Abstract
Background
Canine chronic enteropathies (CE) are believed to be caused by an aberrant immune response towards the intestinal microbiome. Administration of probiotics can alleviate colitis in people. In vitro effects of the probiotic Enterococcus faecium NCIMB 10415 E1707 (EF) previously have been evaluated using canine cells (e.g., whole blood, intestinal biopsies), but data on in vivo efficacy are lacking.
Hypothesis/Objectives
Administration of EF to dogs with food‐responsive CE will improve clinical outcome and decrease the intestinal inflammatory profile.
Animals
Dogs diagnosed with CE were prospectively recruited to receive a hydrolyzed elimination diet plus either a synbiotic product containing EF or placebo for 6 weeks. Both veterinary staff and owners were blinded to the treatment.
Methods
Clinical severity index (CCECAI), clinicopathological data and gene expression using intestinal biopsies (TLR2/4/5/9, IL‐17A, IL‐22, IL‐23p19, RORC, IL‐2, IL‐12p35, TNFα, IL‐4, IFNy, IL‐10, TGFβ, IL‐1β, IL‐18, NLRP3, casp‐1, TFF1, TFF3 and PPARy) before and after 6 weeks of treatment were analyzed using linear mixed modeling.
Results
Of the 45 cases recruited, 12 finished the clinical trial. Seven received the synbiotic and 5 the placebo product. There was no difference between groups or treatments regarding clinical efficacy, histology scores or expression of any of the investigated genes.
Conclusions and clinical importance
Standard dietary treatment induced rapid clinical response in all cases. Because the study was underpowered, it was not possible to determine whether or not EF had an additional effect within the time period of 6 weeks.
Keywords: diarrhea, dog, inflammatory bowel disease, probiotics
Abbreviations
- ACTH
adrenocorticotropic hormone
- ARD
antibiotic‐responsive disease
- ASPA
animal scientific procedure act of the United Kingdom
- casp‐1
caspase 1
- CCECAI
canine chronic enteropathy clinical activity index
- CE
chronic enteropathy
- cPL
canine pancreatic lipase
- EF
enterococcus faecium
- FOS
fructo‐oligosaccharides
- FoxP3
forkhead‐box protein 3
- FRD
food‐responsive disease
- GIT
gastrointestinal tract
- IFNγ
interferon gamma
- IL
interleukin
- MAPK
mitogen‐activated pyruvate kinase
- NFκB
nuclear factor kappa B
- NLRP3
nod‐like receptor with pyrin domain 3
- PI3K
phosphatidyl‐Inositol 3 kinase
- PPARγ
peroxisome proliferator‐activated receptor gamma
- PRR
pathogen‐recognition receptor
- qPCR
quantitative reverse‐transcription real‐time polymerase chain reaction
- RIN
RNA integrity number
- RORC
retinoic acid receptor‐related orphan receptor C
- RVC
Royal Veterinary College, London, United Kingdom
- SRD
steroid‐responsive disease
- TFF
trefoil factor
- TGFβ
transforming‐growth factor beta
- Th cells
T helper lymphocyte cells
- TLI
trypsin‐like immunoreactivity
- TLR
toll‐like receptor
- TNFα
tumor‐necrosis factor alpha
- Tregs
regulatory T‐lymphocytes
- WSAVA
World Small Animal Veterinary Association
Canine chronic enteropathies (CE) are a group of inflammatory intestinal diseases of unknown cause.1, 2 They usually are defined by response to treatment as food‐responsive disease (FRD), antibiotic‐responsive disease (ARD) and (idiopathic) inflammatory bowel disease (IBD), with the latter also being termed steroid‐responsive disease (SRD).2, 3 All of these syndromes manifest in variable degrees and combinations of gastrointestinal signs (e.g., diarrhea, vomiting, weight loss, changes in appetite).
Because microbiome changes are consistently found in dogs with CE,4, 5, 6 modification of the intestinal microbiome in the form of pre‐ and pro‐biotics seems to be an interesting treatment option. Several studies have shown that probiotics can influence key biological signaling pathways in immune cells such as NFκB, MAPK, Akt/PI3K and PPARγ.7, 8, 9, 10, 11 In humans with IBD and mice with experimental colitis, respectively, administration of probiotics has been shown to alleviate intestinal inflammation, prevent relapse or both by induction of a more tolerant microenvironment.12, 13, 14
Knowledge about the usefulness of probiotics in CE in dogs is limited. Only two studies have evaluated the in vivo effect of probiotics in dogs with chronic gastrointestinal diseases.15, 16 One used a combination of different lactobacilli in dogs with FRD compared to placebo (diet only),15 the other a commercially available product for humans, which contains eight different strains of bacteria (lactobacilli, bifidobacteria, streptococci) in comparison to a combination treatment consisting of metronidazole and prednisolone.16 In the FRD study, all dogs improved clinically after treatment, but intestinal mucosal cytokine patterns were not correlated with clinical outcome or probiotic supplementation. In the second study, all dogs also improved clinically, but only in the group treated with probiotics was an increase in FoxP3 positive regulatory T‐lymphocytes (Tregs) as well as an increase in the supposedly protective Faecalibacterium prausnitzii in fecal samples observed.5, 17
Enterococcus faecium NCIMB 10415 E1707 (EF) is commercially available as a probiotic or synbiotic for small animals both in Europe and in the USA (the US strain is SF68). The strain has shown good properties regarding survival in the upper gastrointestinal tract (GIT),18 adhesion to canine intestinal mucus and persistence in fecal samples,19 but has not been tested extensively for its immunomodulatory functions. No information regarding the efficacy of this specific probiotic in dogs with CE is available.
The aim of the present work was to assess the clinical benefit (as measured by the canine chronic enteropathy clinical activity index CCECAI1) of treatment with a synbiotic product (containing EF)1 in dogs with FRD. In addition, selected clinicopathological data, intestinal endoscopic and histology scores and mucosal gene expression for a variety of genes associated with microbial recognition and modulation of the immune system in the intestine were investigated before and after treatment to assess if EF can induce a more anti‐inflammatory environment.
Materials and Methods
Conduct of a Prospective, Blinded, Placebo‐Controlled Clinical Trial
Ethical Approvals and Products Used
The clinical trial was approved by the RVC's Ethics and Welfare Committee (ASPA number 70/7393) and conducted between June 2010 and May 2013. The synbiotic used contained 1 × 109 cfu EF NCIMB 10415 E1707, FOS and gum Arabic1. The placebo consisted of an identical capsule containing maltodextran.
