Abstract
Background
Rotaviral diarrhea (RD) has been associated with the biodiversity of the fecal microbiota in infants; however, the differences in the biodiversity of the fecal microbiota between infants with RD and healthy (H) infants have not been clearly elucidated.
Objectives
This study aimed to reveal the changes in the biodiversity of the fecal microbiota of infants with RD.
Patients and Methods
For this study, 30 fecal samples from 15 RD infants and 15 H infants were collected. The biodiversity of the fecal microbiota from the two groups was compared via polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) and gene sequencing.
Results
The Shannon-Weaver index showed that the biodiversity of the fecal microbiota from the RD infants was significantly lower (P < 0.05) than that from the H infants. All fifteen RD infants were grouped into one cluster and were separated from the H infants by the un weighted-pair group method, with the arithmetic average (UPGMA) clustering algorithm. In addition, when compared with the healthy infants, the communities of the dominant microbes, Lactobacillus and Bifidobacterium, in the fecal microbiota from the RD infants have obviously changed.
Conclusions
With regard to improving the understanding of the differences in the biodiversity of the fecal microbiota between RD infants and H infants, the findings of this study can provide a possible basis to reveal the relationship between RD and intestinal microbiota.
Keywords: Rotaviral Diarrhea, Fecal, Microbiota, Infants, Biodiversity, Denaturing Gradient Gel Electrophoresis, DGGE
1. Background
Rotaviral diarrhea (RD) is the most common cause of gastroenteritis in children (1, 2). Almost all children younger than 5 years old will have suffered from RD, with > 500,000 deaths, > 2 million hospitalizations, and > 25 million clinic visits each year (3, 4). The rotavirus can infect the intestinal epithelium villi cells and lead to watery diarrhea (5); furthermore, RD can cause intestinal dysbacteriosis, and destroy the microbial barrier, making the diarrhea more serious(6, 7).
Previous reports have shown that some diseases, especially intestinal diseases, are associated with the intestinal microbiota, and healthy fecal microbiota are increasingly becoming essential to the maintenance of human health (8, 9). The biodiversity of the fecal microbiota in patients with several diseases, including cow’s milk protein allergy, celiac disease, inflammatory bowel disease, and ulcerative colitis, have been revealed (10-13). In addition, Ma et al. discovered that there was an imbalance in the fecal microbiota in patients aged 2 to 4 years old with viral diarrhea (adenovirus, norovirus, rotavirus, and astrovirus) (14). Although infants are the most susceptible group, the researchers have overlooked studies on the changes of the biodiversity of the fecal microbiota in infants with RD.
2. Objectives
Thirty subjects were included in this study, fifteen of which were untreated RD infants (mean age, 90 days old; range, 90 - 300 days old) from the children’s hospital of Harbin city. The RD infants were diagnosed by the children’s hospital of Harbin city, and the rotavirus (RV) antigens from the fecal samples were detected via specific enzyme immunoassay methods using the RIDASCREEN kit (R-Biopharm, Germany), and confirmed by specific PCR reactions (15). Fifteen of the subjects were healthy (H) infants (mean age, 200 days old; range, 90 - 300 days old) chosen from volunteers in Harbin city. All of the infant subjects were born in natural childbirth, on a breastfeeding diet, and their parents were healthy, without history of illness. In addition, none of the individuals ingested probiotics (including yogurt) or antibiotics within 4 weeks before the sampling.
3. Patients and Methods
3.1. Fecal Sampling
Thirty fecal samples were collected from the RD infants and H infants during November of 2011 in Harbin. All of the fecal samples were transferred into sterile cryotubes and stored at -80°C until further analysis.
3.2. Extraction of Total DNA and PCR Amplification
Approximately 200 mg (wet weight) of the thawed fecal samples were weighed in order to extract the total DNA (13), using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s instructions. The DNA was amplified with the V3 universal primers BA-GC-338f and UN518r for the bacteria, primers Lac1 and Lac2-GC for the Lactobacillus, and primers Bif164-GC-f and Bif662-r for the Bifidobacterium. All of the PCR amplification protocols, as described in previous reports, are shown in Table 1 (16-18).
