Abstract
Background
The gastrointestinal (GI) microbiota has a strong impact on the health of cats and these populations can be altered in GI disease. Little research has been done to associate improvement in diarrhea with changes in GI microbiota.
Objective
To evaluate GI microbiota changes associated with diet change and related improvement in diarrhea in cats with chronic naturally occurring diarrhea.
Animals
Fifteen adult Domestic Shorthair cats with naturally occurring chronic diarrhea.
Methods
Controlled crossover dietary trial for management of diarrhea. Fecal microbiome was assessed using 454‐pyrosequencing. Relationships among fecal score (FS), diet, and microbiome were explored using partial least square method, partial least square method – discriminant analysis, and orthogonal partial least square method with discriminant analysis (OPLS‐DA).
Results
Dominant bacterial phyla included the Firmicutes and Bacteroidetes, followed by Fusobacteria, Proteobacteria, Tenericutes, and Actinobacteria. Orthogonal partial least squares (OPLS‐DA) clustering showed significant microbial differences within cats when fed Diet X versus Diet Y, and with Diet Y versus baseline. Significant correlations were found between the microbiome and FSs. Those bacteria with the strongest correlation with FS included Coriobacteriaceae Slackia spp., Campylobacter upsaliensis, Enterobacteriaceae Raoultella spp., Coriobacteriaceae Collinsella spp., and bacteria of unidentified genera within the families of Clostridiales Lachnospiracea and Aeromonadales Succinivibrionacease, suggesting that increased numbers of these organisms may be important to gut health.
Conclusions and Clinical Importance
Alterations in intestinal microbiota were associated with improvement in diarrhea, but, from our data we cannot conclude if changes in the microbiome caused the improvement in diarrhea, or vice versa.
Keywords: Gastroenterology, Nutrition
Abbreviations
- FS
fecal score
- GI
gastrointestinal
- OPLS‐DA
orthogonal partial least square method with discriminant analysis
- OTU
operational taxonomic unit
- PCA
principal components analysis
- PLS‐DA
partial least square method – discriminant analysis
- PLS
partial least square method
The gastrointestinal (GI) microbiota has a strong impact on the health of cats and dogs,1 and the microbiome can be altered in GI disease.2, 3, 4 Prior research has confirmed alterations in intestinal microbiota in cats with GI disease.5, 6 Documented changes include increases in Clostridium spp., and decreases in Bidifobacterium spp., Lactobacillus spp. and Bacteriodes spp.,6, 7, 8 all of which may be influenced by dietary characteristics including fermentable fiber.3, 9, 10, 11, 12, 13 Studies have shown that dietary changes can result in clinical improvement in diarrhea in cats and dogs.14, 15, 16, 17, 18, 19, 20 In otherwise healthy dogs, dietary‐induced changes were associated with altered microbiota, altered fecal quality, or both.21, 22 However, comprehensive studies are lacking to determine if clinical improvement in cats with diarrhea is associated with changes in the GI microbiome.
Metagenomics is the analysis of genomic patterns of entire communities of microbes, such as the intestinal microbiome. The ability to perform such analyses using 454‐pyrosequencing, which allows accurate and quantitative analysis of DNA, provides a more comprehensive view of the microbiome without the limitations of culture methods.23, 24 Although there is evidence that the intestinal microbiota differs along the GI tract, fecal samples are more readily available for clinical studies, and alterations in fecal microflora have been shown to occur in cats with diarrhea.6, 25, 26 In this study, 16S rRNA sequence data were analyzed using 454‐pyrosequencing to characterize the fecal microbiome in cats with chronic diarrhea before and after response to dietary treatment. Our objectives were to characterize the phylogeny of the feline fecal microbiome, assess the changes induced by the therapeutic diets, and to correlate these microbial community changes with clinical improvement in diarrhea assessed by fecal scores (FS).
Materials and Methods
Animals
Freshly voided fecal samples were collected from adult Domestic Shorthair cats undergoing a controlled, crossover clinical trial for the dietary management of chronic diarrhea. The clinical design and other aspects of this study have been previously reported.20 Briefly, cats with naturally occurring diarrhea lasting at least 3 months were identified among cats at the Nestlé Purina Pet Care Center in Missouri, USA. Diarrhea was defined as a FS of 6 or 7 using a 7‐point scoring system where 1 = extremely dry and firm, 2–3 = normal stools, and 7 = very watery.20 Cats were excluded from the study if they had received any treatment for diarrhea in the 6 weeks before initiation of the study or if they had evidence of intestinal parasites, infectious disease, or a systemic disease that may cause diarrhea.
