Abstract
Background
In wheat, the vernalization requirement is mainly controlled by the VRN genes. Different species of hexaploid and tetraploid wheat are widely used as genetic source for new mutant variants and alleles for fundamental investigations and practical breeding programs. In this study, VRN-A1 and VRN-B1 were analysed for 178 accessions representing six tetraploid wheat species (Triticum dicoccoides, T. dicoccum, T. turgidum, T. polonicum, T. carthlicum, T. durum) and five hexaploid species (T. compactum, T. sphaerococcum, T. spelta, T. macha, T. vavilovii).
Results
Novel allelic variants in the promoter region of VRN-A1 and VRN-B1 were identified based on the change in curvature and flexibility of the DNA molecules. The new variants of VRN-A1 (designated as Vrn-A1a.2, Vrn-A1b.2 – Vrn-A1b.6 and Vrn-A1i) were found to be widely distributed in hexaploid and tetraploid wheat, and in fact were predominant over the known VRN-A1 alleles. The greatest diversity of the new variants of VRN-B1 (designated as VRN-B1.f, VRN-B1.s and VRN-B1.m) was found in the tetraploid and some hexaploid wheat species.
For the first time, minor differences within the sequence motif known as the VRN-box of VRN1 were correlated with wheat growth habit. Thus, vrn-A1b.3 and vrn-A1b.4 were revealed in winter wheat in contrast to Vrn-A1b.2, Vrn-A1b.5, Vrn-A1b.6 and Vrn-A1i. It was found that single nucleotide mutation in the VRN-box can influence the vernalization requirement and growth habit of wheat. Our data suggest that both the A-tract and C-rich segment within the VRN-box contribute to its functionality, and provide a new view of the hypothesised role of the VRN-box in regulating transcription of the VRN1 genes. Specifically, it is proposed that combination of mutations in this region can modulate vernalization sensitivity and flowering time of wheat.
Conclusions
New allelic variants of the VRN-A1 and VRN-B1 genes were identified in hexaploid and tetraploid wheat. Mutations in A-tract and C-rich segments within the VRN-box of VRN-A1 are associated with modulation of the vernalization requirement and flowering time. New allelic variants will be useful in fundamental investigations into the regulation of VRN1 expression, and provide a valuable genetic resource for practical breeding of wheat.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-015-0691-2) contains supplementary material, which is available to authorized users.
Keywords: Cereal, Genetic variation, Flowering time, Vernalization requirement, Wheat, DNA curvature, VRN-box, VRN1 genes, Promoter, New alleles, Anomalous migration
Background
One of the key factors governing the adaptability of wheat to a wide range of environmental conditions is allelic diversity within genes regulating seasonal growth habit (vernalization requirement) and photoperiod response. The vernalization requirement determines the need for a prolonged exposure to low temperature in order for plants to transition to the reproductive phase of development. According to their response to vernalization, wheat plants can be categorised as winter (strong vernalization sensitivity), spring (not sensitive to vernalization), or facultative types (intermediate growth habit). The vernalization requirement and growth habit in wheat is largely controlled by four major loci, which are also critical in determining flowering and maturity times: these include VRN1 (a MADS-box transcription factor), VRN2 (a zinc-finger CCT domain gene - ZCCT), VRN3 (homologous to the Arabidopsis gene FLOWERING LOCUS T) and VRN4 (MADS-box transcription factor) [1–4].
The VRN genes interact epistatically (reviewed by [5]). The products of VRN2 expression downregulate transcription of VRN3 [3] through competitive interaction with other CCT-domain proteins. This occurs in conjunction with the replacement of the HAP2 (HEME ACTIVATOR PROTEIN2) subunit of the HAP2/3/5 complex, which binds with the VRN3 promoter [6]. In turn, VRN3 proteins are transported from the leaves to the shoot apical meristem [7] where they interact with transcription factor FDL2 (FLOWERING LOCUS D-like2) resulting in the formation of a protein complex that activates expression of the VRN1 genes [3, 8] and likely its paralogs FUL2 (FRUITFULL 2) and FUL3 [9]. During vernalization, VRN2 transcription is progressively downregulated by the vernalization conditions (short photoperiod and low temperature) and increasing activation of VRN1, which indirectly suppresses VRN2 [2, 10]. It is supposed that subsequent expression of VRN1 in leaves is important for maintaining the repression of VRN2 transcription in winter wheat after vernalization [9]. Interaction between VRN1, VRN2 and VRN3 form a regulatory positive feedback loop [5, 10] in which VRN3 is the integrator of the vernalization and photoperiod floral pathways [3, 11]. A change in expression of any of the VRN genes in this feedback loop results in modulation of the transcription profiles of the other two genes and a change in flowering time. It is assumed that VRN4 is active in the leaves and operates upstream (or is a part) of the VRN1/VRN2/VRN3 feedback loop, herewith VRN1 is the earliest target (direct or indirect) of VRN-D4 among these three genes [4].
The VRN2 locus includes three tandemly duplicated ZCCT genes. The recessive vrn2 allele is associated with a loss of function for all copies of the ZCCT genes from all homologous genomes which can be caused by missense mutations within the CCT domain coding region [2, 12] or the presence of null alleles [13]. Likely, this is the main reason why accessions carrying recessive vrn2 have so far been described only for T. monococcum [2] and have not been observed in varieties of polyploid wheat [5]. Natural variation for VRN3 has been found only in the B genome [3, 14]. The dominant Vrn-D4 allele is less widespread in wheat, although it is frequent in hexaploid wheat from India and nearby regions [15, 16], and is mainly predominant in accessions of T. sphaerococcum [4]. Thus, the vernalization requirement of an overwhelming number of spring accessions of hexaploid and tetraploid wheat is determined by the allelic diversity of VRN1 genes.
The VRN1 genes have been mapped to the middle of the long arms of the chromosomes 5 homeologous group [1]. Allelic variants at the VRN1 genes are largely responsible for the division of wheat into winter, spring, and facultative types. The presence of the dominant VRN-A1 alleles results in complete elimination of the vernalization requirement (spring type), whereas the presence of either VRN-B1 or VRN-D1 alleles alone is associated with some residual vernalization response and later flowering (facultative type) [17, 18]. VRN1 genes possess no less than two regulatory regions, located in the promoter and first intron respectively [19, 20]. Among dominant alleles of VRN-A1 the predominant carry mutations within the promoter region (in particular the Vrn-A1a/b/d/e and Vrn-Am1a/g alleles), while the dominant alleles of VRN-B1 (Vrn-B1a/b/c) and VRN-D1 (Vrn-D1a and Vrn-D1s alleles) are mostly determined by mutations in the first intron [1, 10, 19–23]. However, a later study identified the dominant VRN-B1 allele (hereafter designated as Vrn-B1(ins)), which has a retrotransposon insertion in the promoter and confers a spring growth habit. Still, this insertion was detected mainly for T. carthlicum (86 % accessions) and only in 3 accessions of T. dicoccum [24]. Similarly, a recent study identified a 174-bp insertion within the VRN-D1 promoter region, which possibly promoted the basal activity level of the VRN-D1 gene (Vrn-D1c allele) [25]. Furthermore, in accessions of T. aestivum were identified from 1 to 4 copies of the VRN-A1 gene [26]. This copy number variation (CNV) polymorphism is an important trait, which influences on vernalization requirement duration and flowering time of wheat [27, 28].
The promoter region of the VRN1 gene contains multiple regulatory sites [1, 29, 30] most important of which are the CArG-box, VRN-box and ACGT-motif. The CArG-box (a common binding site for MADS-box proteins) is located 180 bp upstream of the site of transcription initiation but downstream of the ACGT-motif [1]. However, during subsequent studies it was found that the vernalization requirement is preserved in plants where the CArG-box is fully deleted (vrn-Am1b allele) [31], indicating that the CArG-box is not critical for the determination of the vernalization requirement. Pidal et al. [31] analysed the 5' UTR of the VRN-A1 alleles in spring and winter T. monococcum accessions carrying mutations within the promoter region of VRN-A1. This led to the identification of a sequence located upstream from the CArG-box, containing mutations which were associated with the spring growth habit. Furthermore, the same region is also altered in two dominant, spring Vrn-A1a and Vrn-A1b alleles found in polyploid wheat [19]. This sequence of 16 bp (TTAAAAACCCCTCCCC) was designated as the VRN-box. It is assumed that the VRN-box is involved in determination of the vernalization requirement. Located upstream from CArG- and VRN-boxes are the binding sites for bZIP transcription factors. Overall, within the entire 2.3 kb promoter region of VRN1 several such DNA motifs were found, all with a ACGT core sequence. These consisted of a G-box (CACGTG) and four putative hybrid boxes: A/G, A/C, G/A, G/C, which are binding sites for TaFDL2 (orthologue of FLOWERING LOCUS D-LIKE 2 in wheat) and form a protein complex with TaFT (VRN3) [8]. Thus, interaction of TaFDL2 with TaFT and with the bZIP-binding sites of the VRN1 promoter forms the basis of a mechanism which provides regulation of the transcription of VRN1 by VRN3. Furthermore, activation of VRN1 expression by vernalization is associated with changes in histone methylation in different regions of the promoter [30] and intron-1 [32]. Additionally, recent reports have highlighted the post transcriptional regulation of VRN1 expression involving sequences within the first intron [4, 33].
The less widespread species of hexaploid and tetraploid wheat often attract the attention of researchers as genetic sources of new mutant variants and alleles of agronomically valuable traits. For example, the tetraploid wheat species T. dicoccoides, T. dicoccum and T. carthlicum, and hexaploid wheat T. spelta and T. compactum were where the alleles Vrn-A1d, Vrn-A1e [19], Vrn-B1(ins) [24] and Vrn-D1s [23] respectively, were first identified. And although implementation of these alleles in wheat breeding is difficult to achieve within a short time span, nevertheless, new data provide significant benefits. This research serves to expand our knowledge about the structure and function of VRN genes, facilitates identification and accurate localization of regulatory sites and regions, and furthers our understanding of the ways and mechanisms determining the development of wheat, its vernalization requirement and flowering time.
In the present study the regulatory regions of the VRN1 genes were analysed in tetraploid and hexaploid wheat species in order to evaluate genetic variation and identify of new allelic variants. The phenomenon of anomalously slow migration of DNA fragments in polyacrylamide gels which indicates presence of curved DNA was used as the first step in identification of differences in gene regulatory regions. Focusing our attention on investigation of the VRN-A1 gene, we also analysed VRN-B1 and generalized these data with the results of our previous research on VRN-D1 in the same accessions of hexaploid wheat [23]. This has strengthened the connection between genotype and phenotype of experimental plants, and enabled us to predict the possible effects of new mutant variants in the regulatory regions. Novel variants of VRN-A1 and VRN-B1 identified herein can be useful not only for practical breeding, but also for fundamental investigation of the mechanisms regulating VRN1 gene expression. In particular, new data can contribute to the more accurate localization of regulatory regions within the VRN1 promoter and in the first intron.
