ABSTRACT
Tumor necrosis factor alpha (TNF) plays a critical role in the control of Mycobacterium tuberculosis, in part by augmenting T cell responses through promoting macrophage phagolysosomal fusion (thereby optimizing CD4+ T cell immunity by enhancing antigen presentation) and apoptosis (a process that can lead to cross-priming of CD8+ T cells). M. tuberculosis can evade antituberculosis (anti-TB) immunity by inhibiting host cell TNF production via expression of specific mycobacterial components. We hypothesized that M. tuberculosis mutants with an increased capacity to induce host cell TNF production (TNF-enhancing mutants) and thus with enhanced immunogenicity can be useful for vaccine development. To identify mycobacterial genes that regulate host cell TNF production, we used a TNF reporter macrophage clone to screen an H37Rv M. tuberculosis cosmid library constructed in M. smegmatis. The screen has identified a set of TNF-downregulating mycobacterial genes that, when deleted in H37Rv, generate TNF-enhancing mutants. Analysis of mutants disrupted for a subset of TNF-downregulating genes, annotated to code for triacylglycerol synthases and fatty acyl-coenzyme A (acyl-CoA) synthetase, enzymes that concern lipid biosynthesis and metabolism, has revealed that these strains can promote macrophage phagolysosomal fusion and apoptosis better than wild-type (WT) bacilli. Immunization of mice with the TNF-enhancing M. tuberculosis mutants elicits CD4+ and CD8+ T cell responses that are superior to those engendered by WT H37Rv. The results suggest that TNF-upregulating M. tuberculosis genes can be targeted to enhance the immunogenicity of mycobacterial strains that can serve as the substrates for the development of novel anti-TB vaccines.
IMPORTANCE
One way to control tuberculosis (TB), which remains a major global public health burden, is by immunization with an effective vaccine. The efficacy of Mycobacterium bovis BCG, the only currently approved TB vaccine, is inconsistent. Tumor necrosis factor alpha (TNF) is a cytokine that plays an important role in controlling TB. M. tuberculosis, the causative agent of TB, can counter this TNF-based defense by decreasing host cell TNF production. This study identified M. tuberculosis genes that can mediate inhibition of TNF production by macrophage (an immune cell critical to the control of TB). We have knocked out a number of these genes to generate M. tuberculosis mutants that can enhance macrophage TNF production. Immunization with these mutants in mice triggered a T cell response stronger than that elicited by the parental bacillus. Since T cell immunity is pivotal in controlling M. tuberculosis, the TNF-enhancing mutants can be used to develop novel TB vaccines.
INTRODUCTION
Tuberculosis (TB) is one of the deadliest infectious diseases worldwide (1). It has been estimated that there were 9 million new cases of TB globally in 2013 and that 1.5 million persons died from diseases caused by the tubercle bacillus in that year (1). The propensity of Mycobacterium tuberculosis to persist in an infected host is conducive to the development of persister organisms that are difficult to treat (2). As a result, it takes, on average, 6 to 9 months of multidrug chemotherapy to effectively treat tuberculous infection (3). This requirement causes problems concerning compliance as well as drug toxicity issues, rendering treatment of TB a highly challenging task (1, 3). The emergence of multidrug-resistant and extensively drug-resistant strains of M. tuberculosis presents yet another obstacle to effective TB treatment (1, 3). This hindrance is further complicated by the increased susceptibility to M. tuberculosis of individuals infected with the human immunodeficiency virus (HIV), a pathogen that continues to be a public health threat, as evidenced by the prevalence of HIV/TB coinfection (1, 3). Thus, more effective anti-TB intervention is urgently needed.
Immunization can be an efficacious and cost-effective measure to control infectious diseases (4). For example, the measles vaccine, which is highly efficacious and costs about $17 per disability-adjusted life year, represents a most cost-effective intervention against an infectious agent in developing countries (5). A vaccine of such quality is, however, lacking for the prevention of TB. The difficulty in developing an effective anti-TB vaccine despite the urgent need for one is at least partially due to our lack of understanding of the correlates of protection in tuberculous infection in molecular and biochemical terms (6). The efficacy of Mycobacterium bovis BCG, the only approved TB vaccine in use today, is inconsistent (7).
Proper containment of M. tuberculosis requires the development of optimal innate and adaptive immune responses, and most healthy individuals can control a tuberculous infection upon exposure to the tubercle bacillus (8 – 10). The mechanisms by which an infected host controls M. tuberculosis are, however, not clearly defined (6, 8 – 11). Tumor necrosis factor alpha (TNF), a cytokine with a diverse cellular source, has been shown to play a critical role in mice and nonhuman primates in host defense against M. tuberculosis during both the acute phase and the chronic persistent phase of infection (12 – 14). The enhanced risks for TB observed in individuals receiving anti-TNF therapies for a variety of inflammatory diseases have provided strong evidence that this cytokine plays an important role in mediating host defense mechanisms to prevent reactivation of latent TB (15, 16). Excessive TNF production can, however, result in the development of tissue-damaging immunopathology (12 – 14). Thus, it is generally thought that TNF production during M. tuberculosis infection is tightly controlled in order to attain optimal expression of this cytokine so as to contain the tubercle bacillus without collateral damage (14).
Although the precise mechanisms by which TNF mediates antimycobacterial activity remain to be elucidated, evidence exists that this cytokine can enhance phagosome-lysosome maturation (17), a process that promotes antimycobacterial activity, as well as antigen presentation, the latter process capable of enhancing CD4+ T cell response (18). Additionally, TNF can promote apoptosis in mycobacterium-infected macrophages (19, 20), an event that can lead to cross-priming of CD8+ T cells (21). Since T cell responses to M. tuberculosis and to immunization play an important role in the control of TB and in the development of vaccine-engendered protective immunity, respectively (6, 8 – 11, 22), TNF, via its ability to promote phagosome-lysosome maturation and macrophage apoptosis, can potentially enhance T cell-dependent antimycobacterial host defense mechanisms as well as vaccine immunogenicity and efficacy. A corollary of this notion is the possibility that, as a most adept intracellular pathogen, M. tuberculosis may downregulate host cell TNF production in order to evade the host antituberculous immune mechanisms. Indeed, it has been demonstrated that specific M. tuberculosis genes encode mycobacterial components that can modulate host cell TNF expression, including those that downregulate macrophage production of the cytokine (14, 23 – 25). Of note, certain mutant M. tuberculosis strains deficient in such downregulating elements have been shown to be attenuated for virulence (14, 23 – 25).
Together, the above-described observations prompted us to hypothesize that targeting TNF-downregulating mycobacterial genes can lead to the generation of mutant strains with enhanced immunogenicity that can be exploited to develop effective vaccine candidates. We have developed a genetic screen that has identified a set of mycobacterial genes whose disruption resulted in H37Rv deletion mutants that could stimulate macrophage TNF production at levels higher than that elicited by WT M. tuberculosis. In line with the property of TNF, these mutants, compared to parent bacilli, displayed an enhanced capacity to promote macrophage phagolysosomal fusion and apoptosis. Importantly, mice immunized with these TNF-enhancing mutants engendered CD4+ and CD8+ T cell responses superior to those elicited by WT H37Rv. These studies have provided evidence supporting the notion that targeting TNF-attenuating components in mycobacteria can produce strains with enhanced immunogenicity and therefore that this approach represents a viable approach for the rational design of efficacious TB vaccines.
RESULTS
Generation of a TNF reporter macrophage screening system.
In order to comprehensively identify M. tuberculosis genes that mediate functions that enable the tubercle bacillus to downregulate host cell TNF production, we chose a nonbiased genetic approach, using a platform comprising two components: (i) a macrophage system capable of reporting TNF expression and (ii) an M. tuberculosis cosmid library in the heterologous M. smegmatis strain that enables a gain-of-function screen in infected macrophages. The macrophage was chosen as the surrogate in vitro host to study TNF expression upon interaction with the tubercle bacillus because well-studied robust cell lines exist for this immune cell and are amenable to genetic manipulation (26). In addition, the macrophage is a preferred niche for the tubercle bacillus in vivo and has been used extensively to study phagosome maturation, antigen presentation, and apoptosis (17 – 20), processes that are relevant to the present study. Green fluorescent protein (GFP)-based signal was chosen to assess TNF production as this approach enables an expeditious readout. Further, we have previously used a similar reporter system to study how M. tuberculosis modulates macrophage interleukin-12 (IL-12) production (26).
To generate a TNF-reporter macrophage system, a J774.16 mouse macrophage line was stably transfected with a TNF promoter–humanized recombinant GFP (hrGFP) II-1 fusion (TNFp-hrGFP II-1) (Fig. 1A). The ability of the transfectants to report TNF expression was assessed by fluorescence microscopy as well as by flow cytometric analysis upon treatment with lipopolysaccharide (LPS), a potent TNF inducer (27) (Fig. 1B and D, left panel) and after infection with BCG (Fig. 1C). The latter observation demonstrated that the TNF promoter of this clone could be activated in response to mycobacterial infection, as assessed by the production of GFP signals, a prerequisite for the reporter macrophage system of the proposed genetic screen (Fig. 1C). Stable transfectants were subjected to limiting dilution, and one clone, designated C10, was chosen for further testing based on its level of responsiveness to LPS, as assessed both quantitatively and qualitatively (Fig. 1D, left panel). The functionality of clone C10 in terms of its ability to differentiate mycobacterial isolates with disparate macrophage TNF-inducing capacities was then examined. C10 cells were infected separately with two M. tuberculosis deletion mutants (the ΔrpfAB mutant, deleted for both the rpfA [resuscitation promoting factor A] and rpfB alleles, genes that code for apparent peptidoglycan hydrolases [28], and the ΔsecA2 mutant, deleted for the accessory secretion factor of the Sec-dependent protein export pathway) (23) that have previously been shown to induce macrophage TNF production at levels higher than that elicited by WT bacilli. The results demonstrated that the C10 macrophage clone could discriminate Mycobacterium strains with differential TNF-inducing capacities (Fig. 1D, right panel). C10 was used for the screen to identify TNF-regulating genes throughout this study.
FIG 1 .
