Abstract
Unbiased, high-throughput assays to detect and quantify DNA double-stranded breaks (DSBs) genome-wide in mammalian cells will facilitate basic studies of mechanisms that generate and repair endogenous DSBs. They will also enable more applied studies, such as evaluating on- and off-target activities of engineered nucleases. Here we describe a linear amplification-mediated high-throughput genome-wide sequencing (LAM-HTGTS) method for detecting genome-wide “prey” DSBs via their translocation in cultured mammalian cells to a fixed “bait” DSB. Bait-prey junctions are cloned directly from isolated genomic DNA using LAM-PCR and unidirectionally ligated to bridge adapters; subsequent PCR steps amplify the single-stranded DNA junction library in preparation for Illumina paired-end Miseq sequencing. A custom bioinformatic pipeline identifies prey sequences that contribute to junctions and maps them across the genome. LAM-HTGTS differs from related approaches because it detects a wide range of broken end structures with nucleotide level resolution. Familiarity with nucleic acid methods and next-generation sequencing analysis are necessary for library generation and data interpretation. LAM-HTGTS assays are sensitive, reproducible, relatively inexpensive, scalable, and straightforward to implement with a turnaround time of less than one week.
Introduction
DNA double-stranded breaks (DSBs) are intrinsic to various biological processes such as transcription, are programmed to generate antigen receptor diversification in lymphocytes, and are key substrates for translocations, deletions and amplifications associated with various cancers1,2. There is currently also great interest in defining the range of DSBs across the genome generated by engineered nucleases used for gene editing3. In somatic mammalian cells, a large proportion of DSBs are rejoined by the classical non-homologous DNA end-joining pathway4. Such rejoining often may be accompanied by end-processing—including resections—that can lead to deletion of sequences flanking the break-site5. DSBs that are not immediately rejoined can participate in chromosomal translocations, which frequently result from end-joining of two distinct DSBs2. In this regard, we consider all events in which two separate DSBs are fused as translocations, including those that result in joining two closely linked DSBs in the same chromosome to generate intra-chromosomal deletions2. The frequency of translocations between two sites in the genome is a function of the frequency at which DSB ends at the two sites are available to be translocated and the frequency at which they are physically juxtaposed (“synapsed”)2. The frequency at which DSBs are available is influenced both by their rate of generation and by how long they persist before being rejoined2. Factors that influence DSB generation, persistence, and synapsis are discussed in subsequent sections below and in prior publications6,7.
There have been many methods employed to locate genomic DSBs over the years but each has its limitations (see Table 1). Recently, we developed a Linear Amplification-Mediated High-Throughput Genome-wide Translocation Sequencing (LAM-HTGTS6) and described its application for detecting off-target activities of various types of engineered nucleases and also for a wide-range of other classes of cellular DSBs6-10. Here, we provide a detailed protocol for LAM-HTGTS based on the methods used in these earlier publications.
Table 1.
Various methods to locate DSBs
| Method* | In vivo/ In situ/ In vitro | Assay Type | Comments |
|---|---|---|---|
| BLESS37 | In situ | Maps un-joined broken ends with sequence-specific adapter | Unbiased; high background; narrow time-window (only maps un-joined ends) |
| ChIP-seq18,34,35 | In vivo | Pulls down proteins specifically binding to broken ends or processed ends | Highly depends on the quality of antibody; narrow time-window (only maps un-joined ends); low resolution |
| Digenome-seq44 | In vitro | Cleavage of genomic DNA followed by standard whole genome sequencing | Requires in vivo cleavage confirmation; high skill requirement for bioinformatic analysis |
| DSB-seq36 | In vitro | Maps un-joined broken ends with biotinylated adapter | Unbiased; high background; narrow time-window (only maps un-joined ends) |
| GUIDE-seq27 | In vivo | Randomly incorporates sequence-specific dsDNA fragment into DSB sites | Unbiased; currently limited use for blunt-ended DSBs |
| HTGTS11 | In vivo | Maps translocations with induced DSBs | Relatively higher cost and lower sensitivity compared to LAM-HTGTS |
| IDLV28,45 | In vivo | Randomly incorporates integrase-deficient viral DNA into DSB sites | Low detection frequency; requires high skill; high cost |
| LAM-HTGTS6,8-10 | In vivo | Maps translocations with induced or highly recurrent DSBs | Higher sensitivity on the break-site chromosome; not applicable to limited material |
| TC-seq33 | In vivo | Maps translocations with induced DSBs | Does not resolve junction structures; relatively higher cost and lower sensitivity compared to LAM-HTGTS |
| Whole genome sequencing (WGS)46 | In vivo | Deep genome sequencing | Covers all types of mutations; expensive; high skill requirement for bioinformatic analysis |
Due to space limitations, only typical methods and references were cited here.
Development of LAM-HTGTS
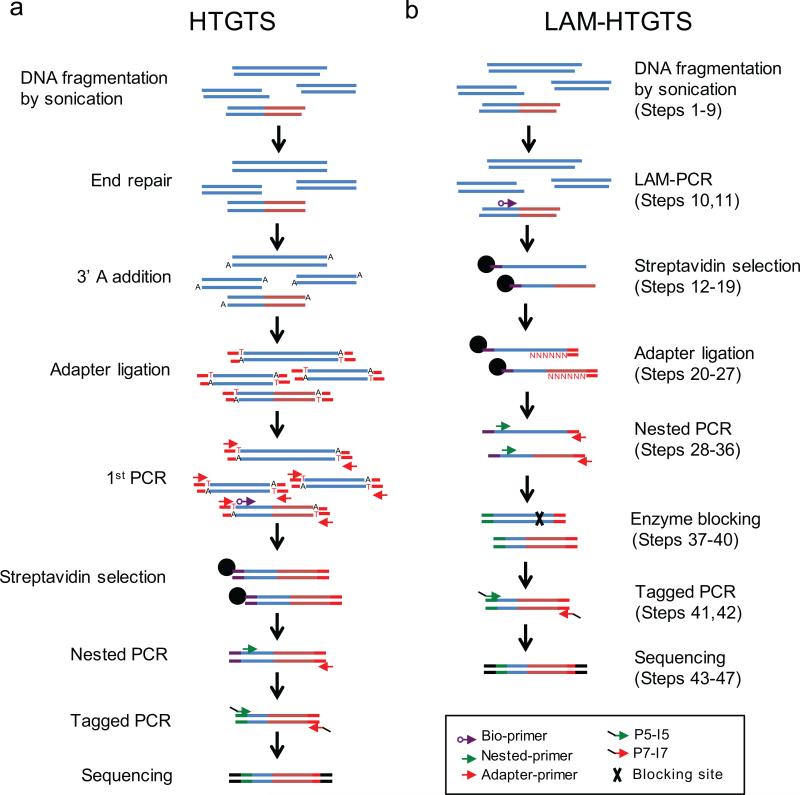
We originally developed an approach called High-Throughput Genome-wide Translocation Sequencing (HTGTS) to identify DSBs genome-wide. HTGTS is based on the ability of DSBs to translocate to a fixed “bait” DSB generated by the yeast I-SceI nuclease, which cleaves at an ectopically integrated 18-bp recognition site in the c-Myc locus of the mouse genome11. Such high-throughput junction cloning11-13, leveraging aspects of whole-genome library construction and next generation sequencing14,15, provides nucleotide-level resolution of translocation junctions that fuse the broken ends of the genome-wide “prey” DSBs to the bait I-SceI DSB (Fig.1a). Prey DSBs represent any broken ends in the cell that join to the bait broken ends; numerous control studies demonstrated that the HTGTS background generated by PCR template switching was very low11. HTGTS not only allows sensitive detection of DSBs genome-wide, but also allows in-depth studies of mechanisms by which these prey DSBs translocate to bait DSBs
Figure 1. Step-by-step overview of HTGTS methods.
(a) The original HTGTS method requires end processing and adapter ligation of sheared genomic DNA fragments prior to PCR amplification, enrichment of biotinylated products, and further amplification steps to increase specificity and to label ends for Miseq sequencing. (b) LAM-HTGTS directly amplifies junctions from sheared genomic DNA using LAM-PCR followed by enrichment and bridge adapter ligation to allow for exponential amplification and Miseq labeling of enriched products.
We have since significantly improved the efficiency of HTGTS and reduced cost, time, and effort by introducing steps to enrich target DNA fragments prior to adapter ligation. This improved method - LAM-HTGTS6 - incorporates LAM-PCR16,17, bridge adapter ligation18, and a customized algorithm to fully characterize sequence reads with respect to the bait DSB employed. LAM-PCR employs a single primer to directly amplify bait-prey junctions from genomic DNA and generate single-stranded DNA (ssDNA) products with diverse ends16,17 (Fig. 1b). The bridge adapter, with a short and nucleotide-variable 3’ overhang, provides a hybridized dsDNA “bridge” to stabilize ligation of the adapter to the diverse ssDNA ends using T4 DNA ligase18 (Fig. 1b). The implementation of these two critical steps improves reproducibility by reducing the number of sample processing steps prior to exponential amplification and increases the junction yield 10-50-fold over the original HTGTS method6.
Overview of the LAM-HTGTS method
The procedure starts with the isolation of genomic DNA from cultured cells using a standard proteinase K digestion method. However, prior to DNA extraction, cells must be cultured for a limited duration to allow for nuclease expression to induce cleavage of the bait break-site and for translocation of bait broken ends to prey DSBs. Prey DSBs can be generated by endogenous mechanisms (e.g. activation-induced cytidine deaminase (AID) or recombination activating gene 1/2 (RAG) cleavage sites, transcriptional start sites, etc.) or ectopic mechanisms (e.g. nuclease-generated DSBs). The numerous approaches available to generate cells with bait DSBs (e.g. transfection, viral transduction, nucleofection) are not described in this procedure but are described elsewhere for commonly used cell lines6,7,9,10.