Results
Identification of three variants of Vrn-A1a
Polyacrylamide gel electrophoresis of PCR fragments of the VRN-A1 promoter region for T. compactum and T. dicoccum accessions revealed three combinations of amplicons, representing three Vrn-A1a allelic variants. These variants were designated as Vrn-A1a.1 (PCR fragments 853 and 944 bp), Vrn-A1a.2 (fragments 853, 924 and 944 bp) and Vrn-A1a.3 (fragments 765 and 853 bp). The Vrn-A1a.1 and Vrn-A1a.3 variants (also known as Vrn-A1a) were previously identified in hexaploid and tetraploid wheat respectively [19]. The new variant, Vrn-A1a.2, differs from Vrn-A1a.1 by the amplification of an additional 924 bp fragment (Fig. 1a). This amplicon was cloned and sequenced (GenBank: KR782255 (T. compactum, PI 352302)). Subsequent analysis revealed a 16 bp deletion and 4 single nucleotide deletions within the MITE insertion when compared to Vrn-A1a.1 (GenBank: AY616458). Yu et al. [34] recently reported that a 222 bp insertion of MITE_VRN in the promoter region of the Vrn-A1a.1 allele processes a 23 bp sequence of microRNA (TamiR1123), which is complementary to the previously identified miR507 [35]. The RNA secondary structure was reconstructed to determine the effect of a 16 bp deletion on the ability to form a stable hairpin loop, which is needed for microRNA processing (Fig. 1b). Results indicated that this deletion not only encompasses 30 % of the sequence, which encodes TamiR1123, but also prevents formation of the hairpin loop structure precluding further processing of microRNA. To investigate the possibility that MITE_VRN is read with the target gene as one transcript with an alternative promoter located within MITE, a similarity search of wheat ESTs using the MITE_VRN sequence was performed. As a result, 9 transcripts from 6 clusters (Ta.98996, Ta.153105, Ta.11542, Ta.127802, Ta.219174, Ta.125868) with identity no less than 82 % and query coverage > 77 % (of these, 6 accessions had coverage of MITE greater than 90 %) were identified. While for 7 accessions MITE was located within the mRNA, for two transcripts the similarity region began from start position of mRNA and correspond of 24 bp position of MITE sequence.
Fig. 1.
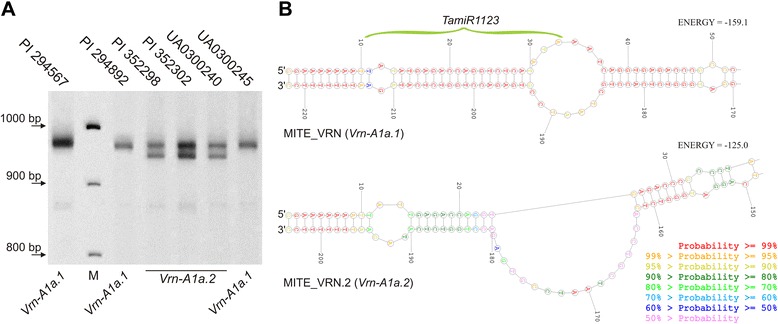
Identification and analysis of the Vrn-A1a allelic variants using PCR with primers VRN1AF/VRN1-INT1R. a Separation of promoter region PCR fragments of Vrn-A1a for T. compactum accessions in polyacrylamide gel (853 bp fragments show a very low amplification). M – DNA molecular size marker. b RNA secondary structures (partial) predicted for MITE_VRN and MITE_VRN.2 from Vrn-A1a.1 and Vrn-A1a.2 respectively. The localization of the hairpin loop structure, which is needed for processing of the TamiR1123 microRNA is indicated
Discovery the novel Vrn-A1i allele
Separation of PCR fragments of the VRN-A1 promoter region, in PAA gels of those fragments identified in agarose gels as the recessive vrn-A1 allele (713 bp), revealed fast migrating amplicons in some accessions of tetraploid wheat. These amplicons migrated closely with 705 bp fragments amplified for reference from the accession of T. monococcum with vrn-Am1 (Fig. 2a). PCR fragments from two accessions of T. turgidum (PI 208912, PI 221422), one T. durum (PI 74830) and a reference accession of T. monococcum (PI 428170) were cloned and sequenced (GenBank: KM016789 (T. monococcum, PI 428170), KM016790 (T. turgidum, PI 208912), KM016791 (T. durum, PI 74830), KM016792 (T. turgidum, PI 221422)). Sequence analysis confirmed the identity of the vrn-Am1 allele for the accession of T. monococcum and revealed a single nucleotide polymorphism within the A-tract (AnTm, n + m > =4) of the VRN-box for accessions of T. turgidum and T. durum (Fig. 2b). This new VRN-A1 allelic variant was designated as Vrn-A1i, according to the nomenclature of vernalization response genes proposed by Yan et al. [19]. The curved DNA migrates more slowly than non-bent DNA of the same length during polyacrylamide gel electrophoresis, as has been demonstrated [36–38]. Usually the anomalously slow electrophoretic mobility is associated with increasing curvature of the DNA molecule, determined by the A-tract bending [39, 40]. The adenine to cytosine mutation leads to a break of the A-tract (A3 instead of A5) within the VRN-box of Vrn-A1i. Furthermore, replacement of adenine with cytosine inside the A-tract is accompanied by an increase in flexibility of the DNA segment through reduction of the rigid local bend, due to the shorter A-tract, and presence of two more flexible nucleotide steps “AC” and “CA” instead of the rigid “AA” steps. Distribution analysis of local bending angles and curvature of the DNA molecule (Additional file 1: Figure S1) shows that the changes in the A-tract result in straightening of DNA fragments, which gives the Vrn-A1i amplicons a characteristic shape. This leads to increasing migration in PAA gels of the Vrn-A1i PCR fragments compared with amplicons of vrn-A1, which migrate anomalously slow as a result of their curvature, as determined by the intact A-tract within the VRN-box.
Fig. 2.
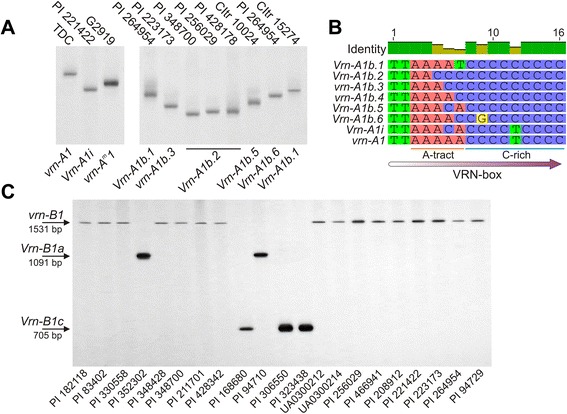
Identification of the VRN-A1 and VRN-B1 alleles during PAGE. a Discrimination of identified herein Vrn-A1i and variants of Vrn-A1b under electrophoresis optimized for low temperature conditions. PCR fragments of the VRN-A1 promoter region were amplified using the primer pair VRN1AF/VRN1-INT1R. b Mutations within VRN-box differentiate Vrn-A1i and variants of Vrn-A1b from vrn-A1 (GenBank: KM047646 (Vrn-A1b.1), KM047641 (Vrn-A1b.2), KM047647 (vrn-A1b.3), KM047651 (vrn-A1b.4), KM047652 (Vrn-A1b.5), KT692944 (Vrn-A1b.6), KM016790 (Vrn-A1i), AY747600 (vrn-A1)). The DNA sequence of the VRN-box of the Vrn-A1b variants as well as VRN-box of Vrn-A1i contains polymorphism within the A-taract, while the VRN-box of Vrn-A1b.5 and Vrn-A1b.6 differed from the VRN-box of Vrn-A1i and vrn-A1b.4 respectively, by the SNP within the C-rich segment. c PCR analysis of the VRN-B1 intron-1 for hexaploid and tetraploid wheat
Discovery of new variants of the Vrn-A1b allele
The next important result obtained during resolution of VRN-A1 promoter region amplicons in PAA gels was the detection of a difference in migration rate for PCR fragments with length near to the DNA marker of Vrn-A1b (Fig. 2a). This was not observed during agarose gel separation or in non-optimized or denaturized PAGE. Several polymorphic amplicons were cloned and sequenced (GenBank: KM047641 (T. dicoccoides, PI 233288), KM047642 (T. dicoccoides, PI 256029), KM047643 (T. dicoccoides, PI 352322), KM047644 (T. spelta, PI 330558), KM047645 (T. spelta, PI 348700), KM047646 (T. turgidum, PI 264954), KM047647 (T. dicoccum, UA0300214), KM047648 (T. durum, PI 655432), KM047649 (T. vavilovii, PI 428342), KM047650 (T. turgidum, PI 223173), KM047651 (T. dicoccoides, PI 466941), KM047652 (T. dicoccum, UA0300212), KT692944 (T. durum, CItr 10024), KT692945 (T. turgidum, PI 220356)). The result of multiple alignments with the reference sequence of the dominant Vrn-A1b allele (GenBank: AY616461) enabled the identification of six variants of Vrn-A1b. All contain three common features in common with Vrn-A1b: a 20 bp deletion located 137 bp upstream of the start codon, the “T” deletion upstream of the VRN-box and a “T- > C” replacement within the C-rich segment of the VRN-box. Five variants (Vrn-A1b.1-b.5) differ from each other by a polymorphism of the A-tract within the VRN-box. These variants were designated as Vrn-A1b.1 (A-tract = “AAAAT”), Vrn-A1b.2 (A-tract mutated to “AACCC”), vrn-A1b.3 (A-tract mutated to “AAACC”), vrn-A1b.4 (A-tract mutated to “AAAAC”) and Vrn-A1b.5 (A-tract mutated to “AAACA”), while Vrn-A1b.6 carries an additional “C- > G” transversion within the C-rich segment compared to vrn-A1b.4 (Fig. 2b). In this nomenclature the Vrn-A1b.1 corresponds to the original Vrn-A1b allele (GenBank: AY616461) previously identified by Yan et al. [19]. It should be noted that PCR fragments of the vrn-A1b.3 and Vrn-A1b.5 as well as vrn-A1b.4 and Vrn-A1b.6 variants migrate in PAA gels at a similar rate, indicating the importance of the length of the A-tract of the VRN-box in generating small observable differences in migration rate. Overall, “A- > C” transversions within the A-tract were accompanied by a reduction in the length of the A tract and curvature of DNA molecules that resulted in a decrease in the anomalously slow migration of PCR fragments of the Vrn-A1b variants during PAGE.
Distribution of the VRN-A1 alleles in wheat
VRN-A1 alleles with mutations in the promoter region and alleles carrying large deletions in the first intron are distributed unevenly among different wheat species. In particular, 74 % carry alleles with mutations in the promoter region, whereas only 26 % carry the 7.2 kb deletion in the first intron (Vrn-A1c (Langdon) allele). No mutations in intron-1 were detected in any hexaploid species or in wild and domesticated emmer wheat, T. dicoccoides and T. dicoccum. The highest distribution of Vrn-A1c was recorded in T. durum (71 %) and T. polonicum (58 %), and Vrn-A1c was detected only one accession within subsets of T. carthlicum and T. turgidum. The dominant Vrn-A1a allele was found in 6 accessions of T. compactum and 4 accessions of T. dicoccum. In each of these species two variants of Vrn-A1a were identified. Vrn-A1a.1 and the newly- identified Vrn-A1a.2 were detected in T. compactum accessions, while Vrn-A1a.1 and Vrn-A1a.3 were recorded in T.dicoccum. The novel Vrn-A1i allele was identified in 5 accessions of T. turgidum and in one sample of T. durum.
