Abstract
Colony biofilms of Bacillus subtilis are a widely used model for studying cellular differentiation. Here, we applied matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) to examine cellular and molecular heterogeneity in B. subtilis colony biofilms. From B. subtilis cells cultivated on a biofilm-promoting medium, we detected two cannibalistic factors not found in previous MALDI MSI studies of the same strain under different culturing conditions. Given the importance of cannibalism in matrix formation of B. subtilis biofilms, we employed a transcriptional reporter to monitor matrix-producing cell subpopulations using fluorescence imaging. These two complementary imaging approaches were used to characterize three B. subtilis strains, the wild type isolate NCIB3610, and two mutants, Δspo0A and ΔabrB, with defective and enhanced biofilm phenotypes, respectively. Upon deletion of key transcriptional factors, correlated changes were observed in biofilm morphology, signaling, cannibalistic factor distribution, and matrix-related gene expression, providing new insights on cannibalism in biofilm development. This work underscores the advantages of using multimodal imaging to compare spatial patterns of selected molecules with the associated protein expression patterns, obtaining information on cellular heterogeneity and function not obtainable when using a single method to characterize biofilm formation.
Keywords: biofilm, mass spectrometry imaging, fluorescence imaging, transcriptional reporter, cell differentiation
Introduction
Biofilms are microbial communities of surface-associated microorganisms embedded in a self-secreted extracellular matrix.1 Research on microbial biofilms improves our understanding of fundamental biological processes, such as cell differentiation2,3 and intercellular communication.4 These efforts have also delivered immediate applications in biomedical and biotechnological settings, including prevention of biofilm growth on indwelling clinical devices5 and wastewater treatment systems.6 Although most naturally occurring biofilms consist of multiple microbial species, single-species biofilms are widely employed in laboratory research as experimental models.7
Bacillus subtilis, a nonpathogenic Gram-positive bacterium, is among the most popular models for studying biofilms.8B. subtilis can develop diverse types of biofilms under different culture conditions, including colony biofilms at the air–solid interface, floating pellicles at the air–liquid interface, and submerged biofilms at the liquid–solid interface.8 In particular, when grown on agar media with a biofilm-inducing composition (minimal salts glycerol glutamate medium, or MSgg medium), B. subtilis forms wrinkled colony biofilms with complex structures.9 During biofilm development, functionally distinct cell subpopulations arise from genetically identical ancestors, following a seemingly ordered differentiation sequence: motile cells–matrix-producing cells–sporulating cells.10 The matrix-producing cells have also been suggested to be the same subpopulation of cannibals, which secrete toxins that can lyse a fraction of their sensitive siblings.11 As the cannibal/matrix-producing cells exhibit resistance to these toxins, they utilize the released nutrients from the cannibalized cells and increase in number, which leads to enhanced matrix production and promotes biofilm formation.11 Phenotypic heterogeneity in B. subtilis biofilms is considered to be a result of spatiotemporal crosstalk between chemical signals and gene expression,10 and yet the ability to characterize such complex interactions requires major advances in currently available analytical approaches.
Imaging techniques have been instrumental in understanding spatial heterogeneity in biofilms. Mass spectrometry imaging (MSI) is a label-free molecular imaging technique that can provide two- or even three-dimensional visualization of metabolite distribution in biological samples, and has been increasingly used in microbiological research.12,13 The most broadly used mass spectrometry (MS)-based imaging techniques in microbiology are matrix-assisted laser desorption/ionization (MALDI) imaging, secondary ionization mass spectrometry (SIMS) imaging, and desorption electrospray ionization imaging.12 Several applications using the aforementioned MSI techniques have been performed successfully to unravel dynamic spatial or temporal chemical information for metabolites in various microbial systems, such as single B. subtilis colonies or coculture systems,14−16Pseudomonas aeruginosa biofilms,17−20 and plant–microbe cocultures.21 With the aid of MSI, in situ visualization of the spatial distribution of individual molecules is enabled without the need for chemical derivatization or immunostaining. The ion images (or m/z images) obtained provide insight into microbe–microbe or microbe–plant interactions and metabolic exchange, and enable discovery of novel natural products.14,15,21,22
Given the complex nature of biofilms, the combination of multiple imaging techniques with complementary figures of merit has potential for unraveling biofilm biology.23 In our previous work, confocal Raman microscopy (CRM) was combined with MALDI18 and SIMS imaging19,20 to study biofilms of the opportunistic pathogen Pseudomonas aeruginosa. CRM can be used to visualize molecular distributions based on characteristic vibrational modes of chemical functional groups, and correlated CRM and MSI has been successfully used to expand chemical coverage, resolve subtly differing compounds, and cross-validate molecular distributions.18−20
Besides measuring small-molecule toxins, we also wanted to examine gene expression patterns to study cellular heterogeneity, and then correlate these results with the presence of specific gene products. While in principle CRM or MSI could provide this information, there are several issues. For example, CRM can detect typical bands of DNA and RNA molecules in bacterial biofilms,18,20 but it does not differentiate the identities of different genetic species. Moreover, MALDI MS of intracellular oligonucleotide species requires complicated sample preparation processes, such as cell lysis and purification,24 which are difficult to adapt for an MSI experiment. On the other hand, fluorescence imaging has long been a primary tool in assisting biologists in monitoring specific gene expression via the use of transcriptional reporters.25 These genetic reporters are constructed by fusing a fluorescence reporter gene with the upstream DNA sequence (promoter) from a gene of interest; expression of the targeted gene can be correlated to the abundance of fluorescence proteins.10,26 For example, using fluorescence reporters of the hag, yqxM, and sspB genes, which are exclusively expressed in motile, matrix-producing and sporulation cells, respectively, revealed that these different cell types exist in distinct spatial locations within Bacillus biofilms.10
For this work we combined MALDI and fluorescence imaging to compare metabolite distributions to spatial patterns of differentiated cell types, revealing molecular mechanisms that are impossible to study when single imaging methods are used alone. Using MALDI MSI to study B. subtilis colony biofilms grown on biofilm-promoting MSgg agar media, we observed two cannibalistic toxins that were absent in previous studies in which the rich medium ISP2 was used to cultivate the same strain. We then combined MALDI MSI and fluorescence stereoscopy to examine one wild type biofilm and two mutant strains with distinct biofilm phenotypes, Δspo0A and ΔabrB. We report correlated changes in metabolite abundance and matrix-related gene expression upon deletion of key transcriptional factors, and discuss these observations in the context of the genetic regulation of B. subtilis biofilm formation.