Table 1. Primers Based on the 16S rDNA Sequences a,b,c,d.
| Target Bacteria | Prime | Sequence (5′ to 3′) |
|---|---|---|
| Total Bacteria | BA-GC-338f ,UN518r | ACTCCTACGGGAGGCAGCAGATT ACC GCG GCTGCT GG |
| Lactobacillus | Lac1, Lac2-GC | AGCAGTAGGGAATCTTCCAATTTCACCGCTACACATG |
| Bifidobacterium | Bif164-GC-f, Bif662-r | GGGTGGTAATGCCGGATGCCACCGTTACACCGGGAA |
| GC d | CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG |
aThe reaction of total bacteria was performed using the following conditions: 92°C for 2 minutes and 30 cycles of 92°C for 1 minute, 55°C for 30 seconds and 72°C for 1 minute, and a final extension at 6°C for 72 minutes.
bThe reaction was performed using the following conditions: 94°C for 2 minutes and 35 cycles of 94°C for 30 seconds, 61°C for 1 minute and 68°C for 1 minute. The reaction was terminated with an extension step of 7 minutes at 68°C.
cThe reaction was performed using the following conditions: 95°C for 5 minutes and 35 cycles of 95°C for 1 minute, 62°C for 20 seconds and 68°C for 40seconds final extension at 6°C for 7 minutes.
dAll GC primers contained a 40 bp GC-clamp sequence at their 5′ end to prevent the complete denaturation of the amplicons.
3.3. Analysis of Fecal Microbiota by Denaturing Gradient Gel Electrophoresis
The denaturing gradient gel electrophoresis (DGGE) scheme was performed by using a DCode apparatus (Bio-Rad, Richmond, CA, USA) at 60°C and employing 8% polyacrylamide gel with a denaturing range of 30% – 55% for the total bacteria, 30% – 50% for the Lactobacillus, and 45% – 55% for the Bifidobacterium. Gel electrophoresis of the total bacteria, Lactobacillus, and Bifidobacterium was run at 20V for 10 minutes, and again at 70V for 18 hours, 70V for 16 hour, and 85V for 16 hour, respectively (16-18). The gels were visualized under UV light after staining them with gene finder (0.5 μg mL-1) and taking photographs.
3.4. Gene Sequencing for the Bands
The bands in the gels were excised and soaked in 50 μL of TE buffer at 4°C overnight to obtain a solution containing the DNA. A PCR program was executed with the same primers without the GC-clamp, and sequenced at the Beijing Genomics Institute (BGI, Beijing, China). The sequences were identified by the BLASTN algorithm in the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/).
3.5. Statistical and Clustering Analysis
The similarities between the two groups were analyzed using the Dice coefficient and the un weighted-pair group method, with the arithmetic average (UPGMA) clustering algorithm using Quantity One software (Bio-Rad) (19). The biodiversity of the bacteria was calculated by the number of bands and by the Shannon–Weaver index of biodiversity (H′), according to previous reports (12, 21). All of the data analyses were performed using the SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA), and statistical significance was established at P < 0.05.
4. Results
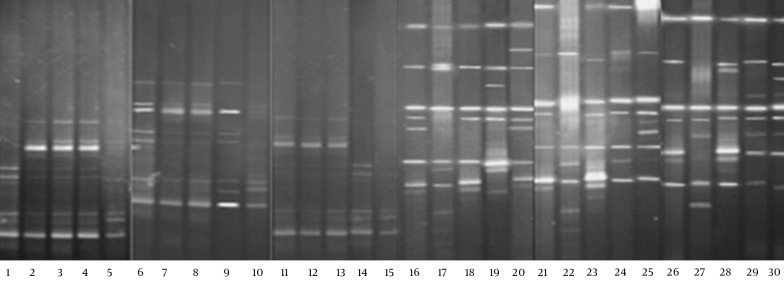
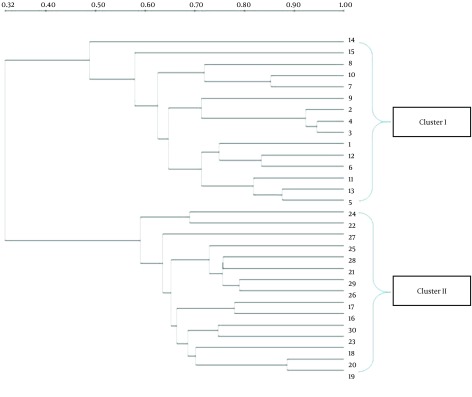
The DGGE images of the 30 fecal samples (fifteen RD infants and fifteen H infants) were obtained by applying PCR-DGGE technology (Figure 1). The 30 fecal samples were divided into two clusters using the UPGMA (Quantity One software); all 15 fecal communities of the RD individuals were placed into cluster I, and separated from all 15 fecal communities of the H infants (cluster II) (Figure 2).
