Abstract
The vitamin D receptor (VDR) is a ligand-activated transcription factor that regulates gene expression in many cell types, including immune cells. It requires binding of 1,25 dihydroxy vitamin D3 (1,25D3) for activation. Many autoimmune diseases show latitude-dependent prevalence and/or association with vitamin D deficiency, and vitamin D supplementation is commonly used in their clinical management. 1,25D3 is regulated by genes associated with the risk of autoimmune diseases and predominantly expressed in myeloid cells. We determined the VDR cistrome in monocytes and monocyte-derived inflammatory (DC1) and tolerogenic dendritic cells (DC2). VDR motifs were highly overrepresented in ChIP-Seq peaks in stimulated monocyte (40%), DC1 (21%) and DC2 (47%), P<E−100 for all. Of the nearly 11 000 VDR-binding peaks identified across the genome in DC1s, 1317 were shared with DC2s (91% of DC2 sites) and 1579 with monocytes (83% of monocyte sites). Latitude-dependent autoimmune disease risk polymorphisms were highly overrepresented within 5 kb of the peaks. Several transcription factor recognition motifs were highly overrepresented in the peaks, including those for the autoimmune risk gene, BATF. This evidence indicates that VDR regulates hundreds of myeloid cell genes and that the molecular pathways controlled by VDR in these cells are important in maintaining tolerance.
Introduction
Genome-wide association studies have identified hundreds of autoimmune risk genes, many shared across diseases.1, 2, 3, 4 The challenge now is to use this genetic road map to better define the pathogenesis of the diseases and develop novel effective therapies and clinically useful biomarkers. The prevalence of many autoimmune diseases increases with distance from the equator in genetically similar populations in many countries.5, 6, 7, 8, 9, 10 These associations have been attributed to reduced exposure to UV radiation, and consequent Vitamin D (Vit D) deficiency. This is supported by evidence of the importance of Vit D in immunomodulation.11, 12, 13, 14, 15, 16, 17
Several recent lines of unrelated investigation provide further strong support for a key role of Vit D in the pathogenesis of autoimmune disorders. Two of the autoimmune-associated genes, CYP27B1 and CYP24A1, control the availability of the ligand for the endogenous Vit D receptor (VDR), 1,25-dihydroxyvitamin D3 (1,25D3): CYP27B1 converts the precursor 25D3 to 1,25D3 and CYP24A1 enhances catabolism of 1,25D3. These genes (CYP27B1/CYP24A1/VDR), singly or collectively, have been identified as risk factors in multiple autoimmune diseases.1, 18, 19, 20, 21, 22, 23 Second, Vit D supplementation has been found to be of therapeutic benefit in animal models of autoimmune disease, including in autoimmune encephalomyelitis, collagen-induced arthritis, type 1 diabetes mellitus, inflammatory bowel disease, autoimmune thyroiditis and systemic lupus erythematosus.24 Further, at a clinical level, higher serum Vit D levels are associated with reduced risk of autoimmune diseases11, 12, 13, 14, 15, 16, 17, 25 and the function of regulatory T cells (Tregs) in patients has been shown to be proportional to serum 25D3 levels.26 As a result of these findings, Vit D has been considered a promising target pathway for therapeutic intervention and clinical trials have commenced (ahead of more detailed knowledge of the microenvironment in which Vit D immunomodulation occurs).
Activation of VDR by 1,25D3 results in the liganded receptor binding to a large number of genes (the VDR cistrome). We propose that a major mechanism by which VDR activation influences autoimmunity is through gene activation in immune cells of myeloid lineage, particularly antigen-presenting dendritic cells (DCs) with consequent activation of tolerizing states in the immune system, especially in DCs.27, 28 We base this on several observations. VDR activation induces a tolerogenic phenotype in antigen-presenting cells, including DCs.29 Within immune cells, CYP27B1 and CYP24A1 are predominantly expressed in DCs30, 31 (it seems likely T cells need an exogenous source of 1,25D3 since they have limited CYP27B1 expression). In mouse models of autoimmune disease treatment with antigen-specific or generic tolerogenic DCs is remarkably successful.32, 33 This has led to clinical trials in humans, with promising results, especially with autologous DCs.34 However, a major limitation is that DCs can be quite plastic, so that the choice of manipulation needs to be further informed by experimental data, and reliable quality control measurements of manipulation are needed. Given that the importance of Vit D in inducing and maintaining a tolerogenic DC phenotype is well established,28, 35 identification of the molecular basis for this may lead to new tools to manipulate and assess DCs.
Although the spectrum of genes regulated by VDR in B cells has been explored, and genes associated with autoimmune diseases are overrepresented in this spectrum,36 the potential role of VDR in DCs and other myeloid cells has yet to be characterized. For the reasons above, we believe these cells to be priority targets.
In this study we identify the VDR cistrome in myeloid (monocytes, inflammatory and tolerogenic DCs) immune cells and map it against genetic loci identified as risk factors for autoimmune states. We used chromatin immunoprecipitation and next generation sequencing to identify the VDR-binding sites in monocytes and two types of in vitro differentiated DCs: inflammatory (stimulated with interferon gamma, DC1) and tolerogenic (stimulated with interferon beta, DC2). The phenotypes of these cells were demonstrated in an earlier study.30 Here we show that there are striking similarities in genomic locations of VDR-binding sites among the myeloid cell subsets, but also many which change dramatically with differentiation state. Differences are also seen in the spectrum of transcription factor (TF) recognition sequences co-located with the VDR peaks. Finally, latitude-dependent autoimmune disease (LAD) risk loci are overrepresented in the genomic vicinity of VDR peaks, pointing to a regulatory architecture that might be exploited for therapeutic purposes.
