Abstract
Epitheliotropic intestinal T-cell lymphoma (EITL, also known as type II enteropathy-associated T-cell lymphoma) is an aggressive intestinal disease with poor prognosis and its molecular alterations have not been comprehensively characterized. We aimed to identify actionable easy-to-screen alterations that would allow better diagnostics and/or treatment of this deadly disease. By performing whole-exome sequencing of four EITL tumor-normal pairs, followed by amplicon deep sequencing of 42 tumor samples, frequent alterations of the JAK-STAT and G-protein-coupled receptor (GPCR) signaling pathways were discovered in a large portion of samples. Specifically, STAT5B was mutated in a remarkable 63% of cases, JAK3 in 35% and GNAI2 in 24%, with the majority occurring at known activating hotspots in key functional domains. Moreover, STAT5B locus carried copy-neutral loss of heterozygosity resulting in the duplication of the mutant copy, suggesting the importance of mutant STAT5B dosage for the development of EITL. Dysregulation of the JAK-STAT and GPCR pathways was also supported by gene expression profiling and further verified in patient tumor samples. In vitro overexpression of GNAI2 mutants led to the upregulation of pERK1/2, a member of MEK-ERK pathway. Notably, inhibitors of both JAK-STAT and MEK-ERK pathways effectively reduced viability of patient-derived primary EITL cells, indicating potential therapeutic strategies for this neoplasm with no effective treatment currently available.
Introduction
Enteropathy-associated T-cell lymphoma (EATL) is a rare aggressive primary intestinal non-Hodgkin lymphoma accounting for 5.4% of peripheral T-cell lymphomas and 10–25% of all primary intestinal lymphomas.1, 2, 3 EATL was included into the World Health Organization classification of hematolymphoid neoplasms for the first time in 2008 and consists of type I and type II.1 Classical or type I EATL is known to associate with celiac disease, HLA-DQ2 and DQ8 haplotypes, and is the more common form in the West.1, 2, 4, 5, 6, 7, 8 On the contrary, type II EATL is more prevalent in Asia and multiple studies have failed to confirm an association with celiac disease.9, 10, 11, 12 Given the distinct clinicopathological features of type II EATL as described below and the lack of association with celiac disease, the term 'enteropathy-associated' in its nomenclature is not appropriate and various names including monomorphic intestinal T-cell lymphoma11 and epitheliotropic intestinal T-cell lymphoma (EITL)13 have been proposed. In this study, we will refer to this neoplasm as EITL henceforth.
EITL has an extremely poor prognosis with a median overall survival of only 7 months.13 No effective treatment or targeted therapies are currently available for this disease. A number of publications have focused on the clinical and pathological characterization of EITL. Together with others, we have comprehensively described the histology of this complex disease.11, 13 In short, EITL displays zonal variation featuring central invasive sheets of monomorphic neoplastic lymphocytes, a peripheral zone of mucosa infiltrated by morphologically atypical intraepithelial lymphocytes (IELs) and a distant zone featuring mucosa with normal villous architecture but increased number of morphologically normal IELs. The putative cell of origin is an intestinal IEL and clonality analyses have shown that the IELs in the distant mucosal zone share clonal relationship with the invasive lymphoma.13, 14 The CD8+ CD56+ phenotype and extensive nuclear expression of megakaryocyte-associated tyrosine kinase are features that are distinct from classical EATL,13, 15 with the majority of neoplastic cells expressing CD8αα homodimers.13 Opinion is still divided whether these cells display mainly T-cell receptor (TCR) αβ or γδ phenotypes.11, 13
Cytogenetic studies have demonstrated gains and translocations of MYC (8q24)10, 13, 16 in EITL, although they may also be seen in classical EATL.17 Conversely, gains of 1q32.2-q41 and 5q34-q35.2 are more common in EATL compared with EITL, whereas 9q31.3 gain and 16q21.1 loss can be observed in both classical EATL and EITL.1, 3 Recently, it was reported that an activating STAT5B p.N642H mutation is common in T-cell lymphoma derived from γδ T-cells, including some cases of EITL.18 We used next-generation sequencing techniques in a multicenter study to describe the frequent genetic changes in EITL. We provide here the first whole-exome sequencing (WES) study of this disease and, in the largest series published to date, demonstrate that STAT5B-activating mutations are found in EITL tumors of both TCRαβ and TCRγδ origin. We present multiple lines of evidence that JAK-STAT and G-protein-coupled receptor (GPCR) signaling pathways are highly activated in EITL, and, most importantly, inhibition of these pathways by targeted therapy effectively reduces the viability of primary EITL cells.
Materials and methods
Patients and samples
EITL was defined as a primary intestinal T-cell lymphoma with characteristic morphology, typically with cytotoxic, CD3+ CD8+ CD56+ EBER− phenotype. Five snap frozen primary tumors and matched whole blood samples were obtained from the SingHealth Tissue Repository. The patients were recruited from 2010 to 2014 and all provided signed informed consent in accordance with the Declaration of Helsinki. An additional 41 archival formalin-fixed paraffin-embedded samples collected from 2003 to 2014 were obtained from the following centers: National Cancer Centre Singapore, Singapore General Hospital, National University Hospital and Tan Tock Seng Hospital in Singapore, Guangdong General Hospital and Sun Yat-sen University Cancer Center in China, University of Malaya and Hospital Raja Permaisuri Bainun in Malaysia, Samsung Medical Centre in Korea, Chi Mei Medical Center in Taiwan and Mahidol University in Thailand. Fourteen of those were also used in our previous study with the same identifiers.13 Clinicopathological features are listed in Supplementary Table 1. Tumor cell content was assessed by hematoxylin and eosin staining. The study was approved by SingHealth Centralised Institutional Review Board (2004/407/F).
Sequencing and mutation analysis
Detailed methods are described in the Supplementary Information. Briefly, four tumor-normal pairs of EITL were whole-exome sequenced using SureSelectXT Human All Exon V4+UTRs capture (Agilent Technologies, Santa Clara, CA, USA) and HiSeq 2000 Sequencing System (Illumina, San Diego, CA, USA). All candidate variants were visually inspected in Integrative Genomics Viewer.19, 20 Only Integrative Genomics Viewer-true mutations were considered somatic and variants with frequency ⩾15% were further validated by Sanger sequencing (Supplementary Table 2).
