Abstract
A periplasmic flagellar chaperone protein, FlgA, is required for P-ring assembly in bacterial flagella of taxa such as Salmonella enterica or Escherichia coli. The mechanism of chaperone-mediated P-ring formation is poorly understood. Here we present the open and closed crystal structures of FlgA from Salmonella enterica serovar Typhimurium, grown under different crystallization conditions. An intramolecular disulfide cross-linked form of FlgA caused a dominant negative effect on motility of the wild-type strain. Pull-down experiments support a specific protein-protein interaction between FlgI, the P-ring component protein, and the C-terminal domain of FlgA. Surface plasmon resonance and limited-proteolysis indicate that flexibility of the domain is reduced in the covalently closed form. These results show that the structural flexibility of the C-terminal domain of FlgA, which is related to the structural difference between the two crystal forms, is intrinsically associated with its molecular chaperone function in P-ring assembly.
The bacterial flagellum is a macromolecular assembly composed of about 30 different proteins with copy numbers ranging from several to tens of thousands1,2. Salmonella enterica serovar Typhimurium (S. enterica) is a motile, Gram-negative bacterium with peritrichous flagella that have been studied intensively by genetic, biochemical, and structural approaches. The bacterial flagellum consists of three major substructures: the basal body, the hook, and the filament. The basal body contains a rotary motor, composed of the MS-ring, the LP-ring, and the drive shaft, called the rod, which traverses the inner- and outer membranes of Gram-negative cells. Connected to the rod is the hook. It transmits motor torque to the filament, which forms a long helical coil that functions as a “propeller” outside the cell. The LP-ring is a molecular bushing that stabilizes high-speed rotation of the flagellum. It is a chemically stable substructure composed of about 26 copies each of FlgH and FlgI, associated with the bacterial outer membrane and peptidoglycan (PG) in the cell wall, respectively3,4. FlgH and FlgI, which comprise the L- and P- rings, respectively, are synthesized in the bacterial cytoplasm and exported to the periplasm by the Sec-dependent pathway5. Except for the bacterial Phylum Firmicutes, for which FlgH and FlgI are not necessary, due to the thick PG layer that holds the rod, the LP-ring is required by most Gram-negative bacteria to form functional flagella6. The L- and P-rings are anchored to the lipopolysaccharide layer of the outer membrane and the PG periplasmic layer3, respectively. After secretion into the periplasm by the Sec-dependent pathway, FlgH is subjected to the outer membrane sorting system Lol7,8, because it has a canonical cysteine residue that is modified with a lipid moiety for outer-membrane localization9.
In the cytoplasm, specific additional chaperones control flagellar assembly10,11,12. FlgA is a periplasmic flagellar protein that chaperones P-ring formation5,13,14. FlgA possesses a typical signal sequence at its N-terminus, recognized by the Sec-dependent pathway, suggesting that FlgA functions as a P-ring assembly chaperone in the periplasm15. Direct evidence of FlgA binding to FlgI was previously demonstrated by genetic and biochemical analyses16. P-ring formation is a key step enabling the bacterial flagellum to pass through the outer membrane17. L-ring formation depends on a pre-formed P-ring, without which L-ring assembly is severely impaired13. To gain insight into the regulatory mechanism of P-ring assembly by FlgA, two different atomic structures of FlgA from S. enterica, the open and closed forms were solved. The structures of these distinct forms reveal structural flexibility that is essential for FlgA function and P-ring formation. Our experiments on the non-flagellated strain of S. enterica SJW1446 show in greater detail, how flexibility of FlgA enables its chaperone function during P-ring assembly.
Results
Open and Closed Conformations of the Chaperone Protein, FlgA
S. enterica FlgA was expressed as a C-terminal, hexa-histidine-tag-fused precursor and was purified from the periplasm of E. coli cells. Purification, crystallization, and diffraction data collection have been described previously18. We solved two crystallographic structures of FlgA. Atomic models were built from amino acid residues Q1 through L198 (numbering corresponds to the mature protein) for the open and closed forms (Fig. 1A,B) and refined at resolutions of 1.95 Å and 2.3 Å, respectively (Table 1). For the open form, a segment from T44 thorough A47 was not traced in the final model due to the poor quality of the electron density map in this region.
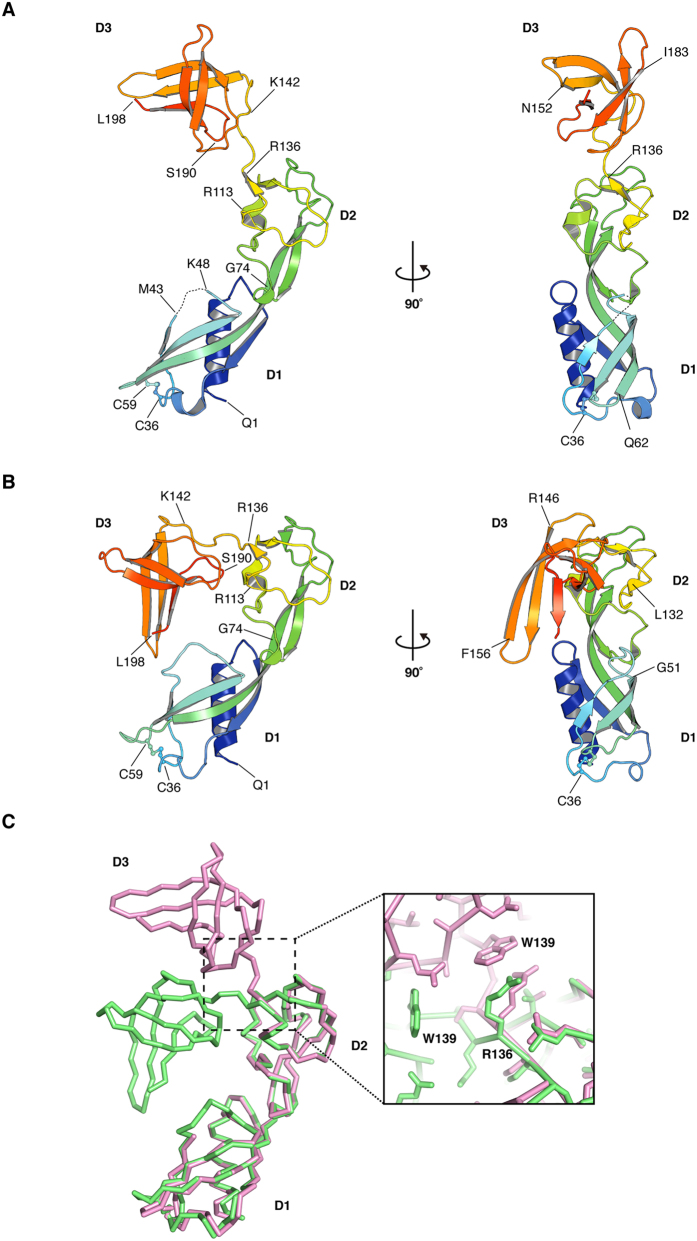
Figure 1. Overall structure of S. enterica FlgA.