Experimental Procedures
Strain Construction and Cultivation
The B. subtilis wild isolate NCIB3610, B. subtilis lab strains PY79 and 168, and their derivatives used in this study are listed in Table S1. Chemicals were purchased from Fisher Scientific (Pittsburgh, PA) or Sigma-Aldrich (St. Louis, MO) unless indicated otherwise. B. subtilis was grown in Luria broth (LB) medium for routine cultivation, or MSgg medium (5 mM potassium phosphate (pH 7), 100 mM MOPS (pH 7), 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 50 μM FeCl3, 1 μM ZnCl2, 2 μM thiamine, 0.5% glycerol, 0.5% glutamate, 50 μg mL–1 tryptophan, 50 μg mL–1 phenylalanine) and ISP2 medium (4 g L–1 yeast extract, 10 g L–1 malt extract, 4 g L–1 glucose) for biofilm formation. When necessary, antibiotics were supplemented at the following concentrations: MLS (1 μg mL–1 erythromycin, 25 μg mL–1 lincomycin); spectinomycin (100 μg mL–1); kanamycin (10 μg mL–1); chloramphenicol (5 μg mL–1). An established protocol was used to form colony biofilms on MSgg agar plates.9 For every strain, three freshly streaked B. subtilis colonies (biological triplicates) on an LB agar plate were inoculated separately into 3 mL of LB liquid media for cultivation at 37 °C and 250 rpm. Then, 1 μL of overnight cell culture from each replicate was spotted onto a single MSgg plate supplemented with 1.5% agar and allowed to grow under static conditions at 37 °C for 24 h. To be compatible with MALDI MSI analysis, 5 mL of MSgg agar medium was dispensed into 10 cm diameter Petri dishes to form thin-layer agar. Petri dishes were sealed with laboratory paraffin film (Parafilm M, Bemis, Neenah, WI) to minimize water evaporation during biofilm cultivation.
Mutations in the wild isolate strains were created by transferring genetic modifications from laboratory strains using SPP1 phage transduction.27 Briefly, 0.1 mL serial dilutions of SPP1 phage stock were added to 0.2 mL of overnight cultures of laboratory donor strains grown in triptone yeast (TY) broth (LB supplemented with 10 mM MgSO4 and 100 μM MnSO4) and incubated for 15 min at 37 °C. TY soft agar, 3 mL (TY supplemented with 0.5% agar), was added to the mixture and poured on top of freshly poured TY plates (TY supplemented with 1.5% agar). The plates were incubated at 37 °C overnight; the one containing near confluent plaques was harvested by adding 5 mL of TY to the plate and scraping top agar layer into a 50 mL conical tube. The tube was vortexed and centrifuged at 5000g for 10 min. The supernatant fluid was treated with 25 μg mL–1 DNase I (New England Biolabs, Ipswich, MA) for 30 min at 20 °C and passed through a 0.45 μm syringe filter before being stored at 4 °C. After being grown to high density, 0.9 mL of recipient cell culture was mixed with 10 μL of SPP1 donor phage stock and 9 mL of TY. The mixture was then statically incubated at 37 °C for 30 min and centrifuged at 5000g for 10 min. The supernatant was discarded and the cell pellet was resuspended in the remaining volume of TY; 100 μL of cells were then plated onto LB, fortified with 1.5% agar, 10 mM sodium citrate, and appropriate antibiotics.
Fluorescence Microscopy and Flow Cytometry
Fluorescence images of B. subtilis colony biofilms were captured using an Axio Zoom.V16 fluorescence microscope (Zeiss, Oberkochen, Germany). The ZEN 2012 software package (Zeiss) was used to control the camera and image processing. The fluorescence Filter Set 47 (Zeiss) was used to visualize cyan fluorescent protein (CFP) fluorescence. All fluorescence pictures were taken with an exposure time of 500 ms and default color balance settings, and processed identically.
Colony biofilms were harvested from the MSgg agar surface and placed in 1 mL of PBS and dispersed with 12 pulses of mild sonication with 1 s duration and 30% amplitude on a Fisher Scientific Sonic Dismembrator Model 500. Cell fixation was performed by mixing 300 μL of cell suspension with 1 mL of 4% paraformaldehyde for a 7 min incubation time at 20 °C. Fixed cells were washed in PBS buffer three times before another 12 pulses of sonication with 1 s duration and 50% amplitude to obtain single cells. An appropriate dilution of the cell suspension was analyzed on a LSR II Flow Cytometer (BD, Franklin Lakes, NJ) by recording 10 000 events for CFP fluorescence (405 nm laser excitation coupled with a 440/40 bandpass filter).
