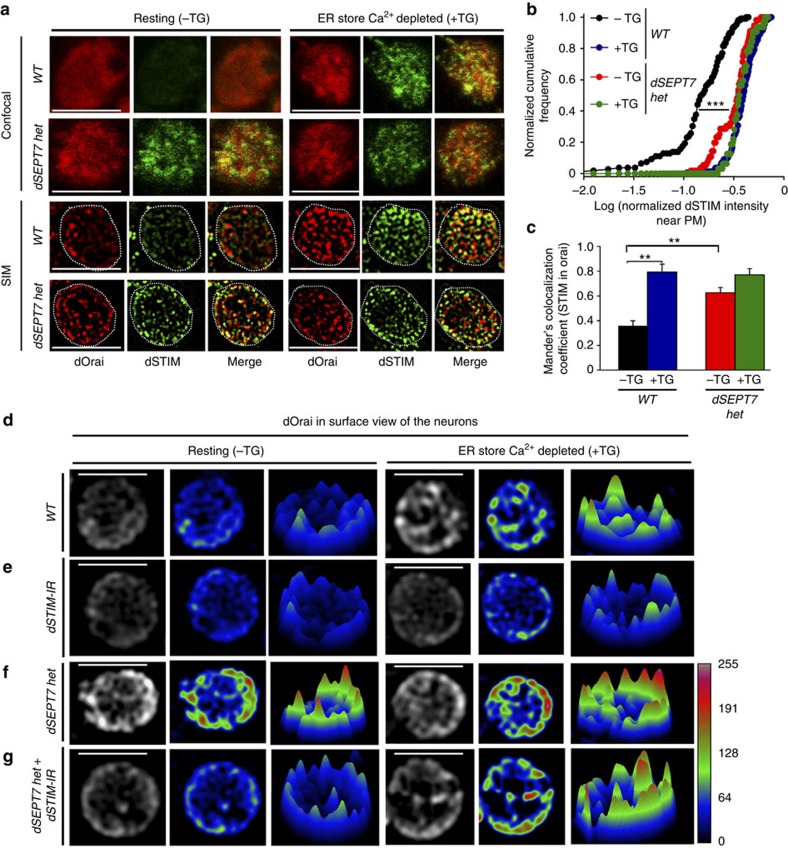
To begin understanding how lower levels of dSEPT7 reduce the dependence of dOrai channel opening on dSTIM, we analysed the extent of endogenous dSTIM co-localization with dOrai near the PM in WT and dSEPT7 het neurons, using previously validated antibodies (Supplementary Fig. 5a,b)40,41. For this purpose, optical sections of ∼300 nm, identified by dOrai immunostaining and representing the PM and subcellular regions in its close proximity, were selected, and dSTIM fluorescence in these sections was quantified (Fig. 5a, confocal). Normalized dSTIM fluorescence detected in these optical sections was significantly higher in store-depleted cells when compared with the resting untreated cells (Fig. 5a, upper panels; Fig. 5b). This resulted in increased co-localization of dSTIM with dOrai after TG treatment (Fig. 5a, +TG, and Fig. 5c). Interestingly, significantly higher intensity of dSTIM was detected near the PM in resting neurons with reduced dSEPT7, and this PM localization was not enhanced significantly after store depletion (Fig. 5a, confocal—lower panels, Fig. 5b,c). Recruitment of STIM to the peripheral ER after store depletion is generally accompanied by clustering and has been visualized by expression of tagged STIM constructs in non-excitable cells3,12. In WT Drosophila neurons, dSTIM near the PM appeared mildly clustered after store depletion (Fig. 5a, confocal, +TG). These clusters were better resolved and visualized by structured illumination microscopy (SIM) in store-depleted neurons as well as in dSEPT7 het neurons at rest and after store depletion (Fig. 5a, SIM).
Figure 5. dSEPT7 reduction results in reorganization of dSTIM and dOrai in resting neurons.
(a) Representative images of immunostained endogenous dSTIM and dOrai, in an optical section of a surface view of the PM and subcellular regions in close proximity of the PM, in either resting (−TG) or store-depleted (+TG) cells of the indicated genotypes. Scale bar, 5 μM. Images shown in the top two panels were acquired using confocal microscopy and those in the bottom two panels were acquired using structured illumination microscopy (SIM). (b) A K–S plot comparing normalized dSTIM intensities near the PM in the indicated genotypes either in resting conditions or after TG treatment. The distributions for TG-treated wild-type cells is shifted significantly to the right when compared with the resting cells, indicating a greater proportion of cells with higher normalized dSTIM intensities. The distribution for resting cells with reduced dSEPT7 is shifted significantly to the right when compared with the resting wild-type cells, indicating a higher proportion of cells with higher normalized dSTIM intensities. ***P<0.0001, K–S test, WT (−TG) compared with either WT (+TG) or dSEPT7 het (−TG). Normalized dSTIM intensity near the surface=dSTIM intensity in the optical section representing the cell surface/total dSTIM intensity for the cell. Images acquired using confocal microscopy were used for the analysis in b,c. (c) Bar graph quantifying the Mander’s co-localization coefficients for the amount of STIM intensity co-localizing with Orai intensity in the indicated genotypes and treatment conditions. **P<0.001, Mann–Whitney U test with Bonferroni correction. (d–g) Representative images of dOrai protein organization on the surface of resting and TG-treated cells of the indicated genotypes. dOrai was visualized by optimal image acquisition settings with confocal microscopy. Original grey scale images were deconvoluted and pseudo-coloured for better representation of high and low dOrai intensities with warmer colours representing higher intensities. A surface plot depicting the spatial distribution of dOrai intensities accompanies each image. Calibration bar represents the grey scale intensities corresponding to each colour. Scale bar, 5 μm.
The appearance of dOrai clusters concomitant with dSTIM was investigated next. A majority of WT Drosophila neurons (∼65%) exhibit diffuse distribution of endogenous dOrai proteins at the PM at rest (Fig. 5a, resting). This diffuse distribution changed to visibly distinct high-intensity clusters in ER store-depleted neurons treated with TG (Fig. 5a, +TG and Fig. 5d, +TG). dOrai clusters, obtained after store depletion by TG, were also observed through SIM (Fig. 5a, SIM). In a fraction (∼35%) of resting WT neurons, dOrai clusters were observed (Fig. 5d, resting), but their normalized intensity was significantly lower as compared with dOrai clusters observed in store-depleted neurons (Supplementary Fig. 6b,c,f). As with dSTIM, resting neurons with reduced dSEPT7, exhibit greater numbers of high-intensity dOrai clusters, and the intensity of these clusters did not change significantly on store depletion (Fig. 5f; Supplementary Fig. 6b,c,g). Thus, reduced dSEPT7 levels pre-dispose resting neurons towards a state that could allow spontaneous Ca2+ entry through dOrai. Higher-resolution SIM images confirmed these observations, where, in resting neurons with reduced dSEPT7, dOrai clusters co-localized with visibly distinct dSTIM clusters (Fig. 5a, SIM).