Figure 1. DGGE Fingerprints of Fecal Microbiota from RD Infants and H Infants.
Lanes 1 – 15, RV infants; lanes 15 – 30, H infants.
Figure 2. Clustering of DGGE Profiles of Fecal Microbiota from RD Infants and H Infants Using Dice’s Coefficient and the UPGMA.
Lanes 1 – 15: RV infants; lanes 15 – 30: H infants.
The number of DGGE bands, Shannon-Weaver, and Dice similarity coefficients (both intragroup and intergroup) were calculated, and the details of the results are given in Table 2. The results showed that the biodiversity of the fecal microbiota from the RD infants (Mean ± SD = 6.6 ± 1.92, Shannon-Weaver = 2.03 ± 0.12) was significantly lower (P < 0.05) than that from the H individuals (Mean ± SD = 8.8 ± 0.94, Shannon-Weaver = 2.24 ± 0.34). The index of similarity coefficient based on the DGGE profiles ranged from 30.9% to 94.6% (average 68.54 ± 13.33%) in the RD infants, from 46.7% to 78.9% (average 65.47 ± 8.34%) in the H infants, and averaged 33.16 ± 8.23% between the two groups. The UPGMA analysis (Figure 2) also revealed that the intragroup similarities within the RD and H groups were significantly higher (P < 0.05) than those calculated between the two groups.
Table 2. Microbiota Diversity and Similarity of the RD and H Groups a,b,c.
| Group | Microbiota Diversity (Mean ± SD) | Microbiota Similarity (Mean H′ or H max′ ± SD) | ||
|---|---|---|---|---|
| DGGE bands | Shannon-Weaver | Intragroup | Intergroup | |
| RV | 6.6 ± 1.92 | 2.03 ± 0.12 | 68.54 ± 13.33 | 33.16 ± 8.23 |
| H | 8.8 ± 0.94 | 2.24 ± 0.34 | 65.47 ± 8.34 | NA |
| p | 0.00022 | 0.0038 | NA | NA |
Abbreviation: NA, not available.
aNumber of DGGE bands produced by each sample.
bDice similarity coefficients comparing DGGE band profiles within individuals of each group.
cDice similarity coefficients comparing DGGE band profiles between each RD infant and H infant.
Gene sequencing technology was used to define the microbiota based on the sequence similarity to their closest neighbor in the NCBI. The results of the sequence alignment are displayed in Appendix 1, and showed that Escherichia coli, Bacteroides vulgatus, Enterococcus faecium, Clostridium, E. fergusonii, C. sardiniense, and Peptostreptococcus anaerobius were the dominant fecal microbiota of the RD infants. Moreover, Bacteroides, Bifidobacterium, Lactobacillus, Proteobacteria bacterium, Clostridium, uncultured bacterium, and E. coli were the dominant fecal microbiota of the H infants.
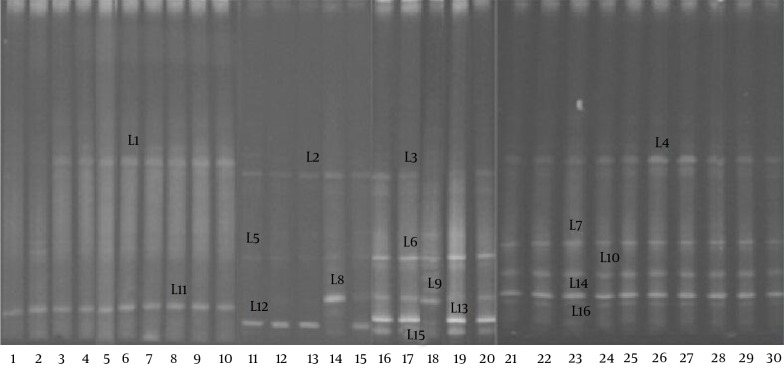
The DGGE profiles (Figure 3) showed that the biodiversity of the Lactobacillus group in the fecal microbiota of the RD infants (mean 2.1 bands) was significantly lower (P < 0.05) than that of the H group (mean 4.6 bands). The gene sequencing results of the bands in the DGGE profiles (Table 3) indicated that a significant reduction in the L. helveticus, L. acidophilus, and L. fermentum was found in the fecal microbiota of the RD infants.
Figure 3. DGGE Profiles of the 16S rRNA Gene Fragments of the Lactobacillus spp.
Lanes 1 – 15, RD infants; lanes 15 – 30: H infants.