The chain is color-coded from blue to red from the N- to the C-terminus. Residues are shown as one-letter codes with numbering. The open structure of FlgA with the main secondary structure annotated with two rotations by 180° difference (A) and the closed structure of FlgA (B). Figures were prepared with PyMOL (The PyMOL Molecular Graphic System, Schrödinger, LLC. http://www.pymol.org.). (C) Comparison of FlgA structures in ribbon. The open form in magenta and the closed form in lime are aligned relative to their D2 domains. A close-up view of residues around R136 is also shown.
Table 1. Refinement statistics for the open and closed forms of S. enterica FlgA.
| Open | Closed | Closed | |
|---|---|---|---|
| Native | SeMet | Native | |
| Space group | C222 | C2221 | P21 |
| Unit-cell parameters (Å,°) | a = 107.52, | a = 53.17, | a = 53.93, |
| b = 131.77, | b = 162.50, | b = 103.32, | |
| c = 49.36 | c = 103.49 | c = 85.50, | |
| β = 107.26 | |||
| Number of molecule in the asymmetric unit | 1 | 2 | 4 |
| Resolution (Å) | 19.8–1.95 | 24.5–2.70 | 25.0–2.30 |
| Rwork | 20.4 | 24.4 | 25.1 |
| Rfree | 23.4 | 27.7 | 28.1 |
| Number atoms | |||
| Protein | 1497 | 2990 | 5980 |
| Water | 165 | 45 | 186 |
| Ligands | 5 (Glycerol) 1 (Chloride ion) | – | – |
| B-factors | |||
| Protein | 35.9 | 74.0 | 60.5 |
| Water | 44.7 | 51.3 | 43.8 |
| Ligands | 73.5 (Glycerol) 57.1(Chloride ion) | – | – |
| RMSD | |||
| Bond length (Å) | 0.015 | 0.004 | 0.004 |
| Bond angles (˚) | 1.496 | 1.044 | 0.969 |
| Ramachandran plot (%) | |||
| Favored | 99.50 | 96.94 | 95.66 |
| Allowed | 0.50 | 3.06 | 4.34 |
| Outliers | 0.00 | 0.00 | 0.00 |
| PDB-id | 3TEE | 3VJP | 3VKI |
Values in parentheses are for the highest resolution shell.
FlgA can be divided into three domains, denominated as D1 (residues 1–74), D2 (residues 75–142), and D3 (residues 143–198). The N-terminus begins with an amphipathic α-helix (α1), followed by a four-stranded, anti-parallel β-sheet. The first three β-strands, β1, β2, and β3, are within D1. The long, fourth β-strand, β4, stretches from D1 into D2. Domain D2 is composed of four short, anti-parallel β-strands linked by loops. Domain D3 is predominantly made of a five-stranded β-sheet (β8, β9, β10, β11, β12) that forms, a short β-barrel (Supplementary Fig. S1). The first four strands are anti-parallel, while the fifth C-terminal strand is parallel to the fourth. FlgA has two cysteine residues (C36 and C59) that form a disulfide bridge in both crystal structures and the side chain and the bond are visible in a ball-and-stick representation (Fig. 1A,B). These cysteines are not conserved throughout the FlgA family (Supplementary Fig. S1) and we found that the disruption of this disulfide bond does not affect motility of SJW1446 (Supplementary Fig. S2).
While the structures of corresponding domains are similar in the open and closed forms, the D1 domains of the two forms align with an RMSD of 0.64 Å, while the D2 and D3 domains align with RMSDs of 0.35 Å and 0.33 Å, respectively (Fig. 1C). The conformation of FlgA is different in the two forms (Supplementary Movie S1). In the closed form, D3 is closer to D1 and forms a compact structure. This occurs largely because of a bend of about 70° around the Cα atom of R136 (Fig. 1C).
Comparison with the FlgA of a thermophile
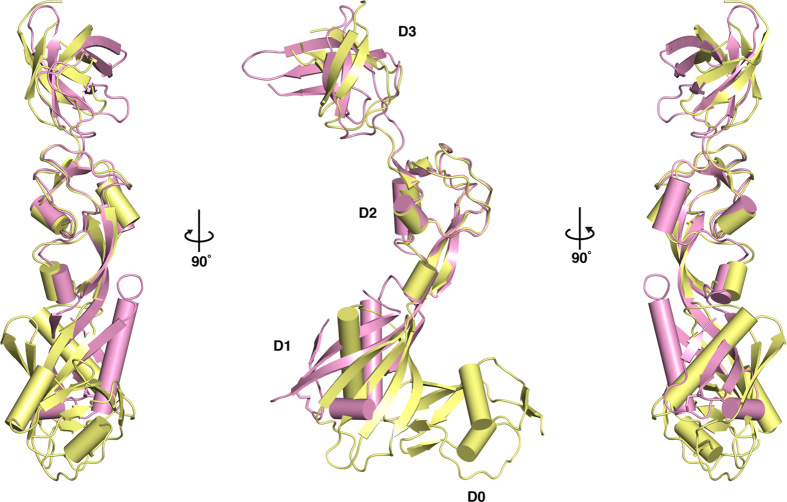
The crystal structure of Thermotoga maritima FlgA, (T. maritima) containing residues 17–283 (out of 286), has been deposited in PDB by a structural genomics consortium. The structure contains four domains: D0 to D3. The N-terminal domain D0, which does not exist in S. enterica, is composed of residues 17–84. There is a slight diversity in the length of these terminal regions among FlgA proteins (Supplementary Fig. S1). Except for the D0 domain, FlgA from both S. enterica and T. maritima share similar domain organization. The overall structures of both FlgAs superimpose with a root-mean-square deviation (RMSD) of 2.7 Å19. The domains common to both FlgAs superimpose with RMSDs of 2.8, 1.2 and 1.3 Å, respectively19. FlgA from T. maritima is a more extended structure, or more open, than that of S. enterica (Fig. 2)
Figure 2. Structural comparison of FlgA proteins from S. enterica and T. maritima.