Study Design
The study protocol was approved by the Nestlé Purina Institutional Animal Care and Use Committee. Sixteen cats were selected and were individually housed. Once cats were assigned to housing, the units were arbitrarily divided into 2 equal‐sized groups. In order to minimize effects caused by differences in the cats’ previous diets and to adapt all cats to a canned food diet, all cats entered into the study were fed the same canned maintenance diet1 during a 2‐week baseline period (Baseline). This period was considered sufficient because prior studies documented that if cats with diarrhea respond to dietary change, they usually do so within 1–2 weeks.15, 16, 19 Two cats refused to eat the canned diet and were replaced during the baseline period. One group of cats then was fed Diet X2 whereas the other group was fed Diet Y3 as their sole diet for 1 month (Period 1), after which they were switched to the alternate diet for an additional month (Period 2). Cats were individually fed once daily based on daily energy requirements and water was available ad libitum. Data collection occurred during the last week on each diet. FSs were assigned daily by 1 of 3 technicians trained in fecal scoring, with each feces being scored separately. During each of the last 3 days of each period, freshly voided fecal samples were collected, mixed with 10% w/w glycerol, flushed with CO2, and frozen at −80°C until analysis. Collection and processing of fecal samples were performed within 15 min of defecation.
Extraction of DNA and Metagenomic 454‐Pyrosequencing
Genomic DNA was extracted from fecal samples using the method of Tsai and Olson.27 Briefly, fecal samples were thawed in ice, weighed and suspended in phosphate‐buffered saline (0.85% NaCl, 120 mM NaH2PO4, pH = 8.0) then centrifuged at 5525 × g for 10 min. After discarding the supernatant, the pellets were resuspended in 0.5 mL lysis solution (0.15 M NaCl, 0.1 M EDTA, pH = 8.0) containing 15 mg/mL lysozyme and incubated for 30 min with constant agitation in a 37°C shaking water bath before, and another 30 min after, addition of 0.5 mL of sodium Tris sodium‐dedocyl‐sulfate solution (0.1 M NaCl, 0.48 M Tris HCl [pH = 8.0], 10% sodium‐dedocyl‐sulfate, pH = 8.0). Three freeze‐thaw cycles were performed to break open the bacterial cell walls. After the third thaw, Proteinase K4 was added to each sample to a final concentration of 50 μg/mL and samples were again incubated in a 37°C shaking water bath for 30 min with constant agitation, then centrifuged at 17,500 × g for 20 min in a refrigerated microcentrifuge. Without disturbing the pellet, 850 μL of the supernatant was removed and placed into a clean tube on ice. Samples remained on ice for the remainder of the processing. Samples were mixed with an equal volume of phenol (pH = 7.9), then centrifuged for 6 min at 16,500 × g. An equal volume of phenol/chloroform/isoamyl alcohol (25 : 24 : 1) was mixed with 700 μL supernatant, then again centrifuged for 6 min at 16,500 × g. This supernatant (550 μL) was mixed with an equal volume of chloroform/isoamyl alcohol (24 : 1) then centrifuged at 16,500 × g for 6 min. A 400 μL aliquot of this supernatant was mixed with 400 μL ice cold isopropanol and 96 μL 10.5 M ammonium acetate (1.125 M final concentration), then frozen at −80°C freezer overnight or until processing (up to 2 weeks).
Samples were thawed on ice for ~5–10 min before further processing. DNA pellets were obtained by centrifuging at 17,500 × g for 20 min in a refrigerated microcentrifuge. The supernatant was removed and pellets were washed with 500 μL 70% ethanol, then dried. The pellets were resuspended in 200 μL Tris‐EDTA buffer (10 mM Tris HCl [pH = 8.0], 1 mM EDTA, pH = 8.0). DNA was quantified by Quant‐It.5 All samples were then stored at −80°C until further analysis.