Genomic DNA is sheared by sonication and the bait-prey junctions are then amplified by LAM-PCR16, using directional primers lying on one or the other side of the bait break-site (or sites). LAM-PCR with a single 5’ biotinylated primer amplifies across the bait sequence into the unknown prey sequence (Fig. 1b). Junction-containing ssDNAs are enriched via binding to streptavidin-coated magnetic beads (Fig. 1b). After washing, bead-bound ssDNAs are unidirectionally ligated to a bridge adapter18. Adapter-ligated, bead-bound ssDNA fragments are then subjected to nested PCR to incorporate a barcode sequence necessary for de-multiplexing (Fig. 1b). Following an optional blocking digest to suppress the potentially large number of uncut and/or perfectly rejoined or minimally-modified bait sequences (Figs. 1b and 2a,b), a final PCR step fully reconstructs Illumina Miseq adapter sequences at the ends of the amplified bait-prey junction sequence (Figs. 1b and 2c). Samples are then separated on an agarose gel, and a resulting population of 0.5-1 kb fragments are collected and quantified prior to Miseq paired-end sequencing, with a typical 2x 250bp HTGTS library sampling ~1×106 sequence reads.
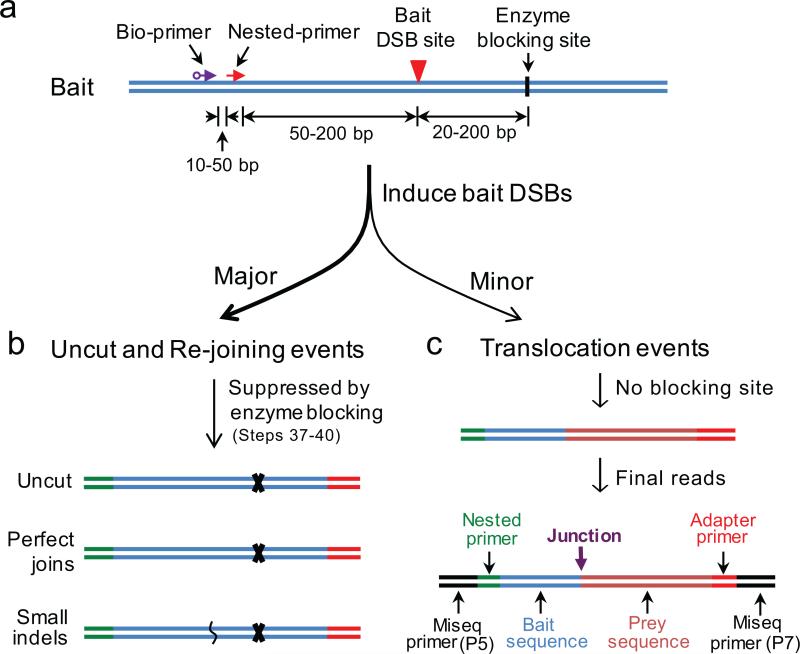
Figure 2. Primer design and enzyme cutting site strategies for LAM-HTGTS.
(a) Strategies for designing biotinylated primer (bio-primer), nested primer and choosing enzyme blocking site for LAM-HTGTS with a given bait DSB site. Note that primers can be placed downstream of the bait DSB site and enzyme blocking sites should be correspondingly upstream of the bait DSB—keeping the relative distances the same—to clone from the other side of the bait DSB. (b) The major events after induction of bait DSBs are uncut, perfect joins and small insertion/deletions (indels) around the bait DSB sites, which are greatly suppressed during the enzyme blocking steps. (c) The minor events after induction of bait DSBs are translocations between bait DSBs and genome-wide DSBs, which lose the enzyme blocking sites and can be readily amplified for Miseq sequencing. The final amplified products will contain the following sequence components in the order listed: Illumina P5-I5, nested primer (with barcode), bait, insertions if any, prey, adapter, Illumina P7-I7. The bait is composed of the nested primer binding sequence leading up to the targeted DSB site. The prey represents the unique genome alignment with the junction representing the resulting join between the bait and prey sequences.
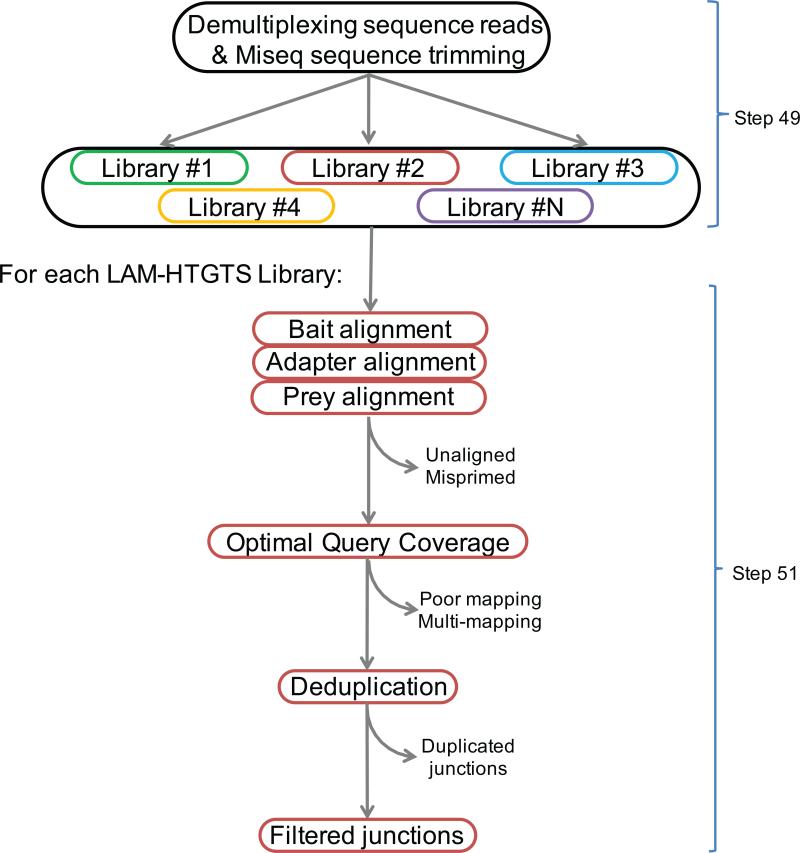
We generated a custom bioinformatic pipeline that can be used to characterize the bait-prey junctions from the library of sequence reads and should be sufficient for most LAM-HTGTS applications using long paired-end sequence reads. The pipeline is available at http://robinmeyers.github.io/transloc_pipeline/ and consists of both third-party stand-alone tools (e.g. aligners) as well as custom programs built in Perl and R, enabling the processing of sequence reads directly off the sequencer into fully annotated translocation junctions in as few as two commands (Fig. 3). Briefly, library pre-processing steps consist of deconvoluting the barcoded libraries and trimming Illumina primers. The main processing pipeline is made up of three major steps: 1) local read alignment, 2) junction detection, and 3) results filtering. We use bowtie 2 to perform read alignments19. The junction detection algorithm is based on the Optimal Query Coverage (OQC) algorithm from the YAHA read aligner and breakpoint detector20. The OQC attempts to achieve the following objective: to optimally infer the full paired-end query sequence from one or more alignments to a reference sequence. The optimal set is determined by using a best-path search algorithm, which enables the detection of not only simple bait-prey junction reads, but also un-joined bait sequences, as well as reads harboring multiple consecutive junctions. The algorithm allows for overlapping alignments, which is required for micro-homology analyses and naturally extends to paired-end reads. The final characterization is an ordered set of alignments termed the Optimal Coverage Set (OCS). The library of resulting OCSs is subjected to a number of filters; the combination of filters and filter parameters used will depend largely on the application. Description of the filters currently employed can also be found at http://robinmeyers.github.io/transloc_pipeline.
Figure 3. Flow chart of bioinformatic pipeline for translocation junction identification.
Multiple HTGTS libraries with different barcodes can be sequenced in the same Miseq flow cell. De-multiplexing separates sequencing reads for each library, followed by sequence read processing which takes into account bait, prey, and adapter alignments to optimally define the sequence read. Uniquely mapped bait-prey junctions are retained as filtered junctions while identical junctions are separated.
Advantages of LAM-HTGTS
To our knowledge, no method, including LAM-HTGTS, is capable of detecting all individual DSBs that occur in a population of cells over a period of time (see Limitations of LAM-HTGTS section). However, thus far, LAM-HTGTS can detect all known classes of recurrent DSBs across the genome, including DSBs introduced by on- or off-target activities of antigen receptor diversification enzymes8,9 and by on- and off-target activities of engineered nucleases6. The assay also detects individual DSBs that occur at lower frequency but are associated with a specific cellular process across the genome, such as active transcription start sites11,21 and sets of DSBs spread across long gene bodies in neural stem and progenitor cells that generate fragile sites7. Finally, the assay also detects low-level wide-spread breaks, such as those generated by ionizing radiation6. In this regard, we also showed that, beyond the I-SceI nuclease, we could employ LAM-HTGTS bait DSBs generated via engineered nucleases such as Cas9 nucleases or TALENs6. Moreover, we also could sensitively employ endogenous DSBs generated by the RAG endonuclease during V(D)J recombination in developing lymphocytes9 or IgH CSR DSBs initiated by AID in activated B lymphocytes as LAM-HTGTS bait DSBs to detect other local or genome-wide DSBs9. The exceptional sensitivity of the method was evidenced by the ability to employ endogenous RAG generated DSBs as bait to discover huge numbers of RAG off-target DSBs locally and across the genome that were not detected by any prior method despite substantial effort put into searching for them9.
Detection of engineered nuclease off-target activity with LAM-HTGTS
Engineered nucleases – including meganucleases20, zinc finger nucleases22, TALENs23,24, and Cas9 nucleases23-26– enable precise targeting of virtually any desired genomic location, However, comprehensive analyses of the collateral damage associated with these nuclease activities had been lacking3. We have previously demonstrated the ability of LAM-HTGTS to reproducibly detect a wide range of off-target nuclease-specific DSB activities, including many predicted but previously undetected sites in addition to new unpredicted sites6. Published examples of how LAM-HTGTS can be used to study nuclease activities include:
1. Characterizing activity of new nucleases
One key to the success of LAM-HTGTS in identifying DSBs genome-wide was the finding that recurrent DSBs can dominate translocation landscapes in mouse and human cells genome-wide regardless of chromosomal location due to cellular heterogeneity in 3-D genome organization2,6,13. LAM-HTGTS can be adapted to use a well-defined engineered nuclease DSB to generate a universal donor bait DSB that can detect the genome-wide prey DSB activities of a co-expressed “uncharacterized” engineered nuclease (Box 1)6. For each uncharacterized nuclease, the frequency at which DSBs occur can be normalized to universal baits, allowing on-target cutting efficiencies of different nucleases to be compared, which is useful for choosing an appropriate nuclease for targeting a desired locus or for a particular application.