Results
Expression of CYP27B1 and CYP24A1 in immune cells
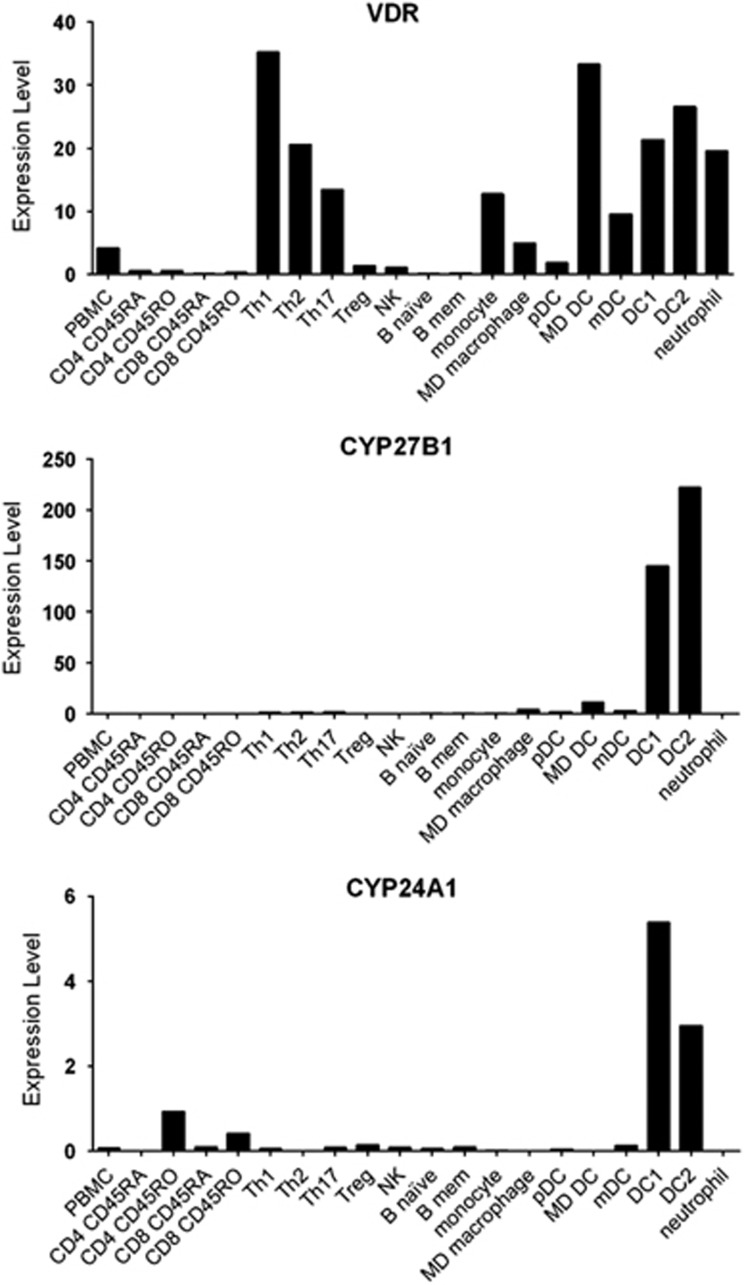
The association of CYP27B1 and CYP24A1 with LADs implicates Vit D in their development. To address this notion, we identified the immune cell subsets able to respond to 25D3 on the basis of expression of CYP27B1 and VDR, and in which CYP24A1 expression was high. Although many cell subsets expressed VDR, and so could respond to 1,25D3, only myeloid cells exhibited high expression of CYP27B1 and CYP24A1 (Figure 1).
Figure 1.
RNA-Seq determined expression of CYP27B1, CYP24A1 and VDR in immune cell subsets. Y-axis is expression level.30 Results are from our data, confirming that of others.31 MD, monocyte derived; mDC, myeloid DC; pDC, plasmacytoid DC.
VDR binding in monocytes, DC1 and DC2
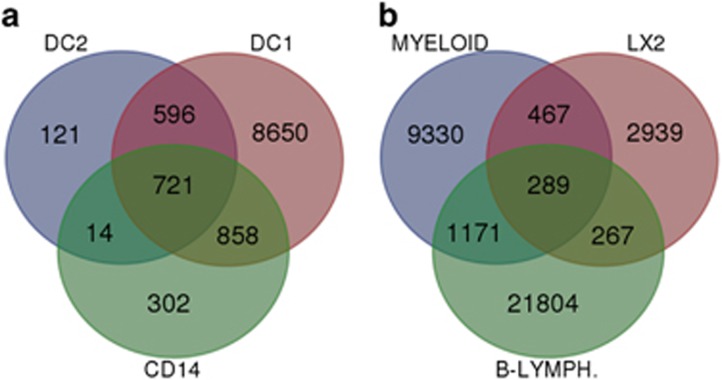
We then sought the genes regulated by VDR in myeloid cells using VDR ChIP-Seq (chromatin immunoprecipitation coupled with high-throughput deep sequencing) to identify the VDR-binding sites in both unstimulated and calcipotriol (synthetic derivative of 1,25D3—used to maximize genomic VDR binding) stimulated monocytes, monocyte-derived inflammatory (DC1) and tolerogenic (DC2) DCs. The canonical Vit D response element (VDRE) motif was highly overrepresented in VDR ChIP-Seq binding peaks in stimulated monocytes (40%), DC1 (21%) and DC2 (47%), P<E−100 for all cell types (Figure 2). Of the nearly 11 000 VDR-binding peaks identified across the genome in DC1s, 1317 were shared with DC2s (91% of DC2 sites) and 1579 with monocytes (83% of monocyte sites) (Figure 3a). The highly significant overrepresentation of the VDRE motif in the binding peaks and the level of overlap between the cell subset cistromes indicate that the data are of high quality.