Generation of a C10 TNF reporter macrophage screen system. (A) The TNF promoter (TNFp) used for the generation of the GFP reporter was derived from a fragment released by KpnI and BamHI digestion of the “Pro-UTR” construct (kindly provided by Jiahuai Han [27]). The released TNFp was cloned into the modified phrGFP II-I vector (m-phrGFP II-1), whose CMV promoter has been deleted, upstream from the hrGFP II-1 to generate the TNFp-rhGFP II-1 fusion reporter construct contained in the m-phrGFP II-1 vector (designated TNFp-hrGFP II-1). (B) The TNFp-hrGFP II-1 fusion construct was stably transfected into J774.16 macrophages. G418 (1 mg/ml)-resistant transfectants were selected for and observed for GFP signals upon LPS treatment (1 µg/ml) by fluorescence microscopy. (C) J774.16 macrophages stably transfected with TNFp-hrGFP II-1 respond to BCG infection (multiplicity of infection of 10:1). (D) Limiting dilution of stably transfected GFP-positive macrophages yielded clone C10, which responded well to LPS to generate GFP signal quantitatively and qualitatively (left panel). Importantly, C10 can differentiate between M. tuberculosis mutant strains with an enhanced capacity to induce macrophage TNF production (the ΔrpfAB and ΔsecA2 mutants) relative to WT bacilli (right panel). 10:1, multiplicity of infection of 10 bacilli to 1 macrophage.
Identification of M. tuberculosis genes involved in downregulating macrophage TNF production.
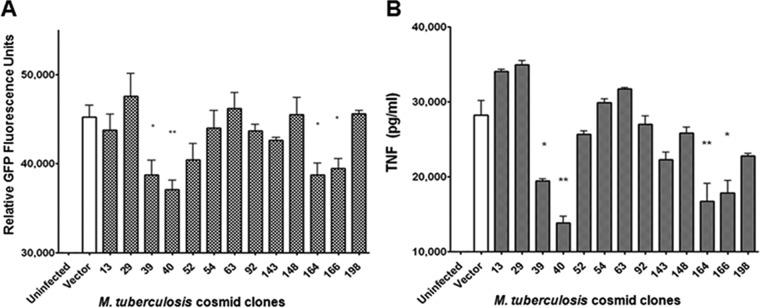
Certain relatively avirulent mycobacterial strains stimulate a higher level of expression of TNF in infected macrophage cultures than virulent pathogenic strains (29 – 31). Relevant to the present study, it has been shown that the ability of the relatively avirulent M. smegmatis species to induce TNF production significantly exceeds that of the virulent M. tuberculosis H37Rv strain in both infected human and infected mouse macrophage cultures (29, 30). In addition, virulent M. tuberculosis, upon deletion of specific genes that lead to deficiency in certain bacterial components, exhibits an increased capacity to induce macrophage TNF production relative to the WT parental strain (23 – 25). These observations have led us to posit that the relatively avirulent M. smegmatis species could be used as the substrate for a nonbiased gain-of-function (attenuation of TNF production) screen to identify M. tuberculosis genes that mediate TNF-regulating functions. Accordingly, we initiated experiments to generate an H37Rv cosmid library in the heterologous M. smegmatis species for this screen. pYUB412-based H37Rv cosmids derived from a library built in Escherichia coli (32) were transformed into M. smegmatis mc2155 (33), resulting in the generation of 105 clones that cover ~50% of the M. tuberculosis genome. The pYUB412-based cosmids exist extrachromosomally in E. coli but integrate into the chromosome upon transformation into M. smegmatis (32). All 105 clones were screened by the C10 reporter cells, whose capability of differentiating mycobacteria with disparate capacities to induce macrophage expression of TNF had been demonstrated (Fig. 1D, right panel). Of these, four clones (clones 39, 40, 164, and 166), which together represented a total of 129 genes, consistently displayed TNF-downregulating activity, eliciting GFP signal levels that were significantly lower than that engendered by WT mc2155 harboring the empty pYUB412 vector containing no H37Rv DNA (Fig. 2A). Enzyme-linked immunosorbent assay (ELISA) analysis of the supernatants of C10 cultures infected with each of the 4 clones for levels of TNF production confirmed their ability to downregulate macrophage production of the cytokine (Fig. 2B). Thus, the relative intensities in GFP signal correspond to relative levels of TNF present in cell supernatants, demonstrating the fidelity of the C10 clone results in reporting TNF expression by macrophages upon infection with M. tuberculosis (Fig. 2).
FIG 2 .
Screening of an M. tuberculosis cosmid library generated in heterologous host M. smegmatis strain mc2155 yielded M. tuberculosis cosmid clones with a TNF-downregulating phenotype. Clone C10 macrophages were infected at a multiplicity of infection (MOI) of 10 with M. smegmatis containing either empty vector (pYUB412 containing no H37Rv DNA) or M. tuberculosis cosmid clones (a total of 105 clones covering approximately half of the M. tuberculosis genome were screened). From the original transformation reaction, three independent colonies were picked for each cosmid clone. Therefore, three sets of the 105 cosmid clone library (named “a,” “b,” and “c”) were stocked. The screen of the 105 cosmid clones (from set “a”) was conducted in triplicate in 96-well plates, and the experiments were repeated 2 to 3 times. Clones with the TNF-downregulating phenotype underwent a second round of C10 screening with a similar setup. Clones that consistently induced GFP signal at a level lower than that generated by M. smegmatis transformed with empty vector were identified. To stringently test the TNF-downregulating phenotype of these second-round hits, corresponding independent clones from set “b” were tested. This third round of screening identified four clones (clones 39, 40, 164, and 166) with a consistent TNF-downregulating phenotype as assessed by GFP fluorescence signal (A) or ELISA measurement of the level of TNF in the supernatants of infected C10 cultures (B). There was a 100% concordance between the C10 fluorescence signal readout and the ELISA-based quantification of TNF production. Values are the means of the results of triplicates ± standard deviations (SD) and are representative of three separate experiments. *, P < 0.05; **, P < 0.005.
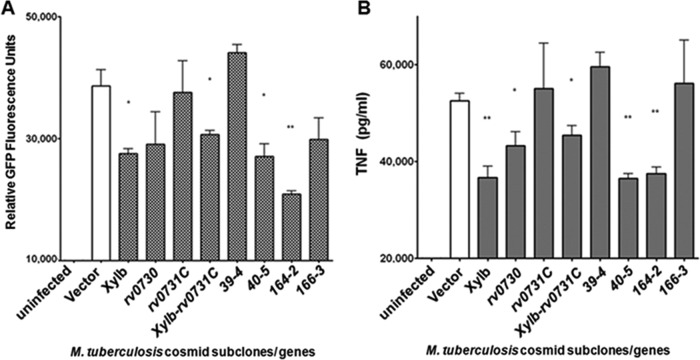
To deconvolute the M. tuberculosis genes within each positive hit, we employed a subcloning strategy. Expressing delimited H37Rv DNA fragments in M. smegmatis under the control of the hsp60 promoter via the integrating vector pMV361 (32), we have identified M. tuberculosis genes in a set of subclones that induced GFP signal at levels higher than that observed with WT mc2155 transformed with pMV361 containing no tuberculous DNA (Fig. 3A). One subclone, designated xylB-Rv0731c, represented the overlapping regions of two contiguous TNF-downregulating clones (clones 39 and 40; see Table S1 in the supplemental material) identified by the original screen of the M. tuberculosis cosmid library constructed in mc2155 (Fig. 2). That TNF-downregulating clones 39 and 40 shared a three-gene overlapping region led us to directly clone the xylB-rv0730-rv0731c fragment into pMV361 to assess its TNF-modulating capacity. rv0729 (xylB) is a putative d-xylulose kinase that can participate in the synthetic pathway of arabinose (34, 35). rv0730 has been annotated to encode a GCN5-related N-acetyltransferase (GNAT family of N-acetyltransferases) that can potentially cross-link peptidoglycans (36). rv0731c has been annotated as an S-adenosylmethionine-dependent methyltransferase which may participate in mycolic acid synthesis (37). Therefore, all three genes located in the overlapping regions of clones 39 and 40 have been annotated to mediate functions that can have an impact on carbohydrate (xylB and rv0730) and lipid (rv0731c) metabolism.
FIG 3 .
Deconvolution of TNF-downregulating M. tuberculosis cosmid clones by subclone analysis. Subclones of the original 4 TNF-downregulating hits (clones 39, 40, 163, and 166) as well as individual genes (xylB, rv0730, and rv0731c) were cloned into pMV361 under the control of the hsp60 promoter and transformed into M. smegmatis mc2155. The cosmid subclones in M. smegmatis were stored in triplicate (sets “a,” “b,” and “c”), and screening for the TNF-regulating activities of the various subclones was conducted as described in the Fig. 2 legend. The subclones underwent two rounds of C10 screening, and the third confirmatory round of screening tested corresponding subclones picked from independent set “b” of the stored stock. C10 were infected at an MOI of 10 in triplicate. Subclones or individual genes contained in the original 4 cosmid hits were assessed for their capacity to induce macrophage TNF expression based on GFP fluorescence signal (A) and direct ELISA measurement of the level of this cytokine in culture supernatants (B). Values are the means of the results of triplicates ± SD and are representative of three separate experiments. *, P < 0.05; **, P < 0.005.
Analysis of clone 40 revealed yet another subclone, named 40-5, that could mediate TNF-downregulating activity in infected C10 cultures (Fig. 3). Subclone 40-5 encompasses rv0755A (a fragment of a putative transposase), thrV (anticodon tRNA-Thr), and rv0756c (hypothetical protein). Subclone 164-2, which contains three genes from the acid-responsive mymA operon (38, 39), rv3087, rv3088, and rv3089, also displayed significant TNF-downregulating capacity relative to the controls, as assessed by GFP expression (Fig. 3A). Both M. tuberculosis rv3087 and rv3088 have been annotated as encoding putative triacylglycerol (TAG) synthases, and rv3089 has been annotated as encoding FadD13, a potential fatty acyl-coenzyme A (acyl-CoA) synthetase. Bioinformatic analysis thus suggests that all three genes contained in subclone 164-2 can mediate functions that can potentially participate in lipid biosynthesis and metabolism.
In sum, of the nine genes located in the TNF-downregulating subclones, six have been annotated with functions that may participate in lipid (rv0731c, rv3087, rv3088, and rv3089) or carbohydrate (rv0729 and rv0730) metabolic and biosynthetic pathways. Since lipids and carbohydrates are major constituents of the M. tuberculosis cell envelope (40, 41), one possible way that these mycobacterial TNF-downregulating genes might modulate C10 production of the cytokine is by modifying the chemical composition of the outer surface of the tubercle bacillus, thereby altering the interaction between M. tuberculosis and macrophages. The TNF-downregulating property of subclones 40-5 and 164-2, xylB-rv0730-rv0731c, and individual genes xylB and rv0370, as assessed by GFP signal of infected C10 macrophages (Fig. 3A), was confirmed by direct ELISA quantification of the amount of the cytokine in the corresponding culture supernatants (Fig. 3B). The only discordance was apparent in the analysis of rv0730, which displayed lower but statistically nonsignificant C10 fluorescence signal but was found to produce significant TNF downregulation based on ELISA analysis. The latter results, similarly to those presented in Fig. 2, again illustrate the reliability of the C10 reporter system.