2. Sensitive detection of DSBs within chromosomes
In the absence of highly recurrent prey DSBs, relative proximity of bait and prey DSBs becomes a more dominant influence in the frequency at which they translocate6. Thus, treating cells with γ-irradiation to generate wide-spread random DSBs across all chromosomes leads the length of each chromosome to become a translocation “hotspot” for the joining of DSBs generated within it due to proximity effects of sequences within a cis chromosome2,6,13. Within a cis chromosome, translocation frequency is further enhanced between sequences within megabase or sub-megabase topologically-associated domains (TADs) due to further increased interaction frequencies2,12,13. These latter properties allow the sensitivity of LAM-HTGTS DSB detection to be extended by employing bait DSBs on different chromosomes or regions of chromosomes to detect DSBs in proximal regions in cis with increased sensitivity6,7. Due to these spatial proximity effects, off-target breaks on the same chromosome as the bait DSB are more likely to be captured by LAM-HTGTS2,6,13. Thus, the sensitivity of LAM-HTGTS can be enhanced (by 5 fold or more) by placing bait DSBs in or along each chromosome to look for off-targets in cis within the chromosome; we recently employed this approach to identify endogenous recurrent DSB clusters in neural stem and progenitor cells7 (see “Detecting endogenous DSB and joining with LAM-HTGTS” section below).
3. Detection of wide-spread low-level breaks
Unlike other assays, such as GUIDE-seq27 or IDLV28, which only describe recurrent DSB activity, LAM-HTGTS is able to interpret changes in the distribution and proportion of translocations along the chromosome harboring the bait DSB as a measure of the amount of genome-wide DSBs that are of low-level recurrence at any particular site (i.e. wide-spread, low-level); this assay property is due in large part to the spatial proximity effects described in example 2. In this context, universal bait LAM-HTGTS revealed that introducing certain TALENs generated an effect reminiscent of treating cells with ionizing radiation6.
4. Characterizing damage at DSBs
Collateral damage is a key problem for nuclease off-target activities. In addition to providing relative frequencies of on- vs off-target DSBs, LAM-HTGTS can also provide an estimate of the frequency of deletions and translocations occurring between on-target and off-target DSBs and also between different off-target DSBs. Given equal DSB frequencies, deletions and translocations occur more frequently for different DSBs that occur on the cis chromosome. GUIDE-seq27 and IDLV28 have not thus far been reported to have been used for an in depth analysis of this kind.
5. Detection of a wide range of broken ends
A unique feature of LAM-HTGTS is that it detects a wide range of broken ends that can be generated by various classes of nucleases, including blunt ends for Cas9 nucleases, nucleotide overhangs for meganucleases, FokI-domain containing nucleases, paired nickases, and also hairpin-sealed ends from RAG-mediated cleavage6,9,11,13. In this context, LAM-HTGTS detected hundreds of off-targets for two tested TALENs as well as robust low-level wide-spread DSB activity6 and further showed that the vast majority of the many TALEN off-targets resulted from homo-dimers recognizing a palindromic cleavage site, as opposed to desired heterodimers recognizing two different sites6. The versatility of LAM-HTGTS DSB detection, thus, should allow characterization of new classes of designer nucleases such as the recently described Cpf1 CRISPR effector family29.
6. Detection of off-target sites on homologous chromosomes
A key feature of LAM-HTGTS not reported for other nuclease off-target assays is the ability to readily detect a major class of “off-targets” that result from targeting of the same “on-target” site on homologous chromosomes. This ability is not trivial because these events can lead to dicentric chromosomes that could promote additional DSBs, translocations, and potentially oncogene amplifications via breakage-fusion-bridge mechanisms6.
Detecting endogenous DSB and joining with LAM-HTGTS
HTGTS and LAM-HTGTS can both be used to detect DSBs generated from the cellular environment (e.g. ionizing radiation, chemotherapeutics, viral integration, etc.)6,9,13. Both methods also detect DSBs generated via endogenous sources such as transcription-associated DSBs and DSBs associated with replication stress7,11,21, programmed DSB-inducing activities in lymphoid cells8-13,30, and likely could be applied to detect endogenous DSBs that arise from other sources such as oxidative DNA damage. More generally, LAM-HTGTS reveals the various classes of DSBs across the genome that can contribute to inter- or intra-chromosomal translocations and deletions, including sources of DSBs that contribute to known oncogenic translocations8,9.
LAM-HTGTS-based studies employed endogenous AID-initiated DSBs in endogenous switch (S) regions as bait in B cells activated for the IgH CSR10. The design of these studies allowed the fate of 14 different AID-target DSBs within a 150 bp region to be followed via a single bait-site LAM-HTGTS primer; these bait-site DSBs joined mainly to targeted S regions 100-200 kb downstream10. S regions are long (up to 10 kb) and highly repetitive which limited prior CSR junction studies to standard PCR-based assays that generally yielded only dozens of junctions, all of which occurred at the S region borders thus were not fully representative of the dominant core S region driven CSR31. However, the LAM-HTGTS assay provided tens of thousands of junctions spread over the entire length of the repetitive S region, offering hugely expanded data sets and far more mechanistic detail than previously could be generated. In addition, the assay was substantially less expensive and time-consuming10. The CSR studies also revealed how LAMHTGTS could be used as a sensitive joining and end-resection assay with respect to rejoining of single DSBs, revealing differential effects of a broad range of DNA damage response factors on the resection process10.
LAM-HTGTS has been applied to study the on-target and off-target activities of the RAG V(D)J recombination specific endonuclease using endogenous RAG-generated DSBs as bait9. While prior studies detected only a handful of off-target RAG generated DSBs32, the LAM-HTGTS studies identified thousands of RAG off-target sites, which are tightly restricted within chromosomal loop domains, strongly suggesting a linear RAG tracking model to explain the generation of most RAG off-target events9.
Finally, recent LAM-HTGTS studies using neural stem and progenitor cells employed bait DSBs on multiple chromosomes in combination with mild replication stress (induced by aphidicolin) to identify, with enhanced sensitivity, 27 recurrent DSB clusters (RDCs) across the genome7. All 27 RDCs occurred in the gene bodies of very long transcribed genes that mostly were late replicating. Moreover, nearly all of these RDC genes were associated with synapse function, neural cell adhesion and/or mental disorders. A number of RDC genes also were associated with various cancers including brain and prostate cancers7.
Comparison of HTGTS with other related methods
Several other DSB detection assays were developed about the same time as the HTGTS method11 or LAM-HTGTS6 that either leveraged chromosomal translocation cloning or in vivo tagging of broken ends27,28,33. Such methods provide higher resolution than ChIP-seq18,34,35 and lower background than DSB-seq36 and BLESS37. Thus, we limit comparison below to these more recently developed translocation-based or in vivo tagging-based methods. However, we do note a recent report applies BLESS for Cas9 off-target detection using strict custom optimization to address the background38.
TC-seq33 has many overlapping features and applications with HTGTS11, including the use of an I-SceI bait DSB approach to detect prey DSBs. However, TC-seq as described did not allow junction structures to be defined at nucleotide resolution, and thus did not allow precise mapping of I-SceI off-targets33. Also, TC-seq studies reported thus far have not employed endogenous DSBs or engineered nuclease-generated DSBs as bait. However, it seems likely that TC-seq could be readily adapted for use in the various contexts outlined for LAM-HTGTS.
GUIDE-seq27 tags engineered nuclease-induced DSBs with blunt-ended, 5’ and 3’ endphosphorothioated, double-stranded DNA (dsDNA) oligos via end-joining; tagged DSBs are then amplified from the inserted dsDNA fragment and mapped genome-wide. GUIDE-seq is very similar to the IDLV DSB detection assay28 but with higher efficiency than IDLV for DSB detection. In its published form, GUIDE-seq DSB detection was dependent on in vivo blunt end-joining mechanisms due to the type of dsDNA oligo tags employed and, thus, would be limited to detecting only these broken end structures in the cell. Hence, DSBs from other types of engineered nucleases or endogenous DSBs with 5’ or 3’ overhangs may not be readily detected by GUIDE-seq. Despite this blunt end-joining limitation, GUIDE-seq is capable of identifying recurrent Cas9 DSBs throughout the genome. Indeed, GUIDE-seq identified the same major off-targets as LAM-HTGTS for the 2 common guides tested. However, while LAM-HTGTS and GUIDE-seq also identified some of the same lower level off-targets, they each uniquely identified other low off-targets. Those differences could be attributable to the different cell lines tested, but could reflect differences in the abilities of the two assays to detect certain DSBs. While LAM-HTGTS can be made more sensitive by using more material and by using baits on different chromosomes, it is not clear if the same applies to GUIDE-seq as the background relative to off-target detection has not been described.
Like HTGTS and TC-Seq, GUIDE-seq requires end-processing and adapter ligation prior to selecting informative sequences27; such an approach was found to present significant financial burden11,33 (Fig. 1a). In contrast, LAM-HTGTS directly amplifies relevant sequences from sheared genomic DNA without prior end-modification, A-tailing, and adapter-ligation, making it approximately 5 times less expensive than the HTGTS method (Fig. 1b).
Limitations of LAM-HTGTS
Although LAM-HTGTS can compare relative recurrent DSB frequencies, currently LAM-HTGTS and all other related assays cannot readily quantify absolute cutting rates due to their inability to differentiate uncut sequences from cut sequences that have perfectly rejoined or rejoined bait sites in regions of very limited sequence diversity10.
LAM-HTGTS only detects DSBs that translocate. However, this potential limitation has thus far not been an issue as HTGTS has been documented to detect all different classes of DSBs, including many that were basically undetectable by other methods6-13,30 (see Advantages of LAM-HTGTS).