Overall structures of S. enterica (PDB-id: 3TEE, magenta) and T. maritima (PDB-id: 3FRN, paleyellow) are shown as ribbon representations with cylindrical helices by superimposing the D2 domains.
Structural similarity of domain D2 to antifreeze protein
Antifreeze protein Type III (PDB-id: 6AME) has been identified as the closest structural homolog to the D2 domain with an RMSD of 1.7 Å over 60 aligned residues, and a sequence identity of 15% with the highest Z-score of 8.7. The structure of the C-terminal antifreeze-like (AFL) domain of human sialic acid synthase (SAS, PDB-id: 1WVO) also showed structural similarity to the D2 domain of FlgA with an RMSD of 1.5 Å over 61 aligned residues and a sequence identity of 15% with the highest Z-score of 8.320. The AFL domain in human SAS seems to play an important role in substrate binding21,22. In spite of the rather poor alignment statistic, the topology of both structures is very similar (Supplementary Fig. S3).
Role of Domain D3 in FlgA Function
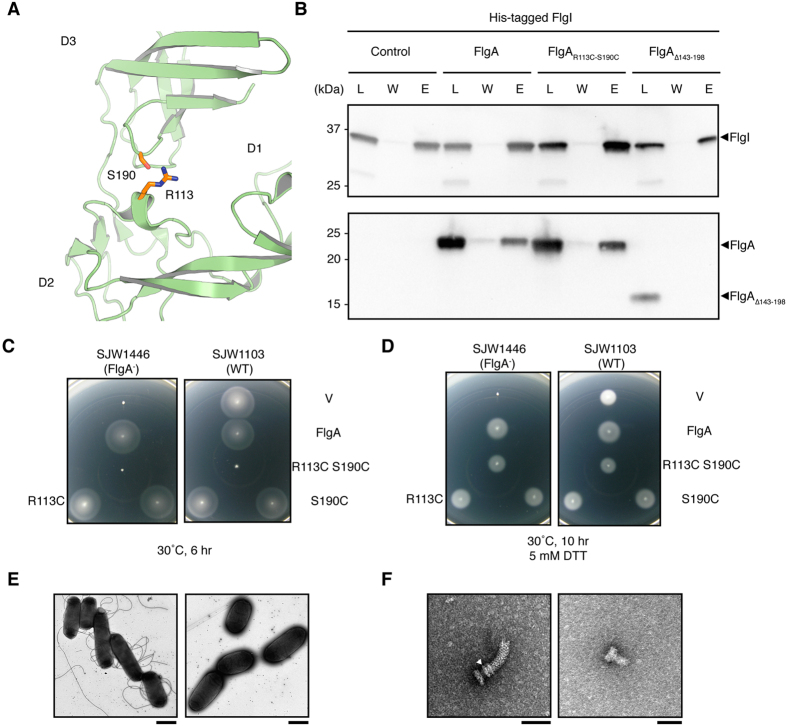
S. enterica strain SJW1446, which is non-flagellated due to a defect in the flgA gene14 that results in deletion of residues V141-G144 (∆VKAG), was used as a host cell for genetics. To examine the role of D3, we constructed a deletion variant of FlgA, FlgA∆143–198, lacking residues 143–198 that form domain D3 (Fig. 3A). This deletion variant neither complemented the flgA (∆141–144) allele of SJW1446, nor had any effect on motility of the wild-type strain SJW1103 (Fig. 3B). However, FlgA∆143–198 was secreted properly into the periplasm as well as wild-type FlgA (Fig. 3C). Degradation products of FlgA were also detected around the molecular size of FlgA∆143–198. In the periplasm, FlgA might be susceptible to proteolytic digestion. The hook-cap protein, FlgD, accumulated in the periplasm of both SJW1446 harboring the empty vector and the mutant strain producing FlgA∆143–198 (Fig. 3C), indicating the failure of LP-ring assembly. Neither FlgD nor FliC was found in the culture supernatant from cells harboring FlgA∆143–198 (Fig. 3C). FlgA∆143–198 did not exert a significant negative dominant effect on motility of SJW1103 (which also possesses a copy of the wild-type gene) suggesting that it failed to bind FlgI due to its lack of the D3 domain (Fig. 3B).
Figure 3. Domain organization of S. enterica FlgA.