Inclusion and Exclusion Criteria
Dogs with a history typical for chronic enteropathy (≥3 weeks of vomiting, diarrhea or both, with or without weight loss) were included. The diagnosis of chronic enteropathy was confirmed based on established criteria (no clinically relevant abnormalities on routine hematology, serum biochemistry; trypsin‐like immunoreactivity [TLI], canine pancreatic lipase [cPL] and adrenocorticotropic hormone [ACTH]‐stimulation test results within the reference range; no abnormalities on abdominal imaging [radiographs, abdominal ultrasound examination or both]). Histopathologic findings of intestinal mucosal biopsies had to show lymphoplasmacytic, eosinophilic or mixed inflammation with or without architectural changes.20, 21 Exclusion criteria were the presence of concurrent diseases or treatment with antimicrobials, anti‐inflammatory drugs or both 7 days before presentation.
Study Design
Dogs were seen at three separate visits: visit 1 (recruitment), visit 2 (14 ± 3 days after starting the trial medication) and visit 3 (42 ± 3 days after starting the trial medication; see Fig 1). The procedures performed at each visit are listed in Table 1. Patients were randomized (using random permutated blocks of n = 12 that had been designed before the start of the study2) to receive either the synbiotic EF product or placebo in conjunction with a hydrolyzed protein diet3. All members of staff involved in the trial (apart from the study monitor, a head nurse) were blinded to the dogs' medications.
Figure 1.
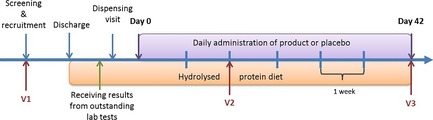
Outline of the clinical trial. Dogs were seen at three separate visits: Visit (V) 1 = recruitment and diagnosis, V2 = after 2 weeks of treatment with either Enterococcus faecium or placebo, V3 = after 6 weeks of treatment. During the duration of the trial dogs were all fed the same hydrolysed protein diet.
Table 1.
Outline and procedures for each dog at each visit of the clinical trial
| Visit | History | PE | CCECAI | CBC/BC | UA | TLI/cPLI | Folate/ cobalamin | ACTH stim | Faecal | Endoscopy and biopsy |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | X | X | X | X | X | X | X | X | X | X |
| 2 | X | X | X | Only albumin | ||||||
| 3 | X | X | X | X | X |
Visit 1 = before start of treatment, visit 2 = 2 weeks after start of treatment for food‐responsive chronic enteropathy with either Enterococcus faecium or placebo, visit 3 = 6 weeks after start of treatment. PE, physical exam; CCECAI, canine chronic enteropathy clinical activity index; CBC, complete blood count; BC, serum biochemistry; UA, urinalysis; ACTH stim, adrenocorticotropin stimulation test; faecal, faecal flotation/sedimentation.
Assessment of Clinical Severity
Clinical severity was assessed using the CCECAI at all three visits.1
Endoscopy and Endoscopic Scoring of Lesions
Gastroduodenoscopy and colonoscopy were performed in all dogs according to standard procedures. Macroscopic endoscopic scoring was performed using the World Small Animal Veterinary Association (WSAVA) approved endoscopic examination report.20 Multiple biopsies (15–20) from the duodenum and colon were taken with a single‐use endoscopic biopsy forceps, and 10–15 from each site were transferred immediately into 2% neutral‐buffered formalin. The remaining biopsies (minimum of 2) were transferred to RNAlater4 , stored at 4°C for 24 h and then at −80°C until further processing.
Histopathology
Initial histopathological assessment was performed at the pathology department of the RVC as part of the routine diagnostic procedure. After finalizing the study, all slides were reviewed by 1 of the authors (HB), and WSAVA standardized classification was applied to all slides.20, 21 As described previously,22 9 histologic variables, five representing morphological changes and four representing inflammatory changes were scored as absent = 0, mild = 1, moderate = 2, or severe = 3 according to the WSAVA guidelines.20, 21 The total histology score was recorded. At this stage of histologic analysis, HB was blinded to original diagnosis, visit number and treatment group.
Gene Expression from Intestinal Tissues
RNA Extraction and cDNA Synthesis
A single biopsy stored in RNAlater was thawed and homogenized in lysis buffer5 using 5 mm stainless steel beads5 and a tissue grinder6. Total RNA was extracted using a commercial kit7 including an on‐column DNAse treatment, and eluted in 30 μL nuclease‐free water. RNA quantity and quality was assessed using an automated analyzer8. Reverse transcription of 1 μg of RNA was performed with a cDNA synthesis kit9, which uses a mixture of oligo‐dT and random nonamer primers.
Generation of Positive Controls and Selection of Reference Genes
Canine sequences for the genes of interest and reference genes were cloned as described before.23 Primers were designed based on publically available gene sequences (Genbank, ENSEMBL) using the Primer3 software (see Table 2).24 A total of 16 genes were assessed for stability using suitable software10 with a commercially available canine reference gene array11. Based on these data, four reference genes were selected for canine duodenum and five for the colon, respectively (Table 2). Primers were optimized using standard PCR12. Amplicons were visualized on a 1.5% agarose gel, purified13 and cloned into a holding vector14 for subsequent sequencing15 and for qPCR standards (see below).
Table 2.