Metagenomic 16S rRNA pyrosequencing was performed by Core for Applied Genomics and Ecology.6 The V1–V2 region of the 16S rRNA gene was amplified using bar‐coded fusion primers with the Roche‐454 A or B titanium sequencing adapters, followed by a unique 8‐base barcode sequence (B) and finally the 5′ ends of primer A‐8FM (5′‐CCATCTCATCCCTGCGTGTCTCCGACTCAGBBBBBBBBAGAGTTTGATCMTGGCTCAG) or primer B‐357R (5′‐CCTATCCCCTGTGTGCCTTGGCAGTCTCAGBBBBBBBBCTGCTGCCTYCCGTA‐3′). All PCR reactions were quality controlled for amplicon saturation by gel electrophoresis; band intensity was quantified against standards using GeneTools7 image analysis software. For each region of a 2‐region picotiter plate, amplicons from 48 reactions were pooled in equal amounts and gel purified. The resulting products were quantified using PicoGreen8 and a Qubit fluorometer9 before sequencing using a Roche‐454 GS‐FLX Pyrosequencer.10
Data Processing Pipeline
The raw data from 454‐pyrosequencing were processed using QIIME.28 Data were filtered to remove low‐quality reads not meeting the following quality criteria: (1) a complete barcode sequence immediately followed by a forward primer sequence, with no mismatch in either barcode or primer sequence; (2) read lengths between 200 and 1,000 bases; (3) average quality score of 25 or higher in a sliding window of 50 bases; and (4) maximum homopolymer run of 6. Processed reads were then demultiplexed into barcode‐indexed sample categories. The barcode, forward primer, and reverse primer were subsequently trimmed from each read. This yielded a total of 556,366 reads from 48 samples with an average of 11,591 reads per sample. The average length for the reads was 358 bases.
Reads were clustered into operational taxonomic units (OTU) using a reference‐based UCLUST algorithm at a 97% sequence similarity level.29 The reference data file was obtained from the greengenes website (http://greengenes.lbl.gov, 2011 release). A consensus taxonomic lineage was assigned to each OTU using the ribosomal database project (RDP) naïve Bayesian classifier30 at a minimum confidence interval of 0.8. The RDP classifier was retrained using the greengenes taxonomy. Finally, an OTU table was constructed with proper taxonomic identifications for each OTU.
Statistical Data Analysis
Mean fecal scores (FSm) for each cat were determined by averaging all of that cat's scores recorded during the last 7 days of each study period. FSs were not affected by feeding order (period), so data from both periods were pooled. The relationships between FSm, diet and microbiome were explored using partial least square method (PLS), partial least square method – discriminant analysis (PLS‐DA), and orthogonal partial least square method with discriminant analysis (OPLS‐DA) using SIMCA‐P+11 and MATLAB12 routines. PLS is a chemometrics method used to measure quantitative relationships between 2 data sets where both are matrices, usually comprising spectral data such as calibration samples (X), and a set of quantitative values (Y). For PLS‐DA, the Y matrix contains qualitative values (eg, presence of diarrhea or treatment group). OPLS‐DA is a multivariate method used to eliminate extraneous variance from the X data matrix that is unrelated to class, in this case, FSs. Having removed the extraneous variation (eg, intersubject age, sex), the OPLS‐DA models enhance the predictive ability of the model and simplify interpretation.31, 32 A standard 7‐fold cross‐ validation method was applied to establish the robustness of the models. In OPLS models, R2X, R2Y, and Q2 are reported. The values of R2X and R2Y show how much of the variation in the datasets X and Y, respectively, are explained by the model. The cross‐validation parameter, Q2 (which can range from −1 to +1), represents the predictability of the models and is used to test the validity of the model against overfitting. A positive Q2 value indicates that differences between groups are statistically significant, whereas a negative Q2 value indicates no significant relationship among the data.
Principal components analysis (PCA) and pairwise discriminant analysis were applied to sample classes (diets) with unit variance scaling (each parameter has a mean of 0 and a variance of 1). This approach filters out metagenomic information that is not correlated with the predefined classes whereas the loadings yield information on which bacterial signals are associated with the observed clustering, thus giving a means for metagenomics interpretation. Pairwise OPLS‐DA models were generated with 1 predictive component, and 2 orthogonal components to discriminate among the 3 diets. Models were calculated using family, genus, and species levels. OPLS‐DA plots were generated only to species level.
Results
One cat was withdrawn from the study for unrelated medical reasons. Fifteen cats (13 neutered males, 2 spayed females; mean age, 10 years [range, 6–17 years]) completed all phases of the study, as previously described.20 Most cats had signs consistent with either large bowel (n = 7) or mixed large and small bowel (n = 7) diarrhea. Mean FS improved (P < .01) from baseline (5.5 ± 0.3) after both Diet X (5.0 ± 0.4 and 4.4 ± 0.4 for periods 1 and 2, respectively) and Diet Y (4.0 ± 0.6 and 4.2 ± 0.5 for periods 1 and 2, respectively), with significantly greater improvement after Diet Y (P < .01). Changes in FS in response to diet were not affected by the order in which the diets were fed (P = .65). Individual FS improved at least 1 unit in 40% of the cats while fed Diet X, and in 67% of the cats while fed Diet Y, resulting in normal stools (FS ≤ 3) in 13.3% of cats fed Diet X and in 46.7% of cats fed Diet Y.20
Pyrosequencing of fecal samples detected 8 phyla, 14 classes, 25 orders, 47 families, 96 genera, and 146 species of bacteria. Firmicutes, Bacteroidetes, Fusobacteria, Proteobacteria, Tenericutes, and Actinobacteria were the most abundant phyla in these cats (Table 1). The most prevalent bacterial classes were Bacilli and Clostridia among the Firmicutes and Bacteroidia and Flavobacteria among the Bacteroidetes. Prevotella was the most abundant genus in fecal samples regardless of diet consumed, with mean of 24% across all samples, followed by Fusobacterium (9%), unclassified Fusobacteriaceae (9%), Clostridium (8%), and Streptococcus (6%).