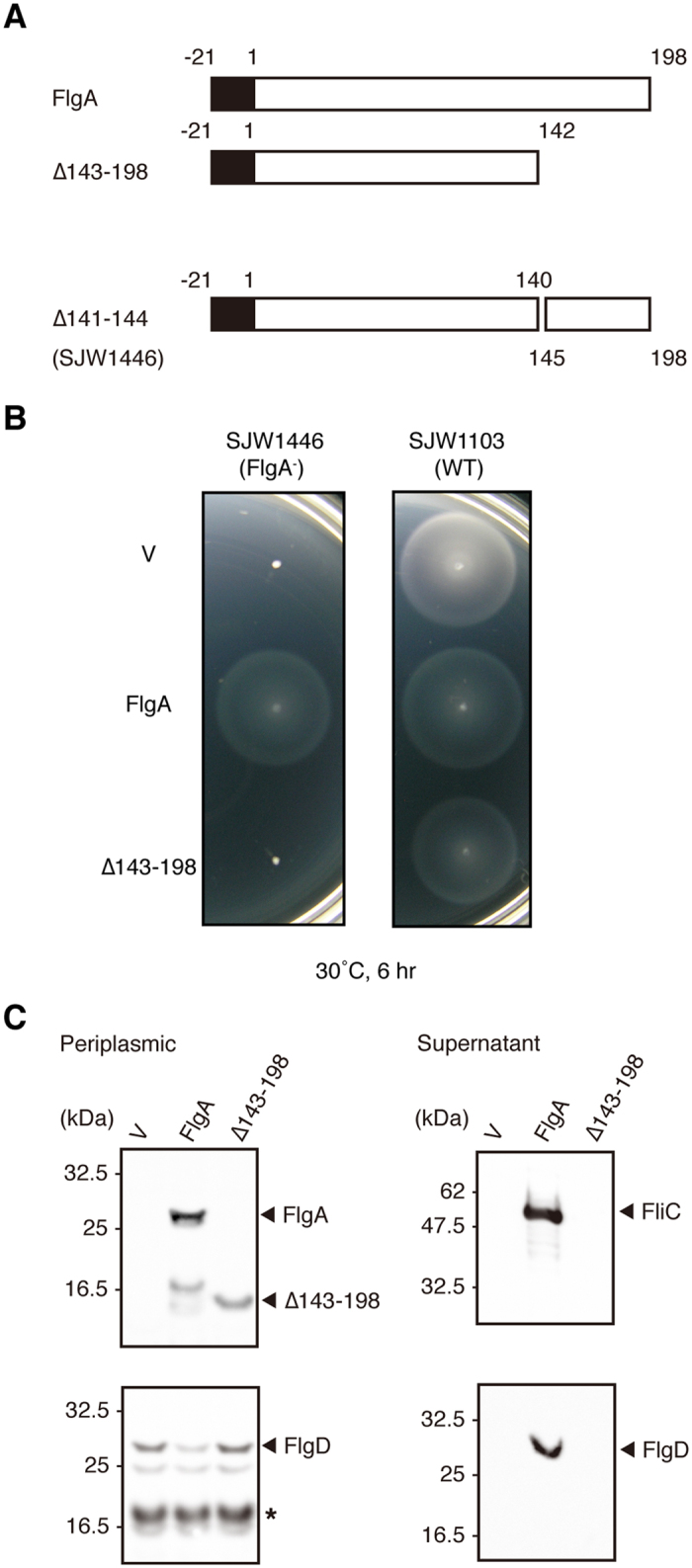
(A) Domain structures of FlgA used in this experiment. (B) Swarming motility of and protein secretion by wild-type (SJW1103, WT) and the flgA deficient strain (SJW1446, FlgA−) with an empty vector (V) or harboring a plasmid expressing either FlgA or FlgA(∆143–198). Swarming motility on soft agar plates. Complementation of the flgA deficient strain SJW1446 with trc-based plasmids encoding either FlgA or FlgA(∆143–198) (left) and the negative dominant effect on wild-type motility of SJW1103 (right). (C) Secretion assay by fractionating proteins (the periplasmic fraction on the left and the supernatant fraction on the right). Protein bands are visualized using antibodies against S. enterica FlgA, FlgD, and FliC. A degradation product of FlgD found in the periplasmic fraction is marked with an asterisk. Molecular marker positions are indicated on the left.
Evidence of the Functional Relevance of Both Conformations
By studying the 3D structures of FlgA, we identified a pair of residues, R113 and S190, which are within a suitable distance for cross-linking with a disulfide bond that keeps the structure of FlgA in the closed conformation (Fig. 4A). To understand how the conformational difference between the open and closed forms of FlgA is related to its function, a disulfide bond was engineered by replacing these residues with cysteines (R113C and S190C, respectively). The disulfide bond forces FlgA to stay in the closed form and constrains the movement of domain D3. Pull-down experiments revealed that FlgA and FlgAR113C-S190C both bound to His-FlgI; however, FlgA∆143–198 could not, suggesting that the C-terminal domain of FlgA is involved in binding to FlgI and that this domain is still intact for binding of FlgAR113C-S190C (Fig. 4B). The complete loss of motility in SJW1446 complemented with the plasmid expressing FlgA∆143–198, is explained by the lack of binding to FlgI (Fig. 3B). Before testing the effect of this double-mutation, the complementation of FlgA with each of the single mutations R113C and S190C on the motility of SJW1446 was tested (Fig. 4C). While each of these single-substitution mutants rescued the functional flagellar assembly of SJW1446, FlgAR113C-S190C strongly suppressed the motility of SJW1103, presumably by blocking flagellar assembly (Fig. 4C). In the presence of dithiothreitol (DTT), which reduces disulfide bonds, the double mutant FlgAR113C-S190C was able to rescue the motility of SJW1446 and had no effect on the motility of SJW1103 (Fig. 4D). The wild-type strain complemented with pHMK339 (FlgA, hereafter called HK1431) produced flagellar filaments, but in contrast with pHMK788 (FlgAR113C-S190C, hereafter called HK1500), no flagellar filament was produced (Fig. 4E). Isolated hook basal-bodies from these strains were morphologically different. A rivet structure, which lacks the LP-ring of the hook basal-body, was isolated from the wild-type strain expressing FlgAR113C-S190C (Fig. 4F). These results indicate that the closed form of FlgA can bind to FlgI, but cannot proceed with subsequent steps, thereby inhibiting FlgI from forming the P-ring.
Figure 4. Effects of an engineered disulfide bond on the chaperone function.
Cysteine substitution creates a novel disulfide bond that mimics the closed conformation of FlgA and eliminates motility. (A) Residues R113 and S190 shown in stick representation in the closed structure of FlgA. Cα atoms of R113 and S190 are 6.3 Å apart in the closed form and 17.5 Å apart in the open form. (B) Immunoblot showing the result of the pulldown assay of FlgA, FlgAR113C-S190C and FlgA(∆143–198) with His-FlgI as a probe. Load (L), wash (W), and elution (E) are shown. Swarming motility assay on tryptone agar plates for complementation of SJW1446 (FlgA−) and negative dominance effect on SJW1103 (WT) transformed with a plasmid expressing the disulfide bond-locked closed form of FlgA. Assays were carried out in the absence (C) and presence (D) of 5 mM DTT. (E) Electron micrographs of cells of HK1431 (left) and HK1500 (right). Scale bars are 1 μm. (F) An isolated hook-basal body from HK1431 (left) and a basal body from HK1500 (right). Scale bars are 50 μm. The P-ring in the basal body is indicated by a white arrowhead.