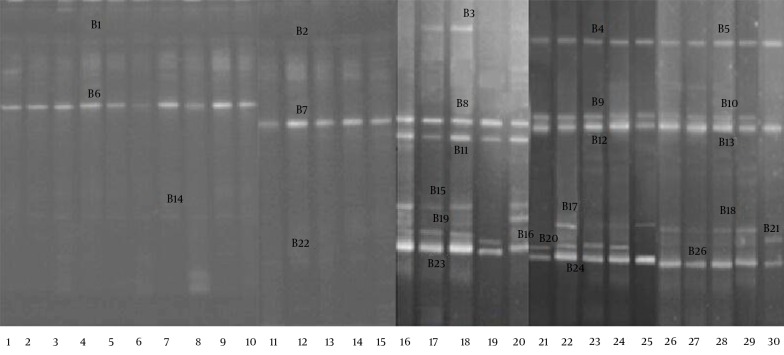
Table 3. The Frequency of the Lactobacillus Species, Identified by Sequencing the DGGE bands Amplified from the Fecal DNA of the RD and H Groups, Using the Lactobacillus and Bifidobacterium Group Specific Primers a,b,c.
| Amplicon ID | Closest relative (Accession number) | Size, bp | H = 15, No. (%) | RD = 15, No. (%) |
|---|---|---|---|---|
| L1, L2, L3, L4 | L. brevis (jx003598) | 1360 | 13 (87) | 13 (87) |
| L5, L6, L7 | L. helveticus (fj749441) | 1462 | 15 (100) | 3 (20) |
| L8, L9, L10 | L. acidophilus (ay763430) | 1475 | 15 (100) | 2 (13) |
| L11, L12, L13, L4 | L. crispatus (jq805668) | 1503 | 14 (93) | 14 (93) |
| L15, L16 | L. fermentum (gq455406) | 1523 | 12 (80) | 0 (0) |
| B1, B2, B3, B4, B5 | B. breve (gu942826) | 294 | 12 (80) | 12 (80) |
| B6, B7, B8, B9, B10 | B. catenulatum (ab846175) | 591 | 15 (100) | 15 (100) |
| B11, B12, B13 | B. infantis (bif16srrge) | 642 | 14 (93) | 0 (0) |
| B15, B17, B18 | B. longum (hq591348) | 1369 | 11 (73) | 0 (0) |
| B16, B21 | B. breve (ay172656) | 1401 | 6 (40) | 0 (0) |
| B14, B19, B20, B22 | B. adolescentis (hq259739) | 1460 | 12 (80) | 5 (33) |
| B23, B24, B25 | B. bifidum (aj311604) | 1469 | 14 (93) | 0 (0) |
aThe marks corresponding to the DGGE bands shown in Figures 3, and 4.
bThe sequence identity was ≥ 99%.
cSignificant differences between RD infants and H infants: P < 0.05.
The DGGE profiles (Figure 4) showed that the fecal microbiota of the RD infants (average 2.1 bands) had a significantly lower level of biodiversity (P < 0.05) than the fecal microbiota of the H infants (average 5.3 bands). The bands were identified with gene sequencing, the details of which are shown in Table 3. The results indicated that the B. infantis, B. longum, B. bifidum, and B. adolescentis in the fecal microbiota of the RD infants were significantly decreased (P < 0.05).
Figure 4. The DGGE Profiles of the 16S rRNA Gene Fragments of the Bifidobacterium spp.
Lanes 1 – 15, RD infants; lanes 15 – 30, H infants.
5. Discussion
The complex intestinal microbial flora harbored by individuals has long been proposed to contribute to intestinal health and disease, and the intestinal microbiota is increasingly considered to be a symbiotic partner in the maintenance of health (9). Studies have shown that many diseases are associated with the intestinal microbiota, and healthy fecal microbiota have become increasingly necessary for the maintenance of human health (8, 21). As a common gastrointestinal disease via viral infection, RD has been considered to be closely related to prominent changes in the gastrointestinal microbiota (22).
Many factors can influence the biodiversity of the fecal microbiota, including genetics, age, gender, feeding, and region (8, 23, 24). Therefore, in order to reduce the differences caused by these factors, the infants chosen for our study were within the same parameters of age range, sex ratio, area, delivery, and feeding. In addition, the infants in this study were between 90 and 300 days old, and during this period the intestinal microbiota are colonizing and forming, and more easily influenced by certain stimulations. The intestinal microbiota of infants is not less complicated when compared with the intestinal microbiota of adults, which provided better feasibility to analyze the differences in the biodiversity of the fecal microbiota between the RD infants and H infants using PCR-DGGE. Our results indicated that the fecal microbial communities of the RD infants were clustered together with higher similarity in the coefficients of the UPGMA; these fecal microbial communities were significantly different from those present in the H infants.