MALDI MSI
The MALDI matrices, 2,5-dihydroxybenzoic acid (DHB), α-cyano-4-hydroxycinnamic acid (CHCA), and Universal MALDI matrix (1:1 mixture of DHB and CHCA), were purchased from Sigma-Aldrich. The matrix application method was modified according to a reported protocol.28 Six square sections (7 mm × 7 mm) containing the biofilms were transferred to a home-built stainless steel substrate that was fitted to MALDI adapters (Bruker Daltonics, Billerica, MA). Prior to MALDI imaging, DHB powder was sprinkled on top of the biofilms using a 53-μm sieve (Hogentogler & Co., Columbia, MD), and then dried in a 37 °C oven (JEIO TECH, Billerica, MA) for 2 h. Dried biofilm samples were stored in a vacuum desiccator until MALDI imaging analysis.
Measurements were performed using an ultrafleXtreme MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) with a frequency tripled Nd:YAG solid state laser (λ = 355 nm). The laser footprint setting was set to “Ultra” at an ∼90 μm diameter. Mass spectrometer calibration was performed using the Peptide Calibration Standard Kit II (Bruker Daltonics). Data acquisition was run in positive reflection mode with pulsed ion extraction, 250-μm laser step size, and a mass range of 500–4800 Da. Biofilms were analyzed with 1000 laser shots fired at 1000 Hz. Tandem MS was conducted on both biofilm surfaces and intact cell extracts (see Additional Methods in the Supporting Information for intact-cell MALDI MS) using the positive reflection LIFT mode of the mass spectrometer. Identification of target ions was based on the comparison of characteristic fragments with reported tandem MS spectra14,15,29 (Table S2 and Figure S2). Spectra were baseline-corrected and analyzed in flexAnalysis 3 (Bruker Daltonics). MALDI imaging was performed with the flexImaging 4 (Bruker Daltonics) and the resulting average mass spectrum was filtered manually in 0.5 Da increments, with individual colors assigned to the specific m/z value. All ion images were normalized to the total ion count (TIC).
Results and Discussion
MALDI MSI of Bacillus Colony Biofilms on MSgg Media
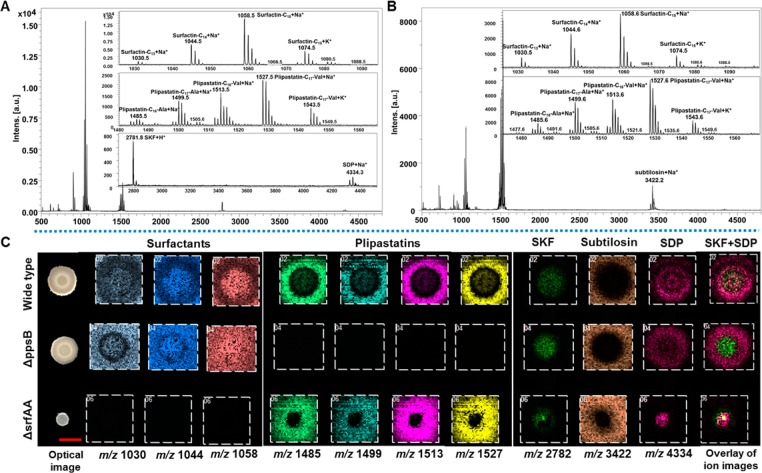
In this work, we focused on the wild type isolate NCIB3610 strain, which is a model for studying B. subtilis colony biofilms.8−10 Elegant previous MALDI MS studies of B. subtilis culture produced exciting data on several molecular distributions,14,15 but only reported images from ISP2 medium, on which relatively unstructured colonies form (Figure S1). To examine whether cell differentiation is associated with the spatial heterogeneity of metabolite distribution in B. subtilis biofilms, we employed MSgg medium, a widely used condition to induce biofilm development.9 When grown on MSgg, NCIB3610 formed characteristic wrinkled colony biofilms (Figure S1), consistent with previous reports.9 As different medium compositions often require customized sample preparation for MALDI MSI of microbial cultures on thin-layer agar,28 we screened appropriate MALDI matrices and matrix application approaches for MSgg. Three commonly used MALDI matrices—DHB, CHCA, and a 1:1 mixture of CHCA and DHB—were tested and sieved to saturate agar samples. In comparison to CHCA and the mixture of CHCA and DHB, the DHB matrix powder provided more homogeneous matrix layers and a broader coverage of target analytes, including surfactins, plipastatins, and subtilosin, as well as two cannibalistic factors, sporulation killing factor (SKF) and sporulation delaying protein (SDP) (Figure 1). Our assignment of these aforementioned known compounds is based on a comparison of our tandem MS results with fragmentation data reported in the literature14,15,29 (Table S2 and Figure S2).
Figure 1.
MALDI imaging analysis of colony biofilms of B. subtilis NCIB3610 and its mutants. (A,B) Single-pixel MALDI-MS spectra of surfactins, plipastatins, subtilosin, SKF and SDP from the inner (A) and outside (B) regions of B. subtilis NCIB3610 biofilms. (C) Selected MALDI images for B. subtilis biofilms. Each column represents (left to right): m/z 1030 (surfactin-C13, [M + Na]+); m/z 1044 (surfactin-C14, [M + Na]+); m/z 1058 (surfactin-C15, [M + Na]+); m/z 1485 (plipastatin-C16-Ala, [M + Na]+); m/z 1499 (plipastatin-C17-Ala, [M + Na]+); m/z 1513 (plipastatin-C16-Val, [M + Na]+); m/z 1527 (plipastatin-C17-Val, [M + Na]+); m/z 2782 (SKF, [M + H]+); m/z 3422 (subtilosin, [M + Na]+); m/z 4334 (SDP, [M + Na]+); and an overlay of ion images (green: SKF; bright pink: SDP). The ion intensity is reflected by the intensity of colors. Each column of ions is displayed using the same intensity scale, optimized per each metabolite and normalized to the TIC. Scale bar = 5 mm.