List of genes investigated by quantitative reverse‐transcriptase real‐time PCR with the respective primers used
| Gene | product size | Forward primer | Reverse primer | Annealing temp | Accession number |
|---|---|---|---|---|---|
| Casp‐1 | 253 | CGACAGACAGCTGGACACAT | ATCTGGGCTTTCACATCTGG | 55 | NM_001003125.1 |
| FoxP3 | 550 | TGG TGC AGT CTC TGG AAC AG | GGC CTT TGG CTT CTC TTC TT | 60 | NM_001168461.1 |
| IFNy | 353 | TTCAGCTTTGCGTGATTTTG | CTGCAGATCGTTCACAGGAA | 54.3 | NM_001003174.1 |
| IL‐10 | 300 | TCTGTTGCTGCCTGGTCCT | TGATGTCTGGGTCGTGGTT | 54.3 | NM_001003077.1 |
| IL‐12p35 | 102 | ATGACGGTCCTGTGCCTTAG | CTGCCTCTTGGGATCCATTA | 55 | NM_001003293.1 |
| IL‐17A | 166 | CCGATCTACCTCACCTTGGA | TCGCAGAACCAGGATCTCTT | 60.2 | NM_001165878.1 |
| IL‐18 | 240 | GAG GAT ATG CCC GAT TCT GA | ATC ATG GCC TGG AAC ACT TC | 55 | NM_001003169.1 |
| IL‐1β | 440 | GCA GTA CCC GAA CTC ACC AG | ACA TTT TCC CCA TTG AGG TG | 55 | NM_001037971.1 |
| IL‐2 | 243 | TCGCACTGACGCTTGTACTT | GCACTTCCTCCAGGTTTTTG | 58.4 | NM_001003305.1 |
| IL‐22 | 254 | TCCAGCAGCCCTATATCACC | TTGGCTTAGCTTGTTGCTGA | 60.2 | XM_538274.2 |
| IL‐23p19 | 198 | ATGGGACATGTGGACCTACC | TAGAGAAGGCTCCCCTGTGA | 55 | XM_538231.3 |
| IL‐4 | 289 | CTCACCTCCCAACTGATTCC | TGCTGCTGAGGTTCCTGTAG | 56.4 | NM_001003159.1 |
| NLRP3 | 207 | GCA ATG CTC TTG GAG ACA CA | AGA GCA GCA TGA CCC CTA GA | 55 | XM_843284.2 |
| PPARy | 413 | GGATTTCTCCAGCATTTCCA | TATGAGACATCCCCACAGCA | 59 | NM_001024632.2 |
| RORC | 289 | TGATGGAGACTGAGCACCTG | ACCAGAACCACTTCCATTGC | 55 | XM_540323.3 |
| TFF1 | 161 | GGA GCA CAG GGT GAT CTA CG | AAC AGC AGC CCT TGT CCT TA | 55 | NM_001002992.1 |
| TFF3 | 105 | ACGAACCTGTGCGAGGTG | GATGCTGGAGTCGAGCAG | 55 | NM_001002990.1 |
| TGFβ | 609 | GCATGTGGAGCTGTACCAGA | TAGTACACGATGGGCAGTGG | 55 | NM_001003309.1 |
| TLR2 | 263 | AGTGGCCAGAAAAGCTGAAA | ATCCAGTTGCTCCTTCGAGA | 54.3 | NM_001005264.2 |
| TLR4 | 200 | ATTCCTTTCCGGACAACTCC | CTGGAGGGAGAGGAGAGGTT | 54.3 | NM_001002950.2 |
| TLR5 | 100 | TGGGCGAGCTCTATGACTCT | CTGAACGTCTGGTCCTGGAT | 55 | NM_001197176.1 |
| TLR9 | 327 | GCCTGGAGTACCTGCTCTTG | AGGCTTTGGTTTTGGTGATG | 55 | NM_001002998.1 |
| TNFα | 715 | TTCTGCCTCAGCCTCTTCTC | GCCCTGAGCCCTTAATTCTC | 60.2 | NM_001003244.4 |
| GAPDHa | 194 | GGAGAAAGCTGCCAAATATG | ACCAGGAAATGAGCTTGACA | 55 | NM_001003142.1 |
| GUSBa | 140 | CATGCTGGTCCAGAGCTACA | CAGGCTTCAGGAAGGAAGTG | 55 | NM_001003191.1 |
| HMBSa | 112 | TCACCATCGGAGCCATCT | GTTCCCACCACGCTCTTC | 54 | XM_546491.3 |
| PPIAa | 159 | GGGGAGAAGTTTGATGACGA | GACCTTGCCAAAGACCACAT | 55 | XM_532723.3 |
| RPL32a | 186 | CCTCAGACCTCTGGTGAAGC | TTCTTGTTGCTCCCGTAACC | 55 | NM_001252169.1 |
| SDHAa | 412 | GGACAGAGCCTCAAGTTTGG | GGCATCCTTCCGTAATGA | 56.3 | XM_535807.3 |
| TBPa | 267 | GGGACCGCAGCAGATTACTA | GCCATAAGGCATCATTGGAC | 60 | XM_849432.2 |
| ATP5Bb | 196 | AGCCCATGGTGGTTACTCTG | GGCAACAGTCAGTCCAGTCA | 55 | XM_531639.3 |
| B2Mb | 99 | ACGGAAAGGAGATGAAAGCA | CCTGCTCATTGGGAGTGAA | 56 | XM_003640047.1 |
| HMBSb | 112 | TCACCATCGGAGCCATCT | GTTCCCACCACGCTCTTC | 54 | XM_546491.3 |
| PLA2R1b | 231 | GAAGCCAGCAATGTGTCTCA | TCACAAGTGCAGGAGGACAG | 55 | XM_545489.3 |
| YESb | 192 | GTCTTTCCATGACGCCATTT | TTTGAAACCGTTCACCCTTC | 55 | NM_001003239.1 |
Reference genes for the duodenum.
Reference genes for the colon.
Quantitative Polymerase Chain Reaction
qPCR reactions were carried out in triplicate at a total reaction volume of 20 μL, using a commercially available reagent mix16 and a primer concentration of 200 nM. Cycling steps were: 95°C for 5 min, then 40 cycles of 94°C for 15 s, 55°C for 10 s, 72°C for 10 s and 80°C for 5 s. Reaction efficiency was determined using 10‐fold dilutions (107 molecules μL−1 to 101 molecules μL−1) of plasmids containing the cloned amplicon. Melting curves were generated to ensure a single amplicon had been produced. Averaged gene expression data for each sample were normalized against the geometric mean of the reference genes.25
Genes of Interest
Several genes of the innate immune system interacting with bacteria (Toll‐like receptors26, 27 [TLRs] 2,4,5, and 9 as well as components of the inflammasome,28 NLRP3, casp‐1, interleukin [IL]‐1β and IL‐18) and the adaptive immune response in the intestine were investigated. For the latter, several signature cytokines and transcription factors of different T helper lymphocyte (Th) cell lines were assessed. These included Th17 cells (IL‐17A, IL‐22, IL‐23p40, RORC),29, 30 Th1 cells (IL‐12p35, IL‐2, TNFα), Th2 cells (IFNγ and IL‐4,31 and Tregs (IL‐10 and TGFβ).32
Expression of the so‐called trefoil factors (TFF), genes associated with epithelial barrier function and repair,33 also was assessed. The nuclear receptor PPARγ, usually responsible for blocking the transcription of inflammatory cytokines34 and supposedly especially up‐regulated in intestinal epithelial cells by enterococci,35 was investigated additionally.
Dendrograms and Heatmaps from Gene Expression Data
To facilitate visualization of gene expression data and changes in gene expression among visits of the clinical trial, heatmaps were created using a statistical package17.