In agreement with previous observations in non-excitable cells12, the intensity of dOrai clusters was significantly lower in dSTIM knockdown cells as compared with WT cells after store depletion (Fig. 5e; Supplementary Fig. 6b,d). dSTIM knockdown also resulted in significant reduction of high-intensity dOrai clusters in resting neurons with reduced dSEPT7, suggesting that clustering of dOrai in resting neurons requires dSTIM (Fig. 5g; Supplementary Fig. 6b).
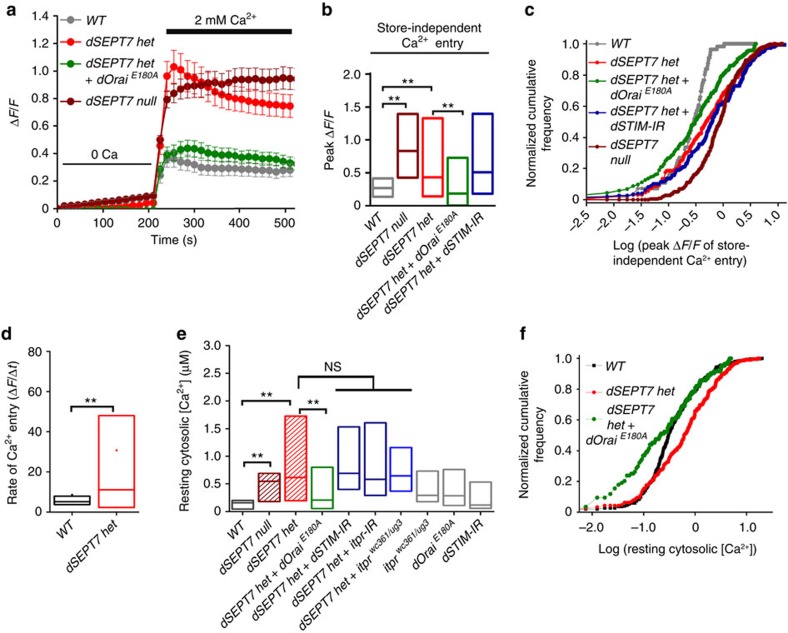
The high levels of dSTIM present near the PM and the resulting dOrai clusters observed in resting neurons with reduced dSEPT7 prompted us to investigate their ability to support Ca2+ entry in the absence of store depletion. WT neurons exhibit a low level of spontaneous Ca2+ entry at rest on addition of Ca2+ to the extracellular medium (Fig. 6a). In dSEPT7 het and dSEPT7 null neurons, spontaneous uptake of extracellular Ca2+ was significantly higher as compared with WT neurons (Fig. 6a–c). Importantly, higher uptake of extracellular Ca2+ by neurons with reduced dSEPT7 was significantly reduced on expression of the ‘dominant-negative’ dOraiE180A transgene (Fig. 6a–c). In agreement with the rescue of flight deficits of dSTIM knockdown animals by introducing dSEPT7 het, store-independent Ca2+ entry remained high in dSEPT7 het+dSTIM-IR neurons (Fig. 6b,c; see discussion). The rate of Ca2+ uptake was significantly higher in dSEPT7 het neurons when compared with WT (Fig. 6d).
Figure 6. dSEPT7 reduction results in a store-independent activation of dOrai channels in resting neurons.
(a) Changes in cytosolic Ca2+ monitored as ΔF/F values during store-independent Ca2+ entry on Ca2+ add-back, after monitoring basal Ca2+ fluctuations in cells plated in ‘0 Ca medium’ for 225 s. Store-independent Ca2+ entry in neurons with reduced (dSEPT7 het) or no dSEPT7 (dSEPT7 null) is higher when compared with WT neurons, and is significantly reduced on expression of dOraiE180A (N≥150 cells). (b) Box plot quantifying the peak ΔF/F obtained during store-independent Ca2+ entry (200–500 s) in the indicated genotypes. **P<0.01, Mann–Whitney U-test with Bonferroni correction. (c) K–S plot analysing the peak ΔF/F obtained during Ca2+ entry in the indicated genotypes. The distribution for neurons with reduced (dSEPT7 het) or no dSEPT7 (dSEPT7 null) is significantly shifted to the right compared with wild-type neurons, indicating a higher proportion of cells with higher peak ΔF/F values. The distribution for dSEPT7 het cells is significantly shifted to the left after expression of dOraiE180A, indicating a greater proportion of cells with lower peak ΔF/F values. P<0.001, K–S test for ‘dSEPT7 het’ or ‘dSEPT7 null’ compared with WT and ‘dSEPT7 het’ compared with ‘dSEPT7 het+dOraiE180A’. (d) Rate of Ca2+ entry in the indicated genotypes. Rate of Ca2+ entry expressed as ΔF/Δt, where ΔF=F255−F225 and Δt=30 s. **P<0.01, Mann-Whitney U test. (e) Resting cytosolic [Ca2+] in the indicated genotypes measured using Indo-1AM (Methods). Higher resting cytosolic [Ca2+] in neurons with reduced or no dSEPT7 (median ∼500 nM) is restored to WT levels (median ∼200 nM) on expression of dOraiE180A. The horizontal line in the middle of the box represents the median. **P<0.001, Mann–Whitney U-test with Bonferroni correction. (f) K–S plots analysing the distribution of resting cytosolic [Ca2+] in the indicated genotypes. The distribution for neurons with reduced dSEPT7 is significantly shifted to the right compared with WT, indicating a greater proportion of cells with higher resting cytosolic [Ca2+]. This distribution was significantly shifted to the left on expression of dOraiE180A, indicating a higher proportion of cells with lower resting cytosolic [Ca2+]; P<0.001, K–S test.