Structural flexibility of FlgA
In solution, both FlgA and FlgAR113C-S190C existed as monomers exclusively (Supplementary Fig. S5). Disulfide bond formation in FlgAR113C-S190C was suggested by gel-shift assay and size-exclusion chromatography. In SDS-PAGE, FlgAR113C-S190C migrated slightly more than FlgA in the absence of the reducing reagent, DTT, indicating that it folded into a more compact structure than FlgA (Supplementary Fig. S6). In addition, the elution of FlgAR113C-S190C by size-exclusion chromatography was more retarded than that of FlgA, indicating a more compact molecular shape, and normal mobility was restored in the presence of DTT (Supplementary Fig. S6).
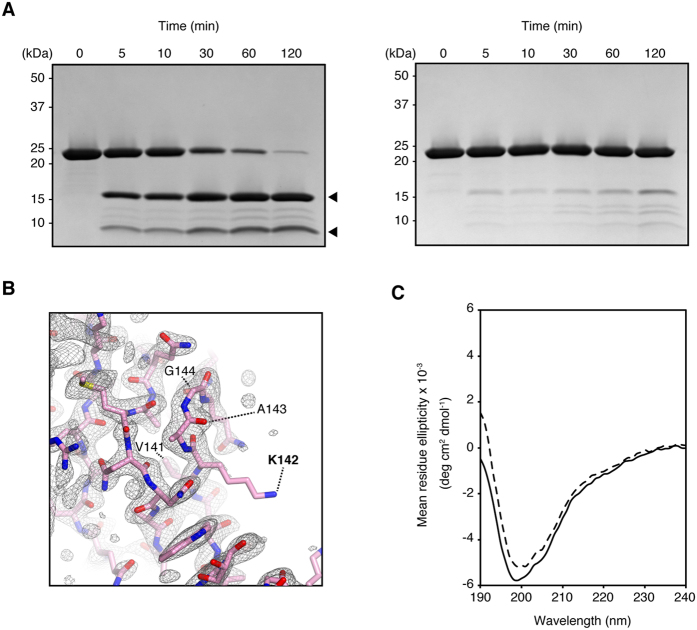
FlgA and FlgAR113C-S190C were subjected to protease digestion with endoprotease Lys-C. In FlgA, major proteolytic fragments appeared shortly after the reaction commenced; however, FlgAR113C-S190C was resistant to digestion (Fig. 5A). The digestion site could reside between residues K142 and A143, judging from the size of the fragments, which overlaps the flgA (∆141–144) allele of SJW1446 (Fig. 5B). This short segment connects domains D2 and D3 and is accessible to the solvent. The strain, SJW1446, harboring pHMK714 (∆141–144), was not motile even upon overexpression of FlgA∆141–144 from the plasmid (Supplementary Fig. S4). The resulting flagellar growth defect was ascribed to the loss of this short segment. FlgA∆141–144 could be quite unstable so that it is not detected in the periplasm (Supplementary Fig. S4). Even If the deletion is acceptable to the protein, the loss of this linker might restrict the movement of D3 relative to D1, thereby suppressing the chaperone function of FlgA for FlgI. In solution, FlgA and FlgAR113C-S190C showed similar CD spectra (Fig. 5C). Calculated secondary structure contents of α and β structures were 19% and 24% for FlgA, and 19% and 27% for FlgAR113C-S190C, respectively. No significant secondary structure transition was found in both proteins. A structural difference between FlgA and FlgAR113C-S190C is solely limited to the D3 positions in the structures.
Figure 5. Analysis of protein stability by proteolytic digestion.
Limited proteolysis of FlgA (left) and FlgAR113C-S190C (right) with Lys-C endoprotease (A). A focused view around residues V141–G144 that are lacking in FlgA of SJW1446. A Sigma-A weighted 2mFo-DFc map generated by phenix_composite_map was shown as mesh by PyMOL with contour level of 1.0. The open form of FlgA was shown as ribbon. These residues including the protease digestion site are labeled (B). CD spectra of the proteins, FlgA (solid line) and FlgAR113C-S190C (dashed line), were measured (C) from 190 to 240 nm.
Quantitative analysis of the interaction of FlgA with its cognate partner, FlgI
The binding of both FlgA and FlgAR113C-S190C to their substrate, FlgI, was investigated using Surface Plasmon Resonance (SPR). Kinetic constants for FlgA and FlgAR113C-S190C against immobilized FlgI, a ligand on the CM5 chip (GE Healthcare), were measured by assuming a two-state reaction model, allowing for conformational changes after complex formation (Supplementary Fig. S7), according to the manufacturer’s protocol (Table 2). Affinity constants (KD) of FlgA and FlgAR113C-S190C against the ligand, FlgI, were estimated as almost identical, indicating no significant difference in binding FlgI. On the other hand, we analyzed the binding to their substrate, FlgI, by immobilizing either FlgA or FlgAR113C-S190C to the CM5 chip (Supplementary Fig. S7). In this case, the KD of FlgA increased to over twice that of FlgAR113C-S190C, indicating that the binding of FlgAR113C-S190C to FlgI is slightly stronger than that of FlgA.
Table 2. Kinetic parameters determined by SPR experiments.
| Ligand-Analyte | ka1(M−1s−1) [105] | kd1(s−1) [10−3] | ka2(s−1)[10−3] | kd2(s−1)[10−3] | KD (M) [10−6] |
|---|---|---|---|---|---|
| FlgI-FlgA | 3.41 | 5.89 | 1.24 | 3.31 | 0.126 |
| FlgI-FlgAR113C-S190C | 3.35 | 5.16 | 2.17 | 4.72 | 0.106 |
| FlgA-FlgI | 0.174 | 9.76 | 0.985 | 1.73 | 0.354 |
| FlgAR113C-S190C -FlgI | 0.152 | 5.02 | 0.673 | 0.980 | 0.166 |
Discussion
The present X-ray structural analysis of FlgA, the flagellar P-ring chaperone, showed unique structural features with no structural similarity to other flagellar chaperone proteins that function in the cytoplasm11,23. Here, structural snapshots of FlgA, revealed by X-ray crystallographic studies, suggest that the conformational difference between the two structures of the chaperone may help to explain flagellar P-ring formation (Supplementary Movie S1). Based upon the structural difference induced by crystal packing, we tested whether the closed and open forms were possible in solution. Functional assays exploring the structural difference between FlgA and FlgAR113C-S190C suggested a difference in binding to FlgI. They also showed different phenotypes in S. enterica strains for flagellar P-ring formation; therefore, structural flexibility of the protein is essential and FlgA is able to bind to FlgI via its C-terminal domain (Figs 3, and 4). FlgAR113C-S190C is locked in one conformation, while FlgA adopts intermediate structures upon immobilization, suggesting that FlgA might exist in the periplasm as a flexible intermediate structure, or in equilibrium between the open and closed forms defined here.