Statistical Analyses
Power Calculation
Clinical severity (as assessed by CCECAI) was selected as the primary outcome measure. Calculations were based on a type I error of 0.05 and a type II error of 0.1, which corresponds to a power of 90%. When assuming a difference (delta, Δ) of 1.5 times the sd (σ) to be medically relevant, the necessary number of animals in each group (treatment versus placebo) was determined to be n = 11.
Analysis of Clincopathological, Histological and Gene Expression Data
Statistical analysis was performed using 2 commercially available software packages18 , 19. Normal distribution was assessed by inspection of histograms or D'Agostino and Pearson omnibus normality test. Subsequently, either parametric or non‐parametric tests were performed: A repeated measures model was used to assess the effect of the treatment interval (before versus after 6 weeks of treatment) and treatment type (EF versus placebo). Kruskal‐Wallis or t‐tests were performed to compare clinical and clinicopathological data, endoscopy and histology scores on visit 1 between treatment groups. Significance was set at P < 0.05.
Results
Patients
A total of 51 possible cases were identified, 45 cases were recruited, and 28 completed visit 1. The remaining 17 dogs dropped out for several reasons: Owners changing their mind about participation in the study without giving specific reasons (n = 5), owners not wanting endoscopy performed (n = 3), additional medical problems were discovered before endoscopy was performed (n = 3), with 1 dog developing pronounced neurological symptoms that were deemed more important than the chronic GIT signs, 1 dog collapsing after sedation for abdominal ultrasound examination and 1 dog being diagnosed with concurrent hypothyroidism, identification of other diseases causing the GIT signs (n = 2, with 1 dog having metastatic gastric carcinoma and the other a pyloric sarcoma), owners not wanting to complete a 6‐weeks food trial (n = 2), owners wanting to perform a food trial before endoscopy (n = 1) and 1 dog that had an aggressive temperament. Of the remaining 28 dogs, 16 were withdrawn after visit 1 and did not complete the study. Reasons were: failure to respond to dietary treatment (n = 6), owner not wishing to continue with the study without giving specific reasons (n = 4), owner objecting to a second endoscopy procedure (n = 2), diagnosis of another condition responsible for the clinical signs (n = 2; 1 dog with colonic mucinous adenocarcinoma and 1 dog with pancreatitis), dog not eating the hydrolyzed protein diet (n = 1), and 1 dog being too difficult to handle for a second period of hospitalization. Thus, a total of 12 dogs finished the clinical trial, including Labrador Retrievers (n = 6), Golden Retrievers (n = 2) and 1 dog each of the following breeds: Bracco Italiano, English Setter, Miniature Schnauzer and Standard Poodle. Six dogs were intact males, two dogs castrated males, and four dogs were spayed females. Their median age was 40 months (range, 12–84 months). Seven dogs had been randomly assigned to receive the synbiotic product and 5 to receive placebo.
Clinical and Clinicopathological Data
The majority of dogs suffered from mixed small and large intestinal clinical signs (n = 6), 3 had solely small intestinal clinical signs, 2 only chronic vomiting and 1 only large intestinal diarrhea. Clinical signs at visit 1 were scored mild to moderate with a median CCECAI of 4 (range, 1–6) in the synbiotic group and 5 (range, 2–7) in the placebo group, without a significant difference between groups. In addition, there was no effect of probiotic treatment on CCECAI at visit 2 or 3 compared to visit 1 (P = 0.72). For both groups combined, CCECAI was significantly decreased at visits 2 and 3 compared to visit 1 (P < 0.001), but not between visits 2 and 3 (Fig 2).
Figure 2.
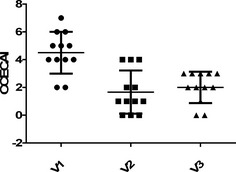
Canine chronic enteropathy clinical acvtivity index (CCECAI) in 12 dogs participating in the clinical trial at visit 1 (V1 = before treatment), visit 2 (V2 = 2 weeks after start of treatment) and visit 3 (V3 = 6 weeks after start of treatment).
There were no significant differences in hematological and biochemical variables (including TLI, cPL, folate and cobalamin) between treatment groups at visit 1. Serum folate concentrations showed a significant increase with treatment at visit 3 compared to visit 1 (P = 0.012). In addition, there was a significant difference in serum folate concentrations between the 2 treatment groups at visit 3 (P = 0.012): dogs treated with placebo had higher serum concentrations of folate (mean, 21.42 μg L−1; SD, 2.56 μg L−1) than dogs treated with the synbiotic (mean, 17.02 μg L−1; SD, 1.68 μg L−1).
Endoscopic Findings
Endoscopic WSAVA scores were low in all areas examined at the initial visit: esophagus median 0 (range, 0–6), stomach median 0 (range, 0–16), duodenum median 1.5 (range, 0–14) and colon median 0 (range, 0–6). Endoscopy scores were even lower (but not significantly) at visit 3: esophagus median 0 (range, 0–1), stomach median 0 (range, 0–3), duodenum median 0 (range, 0–6) and colon median 0 (range, 0–1).
Histopathology
Median total WSAVA scoring of histopathological findings was 3 for the duodenum (range, 1–5) and 2 (range, 1–5) for the colon at visit 1. At visit 3, the median was 1 (range, 0–8) and 3 (range, 0–4) for the duodenum and colon, respectively. There was no significant difference in total scores among visits or between treatment groups for both sites. In the duodenum at visit 1, 7 dogs showed a mild increase in intra‐epithelial lymphocytes, 6 dogs showed mild generalized lymphoplasmacytic infiltration, and 2 dogs mild eosinophilic inflammation. Duodenal architectural changes were present in 8 dogs, and included villous stunting (n = 5), mucosal fibrosis (n = 3), lacteal distension (n = 2), crypt distension (n = 2) and epithelial injury (n = 2). Mild lacteal dilatation was identified in 2 cases.
In the colon (visit 1), mild lymphoplasmacytic infiltration was present in six dogs, mild eosinophilic in two dogs, marked eosinophilic in one dog, and mild neutrophilic inflammation in two dogs. Architectural changes in the colon included crypt hyperplasia (n = 6), crypt distention and distortion (n = 4), fibrosis and atrophy (n = 3) and mild surface epithelium injury (n = 1). In 1 dog, the histology of the colon was reported as normal.
Gene Expression
In both duodenum and colon, TLR2 and TLR4 were more abundantly expressed than TLR5, and TLR9 was expressed at the lowest level (see Fig 3). Genes associated with the inflammasome generally were expressed at a much higher level in the colon than in the duodenum, especially casp‐1 (Fig 4).
Figure 3.