Table 1.
The phylum level composition of the fecal microbiota in cats with chronic diarrhea fed different diets. This analysis was computed using both V1–V2 regions
| Phylum | Total | Baseline | Diet X | Diet Y |
|---|---|---|---|---|
| Percent of bacteria | ||||
| Firmicutes | 34.34 | 33.72 | 35.65 | 33.82 |
| Bacteroidetes | 30.05 | 33.54 | 30.16 | 26.13 |
| Fusobacteria | 18.81 | 14.66 | 16.57 | 25.43 |
| Proteobacteria | 7.66 | 9.14 | 7.52 | 6.16 |
| Tenericutes | 6.56 | 6.21 | 7.45 | 6.12 |
| Actinobacteria | 2.57 | 2.72 | 2.65 | 2.33 |
| Cyanobacteria | 0.0034 | 0.0036 | 0.006 | 0.0008 |
| TM7 | 0.0003 | 0.0007 | 0 | 0 |
The OPLS‐DA models showed significant differences in the fecal microbiome of cats when fed Diet Y versus baseline at the family (Q2 = 0.126), genus (Q2 = 0.399), and species (Q2 = 0.61, Fig 1A) level. Likewise, significant but smaller differences were noted between cats fed Diet X versus Diet Y at the family (Q2 = 0.0588), genus (Q2 = 0.127), and species (Q2 = 0.197, Fig 1B) levels. There were no differences after Diet X compared to baseline. OPLS‐DA clustering showed the greatest microbial differences in cats when fed Diet Y versus baseline (Q2 = 0.61).
Figure 1.
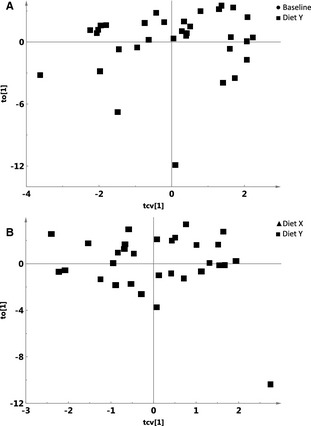
Orthogonal partial least square with discriminant analysis (OPLS‐DA) plot, showing results of multivariate analysis of the effects of dietary changes on fecal microbiota at the species level. Data were visualized by means of component scores plots, where each point represents an individual metagenomic profile of a sample. The score matrix (tcv and to) represent projections onto the latent variables of the OPLS‐DA model. (A) Scores plot showing significant differences (P < .01) between Diet Y versus Baseline; and (B) Scores plot showing significant differences (P < .01) between Diet Y versus Diet X.
Significant changes in bacterial populations occurred in cats fed diet Y versus baseline or diet X (Fig 2). For example, the species Clostridium perfringens, Prevotella copri, Plesiomonas shigelloides, Helicobacter cinaedi, Lactobacillus helveticus, and Bacteroides fragilis and others were decreased and Ruminococcus gnavus, Streptococcus suis and Eubacterium dolichum were increased in cats fed diet Y compared to baseline. Other unidentified species in various genera also changed in cats fed diet Y. Some of these alterations also were found with diet X but not to the same extent, consistent with the observation that Diet X was intermediate to Diet Y regarding improvement in FS.
Figure 2.
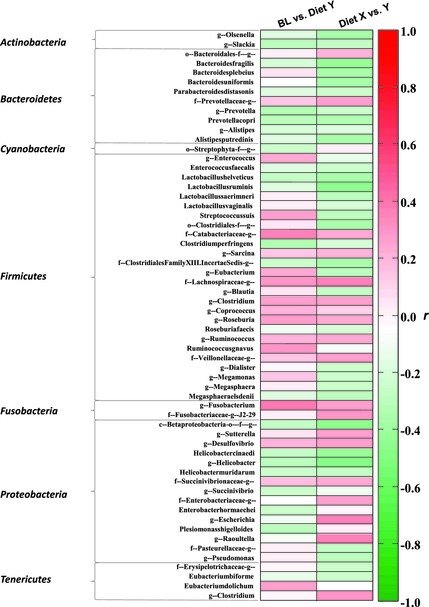
Heat map plot of significant bacterial microbiota. The heat plot (r range from −1 to +1) indicates the abundance of bacteria that were up‐ or down‐regulated in cats after eating each diet. Red corresponds to bacteria that are up‐regulated in Diet Y (high positive r correlation value) and green corresponds to bacteria that are down‐regulated in Diet Y (high negative r correlation value) which resulted from the OPLS‐DA model. The bacterial genera and species were grouped according to their phylum level.