The closed form of FlgA is reminiscent of the PapD structure, an immunoglobulin-like periplasmic chaperone for P pilus biogenesis24,25. In the PapD structure, a deep cleft between domains constitutes the substrate recognition site; however, the cleft between FlgA domains D1 and D3 in the closed form might not be suitable for FlgI binding because the C-terminal domains of both forms of FlgA showed similar affinity for FlgI binding. Upon binding to FlgI, FlgA may adopt the closed structure, since FlgAR113C-S190C showed a strong negative dominant effect on motility of the wild-type strain (Fig. 3). Although a small difference at μM order in the KD values could not be fully explained, the negative dominance effect of FlgAR113C-S190C, it might be effective for P-ring assembly. The KD values are not small enough for simple complex formation (1:1 binding), but might be reasonable for the energy-independent translocation of FlgI in the periplasm. FlgI is released into the periplasm right after secretion by way of the general secretion pathway, and it should be transported to nascent flagellar structures by an unknown sorting mechanism. If FlgA is a pilot for FlgI after complex formation, promoting its localization at the destination, perhaps FlgA appears in the periplasm in the closed form prior to FlgI translocation and then binds to FlgI to recruit it to the flagellar basal body, preventing premature formation of the P-ring.
In case of a pilotin-secretin complex in the Type III secretion system of Shigella flexneri, MxiM and MxiD were intensively studied by X-ray crystallography and nuclear magnetic resonance spectroscopy. Pilotin MxiM binds to the C-terminus of the secretin MxiD to allow proper translocation to the outer membrane in the periplasm with structural rearrangement of secretion26,27. If the C-terminal domain of FlgA binds to FlgI, the rest of the protein would be important for translocation to the periplasm. Domain D2 may target PG through the β-clip fold. Structural similarity of domain D2 to proteins with β-clip folds supports the idea that binding to the PG layer occurs through the hydrophilic surface of domain D228. This hypothesis implies that the role of FlgA in P-ring formation is to assemble FlgI molecules around the nascent rod structure close to the PG layer, probably to the proximal part of the distal rod, composed of FlgG17. It is unknown how FlgA releases bound FlgI prior to P-ring formation. Perhaps FlgA switches conformations from the closed to the open form after interacting with cellular components or other flagellar proteins.
Finally, crystal structures of FlgA will allow future mutagenesis studies to better define its role in FlgI stabilization, polymerization, and localization, as a molecular chaperone in flagellar P-ring formation. In most Gram-negative bacteria, flagellation of cells cannot occur in the absence of FlgA. Thus, this molecule might be a target for novel antimicrobial drugs to inhibit cell flagellation and motility. Further structural information about the FlgA-FlgI complex would be advantageous in the pursuit of new therapeutic drugs.
Our results suggest a stepwise mechanism of P-ring formation mediated by the chaperone. L-ring formation in the outer membrane begins right after P-ring formation. Without a canonical PG binding region, FlgI cannot simply diffuse into the periplasm after secretion by the Sec system, but is probably ushered by FlgA to its destination. In addition to binding FlgI, by preventing inappropriate oligomerization, FlgA might communicate with other flagellar components to determine an exact timing and place for P-ring formation. Further structural approaches could be applied to reveal dynamics subsequent to FlgA-FlgI complex formation.
Experimental Procedures
Bacterial Strains, Plasmids
Bacterial strains and plasmids used in this study are detailed in Supplementary Table S1. DNA manipulations were done according to standard protocols described by Sambrook and Russell (2001). Ampicillin and chloramphenicol were added to final concentrations of 100 and 50 μg ml−1, respectively.
Purification and crystallization of FlgA
Details of expression, purification, and crystallization of S. enterica FlgA have been described in a crystallization note18.
Structure determination of FlgA
X-ray diffraction experiments were performed at beamline BL41XU of Spring-8 with approval of the Japan Synchrotron Radiation Research Institute (JASRI) (proposal no. 2007B1500). Data were indexed and integrated with MOSFLM29 and scaled with SCALA30. Details have been previously described18. In brief, native FlgA crystallized in space group C222 for the open form and in space group P21 for the closed form (Supplementary Table S2). To solve the structure of the open form, a multi-wavelength anomalous dispersion (MAD) experiment of platinum-derivative crystals of FlgA was carried out at 3.0-Å resolution. The initial phase was calculated with SOLVE31 and electron density was modified with DM32 using native data collected at 1.95-Å resolution. Automatic model building was carried out using Arp/Warp33. Iterative model building and refinement were performed with COOT34 and Refmac35. Thereafter, final refinement was accomplished with Buster36. During refinement, manual modification was carried out using “omit map”37. To solve the closed form, a single-wavelength anomalous dispersion (SAD) experiment with selenomethionine-substituted FlgA (SeMet-FlgA) belonging to space group C2221 was carried out at 2.7-Å resolution. The native structure of the closed form crystallized in space group P21 was solved by molecular replacement and refined with Phenix38. In the open form, the C-terminally fused linker region including a hexa-histidine (His6) tag (15 amino acids in length) was partly visible and could be modeled; however, the His6 tag part was omitted from the final model due to poor electron density. For clarity, the C-terminal linker fused to FlgA was omitted from all figures. Refinement statistics are summarized in Table 1. A search for structurally homologous proteins in the Protein Data Bank (PDB) was performed with DALI20.