M. tuberculosis mutants deleted for TNF-downregulating genes display an enhanced capacity to induce TNF production by infected macrophage cultures.
The C10 screen of 105 M. smegmatis clones harboring H37Rv DNA identified a set of M. tuberculosis genes that could mediate functions involved in downregulation of the capacity of mc2155 to induce macrophage TNF production. Because these genes, when knocked into heterologous M. smegmatis, reduced TNF production of infected C10, we reasoned that the respective M. tuberculosis deletion mutants would be strains that would enhance the expression of the cytokine in infected macrophage cultures. To begin testing the possibility that such TNF-upregulating mutants could exhibit enhanced immunogenicity, we chose to focus on the two genes encoding triacylglycerol synthases (rv3087 and rv3088) and the gene encoding fatty-acyl-CoA synthetase (fadD13). The choice to focus on analyzing the Δrv3087, Δrv3088, and Δrv3089 mutants was prompted by the fact that, while the validity of the functional annotation of xylB, rv0730, rv0755A, thrV, and rv0756c has not been tested, results obtained from biochemical analysis of the products of the two genes encoding triacylglycerol synthases (rv3087 and rv3088) and of the gene encoding fatty acyl-CoA synthetase (rv3089) in in vitro systems support the validity of their function assignments (42 – 44). In addition, since the two putative triacylglycerol synthases can possibly affect lipid metabolism and biosynthetic pathways via overlapping or distinct functions, a mutant doubly deleted for rv3087 and rv3088 (the Δrv3087 Δrv3088 mutant) was generated to examine whether the resultant strain displayed an additive or a synergistic TNF-downregulating effect. Finally, since two of the TNF-downregulating genes have been annotated as encoding triacylglycerol synthases, we tested the Δrv3130c strain, a mutant deleted for Tgs1 (encoded by a gene located outside the mymA operon and reportedly the most enzymatically active of the 15 potential M. tuberculosis triacylglycerol synthases [42]), an enzyme that has been shown to play a pivotal role in tuberculous pathogenesis (45).
Indeed, all five H37Rv mutants tested—the Δrv3087, Δrv3088, Δrv3087 Δrv3088, ΔfadD13, and Δtgs1 mutants—demonstrated the ability to enhance the capacity of WT M. tuberculosis to induce macrophage TNF production upon infection, as assessed by ELISA study (Fig. 4). To show that this TNF-modulating effect was specific to the deleted alleles, complementation studies were conducted, and in each case, expression of the complementing gene under the control of the hsp60 promoter via the integrating pMV361 reversed the TNF-upregulating phenotype of the corresponding deletion mutant. These results essentially confirm that the TNF-upregulating phenotype of the various H37Rv knockouts studied was specific to the deleted genes and validate our hypothesis that M. tuberculosis strains with an enhanced capacity to induce macrophage TNF production can be generated by targeting genes that mediate downregulation of the expression of this cytokine in infected macrophages.
FIG 4 .
Analysis of H37Rv mutants deleted for genes harbored in TNF-downregulating subclone 164-2 identified specific M. tuberculosis alleles that mediate functions capable of attenuating C10 macrophage TNF production. Subclone 164-2 harbors rv3087 and rv3088 (both annotated to code for triacylglycerol synthase) and rv3089 (annotated to encode a fatty acyl-CoA synthetase), enzymes that are involved in lipid metabolic and synthesis pathways. J774.16 macrophages were infected at an MOI of 10 with H37Rv, deletion mutants, or complemented strains (designated by the suffix “.c”) or were left untreated. At 16 h postinfection, the amounts of TNF in culture supernatants were quantified by ELISA. All deletion mutants induced significantly higher production of TNF by infected macrophage cultures than H37Rv. The TNF-upregulating phenotype of the deletion mutants can be reversed by complementation with the corresponding WT genes. Values are the means of the results of triplicates ± SD and are representative of three separate experiments. *, P < 0.05; **, P < 0.005; ***, < 0.001.
TNF-enhancing M. tuberculosis mutants affect endosomal processing.
Ample experimental evidence supports the notion that virulent M. tuberculosis strains employ a wide array of strategies to arrest phagosomal maturation, thereby avoiding the hostile environment of the lysosome (46 – 49). One of the consequences of the ability of the tubercle bacillus to prevent phagolysosomal fusion could be suboptimal presentation of antigens to T cells. It has been previously demonstrated that TNF promotes phagolysosomal fusion (17) and, as a result, could enhance CD4+ T cell responses by optimizing antigen processing and presentation via the major histocompatibility complex class II (MHC-II) pathway (18). It is therefore possible that M. tuberculosis TNF-enhancing mutants can promote phagolysosomal fusion in infected macrophages, which, in turn, can lead to augmentation of T cell immunity. To begin testing this possibility, the level of fusion with lysosomes of phagosomes containing WT H37Rv was compared to the level seen with those harboring TNF-enhancing mutants. J774.16 macrophages were infected with WT M. tuberculosis H37Rv and the various TNF-enhancing deletion mutants labeled with fluorescein-5-isothiocyanate (FITC). Rab5 and LAMP1 (lysosome-associated membrane protein 1), the markers for early and late endosomal compartments, respectively, were tracked via the use of fluorescently tagged specific antibodies (Abs) against these surface molecules. Colocalization of FITC-tagged bacilli and the fluorescently labeled Rab5 and LAMP1 was assessed by confocal fluorescence microscopy in a blind fashion. The results revealed that WT H37Rv preferentially localized to the early endosomal compartment. The levels of localization of all five TNF-enhancing M. tuberculosis mutants to the Rab5-positive early endosomal compartments were significantly lower than those seen with WT H37Rv (Fig. 5A and C). Conversely, all five mutants localized to the LAMP1-positive late endosomal vesicles at a level higher than that observed with the WT parental strain (Fig. 5B and D). Together, these results lend support to the notion that targeting TNF-downregulating M. tuberculosis genes is a viable strategy to generate TNF-enhancing strains that, relative to WT bacilli, are attenuated for their capacity to block phagosome-lysosome fusion and, therefore, should promote antigen presentation, thereby augmenting the CD4+ T cell response.
FIG 5 .
M. tuberculosis mutants with the TNF-upregulating property enhance phagolysosomal fusion in infected macrophages. (A and B) J774.16 cells were plated on 8-well chamber slides and allowed to adhere overnight. The next day, macrophages were infected with M. tuberculosis H37Rv or with various TNF-enhancing mutants or were left untreated. Cells were washed after 4 h to remove extracellular bacteria. After an additional 4 h, samples were fixed and reacted with antibodies against Rab5 or LAMP1, followed by staining with a secondary fluorescent antibody. Compared to WT H37Rv, all TNF-enhancing mutants had decreased colocalization with Rab5-positive (Rab5+) endosomes (A) but preferentially colocalized with LAMP1+ endosomes (B). This result indicated that the capacity of the TNF-enhancing M. tuberculosis mutants to promote phagosome maturation is superior to that of WT H37Rv. Values are the means of the results of triplicates ± SD and are representative of three separate experiments. *, P < 0.05; **, P < 0.005; ***, < 0.001. (C and D) Representative photomicrographs from confocal microscopic study depicting colocalization of FITC-labeled M. tuberculosis Δrv3087 Δrv3088 deletion mutant bacilli (green) with the Rab5+ (red) (C) or LAMP1+ (red) (D) endosomal compartments. Yellow coloring represents colocalization.
TNF-enhancing M. tuberculosis mutants promote macrophage apoptosis upon infection.
Apoptosis of M. tuberculosis-infected macrophages is beneficial to the host in that the process can kill tubercle bacilli and the apoptotic vesicles can be usurped to enhance T cell responses by cross-priming CD8+ T cells (21, 50). TNF can promote apoptosis in mycobacterium-infected macrophages (19, 20). We therefore proposed that M. tuberculosis bacteria that have been genetically engineered to upregulate TNF expression in infected macrophages represent strains with enhanced immunogenicity due to the effect of this cytokine on apoptosis. Adept in evading the host immune response, the tubercle bacillus has evolved means to counter this host defense mechanism. Indeed, antiapoptotic M. tuberculosis factors have been shown to impart tuberculous virulence (23, 51, 52). Relatively avirulent mycobacteria are superior to virulent strains in inducing macrophage apoptosis, and this process is dependent on the presence of TNF (53), a well-established inducer of the extrinsic pathway of apoptosis (50). Further, compared to avirulent mycobacteria, virulent M. tuberculosis has been shown to downregulate the expression of proapoptotic genes in infected macrophages (20). These observations have prompted us to speculate that the M. tuberculosis Δrv3087, Δrv3088, Δrv3087 Δrv3088, ΔfadD13, and Δtgs1 deletion mutants, which exhibited an enhanced capacity to induce macrophage TNF production, can be more proapoptotic than WT H37Rv. Macrophage apoptosis was monitored by assessing the activity of effector caspases 3 and 7 using the caspase inhibitor-based FLICA (fluorochrome-labeled inhibitors of caspases) assay, in conjunction with laser scanning cytometry. The results depicted in Fig. 6 reveal that, indeed, TNF-enhancing M. tuberculosis strains stimulated significantly higher levels of activated caspases than the WT strain, demonstrating the proapoptotic property of these mutants.
FIG 6 .
M. tuberculosis mutants with the TNF-upregulating property enhance apoptosis of infected macrophages. (A)J774.16 cells were plated overnight on a 96-well plate (MatriPlate; Brooks Life Science System). The next day, cells were infected with H37Rv or TNF-enhancing mutants or left untreated. After 16 h, samples were analyzed using the caspase inhibitor-based FLICA (fluorochrome-labeled inhibitors of caspases) assay. TNF-enhancing mutants induced significantly more apoptosis of infected macrophages than the WT bacilli. Values are the means of the results of triplicates ± SD and are representative of three separate experiments. *, P < 0.05; **, P < 0.005; ***, < 0.001. (B to H) Representative photomicrographs captured by laser scanning cytometric analysis of J774.16 cells stained with FLICA (green) and DAPI (4′,6-diamidino-2-phenylindole) (blue).