Most bowel diseases can lead to a reduction in the biodiversity of the fecal microbiota (10, 12, 13). In our study, the biodiversity of the fecal microbiota of the RD infants was significantly decreased, and the composition had also changed. The Bacteroides species are the main group in human colonic microbiota, and have a beneficial effect on the prevention of intestinal colonization (25). bacteroides was discovered in the dominant fecal microbiota of the H infants, while B. vulgatus appeared as the dominant fecal microbiota of the RD infants. Therefore, we inferred that the composition of the Bacteroides changed due to the rotavirus infection.
Lactobacillus and Bifidobacterium play important roles in the intestinal microbiota, and have many benefits to human health (26, 27). Lactobacillus can synthesize peptidoglycan, with functional anti-inflammatory activity (28), and Bifidobacterium has a key effect on carbohydrate metabolism in the large bowel, suppressing enteritis in animal models (29). Some researchers have used Lactobacillus and Bifidobacterium for the prevention and treatment of certain intestinal diseases (7, 30, 31). However, Lactobacillus and Bifidobacterium vary in genus, and a further selection of these species was necessary. Therefore, the differences in these two groups of fecal microbiota between the RD infants and H infants were discussed. These results suggested that L. helveticus, L. acidophilus, L. fermentum. B. infantis, B. longum, B. bifidum, and B. adolescentis were considered to be more conducive to mediating the imbalance of the intestinal microbiota of the RD infants.
Enterococcus faecium, as a pathogenic bacteria, can cause serious nosocomial infections, and is found to be associated with intestinal diseases (13, 32). In addition, E. coli and Clostridium have been proven to play important roles in diarrhea and inflammatory processes (33, 34). When compared with the H infants, E. faecium, E. coli, and Clostridium in the fecal microbiota of the RD infants showed obvious advantages.
PCR-DGGE is one of the main ways to assess the biodiversity of the fecal microbiota, in which, the uncultured microorganisms can be identified by the traditional culture-based method (35). In our study, we first analyzed the biodiversity of the dominant fecal microbiota in infants, and found that the harmful microbes were increased in the fecal microbiota of the RD infants. Contrarily, the beneficial microbes were reduced, especially the Lactobacillus and Bifidobacterium. In order to improve the understanding of the differences in the members of these two groups, the biodiversity of the Lactobacillus and Bifidobacterium were revealed via PCR-DGGE using specific primers. However, some researchers have suggested that the DGGE should not be used for quantitative biodiversity analysis (36).
As a supplement, real-time PCR will be used to measure the contents of the microbiota accurately for further research, and linked with metabonomics to explore the connections between the intestinal flora and metabolomics. Moreover, using the PCR-DGGE techniques, we only analyzed the dominant microbiota, and the total biodiversity information of the fecal microbiota cannot be measured. Therefore, we speculate that there should be more differences in the biodiversity of the fecal microbiota between the RD infants and H infants, and our results have provided an important idea and reference for further studies. The next research focus of our team will be to compare the differences in the biodiversity of the fecal microbiota between the two groups using an Illumina MiSeq platform to obtain comprehensive information.
In conclusion, in this study, we showed a comprehensive view of the fecal microbiota in RD infants by using PCR-DGGE and gene sequencing, which revealed the differences in the biodiversity of the dominant microbiota, Lactobacillus and Bifidobacterium in the fecal microbiota of RD infants and H infants. These results suggested that the intestinal microbiota of RD infants exhibited important changes, providing significant information about the relationship between RD and intestinal microbiota.
Appendix 1. The Sequencing Results of Every Band in the DGGE Profiles a.