LAM-HTGTS requires joining of prey DSBs to a known bait DSB and, therefore, cannot be used on previously isolated genomic DNA without a priori knowledge of a recurrent DSB that can serve as bait, such as AID-initiated or RAG-initiated DSBs in B lymphocytes9,10.
Also, LAM-HTGTS only reveals information about the genomic prey DSBs that join to bait DSBs and does not reveal information about prey DSBs that persist as DSBs (See Advantages of LAM-HTGTS). We note, however, that studies using γ-H2AX and 53BP1 foci as markers for DSBs indicate that most DSBs are resolved well within our recommended culture times39 (see Sample requirements).
Translocations are rare events in contrast to rejoining events observed as local insertions and/or deletions (Fig. 2) and are estimated to occur in 1 out of 300 cells by live cell microscopy40 and 1 out of 200-1000 cells on average across HTGTS libraries (can widely vary based on multiple conditions; see Sample requirements) which may constrain the utility of LAM-HTGTS in certain contexts where input DNA is limited.
Recurrent DSBs in highly repetitive regions might also be misrepresented due to difficulties in mapping the sequence reads and due to the potential for mis-priming from incompletely extended PCR products; such problems are universal for any amplification-based high-throughput sequencing method. Notably, however, LAM-HTGTS has, for example in the case of IgH CSR, been useful for solving such potentially confounding issues10.
EXPERIMENTAL DESIGN
Sample requirements
After induction of the recurrent DSBs, cells should be cultured for a sufficient length of time to enable the formation of translocations. We typically culture cells for 48-72 hours after nuclease transfection or induction. Generally, DSBs can be efficiently repaired within 8 hours based on studies of γ-H2AX and 53BP1 foci39, thus 48-72 hours should be sufficient for broken ends to form translocations. Genomic DNA can be isolated using any published method that generates fully dissolved DNA with an absorbance 260/280 ratio higher than 1.8. The amount of starting material required to generate robust HTGTS libraries will be context-dependent, but for initial LAM-HTGTS studies we recommend starting with 20-100 μg genomic DNA at 0.5-1×106 Miseq sequence read depth; based on our findings with bait DSBs generated by I-SceI, Cas9, or TALENs6,8, this should be sufficient to identify thousands of translocations if DSB generation is efficient. However, the final yield of identified junctions may vary considerably depending on the context of the experiment, genetic backgrounds (e.g. between repair deficient versus wild-type), and most notably, the ability to generate sufficient bait DSBs in a particular cell type. We generally perform preliminary libraries to confirm that our HTGTS junction yields for a given experimental setting will be sufficient to achieve the goals of the experiment. Means to increase the number of junctions detected per amount of DNA can include increasing nuclease expression levels for greater bait DSB cleavage, longer culturing periods (though potentially at the cost of affecting junction bias due to selective forces), and deeper sequencing of the library.
Controls
Artifactual background effects can vary depending on the position of and priming strategy used for the bait DSB site; therefore, proper controls must be included to enable full interpretation of the data. To evaluate the primers and determine the level of background, it is essential to generate a control library with the genomic DNA from untreated cells (i.e. no bait DSB). Generally, experimental libraries should generate at least 10-fold more junctions than these uncut control libraries using the same set of primers.
Choice of bait DSB region
Each bait DSB provides two broken ends and, thus, there are two potential bait DSB strategies: either a (+) or (−) chromosomal orientation. Bait sequence within 1kb of the targeted DSB should be analyzed to avoid potential repeat sequences, as determined by repeat masker (www.repeatmasker.org), which can be prone to junction artifacts due to mispriming. It is recommended to clone the bait sequence region from the target cells of interest and sequence for potential polymorphisms which could disrupt nuclease cutting or priming. Finally, it is also suggested, but not required, to identify a rare restriction enzyme site downstream of the bait DSB to suppress detection of uncut or perfectly rejoined sequence and to enhance detection of translocations (Fig. 2a,b; see ‘Blocking enzyme’ section below).
Primer design
Bait sequence length leading up to the bait DSB can be varied but is constrained by the positions of the primers used and sequencing length limitations. LAM-HTGTS uses a nested priming strategy with extension times to amplify 1kb of sequence per cycle. For 2×250bp Illumina Miseq, the outer biotinylated locus primer can be positioned up to 400 bp away from the bait DSB, whereas the nested locus primer (nested primer) must be placed within 200 bp (ideally 80-150 bp) of the bait DSB to allow for optimal contiguous junction mapping across bait and prey sequences (Fig. 2a). Shorter bait sequences limit the number of junctions identified due to resection of the bait sequence beyond the sequencing primer. Longer bait sequences limit the sequence from the forward paired read that is available to uniquely map the translocation partner. This limitation may be partially mitigated if the alignment extends to the reverse paired read. The length of primers range from 20-25 bp, with an optimal melting temperature around 58°C and 60°C for bio-primer and nested primer, respectively. To multiplex LAM-HTGTS libraries from the same bait we typically include a user-defined barcode sequence (0-10 bp) positioned between the nested primer sequence on the 3’ end and a portion of the Illumina-specific sequence on the 5’ end of the primer.
Blocking enzyme (optional)
Translocations are rare cellular events compared to cut and perfectly rejoined or local processing of the bait DSB. Thus, to enhance detection of genome-wide DSBs when the bait DSB-positive cell population and/or cutting levels at the bait DSBs are low, it is suggested to block the amplification of uncut or perfectly rejoined sequence after adapter ligation and nested PCR by using rare restriction enzymes that will cleave downstream of the bait DSB and block PCR amplification due to loss of adapter priming (Figs. 1b and 2a,b). To minimize junction loss at the break-site, the blocking enzyme site should be located as close as possible to the downstream side of the bait DSB. Since restriction enzymes have wide-ranging numbers of substrate sites genome-wide, primarily determined by the length of their recognition sequences, enzymes with six or greater base pair recognition are required. Since the blocking step uses PCR amplified DNA, virtually any rare cutting restriction enzyme that has been employed previously for molecular cloning or Southern analysis can be used. Blocking will only suppress but not eliminate all of the uncut or perfectly rejoined fragments since cutting will not be 100% efficient and some uncut or perfectly rejoined sequences would still be observed. It should be noted that the choice of blocking enzyme should not conflict with nested primers and the bait sequence leading up to and including the break-site. The particular blocking enzyme used will reduce the number of prey junctions harboring the same enzymatic site; thus blocking uncut or perfectly rejoined amplification can be omitted in circumstances where the majority of cells are efficiently cutting at their on-target site. Moreover, deeper sequencing can largely compensate for the omission of enzyme blocking, particularly for lower cutting efficiency at bait DSBs.
DNA polymerase
Any thermo-stable DNA polymerase engineered for PCR should be appropriate for use in LAM-HTGTS. We tested both Taq (Qiagen) and Phusion (Thermo Scientific) to prepare LAM-HTGTS libraries; they showed similar genome-wide profiles and back-to back comparison showed no major difference between the HTGTS libraries generated by these two thermal polymerases. Taq is economical, but its short half-life requires the addition of more polymerase half-way through the 100-cycle PCR6,16,17. The proofreading activity of Phusion enhances the amount of amplified DNA fragments and can increase fidelity across secondary DNA structures. Nonetheless, the proofreading activity also can degrade primers and ssDNA products in the LAM-PCR step. To minimize this, a higher concentration of dNTPs are used (3-fold higher than with Taq).
DNA fragmentation
Although genomic DNA can be used for LAM-PCR directly, the elongation time needs to be very limited (5 seconds17) to suppress the formation of very long amplicons. Furthermore, the reduced accessibility of the biotinylated primer to the denatured long filaments of genomic DNA also reduces the efficiency of LAM-PCR. Shearing DNA into ~1 kb fragments minimizes the accessibility problem, and an extended elongation time (1.5 min in this protocol) suppresses PCR-mediated recombination41. Fragmentation of genomic DNA by sonication is preferred over enzymatic digestion, which requires the presence of a nearby restriction site to capture any given translocation. With sonication, coverage across the genome is less biased leading to more comprehensive genome-wide coverage of potential recurrent DSBs.
Bridge adapter
Standard library preparation protocols for genome-wide sequencing typically require end-polishing and 3’ A-tailing of dsDNA11. To ligate adapters to the ssDNA generated in the LAM-PCR, we use a bridge-adapter ligation strategy18, which introduces a single-stranded “bridge” oligo to stabilize both the adapter and the 3’ end of the unknown prey sequence and improve ligation efficiency; the 3’ ends of the adapter and bridge oligo are amino-modified to suppress adapter-to-adapter ligation. Compared with T4 RNA ligase, the T4 DNA ligase-mediated bridge ligation for ssDNA has higher efficiency, less bias, and lower background18,42.
Sequencing and pre-processing
HTGTS libraries are prepared such that the barcode and bait sequence are always sequenced on read 1 (P5 Illumina adapter) and the adapter end sequenced on read 2 (P7 Illumina adapter) (Fig. 2c). HTGTS libraries are pooled, with the number of libraries per pool depending on the desired number of sequence reads per library, before loading on the flow cell. Pre-processing parameters should be selected depending on the length and uniqueness of the library barcodes.
Alignment and OQC
Reads are aligned to the full reference genome, the bait sequence, and the adapter sequence. Read 1 (R1) and read 2 (R2) are aligned independently and the top scoring alignments from each are passed to the junction detection algorithm. For OCS determination, all R1 and R2 alignments, as well as R1/R2 properly-aligned pairs, are conceptualized as nodes on a directed acyclic graph. The graph may be initialized and guaranteed acyclic by ordering nodes with their query start coordinate and using the following edge rules: an R1 node may only follow other R1 nodes with a smaller query start coordinate; an R1/R2 properly-aligned pair may only follow R1 nodes with smaller query start coordinate; an R2 node may only follow R1/R2 nodes or R2 nodes with a smaller query coordinate. Importantly, an R2 node may not immediately follow an R1 node as this would indicate the junction occurs between the reads. This event may occur, but cannot be fully characterized and inspected as an artifact, and thus is not considered. For each node, the scores of its edges to previous nodes explored are calculated, and the edge with the highest score is retained. Edges are scored by summing the alignment score of the new node with the previous node's score and subtracting any penalties. The OCS is the set of nodes that give the highest scoring path through the graph.