Figure 2.
The vitamin D response element (VDRE) motif is highly enriched in the identified VDR cistromes of DC1, DC2 and monocyte cell subsets. These response elements typically consist of two conserved hexameric half-sites separated by a three nucleotide spacer.
Figure 3.
Venn diagram of overlapping VDR-binding sites in (a) the three myeloid subsets DC1, DC2 and monocyte (CD14) and (b) overlap of the myeloid cell VDR-binding sites with the VDR-binding sites of a hepatic stellate cell line (LX2) and a B-lymphocyte cell line. Overlap of DC2 and DC1 was 91% of DC2 peaks; monocyte and DC1 was 83% of monocyte peaks; DC2 and monocyte was 51% of monocyte peaks; LX2 and myeloid was 19% of LX2 peaks; myeloid and B-lymph was 14% of myeloid peaks; and LX2 and B-lymph was 14% of LX2 peaks. In each case, the cell subset with the largest number of identified VDR peaks is used as the reference to compare overlaps.
Comparison of VDR binding in other cell types
There was overlap of the myeloid VDR cistromes with hepatic stellate cells (LX2) and B cells (Figure 3b), though to a lesser degree than between the myeloid cell subsets. Overlap between each of the myeloid cell subsets with LX2s and B cells were similar. Overlap between the immune cell subsets, B cells and myeloids was greater than with stellate cells.
Representation of autoimmune disease risk genes in the myeloid VDR cistrome
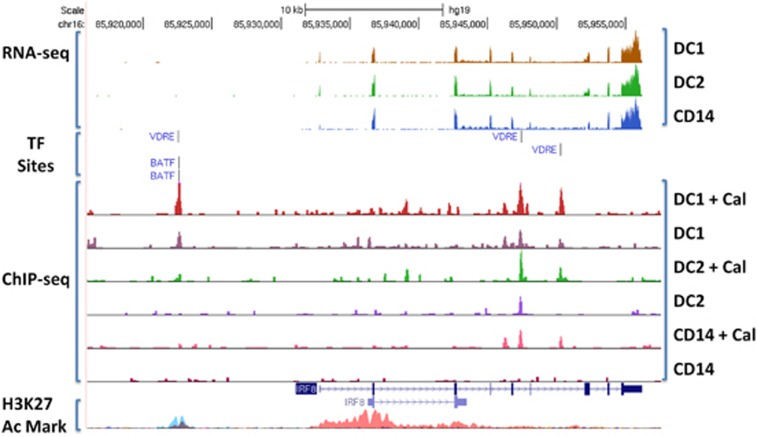
Subsequently, we matched the genes identified as VDR binding in myeloid cell subsets with known risk factor single nucleotide polymorphisms for autoimmune disease. Using HOMER,37 the overrepresentation of autoimmune disease risk factor single nucleotide polymorphisms (from individual GWAS from the NIH GWAS catalog) within 5000 bp of VDR-binding peaks was found to be very high (Table 1). Of the 2293 GWAS on the GWAS catalog, only 15 studies had an excess of single nucleotide polymorphisms from the VDR cistrome at P<10−4 (not corrected for multiple testing). Of these 15 studies, 10 were of autoimmune diseases, 9 of the 10 with evidence of latitude affects and serum 25D3 levels on risk. The autoimmune disease GWAS overrepresented included multiple sclerosis (MS), inflammatory bowel disease, rheumatoid arthritis and systemic lupus erythematosus. Four of the enriched GWAS were for conditions affected by cholesterol, which competes with 25D3 for their mutual precursor squalene. The final disease was attention deficit hyperactivity disorder, known to be affected by latitude and Vit D deficiency. A typical example of VDR-binding peak location, effect of calcipotriol on the peak, co-location with other TF motifs and gene expression in the three myeloid subsets for an LAD risk gene is shown in Figure 4. The MS risk genes that contained VDR-binding peaks and VDRE motifs for each myeloid cell subset examined are shown in Table 2.
Table 1. Risk genes for latitude-dependent autoimmune diseases are highly overrepresented in the vitamin D receptor cistrome.