A predicted outcome of such constitutive Ca2+ entry would be higher basal cytosolic Ca2+ levels in neurons with reduced dSEPT7. Indeed, resting cytosolic [Ca2+] was higher in dSEPT7 het and dSEPT7 null neurons when compared with WT neurons (Fig. 6e,f). Expression of dOraiE180A in dSEPT7 het neurons significantly reduced resting cytosolic [Ca2+] to levels comparable to WT neurons (Fig. 6e,f). Thus, reduction of dSEPT7 promotes Ca2+ entry through dOrai in resting neurons, which is independent of ER store depletion.
dOrai clusters were also reduced in neurons with IP3R knockdown after store depletion (Supplementary Fig. 6a,b,e), suggesting a role for the IP3R in dSTIM-mediated clustering and activation of dOrai. However, knockdown of the IP3R did not reduce dOrai clusters observed in dSEPT7 het neurons at rest (Supplementary Fig. 6a,b). Resting cytosolic [Ca2+] also remained high in neurons with reduced dSEPT7 after either IP3R knockdown or dSTIM knockdown (Fig. 6e) and in IP3R mutant neurons with reduced dSEPT7 (Fig. 6e, dSEPT7 het+itprwc361/ug3). These data are consistent with the observation that dSEPT7 reduction allowed extracellular Ca2+ uptake in neurons with either reduced IP3R (Fig. 2f) or dSTIM (Fig. 4c), but not dOrai (Fig. 4e) and support a model where higher cytosolic [Ca2+] in neurons with dSEPT7 reduction results from store-independent Ca2+ entry through dOrai.
The mechanism by which dSEPT7 supports greater dSTIM localization to the PM and Orai channel opening in the absence of store Ca2+ depletion needs further elucidation. In preliminary experiments, we observed a fraction of endogenous dSEPT7 closely apposed to the ER, marked by GFP-tagged protein disulphide isomerase (Supplementary Fig. 5c, d, upper panels) and in an optical section of the cell surface where it co-localized with the cortical ER, (Supplementary Fig. 5d, lower panels). In addition, dSEPT7 and dOrai co-localized at the periphery of neuronal cell bodies (Supplementary Fig. 5e, upper panels) as well as in neuronal projections (Supplementary Fig. 5e, lower panels; arrows), in agreement with similar PM association found for SEPT7 in mammalian neurons42,43. The intracellular distribution of dSEPT7 near the ER and in the proximity of the PM suggests a direct role in the organization and function of dSTIM and dOrai.
Discussion
In this study, we show that dSEPT7 prevents dOrai-mediated spontaneous Ca2+ entry in neurons. Consequently, lower dSEPT7 levels lead to Ca2+ store-independent constitutive opening of dOrai channels in resting neurons. Thus, dSEPT7 functions as a ‘molecular brake’ on dOrai channel activation in unstimulated neurons. Consistent with the negative regulatory function, dSEPT7 overexpression resulted in lower SOCE in neurons (Fig. 2h).
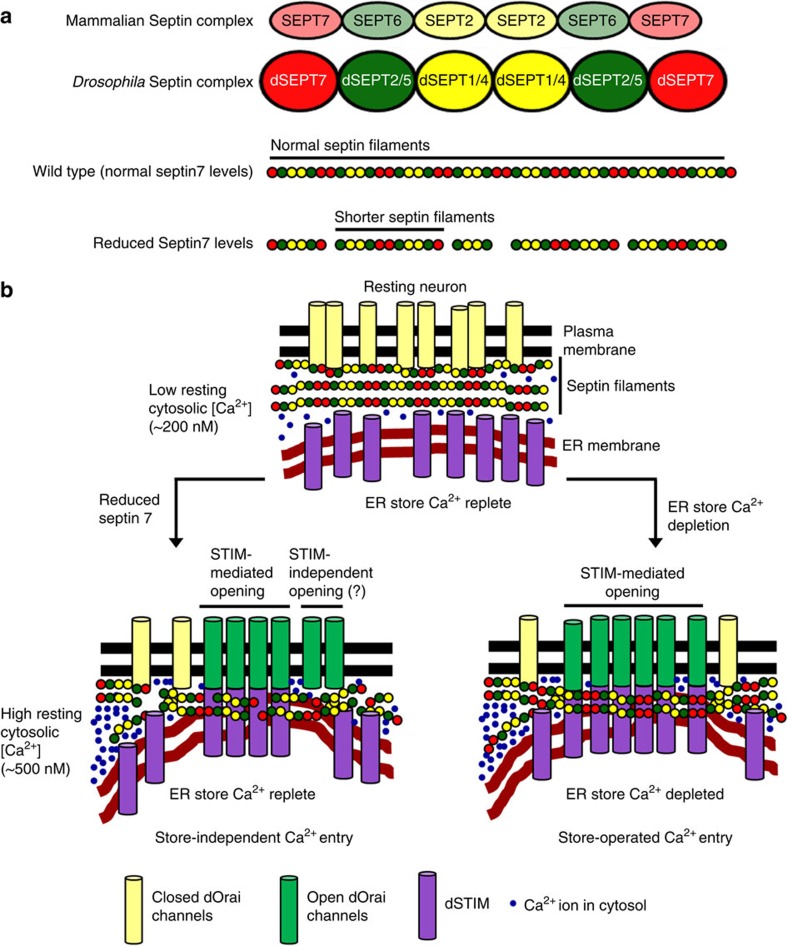
dSEPT7 forms a complex with dSEPT1 and dSEPT2 (ref. 33) similar to mammalian SEPT7, which forms linear hexamers with SEPT2 and SEPT6 (ref. 44; Fig. 7a). SEPT7 occupies terminal positions in this hexamer44. These hexamers are further linked to form non-polar linear septin filaments26,45 (Fig. 7a). We propose a model where dSEPT7-containing septin filaments, in resting neurons with normal dSEPT7 levels, maintain a ‘check’ on dSTIM recruitment to the ER–PM regions (Fig. 7b, resting neuron). Reducing dSEPT7 removes this block in neurons with replete stores and results in dSTIM recruitment to the ER–PM regions, with concomitant formation of dOrai clusters and opening of Orai channels (Fig. 7b, reduced Septin 7). STIM recruitment to the peripheral ER followed by clustering of Orai has been visualized by expression of tagged molecules in most cell types after store depletion12,25. However, Orai clusters are not necessary for opening of Orai channels12,46,47. In agreement with these previous findings from non-excitable cells, lower dSTIM levels also reduce dOrai clusters in resting neurons with reduced dSEPT7 (Fig. 5g; Supplementary Fig. 6b). Higher spontaneous Ca2+ entry and basal cytosolic [Ca2+] in dSEPT7 het neurons with dSTIM knockdown (Fig. 6b,c) suggest the presence of non-clustered but active dOrai channels, and indicate that dSEPT7-containing septin filaments help in organizing membrane domains that favour the inactive state of dOrai channels. Reducing dSEPT7 possibly leads to the presence of shorter septin filaments (Fig. 7a,b), which we speculate help reorganize PIP2 in the PM25 and formation of membrane domains48,49 that favour dOrai channel opening by residual dSTIM proteins. Thus, reduction of dSEPT7 at the PM probably mimics a membrane state that is triggered by septin filament reorganization after store depletion and is permissive for Orai activation (Fig. 7b).
