Abstract
Purpose of Review
The recent publication of The Cancer Genome Atlas molecular taxonomy of primary prostate cancer highlights the increased understanding of the genomic basis of human prostate cancer, but also emphasizes the complexity and heterogeneity of prostate cancer.
Recent Findings
7 molecular subclasses have been defined on the basis of early genomic alterations, which are largely mutually exclusive.
Summary
We review the recent advances in the genomic understanding of human prostate cancer, with focus on molecular subclassification. Broadly, prostate cancer can be classified based upon whether specific genomic rearrangements, such as the TMPRSS2-ERG fusion occur or whether specific alterations such as SPOP and FOXA1 mutations occur. The molecular drivers remain to be identified in a further quarter of human prostate cancers. Depending upon the molecular subclassification and the coincident genomic alterations, specific clinical insights can be gained from this information, including associations with pathologic factors, race, and prognosis, as well as the possibility for future precision therapies.
Keywords: Prostate Cancer, Genomics, Molecular Classification
Introduction
Great progress in understanding the molecular basis of Prostate Cancer (PCa) and the genomic alterations underlying the disease has occurred over the past decade. Next-generation sequencing has allowed the classification of prostate cancers at multiple strata of molecular information, incorporating data at genomic, transcriptomic, epigenetic, and proteomic levels. Distinct molecular subclasses have emerged, with the potential to transform PCa from a poorly-understood, heterogeneous disease with a highly variable clinical course to a collection of homogenous molecular subtypes with relevant clinical implications.
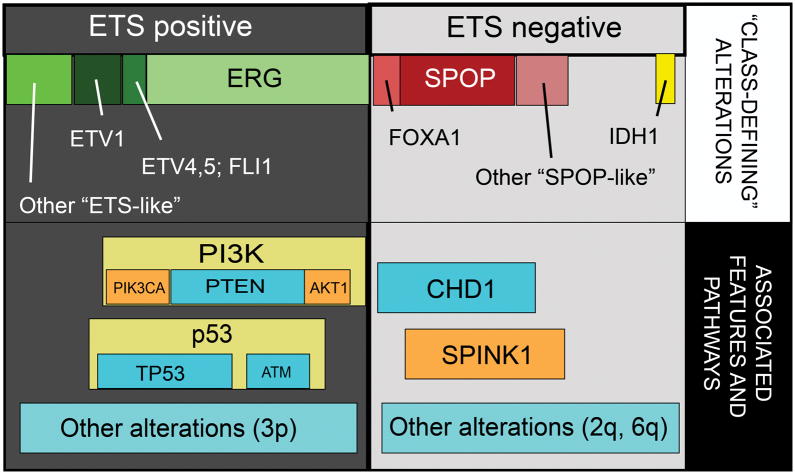
In early genomic and transcriptomic analyses, prostate tumors were able to be stratified based on mRNA expression signatures and patterns of somatic copy number alterations (SCNAs). Several of these studies showed potential for utilization as prognostic biomarker signatures 1–4. Recently published data from The Cancer Genome Atlas (TCGA) support that major molecular subclasses of localized prostate cancer can be divided into ETS-rearrangement prostate cancer (PCa with rearrangements and overexpression of ERG, ETV1, ETV4, or other ETS family transcription factors), SPOP/CHD1 altered cancers, and several smaller categories (Figure) 5. ETS-rearranged tumors are generally enriched in genomic alterations in the PI3K and p53 signaling pathways, while other specific SCNAs predominate in SPOP-mutant cancers (Figure). SPINK1, a secreted serine peptidase inhibitor which is overexpressed in a subset of ETS-negative cancers (including SPOP-mutant cancers) and associated with poor prognostic features, is another marker commonly used for disease classification 6–8. Data such as these will continue to evolve and form the basis for the future molecular classification of prostate cancer. Furthermore, ongoing efforts to establish the timeline of these genomic events and define cancer-initiating lesions versus subsequent alterations perhaps promoting disease progression will be critical for predicting prostate cancer progression and aggressiveness at the molecular level. A molecular definition of progression based on these ideas could prove an invaluable tool for patients on active surveillance or for risk-stratification of intermediate risk patients. In this review, we will summarize the current data and recent findings regarding molecular subtyping of prostate cancer, and explore the potential clinical utility of these disease classification tools.