| Study (GWAS catalog no.) | Total risk SNPs | DC1 peaks overlap | DC1 P-value | DC2 peaks overlap | DC2 P-value | CD14 peaks overlap | CD14 P-value | Latitude effect? | Vit D deficiency? |
|---|---|---|---|---|---|---|---|---|---|
| Inflammatory bowel disease (23128233) | 110 | 29 | 4.8E−18 | NS | NS | Khalili et al.7 | Del Pinto et al.12 | ||
| Ulcerative colitis (21297633) | 47 | 13 | 3.7E−09 | NS | 2 | 2.11E−02 | Khalili et al.7 | Del Pinto et al.12 | |
| Multiple sclerosis (21833088) | 81 | 16 | 1.2E−08 | 3 | 3.30E−03 | 3 | 6.88E−03 | Simpson et al.10 | Nieves et al.13 |
| Crohn's disease (21102463) | 69 | 14 | 6.9E−08 | NS | 5 | 2.07E−05 | Khalili et al.7 | Jørgensen et al.14 | |
| HDL cholesterol (20686565) | 47 | 10 | 3.2E−06 | 2 | 1.28E−02 | NS | Grimes et al.53 | NA | |
| Mean platelet volume (22139419) | 29 | 8 | 3.9E−06 | 4 | 3.82E−06 | 2 | 8.37E−03 | Icli et al.54 | Cumhur Cure et al.55 |
| Celiac disease (20190752) | 40 | 9 | 6.2E−06 | 4 | 1.42E−05 | 3 | 9.20E−04 | NA | Tavakkoli et al.15 |
| Fibrinogen (23969696) | 23 | 7 | 7.8E−06 | NS | NS | Grimes et al.53 | NA | ||
| HDL cholesterol (24097068) | 71 | 11 | 2.6E−05 | 2 | 2.77E−02 | NS | Grimes et al.53 | NA | |
| Rheumatoid arthritis (20453842) | 28 | 7 | 3.2E−05 | NS | NS | Vieira et al.8 | Hong et al.16 | ||
| ADHD (23527680) | 21 | 6 | 5.3E−05 | NS | NS | Arns et al.56 | Humble et al.57 | ||
| Multiple sclerosis (19525955) | 8 | 4 | 8.3E−05 | 1 | 2.87E−02 | NS | Simpson et al.10 | Nieves et al.13 | |
| Crohn's disease (23850713) | 8 | 4 | 8.3E−05 | 1 | 2.87E−02 | NS | Khalili et al.7 | Jørgensen et al.14 | |
| Multiple sclerosis (19525953) | 15 | 5 | 1.0E−04 | NS | NS | Simpson et al.10 | Nieves et al.13 | ||
| Systemic lupus erythematosus (23273568) | 34 | 7 | 1.2E−04 | NS | NS | Walsh and Gilchrist9 | Mok17 |
Abbreviations: ADHD, attention deficit hyperactivity disorder; GWAS, genome-wide association study; HDL, high-density lipoprotein; NA, no supporting evidence; NS, no statistically significant enrichment at P=0.05; Peaks Overlap; number of VDR-binding peaks identified where a GWAS risk SNP is located within 5000 bp.
Figure 4.
Visualization of RNA-seq, VDR ChIP-seq and co-localization of VDRE and BATF-binding sites. Total ChIP-seq peaks for each sample normalized to 1.00e7 to allow for comparison of peak heights. +Cal indicates cells treated for 16 h with calcitriol. RNA-seq experiment described in Shahijanian et al.30 UCSC genome browser and H3K27 Mark ENCODE track used for visualization.
Table 2. MS risk genes with VDR peaks and VDRE motifs.
| MS risk genes | Myeloid cell subsets |
|---|---|
| CARD11, CYP24A1, FOXP1, MAPK1, MIR1208, SLC44A2, SLC9A8, TNFAIP3, ZMIZ1, ZFP36L1 | CD14, DC1, DC2 |
| CD86, ZFP36L2 | CD14, DC1 |
| EOMES, PTGER4, RGS1, RMI2, CD69 | DC1, DC2 |
| CD40, CD83, ELMO1, IQGAP1, IRF8, JDP2, MAF, MALT1, PLEK, SATB1, STAT3, TRAF3, VMP1, IL7R, CTSH | DC1 |
| IFI30 | DC2 |
Autoimmune risk gene TF motifs in VDR peaks
Genes encoding TFs are overrepresented among MS risk genes.38 Of the 26 TFs affecting genetic risk of MS, 14 are predominantly expressed in myeloid cells;38 it is likely they regulate myeloid cell differentiation and/or state. Of these, seven had a VDR ChIP-Seq peak within 5 kb of the MS risk single-nucleotide polymorphism closest to the gene, with six having the canonical VDRE motif within the peak (Table 3, see next section for further detail on binding peaks with and without the VDRE motif).
Table 3. Evidence for MS risk myeloid transcription factors regulated by VDR.
| MS risk myeloid transcription factor | Myeloid cell in which expressed |
VDR |
BATF | |
|---|---|---|---|---|
| VDR Peak | VDR motif | BATF motif | ||
| BCL10 | CD14 | No | No | No |
| BATF | DC1, DC2 | No | No | No |
| HHEX1 | CD14 | No | No | No |
| IRF8 | All | Yes | Yes | Yes |
| MAF | CD14, DC1 | No | No | No |
| NFKB1 | DC2 | Yes (DC1 only) | No | Yes |
| POU5F1B | DC1, DC2 | No | No | No |
| STAT3 | DC1, DC2 | Yes | Yes | Yes |
| STAT4 | DC2 | Yes | Yes | Yes |
| ZBTB46 | CD14, DC2 | No | No | No |
| ZFP36L1 | CD14 | Yes | Yes | Yes |
| ZNF438 | All | Yes | Yes | Yes |
| ZMIZ1 | CD14, DC1 | Yes | Yes | Yes |
| ZFP36L2 | All | No | No | No |
Of TF motifs overrepresented in the VDR ChIP-Seq-binding peaks, one MS risk factor, BATF, was highly overrepresented; this was seen in DC1s and DC2s at P<E−100 but not in monocytes. All the seven MS risk gene TFs containing VDR-binding peaks had BATF recognition motifs co-localized with the peak (genes NFKB1, IRF8 (Figure 4), ZMIZ1, STAT3, STAT4, ZNF438).
Other TF recognition motifs in VDR-binding peaks with/without the VDRE motif
We characterized the presence of other TF motifs in all the VDR-binding peaks, those containing the VDRE motif and those without (Supplementary Table S1). Eight of the TF motifs were for genes expressed predominantly in myeloid cell subsets, although there were some striking differences, pointing to interactions between VDR and these TFs in orchestration of myeloid development. Six TFs were monocyte specific, 16 DC1 specific and 10 DC2 specific. To determine if these different TFs represented particular cellular roles, we investigated if they were overrepresented in curated pathways (Supplementary Figure S1). IL1 (2.6E−6), IL6 (6.1E−9) and MIF (9.2E−5) (not corrected for multiple testing) signaling pathways associated with inflammation were overrepresented in the DC1 motifs, but not significantly overrepresented in the DC2 motifs.