Correlation between the phagosome maturation and apoptosis phenotypes of the TNF-upregulating M. tuberculosis mutants and enhanced TNF production in infected macrophages.
Apoptosis and phagolysosomal fusion are highly regulated processes that involve complex mechanisms, including those that are TNF independent (46 – 50). Consequently, we initiated studies to probe the TNF specificity of the apoptosis- and phagolysosomal fusion-promoting phenotypes of the TNF-upregulating mutants using the TNF-neutralizing monoclonal antibody (MAb) MP6-XT22. For these studies, the TNF-downregulating rv3089 (fadD13) and rv3130c (tgs1) alleles were evaluated using the corresponding single deletion mutants, while rv3087 and rv3088 were assessed in the context of the Δrv3087 Δ3088 double knockout strain. The results of these experiments have shown that the phagolysosomal fusion-promoting capacity of the three deletion mutants examined, as assessed by colocalization of FITC-labeled bacilli with late endosomal compartments, can be attenuated by the TNF neutralization (Fig. 7A). However, the MP6-XT22-mediated attenuation did not cause the level of colocalization to revert to that observed in TNF-neutralized, WT H37Rv-infected macrophage cultures. This partial TNF dependency of the colocalization of the mutant strains with LAMP1-positive compartments in infected macrophages suggests that TNF-independent mechanisms are operative in regulating phagosome maturation by the Δrv3087 Δrv3088, Δrv3089 (ΔfadD13), and Δrv3130c (Δtgs1) mutants (Fig. 7A). The MP6-XT22 in vitro infection system was also utilized to examine the relationship between the apoptosis-promoting attribute of the various mutant strains and their TNF-enhancing capacity. The results of these experiments have revealed that TNF neutralization resulted in a decrease in the level of apoptosis in macrophages infected with the Δrv3087 Δ3088 mutant, the Δrv3089 (ΔfadD13) mutant, or the Δrv3130c (Δtgs1) H37Rv mutant (Fig. 7B). This decrease, however, reached statistical significance only in cultures infected with the Δrv3087 Δ3088 (P < 0.05) mutant or the Δrv3089 (ΔfadD13) (P < 0.05) mutant and not in those infected with the Δrv3130c (Δtgs1) strain (Fig. 7B). As in the phagosome maturation study, TNF neutralization fell short of attenuating the apoptosis seen in cultures infected with the Δrv3087 Δrv3088 mutant or the Δrv3089 (ΔfadD13) mutant to a level comparable to that observed in MP6-XT22-treated macrophages harboring WT M. tuberculosis (Fig. 7B). Together, these results suggest that the ability of the TNF-upregulating mutants to promote apoptosis and phagosome maturation is partially due to their ability to enhance macrophage production of this cytokine and that the TNF specificity of the phenotypes can be gene dependent.
FIG 7 .
Correlation between the phagosome maturation and apoptosis phenotypes of the various M. tuberculosis mutants and their capacity to enhance macrophage TNF production. (A) For the phagosome maturation study, 1 × 105 cells of J774.16 macrophages per well were cultured in eight-well chambered slides with or without the TNF-neutralizing MP6-XT22 MAb (final concentration, 10 µg/ml) for 16 h. The macrophages, in culture medium with or without MP6-XT22, were then synchronously infected with FITC-stained WT M. tuberculosis H37Rv or the various TNF-upregulating mutants. Macrophages were washed to remove extracellular bacilli after 4 h of infection, and cultures were replenished with medium with or without MP6-XT22. Cells were fixed with 4% paraformaldehyde after incubation for an additional 4 h, permeabilized, and allowed to react with primary antibodies against LAMP1, followed subsequently by staining with fluorescently tagged secondary antibodies. Analysis performed with confocal microscopy has revealed that colocalization of M. tuberculosis mutants with LAMP1 can be significantly attenuated by TNF neutralization but that the attenuation did not attain the level of that observed in MP6-XT22-treated, WT bacillus-infected macrophages. (B) For the apoptosis study, J774.16 cells were similarly cultured, in the presence or absence of MP6-XT22, in wells of 96-well plates (MatriPlate; Brooks Life Science System) at 1 × 105 macrophages per well. Cultures were infected with the WT or with the various mutant strains of M. tuberculosis for 4 h and were then incubated for 16 h after removal of extracellular bacilli before being subjected to analysis for evidence of apoptosis using a FAM-FLICA polycaspase kit. Analysis by laser scanning cytometry (iCys; Thorlabs) has revealed that, relative to the levels seen with the MP6-XT22-treated WT H37Rv-infected cultures, the apoptosis-promoting capacity of the Δrv3087 Δrv3088 and Δrv3089 (ΔfadD13) strains can be partially and significantly attenuated upon TNF neutralization. MP6-XT22 treatment had no significant effects on the apoptosis-enhancing capacity of the Δrv3130c (Δtgs1) strain. Values are the means of the results of triplicates ± SD. *, P < 0.05; **, P < 0.005.
Immunization with TNF-enhancing M. tuberculosis mutants elicited a T cell response superior to that engendered by WT bacilli.
The phagolysosomal fusion- and apoptosis-promoting properties of the TNF-enhancing M. tuberculosis mutants predicted that these strains should exhibit enhanced immunogenicity, in part via optimization of antigen presentation to CD4+ T cells through the MHC-II pathway (17) and cross-priming of CD8+ T cells (21), respectively. To determine the immunogenicity of the TNF-enhancing strains, C57BL/6 mice were immunized subcutaneously with 1 × 106 CFU of WT H37Rv or the various deletion mutants. The vaccination-engendered T cell response was assessed by quantification of the level of gamma interferon (IFN-γ)-producing splenic CD4+ and CD8+ T cells at 1 month postinoculation using the enzyme-linked immunosorbent spot (ELISPOT) assay. The results shown in Fig. 8 demonstrate that all five TNF-enhancing M. tuberculosis deletion mutants (the Δrv3087, Δrv3088, Δrv3087 Δrv3088, ΔfadD13, and Δtgs1 strains) induced a significantly higher level of M. tuberculosis ESAT-6 (6-kDa early secretory antigenic target)-specific IFN-γ-producing CD4+ T cells than the WT H37Rv bacilli. Similarly, all TNF-enhancing mutants elicited a significantly higher level of splenic IFN-γ-positive CD8+ T cells with respect to M. tuberculosis GAPDH (glyceraldehyde 3-phosphate dehydrogenase) than the WT tubercle bacilli (Fig. 8). These immunization studies support the notion that targeting TNF-downregulating M. tuberculosis genes is a viable strategy to produce strains with enhanced immunogenicity that can possibly serve as the substrates for developing effective anti-TB vaccines.
FIG 8 .
TNF-upregulating mutants enhance the IFN-γ response in CD4+ and CD8+ T cells. C57BL/6 mice were immunized subcutaneously with 106 bacilli of either H37Rv or TNF-enhancing mutants. After 30 days, IFN-γ-producing splenic CD4+ and CD8+ T cells were quantified by ELISPOT assay. Mice immunized with TNF-enhancing mutants developed more IFN-γ-producing CD4+ T cells (A) and CD8+ T cells (B) than animals vaccinated with WT H37Rv. **, P < 0.005; ***, < 0.001 (unpaired Student’s t test). Values are the means ± SD of the results from experiments performed in triplicate and are representative of three separate experiments.
DISCUSSION
TNF is arguably the best-established antimycobacterial immunological factor in humans, as reflected by the enhanced susceptibility to M. tuberculosis infection of individuals treated with TNF blockade therapy (15). This relevance in human TB has prompted extensive investigative efforts to study the role of TNF in shaping the immune response during M. tuberculosis infection (13). As one of the most tenacious intracellular pathogens, virulent M. tuberculosis is endowed with components of diverse biochemical properties that can attenuate host cell production of TNF (12 – 14). These mycobacterial countermechanisms are relevant in tuberculous pathogenesis, as evidenced by the observation that ablation of certain TNF-attenuating mycobacterial factors can lead to attenuation of virulence (14, 23 – 25). By virtue of its ability to promote phagosome maturation (17) and apoptosis (19, 20) in M. tuberculosis-infected macrophages, TNF can enhance T cell responses (18, 21). The latter property has implications in TB vaccine development, since T cell immunity plays a significant role in engendering immunization-induced protective responses (22). Our objectives in this study were first to identify genetic loci involved in the attenuation of host TNF production, using a TNF reporter macrophage system to screen an M. tuberculosis cosmid library, and then to test whether disruption of these genes resulted in strains of tubercle bacilli that can elevate host TNF production and enhance anti-TB immunity. Focusing on a subset of TNF-enhancing mutants, the results of these proof-of-concept experiments have provided strong evidence supporting the notion that targeted disruption of M. tuberculosis genes that mediate downregulation of the production of TNF by infected macrophages can lead to the generation of mutant strains with enhanced immunogenicity that can therefore serve as the substrates for vaccine development.
Screening approximately half of the H37Rv genome yielded a set of genes with TNF-downregulating attributes, the majority of which have been annotated to encode functions related to metabolism of lipids or carbohydrates, macromolecules that are major components of the M. tuberculosis cell envelope (40, 41). It is thus possible that the cell surface of these TNF-enhancing mutants, which presents an interface for the interaction between the bacilli and host cells, is chemically distinct from that of WT H37Rv. The altered chemical nature of the mutant envelope can also lead to changes in its architecture that, in turn, can result in unmasking or concealing of components that interact with host cells. The altered cell envelope of the mutants can thus result in aberrant interaction of the bacillus with macrophages, perhaps via specific surface molecules such as the pattern recognition receptors (54 – 56), thereby leading to differential expression of TNF. Indeed, precedents exist that indicate that M. tuberculosis lipid and carbohydrate moieties can alter macrophage TNF production and that this can lead to modulation of virulence (14, 23 – 25). Relevant to this notion, it has been reported that inactivation of the acid-responsive mymA operon (38, 39), which harbors three of the TNF-enhancing genes, rv3087, rv3088, and rv3089 (fadD13), results in the production of anomalous mycolic acid species as well as in aberrant cell colony morphology and envelope architecture (57 – 59). The commonality of rv3087 and rv3088 as encoding putative triacylglycerol synthases (Tgs) and, in addition, displaying Tgs activity in vitro may shed light on the mechanisms by which the members of this class of molecules modulate macrophage TNF production. Indeed, disruption of rv3130c (tgs1), encoding the triacylglycerol synthase with the most robust enzymatic activity among the 15 putative M. tuberculosis Tgs (42), resulted in an H37Rv mutant that also upregulated macrophage TNF production. Rv3130c is perhaps the best-studied M. tuberculosis Tg (42, 45, 60); it is the Tgs that is the most highly induced upon hypoxia and NO treatment, conditions which are likely encountered inside in the host (42). Deletion of Rv3130c resulted in nearly complete loss of triacylglcerol (TAG) synthesis (60), and there is evidence that this Tgs plays an important role in regulating energy storage in the form of TAG, which may be required for tuberculous persistence and reactivation (45). Finally, TAG can also serve as a source of fatty acids for phospholipid synthesis, thus influencing the properties of membrane lipid bilayers and their associated components (61). Of note, while the double deletion mutant (the Δrv3087 Δrv3088 mutant) deficient for the two putative Tgs's Rv3087 and Rv3088 displayed an apoptosis phenotype that was stronger than that of the single tgs knockouts, its effect on phagosome maturation is comparable to that exhibited by the single deletion mutant. The latter observation suggests a complex role of Tgs in modulating host cell-bacterium interactions. It is also possible that Rv3087 and Rv3088 might possess yet-to-be-elucidated biochemical functions other than that of triacylglycerol synthase. Thus, despite the putative assigned functions shared among Rv3087, Rv3088, and Tgs1, the mechanisms by which these enzymes mediate TNF-enhancing properties remain to be determined.