Motility assay and protein secretion experiments
The S. enterica flgA mutant strain SJW1446 was used as the host for trc-based plasmids for complementation assays on semi-solid agar plates (Table S1). The S. enterica wild-type strain SJW1103 was used as the host for the negative dominance assay as well. Assays were performed at 30 °C. Periplasmic fractions were prepared as described previously39. Secreted proteins were detected by Western blotting. Fractionated proteins were subjected to SDS-PAGE and then electroblotted to nitrocellulose membranes, where they were visualized using a photoimager LAS3000 (Fuji film, Tokyo, Japan) or an imaging system ChemiDoc XRS+ (Bio-Rad, Hercules, CA, USA) with antibodies against S. enterica FlgA, FlgD, and FliC (MBL, Nagoya, Japan).
DNA sequencing
Genomic DNA was isolated from S. enterica SJW144613 using Gentra Puregene Yeast/Bacteria kit (QIAGEN, Gaithersburg, MD). The flgA gene was amplified by PCR using genomic DNA as a template. The gene was then cloned into the NdeI/BamHI site in pHMK11 to create pHMK714. The flgA DNA sequence was confirmed with an ABI3700 using a BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA).
Pull-down Assays
Pull-down assays of FlgA-FlgI complex formation with TALON metal affinity chromatography were carried out following the manufacturer’s instructions (Clontech, Mountain View, CA). Cell extracts prepared from E. coli Origami2 (DE3), harboring the N-terminally His-tagged FlgI (His-FlgI, pHMK385) co-expressed without FlgA (pHMK14) or FlgA proteins (pHMK720, pHMK844 or pHMK869) were loaded onto a TALON spin column. Following extensive washing with a buffer containing 10 mM imidazole, bound proteins were eluted with 300 mM imidazole and separated by SDS-PAGE, transferred to a PVDF membrane, and probed with custom antibodies raised against FlgA and FlgI (MBL, Nagoya, Japan). Membrane images were acquired with a ChemiDoc XRS+ system (Bio-Rad, Hercules, CA).
Transmission electron microscopy (TEM) observation
Cells were cultured until late log phase in LB medium supplemented with Amp at 30 °C and spun down to remove culture media. After washing with phosphate-buffered saline (pH 7.4), cells were re-suspended in the buffer and applied to freshly glow-discharged carbon grids for negative staining with 1% (w/v) uranyl acetate. Images were collected on a JEM-1230R (Jeol, Tokyo, Japan) operated at 100 keV, equipped with a 2k × 2k Gatan slow-scan, charge-coupled device camera and processed with Digital Micrograph 3.1 (Gatan, Pleasanton, CA). Flagellar basal bodies were isolated as described previously with several modifications40. Sucrose density gradient centrifugation (20–50%(w/v)) was employed instead of a CsCl gradient.
Limited proteolysis of FlgA
Purified FlgA and FlgAR113C-S190C were mixed with endoprotease Lys-C (Roche Molecular Diagnostics, Indianapolis, IN) at a weight ratio of 250:1 (protein:protease) in 10 mM HEPES (pH 7.0), 100 mM NaCl. Reaction mixtures at each time interval were stopped by mixing with SDS loading buffer and heating at 95 °C for 5 min.
Circular dichroism (CD) spectroscopy
Protein concentrations of FlgA and FlgAR113C-S190C were determined with an Ultrospec3100pro (GE Healthcare) using extinction coefficients at 280 nm of 0.903 and 0.910, respectively. CD spectra of FlgA (0.13 mg/ml) and FlgAR113C-S190C (0.12 mg/ml) were measured in 10 mM sodium phosphate buffer (pH 7.0) with a J-820 spectrophotometer (Jasco, Tokyo, Japan) at wavelengths from 190 to 240 nm at 25 °C. Data were analyzed with Spectra Manager ver. 1.55 (Jasco, Tokyo, Japan). Secondary structure contents of proteins were estimated with K2D341.
Surface Plasmon Resonance (SPR) analysis
All SPR experiments were carried out using a Biacore T200 (GE healthcare) at 25 °C. Proteins (His-FlgI, FlgA, FlgAR113C-S190C were purified as described above. For FlgA and FlgAR113C-S190C, the N-terminal-fused histidine tags were removed with thrombin (GE Healthcare) prior to gel filtration. Ligand proteins were immobilized on CM5 sensor chips followed by the manufacture’s standard amine coupling protocol. Analytes were diluted with a running buffer [10 mM HEPES, 150 mM NaCl, 0.05% (v/v) Surfactant P20, (pH 7.4)]. Regeneration of the ligands was conducted by injecting 10 mM Glycine-HCl (pH 1.5). Data were processed with Biacore T200 Evaluation Software v1.0 (GE Healthcare).
Structures reported here are explained in interactive 3D at http://Proteopedia.Org/w/Samatey/2.
Additional Information
Accession codes: Atomic coordinates and structure factors of the native forms (open and closed) and the SeMet-labeled form (closed) have been deposited in the Protein Data Bank (PDB; http://www.pdb.org) as accession numbers 3TEE, 3VKI and 3VJP, respectively.
How to cite this article: Matsunami, H. et al. Structural flexibility of the periplasmic protein, FlgA, regulates flagellar P-ring assembly in Salmonella enterica. Sci. Rep. 6, 27399; doi: 10.1038/srep27399 (2016).
Supplementary Material
Acknowledgments
We thank K. Imada for invaluable advice with data collection and N. Shimizu, K. Hasegawa, and M. Yamamoto at SPring-8 for technical help in the use of beamline BL41XU. Y. Furukawa supported the early stages of this work and T. Miyata gave useful advice on the flagellar basal body isolation. T. Sasaki supported the electron microscopy. We thank E. Martz at the University of Massachusetts Amherst for making the illustration of the proteopedia web pages and the videos. We thank S. Silver at the University of Illinois for critical reading of the manuscript. This work was supported by direct funding provided by OIST and JSPS KAKENHI Grant Number 25000013 to K.N.
Footnotes
Author Contributions H.M., K.N. and F.A.S. designed the project. H.M., Y.-H.Y., V.A.M. and F.A.S. collected the X-ray data, solved the crystal structure. All authors discussed the results. H.M., K.N. and F.A.S. wrote the manuscript with the help of other authors. F.A.S. was responsible for the overall project strategy and management.