Using OPLS‐based evaluations, significant correlations between microbiome and FS were found for cats fed Diet Y at the genus (Q2 = 0.464) and species (Q2 = 0.115) levels, as well as for cats fed Diet X at family (Q2 = 0.232), genus (Q2 = 0.736), and species (Q2 = 0.717) levels. Prediction plots generated from the OPLS data at the species level, wherein FSs were predicted based on the microbiome, showed strong correlations between predicted and actual FS (r 2 = 1.0 and 0.6, P < .005, for Diets X and Diet Y, respectively). OPLS regression identified 63 different bacteria with significant negative or positive correlations (r > ±0.2; P ≤ .05) to FS in cats consuming Diets X or Y, although they differed between diets (Table S1: on‐line supplement) Those organisms most strongly correlated (r > ±0.70) with FS were within cats while fed Diet Y, and included Coriobacteriaceae Slackia spp. (r = −0.83), Campylobacter upsaliensis (r = −0.79), Enterobacteriaceae Raoultella spp. (r = −0.72), and bacteria of unidentified genera within the families of Clostridiales Lachnospiracea (r = −0.76), and Aeromonadales Succinivibrionacease (r = −0.71).
Discussion
To our knowledge, this study is one of the first to apply “next generation” pyrosequencing to characterize the hindgut microbiome of cats with chronic diarrhea, and to show changes in the microbiome with improvement in diarrhea associated with dietary management. The data showed strong correlations between FS after consumption of therapeutic diets and several organisms, generating a model that accurately predicted the FS of these cats based on the microbiota. Those bacteria with the strongest correlation with FS‐included Coriobacteriaceae Slackia spp., Campylobacter upsaliensis, Enterobacteriaceae Raoultella spp., Coriobacteriaceae Collinsella spp., and bacteria of unidentified genera within the families of Clostridiales Lachnospiracea and Aeromonadales Succinivibrionacease, suggesting that increased numbers of these organisms may be important to gut health.
In this study, we detected the presence of 8 bacterial phyla. Dominant bacterial phyla included the Firmicutes and Bacteroidetes, both of which comprised 30–34% of all sequences. Firmicutes includes, among many others, the commonly recognized genera Lactobacillus, Clostridium, and Enterococcus, whereas Bacteroidetes includes Helicobacteria, Escherichia, Pasturella, and Pseudomonas among its genera. These were followed in abundance by Fusobacteria (18.8%), Proteobacteria (7.7%), Tenericutes (6.6%), and Actinobacteria (2.6%). The proportions were similar across all diets with the exception that Fusobacteria were higher in cats fed Diet Y. Although the specific abundance of Firmicutes in this study is less, and the abundance of Bacteroidetes and Tenericutes is more than reported by others, our results are consistent with most other studies using nonculture methods of evaluation, indicating that the dominant phyla in cat feces are Firmicutes, Bacteriodetes, Proteobacteria, Fusobacteria, and Actinobacteria.24, 25, 26, 33, 34 In contrast, Tun et al. reported that Bacteroidetes were the most prevalent in cats (68%), followed by Firmicutes (13%) and Proteobacteria (6%).35 The reported differences in specific abundance among these phyla may be related to differences in diets consumed, or differences in methods and the use of bacterial primers that target certain hypervariable regions of the 16S rRNA. The selection of bacterial primers of the 16S rRNA is an important factor in any metagenomics study because some primers could underestimate or overestimate certain classes of bacteria.24, 26, 36 The large number of Tenericutes relative to prior papers may be related to recent changes in taxonomy, with many genera in the Tenericutes phylum previously classified as Firmicutes.37, 38 However, the differences between results from this study and prior studies may reflect true differences in bacterial populations associated with naturally occurring chronic diarrhea.
The microbiota of cats at baseline with active diarrhea and of cats after being fed Diet X with partial resolution of diarrhea were very similar. However, samples collected after Diet Y, which was associated with greater improvement in diarrhea, showed significant differences compared to baseline and compared to Diet X. Mostly, these changes were in the same direction; ie, the abundance was increased or decreased compared to both baseline and Diet X. Among the few exceptions, Eubacterium spp., Enterococcus spp. and Streptococcus suis all were increased after Diet Y compared to baseline but decreased compared to Diet X.