Figure 7. A model explaining activation of Orai upon dSEPT7 reduction in neurons.
(a) Arrangement of mammalian and Drosophila septins in septin complexes (hetero-hexamers) that are the basic building blocks of septin filaments. Reduction of dSEPT7 is predicted to result in breaks in the linear septin filaments present in wild-type cells, leading to the formation of shorter septin filaments. (b) Septin filaments closely associate with the PM and near the ER in resting neurons. ER store Ca2+ depletion results in a reorganization of these filaments, followed by movement of STIM to the peripheral ER, dSTIM/dOrai coupling and Ca2+ entry through dOrai (SOCE). dSEPT7 reduction leads to formation of shorter septin filaments that support dSTIM recruitment to the peripheral ER in resting neurons and promote Orai channel opening. This store-independent activation of dOrai results in elevation of cytosolic [Ca2+].
Depletion of ER store Ca2+ is the primary stimulus for STIM translocation to the ER–PM regions3, although recent findings indicate multiple other proteins additionally regulate this process50,51. Overexpression of one such regulator, STIMATE resulted in STIM clustering and recruitment to the ER–PM regions in mammalian cells with replete ER store Ca2+ levels50. Importantly, it also resulted in store-independent Ca2+ entry through Orai channels50. In neurons with reduced dSEPT7, we observed increased localization of dSTIM clusters near the ER–PM regions (Fig. 5a,b). These dSTIM clusters may arise due to a small drop in ER calcium, undetected by TG-mediated passive store release in our experiments. Because partial depletion of dSTIM with an RNAi continued to support store-independent Ca2+ entry and higher resting cytosolic [Ca2+] in dSEPT7 het neurons to the same extent as dSEPT7 het neurons with WT levels of dSTIM (Fig. 6c,e), we predict that Orai-mediated Ca2+ entry observed on dSEPT7 reduction maybe a cumulative effect of dOrai activation by residual dSTIM proteins and other non-canonical modes of Orai activation, which remain to be elucidated.
Septins are ubiquitous proteins, but the levels of individual septin subunits and their functions vary across cell types26. For example, dSEPT7 can function differently during cytokinesis and cellularization, based on the cell type under study28. Interestingly, the role of mammalian SEPT7 in cytokinesis is also cell-type specific52. Thus, the functional consequences of modulating septin levels in cells depends on the subgroup of septin depleted, its effect on septin filament organization and ultimately the cell-type under investigation. In this context, our findings that the SEPT2 subgroup of septins (Table 1; Fig. 7a) function as positive regulators of SOCE, whereas dSEPT7 acts as a ‘molecular brake’ on Orai activation, are not unprecedented. The regulation of Orai function by septins will require a better understanding of the nature of septin complexes and filaments present in resting neurons and in neurons after store depletion.
SOCE is required in neurons of the Drosophila flight circuit for transcriptional regulation of genes required during maturation of the flight circuit40. We have earlier demonstrated that raising cytosolic [Ca2+] by reducing sarco-endoplasmic reticulum Ca2+ATPase function with a dominant-negative mutant allele CaP60Akum170/+ (ref. 53), partially restores flight bouts in itpr mutants54, and the length of these flight bouts are further enhanced by introducing dominant hypermorphic alleles of dOrai21. A similar increase in resting cytosolic [Ca2+] is observed in Drosophila neurons with reduced dSEPT7; this is very likely responsible for the rescue of itpr mutant flight deficits obtained on dSEPT7 reduction. Thus, Septin 7 functions as a novel regulator of neuronal Ca2+ homeostasis with an impact on physiological and behavioural phenotypes.
The ability of SEPT7 to control cytosolic Ca2+ levels in Drosophila neurons through Orai needs testing in mammalian neurons because deranged Ca2+ homeostasis has been linked to several neurodegenerative diseases55,56. Our findings could thus provide insights into new therapeutic targets for regulating neuronal Ca2+ homeostasis.
Methods
Fly strains
All flies were reared on corn flour agar medium supplemented with yeast at 25 °C. Canton-S was used as the WT strain. Single point mutations in the Drosophila itpr gene have been described earlier30. The genetic deficiency allele pnutxp27(BL5687) and pan-neuronal elavC155GAL4 (BL458) were obtained from the Bloomington Stock Centre, Indiana, USA. An RNAi line (referred to as ‘itpr-IR’) for the itpr gene (1063) was obtained from the National Institute of Genetics (NIG), Japan. RNAi lines for dSTIM(v47073), dOrai(v12221), dSEPT7 (v11791), dSEPT1 (v101445) and dSEPT4 (v7742) were obtained from the Vienna Drosophila RNAi Centre (VDRC), Vienna, Austria. The RNAi strains obtained from VDRC and NIG contain, short regions (∼200 bp) of the target gene cloned as inverted repeats. These inverted repeats are introduced as transgenes under control of the upstream activator sequence (UAS)57. When expressed in neurons using the pan-neuronal elavC155GAL4, a resulting hairpin RNA is formed in vivo and recognized by the existing RNAi machinery within the cell. This subsequently generates smaller double-stranded RNAs of 21–22 nucleotides in length, which efficiently target and knockdown messenger RNAs from the gene of interest. Gene sequences, cloned as inverted repeats for each gene, are publicly available from the VDRC (http://stockcenter.vdrc.at/control/main) and NIG (www.nig.ac.jp/nig/) stock centres. The hypomorphic dOrai allele (dOrai3)40, UASdSTIM (referred to as dSTIM+)22, UASdOraiE180A (referred to as dOraiE180A)40 and a transgenic strain expressing Sep2-GFP under the control of the endogenous Sep2 promoter39 have been described earlier. A full-length complementary DNA of the dSEPT7 (or pnut) gene (BDGP LD37170 (ref. 58)) was subcloned into the pUASTattB vector between the EcoR1 and Xba1 sites, and microinjected into embryos to generate the UASpnut+(UASdSEPT7+) transgenic line. The pan-neuronal elavC155GAL4 was used for expression of the gene-specific RNAi or the dSEPT7+ transgene in all neurons of the fly. ‘Gene-IR-control’ refers to animals containing the RNAi construct, but no GAL4, resulting in no expression of the RNAi. All fly strains were generated using standard protocols for fly genetics.