Prevalence screening was performed for GNAI2, JAK3 and STAT5B genes in 42 tumor samples using amplicon deep sequencing. Only common hotspots reported in COSMIC v.72 and regions identified through WES were sequenced for each gene (Supplementary Table 3). CREBBP mutation frequency was determined in a partial prevalence cohort (16 cases) using Single Primer Enrichment Technology (NuGEN Technologies, San Carlos, CA, USA).
SNP genotyping array and allele-specific copy number analysis of tumors
Genomic DNA from four tumor-normal pairs was hybridized to Genome-Wide Human Single-Nucleotide Polymorphism (SNP) Array 6.0 (Affymetrix, Santa Clara, CA, USA) chips. Allele-specific copy number analysis of tumors was performed as described previously.21
Stable cell line construction and western blot
Full-length GNAI2 cDNA was amplified using AccuPrime Pfx DNA Polymerase (Invitrogen, Carlsbad, CA, USA) and cloned into retroviral vector MIGR1 (Addgene; plasmid no. 27490)22 at XhoI and EcoRI sites. GNAI2 mutations (c.535C>T for p.R179C and c.535C>A for p.R179S) were generated by QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) and confirmed by Sanger sequencing.
HuT78 and Jurkat cell lines were from ATCC (Manassas, VA, USA) and maintained in Iscove's modified Dulbecco's medium (Invitrogen) supplemented with 20% fetal bovine serum (HyClone; GE Healthcare, Little Chalfont, UK) and RPMI-1640 (Invitrogen) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare), respectively. The cultures were routinely checked for mycoplasma contamination. Stable cell lines were generated by retroviral infection and sorting of green fluorescent protein-positive cells using BD FACSAria III Cell Sorter (BD Biosciences, Franklin Lakes, NJ, USA).
Protein extracts were prepared in RIPA cell lysis buffer (150 mm NaCl, 50 mm Tris-HCl, 0.5% deoxychlorate sodium, 200 mm NaF, 200 mm PMSF, 1% NP-40 and 1 mm EDTA) in the presence of freshly added protease inhibitor (Roche, Basel, Switzerland). HuT78 lines underwent serum starvation for 24 h before collection of protein lysates. Total protein extracts were separated on sodium dodecyl sulfate-polyacrylamide gel and transferred to Trans-Blot Turbo Mini PVDF membranes (Bio-Rad, Hercules, CA, USA). The used antibodies are listed in Supplementary Table 4. Signals were visualized using ChemiDoc MP System (Bio-Rad).
Ex vivo cell viability assay in primary EITL cells
Primary EITL cells from patient 065T were subcutaneously injected into the NOD scid gamma (NSG) mouse (P1; The Jackson Laboratory, Bar Harbor, ME, USA), followed by further passaging of the subcutaneously formed tumor cells by subcutaneous and intraperitoneal injection (P2). Eight-week-old female animals were used in each passage. P2 mouse presented with highly swollen peritoneum, and intestinal and peritoneal tumors. All samples, including peritoneal fluid cells from P2, were histologically characterized by a senior hematopathologist (SYT) and diagnosed EITL. Owing to the consistent phenotype between patient's primary tumor and P2 peritoneal fluid cells, we will refer to the latter as primary EITL cells henceforth. Sanger sequencing confirmed the presence of all somatic mutations identified in the original tumor (Supplementary Table 5) also in the primary cells. Animal experiments were in compliance with ethical regulations of the SingHealth Institutional Animal Care and Use Committee.
Primary EITL cells were maintained in RPMI-1640 (Invitrogen) supplemented with 10% human serum (Sigma-Aldrich, St Louis, MO, USA) and 500 IU/ml interleukin-2 (Miltenyi Biotec, Bergisch Gladbach, Germany). To test the feasibility of targeted therapy in EITL, 5 × 103 cells were seeded in 96-well plates in three technical replicates and treated with Stattic (CAS 19983-44-9; Selleck Chemicals, Houston, TX, USA) and MEK inhibitor PD0325901 (CAS 391210-10-9; Axon Medchem, Groningen, The Netherlands) for 72 h, and Tofacitinib (CP 690550; Selleck Chemicals), Trametinib (GSK 1120212; Axon Medchem) and Gefitinib (ZD 1839; Axon Medchem) for 96 h at indicated concentrations. Cell viability was measured using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA). The efficacy of inhibitors was further evaluated by their effect on downstream targets using western blot. Cell lysates for PD0325901, Trametinib and Tofacitinib were collected 2 h and for Stattic 24 h post-treatment. Dose–response curves were plotted using the Prism software (GraphPad, La Jolla, CA, USA). All drug treatment experiments were repeated at least two times with each inhibitor.
Accession numbers
Sequencing data from WES, Single Primer Enrichment Technology and amplicon deep sequencing has been deposited in the European Nucleotide Archive with the accession number PRJEB9720. The Affymetrix Genome-Wide Human SNP Array 6.0 and GeneChip Human Genome U133 Plus 2.0 Array data are available in the Gene Expression Omnibus23 with the accession number GSE70654.
Results and discussion
Identification of somatic variants by WES
To identify somatic mutations in EITL, we performed WES of four tumors with matching whole blood samples (Figure 1a). To maximize variant detection from a limited sample size, three different variant callers were applied—Genome Analysis Toolkit,24 MuTect25 and Strelka.26 The mean depth of coverage was 126 × and on average 88% of targeted bases were covered by at least 20 reads (Supplementary Table 6). Sanger validation of 296 candidate variants showed a false-positive rate of 3.72%. EITL samples presented a high mutation burden with an average of 85 mutations per sample (range 59–137). A total of 340 nonsynonymous somatic mutations were identified in 313 genes. Seven of those genes also harbored silent somatic mutations (Supplementary Table 5). The nonsynonymous variants comprised 284 missense and 16 nonsense mutations, 13 frameshift indels, 25 splice site mutations and 2 mutations resulting in loss of stop codon (Supplementary Figure 1a). As observed in most other cancers,27, 28 EITL mutation spectrum was characterized by the predominance of C>T transitions (Supplementary Figure 1b).