The absence of canonical VDR motifs in many peaks has been observed in other ChIP-Seq data sets, and implicates tethering of VDR to other TFs (and their target DNA motif).39, 40 In each cell type, the TFs overrepresented in the VDR-binding peaks with and without the VDRE motif largely did not overlap. The overlapping TFs were AP-1, SFPI1, JUND for monocyte; BATF, EGR2, HNF4A, RELA and SPI1 for DC1 and CEBPB and SPI1 for DC2.
The tolerogenic cistrome
The 121 genes containing VDR-binding peaks in DC2s, but not the DC1s or monocytes, are defined here as the tolerogenic cistrome (Supplementary Table S2). Notably, mRNA expression of PGE synthase, known to modulate tolerance through the PGE2 signaling network,41 was the only gene (from the 121 defining the tolerogenic cistrome) differentially expressed between the cell subsets based on RNA-Seq data.
Discussion
We present evidence that the VDR binds at thousands of sites in the genome of human monocytes and DCs, of which about half have the canonical VDRE motif. Furthermore, we show a higher degree of overlap of VDR-binding sites between myeloid subsets than between myeloids, hepatic stellate cells and B lymphocytes, and that the binding sites can change depending on the myeloid cell differentiation state. The recognition motifs for other TFs at these sites are highly non-random, with an overrepresentation of TFs known to interact with VDR from other studies, and which are known to control immune responses. Also overrepresented within 5 kb of these sites are genes associated with LADs, especially with MS. These data indicate that VDR interacts with key TFs in complexes to control the transcriptome and thereby the differentiation state of myeloid cells. Our findings suggest that perturbed VDR binding at autoimmune disease risk gene variants may contribute to the susceptibility to LADs.
Genes nearest to VDR-binding sites are enriched for immune function and risk of LADs. Since the myeloid cells provide a crucial arm of the immune response, controlling processes such as antigen presentation, removal of cellular debris and other sources of antigen, licensing of T and B cells, controlling the cytokine environment at sites of inflammation (trauma, infection) and in the primary and secondary lymphoid organs, it is not surprising that a large proportion of the genes known to be associated with autoimmune risk are predominantly expressed in myeloid cells.42 The identification of these genes in this study, together with future evaluation of the effects of the risk genetic variants on VDR and other TF regulation of the risk genes, should enable a more directed understanding of the transcriptomic interactions important in the dysregulation of the immune response that leads to autoimmune disease.
A number of papers have recently characterized the transcriptome of myeloid subsets, in conjunction with analysis of genetic variation.43, 44, 45 Autoimmune genes were overrepresented in those genes whose expression changed with differentiation, notably on tumor necrosis factor-α stimulation.45 The enzyme catalyzing activation of 25D3 to 1,25D3, Cyp27B1, is one of the most upregulated with the majority of the forms of monocyte stimulation employed in these studies. It remains to be determined if this results in increased VDR activity and subsequent restoration of homeostasis, or if a more complex form of control of myeloid cells is initiated. Given the complexity of the VDR cistrome in inflammatory as well as tolerogenic DCs, the latter seems more likely.
The changes in distribution of VDR-binding sites in different myeloid subsets, and the different spectrum of TF motifs overrepresented in these sites, indicate the architecture of myeloid differentiation is heavily influenced by the VDR activity. Specifically, we have compelling evidence that VDR and BATF may share an intersecting genomic circuit that regulates development of DCs. This extends from our previous demonstration that VDR ligands inhibit hepatic stellate activation by TGFβ1 and abrogate liver fibrosis, whereas VDR knockout mice spontaneously develop hepatic fibrosis.46 Mechanistically, we showed that TGFβ1 signaling causes a redistribution of genome-wide VDR-binding sites, which facilitates VDR binding at SMAD3 profibrotic target genes in these cells. In the presence of VDR ligands, VDR binding to the coregulated genes reduces SMAD3 occupancy at these sites, inhibiting fibrosis. We propose that VDR and BATF may interact in a similar fashion to control monocyte maturation.