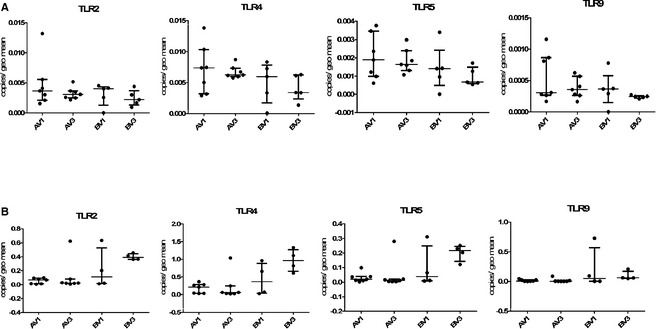
TLR expression in the duodenum (panels A) and colon (panels B) of 12 dogs participating in the clinical trial. V1 = pre‐treatment visit, V3 = after 6 weeks of dietary treatment and either synbiotic (labelled A on the x‐axis) or placebo (labelled B on the x‐axis) administration.
Figure 4.
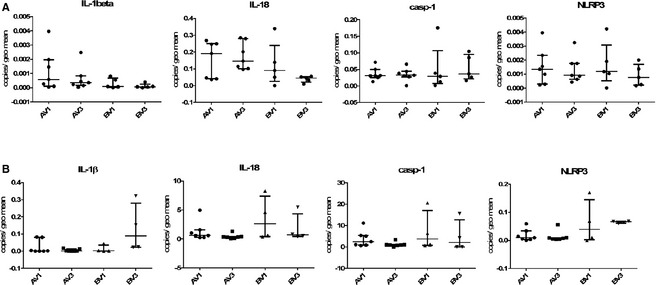
Expression of genes related to the inflammasome in the duodenum (panels A) and colon (panels B) of 12 dogs participating in the clinical trial. V1 = pre‐treatment visit, V3 = after 6 weeks of dietary treatment and either synbiotic (labelled A on the x‐axis) or placebo (labelled B on the x‐axis) administration.
There was no expression of IL‐17A in any duodenal sample, although RORC was expressed at a moderate level (Fig 5). In the colon, IL‐17A gene expression was detectable, but still negligible. Moderate levels of IL‐22 and IL‐23p19 and high levels of RORC mRNA expression were present in the colon (see Fig 6).
Figure 5.
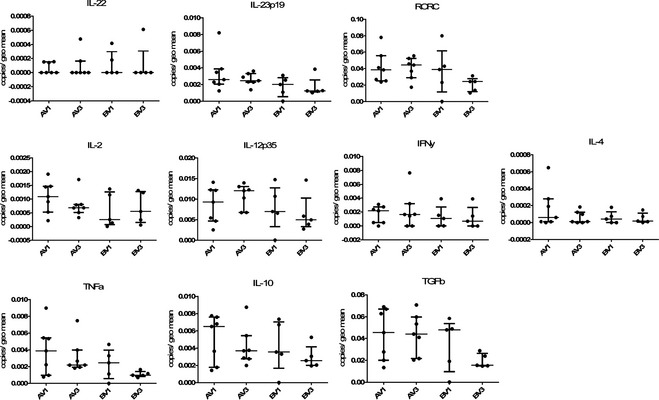
Relative gene expression of cytokines related to differen T helper cell lines in the duodenum of dogs with chronic enteropathy participating in the clinical trial. The upper panel shows factors related to Th17 cells (IL‐[Interleukin] 22, IL‐23p35 and the transcription factor RORC), the middle panel Th1 (IL‐2, IL‐12p35) and Th2 (IFNγ, IL‐4) cytokines, and the lower panel TNFα, which can be classed as innate cytokine or Th1 related, as well as IL‐10 and Transforming growth factor β (TGFβ) as representatives of cytokines produced by regulatory T cells. On the x‐axis groups represent different visits (V1 = before treatement, V3 = after 6 weeks of treatment) and different treatment groups: A = hydrolyzed protein diet plus synbiotic product containing Enterococcus faecium; B = hydrolyzed protein diet plus placebo.
Figure 6.
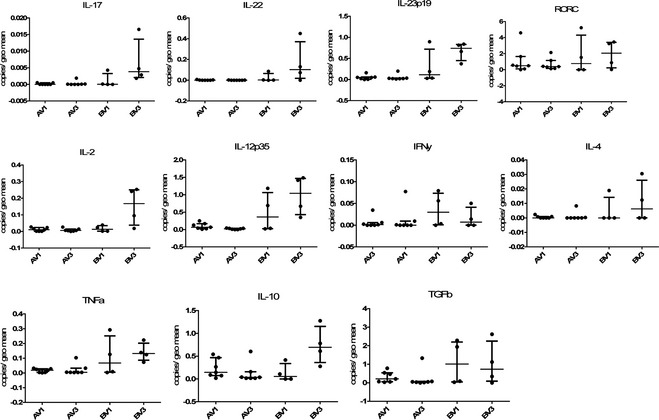
Relative gene expression of cytokines related to differen T helper cell lines in the colon of dogs with chronic enteropathy participating in the clinical trial. The upper panel shows factors related to Th17 cells (IL‐[Interleukin] 17, IL‐22, IL‐23p35 and the transcription factor RORC), the middle panel Th1 (IL‐2, IL‐12p35) and Th2 (IFNγ, IL‐4) cytokines, and the lower panel TNFα, which can be classed as innate cytokine or Th1 related, as well as IL‐10 and Transforming growth factor β (TGFβ) as representatives of cytokines produced by regulatory T cells. On the x‐axis groups represent different visits (V1 = before treatement, V3 = after 6 weeks of treatment) and different treatment groups: A = hydrolyzed protein diet plus synbiotic product containing Enterococcus faecium; B = hydrolyzed protein diet plus placebo.
As observed for other genes, expression levels of Th1 and Th2 cell‐related cytokines generally were higher in the colon than in the duodenum. The IL‐12p35 subunit was most abundantly expressed across all samples, followed by IFNy and TNFα. IL‐2 was expressed at low levels and IL‐4 showed the lowest expression (see Figs 5, 6).
Of the genes associated with Tregs, TGFβ showed the highest expression levels, followed by IL‐10, whereas FoxP3 was undetectable (Figs 5, 6).
TFF1 expression was very low in duodenal samples, whereas TFF3 was highly expressed. The nuclear receptor PPARγ showed decreased gene expression at visit 3 compared to visit 1 in the duodenum (possibly more pronounced in the placebo‐treated group than the synbiotic‐treated dogs), but these differences were not statistically significant.
For all of the examined genes, there was no significant difference in expression with regard to time (before versus after treatment) or treatment (synbiotic versus placebo).