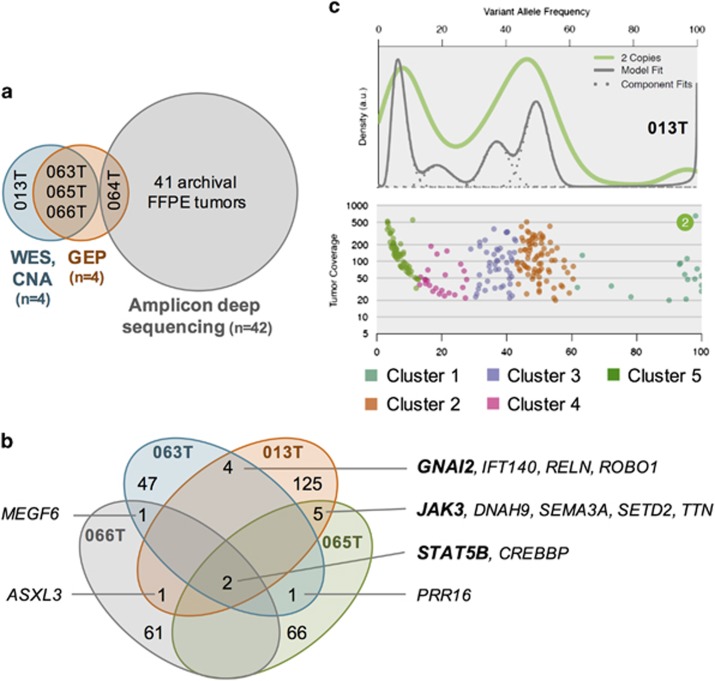
Figure 1.
Graphical overview of the study, and mutation and subclonality profile of EITL tumors. (a) Types of analyses performed and the number of samples used for each experiment. Snap frozen cases are listed by their respective IDs. (b) Venn diagram illustrating the recurrent mutations in the discovery cohort. (c) The subclonal architecture of EITL sample 013T derived from SciClone analysis. The upper plot demonstrates kernel density estimation and the lower one variant allele frequencies plotted against read depth (⩾20 × coverage) in copy-neutral loss of heterozygosity-free regions. CNA, copy number analysis; FFPE, formalin-fixed paraffin-embedded; GEP, gene expression profiling.
A total of 14 genes were found recurrently mutated (Figure 1b and Supplementary Table 5). Among those were the calcium ion-binding gene multiple EGF-like-domains 6 (MEGF6), regulator of cell size proline-rich 16 (PRR16) and the giant muscle protein Titin (TTN), genes with functions in neuronal migration, axon guidance and chemoattraction—Reelin (RELN), Roundabout 1 (ROBO1) and Semaphorin 3 A (SEMA3A)—and with functions in the movement and transportation of cilia—Dynein 9 (DNAH9) and Intraflagellar Transport 140 (IFT140). In addition, mutations were identified in the histone acetyltransferase CREB-binding protein (CREBBP) and two (putative) histone methyltransferases—additional sex comb-like transcriptional regulator 3 (ASXL3) and SET domain-containing 2 (SETD2)—suggesting that epigenetic dysregulation may have a role in EITL.
Interestingly, three of the recurrently mutated genes presented with known activating hotspot variants. These included two key members of the JAK-STAT pathway, a central signaling cascade in immune cells—janus kinase 3 (JAK3) and signal transducer and activator of transcription 5B (STAT5B)—and G protein alpha inhibiting activity polypeptide 2 (GNAI2), a member of the family of Gα proteins. Remarkably, STAT5B was mutated in all four cases, suggesting a critical role in EITL formation. Three samples possessed the common p.N642H mutation, whereas the fourth harbored a complex substitution event in the beginning of exon 17, resulting in two amino-acid changes—p.V712E and p.S715L (see Supplementary Note and Supplementary Figure 2). Both JAK3 and GNAI2 were mutated in a remarkable 2/4 cases. JAK3 alterations occurred at two distinct activating positions (p.V674A and p.M511I), whereas GNAI2 was mutated in the same codon in both samples (p.R179C and p.R179S).
To understand the clonal architecture of EITL tumors, we used SciClone29 to identify populations of cells containing specific mutations. All four discovery cohort samples demonstrated notable intratumor heterogeneity with five subclones present in each tumor sample (Figure 1c, Supplementary Figure 3 and Supplementary Table 7). As only copy-neutral loss of heterozygosity-free segments were analyzed, we were not able to determine whether the JAK3 and STAT5B mutations were located in different subclones. However, respective corrected variant allele frequencies do indicate that these mutations are located within two separate clones (Supplementary Table 5).
Prevalence screening of GNAI2, JAK3, STAT5B and CREBBP
The high frequency of STAT5B, JAK3 and GNAI2 mutations in a small discovery cohort of only four samples was intriguing, prompting us to further analyze the prevalence of these mutations in a larger series of EITL samples. Forty-two additional EITL tumors (1 snap frozen and 41 archival samples) were screened for GNAI2, JAK3 and STAT5B mutations using amplicon deep sequencing. A total of 57 identified mutations were covered by an average of 20 004 reads (range 104–175 515; Supplementary Table 8). As the amplicons covered only common mutation hotspots reported in COSMIC v.72 and positions identified in the discovery cohort, it is likely that our results are lower-bound estimates of the true mutation frequency of these three genes.