Flight assays
Flight assays were performed as described earlier40. In brief, single flies were aged post-eclosion for 3 days, anaesthesized on ice for 15 min before recording flight durations. Each fly was tethered in the neck region between the head and thorax to a thin-metal wire. Flight was monitored for 30 s after delivery of a gentle mouth blown air-puff. Flight duration for each fly was recorded manually. Statistical significance between genotypes was computed with the Mann–Whitney U-test on the Origin 8.0 software (Micro Cal).
Primary neuronal culture
Primary neurons were cultured from the third instar larval CNS (CNS comprising the brain and ventral ganglion) as described previously21,40. In brief, larval CNS was dissected and subjected to enzymatic digestion using 0.75 μg μl−1 collagenase and 0.40 μg μl−1 dispase for ∼40 min. Single dissociated neurons were spun down, followed by plating in growth medium comprising of DMEM/F12-1065 (Gibco) supplemented with 20 mM HEPES (pH 7.2), 50 U ml−1 penicillin (Invitrogen), 50 μg ml−1 streptomycin (Invitrogen) and 10 μg ml−1 Amphotericin B (Invitrogen). Cells were incubated at 25 °C with 5% CO2 for 14–16 h. All chemicals for cell culture were obtained from Sigma-Aldrich unless mentioned otherwise. This protocol primarily supports growth of neuronal cells, and the glial cells remain at <1% of the total cell population59.
Calcium imaging in primary neurons
For the measurement of store Ca2+ release and SOCE, cells were washed with M1 medium21, following which they were loaded with 2.5 μM of Fluo-4 acetoxymethylester (AM; Life Technologies) with 0.002% Pluronic-F-127 (Sigma-Aldrich) in M1 medium for 30 min in the dark. Following dye loading, cells were washed in M1 and placed in nominally ‘0’ Ca2+ M1 (containing 12 nM-free Ca2+) supplemented with 0.5 mM EGTA and imaged for Ca2+ changes. Imaging and quantification were performed as described previously21. Images were acquired every 15 s (for passive store depletion induced SOCE) or every 10 s (for CCh-stimulated SOCE) for a total duration of 10 min (600 s) on a Nikon TE2000 inverted wide-field microscope equipped with × 60/1.4 numerical aperture (NA; oil) objective lens, using the 488-nm excitation and 520-nm emission filter sets. Store depletion was induced by adding 10 μM TG (Life Technologies) or 20 μM Carbamoylcholine chloride (CCh; Sigma) after the first frame of acquisition. CCh-containing medium was exchanged with ‘0 Ca’ medium after 200 s, and the cells were imaged for another 100 s before Ca2+ add-back. CaCl2 was added after the 20th frame of acquisition (or 300 s). CCh measurements were performed with overexpression of muscarinic acetylcholine receptor in neurons of all genotypes. For the measurement of resting cytosolic Ca2+, a previously described protocol21 was followed. In brief, cells were incubated with 5 μM Indo-1 AM and 0.002% Pluronic F-127 in M1 media for 45 min at room temperature in the dark. Cells were washed twice with M1 before and after dye incubation and finally covered with M1 for imaging. Data acquisition was performed by 365-nm excitation and 410/485 dual-emission filter sets at 5-s interval for five frames, after which 10 μM ionomycin (Calbiochem, USA) was added to obtain the maximum florescence intensity and imaged for 10 more frames. EGTA (1 mM) and 0.01% Triton-X-100 were added after the 10th frame to obtain the minimum florescence intensity for each cell. Image acquisition for basal calcium measurements was performed on an Olympus IX81-ZDC2 inverted wide-field microscope equipped with focus drift compensating and × 20/0.5 NA (air), × 60/1.35 NA (oil) and × 100/1.4 NA (oil) objective lenses. Excitation of fluorescent Ca2+ indicator dye was performed using specific wavelength illumination from a halogen arc lamp with a TILL Polychrome 5000 monochromator (TILL Photonics, Graefelfing, Germany) for variable bandwidth and intensity. Emitted light was detected through band-pass filter sets (Chroma, Brattleboro, VT). Image acquisition was performed using the Andor iXON 897E EMCCD camera and AndoriQ 2.4.2 imaging software. The time-lapse acquisition mode of the software was used to follow fluorescence changes over time.
Pharmacological perturbation of septin dynamics
For perturbation of septin dynamics, 14–16-h-old primary neurons were washed with M1 medium, following which they were covered with growth medium (DMEM supplemented with 20 mM HEPES and antibiotics) containing either dimethylsulphoxide (0.1%, solvent control) or 100 μM FCF (Sigma-Aldrich) and incubated at room temperature for either 20 or 45 min. This was followed by three consecutive washes with M1 medium, after which the cells were loaded with Fluo-4-AM for 30 min for calcium measurements or fixed for immunostaining.
Data analysis
For quantifying the changes in fluorescence, images were processed using the Image Pro plus software, V1.33 (Media Cybernetics). Region of interest (ROI) was drawn around each cell, and the fluorescence intensity at each time points was determined and plotted using the Origin 8.0 software as follows: ΔF/F=(Ft−Fbasal)/Fbasal for each time point, where Ft is fluorescence at the time point and Fbasal is the fluorescence of the cell when starting the experiment. Peak values of ΔF/F were obtained for every cell, and a box chart representing the data spread was plotted. The rectangular boxes represent the spread of data points between 25 and 75% of cells and the horizontal line within each box is the median. The smaller box inside each box represents the mean. Significance was computed for different data sets using Mann–Whitney U-test for nonparametric distributions. The distribution of peak ΔF/F values or resting cytosolic [Ca2+] for different genotypes and conditions were also analysed using Kolmogorov–Smirnov (K–S) test. For K–S tests, the cumulative frequency of the peak ΔF/F values or resting cytosolic [Ca2+] was normalized to the total number of cells and plotted against the log of the peak ΔF/F values or resting cytosolic [Ca2+]. Significance was computed based on the maximum difference between the two distributions. Rate of Ca2+ entry during SOCE was calculated by computing the rate of change in fluorescence intensity (ΔF) between the 315 and 285 s time points and expressed as ΔF/Δt, where Δt=30 s.