The variants of Vrn-A1b are widespread in tetraploid and hexaploid wheat species: 80 and 77 % from alleles carrying mutations in promoter for tetraploid and hexaploid wheat respectively (Additional file 2: Table S1, Additional file 3: Table S2). The different variants of Vrn-A1b were represented in all accessions of T. spelta and T. dicoccoides with a mutant VRN-A1 promoter. Among all five variants of the Vrn-A1b allele, the canonical variant Vrn-A1b.1 was observed only in 8 % cases. Most (92 %) carried the variants described herein - Vrn-A1b.2-b.6. In tetraploid wheat Vrn-A1b.2 was found in all spring accessions of T. dicoccoides and in 42 % accessions of T. dicoccum. The vrn-A1b.3 allele predominates in T. turgidum (40 % accessions). Vrn-A1b.6 was identified in all accessions of T. polonicum with Vrn-A1b and in samples of T. durum and T.turgidum. The original Vrn-A1b allele (Vrn-A1b.1 variant) was found only in two accessions of T. durum and 3 accessions of T. turgidum. In hexaploid wheat new variants of Vrn-A1b are represented in almost all analysed species with exception of T. sphaerococcum. The greatest diversity in Vrn-A1b was detected in T. spelta, where the Vrn-A1b.2, vrn-A1b.3 and Vrn-A1b.6 variants were identified. Two accessions of T. compactum (PI 262666 and PI 41023) possessed both vrn-A1 and Vrn-A1b.6 alleles.
The dominant allele Vrn-A1e, previously identified in T. dicoccum [19], was detected in two accessions of T. carthlicum and one accession of T. dicoccum. PCR fragments of the Vrn-A1e and Vrn-A1f alleles have approximately the same length of 658 bp. Hence, Vrn-A1e was confirmed by sequencing (GenBank: KT361213). Moreover, predicted DNA curvature for the PCR fragments of Vrn-A1f and Vrn-A1e allows us to assume that fragments of Vrn-A1f will migrate slower in PAA gels relative to Vrn-A1e due to the presence of two additional bends generated by the A-tract within the VRN-box and T-tract within CArG-box. Finally, Vrn-A1f likely occurs in wheat of the Timopheevii section [29].
Interestingly, the new variants of VRN-A1 identified here using changes in gel retardation (following changes in curvature and flexibility of DNA molecules) are more common in wheat species than the previously known VRN-A1 alleles. This indicates that probably these new variants are actively involved in natural selection as useful adaptations to environmental conditions.
Discovery of new VRN-B1 variants
Results of VRN-B1 promoter region analysis found that almost all accessions of T. carthlicum (with the exception of PI 168672) carry a retrotransposon insertion (Vrn-B1(ins) allele). Under certain electrophoresis conditions the difference in the migration rate of PCR fragments with the intact VRN-B1 promoter was revealed (Fig. 3a). These variants were designated as VRN-B1.f, VRN-B1.m and VRN-B1.s (according to the migration rate of relevant PCR fragments: “f” = fast, “m” = medium, “s” = slow). The difference in migration rates between amplicons that enabled the detection of alternative VRN-B1 promoter variants was observed only by polyacrylamide gel separation and increased with reduced conformational dynamics and greater stability of DNA molecules (Fig. 3a).
Fig. 3.
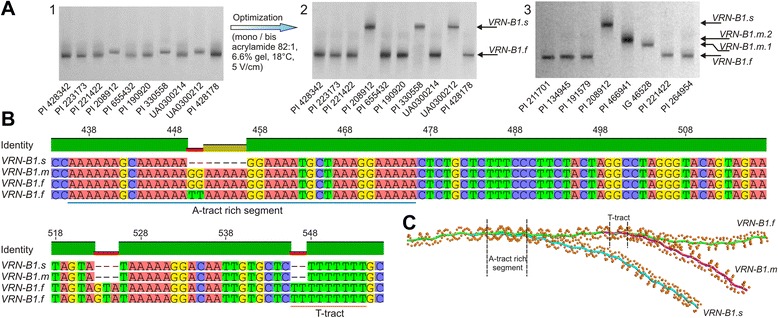
Mutations within the promoter of VRN-B1 indicate tree sequence variants. a Identification of the VRN-B1 variants during PCR with primer pair Pr1/Pr2. Separation of PCR fragments under conditions of low conformational dynamics of DNA molecules contribute to the clear discrimination of the alternative promoter variants. 1 – non-optimized PAA electrophoresis at room temperature without cooling, in 10 % gel, mono/bis-acrylamide ratio 29:1, under 5 V/cm; 2 – the same amplicons resolved in PAGE optimized for low temperature conditions; 3 – support for the contribution of localized DNA bends as determined by the 7 bp deletion within the A-tract rich segment in anomalously migrating PCR fragments of VRN-B1.s, the 7 bp deletion distinguishes VRN-B1.s from VRN-B1.m, observed splitting of VRN-B1.m on two variants. b Comparison of DNA sequences encompassing the center of PCR fragments detecting VRN-B1 variants (GenBank: KR782252 (VRN-B1.s), KT361212 (VRN-B1.m.2), KR782253 (VRN-B1.f from T. turgidum), KR782254 (VRN-B1.f from T. polonicum)). Region showed includes three deletions in VRN-B1.s, two located within the A-tract rich segment and T-tract cause local bends and lead to increased curvature of the DNA molecule. Positions are indicated relative to the start of the VRN-B1.f PCR fragment. c Superposition of predicted DNA paths of PCR fragments of the VRN-B1 promoter variants. Sites of local DNA bends caused by the 7 and 2 bp deletions within the A-tract rich segment and T-tract are indicated
For a more detailed investigation, the amplicons of VRN-B1.f, VRN-B1.s and VRN-B1.m were cloned and sequenced (GenBank: KR782252 (T. turgidum, PI 208912), KR782253 (T. turgidum, PI 191579), KR782254 (T. polonicum, PI 134945), KT361212 (T. dicoccoides, PI 466941)). DNA sequence alignment of these alternative PCR fragments identified 8 SNPs (T102C, G174A, A297C, T341G, +360C, +370C, C558T, C729G) and three short deletions at 7 bp (“GGAAAAA”), 3 bp (“GTA”) and 2 bp (“TT”) for VRN-B1.s relative to VRN-B1.f, where VRN-B1.f corresponds to the vrn-B1 allele with an intact promoter. VRN-B1.m differs from VRN-B1.s by the absence of 7 bp deletion. Furthermore, it was found that SNPs C654G and A693G, identified in the related DNA sequence of VRN-B1.f, are associated with VRN-B1.m. Although altogether the mutations of VRN-B1.s result in a reduction in amplicons length by just 10 bp, the VRN-B1.s PCR fragments (total length 958 bp) migrate through PAA gels much slower than amplicons of VRN-B1.f (968 bp). The difference, seen during PAGE with conditions optimized for low temperature, appeared greater than 100 bp when measured relative to the DNA molecular weight marker. Similarly, amplicons of VRN-B1.m (965 bp) migrate faster than amplicons of VRN-B1.s (958 bp), but slower than VRN-B1.f (968 bp). The 7 bp and 2 bp deletions were located within A-tracts, which in turn were located near the center of the fragment (Fig. 3b). Both of these factors contribute to a change in DNA curvature, leading to the observed anomalously slow migration of VRN-B1.s and VRN-B1.m PCR fragments in PAA gels compare to VRN-B1.f amplicons (Fig. 3c). The 2 bp deletion reduces the length of the T-tract from 9 to 7 bp. The more distal 7 bp deletion located within the A-tract rich segment encompasses a −549 to −509 bp region counting from the start codon of the native VRN-B1 (VRN-B1.f). This region includes 6 A-tracts, which are separated by dinucleotide steps (“GC” and “GG”). The polymorphism of a dinucleotide step separating the 2-nd and 3-rd A-tracts for VRN-B1.f accessions was detected. Comparison of this region within 15 accessions available from GenBank showed that T. dicoccoides and T. turgidum include the dinucleotide “GG”, while T. aestivum, T. durum, T. polonicum and T. carthlicum carry “TT” (Fig. 3b).
To determine what contribution the 7 bp deletion within the A-tract rich segment makes to the change of global DNA curvature and observed retardation of the VRN-B1.s amplicons in PAA gels, the PCR products of accessions PI 208912 (VRN-B1.s), PI 466941 (VRN-B1.m), IG 46528 (the sequence of which (GenBank: KM586665) corresponds to VRN-B1.m) and PI 191579 (VRN-B1.f) were analysed in same electrophoretic conditions (Fig. 3a). The results showed that the contribution of the 7 bp deletion in anomalous migration of the VRN-B1.s PCR fragments is less significant predominant (5−10 %) than the contribution of a 2 bp reduction in the T-tract. Thus both these contributions are equally important to electrophoretic retardation of the VRN-B1.s amplicons. Additionally, it was found that amplicons from IG 46528 (VRN-B1.m.1) show no significant difference in migration compared to amplicons from PI 466941 (VRN-B1.m.2), despite both accessions carrying VRN-B1.m. Sequence alignment showed that PCR fragment of VRN-B1.m.1 contains “T” deletion at position 230 bp and G341T transversion eliminating of the A-tract (“ATG” instead “ATT”), that explains observed difference in migration rate between amplicons of VRN-B1.m.1 and VRN-B1.m.2, where VRN-B1.m.1 migrates faster than VRN-B1.m.2.
Distribution of VRN-B1 alleles
Distribution analysis of the VRN-B1 variants showed that VRN-B1.f is widespread in winter and spring accessions of hexaploid and tetraploid wheat species, while VRN-B1.s is identified predominantly in accessions which, according to passport or experimental date, have a spring growth habit. Distribution of variants VRN-B1 in the different hexaploid wheat species is unequal. VRN-B1.s is found only in accessions of T. compactum and T. spelta. A sharp decrease in VRN-B1 promoter variant diversity, likely the result of a genetic bottleneck and founder effects due to speciation, is observed for such species as T. macha, T. vavilovii and T. shaerococcum, where all analysed accessions were characterized by the VRN-B1.f. Conversely, in tetraploid wheat all species (except T. carthlicum containing a TE insertion in VRN-B1 promoter) were characterized by the more uniform distribution of VRN-B1 variants with VRN-B1.f predominant. VRN-B1.m was identified in only two accessions of T. dicoccoides (PI 352322, PI 466941). Interestingly, all accessions of T. polonicum with Vrn-A1c are associated with VRN-B1.s, while accessions carrying Vrn-A1b are characterized by the VRN-B1.f. Similarly, VRN-B1.s was found in accessions of T. spelta with Vrn-A1b.2.