OCSs with large gaps between bait and prey alignments should be removed since they represent unverifiable (artifactual or biological) events. Bait alignments that minimally extend past the priming site should be removed as these represent potential mispriming events. The prey alignment must have a uniquely high alignment score relative to other overlapping alignments. The pipeline allows duplicate junction detection and filtering, since they may arise from either cellular or PCR replication and not independent events. Duplicate junctions may also arise independently, however, particularly in very dense clusters of junctions. Therefore, in the case of apparently low diversity libraries (i.e. many reads contain identical bait-prey junctions) interpretation needs to take into account both biological (e.g. predicted) and technical (e.g. amplification bias/artifactual) sources of the assay.
MATERIALS
Reagents
- Mammalian cells of interest in which recurrent DSBs that are ectopically induced or from known endogenous sites can be used as LAM-HTGTS bait. We have successfully applied LAM-HTGTS to human 293T (ATCC CRL-3216) and A549 (ATCC CCL-185) cell lines, mouse Abelson virus-transformed pro-B and CH12F3 cell lines, mouse bone marrow and splenic B cells, in vitro differentiated T cell precursors, and cultured primary mouse neural stem and progenitor cells.
- CAUTION: The cell lines used in your research should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
Nuclease-free Milli-Q water (H2O, 0.22 μm filter, autoclaved)
- 10% SDS solution (w/v, Thermo Scientific, cat. no. 24730-020)
- CAUTION: SDS is toxic. Wear gloves and avoid inhalation.
Proteinase K (Thermo Scientific, cat. no. 25530-031)
Isopropanol (Fisher Scientific, BP26184)
Ethanol (Pharmco-AAPER, cat. no. 111000200)
Hydrochloric acid (HCl, Fisher Scientific, cat. no. A144-500LB)
2.5-N Sodium hydroxide solution (NaOH, Fisher Scientific, SS414-1)
Phusion High-Fidelity DNA Polymerase (Thermo Scientific, cat. no. F530)
5x Phusion HF buffer (Thermo Scientific, cat. no. F518)
dNTPs (Fisher Scientific, cat. no. 28406522, 2840502, 2840532, 2840512), four dNTPs are mixed equally and diluted with H2O to 2.5 mM each, stored at −20 °C for up to 3 months
Oligos and primers (synthesized by Integrated DNA Technologies, check Table 2 for sequences), modified primers are synthesized at 100 nmol scale with standard desalting
NaCl (American Bioanalytical, cat. no. AB01915)
EDTA (Sigma Life Sciences, cat. no. E5134)
Tris base (Roche, cat. no. 11814273001)
Dynabeads MyONE C1 streptavidin beads (Life Technologies, cat. no. 65002)
T4 DNA ligase (Promega, cat. no. M1808)
10x T4 DNA ligase buffer (Promega, cat. no. C126A)
Hexammine cobalt (III) chloride (Sigma Life Sciences, cat. no. H7891)
PEG8000 (Sigma Life Sciences, cat. no. P2139)
Agarose (Lonza, cat. no. 50004)
1-kb DNA ladder (Thermo Scientific, cat. no. SM0311)
6x DNA loading buffer (Thermo Scientific, cat. no. R0611)
50x TAE buffer (Thermo Scientific, cat. no. B49), dilute to 1x (40 mM Tris, 20 mM acetic acid, 1 mM EDTA) before use.
Miseq 500V2 kit (Illumina, cat. no. MS-102-2003)
- Ethidium bromide (Life Technologies, cat. no. 15585011)
- CAUTION: Ethidium bromide is toxic. Wear gloves.
QIAquick Gel Extraction kit (Qiagen, cat. no. 28706), including buffer QG and PE
Table 2.
Primers for LAM- HTGTS
| Use | Name | Sequences |
|---|---|---|
| Bridge adapter (step 20) | Adapter-upper* | GCGACTATAGGGCACGCGTGGNNNNNN-NH2 |
| Adapter-lower* | /5-Phosphorylation/CCACGCGTGCCCTATAGTCGC-NH2 | |
| Nested PCR (step 28) | I5-Nested** | ACACTCTTTCCCTACACGACGCTCTTCCGATCT BARCODE NESTEDPRIMER |
| I7-Blue | CTCGGCATTCCTGCTGAACCGCTCTTCCGATCTGACTATAGGGCACGCGTGG | |
| Tagged PCR (step 41) | P5-I5*** | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT |
| P7-I7*** | CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTC |
End modifications can be produced by Integrated DNA Technologies, the synthesis code for “/5-Phosphorylation/” is “/5Phos/”, for “-NH2” is “/3AmMO/”, and for “/5-biotin/” is “/5BiosG/”, “N” means random nucleotide.
“Nested primer” is the locus-specific nested primer (in bold), and “barcode” means the DNA sequence to differentiate samples with the same locus-specific nested primer (underlined), thus these samples can be sequenced in the same Miseq run. Barcodes can be any non-tandem DNA sequences between 0 and 10 bp, or use the Miseq index following the manufacturer's instructions.
Sequences from the Miseq primers are marked in italics. Note that I5 and I7 primers share 14-bp homologies at the 3’ end (compare the italic sequences of I5-nested to that of I7-Blue), thus the Tm in step 42 is 62°C to reduce cross-template amplification. Alternatively, P5-I5 can be further shortened from the 3’ end to avoid annealing to the 3’ region of I7 with same sequences.
Equipment
Bioruptor (Diagenode, cat. no. B01010002), including 1.5-ml tube holder
Vortex-Genie 2 (VWR Scientific)
Precision barrier tips (Denville Scientific, cat. no. P1126, P1122, P1096-FR)
1.5-ml TPX microtubes (Diagenode, cat. no. C30010010)
1.5-ml microtubes (Sarstedt, cat. no. 72.690)
0.2-ml PCR tubes (Thermo Scientific, cat. no. AB-045)
Gel image acquisition system (Alpha Innotech, FluorChem SP)
Magnet stand (Life Technologies, cat. no. 12321D)
PCR machine (MJ Research, cat. no. PTC-200)
Rotary mixer (Labindustries, cat. no. 400-110)
Water Bath (Fisher Scientific, cat. no. 15-462-15Q)
Miseq sequencer (Illumina)
Centrifuge (Eppendorf, cat. no. 5415D)
NanoDrop 2000 spectrophotometer (Thermo Scientific)
Electrophoresis system (Fisher Scientific, cat. no. FB-SBR-2025)
0.22 μm Syringe filter (Fisher Scientific, cat. no. SLGP033RB)
Bioinformatics tools and source codes
Standard PC with at least 8Gb RAM
Translocation pipeline source code: http://robinmeyers.github.io/transloc_pipeline/
ea-utils package: https://code.google.com/p/ea-utils/
SeqPrep package: https://github.com/jstjohn/SeqPrep
Bowtie2 package: http://sourceforge.net/projects/bowtie-bio/files/bowtie2/
Samtools: http://samtools.sourceforge.net/
Reagent setup
- Proteinase K stock: dissolve 0.1 g proteinase K powder in 5 ml H2O to make 20 mg/ml stock, aliquot into 0.5 ml per tube and store at −20 °C for up to 3 months.
- CAUTION: Proteinase K is toxic. Wear gloves.
5-M NaCl: dissolve 292.5 g NaCl in H2O, adjust the total volume to 1 L. Autoclave and store at room temperature (RT; 20-25 °C) for up to 1 year.
0.5-M EDTA (pH 8.0): dissolve 186.12 g EDTA•Na2•2H2O in H2O, adjust the pH to 8.0 using 2.5-N NaOH and then the total volume into 1 L. Autoclave and store at RT for up to 1 year.
1-M Tris-HCl (pH 7.4): dissolve 121.14 g Tris base in H2O, adjust the pH to 7.4 using HCl and then the total volume into 1 L. Autoclave and store at RT for up to 1 year.
- Cell lysis buffer: 200-mM NaCl, 10 mM Tris-HCl (pH 7.4), 2 mM EDTA (pH 8.0), and 0.2% SDS; store at RT for up to 6 months; proteinase K is added (final concentration at 200 ng/ml) before use.
- CRITICAL: Prepare fresh aliquot with proteinase K every time before use.
TE buffer: 10 mM Tris-HCl (pH 7.4), 0.5 mM EDTA (pH 8.0); store at RT for up to 6 months.
50% (w/v) PEG8000: dissolve 5 g PEG8000 in H2O at 56°C, adjust the total volume to 10 ml. Filter through 0.22 μm syringe filter, aliquot into 1 ml per tube and store at −20°C for up to 1 year.
20-mM hexammine cobalt (III) chloride: dissolve 0.53 g hexammine cobalt (III) chloride in H2O, adjust the total volume to 100 ml. Store at RT for up to 3 months.
2x B&W buffer: 2-M NaCl, 10 mM Tris-HCl (pH 7.4), 1 mM EDTA (pH 8.0). Dilute with H2O to make 1x B&W buffer. Store at RT for up to 1 year.
Annealing buffer: 25-mM NaCl, 10 mM Tris-HCl (pH 7.4), 0.5 mM EDTA (pH 8.0). Store at RT for up to 1 year.
- 50 μM bridge adapter: dissolve the two DNA oligos (see Table 2) in annealing buffer to a final concentration 400 μM. Mix equal volumes of the two dissolved oligos in a new 1.5-ml microtube, put the tube in 1 L boiling water with a foam floating tube rack, boil for 5 min, then cool down slowly in water to ~30°C on the bench (adapter concentration is 200 μM). Alternatively, the oligos can be annealed on a PCR thermoblock43. Dilute 4-fold (concentration is 50 μM) with H2O, aliquot 100 μl per tube and store at −20°C for up to 2 months.
- CRITICAL: Thaw the adapter on ice before use.
PROCEDURE
Genomic DNA isolation (Timing: 1 day)
- Resuspend 1× 107 mammalian cells (previously treated to generate bait and prey DSBs) in 500 μl of cell lysis buffer and incubate at 56°C overnight (10-16 hours).
- CRITICAL STEP: When performing LAM-HTGTS for the first time with a new cell type or a new set of LAM-HTGTS primers, a control sample (cells without cleavage at the presumed bait DSB sites) should be processed in parallel.