The M. tuberculosis FadD family members, which play an important role in activating fatty acids, a critical first step for the generation of acyl-CoA via an acyladenylate intermediate for lipid biochemical reactions, can be subdivided into two classes—the fatty acyl-CoA synthetases (fatty acyl-CoA ligases [FACL]) and the fatty acyl-AMP ligases (FAAL) (62). In vitro biochemical analyses using recombinant protein and in silico molecular modeling have shown that FadD13 preferentially activates long-chain fatty acids (C24/C26 versus C16/C10) via FACL activity (43, 44) and may thus play a role in the biosynthesis of the C60-90 mycolic acids (63, 64), whose derivative trehalose dimycolate (TDM) induces host cell TNF production through interaction with cell surface receptors MARCO and Mincle (54, 55, 65). Existing knowledge of the functions of FadD and Tgs suggests that these proteins contribute to modulating the chemical composition and architecture of the mycobacterial cell envelope. Characterization of the cell envelope-related mechanisms by which the lipid mutants upregulate TNF production will likely help efforts to gain insight into processes underlying TB pathogenesis and host defense and may lead to the discovery of bacterial surface-associated molecules that can be exploited for use in the development of TB vaccines.
Importantly, disruption of genes encoding enzymes known to regulate lipid metabolism (rv3087, rv3088, rv3089 [fadD13], and rv3130c [tgs1]) individually (or doubly for rv3087 and rv3088) in WT H37Rv produces deletion mutants that upregulate macrophage TNF production relative to the levels seen with parental bacilli, and complementation of the deletion mutants with the corresponding WT genes reverses the TNF phenotype, proving gene specificity. Relative to WT bacilli, these mutant strains are bestowed with an increased capacity to promote macrophage phagolysosomal fusion and apoptosis, lending support to our hypothesis that TNF-enhancing M. tuberculosis mutants can be more immunogenic than the WT bacillus. Indeed, results of the immunization studies support this notion.
Worthy of note, the results of the TNF neutralization experiments have provided evidence suggesting that the phagolysosomal fusion- and apoptosis-promoting phenotypes observed in macrophages infected with the gene deletion mutants of M. tuberculosis (the Δrv3087 Δ3088, Δrv3089 [ΔfadD13], and Δrv3130c [Δtgs1] mutants) are not totally TNF dependent. Given the complexity of the mechanisms that regulate apoptosis (50) and phagosome maturation (46 – 49) in M. tuberculosis-infected macrophages, including those that involve TNF-independent pathways, the lack of absolute correlation between the capacities of the TNF-upregulating mutants and their proapoptosis and prophagolysosomal fusion phenotypes is not surprising. It is possible that the deletion mutants investigated in the present study can influence the expression of cytokines and/or other host factors, in addition to TNF, which can modulate apoptosis and phagosome maturation. The idea of the existence of such a possibility is supported by the observations derived from the study of a phenolic glycolipid (PGL)-deficient mutant of HN878, a virulent M. tuberculosis strain of the Beijing family, which upregulates macrophage production of at least three cytokines, TNF, IL-6, and IL-12 (24), demonstrating that alteration of a single mycobacterial product could have diverse effects on macrophage responses upon infection. The effect on macrophage expression of cytokines other than TNF and/or other host factors upon infection by the mutants examined in the present study remains to be examined. Detailed characterization of the events that ensue following interactions of the TNF-upregulating M. tuberculosis strains with macrophages should shed light on the precise mechanisms underlying the apoptosis and phagolysosomal fusion phenotypes of these mutants.
One major goal of the development of vaccines against pathogens is the attainment of a long-term robust memory response (22, 66). Observations derived from various immunological models have provided evidence suggesting that certain inflammatory cytokines, including TNF, may promote CD4 T cell activation and proliferation during the priming phase of the immune response, which may lead to enhancement of T cell memory (67 – 69). Indeed, it has been reported recently that a vaccination protocol using BCG-infected macrophages in conjunction with IL-1, IL-6, and TNF administration engenders protection in mice that is superior to that elicited by the regimen without the cytokines or by BCG alone upon challenge with virulent bacilli 8 months postimmunization (70). Interestingly, the enhanced protection displayed by mice vaccinated with BCG-infected macrophages concomitant with IL-1, IL-6, and TNF is associated with an augmented CD4 and CD8 T cell memory response. These results suggest that TNF might modulate the initial phase of T cell activation, which, in turn, can promote the development of robust, long-lasting T cell memory. Whether immunization with the M. tuberculosis mutants evaluated in the present study, which possess the capacity to enhance TNF production in infected macrophages, can elicit a rigorous and long-term memory T cell response that can be recalled to mediate protection against challenge with virulent tubercle bacilli remains to be determined.
In sum, results generated from this study have provided strong evidence that targeting mycobacterial TNF-downregulating genes is a viable approach to augment the immunogenicity of potential vaccine candidates. While that was a major goal of the study, effort expended in this work has generated a set of tools that should prove useful beyond the present research. The C10 TNF reporter macrophage clone has proven effective in identifying TNF-modifying M. tuberculosis genes and will be used to screen the remaining half of the cosmid library, which is currently under construction. This cosmid library proved invaluable for the present study, but its utility can be exploited to identify genes responsible for any defined phenotype that can be assessed by an effective in vitro system. The set of TNF-enhancing mutants can be exploited for studies beyond vaccine development; for example, understanding the biology of these strains will likely be informative regarding the roles of TNF in shaping the host response during tuberculous infection in a system where the level of TNF expression is locally manipulated by the tubercle bacillus.
MATERIALS AND METHODS
Bacterial strains, growth media, and preparation for macrophage infection.
M. smegmatis mc2155 was cultured as previously described (32), with modification. Starter cultures of M. smegmatis clones of the H37Rv cosmid library were initiated by inoculating 5 µl of individual clones from frozen stocks into 1 ml of Middlebrook 7H9 medium supplemented with 0.5% glycerol, 10% oleic acid-albumin-dextrose-catalase (OADC) enrichment (Becton, Dickinson), 0.05% tyloxapol (Sigma), and 50 µg/liter hygromycin (Roche). Strain mc2155 transformed with pYUB412 with no H37Rv genomic DNA (empty vector) was similarly cultured. WT bacteria were grown without antibiotics. Bacteria from starter cultures were reseeded into fresh media and grown to mid-log phase, pelleted, washed in phosphate-buffered saline (PBS)–tyloxapol (Sigma), and sonicated to disrupt clumps. A portion of the bacterial suspension was used for measurement of the optical density at 590 nm (OD590). The remaining bacterial preparation was adjusted to a concentration of 1 × 108 CFU in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and 1× nonessential amino acids (complete DMEM) for use in infection of C10 macrophages.
M. tuberculosis H37Rv and various deletion mutants were cultured at 37°C with shaking in Middlebrook 7H9 broth containing 0.2% glycerol, 0.05% Tween 80, and 10% OADC enrichment medium (Becton, Dickinson). Deletion mutants and complemented strains were grown in similarly supplemented 7H9 medium containing hygromycin (Roche) (50 µg/ml) and kanamycin (Roche) (20 µg/ml), respectively. In preparation for macrophage infection, various M. tuberculosis strains were grown to mid-log phase. Bacteria were washed in complete DMEM and sonicated, and the OD590 was measured. The bacterial suspension was adjusted to appropriate titers for infection of J774.16 macrophages.
Generation of M. tuberculosis gene deletion mutants and complemented strains.
The various gene deletion mutants were generated in the WT H37Rv strain using the mycobacteriophage-mediated specialized transduction method as previously described (32, 71). Complementation of the WT gene of interest (driven by the hsp60 promoter) into the attB site of the corresponding M. tuberculosis knockout strain was achieved using the integrating vector pMV361 as previously described (32).
M. tuberculosis cosmid library construction and subcloning analysis of TNF-downregulating hits.
The plasmid pYUB412 was linearized with XbaI, dephosphorylated with shrimp alkaline phosphatase (Promega), and then digested with BclI to obtain 3,789-bp and 4,773-bp fragments. M. tuberculosis H37Rv genomic DNA was prepared from a 20-ml culture (OD600, ~3) grown in Middlebrook 7H9 supplemented with 10% OADC, 0.2% glycerol, and 0.05% Tween 80 as previously described (32), with the following modification: the genomic DNA, once precipitated in isopropanol, was spooled and immersed in ethanol and was then air-dried instead of being spun down. The genomic DNA was partially digested with Sau3A for 1 h at 37°C to obtain DNA fragments of around 20 to 30 kbp in size and then ligated overnight at 4°C to the pYUB412-derived 3,789-bp and 4,773-bp fragments using T4 ligase (Promega). The ligation reaction mixture (5 µl) was mixed with 25 µl of MaxPlax Lambda Packaging Extract (Epicentre, Madison, WI) at room temperature (RT) for 1 h. Another 2 µl of ligation reaction mixture was added to the packaging reaction, and the mixture was incubated for an additional hour at RT. LB medium (0.1 ml) was added to the packaging reaction along with 200 µl of HB101 cells grown in LB supplemented with 0.3% maltose and 10 mM MgSO4. The resultant mixture was incubated at 37°C for 30 min with no shaking and was then diluted with 0.5 ml of LB media and incubated for an additional 1 h at 37°C with shaking. The packaging reaction mixture was plated on LB-carbenicillin (LB-Carb; 100 µg/ml) plates; individual colonies were picked and grown in LB-Carb. Cosmid DNA was extracted from individual E. coli colonies and was then subjected to sequencing at the two ends of the H37Rv DNA insertion. In parallel, DNA of individual cosmids was analyzed by restriction mapping to validate the finding that the cosmid harbors the M. tuberculosis H37Rv genes (predicted by the sequence analysis of the two ends) in the correct contiguous sequence. pYUB412-based cosmid DNAs procured from E. coli clones were used to transform mc2155 to generate the H37Rv cosmid library in the heterologous M. smegmatis strain. This effort yielded 105 M. smegmatis clones whose genomes harbor distinct M. tuberculosis DNA fragments, covering about 50% of the H37Rv genome in total. Three individual colonies from the transformation reaction were picked and stored for each clone. As a result, the final product consisted of three sets (“a,” “b,” and “c”) of the 105 M. smegmatis H37Rv cosmid clones. Once a TNF-regulating clone of M. smegmatis harboring H37Rv DNA was obtained from the C10 macrophage screen, the validity of the hit was confirmed by studying the same clone from another set from the stored library.