Figure. Molecular subclasses of clinically localized prostate cancer.
Prostate cancers can be classified into those with rearrangements in ETS family transcription factors (like ERG, ETV1, ETV4, and FLI1), and those negative for ETS factors. ETS negative prostate cancers show recurrent mutations in SPOP, FOXA1, and IDH1. Alterations in PI3K and p53 signaling are common in ETS positive cancers, while deletions of CHD1 and overexpression of SPINK1 are specific to ETS negative cancers.
ETS family members
In 2005, a series of landmark papers reported fusions of the 5′ untranslated region (UTR) of the androgen-regulated TMPRSS2 gene with the ETS family transcription factor family members—most notably, ERG and ETV1 9–12. This discovery provided the framework for the molecular organization of prostate cancers into those with ETS rearrangements and those without. The most common ETS family rearrangement is the TMPRSS2:ERG fusion, which has now been identified in approximately half of prostate cancers and accounts for 90% of ETS family fusions9, 13, 14. Fusions of other ETS family members, including ETV1, ETV4, ETV5, and FLII have since been identified 2, 10, 15, 16. These rearrangements result in overexpression of the ETS family transcription factors which confer a neoplastic phenotype 17. Rearrangements involving ETS family members appear to be largely mutually exclusive and even in rare instances where more than one fusion event was detected in a single tumor focus, clonal heterogeneity and convergent phenotypic evolution are thought to explain these events 2, 5, 10, 12. Several 5′ partners have also subsequently been identified, most notably a fusion product involving the androgen-regulated SLC45A3 with the ETS family member ELK4 in 5–10% of prostate cancers, and less commonly, NDRG1 5, 18, 19. Interestingly, other mechanisms of ETS overexpression have been proposed in patients with full length ETS transcripts and no detectable fusions, including cryptic translocations to heavily-transcribed areas of the genome, and by epigenetic mechanisms 5.
ETS rearrangements have been detected in high grade prostatic intraepithelial neoplasia (HgPIN) and seem to be an early event in PCa carcinogenesis 20–22. ERG rearrangements when detected in HgPIN have also been detected in the adjoining prostate cancer, and are thus theorized to precede other mutations 22–24. Additionally, ERG-rearranged HgPIN is rarely identified distant from ERG-rearranged cancer foci in prostatectomy specimens, suggesting that ERG is important for the transition from HgPIN to cancer 21, 23. Indeed, ERG rearrangements in prostate biopsy specimens containing HgPIN have been shown to be predictive of the development of PCa (53% versus 35%) 25. Mice engineered to overexpress ERG or ETV1 under androgen regulation develop pre-neoplastic prostate lesions, and ERG overexpression accelerates prostate cancer pathogenesis when combined with deletions in PTEN 12, 26, 27.
ETS-positive prostate cancers have been demonstrated to have distinct molecular and clinico-pathological features. These rearranged cancers show a distinct gene expression signature from ETS-negative cancers and also show characteristic SCNAs with a notable pattern of genomic rearrangements involving chains of balanced translocations—a phenomenon described as “chromoplexy” 5, 17, 28–32.
The prevalence of ETS rearrangements has ranged from 27% to 79% in radical prostatectomy and biopsy sample series; these generally represent PSA-screened patients. Given the high frequency of TMPRSS2-ERG fusions in human prostate cancer, numerous studies have investigated the prognostic implications of these rearrangements with often-conflicting results. ETS-rearranged PCa has been found to be associated with more aggressive and more indolent disease, likely reflecting a number of confounding factors including multifocality and intra-prostatic molecular heterogeneity, sampling issues, and the heterogeneity of PSA screening practices and treatment patterns, study cohorts, and design, and outcome measurement33, 34. Supporting evidence for the aggressiveness of ETS-rearranged prostate cancers is largely derived from two studies from watchful waiting cohorts of men diagnosed with PCa on transurethral resection of the prostate (TURP). In both studies, men with TMPRSS2-ERG fusion-positive cancers had an increased risk of death from PCa 35, 36. Additionally, ERG-positive cancers in patients managed with active surveillance have been shown to be associated with an increased risk of progression 37. More recently, TMPRSS2-ERG fusions have been found to be associated with younger age at time of diagnosis and low grade PCa 38.