Multiplex PCR was used to investigate VRN-B1 intron-1. The forward primer Ex1/C/F was previously designed to amplify Vrn-A1c [20]. However, sequence alignment indicated that this primer anneals to VRN1 genes from all three genomes (A, B and D). Accordingly, the primer combination Ex1/C/F, Intr1/B/R3 and Intr1/B/R4 was optimised for investigation of VRN-B1. This new multiplex PCR provides four DNA-markers in PCR fragments with lengths of 1531, 1091, 1055 and 705 bp, to discriminate vrn-B1, Vrn-B1a, Vrn-B1b and Vrn-B1c alleles respectively (Fig. 2c). The dominant alleles of Vrn-B1 with mutations in intron-1, prevailed in hexaploid wheat species. In particular, the dominant Vrn-B1a, Vrn-B1b or Vrn-B1c alleles were not detected in tetraploid wheat T. turgidum, T. polonicum, T. carthlicum. Only one accession T. durum carries Vrn-B1a, while 2 and 5 accessions in T. dicoccum and T. dicoccoides respectively carry Vrn-B1c. In hexaploid wheat, the Vrn-B1c was identified in T. spelta (33 %), T. macha (one accession) and T. sphaerococcum (one accession), while Vrn-B1a was detected only in T. compactum (50 %).
Polymorphism of the VRN-box is associated with vernalization requirement and flowering time
According to passport data, almost all accessions with vrn-A1b.3 have a winter growth habit with the exception of two accessions of T. spelta (PI 306550, PI 323438). Nevertheless, DNA-marker analysis has shown that the vernalization insensitive phenotype of these spring accessions of T. spelta is determined by the dominant Vrn-B1c allele. Some of the accessions with vrn-A1b.3 were grown in a glasshouse without vernalization under a long photoperiod (>16 h) and were then genotyped by VRN markers (Tables 1 and 2). The accessions PI 428342, PI 655432 and UA0300214 (detected only vrn-A1b.3, VRN-B1.f) failed to reach stem elongation 120 days (4 months) after planting, when the experiment was terminated. Only PI 223173 (vrn-A1b.3, VRN-B1.s) showed a later flowering time (128 days). This confirms that accessions with vrn-A1b.3 are characterized by a winter growth habit and retain the vernalization requirement. Similarly, the accession of T. dicoccoides (PI 466941) with vrn-A1b.4 according to passport data has a winter growth habit and this was confirmed here experimentally (failed stem elongation after 120 days). In contrast, all accessions with Vrn-A1b.2 and Vrn-A1b.5 according to passport data have a spring growth habit which was also confirmed during the experimental glasshouse planting carried out in the present study. Nevertheless, almost all accessions with Vrn-A1b.2 have VRN-B1.s and/or carry dominant alleles of the other VRN1 genes (VRN-B1/D1). The accessions PI 428178 (T. macha) and PI 190920 (T. dicoccum) were only exceptions.
Table 1.
PCR primers details
| Primers | Primer sequence (5'-3') | Primer design | Annealing temp. °C | Amplified region | Allelic variant | PCR product size (bp) |
|---|---|---|---|---|---|---|
| VRN1AF | gaaaggaaaaattctgctcg | Yan et al. [19] | 60 |
VRN-A1
promoter |
vrn-A1 | 713 |
| VRN1-INT1R | gcaggaaatcgaaatcgaag | Vrn-A1a.1 | 853, 944 | |||
| Vrn-A1a.2 | 853, 924, 944 | |||||
| Vrn-A1a.3 | 765,853 | |||||
| vrn-A m 1 | 705 | |||||
| Vrn-A1b | 691 | |||||
| Vrn-A1d | 685 | |||||
| Vrn-A m 1g | 681 | |||||
| Vrn-A m 1a | 671 | |||||
| Vrn-A1e | 659 | |||||
| Vrn-A1f | 658 | |||||
| vrn-A m 1b | 656 | |||||
| Vrn-A1-intr_F | ccgtcgaaaggatcgctactg | This study | 60 |
VRN-A1
intron-1 |
vrn-A1 | 541 |
| Vrn-A1-intr_R1 | cttgtccccgtgagctacttac | 60 | ||||
| Ex1/C/F | gttctccaccgagtcatggt | Fu et al. [20] | 58 |
VRN-A1
intron-1 |
Vrn-A1c (Langdon) | 522 |
| Intr1/A/R3 | aagtaagacaacacgaatgtgaga | 58 | Vrn-A1c (IL369) | 2188 | ||
| Pr1 | tacccctgctaccagtgcct | Shcherban et al. [22] | 60 |
VRN-B1
promoter |
VRN-B1.f | 968 |
| Pr2 | ggccaaccctacaccccaag | 60 | VRN-B1.s | 958 | ||
| VRN-B1.m | 965 | |||||
| Pr1 | tacccctgctaccagtgcct | Shcherban et al. [22] | 60 |
VRN-B1
promoter |
Vrn-B1(ins) | 1256 |
| PrB/Ins/r2 | tgcggtacgggttatccag | This study | 60 | |||
| Ex1/C/F | gttctccaccgagtcatggt | Fu et al. [20] | 61 |
VRN-B1
intron-1 |
Vrn-B1a | 1091 |
| Intr1/B/R3 | ctcatgccaaaaattgaagatga | 61 | Vrn-B1b | 1055 | ||
| Vrn-B1c | 705 | |||||
| Ex1/C/F | gttctccaccgagtcatggt | Fu et al. [20] | 61 |
VRN-B1
intron-1 |
vrn-B1 | 1531 |
| Intr1/B/R4 | caaatgaaaaggaatgagagca | 61 | ||||
| FT-B-INS-F | cataatgccaagccggtgagtac | Yan et al. [3] | 63 |
VRN-B3
promoter |
Vrn-B3a | 1765 |
| VRN4-B-NOINS-R | ctatccctaccggccattag | 63 | ||||
| VRN4-B-NOINS-F2 | gctgtgtgatcttgctctcc | Yan et al. [3] | 63 |
VRN-B3
promoter |
vrn-B3 | 691 |
| VRN4-B-NOINS-R | ctatccctaccggccattag | 63 | vrn-B3b | 1581 |
Table 2.
Genotype and days to flowering (DTF) of hexaploid and tetraploid wheat accessions grown >120 days under 16 h photoperiod without vernalization
| Species | ID | VRN genotype | DTF (±2 days) |
|---|---|---|---|
| T. sphaerococcum | PI 182118 | vrn-A1/vrn-B1 (VRN-B1.f)/vrn-D1/vrn-B3 | 53 |
| PI 83402 | vrn-A1/vrn-B1 (VRN-B1.f)/Vrn-D1a/vrn-B3 | 72 | |
| T. spelta | PI 168680 | vrn-A1b.3/Vrn-B1c (VRN-B1.f)/vrn-D1/vrn-B3 | 67 |
| PI 330558 | Vrn-A1b.2/vrn-B1 (VRN-B1.s)/Vrn-D1s/vrn-B3 | 59 | |
| PI 348428 | Vrn-A1b.2/vrn-B1 (VRN-B1.s)/Vrn-D1s/vrn-B3 | 61 | |
| PI 348700 | Vrn-A1b.2/vrn-B1 (VRN-B1.s)/Vrn-D1s/vrn-B3 | 60 | |
| T. compactum | PI 211701 | vrn-A1/vrn-B1 (VRN-B1.f)/Vrn-D1s/vrn-B3 | 77 |
| T. vavilovii | PI 428342 | vrn-A1b.3/vrn-B1 (VRN-B1.f)/vrn-D1/vrn-B3 | - |
| T. dicoccum | UA0300212 | Vrn-A1b.5/vrn-B1 (VRN-B1.s)/vrn-B3 | 46 |
| UA0300214 | vrn-A1b.3/vrn-B1 (VRN-B1.f)/vrn-B3 | - | |
| T. dicoccoides | PI 256029 | Vrn-A1b.2/vrn-B1 (VRN-B1.s)/vrn-B3 | 66a |
| PI 466941 | vrn-A1b.4/vrn-B1 (VRN-B1.m.2)/vrn-B3 | - | |
| T. turgidum | PI 208912 | Vrn-A1i/vrn-B1 (VRN-B1.s)/vrn-B3 | 77 |
| PI 221422 | Vrn-A1i/vrn-B1 (VRN-B1.f)/vrn-B3 | 102 | |
| PI 223173 | vrn-A1b.3/vrn-B1 (VRN-B1.s)/vrn-B3 | 128 | |
| PI 264954 | Vrn-A1b.1/vrn-B1 (VRN-B1.f)/vrn-B3 | 68 | |
| T. durum | PI 94729 | Vrn-A1c/vrn-B1 (VRN-B1.f)/vrn-B3 | 54 |
| PI 74830 | Vrn-A1i/vrn-B1 (VRN-B1.s)/vrn-B3 | 76 | |
| PI 655432 | vrn-A1b.3/vrn-B1 (VRN-B1.f)/vrn-B3 | - | |
| T. carthlicum | PI 532505 | vrn-A1/Vrn-B1(ins)/vrn-B3 | 61 |
aPI 256029 with Vrn-A1b.2 was reach stem elongation 7 days earlier than PI 264954 with Vrn-A1b.1
Two accessions of tetraploid wheat with Vrn-A1i (PI 208912 and PI 74830), according to passport data, have a spring growth habit, while four others were characterized by a winter growth habit. Nevertheless, in growing these “spring” and “winter” (PI 221422) accessions, all of them were able to flower without vernalization (Table 2), however flowering was significantly later than accessions with dominant VRN-A1 or VRN-B1 alleles. It should be noted that the “winter” accession (PI 221422) flowered 25 days later than “spring” (PI 208912, PI 74830), herewith they all have recessive vrn-B1 and vrn-B3 alleles. It is assumed that the decreased vernalization sensitivity and facultative growth habit is typical of accessions possessing Vrn-A1i.
The Vrn-A1b.5 accession (UA0300212) flowered 22 days early than Vrn-A1b.1 (PI 264954). Similarly PI 208912 and PI 74830 flowered 25 days earlier than PI 221422, while all of them carried Vrn-A1i (Table 2). The Vrn-A1b.5 has the same polymorphism of the A-tract within VRN-box as Vrn-A1i, but it also contains common mutations characterizing all variants of the Vrn-A1b allele. Interestingly, in all of these cases the earlier accessions (UA0300212, PI 208912, PI 74830) carried VRN-B1.s. Moreover, the VRN-B1.s was identified in the PI 223173 accession, which later flowered without vernalization, in contrast to other accessions also carrying vrn-A1b.3 and VRN-B1.f. This suggests an association between VRN-B1.s, a decrease of vernalization sensitivity and early flowering. It should be noted that the accession of T. dicoccoides (PI 466941) with vrn-A1b.4 and VRN-B1.m failed to achieve stem elongation before the experiment was terminated, in contrast to accession PI 223173 with vrn-A1b.3 and VRN-B1.s. Furthermore, it was found that accession UA0300212 (Vrn-A1b.5, VRN-B1.s) flowered 30 days earlier than PI 208912 and PI 74830 (Vrn-A1i, VRN-B1.s). This difference approximately corresponds to the difference between PI 264954 (Vrn-A1b.1, VRN-B1.f) and PI 221422 (Vrn-A1i, VRN-B1.f), which was 34 days (Table 2). In both cases, the compared accessions do not carry the known dominant VRN-B1 and VRN-B3 alleles, they have the same promoter variants of VRN-B1 and differ only at the VRN-A1 alleles with a common feature: “T- > C” SNP within the C-rich region of the VRN-box (Fig. 2b).