Heatmaps of Gene Expression
When assessing all duodenal or colonic samples, there was no clear effect of visit or treatment on differences in gene expression, both by repeat linear mixed modeling including all gene expression data or by subjective analysis of the clustering created by the heatmaps and dendrograms (see Fig 7). Similarly, there was no effect of visit or treatment on differences in gene expression in the colon by linear mixed modeling, and the heatmap also showed no clustering (see Fig 7). Gene expression responses in both tissue types were very variable among individual dogs.
Figure 7.
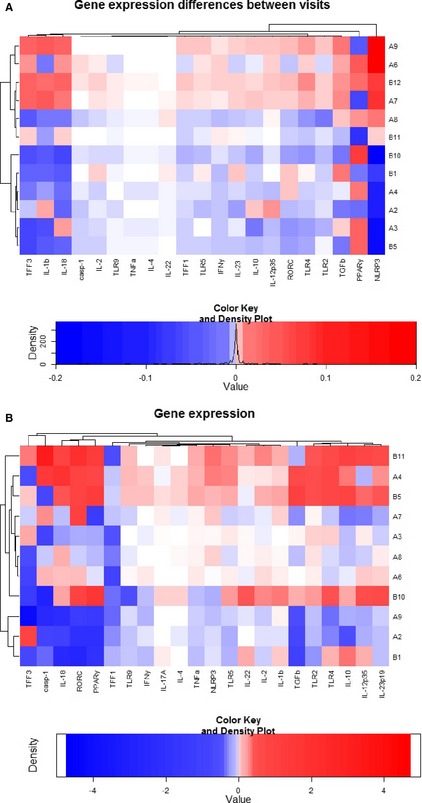
Heatmap and dendrograms depicting the difference in duodenal (A) and colonic (B) gene expression before and after treatment of 12 dogs with food‐responsive chronic enteropathy. Examined genes are listed on the x‐axis, dog IDs in numbers and with a reference to treatment (A = synbiotic; B = placebo) are shown on the y‐axis. Both clustering of genes and dogs was allowed in these heatmaps based on gene expression similarities. Red colour indicates an increase in gene expression from visit 1 (before treatment) to visit 3 (6 weeks after treatment), blue indicates a decrease as depicted by the colour key.
Discussion
The objective of the current study was to test the in vivo effects (clinical outcome and changes in gene expression in intestinal tissues) of the PO administration of a synbiotic containing the probiotic EF in dogs with FRD. Although the number of initially recruited dogs would have likely been sufficient to show a difference based on the power calculation, unfortunately, the drop‐out rate was considerable. Hence, no additional clinical benefit of the administration of the synbiotic could be demonstrated or excluded. Also, no significant differences in expression of the examined genes were observed between visit 1 and 3 or between treatment groups in the duodenum or colon.
The study population was consistent with the literature regarding breed distributaion and age of dogs with CE.5, 36, 37 Especially, dogs with FRD seem to be substantially younger than dogs suffering from other forms of CE.1, 38, 39 There was no significant correlation between CCECAI and histopathological scores, a finding that has been demonstrated before.36, 38 However, it is possible that the dietary trial was not conducted for a sufficient period of time for morphological changes to resolve.
Interestingly, in all dogs, serum folate concentrations increased above the reference range with dietary treatment alone. This was more pronounced in the placebo‐treated group than in the synbiotic group. Because exocrine pancreatic insufficiency had been eliminated in these dogs, this finding could be indicative of a change in bacterial composition induced by the dietary intervention, which could have been ameliorated by probiotic treatment. Complementary investigations into the composition of the intestinal microbiome before treatment unfortunately was not performed. Macroscopic endoscopy scores and WSAVA histology scores did not change significantly with treatment, which is consistent with the literature.1, 36, 40, 41
The gene expression study showed that certain receptors of the innate immune system are differentially expressed in the canine intestine (TLR2 and 4 are more abundant than TLR5 and 9). This, as well as the fact that TFF1 is expressed at low levels, whereas TFF3 is abundantly present, has been reported before.39, 42, 43, 44 The general notion that there is no Th cell lineage‐associated cytokine bias in biopsies from dogs with FRD also could be confirmed45 and was shown to be true for both the duodenum and the colon.
Most genes investigated were expressed at a higher level in the colonic mucosa compared to the duodenum. This is unlikely due to a better preservation or quality of colonic samples, because RNA quality and quantity were assessed for all biopsies and quantification of gene expression was performed relative to several reference genes that were assessed for their stability across samples. The increased gene expression levels in the large intestine could be associated with the larger number of bacteria present in the lumen compared to numbers in the small intestine. However, this also did not change with treatment for FRD or probiotic supplementation.
Limitations of the study included the fact that (despite all efforts) it remained underpowered. Therefore, results regarding the clinical benefit of EF in canine CE, as well as possible effects on gene expression, must be interpreted with caution. The clinical trial was designed to only include dogs with FRD and not other forms of CE. Including more severely affected dogs (with ARD or SRD) needing a variety of treatments would have required even larger numbers of dogs in each treatment group and might have made it more difficult to assess clinical improvement. Even if the desired 11 dogs had been recruited per treatment group, the study might have been underpowered. The choice of the appropriate data set from the literature to use for a power calculation is challenging, because usually a decrease of clinical indices of approximately 75% is considered consistent with remission.46 However, this has been demonstrated mostly in dogs with idiopathic IBD (i.e., SRD), which seem to have much higher disease activity compared to FRD dogs.1 Because the expected probiotic effect was unknown, power calculation was difficult to conduct.
Also, the dose of EF chosen to be administered to the dogs of this clinical trial was recommended by the manufacturer (1 × 109 CFU once daily). However, limited data are available to suggest the appropriate amount of that particular EF strain needed to induce clinical, immunological or microbiome alterations. Administration of 2–3 × 109 CFU/day of a different EF strain resulted in its persistence in feces for 3 months after cessation of the treatment.19 In addition, culture of feces before and after treatment with EF was not performed in this study, which potentially could have been used to ensure owner compliance of capsule administration. However, because EF can be part of the normal fecal flora and the probiotic strain cannot be differentiated from the naturally‐occurring EF strains, the additional benefit of fecal culture is questionable.
In conclusion, this study did not have enough power to enable a statement regarding a potential additional effect of EF (given as a synbiotic) on clinical signs or gene expression in intestinal biopsies in dogs with FRD within a time period of 6 weeks. More studies using a larger number of dogs or dogs with different forms of CE are needed to assess the benefit of probiotics in CE of dogs. Testing of different probiotic strains for their clinical or anti‐inflammatory properties in comparison with EF also should be considered.