Many of the organisms frequently identified using culture methods, such as Lactobacillus spp., Bifidobacterum, C. perfringens, and Escherichia coli, showed weak or no correlation with fecal quality in this study. C. perfringens, however, was significantly decreased in cats fed Diet Y compared to baseline and compared to Diet X. Previous work had shown a positive correlation between increased dietary protein and Clostridium, including C. perfringens.13, 39 In this study, Diet Y contained more protein than Diet X, so factors other than dietary protein also affect fecal C. perfringens. Some Lactobacillus species, such as L. helveticus, also were increased in cats fed Diet Y. Desulfovibrio spp., an organism previously shown to be increased in cats with inflammatory bowel disease,6 also was increased in cats fed Diet Y compared to baseline and Diet X. However, there was no correlation between this organism and FS in the cats in this study.
There were some limitations to this study. It was a clinical study evaluating cats with chronic, naturally occurring diarrhea, and only 15 cats were available for the study. The use of a crossover design increased the power of the study and helped overcome the inherent variation in individual differences in microbiota. Cats were given 3 weeks to adapt to each diet before beginning the sampling period, which previously has been documented as sufficient for a stable response to dietary change in cats with diarrhea,15, 16, 19 but there was not a washout or return to baseline between dietary treatments. When planning the study, it was assumed that there would be no carryover effect because cats with diet‐responsive diarrhea usually respond within 1–2 weeks and a 3‐week adaptation time during each phase was allowed before initiating sample collections.16, 20 The lack of washout did not appear to impact the results because the order in which the diets were fed did not have a significant effect on the results. Cats had poor FS at the beginning of the study which improved after consuming the therapeutic diets, but based on the study design it is not possible to differentiate the effects of diet on the microbiome from the effects of improvement in diarrhea.
Conclusions
In this study, we quantified variations in the composition of hindgut microbiota in cats with chronic diarrhea and after clinical improvement while being fed 2 therapeutic diets. The bacterial phyla Firmicutes and Bacteroidetes each comprised 30–34% of all sequences in this study, which is less for Firmicutes and more for Bacteroidetes compared to most other studies. It is not known if the differences are because of methodology or because of health or dietary effects. Those bacteria with the strongest correlation with FS included Coriobacteriaceae Slackia spp., Campylobacter upsaliensis, Enterobacteriaceae Raoultella spp., Coriobacteriaceae Collinsella spp., and bacteria of unidentified genera within the families of Clostridiales Lachnospiracea and Aeromonadales Succinivibrionacease, suggesting that increased numbers of these organisms may be important to gut health. Finally, from our data we cannot conclude if these changes in the microbiome caused the improvement in diarrhea or were a result of either the diet or the improved fecal quality.
Supporting information
Table S1. Fecal microbiota, determined by 454 pyrosequencing that were significantly (P < 0.05) correlated with fecal scoresa from cats with naturally occurring chronic diarrhea after being fed therapeutic diet Diet Xb or Diet Yc for 4 weeks. Bacteria are sorted according to their phylum.
Acknowledgments
The authors thank the staff veterinarians and technicians at the Nestlé Purina Petcare Center for assistance with data collection and animal care. This study was supported by the Nestlé Research Center – St. Louis.
Conflict of Interest: This project was funded and conducted by Nestlé Purina PetCare, St. Louis, MO 63164. All authors are full‐time employees of this company.
This research was conducted at the Nestlé Purina Pet Care Center in Missouri and the Nestlé Research Center in St. Louis, MO between 2008 and 2009.
This study was presented in abstract form at the 2013 ACVIM Forum, Seattle, WA.