Discussion
Detection of the identified VRN-A1 and VRN-B1 alleles
The difference in curvature and flexibility of DNA molecules caused by the mutations within the VRN-box of VRN-A1 allelic variants first described herein, or by deletions within A-tracts for promoter variants of VRN-B1, leads to modulation in the rate of anomalously migrating PCR fragments. More than 25 years ago it was shown that anomalous migration of intrinsically bent DNA molecules in PAA gels allows the detection of small sequence-specific DNA structure variations [41]. In our previous study the haplotypes of VRN-D1 and Ppd-A1 haplogroups were discriminated based on anomalous migration [23, 42]. In the present study we found that the sensitivity of PCR fragments migration rate in PAA gels to curvature of DNA molecules provides a good basis for the development of a simple and fast method to discriminate between the new VRN-A1 and VRN-B1 allelic variants. Nevertheless, identification of the new VRN-A1 and VRN-B1 variants requires very specific conditions of PAGE. Hence, the electrophoretic conditions were optimized to obtain the minimal loss of resolution during electrophoresis run under low and room temperature. For example, as seen in Fig. 3 during a typical neutral PAGE the difference between VRN-B1 variants is insignificant or fully absent, whereas optimization of PAGE results in a clear discrimination between the two. Overall, the retardation of anomalously slow migrating PCR fragments and sharper bands definition are especially important for identification of the Vrn-A1b variants, where Vrn-A1b.1/Vrn-A1b.6 and vrn-A1b.3/Vrn-A1b.2 show a small difference in migration rate, and also are the predominant variants of VRN-B1. Adherence to recommendations proposed herein promotes the clear discrimination of Vrn-A1b and VRN-B1 variants and prevents potential errors in the analysis of genetic material for vernalization experiments. For example, electrophoresis via agarose gels or PAA gels under standard conditions does not allow for the detection of differences between variants of Vrn-A1b. Thus, vrn-A1b.3 and vrn-A1b.4, associated with vernalization requirement and winter growth habit can be erroneously taken as the known dominant Vrn-A1b allele, which is associated with a spring growth habit.
Polymorphism of the VRN-B1 promoter region results in a change of curvature of the DNA molecule
Analysis of the VRN-B1 promoter region identified three sequence variants (VRN-B1.f, VRN-B1.s and VRN-B1.m), which could be resolved during separation in PAA gels under low conformational dynamics of DNA molecules. Compared to VRN-B1.f (968 bp), the VRN-B1.s (958 bp) contains three short deletions at 2, 3 and 7 bp, while VRN-B1.m (965 bp) differs from VRN-B1.s by the absence of a 7 bp deletion. Nevertheless, VRN-B1.s migrates slower than VRN-B1.m which is in turn slower than VRN-B1.f. As was described previously the curved DNA molecules migrate slower in polyacrylamide but not in agarose gels [36, 41] at a rate mostly determined by the length and localization of A-tracts [38–40]. Anomalously slow migration of the PCR fragments of VRN-B1.s and VRN-B1.m is determined by the localization of 2 and 7 bp deletions within A-tracts. In particular, a 2 bp deletion reduces the length of the 9 bp T-tract. As it was previously shown by Koo et al. [39] and our recent study on discrimination of haplotypes of VRN-D1 [23], with decreasing length of the A-tract DNA curvature increases, while an A-tract length of more than 7 nt, reduces curvature through antiphase compensation of equivalent bends in opposite directions. Hence, a reduction in length of the T-tract from 9 to 7 nucleotides leads to an increase in local curvature of this DNA segment. This explains the contribution of the 2 bp deletion to the anomalous slow migration of VRN-B1.s and VRN-B1.m. The 7 bp deletion is located within A-tract rich segment, which includes 6 A-tracts separated by dinucleotide steps. Thus, a bend produced by the 1st tract is compensated by the bend of the 2nd tract, which is oriented in the opposite direction. Similarly, the 4th tract compensates for the bend of the 3rd tract, and the 6th tract for the 5th. As a result, curvature in this region is insignificant. However in VRN-B1.s the 4th tract is not compensated by an opposite bend of the 3rd tract due to the absence, through the 7 bp deletion encompassing this segment. This leads to increased curvature of the DNA molecule and retardation of VRN-B1.s amplicons in the PAA gel. The effect of the change of local curvature on the slowdown of VRN-B1.s amplicons is significantly enhanced due to both DNA bends, caused by the 7 and 2 bp deletions, localized the center of the fragment. In a previous study, Shcherban et al. [43] noted variability in the VRN-B1 promoter region for some accessions of T. dicoccoides including 7, 3 and 2 bp deletions, which carry as designated here the VRN-B1.m and VRN-B1.s variants. However, the present study is the first to examine the distribution of these VRN-B1 variants among wheat species and their probable phenotypic association, and to describe a method for their detection. The combination of the dominant VRN-A1 alleles with VRN-B1.s was associated with early flowering (Table 2). Although it seems unlikely that mutations characterizing of VRN-B1.s have an effect on regulatory sites since they are located a long distance away from the transcription start site, these mutations could exert an influence through a distal enhancer or silencer. Also, it cannot be ruled out that the significant bending of the DNA molecule inside the VRN-B1.s promoter region, confirmed here by anomalously slow migration in PAA gels, facilitates an interaction between a remote enhancer and regulatory sites within the VRN1 promoter; thus activating transcription via DNA looping. Additionally, an insertion in the promoter region of Vrn-D1c, associated with increasing transcript levels, is located more than 60 bp upstream of the A-tract rich region and 600 bp upstream of the transcription start site [25]. Nevertheless, while the association between promoter sequence variants of VRN-B1 and phenotypic traits requires further investigations these variants are highly valuable as polymorphic, codominant DNA markers in genetic analysis.
Study of VRN-B1 intron-1
A number of marker systems have already been proposed for identification of the dominant VRN-B1 alleles carrying large deletions within intron-1 [20, 44]. The multiplex PCR system optimized in the present study provides DNA markers almost 150 bp shorter than the existing analogues to discriminate between the intact intron-1 and the three dominant Vrn-B1a, Vrn-B1b and Vrn-B1c alleles. Furthermore, the use of the forward primer Ex1/C/F simultaneously in the two separate PCR assays for identification of alleles of the VRN-B1 gene and dominant allele Vrn-A1c [20] is economically advantageous. Distribution analysis of the VRN-B1 dominant alleles, showed that the Vrn-B1c allele is more commonly found in spring accessions of hexaploid and tetraploid wheat species than Vrn-B1a or Vrn-B1b (which was not detected), while an insertion retrotransposon in the promoter region of the VRN-B1 gene is likely specific to tetraploid wheat T. carthlicum, where it is the main determinant of spring growth habit (Additional file 3: Table S2). The majority of the spring wheat germplasm surveyed here carried the dominant VRN-B1 alleles in combination with dominant VRN-A1. Interestingly, accessions of T. compactum with dominant VRN-B1 are characterized by the Vrn-B1a allele, while some accessions of T. spelta carry Vrn-B1c exclusively.
Less frequent VRN alleles in hexaploid and tetraploid wheat
All accessions of T. sphaerococcum carry the recessive vrn-A1 allele (with intact of VRN-A1 promoter and intron-1) and according to passport data, 16 accessions have a spring growth habit and two samples have a facultative or winter growth habit. However, Vrn-B1c was identified in only one accession and Vrn-D1a in just two [23]. Thus, the spring growth habit of the predominant number of T. sphaerococcum accessions is mostly determined by genes non-allelic to VRN1 or VRN-B3. This is consistent with the assumption that the spring growth habit of T. sphaerococcum accessions is largely controlled by the dominant Vrn-D4 allele [4].
The newly described allele Vrn-A1a.2 can indicate the presence of an additional copy of the VRN-A1
Yan et al. [19] identified two variants of Vrn-A1a, named here Vrn-A1a.1 and Vrn-A1a.3. They assumed that Vrn-A1a.1 is specific to hexaploid wheat, while Vrn-A1a.3 is specific to tetraploid wheat. In the present study Vrn-A1a.3 was detected only in tetraploid wheat, whereas Vrn-A1a.1 was identified in both hexaploid and tetraploid wheat species. A new allele, Vrn-A1a.2, first identified here was detected only in accessions of hexaploid wheat.
Southern blot hybridization using a probe matched to the VRN-A1 region between exons 4 and 8 [19] showed that isogenic lines carrying Vrn-A1a.1 and Vrn-A1a.3 (TDD and Anza respectively) have the same hybridization fragments with a similar intensity as lines possessing the recessive vrn-A1 allele (TDC, Winter-Anza). These data indicate that accessions with Vrn-A1a have the same copy number of VRN-A1 genes as accessions with vrn-A1, such as the TDC line. Recently, using a VRN1 copy number Taqman assay, it was found that TDC contains one copy of VRN-A1 [4]. Identification in the Vrn-A1a.2 of additional copy of the promoter region and exon-1, comparing with Vrn-A1a.1 and Vrn-A1a.3 allows us to assume that accessions with Vrn-A1a.2 contain no less than 2 copies of VRN-A1 (i.e. one copy more than Vrn-A1a.1 and Vrn-A1a.3), differing from each other by the 16 bp deletion during MITE_VRN insertion in the promoter region.
The original MITE from the Vrn-A1a.1 allele, when transcribed to RNA forms a highly stable hairpin loop structure (Fig. 1b) that facilitates later processing to form a microRNA - TamiR1123 [34]. However the mutant variant of MITE identified in this study (designated Spring.2 or MITE_VRN.2) from the Vrn-A1a.2 allele has a 16 bp deletion encompassing 8 bp within the DNA sequence of TamiR1123 which leads to a change in RNA secondary structure preventing the loop in the pri-miRNA (Fig. 1b). The 16 bp deletion is flanked by a nucleotide repeat (“CAGGT” in this case) that is characteristic of deletions within VRN1 genes [45] and suggests that the doublestranded DNA break repair mechanism of non-homologous end-joining, mediated by the single-strand annealing-like mechanism [46] can be invoked to explain their emergence. Features of sequence and target site duplication characterize MITE_VRN as a Mutator-like element (MULE) [34]. It is known that MULEs can harbor promoters of genes for transcription [47]. Furthermore, most miRNA genes in plants contain the TATA box motif and are transcribed by RNA polymerase II [48, 49]. In any case, this promoter must be located upstream of the sequence encoding TamiR1123 since the transcription level of TamiR1123 positively correlates with the transcription level of Vrn-A1a.1 in the TDD line [34]. Hence, the Vrn-A1a.2 promoter must be intact, since the 16 bp deletion within MITE is located downstream of the start position of TamiR1123. This is true if TamiR1123 and Vrn-A1a are transcribed as a single product [34]. Results of the BLAST analysis of wheat ESTs assume that mRNA can be read with an alternative promoter located within MITE upstream of the 24 bp site, indicating that MITE_VRN and the target gene in reality can be read as a single transcript. The possibility of direct or indirect induction of Vrn-A1a by TamiR1123 to explain the positive correlation in their transcript levels seems unlikely since Vrn-A1a is not the only source of TamiR1123 - it is also present in winter accessions [34].