Add 500 μl isopropanol directly into the microtube, and mix immediately by inverting the microtube until the genomic DNA can be seen to form a pellet.
Using a pipette, transfer the DNA pellet to a new microtube containing 1 ml 70% ethanol. Centrifuge at 13,000× g for 5 mins at 4°C.
Discard the supernatant completely; dissolve the pellet in 200 μl TE at 56°C for at least 2 hours.
Check the concentration of a 1 μl aliquot with a NanoDrop; the A260/280 should be above 1.8.
Sonication (Timing: 1 hr)
-
6.Transfer 20-100 μg genomic DNA from step 5 into a 1.5-ml TPX microtube, adjust the final volume to 200 μl with H2O, mix by vortexing and then incubate on ice for 5 min.
- CRITICAL STEP: Make sure the DNA is dissolved completely before proceeding to sonication.
-
7.
Fix the tube in 1.5-ml Bioruptor tube holder, fill in empty spaces of the holder with 1.5-ml TPX microtubes containing 200 μl H2O each.
-
8.Turn on the water bath to cool the Bioruptor system to 4°C, then set the Bioruptor as below to fragment the genomic DNA:
Setting Value Energy output Low Working time 25 seconds Resting time 60 seconds Sonication cycles 2 cycles - CRITICAL STEP: Sonication settings need to be optimized for different sonication equipment.
-
9.After sonication, run 1 μl fragmented DNA on a 1% agarose gel (w/v) in 1× TAE buffer; the DNA smear should range from 0.2-2 kb with a peak at approximately 750 bp.
- PAUSE POINT: Fragmented DNA can be stored at −20°C for months or 4°C for one week.
- CRITICAL STEP: Insufficient sonication or over-sonication of genomic DNA results in lower junction yield.
LAM-PCR (Timing: 6 hr)
-
10.Set up eight 50-μl LAM-PCR reactions for each sample as below:
Reagents Volume (μl) Final 5x Phusion HF buffer 10 1x dNTPs (2.5 mM each) 1.5 75 μM Bio-primer (1 μM) 0.5 10 nM Phusion polymerase (2 U/μl) 0.5 1 U sonicated DNA (from step 8) 25 1-10 μg H2O 12.5 - Total 50 - CRITICAL STEP: The amount of sonicated DNA for each 50-μl LAM-PCR reaction should be 1-10 μg, optimally around 5 μg; eight 50-μl PCR reactions are recommended when using 20-80 μg input genomic DNA and 16 reactions are recommended when using >80 μg input genomic DNA.
-
11.Set the PCR machine to amplify the DNA fragments as below:
Cycle number Denature Anneal Extend 1 98°C, 2 min 2-81 95°C, 30 s 58°C, 30 s 72°C, 90 s 82 72°C, 2 min - PAUSE POINT: Amplified ssDNA fragments can be stored for up to one week at −20°C; longer storage is not recommended because Phusion polymerase may cause slight degradation of ssDNA products.
- CRITICAL STEP: Do not leave PCR products in the PCR machine for too long (> 4 hours) after the PCR amplification is complete since Phusion may resect the 3’ ends of the ssDNA products.
Streptavidin purification (Timing: 3 hr)
-
12
. Pool the 8 PCR products from step 11 in a new 1.5-ml microtube (total volume ~400 μl), add 100 μl 5-M NaCl (1 M final) and 5 μl 0.5-M EDTA (pH 8.0; 5 mM final).
-
13.Transfer 40 μl Dynabeads C1 streptavidin beads (400 μg) to another new 1.5-ml microtube, add 600 μl 1x B&W buffer and mix by pipetting.
- CRITICAL STEP: Before pipetting streptavidin beads, fully resuspend the beads by vortexing for at least 30 seconds.
-
14.
Capture the beads on a magnet stand for 1 min, and discard the supernatant.
-
15.
Resuspend the beads in 600 μl 1x B&W buffer, capture the beads on the magnet stand for 1 min, discard the supernatant.
-
16.Resuspend the beads with pooled PCR products from step 12, incubate the mixture on a rotary mixer at RT for at least 2 hours;
- CRITICAL STEP: 2 hours are sufficient for the beads to capture most of the biotinylated PCR products, however, 4-hour incubation time is recommended.
- PAUSE POINT: Binding mixture can be incubated at RT overnight.
-
17.
Capture the DNA-beads complex on the magnet stand and wash the DNA-beads complex with 600 μl 1x B&W buffer (as described in step 15) three times;
-
18.
Resuspend the beads in 1 ml H2O, capture the beads on the magnet stand for 1 min, discard the supernatant.
-
19.
Resuspend the beads in 45 μl H2O.
On-beads ligation (Timing: 5 hr)
-
20.Set up a 100-μl ligation reaction as below:
Reagents Volume (μl) Final DNA-beads complex (from step 19) 45 – 10x T4 ligation buffer 10 1x hexammine cobalt (III) chloride (20 mM) 5 1 mM Bridge adapter (50 μM) 5 2.5 μM T4 DNA ligase (3 U/μl) 5 15 U 50% PEG8000 30 15% Total 100 - CRITICAL STEP: Thaw the bridge adapter on ice; combine and mix well all the reagents except 50% PEG8000, then add 30 μl 50% PEG8000 using cut tips for more accurate pipetting of the viscous solution; mix thoroughly by pipetting.
-
21.
Aliquot ligation mixture evenly into two PCR tubes (50 μl each).
-
22.Set PCR machine as below using a heated lid to incubate the ligation for 4 hours:
Temperature Time 25°C 1 hour 22°C 2 hours 16°C 1 hour - PAUSE POINT: the ligation reactions can be optionally incubated at 16°C overnight instead of for 1 hour.
- CRITICAL STEP: To improve the ligation efficiency, resuspend the mixture after 2 hours of incubation. Do not spin the mixture before incubation since the settlement of DNA-beads greatly reduces the ligation efficiency.
-
23.
Add 50 μl 2x B&W buffer into each PCR tube, transfer and combine the mixture in a new 1.5-ml microtube.
-
24.
Add 50 μl 1x B&W buffer into each old PCR tube to collect residual ligation products, transfer the residual ligation products into the new microtube from step 23.
-
25.
Capture the on-beads ligation products on the magnet and wash the DNA-beads complex with 600 μl 1x B&W buffer (as described in step 15) twice;
-
26.
Resuspend the on-beads ligation products in 1 ml H2O, capture the beads on the magnet stand for 1 min, discard the supernatant.
-
27.Resuspend the on-beads ligation products in 200 μl H2O.
- PAUSE POINT: The on-beads ssDNA can be stored for up to one week at −20°C; longer storage is not recommended because the streptavidin beads gradually lose binding activity in H2O.
Nested PCR (Timing: 2 hr)
-
28.Set up eight 50-μl PCR reactions for each DNA sample as below:
Reagents Volume (μl) Final 5x Phusion HF buffer 10 1x dNTPs (2.5 mM each) 4 200 μM I5-nested (10 μM) 2 400 nM I7-blue (10 μM) 2 400 nM Phusion polymerase (2 U/μl) 0.5 1 U DNA-beads complex (from step 27) 25 - H2O 6.5 - Total 50 -
29.Set the PCR machine to amplify the DNA fragments as below:
Cycle number Denature Anneal Extend 1 95°C, 5 min 2-16 95°C, 60 s 60°C, 30 s 72°C, 60 s 17 72°C, 6 min - PAUSE POINT: Amplified DNA products can be stored at −20°C for months. CRITICAL STEP: Do not spin the PCR mixture before amplification because the settlement of DNA-beads greatly reduces the amplification efficiency.
-
30.
Pool the eight PCR products from step 29 together in a new 1.5-ml microtube, centrifuge at 15,000 ×g for 5 min at RT.
-
31.
Transfer the supernatant to a new 1.5-ml microtube, add 1.2 ml buffer QG included in the QIAquick Gel Extraction kit.
-
32.
Spin the mixture through a QIAquick Gel Extraction column at 15,000 ×g for 1 min at RT, discard the flow through.
-
33.
Add 800 μl buffer PE to the column, spin at 15,000 ×g for 1 min at RT, discard the flow through.
-
34.
Spin the column at 15,000 ×g for 2 min at RT, transfer the column to a new 1.5-ml microtube.
-
35.
Add 30 μl H2O to the column, spin at 15,000 ×g for 1 min at RT. Repeat this step once more (60 μl total elution volume).
-
36.
Check the concentration of a 1 μl aliquot with a NanoDrop.
(OPTIONAL) Enzyme blocking (Timing: 1 hr 30 mins)
-
37.Set up a 100-μl blocking reaction as below:
Reagents Amount DNA products (from step 35) 60 μl 10x enzyme buffer 10 μl Blocking enzyme 5 U H2O 30 μl - CRITICAL STEP: Several Blocking enzymes can be used together to improve the blocking efficiency.
-
38.Aliquot blocking mixture equally into two PCR tubes (50 μl each), incubate at recommended temperature by enzyme manufacturer in water bath for 1 hour.
- PAUSE POINT: Blocked DNA products can be stored at −20°C for months after heat inactivation (65°C for 15 mins or following the manufacturer's instruction) of the blocking enzyme.
-
39.
Add 300 μl buffer QG into the blocking mixture, recover the blocked DNA products with a Qiagen column as described in steps 32-35, and elute the products with 60 μl H2O as in step 35.
-
40.Check the concentration of a 1 μl aliquot with a NanoDrop.
- PAUSE POINT: Purified DNA products can be stored at −20°C for months.
Tagged-PCR (Timing: 1 hr)
-
41.Set up four 50-μl PCR reactions for each DNA sample from step 39 (or step 35 if blocking was omitted) as below:
Reagents Volume (μl) Final 5x Phusion HF buffer 10 1x dNTPs (2.5 mM each) 4 200 μM P5-I5 (10 μM) 2 400 nM P7-I7 (10 μM) 2 400 nM Phusion polymerase (2 U/μl) 0.5 1 U DNA products (from step 39 or step 35 if blocking is omitted) 15 - H2O 16.5 - Total 50 -
42.Set the PCR machine as below to amplify the DNA fragments:
Cycle number Denature Anneal Extend 1 95°C, 3 min 2–n 95°C, 30 s 62°C, 30 s 72°C, 60 s n+1 72°C, 6 min “n” can be 11 to 16, depending on the amount of template DNA- PAUSE POINT: Amplified DNA products can be stored at −20°C for months.