For computation of resting cytosolic [Ca2+], fluorescence values obtained at each time point were converted to ratios of 410/485 emission. The absolute concentration of free calcium was calculated using the Grynkiewicz equation: [Ca2+]i (μM)=Kd × (Fbasal−Fmin)/(Fmax−Fbasal), where Fbasal is the basal cytosolic fluorescence (at the first frame or start of imaging), Fmax is the peak fluorescence obtained after adding 10 μM ionomycin (to allow the cytosolic Ca2+ to equilibrate to external Ca2+) and Fmin refers to the fluorescence on chelating all free cytosolic Ca2+ with EGTA. The published Kd value of 1.16 μM for Indo-1 in Drosophila cells was used60.
Immunocytochemistry and confocal microscopy
Primary neuronal cultures, with or without TG treatment for 5 min, were washed with PBS (pH=7.5), followed by fixation with 3.5% paraformaldehyde in PBS for 20 min at room temperature. Fixed neurons were washed once with a wash buffer (1/10th dilution of blocking buffer) and permeabilized with three 10-min washes of wash buffer, followed by blocking with blocking buffer (5% bovine serum albumin (w/v), 0.5% Triton X (v/v), 0.5% glycerol (v/v) in PBS) for 1 h. The cultures were incubated overnight with primary antibodies at the appropriate dilution in wash buffer. The following day cells were washed three times, for 10 min each, with wash buffer at 4 °C. Incubation with the appropriate secondary antibody was in wash buffer for 30 min at 4 °C, 45 r.p.m., followed by three 15-min washes with wash buffer at room temperature. Cells were finally covered with 60% glycerol (v/v) for imaging in an inverted Olympus FV1000 confocal microscope. Images were acquired through a × 60/oil objective with × 6 optical zoom, with the Fluoview 2.1 C software and FV10-SPD detectors, under optimal settings. Entire cell was imaged using several sequential optical sections of 300-nm thickness. For immunostaining of Sep2-GFP, rabbit anti-GFP antibody (1:10,000; catalogue no. A6455, Life Technologies) was used. dSEPT7 (or Pnut) was detected with a mouse anti-Pnut antibody27 (1:4; catalogue no. 4C9H4, DSHB, University of Iowa, Iowa). A dOrai-specific antibody generated in rats was used at a dilution of 1:1,000 and has been described earlier40. Two anti-dSTIM mouse antibodies41 (8G1) and (3C1) mixed 1:1 were used at a dilution of 1:20. Secondary antibodies used at a dilution of 1:400 are as follows: anti-rabbit Alexa Fluor 488 (catalogue no. A1108; Life Technologies), anti-mouse Alexa Fluor 568 (catalogue no. A1104; Life Technologies) and anti-rat Alexa Fluor 633 (catalogue no. A21094; Life Technologies).
Particle intensity analysis
For the analysis of dOrai particle intensities, raw confocal images (grey scale) were exported as TIFF files. A ROI was drawn around each cell and a mask image was generated using ROIs for all cells in a field. Subsequent analysis of the intensity of each particle was carried out in Matlab R2012 (Licence number 828962; host ID: A4BADB34D85C). The raw particle intensities were normalized to the mean intensity of dOrai for each cell to account for slight variations in antibody staining. The normalized dOrai particle intensities are quantified for each genotype and treatment condition (Supplementary Fig. 6b). Normalized intensities of particles between 3 and 8 pixels were used for analysis. Statistical significance was computed using the Mann–Whitney U-test and the K–S test. For representation, images were deconvoluted with the deconvolve three-dimensional (3D) plugin61 of ImageJ 1.41o (National Institute of Health, USA)62. The diffraction PSF 3D plugin in ImageJ was used to generate a theoretical point spread function (PSF) that was next used in the ‘Deconvolve 3D’ plugin to generate the deconvoluted image with a single iteration. The deconvoluted image was pseudo-coloured and a surface plot generated using ImageJ. For analysing the extent of co-localization, Pearson’s Correlation Coefficients and Mander’s co-localization coefficients were calculated using ImageJ.
Quantification of STIM intensities near the surface
Untreated or TG-treated cells were fixed and the endogenous STIM and Orai proteins labelled using the respective antibodies in the same cell. Confocal images were obtained using simultaneous acquisition in both channels (STIM, excitation wavelength: 488 nm and Orai, excitation wavelength, 633 nm), using an Olympus FV1000 Confocal microscope as described earlier. For computation of STIM intensities near the PM, an optical section that represents the PM and intensities approximately within 300 nm from the coverslip were selected using immunostaining against Orai. The same optical section was selected from the other channel (showing STIM immunostaining) and the STIM intensities in this optical section computed using ImageJ. This STIM intensity near the PM was normalized to the total STIM fluorescence for the whole cell and expressed as ‘normalized STIM fluorescence near the PM’. Total STIM intensity was not significantly different between resting and TG-treated cells. Single-stack images from each of these two channels (showing the PM Orai staining and STIM intensity near the PM) were used to analyse STIM/Orai co-localization using ImageJ.
Structured illumination microscopy
SIM was performed on a Nikon N-SIM microscope with a × 100 (oil, NA 1.49) CFI SR APOCHROMAT objective using the 488- or 633-nm laser lines. Cell footprints were imaged as a single-plane image. The super-resolution image obtained using 3D SIM was reconstructed, using the Nikon NIS ELEMENT Software version 4.30. Sample preparation was similar to that used for confocal imaging. Optimum intensity settings were maintained to obtain the super-resolution images.