Acknowledgments
The authors thank the owners of the dogs participating in this study for taking part, as well as the help of the staff of the Queen Mother Hospital for Animals and the Clinical Investigation Centre of the RVC for their support in recruiting cases and acquiring all necessary samples and data. Additional thanks go to Dr. Victoria Offord (Bioinformatics Unit of the RVC) for her help with using the statistical programme “R” and creating the heat maps. The study was supported by the BBSRC with a studentship with Probiotics International Ltd., Somerset, UK. The RVC manuscript approval number is CSS_00823.
Conflict of Interest Declaration: Authors disclose no conflict of interest.
Off‐label Antimicrobial Declaration: Authors declare no off‐label use of antimicrobials.
The work presented in this manuscript was performed at the Royal Veterinary College, London, UK.
This paper was presented as an oral abstract at the ECVIM Congress 2013 in Liverpool, UK.
Footnotes
Synbiotic D‐C; Probiotics International/ Protexin, Somerset, UK
Microsoft Office, Excel, Microsoft Cooperation, Redmond, USA
Purina Veterinary Diet canine HA Hypo Allergenic, Purina, Nestle, York, UK
RNAlater (Ambion, UK)
Qiagen (Manchester, UK)
Mixer Mill MM300 tissue grinder (Retch, Leeds, UK)
RNeasy micro kit (Qiagen)
Eukaryote Total RNA Nano chip, Agilent Bioanalyzer, Agilent Technologies, Wokingham, UK)
iScript cDNA synthesis kit (Bio‐Rad, Hemel Hampstead, UK)
qBase/GeNorm software (Biogazelle, Gent, Belgium)
PrimerDesign Ltd., Southampton, UK)
Immolase polymerase, Bioline, London, UK)
Wizard SV Gel and PCR clean‐up system (Promega)
pGemT‐Easy vector (Promega)
Source Bioscience, Cambridge, UK
SsoFast Evagreen Supermix (Bio‐Rad, Hemel Hampstead, UK)
The R project for statistical computing; www.r-project.org; “gplots” package; function heatmap.2
SPSS (IBM SPSS statistics, version 19.0)
Graph Pad Prism Version 5 (Graph Pad Software Inc., La Jolla, California, USA)
References
- 1. Allenspach K, Wieland B, Gröne A, Gaschen F. Chronic enteropathies in dogs: evaluation of risk factors for negative outcome. J Vet Intern Med 2007;21:700–708. [DOI] [PubMed] [Google Scholar]
- 2. Jergens AE, Moore FM, Haynes JS, Miles KG. Idiopathic inflammatory bowel disease in dogs and cats: 84 cases (1987–1990). J Am Vet Med Assoc 1992;201:1603–1608. [PubMed] [Google Scholar]
- 3. German AJ, Hall EJ, Day MJ. Chronic intestinal inflammation and intestinal disease in dogs. J Vet Intern Med 2003;17:8–20. [DOI] [PubMed] [Google Scholar]
- 4. Suchodolski JS, Xenoulis PG, Paddock CG, et al. Molecular analysis of the bacterial microbiota in duodenal biopsies from dogs with idiopathic inflammatory bowel disease. Vet Microbiol 2010;142:394–400. [DOI] [PubMed] [Google Scholar]
- 5. Suchodolski JS, Markel ME, Garcia‐Mazcorro JF, et al. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS ONE 2012;7:e51907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xenoulis PG, Palculict B, Allenspach K, et al. Molecular‐phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiol Ecol 2008;66:579–589. [DOI] [PubMed] [Google Scholar]
- 7. van Baarlen P, Troost FJ, van Hemert S, et al. Differential NF‐kappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc Natl Acad Sci USA 2009;106:2371–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Di Caro S, Tao H, Grillo A, et al. Effects of Lactobacillus GG on genes expression pattern in small bowel mucosa. Dig Liver Dis 2005;37:320–329. [DOI] [PubMed] [Google Scholar]
- 9. Foligne B, Zoumpopoulou G, Dewulf J, et al. A key role of dendritic cells in probiotic functionality. PLoS ONE 2007;2:e313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thomas CM, Versalovic J. Probiotics‐host communication: modulation of signaling pathways in the intestine. Gut Microbes 2010;1:148–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shida K, Nanno M, Nagata S. Flexible cytokine production by macrophages and T cells in response to probiotic bacteria: a possible mechanism by which probiotics exert multifunctional immune regulatory activities. Gut Microbes 2011;2:109–114. [DOI] [PubMed] [Google Scholar]
- 12. Bellavia M, Rappa F, Lo Bello M, et al. Lactobacillus casei and bifidobacterium lactis supplementation reduces tissue damage of intestinal mucosa and liver after 2,4,6‐trinitrobenzenesulfonic acid treatment in mice. J Biol Regul Homeost Agents 2014;28:251–261. [PubMed] [Google Scholar]
- 13. Jeon SG, Kayama H, Ueda Y, et al. Probiotic Bifidobacterium breve induces IL‐10‐producing Tr1 cells in the colon. PLoS Pathog 2012;8:e1002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Linskens RK, Huijsdens XW, Savelkoul PH, et al. The bacterial flora in inflammatory bowel disease: current insights in pathogenesis and the influence of antibiotics and probiotics. Scand J Gastroenterol Suppl 2001;234:29–40. [DOI] [PubMed] [Google Scholar]
- 15. Sauter SN, Benyacoub J, Allenspach K, et al. Effects of probiotic bacteria in dogs with food responsive diarrhoea treated with an elimination diet. J Anim Physiol Anim Nutr (Berl) 2006;90:269–277. [DOI] [PubMed] [Google Scholar]
- 16. Rossi G, Pengo G, Caldin M, et al. Comparison of microbiological, histological, and immunomodulatory parameters in response to treatment with either combination therapy with prednisone and metronidazole or probiotic VSL#3 strains in dogs with idiopathic inflammatory bowel disease. PLoS ONE 2014;9:e94699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti‐inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 2008;105:16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marcináková M, Klingberg TD, Lauková A, Budde BB. The effect of pH, bile and calcium on the adhesion ability of probiotic enterococci of animal origin to the porcine jejunal epithelial cell line IPEC‐J2. Anaerobe 2010;16:120–124. [DOI] [PubMed] [Google Scholar]