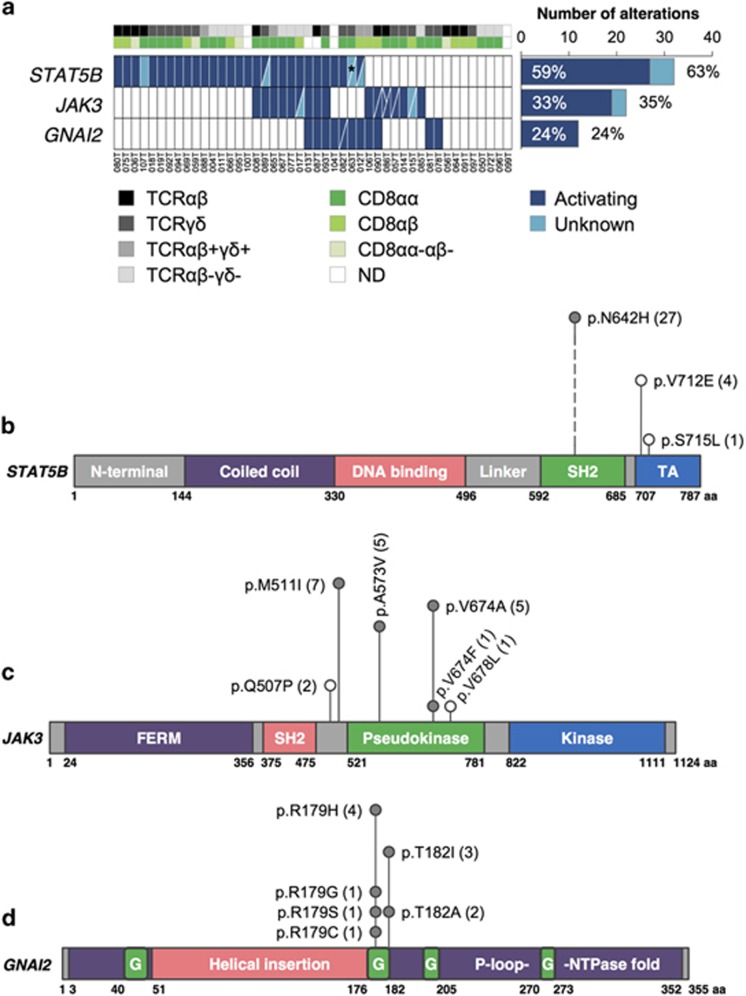
Figure 2 summarizes the results from both the discovery and prevalence cohorts. STAT5B mutations were detected in 25/42 (60%) cases, with 24/42 (57%) carrying known activating mutations (p.N642H) in the Src homology 2 dimerization domain (Figure 2b). JAK3 was mutated in 14/42 (33%) samples with 13/42 (31%) harboring activating mutations (p.M511I, p.A573V and p.V674A/F), the majority located in the pseudokinase domain (Figure 2c). We identified two samples carrying two separate JAK3-activating mutations and a remarkable three activating mutations were identified in a third case indicating profound alteration of JAK3 function in these patients. Notably, none of these samples contained an activating STAT5B mutation (Figure 2a). Collectively, the JAK-STAT pathway was altered in 32/42 (76%) cases. It is remarkable that a large subset of tumors harbor co-occurring activating mutations in two members of the same pathway. A similar phenomenon was recently described for JAK1 and STAT3 in systemic ALK-negative anaplastic large T-cell lymphomas.30 Moreover, these mutants were demonstrated to act in synergy to constitutively activate downstream STAT3 signaling. Further studies for mutated JAK3 and STAT5B are thus warranted and might shed light on EITL tumorigenesis.
Figure 2.
Frequently mutated genes in EITL identified by WES and amplicon deep sequencing. (a) Mutation frequencies of STAT5B, JAK3 and GNAI2 correlating to CD8 dimer and TCR type. Distribution of mutations in (b) STAT5B, (c) JAK3 and (d) GNAI2 genes. The frequency of each alteration is denoted within parentheses after its label. Filled circles indicate known mutation hotspots and empty circles mutations with unknown functional consequence. *A total of four nonsynonymous missense mutations identified resulting in the change of two amino acids. G, GTP-binding; ND, not done; SH2, Src Homology 2; TA, transcriptional activation domain; TCR, T-cell receptor.
GNAI2 mutations were observed in 9/42 (21%) samples. Two major hotspots were identified—p.R179 and p.T182—both located in the GTP-binding domain (Figure 2d) and are known activating variants.31, 32, 33 A single case was found to contain both mutations. Although exhibiting a lower mutation incidence compared with STAT5B and JAK3, it has been suggested that the lower frequencies for G protein α-subunits could be explained by mutations in receptors coupled to these G proteins.34 On that note, both the GNAI2 wild-type discovery cohort samples harbored somatic mutations in GPCRs (Supplementary Table 5). It would be therefore interesting to analyze the mutational prevalence of GPCRs and other members of Gαi signaling in a larger cohort to determine the frequency at which this cascade is altered in EITL.
Alterations in all three genes were found to be not mutually exclusive (JAK3 vs STAT5B: P-value=0.2527; GNAI2 vs STAT5B: P-value=0.5118; GNAI2 vs JAK3: P-value=0.9998; one-sided Fisher's exact test by CoMEt35). Owing to the limited number of events, we were not able to analyze the subclonality of amplicon deep sequencing data; however, corrected variant allele frequencies do suggest that multiple mutations in all three genes can be found both within the same as well as different subclones (Supplementary Table 8).
We also analyzed the mutation frequency of CREBBP in a partial prevalence cohort of 16 samples. Mutations were identified in 2/16 (13%) cases, and in total, CREBBP was mutated in 6/20 (30%) samples (Supplementary Figure 4 and Supplementary Table 9). Inactivating CREBBP mutations have been reported in a number of malignancies, including B-cell lymphomas,36, 37 and the mutations have been described as early events associated with inferior outcome.38, 39 Although in our series each identified variant was predicted to be disruptive, all mutations occurred in samples with coinciding STAT5B and/or JAK3 mutations, suggesting that CREBBP mutations might not be the major initiating event and further support the importance of activated JAK-STAT pathway in EITL.
Correlation of mutations with pathological features
The identified STAT5B mutation frequency is nearly twice that of a previously published study of EITLs with TCRγδ expression.18 As the latter relied on Sanger sequencing, this difference may at least, in part, be explained by the lower sensitivity of Sanger sequencing. However, our observations do not support the hypothesis that STAT5B-activating mutations only occur in the TCRγδ lineage of EITLs as activating variants were equally identified in TCRαβ, TCRγδ, double-negative and aberrant double-positive cases (20%, 36%, 32% and 12% in each subset, respectively; Figure 2a and Supplementary Table 10). A similar observation was also made for the JAK3- and GNAI2-activating mutations.
We further segregated the samples by the expression of CD8 dimer. Activating mutations of each gene were present in tumors with CD8αα, CD8αβ and double-negative T-cells, with more than half of samples with STAT5B-activating variants being of the CD8αα type (61% CD8αα, 26% CD8αβ and 13% double negative; Supplementary Table 10). This is consistent with the notion that EITL is predominantly a tumor of CD8αα IELs.13 Importantly, there is no phenotypic variation between the mutant cases with different TCR and/or CD8 expression (Supplementary Figures 5 and 6).
Copy number alterations in EITL
To characterize the chromosomal abnormalities in EITL, we profiled all samples from the discovery cohort on a genome-wide SNP array. All tumors were approximately diploid. We identified each of the previously reported frequent chromosomal change (loss of 16q12.1 and gains of 9q31.3 and 8q24)1, 3, 10, 13 recurrently altered in 2/4 samples (Supplementary Table 11). This includes amplification of the MYC gene (four copies in two samples). Importantly, no gains of 1q32.2-q41 and 5q34-q35.5, alterations more common in classical EATL compared with EITL, were identified. Other recurrent losses included 8p, 11q and 12p, and gain of 7q (detailed regions in Supplementary Table 11).