Subclones of the original TNF-regulating hits were transformed into M. smegmatis mc2155 via the use of the integrating pMV361. Similarly to the storage of stocks for the original 105 M. smegmatis H37Rv cosmid clones described above, three individual colonies from the transformation reaction were picked and stored for each subclone, resulting in three sets (“a,” “b,” and “c”) of subclones derived from the original four TNF-regulating cosmid hits. This allowed confirmation of the TNF-regulating property of a hit by examining whether this attribute was reproducible using the same subclone from another set of the stored stocks.
Transfection of J774.16 macrophages with a TNF promoter-GFP construct.
The TNF promoter, whose function has been previously characterized using chloramphenicol acetyltransferase (CAT) as the reporter, was derived from the construct Pro-UTR (a kind gift of Jiahuai Han), harboring the TNF promoter (TNFp) in tandem with the CAT coding sequence and the 3′ untranslated region (3′ UTR) of the TNF allele (27). Digestion of pro-UTR with KpnI and BamHI released a fragment containing the TNFp. The cytomegalovirus (CMV) promoter was deleted from phrGFP II-1 (Stratagene) to yield the modified phrGFP II-1 (m-phrGFP II-1) and then digested with KpnI and BamHI. The TNFp fragment was ligated into m-phrGFP II-1 (upstream of hrGFP II-1), yielding the TNFp-hrGFPII-1 fusion construct, which was transformed into E. coli HB101. Transformants were selected for resistance to kanamycin (50 µg/ml). J774.16 mouse macrophages were transfected with the m-phrGFP II-1 vector containing the TNFp-hrGFP II-1 fusion by nucleofection using an Amaxa Nucleofector (Lonza, Cologne, Germany) and were subjected to selection using G418 (1 mg/ml). G418-resistant transfectants were stimulated with lipopolysaccharides (LPS) (Sigma) (1 µg/ml; Escherichia coli 0127:B8) and examined by fluorescence microscopy. GFP-positive colonies were picked and transferred individually into wells of 24-well plates. Expanded GPF-positive colonies underwent a second round of inspection by fluorescence microscopy. Colonies with positive GFP signals were collected, expanded, and stored for further selection.
Macrophage infections and assessment of TNF expression.
J774.16 macrophages were grown as previously described (72). C10 macrophages were cultured in 100-mm-diameter Optilux petri dishes (BD Falcon) in complete DMEM. Day 3 C10 cultures (80% to 90% confluent) were used to seed wells of 96-well tissue culture plates (Becton, Dickinson) at 105 cells per well and were allowed to adhere overnight at 37° in a 5% CO2 atmosphere prior to infection. The macrophages were subjected to infection with individual M. smegmatis bacteria harboring H37Rv M. tuberculosis DNA (prepared as described above) at a multiplicity of infection (MOI) of 10:1 (10 bacilli to 1 macrophage). Uninfected C10 or cultures infected with M. smegmatis transformed with the pYUB412 empty vector served as controls. After 4 h of infection, the cultures were washed twice with warm supplemented DMEM, replenished with the same medium containing 10 µg/ml of gentamicin. At 16 h later, the culture medium was replaced by warm PBS and GFP expression was quantified by the use of a Walac Viktor II plate reader (PerkinElmer) (excitation wavelength, 488 nm; emission wavelength, 530 nm). Culture supernatants were collected for direct measurement of TNF levels by ELISA (eBioscience) following the manufacturer’s instructions.
M. tuberculosis H37Rv and the various deletion mutants and complemented strains were prepared as described above for infection of C10 macrophages. The C10 cultures were infected with WT H37Rv, deletion mutants, and complemented strains at an MOI of 10:1. At 4 h after initiation of the infection, cells were washed three times with prewarmed complete DMEM to remove extracellular bacteria and incubated for an additional 16 h in growth media. Samples were then analyzed for GFP fluorescence in a Viktor II plate reader as described above. For measurement of TNF in culture supernatants, samples were aspirated and filtered to remove infectious material and then subjected to analysis by ELISA.
Apoptosis studies.
J774.16 cells were cultured in complete DMEM and plated at 1 × 105 cells per well on a glass-bottom 96-well plate (MatriPlate; Brooks Life Science System). Cells were allowed to adhere overnight, and macrophage cultures were infected the next day with mid-log-phase WT H37Rv, deletion mutants, and complemented strains (prepared as described above) at an MOI of 10. After 4 hours, cells were washed with prewarmed culture media and incubated for an additional 16 h. Samples were labeled using a FAM-FLICA (6-carboxyfluorescein–fluorochrome-labeled inhibitors of caspases) in vitro poly caspase kit (ImmunoChemistry) following the manufacturer’s instructions and then fixed with 4% paraformaldehyde. Levels of FLICA-positive cells were measured using laser scanning cytometry (iCys; Thorlabs) followed by analysis performed with the accompanying iCys software. In certain experiments, the TNF neutralization MP6-XT22 MAb was added to cultures at a concentration of 10 µg/ml. The dose chosen derived from standardization experiments testing a range of concentrations previously used (17, 73, 74).
Phagolysosomal fusion studies.
J774.16 macrophages were cultured in complete DMEM and seeded at 2 × 105 cells per well in an eight-well chambered slide with glass coverslips (Nunc Lab-TekII chamber slides) and allowed to adhere overnight. Mid-log-phase WT H37Rv, deletion mutants, and complemented strains (prepared as described above) were labeled with fluorescein-5-isothiocyanate (FITC) (Life Technologies) (10 µg/ml) for 4 h at 37°C with shaking and were then washed with PBS containing 0.05% Tween 80 (PBS-T) two times. Macrophage samples were synchronized on ice and then infected with various strains of M. tuberculosis at an MOI of 1 to 5. At 4 h later, cells were washed three times with prewarmed complete DMEM to remove extracellular bacteria. After 4 h of additional incubation, samples were fixed with 4% paraformaldehyde and then permeabilized with 0.01% Triton X-100 (Sigma). After 1 h of blocking with 10% serum from the same source as the secondary antibody, primary antibodies (Abcam; rabbit polyclonal IgG to Rab5 [ab18211] and rabbit polyclonal IgG to LAMP1 [ab24170]) were added at a dilution of 1:100 and incubated overnight at 4°C. The next day, samples were washed three times with PBS-T and fluorescently tagged secondary antibodies (Abcam; polyclonal goat anti-rabbit IgG H&L [ab150086] conjugated to Alexa Fluor 555) were added at a dilution of 1:1,000 and incubated for 1 h at room temperature. After secondary antibodies were washed away with PBS-T, samples were mounted with ProLong Gold antifade reagent (Life Technologies) and then imaged with a Leica SP2 light microscope. At least five images per well were assessed, which amounted to more than 100 bacterium-containing phagosomes per sample. Colocalization between fluorescent bacteria and cell markers was scored blind. When appropriate, the TNF neutralization MP6-XT22 MAb was added to cultures at a concentration of 10 µg/ml. The dose used was based on pilot studies testing a range of concentrations previously used (17, 73, 74).
Mouse immunization and IFN-γ ELISPOT assay.
Animal studies were conducted according to protocols that have been approved by the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine. C57BL/6 female mice (Jackson Laboratories) (8 to 10 weeks old) were immunized subcutaneously with 106 cells of M. tuberculosis H37Rv, deletion mutants, or complemented strains. After 30 days, mice were sacrificed and splenic T cells were isolated by magnetically activated cell sorting (MACS) using a Pan T cell isolation kit (Miltenyi Biotec) according to the manufacturer’s protocol. Detection of IFN-γ-producing T cells was carried out as previously described (75), using a mouse IFN-γ ELISPOT Ready-Set-Go kit (eBioscience) according to the manufacturer’s instructions. Briefly, T cells (1 × 105 and 3 × 105) were seeded in 96-well ELISPOT plates (Millipore) that had been coated previously with IFN-γ capture antibody overnight. Peptides of M. tuberculosis ESAT-6 (the first 20 amino acids of Rv3875) were used to stimulate CD4+ T cells, and that of M. tuberculosis GAP (amino acid GAPINSATAM of Rv0125) was used to stimulate CD8+ T cells in this assay. Splenocytes (2 × 105) from naive uninfected mice were used as antigen-presenting cells (APCs) and were incubated with ESAT-6 (6-kDa early secretory antigenic target) or GAPDH (glyceraldehyde 3-phosphate dehydrogenase) (10 µg/ml) for 1 h at 37°C. After two careful washes were performed, the APCs with or without peptides were added to the ELISPOT wells. After a 36-h incubation at 37°C and 5% CO2, cells were removed, the plates were washed, and the captured cytokine was detected by incubating wells with a biotinylated anti-mouse IFN-γ antibody (clone XMG1.2; eBiosciences) for 2 h at 37°C. Avidin-horseradish peroxidase (eBioscience) was added to the wells for 45 min at 37°C, and spots were detected using AEC substrate solution (Sigma). The substrate reaction was stopped by washing the plate with distilled water. Spots were enumerated using an automated ELISPOT reader (Autoimmun Diagnostika).
Statistical analysis.
Statistical analysis was performed with Prism 5.0 software (GraphPad) using the unpaired t test. A P value of less than 0.05 was considered significant.
SUPPLEMENTAL MATERIAL
The TNF-enhancing M. tuberculosis cosmids (*, overlapping clones).