Footnotes
Baseline maintenance diet: Fancy Feast® Savory Salmon Feast Cat food, Nestlé Purina PetCare Company, St. Louis, MO
Diet X: Hill's® Prescription Diet® i/d® Feline, Hill's Pet Nutrition Inc, Topeka, KS
Diet Y: Purina Veterinary Diets® EN Gastroenteric® brand Feline Formula, Nestlé Purina PetCare Company
Proteinase K: Fisher BioReagents # BP1700‐500, Fisher Scientific, Pittsburg, PA
Quant‐It: Invitrogen Quant‐It kit, dsDNA, broad range, Life Technologies, Grand Island, NY
Core for Applied Genomics and Ecology, University of Nebraska‐Lincoln, Lincoln, NE
GeneTools image analysis software, Syngene, Frederick, MD
PicoGreen: Invitrogen, Life Technologies
Qubit fluorometerg: Invitrogen, Life Technologies
Roche‐454 GS‐FLX Pyrosequencer: 454 Life Sciences, a Roche company, Branford, CT
SIMCA‐P+, verion 12.0.1.0: Umetrics AB, Umeå, Sweden
MATLAB: MathWorks Inc, Natick, MA
References
- 1. Bell JA, Kopper JJ, Turnbull JA, et al. Ecological characterization of the colonic microbiota of normal and diarrheic dogs. Interdiscip Perspect Infect Dis 2008;2008:149694. doi: 10.1155/2008/149694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blaut M. Relationship of prebiotics and food to intestinal microflora. Eur J Nutr 2002;41(Suppl. 1):I11–I16. [DOI] [PubMed] [Google Scholar]
- 3. Blaut M, Clavel T. Metabolic diversity of the intestinal microbiota: Implications for health and disease. J Nutr 2007;137:751S–755S. [DOI] [PubMed] [Google Scholar]
- 4. deVrese M, Marteau PR. Probiotics and prebiotics: Effects on diarrhea. J Nutr 2007;137:803S–811S. [DOI] [PubMed] [Google Scholar]
- 5. Johnston KL, Lamport A, Ballevre O, Batt RM. A comparison of endoscopic and surgical collection procedures for the analysis of the bacterial flora in duodenal fluid from cats. Vet J 1999;157:85–89. [DOI] [PubMed] [Google Scholar]
- 6. Inness VL, McCartney AL, Khoo C, et al. Molecular characterisation of the gut microflora of healthy and inflammatory bowel disease cats using fluorescence in situ hybridisation with special reference to Desulfovibrio spp. J Anim Physiol Anim Nutr (Berl) 2007;91:48–53. [DOI] [PubMed] [Google Scholar]
- 7. Johnston KL, Swift NC, Forster‐van HM, et al. Comparison of the bacterial flora of the duodenum in healthy cats and cats with signs of gastrointestinal tract disease. J Am Vet Med Assoc 2001;218:48–51. [DOI] [PubMed] [Google Scholar]
- 8. Janeczko S, Atwater D, Bogel E, et al. The relationship of mucosal bacteria to duodenal histopathology, cytokine mRNA, and clinical disease activity in cats with inflammatory bowel disease. Vet Microbiol 2008;128:178–193. [DOI] [PubMed] [Google Scholar]
- 9. Krecic MR. Feline inflammatory bowel disease: Treatment, prognosis, and new developments. Compendium 2001;23:964–973. [Google Scholar]
- 10. Zentek J, Marquart B, Pietrzak T, et al. Dietary effects on bifidobacteria and Clostridium perfringens in the canine intestinal tract. J Anim Physiol Anim Nutr (Berl) 2003;87:397–407. [DOI] [PubMed] [Google Scholar]
- 11. Barry KA, Wojcicki BJ, Middelbos IS, et al. Dietary cellulose, fructooligosaccharides, and pectin modify fecal protein catabolites and microbial populations in adult cats. J Anim Sci 2010;88:2978–2987. [DOI] [PubMed] [Google Scholar]
- 12. Middelbos IS, Vester Boler BM, Qu A, et al. Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS ONE 2010;5:e9768. doi:10.1371/journal.pone.0009768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hooda A, Vester‐Boler BM, Kerr KR, et al. The gut microbiome of kittens is affected by dietary protein:carbohydrate ratio and associated with blood metabolite and hormone concentrations. Brit J Nutr 2013. May;109(9):1637–1646. doi:10.1017/S0007114512003479. Epub 2012 Aug 31. [DOI] [PubMed] [Google Scholar]
- 14. Marks SL, Laflamme DP, McAloose D. Dietary trial using a commercial hypoallergenic diet containing hydrolyzed protein for dogs with inflammatory bowel disease. Vet Ther 2002;3:109–118. [PubMed] [Google Scholar]
- 15. Guilford WG, Jones BR, Markwell PJ, et al. Food sensitivity in cats with chronic idiopathic gastrointestinal problems. J Vet Intern Med 2001;15:7–13. [DOI] [PubMed] [Google Scholar]
- 16. Laflamme DP, Long GM. Evaluation of two diets in the nutritional management of cats with naturally occurring chronic diarrhea. Vet Ther 2004;5:43–51. [PubMed] [Google Scholar]