Polymorphism of the VRN-box is associated with modulation of the vernalization requirement and flowering time of wheat
Polymorphism of four promoter regions differentiates the known variants of Vrn-A1b and the intact vrn-A1 allele. This includes polymorphism of the A-tract and C-rich region within the VRN-box and also a 20 bp deletion and “T” deletion downstream and upstream of the VRN-box, respectively. It was determined that deletion of the “T” upstream as well as the 20 bp deletion dowstream of the VRN-box is likely not critical for determination of the spring growth habit, because it is identified only in Vrn-A1b variants and absent in the other dominant Vrn1 alleles with a mutant VRN-box, such as Vrn-A1i, Vrn-Am1g, Vrn-Am1a. Furthermore, vrn-A1b.3 and vrn-A1b.4 carrying these mutations are associated with a winter growth habit. Due to this, we assume that namely polymorphism of the VRN-box determines the decreased vernalization sensitivity of accessions which carry Vrn-A1b. However we cannot rule out the possible influence of mutations outside the VRN-box, which are observed in association with specific mutations within the VRN-box. The VRN-box was proposed by Pidal et al. [31] as an alternative regulatory site for vernalization. Alignments of most known VRN-A1 alleles possessing mutations within the promoter region influencing vernalization requirement (Vrn-A1a, Vrn-A1b, Vrn-Am1a, Vrn-A1d, Vrn-A1e, Vrn-Am1g, Vrn-A1h and Vrn-A1i) have found that all contain mutations within the VRN-box such as SNPs, InDels or its total elimination (Fig. 4b). This fact emphasizes the importance of the VRN-box sequence in the transcriptional regulation of VRN1 and in the vernalization response of wheat.
Fig. 4.
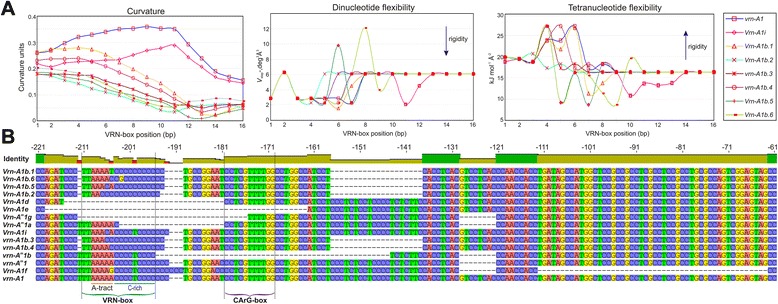
Polymorphism of the VRN-box differentiates the VRN-A1 alleles. a Distribution of local curvature, dinucleotide and tetranucleotide flexibility within the VRN-box for identified here Vrn-A1i and variants of Vrn-A1b. Arrows indicate increasing rigidity of the central dinucleotide steps. b Multiple alignments of VRN-A1 alleles carrying mutations within the promoter region (excluding Vrn-A1a); GenBank: KM047646 (Vrn-A1b.1), KT692944 (Vrn-A1b.6), KM047652 (Vrn-A1b.5), KM047641 (Vrn-A1b.2), AY616462 (Vrn-A1d), AY616463 (Vrn-A1e), DQ146422 (Vrn-A m 1g), AY244509 (Vrn-A m 1a), KM016790 (Vrn-A1i), KM047647 (vrn-A1b.3), KM047651 (vrn-A1b.4), EU875079 (vrn-A m 1b), KM016789 (vrn-A m 1), GQ451816 (Vrn-A1f), AY747600 (vrn-A1). The effect of deletion within Vrn-A1f on vernalization requirement is unclear. Positions are indicated relative to the vrn-A1 transcription start site
Overall, polymorphism of the VRN-box is mostly determined by mutations within A-tract and C-rich regions. Our study assumed that mutations in both of these regions influence Vrn1 expression, detected as a difference in vernalization requirement and flowering time for accessions with a mutant VRN-box of VRN-A1 (Table 2). The 5’-end of the VRN-box includes T-repeat and A-tract sequences. Yan et al. [19] noted that although this sequence has significant similarity to the TATA-box, it seems unlikely that this region plays an essential role in the initiation of VRN1 transcription, because dominant alleles, which carry deletions encompassing this region have similar VRN1 transcription levels to the intact gene. In fact, the region encompassing the A-tracts and T-repeat region (“TTAAAAA”) shows a good correlation with the TATA-box, according to the weight matrix proposed earlier by Bucher [50]. Moreover, this sequence was recognized as the TATA-box when analysing more than 1500 bp of the 5’ UTR of VRN1 genes from B, A or D genomes using multiple independent tools for prediction of plant promoters such as TSSP [51], NSITE-PL [52] and PlantCARE [53]. In the last case, the sequence “TTTAAAAA” was identified as the core promoter element for Zea mays glyceraldehyde-3-phosphate dehydrogenase (gpc2) gene. Nevertheless, the VRN-box within the dominant Vrn-A1d, Vrn-A1e and Vrn-Am1g alleles is fully deleted. Expression of these alleles can be explained by the initiation of transcription via an alternative promoter. It is likely that transcription with either a putative alternative promoter located upstream of the VRN-box or transcription from a promoter inside the intact VRN-box is blocked by a hypothetical repressor which binds to the VRN-box sequence. Since transcription with an alternative promoter can be realized only if the VRN-box was damaged by mutations. The initiators of VRN1 transcription, associated with insertions of genetically mobile elements within the VRN1 promoter region and intron-1, or large deletions within the first intron, is not fully clear.
Usually it is assumed that protein affinity for DNA involves the additive contribution of nucleotides located within the target sequence. Nevertheless, the sequence-dependent features of intrinsic DNA structure and its deformability are components of so-called “indirect readout”, the principal source of non-additivity for specific protein-DNA recognition [54]. For example, TATA boxes containing 3–4 consecutive adenines have a rigid context-independent cooperative structure, which is best described by a nearest-neighbor non-additive model of protein binding [55]. According to this model the TATA-binding protein (TBP)/TATA-box system is a specific protein-DNA complex, where DNA conformation is used in extreme case and interaction is mainly through “indirect readout” [56–59]. In particular, among structural signals for TBP recognizing TATA boxes can be distinguished the helical twist angle in the middle of TATA boxes and recognition of global DNA flexibility [60]. However, in the case of A-tract polymorphism revealed in this study, the additive and non-additive components cannot be clearly separated, because the nucleotide sequence (length) of A-tract (additive component) determines flexibility and spatial structure (shape) of the DNA molecule of this sequence region (non-additive component).
Winter and spring variants of Vrn-A1b, and also vrn-A1 and Vrn-A1i, differing by only one or several SNPs located within A-tract of VRN-box, are strongly associated with differences in vernalization requirement and flowering time. Due to this, it is assumed that polymorphism of the A-tract within the VRN-box contributes greatly to modulation of VRN1 expression. By now it is definitively known that due to the unique structure and ability to affect intrinsic DNA curvature, the A-tracts provide a general genetic mechanism for regulation of the promoter activity by the different transcription factors and fine-tuning expression in a predictable manner, with resolution that can be even finer than that attained through altering transcription factor sites [61]. The function of such a promoter element mainly depends on its intrinsic structure, not its interaction with sequence-specific DNA-binding proteins [62]. In the present study, it was shown that polymorphism of the A-tract within the VRN-box was accompanied by a change in the curvature and flexibility of DNA molecules (Fig. 4a). Hence, it cannot be ruled out that a change in curvature and flexibility of DNA, determined by mutations of the A-tract within the VRN-box, affects the binding strength of the regulatory DNA sequence inside the VRN-box (or within flanking regions) with a hypothetical protein or protein complex that downregulates transcription of VRN1. For example, as was previously shown for the CArG-box, the presence of A-tracts flanking CArG-box regions can facilitate the looping of the DNA on the binding of MADS-domain transcription factors which in turn may contribute to the DNA-binding specificity of higher order complexes [63]. On the other hand, molecular dynamics simulation and experimental data revealed that the minor groove of the A-tract is a highly hydrated site, indicating that these steps have the potential to serve as a hydrogen-bonding site during protein-DNA interaction [64].
The importance of the C-rich segment within the VRN-box also cannot be excluded. The “T- > C” transition was identified only for mutant variants of Vrn-A1b and hence detected for “spring” (Vrn-A1b.1, Vrn-A1b.2, Vrn-A1b.5, Vrn-A1b.6) and “winter” (vrn-A1b.3, vrn-A1b.4) variants. However, previously it has been shown that a 20 bp deletion in the Vrn-Am1a promoter was linked to a spring growth habit [1]. This deletion includes part of C-rich region within the VRN-box (C3 instead C4TC4), which includes the site of the “T- > C” transition to 12 bp downstream. The present study revealed approximately 30 days difference between accessions with the same alleles of VRN-B1 and VRN-B3 genes, but differences in VRN-boxes of VRN-A1 of only one SNP (“T- > C” transition) within the C-rich segment (Vrn-A1b.5 and Vrn-A1i alleles), were found to be significant, with a more later flowering phenotype characterized for accessions with an intact C-rich region (Vrn-A1i allele). Furthermore, the dominant allele Vrn-A1b.6 differs from the recessive vrn-A1b.4 by only one SNP within the C-rich segment. Finally, almost all dominant alleles of Vrn1 with a mutant VRN-box carry mutations within the C-rich segment (Fig. 4b). Only Vrn-A1i has an intact C-rich, region, and this was associated with a later flowering time. These data emphasize the importance of the C-rich sequence within the VRN-box in repression of VRN-A1 transcription. It is known that guanine- and cytosine-rich regions of DNA are able to form a non-canonical nucleic acid substructure conformations comprising four-stranded DNA secondary structures, namely the G-quadruplex and i-motif. These structures are involved in the control of gene expression through regulation of transcription activity [65, 66] or post-transcriptional regulation [67]. Hence, it can be hypothesised that the C-rich region of the VRN-box regulates Vrn1 transcription through formation of quadruplex structures which are destabilized by a “T- > C” transition (for variants of Vrn-A1b) or cannot be formed due to almost full deletion of the nucleotide sequence (for Vrn-Am1a) accompanying transcription activation of VRN1. For example, mutational destabilization of the C-MYC (human oncogene) promoter G-quadruplex leads to greater transcriptional activity due to the destabilization of a DNA-protein complex, where the protein is a transcriptional repressor [68]. Furthermore, there is an opinion that G-tracts have transcriptional properties virtually identical to those of A-tracts [62]. Interestingly, according to biophysical stability studies the formation of G-quadruplex and i-motif conformations leads to destabilization of the proximal duplex regions [69] that can be the basis for interactions between the C-rich segment and A-tract within the VRN-box. Although with less similarity than a quadruplex motif, C-rich region of VRN-box (as well as the 20 bp deletion downstream of the VRN-box for Vrn-A1b alleles) is also associated with a core promoter element, the so-called Y-Patch, a CT-rich region often found in plant TATA box promoters [70]. For example, around 50 % of rice core promoters contain one or more Y-Patches [71].
Conclusions
The VRN-A1 and VRN-B1 genes were analysed in hexaploid and tetraploid wheat species. During investigation the seven novel variants of VRN-A1 and three promoter sequence variants of VRN-B1 were identified. For detection of these variants during PAGE of PCR fragments the electrophoretic conditions were optimized for low and room temperature. Novel allelic variants of VRN-A1 identified herein are more frequent compared to previously known VRN-A1 alleles. It is assumed that polymorphism of the A-tract within the VRN-box of the VRN-A1 gene is associated with modulation of the vernalization requirement and flowering time of wheat. Furthermore, it is apparent that the C-rich segment and the adjacent A-tract, both contribute to the functionality of the VRN-box. VRN-B1.s was associated with early flowering of the spring wheat accessions. In summary, the results of our study not only enrich the genetic source material for wheat breeding with new agronomically valuable alleles, but also provide a set of natural mutants for fundamental investigations essential to achieving a better understanding of the mechanisms of the regulation of VRN1 expression in wheat.