To confirm these identifications, strains with gene deletions in surfactin and plipastatin biosynthesis pathways were analyzed.15 The absence of surfactin and plipastatin ions was observed in the ΔsrfAA and ΔppsB strains, respectively (Figure 1). Furthermore, MALDI imaging revealed region-specific distributions of various metabolites (Figure 1). Surfactins, including surfactin-C13, surfactin-C14, and surfactin-C15, were mainly detected as Na+ and K+ adduct ions. Surfactins were visualized in both agar media and bacteria cells, with ion intensities being higher in the cells than in the agar. In contrast, plipastatins (plipastatin-C16-Ala, plipastatin-C17-Ala, plipastatin-C16-Val, and plipastatin-C17-Val) and subtilosin were preferably located in the agar media outside of the biofilms (Figure 1), and were detected as H+, Na+, and K+ adduct ions. For the cannibalistic factors, SKF was merely detected as H+ ions, but SDP was detected as H+, Na+, and K+ adduct ions. While both SKF and SDP were primarily associated with the biofilms, SKF was preferably located at the center and SDP was detected across the entire biofilm surface (Figure 1).
Notably, SKF and SDP were not detected in previous MALDI imaging studies when the same strain was cultivated on ISP2 medium.14 Considering the importance of cannibalism in matrix formation and biofilm development,11 it is not surprising that the absence of SKF and SDP correlates with the lack of biofilm formation in NCIB3610 cultures on ISP2 (Figure S1).14,15 Moreover, previous studies on SKF and SDP primarily employed the laboratory B. subtilis strain PY79, which is incapable of forming biofilms,9 and MALDI imaging revealed that both SKF and SDP were located evenly on the surfaces of PY79 colonies.14 In contrast, SKF and SDP exhibited distinct spatial patterns on NCIB3610 biofilms (Figure 1), suggesting an association between heterogeneity in metabolite distribution and cell differentiation. Furthermore, a previous study suggested that surfactins were required to activate cannibalism, based on the appearances of biofilms formed by a few mutant B. subtilis strains.11 While our results (Figure S1) agree with the prior reports on the aberrant morphology of ΔsrfAA biofilms,9,11 we still observed SKF and SDP production in the absence of surfactins (Figure 1). Together, the capability to detect SKF and SDP from B. subtilis biofilms using MSgg suggests that in certain systems, it is necessary to use specific cultivation media to replace ISP2, the most commonly used medium in MALDI MSI analysis of microbial agar cultures.
Monitoring Matrix-producing Cells Using a Fluorescence Reporter
The metabolites we characterized via MALDI MSI perform different roles in B. subtilis biofilm development: surfactins can promote surface swarming of B. subtilis cells,30 and they have been proposed as a class of signaling molecules that trigger biofilm initiation.26,31 SKF and SDP reduce the proportion of nonmatrix-producing cells to enhance biofilm formation;11,14 however, the biofilm-related functions of plipastatins and subtilosin remain elusive. Although extracellular matrix production is a key step in biofilm development to encase constituent cells into a structurally integrated community,32,33 matrix components were not detected in this and previous MALDI imaging studies.14,15 The B. subtilis biofilm matrix mainly consists of exopolysaccharide and proteins.8,34 In particular, amyloid fibers formed by the TasA protein are major matrix components,35 providing structural integrity to B. subtilis biofilms.36 The absence of TasA in previous MALDI MSI studies14,15 may be due to the low desorption/ionization efficiency of this amyloid protein from thick biofilms, or its formation into high molecular weight insoluble fibers. In addition, the typical sample preparation steps for analyzing amyloids from thin sections of mammalian tissues37 may not be suitable for studying biofilms, as rinsing and tryptic digestion may cause biofilm flaking and dislocation of small metabolites. Therefore, instead of targeting matrix molecules, we focused on cell-type-specific gene expression to visualize spatial distributions of matrix-producing cells.
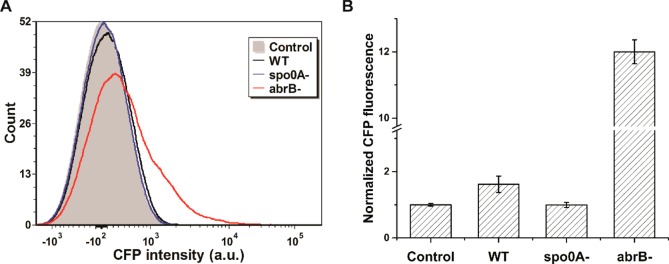
We chose to use a fluorescence transcriptional reporter, PyqxM-CFP, whereby the CFP gene is fused to the promoter of the yqxM-sipW-tasA operon.26 As this TasA-encoding operon is highly expressed in matrix-producing cells, this subpopulation becomes CFP positive during biofilm development in strains harboring the PyqxM-CFP reporter.26 We cultivated colony biofilms of a B. subtilis NCIB3610 strain integrated with PyqxM-CFP,26 and used flow cytometry to measure CFP expression at the single cell level. Compared to the negative control strain without PyqxM-CFP, the reporter strain (herein referred to as WT) exhibited a 62 ± 25% (mean ± SD) increase in CFP fluorescence (Figure 2), confirming the functional reconstitution of the genetic reporter.
Figure 2.