The impact of ETS fusions on aggressive features or outcome following treatment is less clear, with studies showing positive, neutral, and negative association between ETS fusion status and features of aggressive prostate cancer (including increased Gleason grade, stage, or biochemical recurrence). The largest and most recent is a prospective study of over 1100 patients who were treated with radical prostatectomy and for whom ERG rearrangement or overexpression were found to be associated with tumor stage, but not biochemical recurrence or PCa-specific mortality 39. Of uncertain clinical significance is the fact that anteriorly-located tumors are much less likely to contain ETS rearrangements, a pathological finding which is also associated with increased incidence in African-American patients.40, 41 Furthermore, there appears to be racial variation in the incidence of ETS rearrangements, with African-American patients approximately 50% less likely to have ETS family rearrangements overall, but more likely to have non-ERG ETS family rearrangements in low-risk prostate cancers. 40, 41.
From a molecular standpoint, multiple patterns of hypermethylation changes occur within ETS-rearranged prostate cancers, which may in part explain the variable clinical outcomes seen 5. TCGA analysis of primary prostate cancer specimens found that ERG-positive cancers exhibited two patterns of hypermethylation: approximately two-thirds showed moderate hypermethylation, whereas the remainder belonged to a distinct hypermethylation cluster exclusive to ERG-positive tumors. Interestingly, the hypermethylation patterns of the ERG-rearranged prostate cancers were distinct from other ETS family members which showed heterogenous hypermethylation changes 5. The ETS-rearranged family of prostate cancers is also notable for enrichment of genomic alterations in a number of canonical pathways, including PTEN deletions, TP53 alterations, PI3K pathway alterations and specific amplifications in 3p 5. The molecular diversity within this ETS-rearrangement subclassification may make broad attempts at predicting clinical endpoints based upon this subclassification alone not feasible without further information.
In summary, it appears that ERG rearrangement may be associated with poor prognosis and adverse features in population-based studies of watchful waiting cohorts, but series of patients treated with radical prostatectomy have conflicting results regarding aggressiveness and prognosis. A variety of factors, including variation in techniques to detect ERG rearrangement and lack of PSA screening in presently evaluated population cohorts, complicate interpretation across studies. Furthermore, there is marked epigenetic heterogeneity within the ETS fusion tumor subclass, and additionally, the clinical impact of non-ERG ETS rearrangements (ETV1, ETV4, ETV5, and FLI1) is still unclear.
SPOP/CHD1
Recurrent mutations in the SPOP gene are found in 5–15% of tumors, making it the most common point mutation in PCa 42, 43. SPOP encodes the substrate-binding subunit of a Cullin-based E3 ubiquitin ligase, and mutations affect conserved residues in the structurally defined substrate-binding cleft. SPOP mutation appears to occur exclusively in tumors without ETS rearrangement, and constitute a unique subclass of PCa with several distinguishing molecular characteristics 42. SPOP mutations have been identified in HGPIN adjacent to adenocarcinoma, and likely represent early events in the natural history of PCa 42. SPOP-mutant tumors have been found to have recurrent somatic deletions at 5q21 at the CHD1 locus, as well as loss of 2q and 6q 42, 43. CHD1 is an ATP-dependent chromatin-remodeling enzyme, and the genomic locus is deleted in approximately 5–10 % of prostate cancers 44, 45. Prostate cancers with homozygous CHD1 loss display increased genomic rearrangements 44. Intriguingly, SPOP-mutant/CHD1-deleted primary prostate cancers have been recently shown to possess homogenous gene expression patterns, have elevated levels of DNA methylation, and to overexpress SPINK15,
A recent study found no association between SPOP mutation and clinical or pathological parameters 43; however, others have reported that mutations and decreased expression of the SPOP gene are associated with worse progression free survival 46. Functionally, SPOP mutation has been shown to modulate carcinogenesis by preventing the degradation of oncogenic factors including ERG and the androgen receptor 47–51. Concordant with this, SPOP-mutant tumors have been found to have among the highest androgen receptor transcriptional activity 5. Importantly, it has been recently demonstrated that SPOP modulates DNA double strand break (DSB) repair, is associated with genomic instability, and sensitizes to DNA damaging agents such as PARP inhibitors 52.