Methods
Plant material
A total of 178 accessions representing 5 hexaploid and 6 tetraploid wheat species from 57 countries and different eco-geographic areas were investigated (Additional file 2: Table S1, Additional file 3: Table S2). Hexaploid species (genome BBAuAuDD) included were T. spelta L., T. macha Dekap, T. vavilovii Jakubz, T. compactum Host, T. sphaerococcum Percival. Tetraploid species (genome BBAuAu) consisted of T. durum Desf., T. turgidum L., T. polonicum L., T. carthlicum Nevski, T. dicoccum Schrank and T. dicoccoides Körn. Germplasm was obtained from the National Plant Germplasm System (NPGS, USA) and National Center of Plant Genetic Resources (Ukraine).
The experimental plants were grown in a photoperiod-controlled glasshouse under 16 h of natural light (long day conditions). Days to flowering were registered for each accession. The plants that failed to reach stem elongation by the end of the experiment (120 days) were considered to have a winter growth habit.
DNA extraction and PCR amplification
Total DNA from 4-day-old wheat seedlings was extracted following a modified CTAB-method [72]. PCR reactions consisted of: DNA (~40 ng), 20 mM Tris–HCl (pH 8.8), 10 mM (NH2)2SO4, 2.6 mM MgCl2, 1 mM KCl, 0.1 % Triton X-100, 250 μM dNTPs, 1.5 % DMSO, 3 ng/uL each primer, 0.05 U/uL Taq-polymerase. PCR was performed using the following program: initial denaturation at 94 °C (2 min); 30 cycles of amplification: 94 °C (10 s), annealing (10 s), 74 °C (50 s or 1.5 min for amplification of the VRN-B1 intron-1) per cycle, followed by 3 cycles of: annealing (10 s), 74 °C (40 s) and a final elongation step of 72 °C for 3 min. Further details of all primers, including annealing temperatures, are listed in Table 1.
Electrophoresis in polyacrylamide and agarose gels
Amplification products of the VRN-A1 and VRN-B1 first intron, VRN-B3 alleles and variants of Vrn-A1a were separated on 6.6 % nondenaturing polyacrylamide (PAA) gels (mono/bis-acrylamide ratio 82:1) in 1.38 TBE buffer (123 mM ionic strength), at room temperature, under 6 V/cm until the bands had migrated 50–70 % of the length of the gel. To enable detection of the allelic variants of VRN-A1 and VRN-B1 by PCR fragments of the VRN1 promoter region the conditions of polyacrylamide gel electrophoresis (PAGE) were optimized for low and room temperature to obtain the minimal loss of resolution under these conditions (Additional file 4). Thus electrophoresis in 6.6 % polyacrylamide gels at mono/bis-acrylamide ratio 82:1, under low electric field (2.5 V/cm) and high ionic strength (123 mM, x1.38 TBE in gel) was carried out at room temperature, while the best results for a short duration were obtained when electrophoresis was run at low temperature (10–20 °C) with high electric field strength (5–10 V/cm). Visualization of PCR fragments in PAA gels was conducted using a modified silver staining protocol [73]. Agarose gel electrophoresis was performed using 1.5 % agarose in x1 TBE buffer. PCR products were stained with ethidium bromide and viewed under UV light.
Cloning and sequencing of PCR fragments
PCR amplicons were separated on 1.5 % agarose gels, gel-extracted using a GeneJET Gel Extraction Kit (Thermo Scientific), and DNA fragments ligated into pGEM-T (Promega), according to manufacturer’s instructions. Plasmid DNA was transformed into JM109 Escherichia coli competent cells (L2001, Promega). Transformed cells containing the plasmid carrying an insertion of foreign DNA fragment were detected using white-blue selection on growth medium containing ampicillin, X-Gal, and IPTG. Positive colonies were tested for the presence of cloned PCR products by PCR with universal pUC primers (forward and reverse M13 primers), followed by separation and visualisation of PCR products on agarose gels. Plasmid DNA was extracted using GeneJET Plasmid Miniprep Kit (Thermo Scientific), and sequencing PCRs performed using an ABI3700 Bioanalyser (Applied Biosystems), following the manufacturer’s protocol. Fluorescently labelled extension products were precipitated and resuspended in HiDi Formamide (Applied Biosystems).
Nomenclature
The different sequence variants of the Vrn-A1b allele were designated according to a numerical index, i.e. “b.1”, “b.2”, etc. A similar convention was employed for variants of Vrn-A1a, which were be identified by the specific combination of PCR fragments of certain lengths. Sequence variants of the VRN-B1 promoter region were designated by the literal index “f”, “m” and “s”, indicating respectively a “fast”, “medium” or “slow” relative migration rate of the associated PCR fragments in PAA gels. These sequence variants combined with the different VRN-B1 alleles (vrn-B1, Vrn-B1a and Vrn-B1c) with the exception of Vrn-B1(ins). The partial nucleotide sequences of VRN-A1 and VRN-B1 (promoter region) reported in this paper were deposited in GenBank under the accessions: KR782255 (Vrn-A1a.2), KM016789 (vrn-Am1), KM016790-KM016792 (Vrn-A1i), KT361213 (Vrn-A1e), KM047646 (Vrn-A1b.1), KM047641-KM047645 (Vrn-A1b.2), KM047647-KM047650 (vrn-A1b.3), KM047651 (vrn-A1b.4), KM047652 (Vrn-A1b.5), KT692944-KT692945 (Vrn-A1b.6), KR782252 (VRN-B1.s), KT361212 (VRN-B1.m), KR782253-KR782254 (VRN-B1.f).
Data analyses
For estimation of intrinsic curvature and local bend angles, three dimensional DNA models describing the trajectory of the contour path, central axis and phosphates were generated. The calculation of reconstructed DNA trajectory and the evaluation of DNA curvature was performed according to previously published methods (in Methods section of [42]). Structures were visualized by the PyMOL Molecular Graphics System, Version 1.7.2 Schrödinger, LLC. Distribution of flexibility of DNA molecules was evaluated based on the relevant parameters for di- and tetranucleotide steps published earlier [74, 75]. Multiple sequence alignments were generated using Clustal W [76]. The T. aestivum ESTs database (UniGene Build #63) containing 1551792 sequences in 178464 clusters was downloaded from NCBI (ftp://ftp.ncbi.nih.gov/repository/UniGene/Triticum_aestivum/Ta.seq.all.gz). A sequence similarity search was performed using BLAST+ software [77]. RNA secondary structure was predicted using the RNAstructure web service [78].
Acknowledgements
We are grateful to Carly Schramm for comments and English polishing in the manuscript. The analysis of the VRN-B1 promoter region in hexaploid and tetraploid wheat species and sequencing of the Vrn-A1b.6 and VRN-B1.m variants were supported by the Russian Scientific Foundation (Project No. 14-14-00161).
Additional files
“Distribution of curvature and bending angles for PCR fragments of vrn-A1 and Vrn-A1i”. (DOC 1683 kb)
“The VRN-A1 and VRN-B1 alleles of hexaploid wheat accessions”. (DOC 118 kb)
“The VRN1 alleles identified for tetraploid wheat accessions”. (DOC 141 kb)
“Effect of electrophoretic conditions on discrimination of VRN-A1 and VRN-B1 alleles”. “Optimization of PAGE for detection of the VRN-A1 and VRN-B1 alleles”. (DOC 1476 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AM carried out the experiments, developed of method for detection of the new VRN alleles, sequenced of the Vrn-A1b.6 and VRN-B1.m alleles, and wrote the manuscript. RK cloned and sequenced VRN-A1 and VRN-B1 variants and contributed to the preparation of the manuscript. EA revised the manuscript, contributed to the discussion and participated in preparing the manuscript. All authors read and approved the final manuscript.
Declarations
Publication of this article has been funded by the Russian Scientific Foundation (Project No. 14-14-00161).
This article has been published as part of BMC Plant Biology Volume 16 Supplement 1, 2015: Selected articles from PlantGen 2015 conference: Plant biology. The full contents of the supplement are available online at http://www.biomedcentral.com/bmcplantbiol/supplements/16/S1.