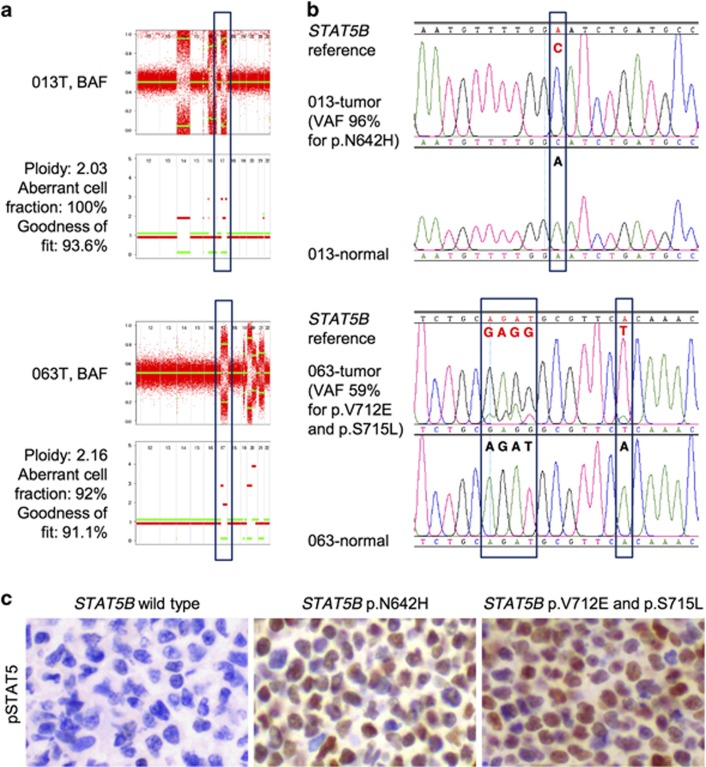
Remarkably, all four samples demonstrated copy-neutral loss of heterozygosity at chromosome 17q where STAT5B is located (Figures 3a and b). This phenomenon has previously been observed in JAK2 and JAK3 in other hematological neoplasms,40, 41 and is thought to arise from somatic recombination events. To the best of our knowledge, this is the first time acquired uniparental disomy has been described for STAT5B and further indicates the pivotal role played by this gene in the development of EITL. Consistent with this, immunohistochemical analysis of samples with mutated STAT5B revealed noticeable upregulation of phosphorylated STAT5 (Supplementary Table 12), an example of which is shown in Figure 3c. It would be interesting to see whether there exists a transition from heterozygosity to homozygosity either histologically from distant mucosa to central tumor zone or clinically from primary diagnosis to relapse representing clonal evolution as observed in other tumors.41, 42
Figure 3.
Acquired uniparental disomy in STAT5B locus. (a) Allele-specific copy number analysis of tumor (ASCAT) close-ups of two STAT5B mutant samples highlighting chromosome 17q. The upper panels demonstrate the raw (red) and segmented (green) B-allele frequencies (BAFs) of SNPs. The lower panels present allele-specific copy number profiles—red lines indicate the higher and green the lower copy number chromosomal haplotypes. The green line is at zero, which indicates loss of heterozygosity. (b) Sanger sequencing electropherograms demonstrating the presence of STAT5B somatic mutation in the same samples. (c) Representative images of pSTAT5 immunohistochemical staining in STAT5B wild-type and mutant cases. Magnification × 400. VAF, variant allele frequency from WES.
We also identified two samples with amplification of the PIK3CG gene (five and four copies, respectively) that was also evident on protein level both for PIK3CG itself and downstream pAKT in one sample (Supplementary Figure 7). The prevalence of such amplification should be studied in a larger cohort as it might open up novel possibilities for targeted therapy. The first PI3K inhibitor is already in use for treatment of certain B-cell malignancies with many more currently in development.43
Pathways and biological functions affected in EITL
To identify key biological functions and signaling pathways affected in EITL, two different approaches were used—Ingenuity Pathway Analysis (IPA) of mutated genes from WES and Gene Set Enrichment Analysis (GSEA) following gene expression profiling of EITL tumors.
Among all mutated genes, major cellular functions such as cell death, survival, development, differentiation, homeostasis and proliferation were found significantly enriched (Supplementary Table 13), many of these affecting lymphocytes or T-cells specifically. In addition, mutated genes were also significantly associated with components involved in T-cell and intestine morphology and genes known to be involved in cancer and lymphoid malignancies. Importantly, enrichment of cell signaling and binding of DNA were also noted. All these suggest that key cellular functions are involved in the development of EITL and correlate well with the nature of this disease.
Further supporting our genomic findings, canonical JAK-STAT and GPCR signaling were among the top-enriched IPA pathways (false discovery rate q-value=0.24 and 0.03, respectively; Supplementary Table 14). In addition, several other aspects of these cascades were also altered indicating the importance of GNAI2, JAK3 and STAT5B mutations described above. These include the role of JAK1 and JAK3 in γc cytokine signaling, Oncostatin M signaling, interleukin-2 and interleukin-9 signaling, cAMP-mediated signaling and Gαi signaling. Molecular mechanisms of cancer was also found enriched, suggesting the involvement of classical cancer genes in this disease. The latter is supported by the finding of at least one mutated tumor suppressor or oncogene in each case (Supplementary Table 5). Because IPA does not consider recurrence of gene alterations, we repeated the analysis with the list of recurrently mutated genes only. Importantly, all the above pathways, except Gαi signaling, were found significantly enriched in EITL (Supplementary Table 15).
To verify the above by an unrelated approach, we performed gene expression profiling of four EITL tumors, two of CD8αα and two of CD8αβ origin. As EITL is suggested to arise from CD8+ IELs,13 we integrated our data with publicly available profile of CD8αα and CD8αβ T-cells from healthy donors.44 There was a total of 1760 genes significantly upregulated in EITL tumors (P<0.05 and fold change ⩾1.5; Supplementary Table 16). GSEA against Molecular Signatures Database gene sets45 was used to identify deregulated pathways. The used gene sets and all statistics from GSEA are shown in Supplementary Table 17. It is noteworthy that GSEA independently confirmed the findings of IPA analysis using mutated genes from WES. Several aspects of GPCR signaling were found significantly enriched in EITL samples including GPCR ligand binding, signaling by GPCR and, importantly, Gαi signaling (Supplementary Figure 8). These clearly suggest a marked deregulation of Gαi signaling in EITL and further indicate the functional importance of discovered GNAI2 mutations.