SPINK1
Using the same Cancer Outlier Profile Analysis (COPA) used to define ETS gene rearrangements, Tomlins et al identified a second subclass of prostate cancers, which overexpress Serine peptidase inhibitor, Kazal type 1 (SPINK1) 28. SPINK1 is commonly overexpressed in SPOP-mutant and other ETS-negative prostate cancers (Figure) 5. SPINK1 outlier expression has been identified in ~10% of prostate cancers, and appears to be mutually exclusive from ERG rearrangements 6. Interestingly, patients harboring these tumors were found to have a shorter time to biochemical recurrence than patients who do not overexpress SPINK1. SPINK1 outlier status, independent of Gleason score, lymph node status, surgical margin status, seminal vesicle invasion, extracapsular extension, and preoperative PSA, has been shown to be a significant predictor of clinical recurrence 6. SPINK1 overexpressing tumors have also been found to be associated with higher Gleason scores and African-American patients 53. SPINK1 is an extracellular secreted protein and therefore is amenable to both therapeutic targeting and non-invasive diagnosis 6, 54, 55. Indeed, studies using antibodies against SPINK1 in mouse prostate cancer xenografts have identified SPINK1 as a likely target in patients harboring SPINK1+/ETS− tumors54.
FOXA1 mutations
Forkhead box A1 (FOXA1) is a pioneering transcription factor of the androgen receptor which is thought to affect prostate cancer oncogenesis and progression through multiple mechanisms 56. The mutations that define the subset of FOXA1-mutant prostate cancers are mostly missense mutations altering the winged-helix DNA binding domain, the effect of which is currently unknown, and occur at a frequency of approximately 4% of primary prostate cancers 5, 29, 42. Additionally, tumors with FOXA1 mutations were found to have similar molecular features to SPOP-mutant tumors, including similar mRNA, SCNAs, and methylation profiles 5 Furthermore, along with SPOP-mutant cancers, FOXA1 mutations were associated with the highest levels of androgen receptor transcriptional activity in TCGA cohort 5. While FOXA1 and SPOP mutations were mostly mutually exclusive, several tumors exhibited concurrent FOXA1 and SPOP mutations within the same dataset, which retained elevated levels of androgen receptor transcription.
IDH1 mutations
The metabolic enzyme, Isocitrate dehydrogenase-1 (IDH1), is recurrently mutated in several human malignancies including acute myeloid leukemia and gliomas, and result in a methylator phenotype 57. Increased production of the oncometabolite 2-hydroxyglutarate via neomorphic activity of IDH1 gained through characteristic mutations is thought to result in the inhibition of Tet Methylcytosine Dioxygenase 2 (TET2), thereby resulting in hypermethylation across the genome 57.
The integration of multiple genomic platforms in primary PCa allowed for the identification of this rare, novel molecular subclass of prostate cancers characterized by IDH1 mutations, most notably at residue R132 5, 58. These cancers were found to be associated with early age of onset, few SCNAs, and similar to IDH1-mutant gliomas and acute myeloid leukemias, vast, genome-wide hypermethylation, although at disease-specific loci. While uncommon, this mutation may be clinically actionable, as clinical trials with IDH1 inhibitors specific to R132 IDH1-mutants are ongoing in acute myelogenous leukemia and other malignancies.59
Conclusion
Identification of early driver genomic events in the oncogenesis of PCa has allowed for a schema for the molecular classification of prostate cancer, which increasingly can inform clinical decision-making and aid in the development of precision therapies. However, even within these broad molecular subclassifications, PCa remains a heterogenous disease, making clinically-relevant observations challenging. Despite the multiplatform, intensely characterized TCGA genomic analysis of a large cohort of primary prostate cancers, molecular drivers could not be identified in 26% of patients with both low and high grade tumors 5. Nevertheless, recent strides in the understanding of the molecular basis of human prostate cancer will continue to improve clinical insights gained through the use of genomics and assist in the development of targeted strategies for the treatment of advanced disease.
Key Points.
Primary prostate cancer can be molecularly classified into at least 7 subclasses based upon mostly mutually-exclusive early genomic alterations
A number of these alterations provide clinically-relevant insights, including associations with race, disease aggressiveness, and tumor location.
Some of these molecular subclasses, including IDH1-mutant PCa, may provide avenues towards precision medicine-based therapies in the instance of advanced disease.
Acknowledgments
We thank Dr. Mark Rubin and Dr. Yu Chen for mentorship and helpful discussions, and members of the Barbieri, Rubin, and Chen labs.