Flow cytometric analysis of B. subtilis colony biofilms with the PyqxM-CFP reporter. B. subtilis strains were cultured on MSgg agar media for 24 h before harvest. Colony biofilms were dispersed into single cells using sonication for analysis via flow cytometry. Control denotes the NCIB3610 strain without the reporter. WT denotes the NCIB3610 strain integrated with a PyqxM-CFP cassette. Two gene deletion strains based on WT are denoted as spo0A- and abrB-. (A) Histogram of the flow cytometry. (B) Mean fluorescence intensities of 10 000 cells. CFP intensities were normalized to the control. Error bars indicate the SDs of three biological replicates. P values were calculated using the independent two-tailed, two-sample t-test for equal sample sizes and equal variance: 0.042 (WT/Control), 0.944 (spo0A-/Control), and 0.013 (abrB-/Control).
We then explored whether mutations in genetic regulators may affect matrix-related gene expression. Formation of B. subtilis colony biofilms is controlled by a complex regulatory network in response to environmental stimuli.8,38 In particular, Spo0A and AbrB are two important transcriptional factors modulating expression of many biofilm-related genes. It has been reported that mutations of the spo0A and abrB genes can result in defective and enhanced biofilm phenotypes, respectively.9,26,39 We disrupted the spo0A or the abrB gene in the B. subtilis reporter strain, and found that these gene deletions exerted opposite effects on CFP intensity. Whereas Δspo0A deletion completely abolished CFP signals, ΔabrB increased CFP signals substantially (Figure 2). These observations correlate well with the biological functions of Spo0A and AbrB. As Spo0A and its phosphorylated forms are required for initiating matrix production,40 the PyqxM promoter remains silent in the absence of Spo0A. AbrB inhibits gene expression from PyqxM, and hence, the PyqxM promoter can be activated upon removal of its genetic repressor AbrB.41 Accordingly, a genetic fluorescence reporter was successfully constructed to monitor gene expression in B. subtilis.
Analysis of Colony Biofilms Using MALDI MSI and Fluorescence Microscopy
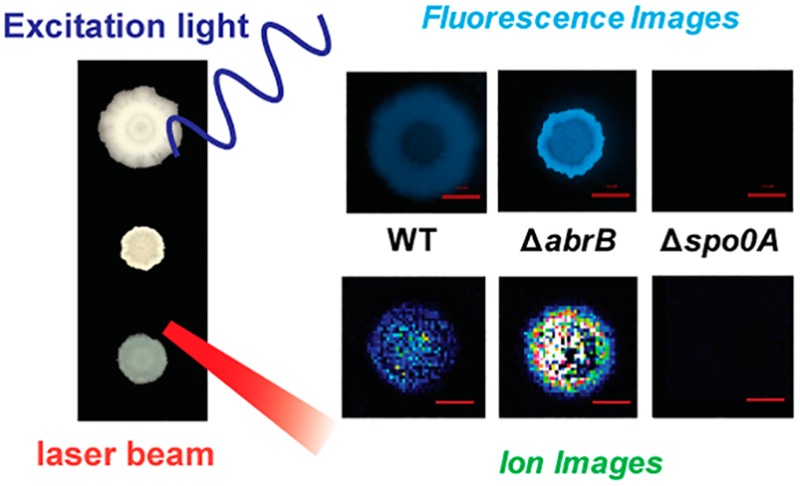
With an optimized MALDI MSI protocol and a functional fluorescence reporter, we sought to combine MS and fluorescence imaging to characterize both chemical and genetic heterogeneity in B. subtilis biofilms. Three B. subtilis strains harboring the PyqxM-CFP reporter were examined: the WT, Δspo0A, and ΔabrB strains. When cultivated on the biofilm-promoting MSgg medium, the WT formed colony biofilms with characteristic wrinkles (Figure S1), and CFP intensity was higher at the edge of biofilms than in the central region (Figure 3). Compared with the wild type NCIB3610 strain, the recombinant NCIB3610 strain integrated with the PyqxM-CFP reporter exhibited no observable changes in biofilm morphology (Figure S1) or metabolite distribution (Figure 1 and Figure 3). On the other hand, the mutant lacking spo0A formed unstructured colonies, and the mutant lacking abrB produced biofilms with hyper-wrinkly structures (Figure S1).
Figure 3.
Fluorescence imaging and MALDI MSI of B. subtilis biofilms of NCIB3610 and its mutants integrated with the PyqxM-CFP reporter. CFP images were acquired using a fluorescence stereoscope before MALDI analysis. Each individual column represents (left to right): m/z 1030 (surfactin-C13, [M + Na]+); m/z 1044 (surfactin-C14, [M + Na]+); m/z 1058 (surfactin-C15, [M + Na]+); m/z 1485 (plipastatin-C16-Ala, [M + Na]+); m/z 1499 (plipastatin-C17-Ala, [M + Na]+); m/z 1513 (plipastatin-C16-Val, [M + Na]+); m/z 1527 (plipastatin-C17-Val, [M + Na]+); m/z 2782 (SKF, [M + H]+), m/z 3422 (subtilosin, [M + Na]+), m/z 4334 (SDP, [M + Na]+), and an overlay of ion images (green: SKF; bright pink: SDP). The ion intensity is reflected by the intensity of colors. Each column of ions is displayed using the same intensity scale, optimized per each metabolite and normalized to the TIC. Scale bar = 2 mm for fluorescence images and 5 mm for optical and ion images.