ACKNOWLEDGMENTS
This work was supported by NIH grants P01AI063537 (J.C., S.A.P., W.R.J.), R01 AI093649 (S.A.P.), R01 AI26170 (W.R.J.), R01 AI098925 (W.R.J.), and T32 AI007501 (HIV, AIDS, and Opportunistic Training Grant) and by grant P30 AI051519 (The Einstein/Montefiore Center for AIDS Research). A portion of this work was supported by the Heiser Foundation (grant no. P11-000255 to Y.C.). Flow cytometry resources were supported by the Einstein Cancer Center (P30 CA013330).
We thank members of the Chan, Porcelli, and Jacobs laboratory for helpful discussion.
Funding Statement
This work was supported by NIH grants P01AI063537 (JC, SAP, WRJ), R01 AI093649 (SAP), R01 AI26170 (WRJ), R01 AI098925 (WRJ), T32 AI007501 (HIV, AIDS, and Opportunistic Training Grant), P30 AI051519 (The Einstein/Montefiore Center for AIDS Research). Portion of this work was supported by the Heiser Foundation Grant #P11-000255 (YC). Flow cytometry resources were supported by the Einstein Cancer Center (P30 CA013330).
Footnotes
Citation Olsen A, Chen Y, Ji Q, Zhu G, De Silva AD, Vilchèze C, Weisbrod T, Li W, Xu J, Larsen M, Zhang J, Porcelli SA, Jacobs WR, Jr., Chan J. 2016. Targeting Mycobacterium tuberculosis tumor necrosis factor alpha-downregulating genes for the development of antituberculous vaccines. mBio 7(3):e01023-15. doi:10.1128/mBio.01023-15.
REFERENCES
- 1.World Health Organization 2014. WHO Report 2014: Global Tuberculosis Control. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Balaban NQ, Gerdes K, Lewis K, McKinney JD. 2013. A problem of persistence: still more questions than answers? Nat Rev Microbiol 11:587–591. doi: 10.1038/nrmicro3076. [DOI] [PubMed] [Google Scholar]
- 3.Fitzgerald DW, Sterling TR, Hass DW. 2015. Mycobacterium tuberculosis. In Mandell, Douglas, and Bennett’s principles and practice of infectious diseases, 8th ed. Elsevier Saunders, Philadelphia, PA. [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC) 1999. Impact of vaccines universally recommended for children—United States, 1990–1998. MMWR Morb Mortal Wkly Rep 48:243–248. [PubMed] [Google Scholar]
- 5.Koff WC, Burton DR, Johnson PR, Walker BD, King CR, Nabel GJ, Ahmed R, Bhan MK, Plotkin SA. 2013. Accelerating next-generation vaccine development for global disease prevention. Science 340:1232910. doi: 10.1126/science.1232910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Modlin RL, Bloom BR. 2013. TB or not TB: that is no longer the question. Sci Transl Med 5:213sr216. doi: 10.1126/scitranslmed.3007402. [DOI] [PubMed] [Google Scholar]
- 7.Mangtani P, Abubakar I, Ariti C, Beynon R, Pimpin L, Fine PE, Rodrigues LC, Smith PG, Lipman M, Whiting PF, Sterne JA. 2014. Protection by BCG vaccine against tuberculosis: a systematic review of randomized controlled trials. Clin Infect Dis 58:470–480. doi: 10.1093/cid/cit790. [DOI] [PubMed] [Google Scholar]
- 8.Cooper AM. 2009. Cell-mediated immune responses in tuberculosis. Annu Rev Immunol 27:393–422. doi: 10.1146/annurev.immunol.021908.132703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flynn JL, Chan J. 2001. Immunology of tuberculosis. Annu Rev Immunol 19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 10.North RJ, Jung YJ. 2004. Immunity to tuberculosis. Annu Rev Immunol 22:599–623. doi: 10.1146/annurev.immunol.22.012703.104635. [DOI] [PubMed] [Google Scholar]
- 11.Ernst JD. 2012. The immunological life cycle of tuberculosis. Nat Rev Immunol 12:581–591. doi: 10.1038/nri3259. [DOI] [PubMed] [Google Scholar]
- 12.Chan J, Flynn J. 2004. The immunological aspects of latency in tuberculosis. Clin Immunol 110:2–12. doi: 10.1016/S1521-6616(03)00210-9. [DOI] [PubMed] [Google Scholar]
- 13.Dorhoi A, Kaufmann SH. 2014. Tumor necrosis factor alpha in mycobacterial infection. Semin Immunol 26:203–209. doi: 10.1016/j.smim.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Flynn JL, Chan J. 2005. What’s good for the host is good for the bug. Trends Microbiol 13:98–102. doi: 10.1016/j.tim.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Keane J, Gershon S, Wise RP, Mirabile-Levens E, Kasznica J, Schwieterman WD, Siegel JN, Braun MM. 2001. Tuberculosis associated with infliximab, a tumor necrosis factor alpha-neutralizing agent. N Engl J Med 345:1098–1104. doi: 10.1056/NEJMoa011110. [DOI] [PubMed] [Google Scholar]
- 16.Wallis RS. 2009. Infectious complications of tumor necrosis factor blockade. Curr Opin Infect Dis 22:403–409. doi: 10.1097/QCO.0b013e32832dda55. [DOI] [PubMed] [Google Scholar]
- 17.Harris J, Hope JC, Keane J. 2008. Tumor necrosis factor blockers influence macrophage responses to Mycobacterium tuberculosis. J Infect Dis 198:1842–1850. doi: 10.1086/593174. [DOI] [PubMed] [Google Scholar]
- 18.Ramachandra L, Simmons D, Harding CV. 2009. MHC molecules and microbial antigen processing in phagosomes. Curr Opin Immunol 21:98–104. doi: 10.1016/j.coi.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balcewicz-Sablinska MK, Keane J, Kornfeld H, Remold HG. 1998. Pathogenic Mycobacterium tuberculosis evades apoptosis of host macrophages by release of TNF-R2, resulting in inactivation of TNF-alpha. J Immunol 161:2636–2641. [PubMed] [Google Scholar]
- 20.Spira A, Carroll JD, Liu G, Aziz Z, Shah V, Kornfeld H, Keane J. 2003. Apoptosis genes in human alveolar macrophages infected with virulent or attenuated Mycobacterium tuberculosis: a pivotal role for tumor necrosis factor. Am J Respir Cell Mol Biol 29:545–551. doi: 10.1165/rcmb.2002-0310OC. [DOI] [PubMed] [Google Scholar]
- 21.Winau F, Weber S, Sad S, de Diego J, Hoops SL, Breiden B, Sandhoff K, Brinkmann V, Kaufmann SH, Schaible UE. 2006. Apoptotic vesicles crossprime CD8 T cells and protect against tuberculosis. Immunity 24:105–117. doi: 10.1016/j.immuni.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Andersen P, Kaufmann SH. 2 June 2014. Novel vaccination strategies against tuberculosis. Cold Spring Harb Perspect Med doi: 10.1101/cshperspect.a018523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurtz S, McKinnon KP, Runge MS, Ting JP, Braunstein M. 2006. The SecA2 secretion factor of Mycobacterium tuberculosis promotes growth in macrophages and inhibits the host immune response. Infect Immun 74:6855–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed MB, Domenech P, Manca C, Su H, Barczak AK, Kreiswirth BN, Kaplan G, Barry CE III. 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 25.Stanley SA, Raghavan S, Hwang WW, Cox JS. 2003. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci U S A 100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dao DN, Sweeney K, Hsu T, Gurcha SS, Nascimento IP, Roshevsky D, Besra GS, Chan J, Porcelli SA, Jacobs WR. 2008. Mycolic acid modification by the mmaA4 gene of M. tuberculosis modulates IL-12 production. PLoS Pathog 4:e1000081. doi: 10.1371/journal.ppat.1000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han J, Huez G, Beutler B. 1991. Interactive effects of the tumor necrosis factor promoter and 3′-untranslated regions. J Immunol 146:1843–1848. [PubMed] [Google Scholar]
- 28.Russell-Goldman E, Xu J, Wang X, Chan J, Tufariello JM. 2008. A Mycobacterium tuberculosis Rpf double-knockout strain exhibits profound defects in reactivation from chronic tuberculosis and innate immunity phenotypes. Infect Immun 76:4269–4281. doi: 10.1128/IAI.01735-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beltan E, Horgen L, Rastogi N. 2000. Secretion of cytokines by human macrophages upon infection by pathogenic and non-pathogenic mycobacteria. Microb Pathog 28:313–318. doi: 10.1006/mpat.1999.0345. [DOI] [PubMed] [Google Scholar]
- 30.Falcone V, Bassey EB, Toniolo A, Conaldi PG, Collins FM. 1994. Differential release of tumor necrosis factor-alpha from murine peritoneal macrophages stimulated with virulent and avirulent species of mycobacteria. FEMS Immunol Med Microbiol 8:225–232. [DOI] [PubMed] [Google Scholar]
- 31.Yadav M, Roach SK, Schorey JS. 2004. Increased mitogen-activated protein kinase activity and TNF-alpha production associated with Mycobacterium smegmatis- but not Mycobacterium avium-infected macrophages requires prolonged stimulation of the calmodulin/calmodulin kinase and cyclic AMP/protein kinase A pathways. J Immunol 172:5588–5597. doi: 10.4049/jimmunol.172.9.5588. [DOI] [PubMed] [Google Scholar]
- 32.Larsen MH, Biermann K, Tandberg S, Hsu T, Jacobs WR Jr.. 2007. Genetic manipulation of Mycobacterium tuberculosis. Curr Protoc Microbiol Chapter 10:Unit 10A.2. doi: 10.1002/9780471729259.mc10a02s6. [DOI] [PubMed] [Google Scholar]
- 33.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR Jr.. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 34.De Groot MJ, van de Vondervoort PJ, de Vries RP, vanKuyk PA, Ruijter GJ, Visser J. 2003. Isolation and characterization of two specific regulatory Aspergillus niger mutants shows antagonistic regulation of arabinan and xylan metabolism. Microbiology 149:1183–1191. doi: 10.1099/mic.0.25993-0. [DOI] [PubMed] [Google Scholar]
- 35.VanKuyk PA, de Groot MJ, Ruijter GJ, de Vries RP, Visser J. 2001. The Aspergillus niger d-xylulose kinase gene is co-expressed with genes encoding arabinan degrading enzymes, and is essential for growth on d-xylose and l-arabinose. Eur J Biochem 268:5414–5423. doi: 10.1046/j.0014-2956.2001.02482.x. [DOI] [PubMed] [Google Scholar]
- 36.Vetting MW, de Carvalho LPS, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. 2005. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys 433:212–226. [DOI] [PubMed] [Google Scholar]