Financial support and sponsorship
S.D.K. is supported by NIH grant T32CA082088-15. C.E.B. is supported by the Prostate Cancer Foundation, and a Urology Care Foundation Rising Star in Urology Research Award.
Footnotes
Conflicts of interest
C.E.B. is co-inventor on a patent issued to Weill Medical College of Cornell University on SPOP mutations in prostate cancer.
References
- 1.Lapointe J, Li C, Higgins JP, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:811. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lapointe J, Li C, Giacomini CP, et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- 4.Setlur SR, Mertz KD, Hoshida Y, et al. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. Journal of the National Cancer Institute. 2008;100:815. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Cancer Genome Atlas Research Network. Electronic address, s. c. m. o., Cancer Genome Atlas Research, N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011. doi: 10.1016/j.cell.2015.10.025. Comprehensive molecular characterization of over 300 clinically localized prostate cancer samples. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomlins SA, Rhodes DR, Yu J, et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell. 2008;13:519. doi: 10.1016/j.ccr.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paju A, Hotakainen K, Cao Y, et al. Increased expression of tumor-associated trypsin inhibitor, TATI, in prostate cancer and in androgen-independent 22Rv1 cells. European urology. 2007;52:1670. doi: 10.1016/j.eururo.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 8.Lippolis G, Edsjo A, Stenman UH, et al. A high-density tissue microarray from patients with clinically localized prostate cancer reveals ERG and TATI exclusivity in tumor cells. Prostate cancer and prostatic diseases. 2013 doi: 10.1038/pcan.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 10.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 11.Tomlins SA, Mehra R, Rhodes DR, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 12.Tindall DJ. Protein Reviews. Vol. 16. New York, NY: Springer New York: Imprint: Springer; 2013. SpringerLink (Online service): Prostate Cancer Biochemistry, Molecular Biology and Genetics; p. XII.p. 522. 41 illus., 38 illus. in color. [Google Scholar]
- 13.Pettersson A, Graff RE, Bauer SR, et al. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21:1497. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomlins SA, Bjartell A, Chinnaiyan AM, et al. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol. 2009;56:275. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 15.Helgeson BE, Tomlins SA, Shah N, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 16.Paulo P, Barros-Silva JD, Ribeiro FR, et al. FLI1 is a novel ETS transcription factor involved in gene fusions in prostate cancer. Genes Chromosomes Cancer. 2012;51:240. doi: 10.1002/gcc.20948. [DOI] [PubMed] [Google Scholar]
- 17.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 18.Maher CA, Kumar-Sinha C, Cao X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rickman DS, Pflueger D, Moss B, et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009;69:2734. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perner S, Mosquera JM, Demichelis F, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 21.Park K, Tomlins SA, Mudaliar KM, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerveira N, Ribeiro FR, Peixoto A, et al. TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006;8:826. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosquera JM, Perner S, Genega EM, et al. Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications. Clin Cancer Res. 2008;14:3380. doi: 10.1158/1078-0432.CCR-07-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furusato B, Tan SH, Young D, et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 2010;13:228. doi: 10.1038/pcan.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park K, Dalton JT, Narayanan R, et al. TMPRSS2:ERG gene fusion predicts subsequent detection of prostate cancer in patients with high-grade prostatic intraepithelial neoplasia. J Clin Oncol. 2014;32:206. doi: 10.1200/JCO.2013.49.8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King JC, Xu J, Wongvipat J, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Setlur SR, Mertz KD, Hoshida Y, et al. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst. 2008;100:815. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demichelis F, Rubin MA. TMPRSS2-ETS fusion prostate cancer: biological and clinical implications. J Clin Pathol. 2007;60:1185. doi: 10.1136/jcp.2007.046557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baca SC, Prandi D, Lawrence MS, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbieri CE, Rubin MA. Genomic rearrangements in prostate cancer. Curr Opin Urol. 2015;25:71. doi: 10.1097/MOU.0000000000000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomlins SA, Bjartell A, Chinnaiyan AM, et al. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. European urology. 2009;56:275. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 34.Brenner JC, Chinnaiyan AM, Tomlins SA. ETS Fusion Genes in Prostate Cancer. In: Tindall DJ, editor. Prostate Cancer: Biochemistry, Molecular Biology and Genetics. New York: Springer New York; 2013. pp. 139–183. vol. Protein Reviews. [Google Scholar]