In addition, we also identified KEGG JAK-STAT signaling pathway as significantly enriched in EITLs. The higher side q-value (Supplementary Table 17) is in this regard not surprising as the activation of JAK-STAT pathway is known to occur through protein phosphorylation and not increased expression of mRNA. More importantly, an outstanding association was identified with genes containing STAT5B binding sites in their promoters, both for the shorter and longer consensus motif. We further confirmed this by GSEA against publicly available gene set of STAT5 target genes.46 There was a significant enrichment between STAT5 targets and genes upregulated in EITL (normalized enrichment score=1.78, false discovery rate q-value<0.001; Supplementary Figure 8d).
Gene expression profiling data was validated by real-time PCR of randomly selected genes (Supplementary Table 18). The values from the two experiments were consistent, although the correlation was not statistically significant because of the small sample size (Supplementary Figure 9). In summary, JAK-STAT and GPCR signaling pathways were demonstrated as significantly enriched in EITL tumors using two different data sets and methodologies.
Targeted therapy inhibits viability of primary EITL cells
While activating mutations in genes encoding α-subunits of G proteins occur in many human cancers, oncogenic alterations in the inhibitory subunits of G proteins exist at a much lower frequency.34 Oncogenic form of GNAI2 (GNAI2 p.R179C/H), referred to as gip2,32 was first identified in endocrine tumors of adrenal cortex and ovary,31 and p.T182A was thereafter also described as a constitutively activated mutant.33 Notably, gip2 was demonstrated to constitutively activate mitogen-activated protein kinase,47 likely through inhibitory effect on intracellular cAMP levels.48
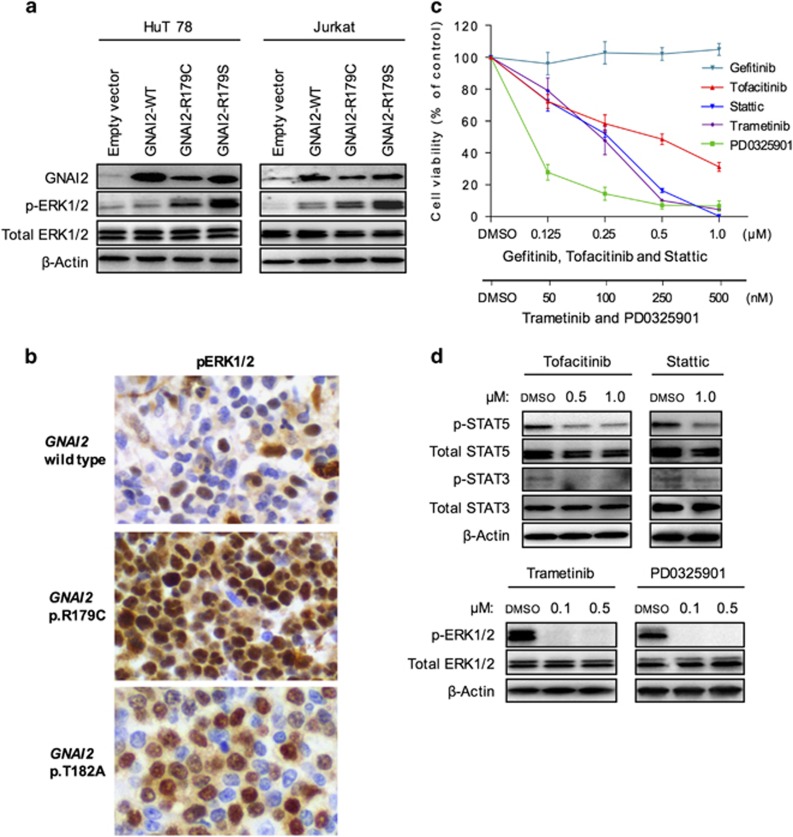
To better understand the above, we created stable cell lines overexpressing wild-type and mutant forms (p.R179C and p.R179S) of GNAI2. A strong upregulation of phosphorylated ERK1/2 level was detected in both mutant cells, whereas the total ERK1/2 protein levels remained unchanged (Figure 4a). The same observation was made in two independent cell lines. Consistent with this, immunohistochemical analysis of GNAI2 mutant cases also revealed upregulation of phosphorylated ERK1/2 (Supplementary Table 12), examples of which are shown in Figure 4b, suggesting that the MEK-ERK signaling pathway is aberrantly activated in these EITL tumors.
Figure 4.
Effective targeted therapy by JAK, STAT and MEK inhibitors in primary EITL cells. (a) Stable cell lines were created by overexpressing GNAI2 wild-type and mutants in HuT78 and Jurkat cells. The expression of relevant proteins was assessed by western blot. (b) Representative images of pERK1/2 immunohistochemical staining in GNAI2 wild-type and mutant tumors. Magnification × 400. (c) Representative ex vivo cell viability assay in primary EITL cells treated with PD0325901 and Stattic for 72 h, and Tofacitinib, Trametinib and Gefitinib for 96 h. All results are normalized to the control (dimethyl sulfoxide (DMSO)) and presented as mean±s.d. The experiment was repeated at least two times with each inhibitor. (d) Immunoblots of indicated proteins in primary EITL cells treated with PD0325901, Tofacitinib and Trametinib for two and Stattic for 24 h at indicated concentrations.
Notably, the identification of activated pathways in EITL offers novel treatment opportunities. JAK3 and STAT5B are both directly targetable by small-molecule inhibitors, and because mutant GNAI2 activates the MEK-ERK pathway, MEK inhibitors may be effective for the treatment of EITL. To further confirm the involvement of JAK-STAT and GPCR signaling pathways in the survival of EITL, we evaluated the effect of two inhibitors of both pathways in primary EITL cells—Tofacitinib (pan-JAK inhibitor), Stattic (STAT3 inhibitor) and MEK inhibitors Trametinib and PD0325901. All four inhibitors reduced cell viability in a dose-dependent manner compared with the vehicle-treated control (Figure 4c). Importantly, the primary cells were resistant to an inhibitor of a non-relevant pathway, the EGFR inhibitor Gefitinib, suggesting that the observed effect was not because of general cytotoxicity. Moreover, all four drugs clearly inhibited the downstream signaling cascades as demonstrated by suppression of STAT3, STAT5 and ERK1/2 phosphorylation (Figure 4d).