Consistent with the flow cytometry results (Figure 2), biofilms of the Δspo0A and ΔabrB strains showed substantially reduced and enhanced CFP fluorescence compared with the WT, respectively (Figure 3). Furthermore, Δspo0A and ΔabrB also exerted different effects on metabolite abundance and distribution. For surfactins, plipastatins, subtilosin, and SDP, the ΔabrB mutant exhibited no observable changes relative to the WT, and the abundance of SKF was enhanced in ΔabrB compared to the WT (Figure 3). On the other hand, while surfactins of the WT were observed both in agar and cells, surfactins of the Δspo0A mutant were mostly confined within the biofilm region (Figure 3). These results are in agreement with previous findings that surfactin secretion is absent upon spo0A deletion.42 Also, the abundances of plipastatins, subtilosin, SKF, and SDP were greatly reduced by Δspo0A deletion (Figure 3), consistent with observations in earlier reports.14 Notably, while the Δspo0A and ΔabrB strains exhibited moderately decreased growth rates compared with WT in liquid LB media (Figure S3A), cell growth was significantly (p < 0.001) impaired on MSgg agar media for both mutants relative to WT (Figure S3B). Therefore, it may be reasonable to attribute enhanced SKF signals in the ΔabrB biofilms to increased biosynthesis, but it was unclear whether reduced metabolite amounts in the Δspo0A biofilms resulted from decreased cell numbers or suppressed biosynthesis (Figure S3B).
Our results confirm the current thought on the regulatory functions of Spo0A and AbrB in biofilm development. Spo0A is a master transcriptional regulator that modulates expression of many genes involved in biofilm formation and sporulation.39,40 A single aspartate residue of Spo0A is subjected to phosphorylation modification, and the cellular concentration of Spo0A∼P (phosphorylated Spo0A protein) determines a specific transcriptomic state.43 For example, intermediate Spo0A∼P concentrations activate matrix production, but high concentrations trigger sporulation. Hence, diverse Spo0A∼P levels in a cell population over the time course of biofilm formation enable temporospatial cellular differentiation.43 Consistent with this model, unstructured colonies were formed by the Δspo0A strain, suggesting cellular differentiation was absent. Also, expression from the PyqxM promoter (Figure 2) was blocked upon the Δspo0A deletion, indicating that the pathway for matrix synthesis is subjected to genetic regulation by Spo0A∼P levels.
For AbrB, it represses a number of genes, including those involved in biofilm formation via direct association with promoter sequences.44 AbrB is negatively regulated by Spo0A∼P via both transcriptional repression45 and allosteric inhibition.46 When environmental signals trigger phosphorylation of Spo0A∼P, cellular AbrB abundancy is reduced, which in turn activates matrix gene expression and biofilm formation.41 In the ΔabrB strain, hyper-wrinkly biofilms were formed (Figure S1), consistent with the role of AbrB in the negative regulation of biofilm development. For the PyqxM reporter, fluorescence intensity was greatly enhanced by ΔabrB (Figure 2 and Figure 3), in agreement with previous reports that the yqxM-sipW-tasA operon is a repression target of AbrB.41 Compared with the WT, SKF abundance was greatly increased in the ΔabrB strain (Figure 3), suggesting that repression of the SKF pathway by AbrB was not fully relieved in the WT strain when cultivated using MSgg. In contrast, production of the other metabolites observed did not exhibit obvious changes between the WT and the ΔabrB mutant, indicating that the activation of corresponding pathways was no longer limited by AbrB in the WT when cultivated on MSgg.
However, caution needs to be taken when interpreting quantitative changes among different strains in the fluorescence and MALDI imaging results, which can be affected by both total cell numbers and protein/metabolite abundance in individual cells. In particular, as cell growth was substantially impaired in the Δspo0A mutant relative to WT on MSgg agar (Figure S3), it was difficult to conclude whether decreased CFP and MS signals (Figure 3) resulted from reduced cell numbers or suppressed biosynthesis, or both, based solely on the imaging results. Several approaches can be used to alleviate this concern. First, if changes in fluorescence/MS imaging signals and cell amounts are in opposite directions, it may be reasonable to speculate modified biosynthetic productivity. In this study, it is highly likely that enhanced CFP signal and SKF abundance in the ΔabrB strain (Figure 3) were due to increased biosynthesis, as there were fewer cells in the mutant colony biofilms compared with WT (Figure S3B). Second, complementary approaches can be used to provide single cell-level information. For example, flow cytometry measures fluorescence intensities of individual cells, and therefore, one may conclude that the Δspo0A and ΔabrB mutations changed CFP transcription from the PypxM promoter (Figure 2). Third, other analytical techniques more amenable to quantitative analysis than MALDI imaging (such as liquid chromatography) can be performed to estimate aggregated averages of metabolite abundance in individual cells. Together, as the TIC normalization method for processing MALDI imaging data does not account for cell number variations, it requires careful assessment when comparing metabolite abundance results between different microbial strains.
It is also important to note that the combined use of MALDI and fluorescence imaging supports insights on cannibalism and matrix production in B. subtilis biofilm development. Previously, matrix-producing and cannibal cells were considered the same population.11 The main evidence for this assertion was obtained using transcriptional reporters, whereby biosynthesis pathways responsible for TasA and SKF production were coordinately activated in the same subpopulation.11 In contrast, by using MALDI and fluorescence imaging we observed that in WT biofilms, SDP-, SKF- and matrix-producing cells exhibited distinct spatial patterns (Figure 3), suggesting further differentiation in the cannibal/matrix-producing subpopulation. Such differentiation may be mediated by AbrB, as the distributions of SDP-, SKF-, and CFP-positive cells were almost overlapping in ΔabrB biofilms (Figure 3). This discrepancy may be a result of the analytical methods used in the other study,11 which dispersed biofilms into single cells before flow cytometric analysis, and so did not yield spatial information. We believe that even if fluorescence microscopy were to be used to monitor the spatial activities of multiple transcriptional reporters, it still cannot match the information-rich data from our combined method. Given the broad bandwidths of emission spectra, only two transcriptional reporters are commonly used in a single B. subtilis strain.10,26 Therefore, visualization of three different subpopulations in B. subtilis biofilms using fluorescence reporters required construction of at least two different strains.10 In contrast, the mass resolution of MALDI allows detection of a myriad of preselected and unanticipated compounds from a biofilm, minimizing the effort in strain creation and reducing sample-to-sample variations as compared to using fluorescence microscopy alone.