- 35.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 36.Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berg KD, Vainer B, Thomsen FB, et al. ERG protein expression in diagnostic specimens is associated with increased risk of progression during active surveillance for prostate cancer. Eur Urol. 2014;66:851. doi: 10.1016/j.eururo.2014.02.058. [DOI] [PubMed] [Google Scholar]
- 38.Steurer S, Mayer PS, Adam M, et al. TMPRSS2-ERG fusions are strongly linked to young patient age in low-grade prostate cancer. Eur Urol. 2014;66:978. doi: 10.1016/j.eururo.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 39.Pettersson A, Graff RE, Bauer SR, et al. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21:1497. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faisal FA, Sundi D, Tosoian JJ, et al. Racial Variations in Prostate Cancer Molecular Subtypes and Androgen Receptor Signaling Reflect Anatomic Tumor Location. Eur Urol. 2015 doi: 10.1016/j.eururo.2015.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khani F, Robinson BD. A “Chicken or Egg” Conundrum: Race, Molecular Subtype, and Tumor Location in Prostate Cancer. Eur Urol. 2015 doi: 10.1016/j.eururo.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 42.Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blattner M, Lee DJ, O’Reilly C, et al. SPOP mutations in prostate cancer across demographically diverse patient cohorts. Neoplasia. 2014;16:14. doi: 10.1593/neo.131704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W, Lindberg J, Sui G, et al. Identification of novel CHD1-associated collaborative alterations of genomic structure and functional assessment of CHD1 in prostate cancer. Oncogene. 2012;31:3939. doi: 10.1038/onc.2011.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang S, Gulzar ZG, Salari K, et al. Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene. 2012;31:4164. doi: 10.1038/onc.2011.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia-Flores M, Casanova-Salas I, Rubio-Briones J, et al. Clinico-pathological significance of the molecular alterations of the SPOP gene in prostate cancer. Eur J Cancer. 2014;50:2994. doi: 10.1016/j.ejca.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Theurillat JP, Udeshi ND, Errington WJ, et al. Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science. 2014;346:85. doi: 10.1126/science.1250255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.An J, Ren S, Murphy SJ, et al. Truncated ERG Oncoproteins from TMPRSS2-ERG Fusions Are Resistant to SPOP-Mediated Proteasome Degradation. Mol Cell. 2015;59:904. doi: 10.1016/j.molcel.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 49.Gan W, Dai X, Lunardi A, et al. SPOP Promotes Ubiquitination and Degradation of the ERG Oncoprotein to Suppress Prostate Cancer Progression. Mol Cell. 2015;59:917. doi: 10.1016/j.molcel.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geng C, Rajapakshe K, Shah SS, et al. Androgen receptor is the key transcriptional mediator of the tumor suppressor SPOP in prostate cancer. Cancer Res. 2014;74:5631. doi: 10.1158/0008-5472.CAN-14-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.An J, Wang C, Deng Y, et al. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6:657. doi: 10.1016/j.celrep.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Boysen G, Barbieri CE, Prandi D, et al. SPOP mutation leads to genomic instability in prostate cancer. Elife. 2015;4 doi: 10.7554/eLife.09207. Defines SPOP mutant prostate cancer as predisposed to genomic instability, with clinical implications as to therapeutic susceptibility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomlins SA, Alshalalfa M, Davicioni E, et al. Characterization of 1577 primary prostate cancers reveals novel biological and clinicopathologic insights into molecular subtypes. Eur Urol. 2015;68:555. doi: 10.1016/j.eururo.2015.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ateeq B, Tomlins SA, Laxman B, et al. Therapeutic targeting of SPINK1-positive prostate cancer. Sci Transl Med. 2011;3:72ra17. doi: 10.1126/scitranslmed.3001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laxman B, Morris DS, Yu J, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin HJ, Zhao JC, Ogden I, et al. Androgen receptor-independent function of FoxA1 in prostate cancer metastasis. Cancer Res. 2013;73:3725. doi: 10.1158/0008-5472.CAN-12-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kang MR, Kim MS, Oh JE, et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125:353. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- 59*.Okoye-Okafor UC, Bartholdy B, Cartier J, et al. New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat Chem Biol. 2015;11:878. doi: 10.1038/nchembio.1930. Suggests that rare IDH1 mutant prostate cancers may be susceptible to targeted therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]