- CRITICAL STEP: the cycle number “n” depends on the concentration determined at step 40 (or step 36 if blocking is omitted). The cycle number “n” can be generally calculated as below:
DNA concentration (ng/μl) from step 40 DNA concentration (ng/μl) from step 36 if blocking is omitted Cycle number (“n”) > 10 > 15 11 7-10 10-15 12-13 < 7 < 10 14-16
Library purification (Timing: 1 hr)
-
43.Pool the four samples from step 42 together, run the entirety of the amplified DNA products on a 1% agarose gel in 1× TAE buffer.
- CRITICAL STEP: Add 40μl 6x DNA loading buffer directly into the pooled products and run them in multiple wells. It's unnecessary to condense products before loading.
-
44.
Excise from the gel the DNA fragments between 500-1000 bp (see Fig. 4).
-
45.
Purify each library with one Qiagen column following the manufacturer's instruction (similarly as performed in steps 31-35), elute with 30 μl H2O.
-
46.
Check the concentration of a 1 μl aliquot with a NanoDrop.
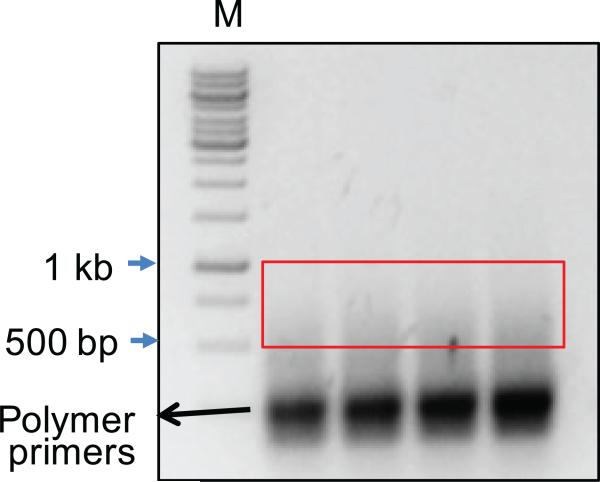
Figure 4. Representative smear of amplified and Illumina sequence tagged products.
Sequences harboring Bait/Prey components can vary in size due to the combination of stochastic shearing of genomic DNA and the juxtaposition of Bait/Prey sequences. Products ranging from 500bp-1kb are excised and purified for Miseq sequencing. Smaller products may also contain sequences with relevant junction information but co-migrate with various artifactual poly-priming intermediates. M = Molecular weight ladder.
High-throughput sequencing (Timing: 2 days)
-
47.
Pool 10-15 LAM-HTGTS libraries equally and apply pooled library DNA to Miseq sequencer for 2x 250 bp sequencing with the 500V2 kit following the manufacturer's instruction.
Sequence read preprocessing (Timing: <1 hour)
CRITICAL
Data is processed using our custom translocation bioinformatics pipeline as outlined in steps 48-51. Further details are available in the program documentation (http://robinmeyers.github.io/transloc_pipeline/).
Sequencing read main processing (Timing: 2-8 hours)
-
50.
Verify the location on disk of both the fasta file and bowtie2 index of the target genome. A custom script for modifying an existing genome is included in the pipeline.
-
51.Execute the main processing command below included with the pipeline. The detailed information for each junction is contained in a .tlx file (See Box 3).
TIMING
Steps 1-5, genomic DNA isolation: 1 day
Steps 6-9, sonication: 1 hour
Steps 10-11, LAM-PCR: 6 hours
Steps 12-19, streptavidin purification: 3 hours
Steps 20-27, on-beads ligation: 5 hours
Steps 28-36, nested PCR: 2 hours
Steps 37-40, (OPTIONAL) enzyme blocking: 1 hour 30 minutes
Steps 41-42, tagged PCR: 1 hour
Steps 43-46, library purification: 1 hour
Step 47, high-throughput sequencing: 2 days
Steps 48-49, sequence read pre-processing: <1 hour
Steps 50-51, sequence read main processing: 2-8 hours
TROUBLESHOOTING
Troubleshooting advice is provided in Table 3
Table 3.
Troubleshooting
| Steps | Problem | Possible reason | Possible solution |
|---|---|---|---|
| 5 | A260/280 is low | Proteinase K digestion is not sufficient for removing all the protein | Extract the DNA with Phenol-Chloroform twice |
| 9 | Average size of DNA smear is too large | Insufficient sonication or incompletely dissolved genomic DNA | Perform 1-2 more sonication cycles. |
| 36 | Very low DNA concentration (<3ng/μl) | Melting temperature (Tm) is not optimal for steps 11, 29 | Test the primer by gradient PCR and choose the right Tm |
| Wrong primer and/or dNTP concentrations for step 10 | Use correct concentrations for step 10 | ||
| Wrong pair of primers for steps 11, 29 | Check the sequences of the primers and make sure the nested primer corresponds with the bio primer | ||
| Bait DSB site is not cutting | Check amplified region to ensure target sequence is present; test another nearby target site | ||
| The bridge adapter is thawed and frozen too many times | Use fresh aliquot of bridge adapter | ||
| Beads were spun to bottom before ligation or nested PCR at steps 22, 29 | Do not spin the mixture before PCR | ||
| Operation error in some step | Re-do on-bead PCR or start over | ||
| Too high DNA concentration (>50ng/μl) | Unspecific priming | Design new primers | |
| Test background in an untreated (uncut bait DSB) library | |||
| 40 | Very low DNA concentration (<2ng/μl) | Blocking enzyme sites on the bait region or the I7-Blue primer | Change blocking enzyme |
| Operation error in some step | Re-do on-bead PCR or start over | ||
| 44 | Very short DNA smear tail | Too few PCR cycles | Increase the PCR cycles |
| Very long DNA smear tail | Too many PCR cycles | Reduce the input DNA amount for step 41 or decrease PCR cycles | |
| 49,51 | Pipeline does not execute | Incorrect metadata file | Check to make sure primer sequences match specified coordinates |
| 49 | No reads | Wrong barcode sequence | Check the sequences of the barcode and primer |
| Multiple samples with identical barcode and primer sequence at step 427 | Run identical barcode/primer samples on separate Miseq runs | ||
| Operation error for step 41 | Re-do step 41 | ||
| 51 | Very few junctions | Poor cutting at the bait DSB | Verify sufficient cutting at bait DSB |
| Primers are not annealing properly | Check amplified region to ensure primers do not overlap with a polymorphic site | ||
| Too high background | Cells are unhealthy or dying | Make sure the treatment doesn't cause too much DNA damages to the cells | |
| Many junctions in uncut-cell control | Repetitive sequence in bait region | Design a new bait DSB site |
ANTICIPATED RESULTS
A typical LAM-HTGTS library should generate several thousand or tens of thousands of unique translocation junctions, or at least >10-fold more than the junction numbers in the library from parallel control cells without bait DSB cleavage. The junction yield is influenced by the level of bait DSB cutting in the cells assayed and the amount of input genomic DNA used for HTGTS; increasing junction yields are more susceptible to saturation bias and optimization of user-defined conditions may be needed. All libraries are expected to have substantial enrichment at frequent DSB sites of the cells, for example, bait DSB break-sites for libraries with engineered nucleases- or AID-generated bait DSBs6,8,10 or bona fide recombination signal sequences for libraries with RAG-induced bait DSBs9. Repeat masked reference genomes can be used for alignment but is not recommended. Junctions in such masked regions, especially telomere, ribosomal, and LINE element repeats, are good indicators of the quality of the libraries. Libraries may need to be generated again if repetitive region junctions comprise more than 20% of the total, indicating relevant junctions are likely under-amplified and may impact downstream analyses.
To monitor the library preparation process, we quantify DNA products at steps 36 and 40. For our libraries with bait DSBs generated by I-SceI8, Cas9:gRNA6, TALENs6, AID10, or RAG9, concentrations ranged from 8-20 ng/μl for step 36 and 5-15 ng/μl for step 40 using 20-100 μg total input genomic DNA, respectively. It is important to optimize the cycle number for the Tagged-PCR to control for over-amplification bias, and generally, the final library DNA concentration should be within 20-40 ng/μl. For control libraries (i.e. no bait DSB), similar, but not lower, concentrations are expected for steps 36 and 40; however, these libraries result in very few junctions. If primers anneal to many sites in the genome, or the bait region contains repetitive sequences, very high DNA concentrations are expected in steps 36 and 40 (i.e. > 50 ng/μl final) with filtered junctions typically containing a high background. In this case, we recommend choosing another bait DSB site/strategy, or reduce the amount of amplified DNA in the above steps if choice of bait site is limited.
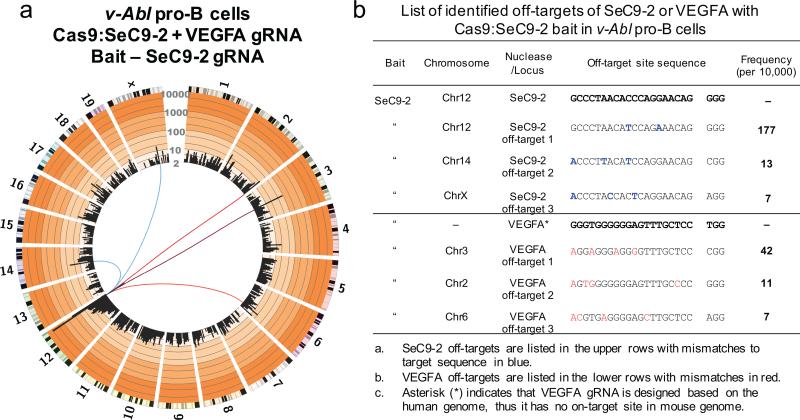
Example data for a universal bait LAM-HTGTS assay is shown in Fig. 5. Data were generated from Abelson virus-transformed (v-Abl) murine pro-B cells co-expressing the universal bait Cas9:SeC9-2 gRNA located in the IgH locus (at the end of chromosome 12) and a VEGFA gRNA designed to target the human VEGFA locus, but which generates 38 additional off-targets in the human genome (Fig. 5a)6. The SeC9-2 universal bait identified 3 SeC9-2 off-targets (1 very close to the bait). Even though VEGFA has no on-target site in the mouse genome, 3 VEGFA off-targets were identified with this universal bait assay (Fig. 5a,b).