- 17. Allenspach K, Wieland B, Grone A, Gaschen F. Chronic enteropathies in dogs: Evaluation of risk factors for negative outcome. J Vet Intern Med 2007;21:700–708. [DOI] [PubMed] [Google Scholar]
- 18. Mandigers PJ, Biourge V, van den Ingh TS, et al. A randomized, open‐label, positively‐controlled field trial of a hydrolyzed protein diet in dogs with chronic small bowl enteropathy. J Vet Intern Med 2010;24:1350–1357. [DOI] [PubMed] [Google Scholar]
- 19. Laflamme DP, Xu H, Long G. Effect of diets differing in fat content on chronic diarrhea in cats. J Vet Intern Med 2011;25:230–235. [DOI] [PubMed] [Google Scholar]
- 20. Laflamme DP, Xu H, Cupp CJ, et al. Evaluation of canned therapeutic diets for the management of cats with naturally occurring chronic diarrhea. J Fel Med Surg 2012;14:669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martineau B, Laflamme DP, Jones W, Jones J. Effect of diet on markers of intestinal health in dogs. Res Vet Sci 2002;72:223–227. [DOI] [PubMed] [Google Scholar]
- 22. Wakshlag JJ, Simpson KW, Struble AM, Dowd SE. Negative fecal characteristics are associated with pH and fecal flora alterations during dietary change in dogs. Intern J Appl Res Vet Med 2011;9:278–283. [Google Scholar]
- 23. Swanson KS, Suchodolski JS, Turnbaugh PJ. Companion animals symposium: Microbes and health. J Anim Sci 2011;89:1496–1497. [DOI] [PubMed] [Google Scholar]
- 24. Suchodolski JS. Companion animals symposium: Microbes and gastrointestinal health of dogs and cats. J Anim Sci 2011;89:1520–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ritchie LE. Molecular Characterization of the Intestinal Bacteria in Healthy Cats and a Comparison of the Fecal Bacterial Flora Between Healthy Cats and Cats with Inflammatory Bowel Disease (IBD). College Station, TX: Texas A&M University; 2008. Masters of Science Thesis. [Google Scholar]
- 26. Ritchie LE, Burke KF, Garcia‐Mazcorro JF, et al. Characterization of fecal microbiota in cats using universal 16S rRNA gene and group‐specific primers for Lactobacillus and Bifidobacterium spp. Vet Microbiol 2010;144:140–146. [DOI] [PubMed] [Google Scholar]
- 27. Tsai YL, Olson BH. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol 1991;57:1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high‐throughput community sequencing data. Nat Methods 2010;7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–2461. [DOI] [PubMed] [Google Scholar]
- 30. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res 2007;6:469–479. [DOI] [PubMed] [Google Scholar]
- 32. Trygg J, Wold S. Orthogonal projections to latent structures (OPLS). J Chemom 2002;16:119–128. [Google Scholar]
- 33. Garcia‐Mazcorro JF, Lanerie DJ, Dowd SE, et al. Effect of a multi‐species synbiotic formulation on fecal bacterial microbiota of healthy cats and dogs as evaluated by pyrosequencing. FEMS Microbiol 2011;78:542–554. [DOI] [PubMed] [Google Scholar]
- 34. Handl S, Dowd SE, Garcia‐Mazcorro JF, et al. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol Ecol 2011;76:301–310. [DOI] [PubMed] [Google Scholar]
- 35. Tun HM, Brar MS, Khin N, et al. Gene‐centric metagenomics analysis of feline intestinal microbiome using 454 junior pyrosequencing. J Microbiol Methods 2012;88:369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 2008;6:e280. doi:10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wolf M, Muller T, Dandekar T, Pollack JD. Phylogeny of Firmicutes with special reference to Mycoplasma (Mollicutes) as inferred from phosphoglycerate kinase amino acid sequence data. Int J Syst Evol Microbiol 2004;54:871–875. [DOI] [PubMed] [Google Scholar]
- 38. Ludwig W, Schleifer KH, Whitman WB. Revised road map to the phylum Firmicutes In: DeVos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB. (eds). Bergey's Manual of Systemic Bacteriology, volume 3, 2nd ed New York: Springer‐Verlag; 2009:1–13. [Google Scholar]
- 39. Lubbs DC, Vester BM, Fastinger ND, Swanson KS. Dietary protein concentration affects intestinal microbiota of adult cats: A study using DGGE and qPCR to evaluate differences in microbial populations in the feline gastrointestinal tract. J Anim Physiol Anim Nutr (Berl) 2009;93:113–121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Fecal microbiota, determined by 454 pyrosequencing that were significantly (P < 0.05) correlated with fecal scoresa from cats with naturally occurring chronic diarrhea after being fed therapeutic diet Diet Xb or Diet Yc for 4 weeks. Bacteria are sorted according to their phylum.