- 37.Boissier F, Bardou F, Guillet V, Uttenweiler-Joseph S, Daffé M, Quémard A, Mourey L. 2006. Further insight into S-adenosylmethionine-dependent methyltransferases: structural characterization of Hma, an enzyme essential for the biosynthesis of oxygenated mycolic acids in Mycobacterium tuberculosis. J Biol Chem 281:4434–4445. doi: 10.1074/jbc.M510250200. [DOI] [PubMed] [Google Scholar]
- 38.Cheruvu M, Plikaytis BB, Shinnick TM. 2007. The acid-induced operon Rv3083-Rv3089 is required for growth of Mycobacterium tuberculosis in macrophages. Tuberculosis (Edinb) 87:12–20. [DOI] [PubMed] [Google Scholar]
- 39.Fisher MA, Plikaytis BB, Shinnick TM. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol 184:4025–4032. doi: 10.1128/JB.184.14.4025-4032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaur D, Guerin ME, Skovierová H, Brennan PJ, Jackson M.. 2009. Chapter 2: biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv Appl Microbiol 69:23–78. doi: 10.1016/S0065-2164(09)69002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daffé M, Draper P. 2012. The envelope layers of Mycobacteria with reference to their pathogenicity, p 131–203. In Cardona PJ (ed), Understanding tuberculosis—analyzing the origin of Mycobacterium tuberculosis pathogenicity. InTech, Rijeka, Croatia. [Google Scholar]
- 42.Daniel J, Deb C, Dubey VS, Sirakova TD, Abomoelak B, Morbidoni HR, Kolattukudy PE. 2004. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol 186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jatana N, Jangid S, Khare G, Tyagi AK, Latha N. 2011. Molecular modeling studies of fatty acyl-CoA synthetase (FadD13) from Mycobacterium tuberculosis—a potential target for the development of antitubercular drugs. J Mol Model 17:301–313. doi: 10.1007/s00894-010-0727-3. [DOI] [PubMed] [Google Scholar]
- 44.Khare G, Gupta V, Gupta RK, Gupta R, Bhat R, Tyagi AK. 2009. Dissecting the role of critical residues and substrate preference of a fatty acyl-CoA synthetase (FadD13) of Mycobacterium tuberculosis. PLoS One 4:e8387. doi: 10.1371/journal.pone.0008387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baek SH, Li AH, Sassetti CM. 2011. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol 9:e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deretic V, Singh S, Master S, Harris J, Roberts E, Kyei G, Davis A, de Haro S, Naylor J, Lee HH, Vergne I. 2006. Mycobacterium tuberculosis inhibition of phagolysosome biogenesis and autophagy as a host defence mechanism. Cell Microbiol 8:719–727. doi: 10.1111/j.1462-5822.2006.00705.x. [DOI] [PubMed] [Google Scholar]
- 47.Lugo-Villarino G, Neyrolles O. 2014. Manipulation of the mononuclear phagocyte system by Mycobacterium tuberculosis. Cold Spring Harb Perspect Med 4:a018549. doi: 10.1101/cshperspect.a018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell DG. 2011. Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol Rev 240:252–268. doi: 10.1111/j.1600-065X.2010.00984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vergne I, Gilleron M, Nigou J. 2014. Manipulation of the endocytic pathway and phagocyte functions by Mycobacterium tuberculosis lipoarabinomannan. Front Cell Infect Microbiol 4:187. doi: 10.3389/fcimb.2014.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Behar SM, Martin CJ, Booty MG, Nishimura T, Zhao X, Gan HX, Divangahi M, Remold HG. 2011. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol 4:279–287. doi: 10.1038/mi.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hinchey J, Lee S, Jeon BY, Basaraba RJ, Venkataswamy MM, Chen B, Chan J, Braunstein M, Orme IM, Derrick SC, Morris SL, Jacobs WR Jr., Porcelli SA. 2007. Enhanced priming of adaptive immunity by a proapoptotic mutant of Mycobacterium tuberculosis. J Clin Invest 117:2279–2288. doi: 10.1172/JCI31947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Velmurugan K, Chen B, Miller JL, Azogue S, Gurses S, Hsu T, Glickman M, Jacobs WR Jr., Porcelli SA, Briken V. 2007. Mycobacterium tuberculosis nuoG is a virulence gene that inhibits apoptosis of infected host cells. PLoS Pathog 3:e110. doi: 10.1371/journal.ppat.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bohsali A, Abdalla H, Velmurugan K, Briken V. 2010. The non-pathogenic mycobacteria M. smegmatis and M. fortuitum induce rapid host cell apoptosis via a caspase-3 and TNF dependent pathway. BMC Microbiol 10:237. doi: 10.1186/1471-2180-10-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bowdish DM, Sakamoto K, Kim MJ, Kroos M, Mukhopadhyay S, Leifer CA, Tryggvason K, Gordon S, Russell DG. 2009. Marco, TLR2, and CD14 are required for macrophage cytokine responses to mycobacterial trehalose dimycolate and Mycobacterium tuberculosis. PLoS Pathog 5:e1000474. doi: 10.1371/journal.ppat.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, Kinoshita T, Akira S, Yoshikai Y, Yamasaki S. 2009. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med 206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jo EK. 2008. Mycobacterial interaction with innate receptors: TLRs, C-type lectins, and NLRs. Curr Opin Infect Dis 21:279–286. doi: 10.1097/QCO.0b013e3282f88b5d. [DOI] [PubMed] [Google Scholar]
- 57.Singh A, Gupta R, Vishwakarma RA, Narayanan PR, Paramasivan CN, Ramanathan VD, Tyagi AK. 2005. Requirement of the mymA operon for appropriate cell wall ultrastructure and persistence of Mycobacterium tuberculosis in the spleens of guinea pigs. J Bacteriol 187:4173–4186. doi: 10.1128/JB.187.12.4173-4186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh A, Jain S, Gupta S, Das T, Tyagi AK. 2003. mymA operon of Mycobacterium tuberculosis: its regulation and importance in the cell envelope. FEMS Microbiol Lett 227:53–63. doi: 10.1016/S0378-1097(03)00648-7. [DOI] [PubMed] [Google Scholar]
- 59.Singh R, Singh A, Tyagi AK. 2005. Deciphering the genes involved in pathogenesis of Mycobacterium tuberculosis. Tuberculosis (Edinb) 85:325–335. [DOI] [PubMed] [Google Scholar]
- 60.Sirakova TD, Dubey VS, Deb C, Daniel J, Korotkova TA, Abomoelak B, Kolattukudy PE. 2006. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology 152:2717–2725. doi: 10.1099/mic.0.28993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alvarez HM, Steinbüchel A. 2002. Triacylglycerols in prokaryotic microorganisms. Appl Microbiol Biotechnol 60:367–376. doi: 10.1007/s00253-002-1135-0. [DOI] [PubMed] [Google Scholar]
- 62.Duckworth BP, Nelson KM, Aldrich CC. 2012. Adenylating enzymes in Mycobacterium tuberculosis as drug targets. Curr Top Med Chem 12:766–796. doi: 10.2174/156802612799984571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bhatt A, Molle V, Besra GS, Jacobs WR Jr., Kremer L. 2007. The Mycobacterium tuberculosis FAS-II condensing enzymes: their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development. Mol Microbiol 64:1442–1454. doi: 10.1111/j.1365-2958.2007.05761.x. [DOI] [PubMed] [Google Scholar]
- 64.Marrakchi H, Lanéelle MA, Daffé M. 2014. Mycolic acids: structures, biosynthesis, and beyond. Chem Biol 21:67–85. doi: 10.1016/j.chembiol.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 65.Matsunaga I, Moody DB. 2009. Mincle is a long sought receptor for mycobacterial cord factor. J Exp Med 206:2865–2868. doi: 10.1084/jem.20092533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pulendran B, Ahmed R. 2006. Translating innate immunity into immunological memory: implications for vaccine development. Cell 124:849–863. doi: 10.1016/j.cell.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 67.Haynes L, Eaton SM, Burns EM, Rincon M, Swain SL. 2004. Inflammatory cytokines overcome age-related defects in CD4 T cell responses in vivo. J Immunol 172:5194–5199. doi: 10.4049/jimmunol.172.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pape KA, Khoruts A, Mondino A, Jenkins MK. 1997. Inflammatory cytokines enhance the in vivo clonal expansion and differentiation of antigen-activated CD4+ T cells. J Immunol 159:591–598. [PubMed] [Google Scholar]
- 69.Vella AT, Mitchell T, Groth B, Linsley PS, Green JM, Thompson CB, Kappler JW, Marrack P. 1997. CD28 engagement and proinflammatory cytokines contribute to T cell expansion and long-term survival in vivo. J Immunol 158:4714–4720. [PubMed] [Google Scholar]
- 70.Singh V, Jain S, Gowthaman U, Parihar P, Gupta P, Gupta UD, Agrewala JN. 2011. Co-administration of IL-1+IL-6+TNF-alpha with Mycobacterium tuberculosis infected macrophages vaccine induces better protective T cell memory than BCG. PLoS One 6:e16097. doi: 10.1371/journal.pone.0016097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bardarov S, Bardarov S Jr, Pavelka MS Jr., Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR Jr.. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 72.Chan J, Xing Y, Magliozzo RS, Bloom BR. 1992. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med 175:1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Algood HM, Lin PL, Yankura D, Jones A, Chan J, Flynn JL. 2004. TNF influences chemokine expression of macrophages in vitro and that of CD11b+ cells in vivo during Mycobacterium tuberculosis infection. J Immunol 172:6846–6857. doi: 10.4049/jimmunol.172.11.6846. [DOI] [PubMed] [Google Scholar]
- 74.Rojas M, Olivier M, Gros P, Barrera LF, García LF. 1999. TNF-alpha and IL-10 modulate the induction of apoptosis by virulent Mycobacterium tuberculosis in murine macrophages. J Immunol 162:6122–6131. [PubMed] [Google Scholar]
- 75.Kozakiewicz L, Chen Y, Xu J, Wang Y, Dunussi-Joannopoulos K, Ou Q, Flynn JL, Porcelli SA, Jacobs WR Jr., Chan J. 2013. B cells regulate neutrophilia during Mycobacterium tuberculosis infection and BCG vaccination by modulating the interleukin-17 response. PLoS Pathog 9:e1003472. doi: 10.1371/journal.ppat.1003472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The TNF-enhancing M. tuberculosis cosmids (*, overlapping clones).