Figure 5. Universal bait detection of off-targets for designed VEGFA gRNA.
(a) Circos plot (circos.ca) on custom log scale showing genome-wide profile of Cas9:SeC9-2 junctions in cycling v-Abl pro-B cells. Bin size is 5 Mb and 8751 unique junctions are shown from 2 independent libraries. Chromosomes are displayed as centromere to telomere in a clock-wise orientation. Blue lines link SeC9-2 off-targets to bait break-site while red lines link VEGFA off-targets to bait break-site. (b) List of identified off-targets for SeC9-2 or VEGFA. The off-targets were identified by MACS2 as described previously6. v-Abl cells (3×106) were nucleofected with a combined total of 3 μg plasmid DNA in SF solution using the DN-100 program (Lonza) and collected 48 hours post nucleofection.
Box 1: Bait DSB site strategy.
Recurrent off-target activity can be detected either by directly cloning from the on-target DSB to the off-target prey DSBs or by co-expressing a second previously established nuclease which can provide the donor bait DSB necessary to compare the joining rates of the on-target and off-target prey DSBs of the candidate nuclease; the latter strategy is referred to as the universal bait approach. Direct bait DSB cloning presents joining events with respect to the on-target site, but suffers from the inability to accurately compare its own on-target activity relative to potential off-target activity. Universal bait DSB cloning provides a tertiary bait DSB that can compare the relative joining rates between predicted on-target sites and empirically derived off-target sites of the candidate nuclease. Furthermore, the off-target sites of the defined universal bait nuclease can also be used as bait DSBs to control for off-target detection frequencies on the initial on-target universal bait chromosome.
Box2: Instructions for creating metadata files.
The metadata file contains the configuration information necessary to process sequence reads for any particular library. Incorrect information may result in errors at various stages of the translocation pipeline or produce incorrect results. Several of the columns are used to define the breaksite locus. The breaksite locus is defined as the sequence between the first nucleotide of the nested primer and the last nucleotide before the specific DSB. The metadata file is a tab-delimited plain text file containing the following header line with each subsequent row describing the design of a single library.
| Library | Researcher | Assemby | Chr | Start | End | Strand | MID | Primer | Adapter | Cutter |
“Library”-the unique name of library, this ID will be used to name most files generated by the pipeline.
“Researcher”- the creator of the library, for record keeping purposes, otherwise leave blank.
“Assembly”-reference genome (e.g. mm9, hg19). The pipeline uses this name to find the reference genome sequence and bowtie2 index on the file system.
“Chr”-the name of the chromosome which contains the break-site locus (e.g. chr15).
“Start”-position of the first nucleotide of the break-site locus
“End”-position of the last nucleotide of the break-site locus
“Strand”- either “+” or “-” based on the orientation of the nested primer
“MID”-barcode sequence if positioned at the start of the forward read, otherwise leave blank
“Primer”-sequence of nested primer;
“Adapter”-sequence of adapter;
“Cutter”-restriction enzyme target sequence if frequent cutter is used to fragment the genomic DNA, otherwise leave blank
Box3: Main processing of sequence reads.
Once the main processing command in step 51 is executed, the pipeline will read the demultiplexed R1 and R2 file according to the information provided in the metadata file and generate result files characterizing the set of translocation junctions. Multiple libraries can be processed in the same batch. The main steps are listed below:
The pipeline will read in the metadata file and call bowtie2 to align the forward and reverse reads against the genome build and the bridge adapter.
The pipeline will pool all alignments for each paired-end read to run through the OQC algorithm.
For each read, the pipeline will return the OCS which is defined by the set of alignments that optimally cover the paired-end query sequences. See the pipeline documentation (http://robinmeyers.github.io/transloc_pipeline/) for the parameters that control this process.
By default, the pipeline will filter OCS-defined reads that do not satisfy certain conditions. For example: reads with insufficient bait sequence length (associated with mispriming events), reads with a bait sequence that extends past the cut site, reads that do not contain a prey junction, or reads with a large gap between bait and prey alignments.
The pipeline will filter out reads with a strong competing prey alignment, indicating that the translocation cannot be uniquely mapped.
The pipeline will identify and filter duplicate junctions.
Using a program included in the pipeline, the user may refilter reads in a manner different from the default filters, depending on the nature of experiment (e.g. keep unjoined bait sequences, keep duplicate junctions, etc).
Acknowledgements
The authors thank members of the Alt lab for discussions about improving LAM-HTGTS and Zach Herbert from the Molecular Biology Core Facilities at Dana-Farber Cancer Institute for discussions on transitioning HTGTS to Illumina Miseq. This work is supported by National Institutes of Health Grants R01AI020047 and R01AI077595 to F.W.A.. R.L.F. was supported by the National Health Institutes of Health NRSA T32AI007512. J.H. is supported by Robertson Foundation/Cancer Research Institute Irvington Fellowship. F.W.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions
J.H., R.M.M., F.W.A., R.L.F. wrote the manuscript with additional comments from J.D. and R.A.P. J.H. and R.L.F. designed and experimentally developed the LAM-HTGTS approach and R.M.M. wrote the translocation pipeline program. J.H., R.L.F, J.D., and R.A.P. performed experiments quoted in the manuscript that employed the LAM-HTGTS approach, and all authors analyzed data that contributed to the development and/or application of the approach.
Competing Financial Interests
The authors declare competing financial interests (see the HTML version of this article for details). A patent application has been filed relating to the current LAM-HTGTS method.
References
- 1.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alt FW, Zhang Y, Meng F-L, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hendel A, Fine EJ, Bao G, Porteus MH. Quantifying on- and off-target genome editing. Trends Biotechnol. 2015;33:132–140. doi: 10.1016/j.tibtech.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv. Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 5.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 6.Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei P-C, et al. Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells. Cell. doi: 10.1016/j.cell.2015.12.039. doi:10.1016/j.cell.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng F-L, et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J, et al. Chromosomal loop domains direct the recombination of antigen receptor genes. Cell. 2015;163:947–959. doi: 10.1016/j.cell.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong J, et al. Orientation-specific joining of AID-initiated DNA breaks promotes antibody class switching. Nature. 2015;525:134–139. doi: 10.1038/nature14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gostissa M, et al. IgH class switching exploits a general property of two DNA breaks to be joined in cis over long chromosomal distances. Proc. Natl. Acad. Sci. U.S.A. 2014;111:2644–2649. doi: 10.1073/pnas.1324176111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Malley RC, Alonso JM, Kim CJ, Leisse TJ, Ecker JR. An adapter ligation-mediated PCR method for high-throughput mapping of T-DNA inserts in the Arabidopsis genome. Nat. Protoc. 2007;2:2910–2917. doi: 10.1038/nprot.2007.425. [DOI] [PubMed] [Google Scholar]
- 15.Williams R, et al. Amplification of complex gene libraries by emulsion PCR. Nat. Meth. 2006;3:545–550. doi: 10.1038/nmeth896. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt M, et al. High-resolution insertion-site analysis by linear amplification–mediated PCR (LAM-PCR). Nat. Meth. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 17.Paruzynski A, et al. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat. Protoc. 2010;5:1379–1395. doi: 10.1038/nprot.2010.87. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Z-X, et al. Mapping genomic hotspots of DNA damage by a single-strand-DNA-compatible and strand-specific ChIP-seq method. Genome Res. 2013;23:705–715. doi: 10.1101/gr.146357.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faust GG, Hall IM. YAHA: fast and flexible long-read alignment with optimal breakpoint detection. Bioinformatics. 2012;28:2417–2424. doi: 10.1093/bioinformatics/bts456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwer B, et al. Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells. Proc. Natl. Acad. Sci. U.S.A. doi: 10.1073/pnas.1525564113. doi:10.1073/pnas.1525564113/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinek M, et al. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim H, Kim J-S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 27.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2014;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol. 2015;33:175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- 29.Zetsche B, et al. Cpf1 Is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu J, Tepsuporn S, Meyers RM, Gostissa M, Alt FW. Developmental propagation of V(D)J recombination-associated DNA breaks and translocations in mature B cells via dicentric chromosomes. Proc. Natl. Acad. Sci. U.S.A. 2014;111:10269–10274. doi: 10.1073/pnas.1410112111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boboila C, et al. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J. Exp. Med. 2010;207:417–427. doi: 10.1084/jem.20092449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teng G, et al. RAG represents a widespread threat to the lymphocyte genome. Cell. 2015;162:751–765. doi: 10.1016/j.cell.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein IA, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barlow JH, et al. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152:620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khair L, Baker RE, Linehan EK, Schrader CE, Stavnezer J. Nbs1 ChIP-seq identifies off-target DNA double-strand breaks induced by AID in activated splenic B cells. PLoS Genet. 2015;11:e1005438. doi: 10.1371/journal.pgen.1005438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baranello L, et al. DNA break mapping reveals topoisomerase II activity genome-wide. Int. J. Mol. Sci. 2014;15:13111–13122. doi: 10.3390/ijms150713111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crosetto N, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Meth. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asaithamby A, Chen DJ. Cellular responses to DNA double-strand breaks after low-dose-irradiation. Nucleic Acids Res. 2009;37:3912–3923. doi: 10.1093/nar/gkp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roukos V, et al. Spatial dynamics of chromosome translocations in living cells. Science. 2013;341:660–664. doi: 10.1126/science.1237150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Judo MS, Wedel AB, Wilson C. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Res. 1998;26:1819–1825. doi: 10.1093/nar/26.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clepet C. Improved full-length cDNA production based on RNA tagging by T4 DNA ligase. Nucleic Acids Res. 2004;32:6e–6. doi: 10.1093/nar/gng158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Meth. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 45.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 46.Veres A, et al. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]