BATF is known to be a core TF in myeloid cell differentiation.47 The BATF motif was found in 24% of VDR-captured sequences for DC1s and 28% for DC2s. It is interesting that BATF is relatively highly expressed in DC1s and DC2s, but not monocytes. Since the overrepresentation of BATF sites at VDR peaks occurs only where BATF protein would be present, this is consistent with the BATF protein actually binding to its cognate motif in the VDR-captured sequence. The VDR ChIP-Seq BATF sites were in putative promoter regions of LAD risk genes likely to control the tolerance/inflammatory development of DCs, including CYP27B1 and CYP24A1. The protein encoded by BATF is a nuclear basic leucine zipper protein that belongs to the AP-1/ATF superfamily of TFs; BATF knockout mice lack Th17 cells and are resistant to encephalomyelitis, a mouse model of MS.47 Its absence from murine cells has been shown to perturb the balance of many TFs, which also produce an altered CD8 T-cell phenotype and functional consequences.48
In mouse models of autoimmune diseases, including encephalomyelitis, treatment with antigen-specific or generic tolerogenic DCs is remarkably successful.32 This has led to clinical trials in humans, with promising results, especially with autologous DCs.34 However, a major limitation is that the functional state of DCs can vary dramatically according to their surrounding cytokine environment or presence of stimulating factors, so that the choice of manipulation needs to be further informed by experimental data, and reliable quality control measurements of manipulation are needed. The importance of Vit D in inducing and maintaining a tolerogenic DC phenotype is well established.28, 35 Knowledge of the precise changes to the DC VDR cistrome and subsequent transcriptome, including with the interactions with other TFs such as BATF, and determination of the effects of the LAD risk gene genotypes on these processes, may allow a better understanding of myeloid cell plasticity and thereby utility for therapy. From this study, a set of genes in cis with VDR peaks in DC2s but not with DC1s were identified, and of these PGE2 synthase was differentially expressed between the two. This protein promotes the synthesis of PGE2, a ligand for the MS risk factor PTGER4, and a protein known to mediate resistance to encephalomyelitis.41
Another translational benefit of a better understanding of VDR function in myeloid cells is that it may provide a more targeted approach to preventing or treating autoimmune disease than simple oral 25D3 supplementation. Many drugs have been designed to manipulate the Vit D3 pathway.49 The two main issues affecting the therapeutic use of VDR agonist ligands are increased intestinal calcium absorption leading to hypercalcaemia and loss of activity due to VDR-mediated upregulation of CYP24A1 leading to ligand catabolism. To overcome this a number of approaches have been adopted, including design of VDR ligands that cause minimal elevation of serum calcium and display relative resistance to Cyp24A1-mediated degradation.49 Further, giving the ligand in intermittent pulses helps overcome both of these issues. Finally, targeting the VDR agonist to the cell type relevant to the therapeutic effect is a logical approach to therapy, but this necessitates a detailed mechanistic knowledge of the disease process. To date, many clinical trials have been undertaken, but only with the natural endogenous ligand (1,25D3) or its immediate precursor (25D3), with mixed results.50
Our results indicate that VDR regulates transcription from thousands of genomic sites, both with and without canonical VDR recognition motifs, in myeloid cell subsets. There is evidence of interaction with many other immune cell TFs, and of control of myeloid cell differentiation through this interaction, including of genes which are autoimmune disease risk factors. Our data compellingly suggest that VDR and other TFs, notably BATF, combine to regulate myeloid cells, that the balance of cell differentiation, tolerant vs activating, is fundamental to autoimmune disease risk and that manipulation of the process may be therapeutically beneficial.
Materials and methods
Cell isolation, differentiation and stimulation
Peripheral blood mononuclear cells were isolated from EDTA-treated whole blood of healthy controls using Ficoll Paque Plus (GE Healthcare, Uppsala, Sweden). CD14+ cells were purified from the peripheral blood mononuclear cells using the Human CD14 Positive Selection Kit (Stem Cell Technologies, Vancouver, BC, Canada) according to the manufacturer's instructions. Cells were plated in X-Vivo15 medium (Lonza, Basel, Switzerland) with 2% AB serum in 25 cm2 flasks and treated with three changes of granulocyte macrophage colony-stimulating factor (70 ng ml−1) and IL-4 (10 ng ml−1) over 6 days. On day 6, cells were treated with lipopolysaccharide (250 ng ml−1) and interferon-γ (10 ngml−1, for DC1 differentiation) or interferon-β (1000 IUml−1, for DC2 differentiation). Cells cultured without addition of calcipotriol were harvested after 48 h. For calcipotriol-treated samples, cells were cultured for 32 h before addition of calcipotriol at a final concentration of 100 nm and cultured for an additional 16 h prior to harvest.
Chromatin immunoprecipitation coupled with high-throughput deep sequencing (ChIP-seq)
The experimental procedure for ChIP was as previously described.51 Briefly, after fixation, nuclei from monocyte, DC1 and DC2 cells were isolated, lysed and sheared with a Diagenode Bioruptor (Denville, NJ, USA) to yield DNA fragment sizes of 200–1000 bp followed by immunoprecipitation using the following antibodies: normal rabbit immunoglobulin G (Santa Cruz, Dallas, TX, USA; sc-2027), VDR (Santa Cruz; sc-1008). ChIP-Seq analysis was performed as previously described.51 Briefly, short DNA reads generated on an Illumina HiSeq 2000 platform were aligned against the human hg19 reference genome and aligned using the Bowtie2 v2.2.3 aligner with default settings.52 Only tags that mapped uniquely to the genome were considered for further analysis. Subsequent peak calling and motif analysis were conducted using HOMER (http://homer.salk.edu/homer/).37 One tag from each unique position was considered to eliminate peaks resulting from clonal amplification of fragments during the ChIP-Seq protocol. Peaks were identified by searching for clusters of tags within a sliding 200 bp window, requiring adjacent clusters to be at least 1 kb away from each other. The threshold for the number of tags that determine a valid peak was selected for a false discovery rate <0.0001, as empirically determined by repeating the peak finding procedure using randomized tag positions. Peaks are required to have at least fourfold more tags (normalized to total count) than input or immunoglobulin G control samples and fourfold more tags relative to the local background region (10 kb) to avoid identifying regions with genomic duplications or nonlocalized binding. Peaks are annotated to gene products by identifying the nearest RefSeq transcriptional start site. Visualization of ChIP-Seq results was achieved by uploading custom tracks onto the University of California, Santa Cruz genome browser.