- 19. Marcináková M, Simonová M, Strompfová V, Lauková A. Oral application of Enterococcus faecium strain EE3 in healthy dogs. Folia Microbiol. (Praha) 2006;51:239–242. [DOI] [PubMed] [Google Scholar]
- 20. Washabau RJ, Day MJ, Willard MD, et al. Endoscopic, biopsy, and histopathologic guidelines for the evaluation of gastrointestinal inflammation in companion animals. J Vet Intern Med 2010;24:10–26. [DOI] [PubMed] [Google Scholar]
- 21. Day MJ, Bilzer T, Mansell J, et al. Histopathological standards for the diagnosis of gastrointestinal inflammation in endoscopic biopsy samples from the dog and cat: a report from the World Small Animal Veterinary Association Gastrointestinal Standardization Group. J Comp Pathol 2008;138(Suppl 1):S1–S43. [DOI] [PubMed] [Google Scholar]
- 22. Procoli F, Mõtsküla PF, Keyte SV, et al. Comparison of histopathologic findings in duodenal and ileal endoscopic biopsies in dogs with chronic small intestinal enteropathies. J Vet Intern Med 2013;27:268–274. [DOI] [PubMed] [Google Scholar]
- 23. Schmitz S, Henrich M, Neiger R, et al. Stimulation of duodenal biopsies and whole blood from dogs with food‐responsive chronic enteropathy and healthy dogs with toll‐like receptor ligands and probiotic Enterococcus faecium . Scand J Immunol 2014;80:85–94. [DOI] [PubMed] [Google Scholar]
- 24. Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000;132:365–386. [DOI] [PubMed] [Google Scholar]
- 25. Vandesompele J, de Preter K, Pattyn F, et al. Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abreu MT. Toll‐like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol 2010;10:133–141. [DOI] [PubMed] [Google Scholar]
- 27. Tyrer P, Foxwell AR, Cripps AW, et al. Microbial pattern recognition receptors mediate M‐cell uptake of a gram negative bacterium. Infect Immun 2006;74:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cario E. Heads up! How the intestinel epithelium safeguards mucosal barrier immunity through the inflammasome and beyond. Curr Opin Gastroenterol 2010;26:583–590. [DOI] [PubMed] [Google Scholar]
- 29. Hirota K, Martin B, Veldhoen M. Development, regulation and functional capacities of Th17 cells. Semin Immunopathol 2010;32:3–16. [DOI] [PubMed] [Google Scholar]
- 30. Kanai T, Mikanmi Y, Sujuno T, et al. RORyt‐dependent IL‐17A‐producing cells in the pathogenesis of intestinal inflammation. Mucosal Immunol 2012;5:240–247. [DOI] [PubMed] [Google Scholar]
- 31. Mosmann TR, Coffman RL. Th1 and Th2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol 1989;7:145–173. [DOI] [PubMed] [Google Scholar]
- 32. Fontenot JD, Gavin MA, Rudensky AY. FoxP3 programs the development and function of CD4 + CD25 + regulatory T‐cells. Nat Immunol 2003;4:330–336. [DOI] [PubMed] [Google Scholar]
- 33. Thim L. Trefoil peptides: from structure to function. Cell Mol Life Sci 1997;53:888–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wahli W, Michalik L. PPARs at the crossroads of lipid signalling and inflammation. Trends Endocrinol Metab 2012;23:351–363. [DOI] [PubMed] [Google Scholar]
- 35. Are A, Aronsson L, Wang S, et al. Enterococcus faecalis from newborn babies regulates endogenous PPARgamma activity and IL‐10 levels in colonic epithelial cells. Proc Natl Acad Sci USA 2008;106:1943–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schreiner NMS, Gaschen F, Gröne A, et al. Clinical signs, histology, and CD3‐positive cells before and after treatment of dogs with chronic enteropathies. J Vet Intern Med 2008;22:1079–1083. [DOI] [PubMed] [Google Scholar]
- 37. Mandigers PJJ, Biourge V, van den Ingh TSGAM, et al. A randomized, open‐label, positively‐controlled field trial of a hydrolyzed protein diet in dogs with chronic small bowel enteropathy. J Vet Intern Med 2010;24:1350–1357. [DOI] [PubMed] [Google Scholar]
- 38. Walker D, Knuchel‐Takano A, McCutchan A, et al. A comprehensive pathological survey of duodenal biopsies from dogs with diet‐responsive chronic enteropathy. J Vet Intern Med 2013;27:862–874. [DOI] [PubMed] [Google Scholar]
- 39. Allenspach K, House A, Smith K, et al. Evaluation of mucosal bacteria and histopathology, clinical disease activity and expression of Toll‐like receptors in German shepherd dogs with chronic enteropathies. Vet Microbiol 2010;146:326–335. [DOI] [PubMed] [Google Scholar]
- 40. Luckschander N, Allenspach K, Hall J, et al. Perinuclear antineutrophilic cytoplasmic antibody and response to treatment in diarrheic dogs with food responsive disease or inflammatory bowel disease. J Vet Intern Med 2006;20:221–227. [DOI] [PubMed] [Google Scholar]
- 41. Gaschen L, Kircher P, Stüssi A, et al. Comparison of ultrasonographic findings with clinical activity index (CIBDAI) and diagnosis in dogs with chronic enteropathies. Vet Radiol Ultrasound 2008;49:656–664. [DOI] [PubMed] [Google Scholar]
- 42. McMahon LA, House AK, Catchpole B, et al. Expression of Toll‐like receptor 2 in duodenal biopsies from dogs with inflammatory bowel disease is associated with severity of disease. Vet Immunol Immunopathol 2010;135:158–163. [DOI] [PubMed] [Google Scholar]
- 43. Burgener IA, König A, Allenspach K, et al. Upregulation of toll‐like receptors in chronic enteropathies in dogs. J Vet Intern Med 2008;22:553–560. [DOI] [PubMed] [Google Scholar]
- 44. Schmitz S, Hill S, Werling D, Allenspach K. Expression of trefoil factor genes in the duodenum and colon of dogs with inflammatory bowel disease and healthy dogs. Vet Immunol Immunopathol 2013;151:168–172. [DOI] [PubMed] [Google Scholar]
- 45. Jergens AE, Sonea IM, O'Connor AM, et al. Intestinal cytokine mRNA expression in canine inflammatory bowel disease: a meta‐analysis with critical appraisal. Comp Med 2009;59:153–162. [PMC free article] [PubMed] [Google Scholar]
- 46. Jergens AE, Crandell J, Morrison JA, et al. Comparison of oral prednisone and prednisone combined with metronidazole for induction therapy of canine inflammatory bowel disease: a randomized‐controlled trial. J Vet Intern Med 2010;24:269–277. [DOI] [PubMed] [Google Scholar]