Western blots
Larval CNS or adult heads of appropriate genotypes were dissected in cold dissection buffer (20 mM HEPES, 100 mM KCl, pH 7.5). Homogenization of 10 brains or adult heads was performed in 100 μl of homogenizing buffer (20 mM HEPES, 100 mM KCl, 1% Triton-X-100, 1 mM PMSF, pH 7.5) and 15 μl of the homogenate was run on a 6% SDS–polyacrylamide gel (for IP3R) or 8% SDS–polyacrylamide gel (for all other proteins). The protein was transferred to a nitrocellulose membrane by standard protocols, and the membrane was incubated in the primary antibody overnight at 4 °C. Primary antibodies were used at the following dilutions: rat anti-dOrai, 1:1,000; mouse anti-β-tubulin monoclonal, 1:5,000 (catalogue no. E7, DSHB); and mouse anti-Pnut, 1:20 (catalogue no. 4C9H4, DSHB). The affinity-purified anti-InsP3R rabbit polyclonal antibody31(IB-9075) was used at a dilution of 1:300. A mouse anti-spectrin antibody (1:50; catalogue no. 3A9, DSHB) was used as a loading control for IP3R. Two anti-dSTIM mouse antibodies (8G1) and (3C1) mixed 1:1 were used at a dilution of 1:20. Secondary antibodies used were anti-mouse HRP (1:3,000; catalogue no. 7076S, Cell Signaling Technologies), anti-rabbit HRP (1:3,000; catalogue no. 32260, Thermo scientific, Rockford) and anti-rat HRP (1:10,000; catalogue no. 012030003, Jackson Immuno Research). Proteins were detected on the blot by a chemiluminiscent detection solution, SuperSignal West Dura Extended Duration Substrate (catalogue no. 34075, Thermo Scientific). Full-size western blots are shown in Supplementary Fig. 7. Images of the blots shown in the main and Supplementary Figs have been cropped for presentation.
Statistical tests
Unless otherwise mentioned, statistical significance was computed between a pair of genotypes or treatment conditions, indicated using brackets in the figures, with the Mann–Whitney U-test or K–S test. If multiple genotypes or treatment conditions were compared, the P value used for determining the significance of the differences observed was calculated after applying Bonferroni correction. Nonparametric tests were used for computation of statistical significance as most of the data are not normally distributed.
Data availability
The data that support the findings of this study are available from the corresponding author on request.
Additional information
How to cite this article: Deb, B. K. et al. Store-independent modulation of Ca2+ entry through Orai by Septin 7. Nat. Commun. 7:11751 doi: 10.1038/ncomms11751 (2016).
Supplementary Material
Supplementary Figures 1 - 7
Acknowledgments
We thank Professor Patrick Hogan (La Jolla Institute for Allergy and Immunology, San Diego) for useful discussions on the manuscript, Balaji Ramalingam for MATLAB codes, Dhruv Raina (inStem) for help with MATLAB analysis of particle intensities, Dr Richa Rikhy (IISER, Pune) for fly strains, NCBS Central Imaging and Flow cytometry Facility (CIFF) for confocal and super-resolution imaging, Suparno Gupta for help with SIM imaging and the Fly Facility, NCBS for generating transgenic fly lines. B.K.D. and T.P. are supported by research fellowships from the Council of Scientific and Industrial research (CSIR), Government of India. This research was funded by core grants from NCBS, TIFR.
The authors declare no competing financial interests.
Author contributions B.K.D. and G.H. designed the experiments and wrote the paper. B.K.D. and T.P. performed the experiments. B.K.D. analysed the data. G.H. contributed reagents and overall guidance to the project.
07/18/2018
The original HTML version of this Article had an incorrect article number of ‘0’; it should have been ‘11751’. This has now been corrected in the HTML version of the Article. The PDF version was correct from the time of publication.
References
- Soboloff J., Rothberg B. S., Madesh M. & Gill D. L. STIM proteins: dynamic calcium signal transducers. Nat. Rev. Mol. Cell Biol. 13, 549–565 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge M. J. Inositol trisphosphate and calcium signalling mechanisms. Biochim. Biophys. Acta 1793, 933–940 (2009). [DOI] [PubMed] [Google Scholar]
- Wu M. M., Buchanan J., Luik R. M. & Lewis R. S. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 174, 803–813 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. L. et al. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature 437, 902–905 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J. et al. STIM is a Ca2+ sensor essential for Ca2+-store- depletion-triggered Ca2+ influx. Curr. Biol. 15, 1235–1241 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J., Fivaz M., Inoue T. & Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc. Natl Acad. Sci. USA 104, 9301–9306 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske S. et al. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 441, 179–185 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang S. L. et al. Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proc. Natl Acad. Sci. USA 103, 9357–9362 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig M. et al. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 312, 1220–1223 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M. et al. Orai1 is an essential pore subunit of the CRAC channel. Nature 443, 230–233 (2006). [DOI] [PubMed] [Google Scholar]
- Yeromin A. V. et al. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature 443, 226–229 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. Y. et al. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell 136, 876–890 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M. & Lewis R. S. Store-operated calcium channels. Pharmacology 95, 1383–1436 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S. et al. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat. Cell Biol. 12, 436–446 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S. et al. Junctate is a Ca2+-sensing structural component of Orai1 and stromal interaction molecule 1 (STIM1). Proc. Natl Acad. Sci. USA 109, 8682–8687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuttell L. et al. Undertaker, a Drosophila junctophilin, links Draper-mediated phagocytosis and calcium homeostasis. Cell 135, 524–534 (2008). [DOI] [PubMed] [Google Scholar]
- Woo J. S. et al. Hypertrophy in skeletal myotubes induced by junctophilin-2 mutant, Y141H, involves an increase in store-operated Ca2+ entry via Orai1. J. Biol. Chem. 287, 14336–14348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng M. et al. Store-independent activation of orai1 by SPCA2 in mammary tumors. Cell 143, 84–98 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palty R., Raveh A., Kaminsky I., Meller R. & Reuveny E. SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell 149, 425–438 (2012). [DOI] [PubMed] [Google Scholar]
- Feng J.-M. et al. Golli protein negatively regulates store depletion-induced calcium influx in T cells. Immunity 24, 717–727 (2006). [DOI] [PubMed] [Google Scholar]
- Venkiteswaran G. & Hasan G. Intracellular Ca2+ signaling and store-operated Ca2+ entry are required in Drosophila neurons for flight. Proc. Natl Acad. Sci. USA 106, 10326–10331 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N. et al. Inositol 1,4,5-trisphosphate receptor and dSTIM function in Drosophila insulin-producing neurons regulates systemic intracellular calcium homeostasis and flight. J. Neurosci. 30, 1301–1313 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S. & Hasan G. Functional complementation of Drosophila itpr mutants by rat Itpr1. J. Neurogenet. 26, 328–337 (2012). [DOI] [PubMed] [Google Scholar]