RNA-Seq
We used our previously generated RNAseq data set of ex vivo and in vitro differentiated immune cell subsets to compare relative gene expression levels across different immune cell populations.30
Acknowledgments
This work was supported by the Australian NHMRC Project Grant 1049936. DB was the Hunt Family MSRA Senior Research Fellow, now NHMRC Senior Research Fellow. RME is an Investigator of the Howard Hughes Medical Institute (HHMI) at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology, and is supported by National Institutes of Health (NIH) grants (DK057978, DK090962, HL088093, HL105278 and ES010337), the Glenn Foundation for Medical Research, the Leona M and Harry B Helmsley Charitable Trust (#2012-PG-MED002). CL and MD were funded by grants from the National Health and Medical Research Council of Australia Project Grants 632886 and 1043199.
Author contributions
DB and CL conceived the study. FS and SC carried out the cell culture, ND the ChIP-Seq experiments, GP the RNA-seq experiments and CL conducted the bioinformatic analysis with support from DB and GP. DB wrote the first draft and prepared figures with GP and CL. All authors contributed to and approved the final document.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Genes and Immunity website (http://www.nature.com/gene)
Supplementary Material
References
- Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011; 476: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 2013; 45: 1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet 2013; 14: 661–673. [DOI] [PubMed] [Google Scholar]
- Cotsapas C, Hafler DA. Immune-mediated disease genetics: the shared basis of pathogenesis. Trends Immunol 2013; 34: 22–26. [DOI] [PubMed] [Google Scholar]
- Ascherio A, Munger KL, Simon KC. Vitamin D and multiple sclerosis. Lancet Neurol 2010; 9: 599–612. [DOI] [PubMed] [Google Scholar]
- Miller DH, Hammond SR, McLeod JG, Purdie G, Skegg DC. Multiple sclerosis in Australia and New Zealand: are the determinants genetic or environmental? J Neurol Neurosurg Psychiatry 1990; 53: 903–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili H, Huang ES, Ananthakrishnan AN, Higuchi L, Richter JM, Fuchs CS et al. Geographical variation and incidence of inflammatory bowel disease among US women. Gut 2012; 61: 1686–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira VM, Hart JE, Webster TF, Weinberg J, Puett R, Laden F et al. Association between residences in U.S. northern latitudes and rheumatoid arthritis: a spatial analysis of the Nurses' Health Study. Environ Health Perspect 2010; 118: 957–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh SJ, Gilchrist A. Geographical clustering of mortality from systemic lupus erythematosus in the United States: contributions of poverty, Hispanic ethnicity and solar radiation. Lupus 2006; 15: 662–670. [DOI] [PubMed] [Google Scholar]
- Simpson S Jr, Blizzard L, Otahal P, Van der Mei I, Taylor B. Latitude is significantly associated with the prevalence of multiple sclerosis: a meta-analysis. J Neurol Neurosurg Psychiatry 2011; 82: 1132–1141. [DOI] [PubMed] [Google Scholar]
- Berlanga-Taylor AJ, Disanto G, Ebers GC, Ramagopalan SV. Vitamin D-gene interactions in multiple sclerosis. J Neurol Sci 2011; 311: 32–36. [DOI] [PubMed] [Google Scholar]
- Del Pinto R, Pietropaoli D, Chandar AK, Ferri C, Cominelli F. Association between inflammatory bowel disease and vitamin D deficiency: a systematic review and meta-analysis. Inflamm Bowel Dis 2015; 21: 2708–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves J, Cosman F, Herbert J, Shen V, Lindsay R. High prevalence of vitamin D deficiency and reduced bone mass in multiple sclerosis. Neurology 1994; 44: 1687–1692. [DOI] [PubMed] [Google Scholar]
- Jorgensen SP, Hvas CL, Agnholt J, Christensen LA, Heickendorff L, Dahlerup JF. Active Crohn's disease is associated with low vitamin D levels. J Crohn's Colitis 2013; 7: e407–e413. [DOI] [PubMed] [Google Scholar]
- Tavakkoli A, DiGiacomo D, Green PH, Lebwohl B. Vitamin D status and concomitant autoimmunity in celiac disease. J Clin Gastroenterol 2013; 47: 515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Q, Xu J, Xu S, Lian L, Zhang M, Ding C. Associations between serum 25-hydroxyvitamin D and disease activity, inflammatory cytokines and bone loss in patients with rheumatoid arthritis. Rheumatology (Oxford) 2014; 53: 1994–2001. [DOI] [PubMed] [Google Scholar]
- Mok CC. Vitamin D and systemic lupus erythematosus: an update. Expert Rev Clin Immunol 2013; 9: 453–463. [DOI] [PubMed] [Google Scholar]
- Bahlo M, Booth DR, Broadley SA, Brown MA, Foote SJ, Griffiths LR et al. Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet 2009; 41: 824–U84. [DOI] [PubMed] [Google Scholar]
- Bailey R, Cooper JD, Zeitels L, Smyth DJ, Yang JH, Walker NM et al. Association of the vitamin D metabolism gene CYP27B1 with type 1 diabetes. Diabetes 2007; 56: 2616–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, Wallace C et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet 2011; 7: e1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012; 491: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014; 506: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, Stahl EA, Trynka G, Raychaudhuri S, Festen EA, Franke L et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet 2011; 7: e1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon M, Fitzgerald O. Vitamin D and its emerging role in immunopathology. Clin Rheumatol 2012; 31: 199–202. [DOI] [PubMed] [Google Scholar]
- Munger KL, Levin LI, Hollis BW, Howard NS, Ascherio A. Serum 25-hydroxyvitamin D levels and risk of multiple sclerosis. JAMA 2006; 296: 2832–2838. [DOI] [PubMed] [Google Scholar]