These primary EITL cells harbor mutations in both JAK3 and downstream STAT5B, which may explain the inferior sensitivity of Tofacitinib compared with Stattic. Further supporting our data, JAK-STAT cascade inactivation either by knockdown of pathway members or targeted therapy also effectively reduces cell viability of cutaneous T-cell lymphoma cells that harbor an activating p.A573V JAK3 mutation.49, 50, 51, 52 Despite the lack of GNAI2 mutation in the primary EITL cells, several other members of the GPCR pathway—the GPCRs HTR1F, GRM5, MRGPRX3 and GPR179, and protein tyrosine phosphatase DUSP4 (Supplementary Table 5)—are all altered in these cells potentially leading to downstream activation of the GPCR pathway. Importantly, DUSP4 is known to inactivate ERK1/2.53 The p.R300W mutation identified in the primary EITL cells is positioned right in the middle of DUSP4 catalytic dual-specificity phosphatase domain. As commonly observed for mutations within the functional domains of enzymes, it is likely that the identified p.R300W mutation causes inactivation of the DUSP4 protein that leads to the activation of ERK proteins and might explain the effectiveness of MEK inhibition in these cells. In addition, GPCR, Gαi and MEK-ERK pathways are all significantly enriched in the original tumor sample (Supplementary Figures 8a and c), further supporting the aberrant activation of these signaling pathways. Taken together, these results indicate that targeted therapy may be useful in the treatment of EITL patients.
Conclusions
This is the first study to characterize comprehensively the molecular changes in EITL and provides novel insights into the pathogenesis of this disease. We have clearly demonstrated alterations in the JAK-STAT and GPCR signaling pathways on multiple levels, including mutations, chromosomal alterations and transcriptional upregulation. The high mutation frequency and location of mutations in key functional domains indicate the importance of these variants and notably may present novel opportunities for targeted therapies or better diagnostics. This is supported by our in vitro data demonstrating response of primary EITL cells to inhibitors of both pathways. Further studies are required to support these findings and to functionally evaluate them in a disease-specific model.
Acknowledgments
This work was supported by the National Medical Research Council of Singapore (TCR12DEC005), Tanoto Foundation as Professorship in Medical Oncology, New Century Foundation Limited, Ling Foundation and Singapore National Cancer Centre Research Fund. We thank Swe Swe Myint and Jia Liang Loh for help with sample preparation; Minghui Lee and Angie Tan from Duke-NUS Genome Biology Facility for gene expression profiling; Qian Qiao Tang, Alisa Noor Hidayah Sairi, Liang Kai Koh, Gerald Chua, Agnieszka Maliszewska and Beng Hooi Phang for logistic and technical support, and all co-workers of Laboratory of Cancer Epigenome. We also thank SingHealth Flow Cytometry unit for cell sorting and the following surgeons for their generous contribution of patient samples: Associate Professor Foong Koon Cheah, Prof. Wai Keong Wong, Dr Peng Chung Cheow, Dr Yaw Chong Goh and Dr Weng Hoong Chan.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Isaacson PG, Chott A, Ott G, Stein H. Enteropathy-associated T-cell lymphoma. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H et al (eds). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edn IARC Press: Lyon, France, 2008, pp 289–291.
- Delabie J, Holte H, Vose JM, Ullrich F, Jaffe ES, Savage KJ et al. Enteropathy-associated T-cell lymphoma: clinical and histological findings from the international peripheral T-cell lymphoma project. Blood 2011; 118: 148–155. [DOI] [PubMed] [Google Scholar]
- Foukas PG, de Leval L. Recent advances in intestinal lymphomas. Histopathology 2015; 66: 112–136. [DOI] [PubMed] [Google Scholar]
- Shaw CK, Chen LL, Lee A, Lee TD. Distribution of HLA gene and haplotype frequencies in Taiwan: a comparative study among Min-nan, Hakka, Aborigines and Mainland Chinese. Tissue Antigens 1999; 53: 51–64. [DOI] [PubMed] [Google Scholar]
- Saito S, Ota S, Yamada E, Inoko H, Ota M. Allele frequencies and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population. Tissue Antigens 2000; 56: 522–529. [DOI] [PubMed] [Google Scholar]
- Liang CK, Chen KH, Hsu WM, Chen KH. Association of HLA type and Mooren's ulcer in Chinese in Taiwan. Br J Ophthalmol 2003; 87: 797–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Oh DH, Lee C, Yang SY. Allelic and haplotypic diversity of HLA-A, -B, -C, -DRB1, and -DQB1 genes in the Korean population. Tissue Antigens 2005; 65: 437–447. [DOI] [PubMed] [Google Scholar]
- Lan Q, Shen M, Garcia-Rossi D, Chanock S, Zheng T, Berndt SI et al. Genotype frequency and F ST analysis of polymorphisms in immunoregulatory genes in Chinese and Caucasian populations. Immunogenetics 2007; 59: 839–852. [DOI] [PubMed] [Google Scholar]
- Chim CS, Au WY, Shek TW, Ho J, Choy C, Ma SK et al. Primary CD56 positive lymphomas of the gastrointestinal tract. Cancer 2001; 91: 525–533. [DOI] [PubMed] [Google Scholar]
- Ko YH, Karnan S, Kim KM, Park CK, Kang ES, Kim YH et al. Enteropathy-associated T-cell lymphoma—a clinicopathologic and array comparative genomic hybridization study. Hum Pathol 2010; 41: 1231–1237. [DOI] [PubMed] [Google Scholar]
- Chan JK, Chan AC, Cheuk W, Wan SK, Lee WK, Lui YH et al. Type II enteropathy-associated T-cell lymphoma: a distinct aggressive lymphoma with frequent gammadelta T-cell receptor expression. Am J Surg Pathol 2011; 35: 1557–1569. [DOI] [PubMed] [Google Scholar]
- Sun J, Lu Z, Yang D, Chen J. Primary intestinal T-cell and NK-cell lymphomas: a clinicopathological and molecular study from China focused on type II enteropathy-associated T-cell lymphoma and primary intestinal NK-cell lymphoma. Mod Pathol 2011; 24: 983–992. [DOI] [PubMed] [Google Scholar]
- Tan SY, Chuang SS, Tang T, Tan L, Ko YH, Chuah KL et al. Type II EATL (epitheliotropic intestinal T-cell lymphoma): a neoplasm of intra-epithelial T-cells with predominant CD8alphaalpha phenotype. Leukemia 2013; 27: 1688–1696. [DOI] [PubMed] [Google Scholar]
- Chuang SS, Liao YL, Liu H, Lin SH, Hsieh PP, Huang WT et al. The phenotype of intraepithelial lymphocytes in Taiwanese enteropathy-associated T-cell lymphoma is distinct from that of the West. Histopathology 2008; 53: 234–236. [DOI] [PubMed] [Google Scholar]