Conclusions
We developed a multimodal imaging method to compare metabolite distribution and gene expression patterns on the surfaces of a widely used B. subtilis biofilm model. Using fluorescence imaging coupled with a transcriptional reporter, we were able to analyze matrix production in biofilm development. This key phenotype had not been examined in previous MALDI MSI studies of B. subtilis, perhaps due to the incompatibility of sample preparation methods between metabolites and protein amyloids. Combining MALDI and fluorescence imaging enabled the detection of distinct populations of cells in a biofilm previously assumed to be comprised of an identical population. The combination of genetic tools, fluorescence imaging, and MALDI MSI is applicable to other bacterial biofilms for examining spatial distributions in a range of complex biofilm communities.
Acknowledgments
We thank Dr. Mayandi Sivaguru and Dr. Austin Cyphersmith at the Carl R. Woese Institute for Genomic Biology Core Facility for their help with the fluorescence stereoscope. We also thank Prof. Roberto Kolter at the Harvard Medical School for kindly providing the B. subtilis strains with the fluorescence transcriptional reporter PyqxM-CFP, and Sage J.B. Dunham for proofreading this manuscript. T. S. would like to acknowledge support from the Institute for Genomic Biology Postdoctoral Fellowship. This research was partly supported by the Department of Energy through a subcontract from Oak Ridge National Laboratory (PTX-UT-Battelle) under Award No. ORNL-4000134575 (JVS), the National Institute of Allergy and Infectious Diseases under Award No. R01 AI113219 (JVS), and the Biosystems Design theme startup fund from the Institute for Genomic Biology (HZ). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.6b00127.
Additional methods and supporting figures showing the colony morphology of B. subtilis strains on MSgg and ISP2 media (Figure S1), representative MALDI LIFT TOF/TOF mass spectra of SKF and SDP (Figure S2), and cell growth of the B. subtilis NCIB3610 strain and its mutants (Figure S3). Two tables showing the strains used in this study (Table S1) and selected metabolites assigned in B. subtilis biofilms by MALDI ToF MS analysis of intact cells (Table S2). (PDF)
Author Contributions
# T.S. and B.L. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Hall-Stoodley L.; Costerton J. W.; Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2 (2), 95–108. 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Stewart P. S.; Franklin M. J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6 (3), 199–210. 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Stoodley P.; Sauer K.; Davies D. G.; Costerton J. W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- Parsek M. R.; Greenberg E. P. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13 (1), 27–33. 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L.; Stoodley P. Evolving concepts in biofilm infections. Cell. Microbiol. 2009, 11 (7), 1034–1043. 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- Singh R.; Paul D.; Jain R. K. Biofilms: implications in bioremediation. Trends Microbiol. 2006, 14 (9), 389–397. 10.1016/j.tim.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Monds R. D.; O’Toole G. A. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009, 17 (2), 73–87. 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Vlamakis H.; Chai Y.; Beauregard P.; Losick R.; Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 2013, 11 (3), 157–168. 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda S. S.; Gonzalez-Pastor J. E.; Ben-Yehuda S.; Losick R.; Kolter R. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (20), 11621–11626. 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H.; Aguilar C.; Losick R.; Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008, 22 (7), 945–953. 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D.; Vlamakis H.; Losick R.; Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol. Microbiol. 2009, 74 (3), 609–618. 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous J. D.; Dorrestein P. C. Imaging mass spectrometry in microbiology. Nat. Rev. Microbiol. 2011, 9 (9), 683–694. 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell L. A.; Heeren R. M. Imaging mass spectrometry. Mass Spectrom. Rev. 2007, 26 (4), 606–643. 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- Liu W. T.; Yang Y. L.; Xu Y.; Lamsa A.; Haste N. M.; Yang J. Y.; Ng J.; Gonzalez D.; Ellermeier C. D.; Straight P. D.; Pevzner P. A.; Pogliano J.; Nizet V.; Pogliano K.; Dorrestein P. C. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (37), 16286–16290. 10.1073/pnas.1008368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. L.; Xu Y.; Straight P.; Dorrestein P. C. Translating metabolic exchange with imaging mass spectrometry. Nat. Chem. Biol. 2009, 5 (12), 885–887. 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debois D.; Hamze K.; Guerineau V.; Le Caer J. P.; Holland I. B.; Lopes P.; Ouazzani J.; Seror S. J.; Brunelle A.; Laprevote O. In situ localisation and quantification of surfactins in a Bacillus subtilis swarming community by imaging mass spectrometry. Proteomics 2008, 8 (18), 3682–3691. 10.1002/pmic.200701025. [DOI] [PubMed] [Google Scholar]
- Lanni E. J.; Masyuko R. N.; Driscoll C. M.; Aerts J. T.; Shrout J. D.; Bohn P. W.; Sweedler J. V. MALDI-guided SIMS: multiscale imaging of metabolites in bacterial biofilms. Anal. Chem. 2014, 86 (18), 9139–9145. 10.1021/ac5020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masyuko R. N.; Lanni E. J.; Driscoll C. M.; Shrout J. D.; Sweedler J. V.; Bohn P. W. Spatial organization of Pseudomonas aeruginosa biofilms probed by combined matrix-assisted laser desorption ionization mass spectrometry and confocal Raman microscopy. Analyst 2014, 139 (22), 5700–5708. 10.1039/C4AN00435C. [DOI] [PubMed] [Google Scholar]