- Smolders J, Thewissen M, Peelen E, Menheere P, Tervaert JW, Damoiseaux J et al. Vitamin D status is positively correlated with regulatory T cell function in patients with multiple sclerosis. PLoS One 2009; 4: e6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adorini L, Penna G. Dendritic cell tolerogenicity: a key mechanism in immunomodulation by vitamin D receptor agonists. Hum Immunol 2009; 70: 345–352. [DOI] [PubMed] [Google Scholar]
- Nagy L, Szanto A, Szatmari I, Szeles L. Nuclear hormone receptors enable macrophages and dendritic cells to sense their lipid environment and shape their immune response. Physiol Rev 2012; 92: 739–789. [DOI] [PubMed] [Google Scholar]
- Szeles L, Keresztes G, Torocsik D, Balajthy Z, Krenacs L, Poliska S et al. 1,25-dihydroxyvitamin D3 is an autonomous regulator of the transcriptional changes leading to a tolerogenic dendritic cell phenotype. J Immunol 2009; 182: 2074–2083. [DOI] [PubMed] [Google Scholar]
- Shahijanian F, Parnell GP, McKay FC, Gatt PN, Shojoei M, O'Connor KS et al. The CYP27B1 variant associated with an increased risk of autoimmune disease is underexpressed in tolerizing dendritic cells. Hum Mol Genet 2014; 23: 1425–1434. [DOI] [PubMed] [Google Scholar]
- Liu SM, Xavier R, Good KL, Chtanova T, Newton R, Sisavanh M et al. Immune cell transcriptome datasets reveal novel leukocyte subset-specific genes and genes associated with allergic processes. J Allergy Clin Immunol 2006; 118: 496–503. [DOI] [PubMed] [Google Scholar]
- Van Brussel I, Lee WP, Rombouts M, Nuyts AH, Heylen M, De Winter BY et al. Tolerogenic dendritic cell vaccines to treat autoimmune diseases: can the unattainable dream turn into reality? Autoimmun Rev 2014; 13: 138–150. [DOI] [PubMed] [Google Scholar]
- Mohammad MG, Hassanpour M, Tsai VW, Li H, Ruitenberg MJ, Booth DW et al. Dendritic cells and multiple sclerosis: disease, tolerance and therapy. Int J Mol Sci 2012; 14: 547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. Dendritic cells as targets or therapeutics in rheumatic autoimmune disease. Curr Opin Rheumatol 2014; 26: 211–218. [DOI] [PubMed] [Google Scholar]
- Pearce EJ, Everts B. Dendritic cell metabolism. Nat Rev Immunol 2015; 15: 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res 2010; 20: 1352–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell GP, Gatt PN, Krupa M, Nickles D, McKay FC, Schibeci SD et al. The autoimmune disease-associated transcription factors EOMES and TBX21 are dysregulated in multiple sclerosis and define a molecular subtype of disease. Clin Immunol 2014; 151: 16–24. [DOI] [PubMed] [Google Scholar]
- Downes M, Liddle C. Look who's talking: nuclear receptors in the liver and gastrointestinal tract. Cell Metab 2008; 7: 195–199. [DOI] [PubMed] [Google Scholar]
- Tuoresmaki P, Vaisanen S, Neme A, Heikkinen S, Carlberg C. Patterns of genome-wide VDR locations. PLoS One 2014; 9: e96105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott NM, Ng RL, Gorman S, Norval M, Waithman J, Hart PH. Prostaglandin E2 imprints a long-lasting effect on dendritic cell progenitors in the bone marrow. J Leukoc Biol 2014; 95: 225–232. [DOI] [PubMed] [Google Scholar]
- Booth DR. The clinical implications from the first hundred known MS susceptibility genes. Adv Clin Neurosci Rehabil 2014; 14: 10–13. [Google Scholar]
- Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 2014; 345: 1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014; 40: 274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MN, Ye C, Villani AC, Raj T, Li W, Eisenhaure TM et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science 2014; 343: 1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N, Yu RT, Subramaniam N, Sherman MH, Wilson C, Rao R et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell 2013; 153: 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 2009; 460: 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, DiIorio MA, Lemieux ME et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol 2014; 15: 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster I, Bernhardt R. Inhibition of cytochromes p450: existing and new promising therapeutic targets. Drug Metab Rev 2007; 39: 481–499. [DOI] [PubMed] [Google Scholar]
- Munger KL, Ascherio A. Prevention and treatment of MS: studying the effects of vitamin D. Mult Scler 2011; 17: 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C et al. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev 2010; 24: 2760–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes DS, Hindle E, Dyer T. Sunlight, cholesterol and coronary heart disease. QJM 1996; 89: 579–589. [DOI] [PubMed] [Google Scholar]
- Icli A, Aksoy F, Nar G, Kaymaz H, Alpay MF, Nar R et al. Increased mean platelet volume in familial hypercholesterolemia. Angiology 2015; 67: 146–150. [DOI] [PubMed] [Google Scholar]
- Cumhur Cure M, Cure E, Yuce S, Yazici T, Karakoyun I, Efe H. Mean platelet volume and vitamin D level. Ann Lab Med 2014; 34: 98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arns M, van der Heijden KB, Arnold LE, Kenemans JL. Geographic variation in the prevalence of attention-deficit/hyperactivity disorder: the sunny perspective. Biol Psychiatry 2013; 74: 585–590. [DOI] [PubMed] [Google Scholar]
- Humble MB, Gustafsson S, Bejerot S. Low serum levels of 25-hydroxyvitamin D (25-OHD) among psychiatric out-patients in Sweden: relations with season, age, ethnic origin and psychiatric diagnosis. J Steroid Biochem Mol Biol 2010; 121: 467–470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.