- Tan SY, Ooi AS, Ang MK, Koh M, Wong JC, Dykema K et al. Nuclear expression of MATK is a novel marker of type II enteropathy-associated T-cell lymphoma. Leukemia 2011; 25: 555–557. [DOI] [PubMed] [Google Scholar]
- Okumura K, Ikebe M, Shimokama T, Takeshita M, Kinjo N, Sugimachi K et al. An unusual enteropathy-associated T-cell lymphoma with MYC translocation arising in a Japanese patient: a case report. World J Gastroenterol 2012; 18: 2434–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AL, Swerdlow SH, Przybylski GK, Surti U, Choi JK, Campo E et al. Intestinal gammadelta T-cell lymphomas are most frequently of type II enteropathy-associated T-cell type. Hum Pathol 2013; 44: 1131–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucuk C, Jiang B, Hu X, Zhang W, Chan JK, Xiao W et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat Commun 2015; 6: 6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G et al. Integrative genomics viewer. Nat Biotechnol 2011; 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013; 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutcutache I, Suzuki Y, Tan IB, Ramgopal S, Zhang S, Ramnarayanan K et al. Exome-wide sequencing shows low mutation rates and identifies novel mutated genes in seminomas. Eur Urol 2015; 68: 77–83. [DOI] [PubMed] [Google Scholar]
- Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood 1998; 92: 3780–3792. [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002; 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013; 31: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012; 28: 1811–1817. [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G et al. Patterns of somatic mutation in human cancer genomes. Nature 2007; 446: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, White BS, Dees ND, Griffith M, Welch JS, Griffith OL et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol 2014; 10: e1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescenzo R, Abate F, Lasorsa E, Tabbo F, Gaudiano M, Chiesa N et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell 2015; 27: 516–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons J, Landis CA, Harsh G, Vallar L, Grunewald K, Feichtinger H et al. Two G protein oncogenes in human endocrine tumors. Science 1990; 249: 655–659. [DOI] [PubMed] [Google Scholar]
- Pace AM, Wong YH, Bourne HR. A mutant alpha subunit of Gi2 induces neoplastic transformation of Rat-1 cells. Proc Natl Acad Sci USA 1991; 88: 7031–7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishina H, Nimota K, Kukimoto I, Maehama T, Takahashi K, Hoshino S et al. Significance of Thr182 in the nucleotide-exchange and GTP-hydrolysis reactions of the alpha subunit of GTP-binding protein Gi2. J Biochem 1995; 118: 1083–1089. [DOI] [PubMed] [Google Scholar]
- O'Hayre M, Vazquez-Prado J, Kufareva I, Stawiski EW, Handel TM, Seshagiri S et al. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat Rev Cancer 2013; 13: 412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Wu HT, Vandin F, Raphael BJ. CoMEt: a statistical approach to identify combinations of mutually exclusive alterations in cancer. Genome Biol 2015; 16: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011; 476: 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 2011; 471: 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, Gentles AJ, Nair RV, Irish JM, Kihira S, Liu CL et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood 2013; 121: 1604–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore A, Jurinovic V, Kridel R, Hoster E, Staiger AM, Szczepanowski M et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol 2015; 16: 1111–1122. [DOI] [PubMed] [Google Scholar]
- Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 2005; 352: 1779–1790. [DOI] [PubMed] [Google Scholar]
- Kawashima-Goto S, Imamura T, Seki M, Kato M, Yoshida K, Sugimoto A et al. Identification of a homozygous JAK3 V674A mutation caused by acquired uniparental disomy in a relapsed early T-cell precursor ALL patient. Int J Hematol 2015; 101: 411–416. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 2011; 471: 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer 2015; 15: 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LJ, Kang YH, Smith MO, Tharmalingham H, Ramamurthy N, Fleming VM et al. Human MAIT and CD8alphaalpha cells develop from a pool of type-17 precommitted CD8+ T cells. Blood 2012; 119: 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011; 27: 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Vogiatzi P, Puhr M, Dagvadorj A, Lutz J, Ryder A et al. Stat5 promotes metastatic behavior of human prostate cancer cells in vitro and in vivo. Endocr Relat Cancer 2010; 17: 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SK, Gallego C, Johnson GL, Heasley LE. MAP kinase is constitutively activated in gip2 and src transformed rat 1a fibroblasts. J Biol Chem 1992; 267: 7987–7990. [PubMed] [Google Scholar]
- van Biesen T, Luttrell LM, Hawes BE, Lefkowitz RJ. Mitogenic signaling via G protein-coupled receptors. Endocr Rev 1996; 17: 698–714. [DOI] [PubMed] [Google Scholar]
- Verma NK, Davies AM, Long A, Kelleher D, Volkov Y. STAT3 knockdown by siRNA induces apoptosis in human cutaneous T-cell lymphoma line Hut78 via downregulation of Bcl-xL. Cell Mol Biol Lett 2010; 15: 342–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffon C, Adams J, van der Fits L, Wen S, Townsend P, Ganesan A et al. The histone deacetylase inhibitors vorinostat and romidepsin downmodulate IL-10 expression in cutaneous T-cell lymphoma cells. Br J Pharmacol 2011; 162: 1590–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Out-Luiting JJ, Tensen CP, Zoutman WH, Vermeer MH. Exploring the IL-21-STAT3 axis as therapeutic target for Sezary syndrome. J Invest Dermatol 2014; 134: 2639–2647. [DOI] [PubMed] [Google Scholar]
- McGirt LY, Jia P, Baerenwald DA, Duszynski RJ, Dahlman KB, Zic JA et al. Whole-genome sequencing reveals oncogenic mutations in mycosis fungoides. Blood 2015; 126: 508–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey KL, Camps M, Rommel C, Mackay CR. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat Rev Drug Discov 2007; 6: 391–403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.