References
- 1.Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci U S A. 2003;100(10):6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, et al. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303(5664):1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci U S A. 2006;103(51):19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kippes N, Debernardi JM, Vasquez-Gross HA, Akpinar BA, Budak H, Kato K, et al. Identification of the VERNALIZATION 4 gene reveals the origin of spring growth habit in ancient wheats from South Asia. Proc Natl Acad Sci U S A. 2015;112(39):5401–5410. doi: 10.1073/pnas.1514883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muterko AF, Balashova IA, Fayt VI, Sivolap YM. Molecular genetic mechanisms of regulation of growth habit in wheat. Tsitol Genet. 2015;49(1):58–71. [Google Scholar]
- 6.Li C, Distelfeld A, Comis A, Dubcovsky J. Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J. 2011;67(5):763–773. doi: 10.1111/j.1365-313X.2011.04630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316(5827):1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 8.Li C, Dubcovsky J. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 2008;55(4):543–554. doi: 10.1111/j.1365-313X.2008.03526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen A, Dubcovsky J. Wheat TILLING mutants show that the vernalization gene VRN1 downregulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet. 2012;8(12):e1003134. doi: 10.1371/journal.pgen.1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L. Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol Biol. 2006;60(64):469–480. doi: 10.1007/s11103-005-4814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trevaskis B, Hemming MN, Dennis ES, Peacock WJ. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007;12(8):352–357. doi: 10.1016/j.tplants.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Distelfeld A, Tranquilli G, Li C, Yan L, Dubcovsky J. Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol. 2009;149(1):245–257. doi: 10.1104/pp.108.129353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X, Tan C, Cao S, Yan L. Molecular differentiation of null alleles at ZCCT-1 genes on the A, B, and D genomes of hexaploid wheat. Mol Breed. 2011;27(4):501–510. doi: 10.1007/s11032-010-9447-8. [DOI] [Google Scholar]
- 14.Chen F, Gao M, Zhang J, Zuo A, Shang X, Cui D. Molecular characterization of vernalization and response genes in bread wheat from the Yellow and Huai Valley of China. BMC Plant Bio. 2013;13:199. doi: 10.1186/1471-2229-13-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwaki K, Nakagawa K, Kuno H, Kato K. Ecogeographical differentiation in East Asian wheat, revealed from the geographical variation of growth habit and Vrn genotype. Euphytica. 2000;111(2):137–143. doi: 10.1023/A:1003862401570. [DOI] [Google Scholar]
- 16.Iwaki K, Haruna S, Niwa T, Kato K. Adaptation and ecological differentiation in wheat with special reference to geographical variation of growth habit and Vrn genotype. Plant Breed. 2001;120(2):107–114. doi: 10.1046/j.1439-0523.2001.00574.x. [DOI] [Google Scholar]
- 17.Stelmakh AF. Genetic effects of Vrn genes on heading date and agronomic traits in bread wheat. Euphytica. 1993;65:53–60. doi: 10.1007/BF00022199. [DOI] [Google Scholar]
- 18.Stelmakh AF. Genetic systems regulating flowering response in wheat. Euphytica. 1998;100:359–369. doi: 10.1023/A:1018374116006. [DOI] [Google Scholar]
- 19.Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J. Allelic variation at the VRN1 promoter region in polyploid wheat. Theor Appl Genet. 2004;109(8):1677–1686. doi: 10.1007/s00122-004-1796-4. [DOI] [PubMed] [Google Scholar]
- 20.Fu D, Szucs P, Yan L, Helguera M, Skinner J, Hayes P, et al. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Gen Genomics. 2005;273(4):54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- 21.Santra DK, Santra M, Allan RE, Campbell KG, Kidwell KK. Genetic and molecular characterization of vernalization genes Vrn-A1, Vrn-B1, and Vrn-D1 in spring wheat germplasm from the Pacific Northwest region of the USA. Plant Breed. 2009;128:576–584. doi: 10.1111/j.1439-0523.2009.01681.x. [DOI] [Google Scholar]
- 22.Shcherban AB, Efremova TT, Salina EA. Identification of a new Vrn-B1 allele using two near-isogenic wheat lines with difference in heading time. Mol Breed. 2012;29(3):675–685. doi: 10.1007/s11032-011-9581-y. [DOI] [Google Scholar]
- 23.Muterko A, Balashova I, Cockram J, Kalendar R, Sivolap Y. The new wheat vernalization response allele Vrn-D1s is caused by DNA transposon insertion in the first intron. Plant Mol Biol Rep. 2015;33(2):294–303. doi: 10.1007/s11105-014-0750-0. [DOI] [Google Scholar]
- 24.Chu CG, Tan CT, Yu GT, Zhong S, Xu SS, Yan L. A novel retrotransposon inserted in the dominant Vrn-B1 allele confers spring growth habit in tetraploid wheat (Triticum turgidum L.) G3 (Bethesda) 2011;1(7):637–645. doi: 10.1534/g3.111.001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Gao M, Wang S, Chen F, Cui D. Allelic variation at the vernalization and photoperiod sensitivity loci in Chinese winter wheat cultivars (Triticum aestivum L.). Front. Plant Sci. 2015;6:470. doi: 10.3389/fpls.2015.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Würschum T, Boeven PH, Langer SM, Longin CF, Leiser WL. Multiply to conquer: Copy number variations at Ppd-B1 and Vrn-A1 facilitate global adaptation in wheat. BMC Genet. 2015;16:96. doi: 10.1186/s12863-015-0258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Díaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum) PLoS One. 2012;7(3):e33234. doi: 10.1371/journal.pone.0033234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li G, Yu M, Fang T, Cao S, Carver BF, Yan L. Vernalization requirement duration in winter wheat is controlled by TaVRN-A1 at the protein level. Plant J. 2013;76(5):742–753. doi: 10.1111/tpj.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Golovnina K, Kondratenko E, Blinov A, Goncharov N. Molecular characterization of vernalization loci VRN1 in wild and cultivated wheats. BMC Plant Biol. 2010;10(168):168–183. doi: 10.1186/1471-2229-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diallo AO, Ali-Benali MA, Badawi M, Houde M, Sarhan F. Expression of vernalization responsive genes in wheat is associated with histone H3 trimethylation. Mol Genet Genomics. 2012;287:575–590. doi: 10.1007/s00438-012-0701-0. [DOI] [PubMed] [Google Scholar]
- 31.Pidal B, Yan L, Fu D, Zhang F, Tranquilli G, Dubcovsky J. The CArG-Box located upstream from the transcriptional start of wheat vernalization gene VRN1 is not necessary for the vernalization response. J Hered. 2009;100(3):355–364. doi: 10.1093/jhered/esp002. [DOI] [PubMed] [Google Scholar]
- 32.Oliver S, Finnegan E, Dennis E, Peacock W, Trevaskis B. Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proc Natl Acad Sci U S A. 2009;106(20):8386–8391. doi: 10.1073/pnas.0903566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao J, Xu S, Li C, Xu Y, Xing L, Niu Y, et al. O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat Commun. 2014;5:4572. doi: 10.1038/ncomms5572. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Yu M, Carver BF, Yan L. TamiR1123 originated from a family of miniature inverted-repeat transposable elements (MITE) including one inserted in the Vrn-A1a promoter in wheat. Plant Sci. 2014;215–216:117–123. doi: 10.1016/j.plantsci.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, et al. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.) Genome Biol. 2007;8(6):R96. doi: 10.1186/gb-2007-8-6-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marini JC, Levene SD, Crothers DM, Englund PT. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stellwagen NC. Anomalous electrophoresis of deoxyribonucleic acid restriction fragments on polyacrylamide gels. Biochemistry. 1983;22(26):6186–6193. doi: 10.1021/bi00295a023. [DOI] [PubMed] [Google Scholar]
- 38.Diekmann S. Analyzing DNA, curvature in polyacrylamide gels. Methods Enzymol. 1992;212:30–46. doi: 10.1016/0076-6879(92)12004-A. [DOI] [PubMed] [Google Scholar]
- 39.Koo HS, Wu HM, Crothers DM. DNA bending at adenine-thymine tracts. Nature. 1986;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 40.Hagerman PJ. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- 41.Diekmann S. The migration anomaly of DNA fragments in polyacrylamide gels allows the detection of small sequence-specific DNA structure variations. Electrophoresis. 1989;10(5–6):354–359. doi: 10.1002/elps.1150100513. [DOI] [PubMed] [Google Scholar]
- 42.Muterko A, Kalendar R, Cockram J, Balashova I. Discovery, evaluation and distribution of haplotypes and new alleles of the Photoperiod-A1 gene in wheat. Plant Mol Biol. 2015;88(1–2):149–164. doi: 10.1007/s11103-015-0313-2. [DOI] [PubMed] [Google Scholar]
- 43.Shcherban AB, Strygina KV, Salina EA. VRN-1 gene- associated prerequisites of spring growth habit in wild tetraploid wheat T. dicoccoides and the diploid A genome species. BMC Plant Biol. 2015;15:94. doi: 10.1186/s12870-015-0473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milec Z, Tomkova L, Sumikova T, Pankova K. A new multiplex PCR test for the determination of Vrn-B1 alleles in bread wheat (Triticum aestivum L.) Mol Breed. 2012;30(1):317–323. doi: 10.1007/s11032-011-9621-7. [DOI] [Google Scholar]
- 45.Cockram J, Mackay IJ, O’Sullivan DM. The role of doublestranded break repair in the creation of phenotypic diversity at cereal VRN1 loci. Genetics. 2007;177(4):2535–2539. doi: 10.1534/genetics.107.074765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puchta H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot. 2005;56(409):1–14. doi: 10.1093/jxb/eri025. [DOI] [PubMed] [Google Scholar]
- 47.Ferguson AA, Jiang N. Mutator-like elements with multiple long terminal inverted repeats in plants. Comp Funct Genomics. 2012;2012(2012):695827. doi: 10.1155/2012/695827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138(4):2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004;18(18):2237–2242. doi: 10.1101/gad.307804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212(4):563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 51.Solovyev VV, Shahmuradov IA. PromH: Promoters identification using orthologous genomic sequences. Nucleic Acids Res. 2003;31(13):3540–3545. doi: 10.1093/nar/gkg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solovyev VV, Shahmuradov IA, Salamov AA. Identification of promoter regions and regulatory sites. Methods Mol Biol. 2010;674:57–83. doi: 10.1007/978-1-60761-854-6_5. [DOI] [PubMed] [Google Scholar]
- 53.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Travers AA. DNA-Protein: Structural interactions. Oxford: IRL Press; 1995. pp. 49–75. [Google Scholar]
- 55.Faiger H, Ivanchenko M, Haran TE. Nearest-neighbor non-additivity versus long-range non-additivity in TATA-box structure and its implications for TBP-binding mechanism. Nucleic Acids Res. 2007;35(13):4409–4419. doi: 10.1093/nar/gkm451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365(6446):520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 57.Nikolov DB, Chen H, Halay ED, Hoffman A, Roeder RG, Burley SK. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc Natl Acad Sci U S A. 1996;93(10):4862–4867. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Juo ZS, Chiu TK, Leiberman PM, Baikalov I, Berk AJ, Dickerson RE. How proteins recognize the TATA box. J Mol Biol. 1996;261(2):239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 59.Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, Burley SK. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13(24):3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bareket-Samish A, Cohen I, Haran TE. Signals for TBP/TATA box recognition. J Mol Biol. 2000;299(4):965–977. doi: 10.1006/jmbi.2000.3797. [DOI] [PubMed] [Google Scholar]
- 61.Raveh-Sadka T, Levo M, Shabi U, Shany B, Keren L, Lotan-Pompan M, et al. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat Gen. 2012;44(7):743–750. doi: 10.1038/ng.2305. [DOI] [PubMed] [Google Scholar]
- 62.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14(11):2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muiño JM, Smaczniak C, Angenent GC, Kaufmann K, van Dijk AD. Structural determinants of DNA recognition by plant MADS-domain transcription factors. Nucleic Acids Res. 2014;42(4):2138–2146. doi: 10.1093/nar/gkt1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yonetani Y, Kono H. Sequence dependencies of DNA deformability and hydration in the minor groove. Biophys J. 2009;97(4):1138–1147. doi: 10.1016/j.bpj.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du Z, Zhao Y, Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008;18(2):233–241. doi: 10.1101/gr.6905408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, et al. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J Med Chem. 2008;51(18):5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 67.Bugaut A, Balasubramanian S. 5’-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40(11):4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A. 2002;99(18):11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.König SL, Huppert JL, Sigel RK, Evans AC. Distance-dependent duplex DNA destabilization proximal to G-quadruplex/i-motif sequences. Nucleic Acids Res. 2013;41(15):7453–7461. doi: 10.1093/nar/gkt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamamoto YY, Ichida H, Abe T, Suzuki Y, Sugano S, Obokata J. Differentiation of core promoter architecture between plants and mammals revealed by LDSS analysis. Nucleic Acids Res. 2007;35(18):6219–6226. doi: 10.1093/nar/gkm685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Civán P, Svec M. Genome-wide analysis of rice (Oryza sativa L. subsp. japonica) TATA box and Y Patch promoter elements. Genome. 2009;52(3):294–297. doi: 10.1139/G09-001. [DOI] [PubMed] [Google Scholar]
- 72.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry Bull. 1987;19:11–15. [Google Scholar]
- 73.Budowle B, Chakraborty R, Giusti AM, Eisenberg AJ, Allen RC. Analysis of the VNTR locus D1S80 by the PCR followed by high-resolution PAGE. Am J Hum Genet. 1991;48(1):137–144. [PMC free article] [PubMed] [Google Scholar]
- 74.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc Natl Acad Sci U S A. 1998;95(19):11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Packer MJ, Dauncey MP, Hunter CA. Sequence-dependent DNA structure: tetranucleotide conformational maps. J Mol Biol. 2000;295(1):85–103. doi: 10.1006/jmbi.1999.3237. [DOI] [PubMed] [Google Scholar]
- 76.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]