References
- Berg H. The rotary motor of bacterial flagella. Annu Rev Biochem 72, 19–54 (2003). [DOI] [PubMed] [Google Scholar]
- Macnab R. M. Type III flagellar protein export and flagellar assembly. Biochim Biophys Acta 1694, 207–17 (2004). [DOI] [PubMed] [Google Scholar]
- Jones C. J., Homma M. & Macnab R. M. Identification of proteins of the outer (L and P) rings of the flagellar basal body of Escherichia coli. J Bacteriol 169, 1489–92 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiba T., Yoshimura H. & Namba K. Monolayer crystallization of flagellar L-P rings by sequential addition and depletion of lipid. Science 252, 1544–6 (1991). [DOI] [PubMed] [Google Scholar]
- Homma M., Komeda Y., Iino T. & Macnab R. M. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J Bacteriol 169, 1493–8 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R. & Ochman H. Stepwise formation of the bacterial flagellar system. Proc Natl Acad Sci USA 104, 7116–21 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita S. & Tokuda H. Sorting of bacterial lipoproteins to the outer membrane by the Lol system. Methods Mol Biol 619, 117–29 (2010). [DOI] [PubMed] [Google Scholar]
- Okuda S. & Tokuda H. Lipoprotein sorting in bacteria. Annu Rev Microbiol 65, 239–59 (2011). [DOI] [PubMed] [Google Scholar]
- Schoenhals G. J. & Macnab R. M. Physiological and biochemical analyses of FlgH, a lipoprotein forming the outer membrane L ring of the flagellar basal body of Salmonella typhimurium. J Bacteriol 178, 4200–7 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibuki T., Shimada M., Minamino T., Namba K. & Imada K. Crystallization and preliminary X-ray analysis of FliJ, a cytoplasmic component of the flagellar type III protein-export apparatus from Salmonella sp. Acta Crystallogr Sect F Struct Biol Cryst Commun 65, 47–50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imada K., Minamino T., Kinoshita M., Furukawa Y. & Namba K. Structural insight into the regulatory mechanisms of interactions of the flagellar type III chaperone FliT with its binding partners. Proc Natl Acad Sci USA 107, 8812–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimov A. G. et al. Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nat Struct Biol 10, 789–93 (2003). [DOI] [PubMed] [Google Scholar]
- Kubori T., Shimamoto N., Yamaguchi S., Namba K. & Aizawa S. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol 226, 433–46 (1992). [DOI] [PubMed] [Google Scholar]
- Ohnishi K., Ohto Y., Aizawa S., Macnab R. M. & Iino T. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J Bacteriol 176, 2272–81 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K., Okada T., Yokoseki T. & Iino T. Sequence analysis of the flgA gene and its adjacent region in Salmonella typhimurium, and identification of another flagellar gene, flgN. Gene 143, 49–54 (1994). [DOI] [PubMed] [Google Scholar]
- Nambu T. & Kutsukake K. The Salmonella FlgA protein, a putativeve periplasmic chaperone essential for flagellar P ring formation. Microbiology 146 (Pt 5), 1171–8 (2000). [DOI] [PubMed] [Google Scholar]
- Chevance F. F. et al. The mechanism of outer membrane penetration by the eubacterial flagellum and implications for spirochete evolution. Genes Dev 21, 2326–35 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H., Samatey F. A., Nagashima S., Imada K. & Namba K. Crystallization and preliminary X-ray analysis of FlgA, a periplasmic protein essential for flagellar P-ring assembly. Acta Crystallographica Section F-Structural Biology and Crystallization Communications 68, 310–313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E. & Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60, 2256–68 (2004). [DOI] [PubMed] [Google Scholar]
- Holm L., Kääriäinen S., Rosenström P. & Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko T. et al. The refined crystal structure of an eel pout type III antifreeze protein RD1 at 0.62-A resolution reveals structural microheterogeneity of protein and solvation. Biophys J 84, 1228–37 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawan J. et al. Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol. J Biol Chem 280, 3555–63 (2005). [DOI] [PubMed] [Google Scholar]
- Ibuki T. et al. Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases. Nat Struct Mol Biol 18, 277–82 (2011). [DOI] [PubMed] [Google Scholar]
- Holmgren A. & Bränden C. I. Crystal structure of chaperone protein PapD reveals an immunoglobulin fold. Nature 342, 248–51 (1989). [DOI] [PubMed] [Google Scholar]
- Kuehn M. J. et al. Structural basis of pilus subunit recognition by the PapD chaperone. Science 262, 1234–41 (1993). [DOI] [PubMed] [Google Scholar]
- Lario P. I. et al. Structure and biochemical analysis of a secretin pilot protein. EMBO J 24, 1111–21 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okon M. et al. Structural characterization of the type-III pilot-secretin complex from Shigella flexneri. Structure 16, 1544–54 (2008). [DOI] [PubMed] [Google Scholar]
- Iyer L. M. & Aravind L. The emergence of catalytic and structural diversity within the beta-clip fold. Proteins 55, 977–91 (2004). [DOI] [PubMed] [Google Scholar]
- Leslie A. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr 62, 48–57 (2006). [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50, 760–3 (1994). [DOI] [PubMed] [Google Scholar]
- Terwilliger T. & Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr 55, 849–61 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. & Main P. Phase combination and cross validation in iterated density-modification calculations. Acta Crystallogr D Biol Crystallogr 52, 43–8 (1996). [DOI] [PubMed] [Google Scholar]
- Perrakis A., Sixma T., Wilson K. & Lamzin V. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr D Biol Crystallogr 53, 448–55 (1997). [DOI] [PubMed] [Google Scholar]
- Emsley P. & Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–32 (2004). [DOI] [PubMed] [Google Scholar]
- Murshudov G., Vagin A. & Dodson E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53, 240–55 (1997). [DOI] [PubMed] [Google Scholar]
- Blanc E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr 60, 2210–21 (2004). [DOI] [PubMed] [Google Scholar]
- Bhat T. N. Calculation of an omit map. Journal of Applied Crystallography 21, 279–281 (1988). [Google Scholar]
- Adams P. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T. & Macnab R. M. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol 181, 1388–94 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis N. R., Sosinsky G. E., Thomas D. & DeRosier D. J. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol 235, 1261–70 (1994). [DOI] [PubMed] [Google Scholar]
- Louis-Jeune C., Andrade-Navarro M. A. & Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins 80, 374–81 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.