- Banerjee S. Loss of flight and associated neuronal rhythmicity in inositol 1,4,5-trisphosphate receptor mutants of Drosophila. J. Neurosci. 24, 7869–7878 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. et al. An siRNA screen for NFAT activation identifies septins as coordinators of store-operated Ca2+ entry. Nature 499, 238–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S. & Cossart P. Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 13, 183–194 (2012). [DOI] [PubMed] [Google Scholar]
- Neufeld T. P. & Rubin G. M. The drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell 77, 371–379 (1994). [DOI] [PubMed] [Google Scholar]
- Adam J. C., Pringle J. R. & Peifer M. Evidence for functional differentiation among Drosophila septins in cytokinesis and cellularization. Mol. Biol. Cell 11, 3123–3135 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L. et al. Phylogenetic and evolutionary analysis of the septin protein family in metazoan. FEBS Lett. 581, 5526–5532 (2007). [DOI] [PubMed] [Google Scholar]
- Joshi R., Venkatesh K., Srinivas R., Nair S. & Hasan G. Genetic dissection of itpr gene function reveals a vital requirement in aminergic cells of Drosophila larvae. Genetics 166, 225–236 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N., Padmanabhan N. & Hasan G. Inositol 1,4,5-trisphosphate receptor function in Drosophila insulin producing cells. PLoS ONE 4, e6652 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M. & Lewis R. S. Potentiation and inhibition of Ca2+ release-activated Ca2+ channels by 2-aminoethyldiphenyl borate (2-APB) occurs independently of IP3 receptors. J. Physiol. 536, 3–19 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field C. M. et al. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J. Cell Biol. 133, 605–616 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H., Peifer M. & Pringle J. R. Localization and possible functions of Drosophila septins. Mol. Biol. Cell 6, 1843–1859 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Soriano V., Nieto-Arellano R. & Paricio N. Septin 4, the Drosophila ortholog of human CDCrel-1, accumulates in Parkin mutant brains and is functionally related to the Nedd4 E3 ubiquitin ligase. J. Mol. Neurosci. 48, 136–143 (2012). [DOI] [PubMed] [Google Scholar]
- Sellin M. E., Sandblad L., Stenmark S. & Gullberg M. Deciphering the rules governing assembly order of mammalian septin complexes. Mol. Biol. Cell 22, 3152–3164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase M., Okada S., Oguchi T. & Toh-e A. Forchlorfenuron, a phenylurea cytokinin, disturbs septin organization in Saccharomyces cerevisiae. Genes Genet. Syst. 79, 199–206 (2004). [DOI] [PubMed] [Google Scholar]
- Hu Q., Nelson W. J. & Spiliotis E. T. Forchlorfenuron alters mammalian septin assembly, organization, and dynamics. J. Biol. Chem. 283, 29563–29571 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman-Gavrila R. V., Hales K. G. & Wilde A. Anillin-mediated targeting of peanut to pseudocleavage furrows is regulated by the GTPase Ran. Mol. Biol. Cell 19, 3735–3744 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak T., Agrawal T., Richhariya S., Sadaf S. & Hasan G. Store-operated calcium entry through Orai is required for transcriptional maturation of the flight circuit in Drosophila. J. Neurosci. 35, 13784–13799 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal T., Sadaf S. & Hasan G. A genetic RNAi screen for IP3/Ca2+ coupled GPCRs in Drosophila identifies the PdfR as a regulator of insect flight. PLoS Genet. 9, e1003849 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y. et al. The GTP-binding protein Septin 7 is critical for dendrite branching and dendritic-spine morphology. Curr. Biol. 17, 1746–1751 (2007). [DOI] [PubMed] [Google Scholar]
- Tada T. et al. Role of septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr. Biol. 17, 1752–1758 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M. et al. Structural insight into filament formation by mammalian septins. Nature 449, 311–315 (2007). [DOI] [PubMed] [Google Scholar]
- Mavrakis M. et al. Septins promote F-actin ring formation by crosslinking actin filaments into curved bundles. Nat. Cell Biol. 16, 322–334 (2014). [DOI] [PubMed] [Google Scholar]
- Yuan J. P. et al. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 11, 337–343 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally B. A., Somasundaram A., Yamashita M. & Prakriya M. Gated regulation of CRAC channel ion selectivity by STIM1. Nature 482, 241–245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calloway N. et al. Stimulated association of STIM1 and Orai1 is regulated by the balance of PtdIns(4,5)P2 between distinct membrane pools. J. Cell Sci. 124, 2602–2610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maléth J., Choi S., Muallem S. & Ahuja M. Translocation between PI(4,5)P2-poor and PI(4,5)P2-rich microdomains during store depletion determines STIM1 conformation and Orai1 gating. Nat. Commun. 5, 5843 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing J. et al. Proteomic mapping of ER–PM junctions identifies STIMATE as a regulator of Ca2+ influx. Nat. Cell Biol. 17, 1339–1347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana A. et al. TMEM110 regulates the maintenance and remodeling of mammalian ER-plasma membrane junctions competent for STIM-ORAI signaling. Proc. Natl Acad. Sci. USA 112, E7083–E7092 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon M. B. et al. Genetic deletion of SEPT7 reveals a cell type-specific role of septins in microtubule destabilization for the completion of cytokinesis. PLoS Genet. 10, e1004558 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S. et al. Analysis of conditional paralytic mutants in Drosophila sarco-endoplasmic reticulum calcium ATPase reveals novel mechanisms for regulating membrane excitability. Genetics 169, 737–750 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S. et al. Compensation of inositol 1,4,5-trisphosphate receptor function by altering sarco-endoplasmic reticulum calcium ATPase activity in the Drosophila flight circuit. J. Neurosci. 26, 8278–8288 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorova P., Popugaeva E. & Bezprozvanny I. Disturbed calcium signaling in spinocerebellar ataxias and Alzheimer’s disease. Semin. Cell Dev. Biol. 40, 127–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Leemput J. et al. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet. 3, 1076–1082 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G. et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156 (2007). [DOI] [PubMed] [Google Scholar]
- Rubin G. M. et al. A Drosophila complementary DNA resource. Science 287, 2222–2224 (2000). [DOI] [PubMed] [Google Scholar]
- Wu C. F., Suzuki N. & Poo M. M. Dissociated neurons from normal and mutant Drosophila larval central nervous system in cell culture. J. Neurosci. 3, 1888–1899 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie R. C. INDO-1 measurements of absolute resting and light-induced Ca2+ concentration in Drosophila photoreceptors. J. Neurosci. 16, 2924–2933 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty R. P. in 11th Aeroacoustics Convference 1–13 (Monterey, CA, USA, 2005).
- Schneider C. A., Rasband W. S. & Eliceiri K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1 - 7
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on request.