- Baig N. F.; Dunham S. J.; Morales-Soto N.; Shrout J. D.; Sweedler J. V.; Bohn P. W. Multimodal chemical imaging of molecular messengers in emerging Pseudomonas aeruginosa bacterial communities. Analyst 2015, 140 (19), 6544–6552. 10.1039/C5AN01149C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni E. J.; Masyuko R. N.; Driscoll C. M.; Dunham S. J.; Shrout J. D.; Bohn P. W.; Sweedler J. V. Correlated imaging with C60-SIMS and confocal Raman microscopy: Visualization of cell-scale molecular distributions in bacterial biofilms. Anal. Chem. 2014, 86 (21), 10885–10891. 10.1021/ac5030914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debois D.; Jourdan E.; Smargiasso N.; Thonart P.; De Pauw E.; Ongena M. Spatiotemporal monitoring of the antibiome secreted by Bacillus biofilms on plant roots using MALDI mass spectrometry imaging. Anal. Chem. 2014, 86 (9), 4431–4438. 10.1021/ac500290s. [DOI] [PubMed] [Google Scholar]
- Cawoy H.; Debois D.; Franzil L.; De Pauw E.; Thonart P.; Ongena M. Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens. Microb. Biotechnol. 2015, 8 (2), 281–295. 10.1111/1751-7915.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masyuko R.; Lanni E. J.; Sweedler J. V.; Bohn P. W. Correlated imaging--a grand challenge in chemical analysis. Analyst 2013, 138 (7), 1924–1939. 10.1039/c3an36416j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C.; Cantor C. R. A high-throughput gene expression analysis technique using competitive PCR and matrix-assisted laser desorption ionization time-of-flight MS. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (6), 3059–3064. 10.1073/pnas.0630494100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southward C. M.; Surette M. G. The dynamic microbe: green fluorescent protein brings bacteria to light. Mol. Microbiol. 2002, 45 (5), 1191–1196. 10.1046/j.1365-2958.2002.03089.x. [DOI] [PubMed] [Google Scholar]
- Lopez D.; Vlamakis H.; Losick R.; Kolter R. Paracrine signaling in a bacterium. Genes Dev. 2009, 23 (14), 1631–1638. 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasbin R. E.; Young F. E. Transduction in Bacillus subtilis by bacteriophage SPP1. J. Virol. 1974, 14 (6), 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Y.; Phelan V. V.; Simkovsky R.; Watrous J. D.; Trial R. M.; Fleming T. C.; Wenter R.; Moore B. S.; Golden S. S.; Pogliano K.; Dorrestein P. C. Primer on agar-based microbial imaging mass spectrometry. J. Bacteriol. 2012, 194 (22), 6023–6028. 10.1128/JB.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx R.; Stein T.; Entian K. D.; Glaser S. J. Structure of the Bacillus subtilis peptide antibiotic subtilosin A determined by 1H-NMR and matrix assisted laser desorption/ionization time-of-flight mass spectrometry. J. Protein Chem. 2001, 20 (6), 501–506. 10.1023/A:1012562631268. [DOI] [PubMed] [Google Scholar]
- Kearns D. B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010, 8 (9), 634–644. 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D.; Fischbach M. A.; Chu F.; Losick R.; Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (1), 280–285. 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda S. S.; Vik S.; Friedman L.; Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005, 13 (1), 20–26. 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Flemming H. C.; Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8 (9), 623–633. 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Marvasi M.; Visscher P. T.; Casillas Martinez L. Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis. FEMS Microbiol. Lett. 2010, 313 (1), 1–9. 10.1111/j.1574-6968.2010.02085.x. [DOI] [PubMed] [Google Scholar]
- Branda S. S.; Chu F.; Kearns D. B.; Losick R.; Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 2006, 59 (4), 1229–1238. 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- Romero D.; Aguilar C.; Losick R.; Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (5), 2230–2234. 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley E. H.; Caprioli R. M. Molecular imaging of proteins in tissues by mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (47), 18126–18131. 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns L. S.; Hobley L.; Stanley-Wall N. R. Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms. Mol. Microbiol. 2014, 93 (4), 587–598. 10.1111/mmi.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon M. A.; Lazazzera B. A. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 2001, 42 (5), 1199–1209. 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- Molle V.; Fujita M.; Jensen S. T.; Eichenberger P.; Gonzalez-Pastor J. E.; Liu J. S.; Losick R. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 2003, 50 (5), 1683–1701. 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- Hamon M. A.; Stanley N. R.; Britton R. A.; Grossman A. D.; Lazazzera B. A. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 2004, 52 (3), 847–860. 10.1111/j.1365-2958.2004.04023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Y.Investigating Microbial Metabolites with Novel Mass Spectrometry Tools. Ph.D. Dissertation, University of California, San Diego, San Diego, 2013. [Google Scholar]
- Fujita M.; Gonzalez-Pastor J. E.; Losick R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 2005, 187 (4), 1357–1368. 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumsakul O.; Takahashi H.; Oshima T.; Hishimoto T.; Kanaya S.; Ogasawara N.; Ishikawa S. Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res. 2011, 39 (2), 414–428. 10.1093/nar/gkq780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch M.; Webb V.; Spiegelman G.; Hoch J. A. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. U. S. A. 1990, 87 (5), 1801–1805. 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banse A. V.; Chastanet A.; Rahn-Lee L.; Hobbs E. C.; Losick R. Parallel pathways of repression and antirepression governing the transition to stationary phase in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (40), 15547–15552. 10.1073/pnas.0805203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.