| Band Number | Species | Closest relative ID in NCBI | Size, bp | Identities, % |
|---|---|---|---|---|
| 1-1,2-4,3-4,4-4,6-4,7-4,8-4, 9-3,11-4,12-4,13-4,14-1 | Clostridium spp. | JN605802 | 664 | 100 |
| 1-2,2-5,3-5,4-5,6-5,7-5,8-5, 9-4,11-5,12-5,13-5,14-2 | C. sardiniense | FJ546743 | 681 | 99 |
| 1-3,2-6,3-6,4-6,5-3,6-5,7-6,8-6,9-5,10-1,11-6,12-6,13-614-3,15-1 | Escherichia fergusonii | HE612113 | 1211 | 100 |
| 1-4,2-7,3-7,4-7,5-4,6-7,7-7,8-7,9-6,10-2,11-7,12-7,13-7, 14-4,15-2 | Enterococcus faecium | JQ778275 | 1473 | 99 |
| 1-5,2-8,3-8,4-8,5-5,6-8,7-8,8-8,9-7,10-311-8,12-8,13-8,14-5,15-3,17-6,22-5,27-7 | E. coli | HQ622348 | 1511 | 100 |
| 2-1,3-1,4-1,6-1,7-1,8-1,9-1, 11-1,12-1,13-1 | P. anaerobius | GQ496449 | 300 | 100 |
| 2-2,2-3,3-2,3-3,4-2,4-3,5-1, 5-2,6-2,6-3,7-2,7-3,8-2,8-3,9-2,11-211-3,12-2,12-3,13-2,13-3 | Bacteroides vulgatus | JF298877 | 540 | 99 |
| 16-1,17-1,18-1,19-1,20-1,21-1,23-1,24-1,25-1,26-1,27-1,28-1,29-1,30-1 | Clostridium spp. | AM774623 | 354 | 99 |
| 16-2.17-2,18-2,19-2,20-3,21-1,22-2,23-2, 24-2,25-2,26-2,28-2,28-3,29-2,30-2 | Uncultured bacterium | JF256482 | 644 | 100 |
| 16-3,17-3,18-3,19-4,20-4,21-3,22-2,23-3,24-3,25-3,26-3,27-2,28-4,29-4,30-5 | Bacteroides spp. | AM117579 | 821 | 100 |
| 16-4,18-4,19-5,20-5,21-4,23-4,24-4,25-4,26-4,27-3,28-5,29-5,30-6 | Bifidobacterium spp. | AM117580 | 845 | 99 |
| 16-6,17-4,18-5,19-6,20-7,21-5,22-3,23-5,24-5,25-6,26-5,28-6,29-7,30-8 | Lactobacillus spp. | JH164960 | 936 | 99 |
| 16-7,17-6,18-6,20-8,21-5,22-5,23-7,24-7, 25-7.26-6,27-5,28-8,29-8,30-8 | B. infantis | HQ591348 | 1369 | 99 |
| 16-5,20-6,25-5,27-4,29-6,30-7 | P. bacterium | AXYJ01000008 | 1222 | 100 |
ai - i′ stands for the i′th band of the i th lane in Figure 1.
Acknowledgments
The authors would like to thank Hong Shao for technical assistance. We would also like to thank all of the subjects who participated in the study.
Footnotes
Authors’ Contribution:Peng Fei and Lin Li had full access to all of the data in the study, and take responsibility for the integrity of the data and the accuracy of the data analysis. Both contributed equally to the study and are co-first authors. Xiaolin Cai and Xinjie Zhang played important roles in the experimental operation process, and can be considered as the co-second authors. Hong Jian Bai was responsible for the collection of the fecal samples. Yu Jun Jiang and Zhen Feng provided some help in the data analysis. Ling Guo, as the corresponding author, provided the idea and experimental design of this study, while participating in and guiding the whole study.
Financial Disclosure:The authors declare that no competing financial interests exist.
Funding/Support:This research was supported by the national natural science foundation of China (31501511), research project of science and technology of the Heilongjiang province (12541026), open fund of the Key laboratory of dairy science of the ministry of education (Northeast agricultural university), and postdoctoral researcher scientific research foundation of the Heilongjiang province (LBH-Q12149). The authors are also thankful for the support of the children’s hospital of Harbin city in China.
References
- 1.Jin H, Wang B, Fang Z, Duan Z, Gao Q, Liu N, et al. Hospital-based study of the economic burden associated with rotavirus diarrhea in eastern China. Vaccine. 2011;29(44):7801–6. doi: 10.1016/j.vaccine.2011.07.104. [DOI] [PubMed] [Google Scholar]
- 2.Dennehy PH. Treatment and prevention of rotavirus infection in children. Curr Infect Dis Rep. 2013;15(3):242–50. doi: 10.1007/s11908-013-0333-5. [DOI] [PubMed] [Google Scholar]
- 3.Chen KT, Fan SF, Tang RB, Huang YF, Lee PI, Chen PY, et al. Hospital-based study of the economic burden associated with rotavirus diarrhea in Taiwan. Vaccine. 2007;25(21):4266–72. doi: 10.1016/j.vaccine.2007.02.056. [DOI] [PubMed] [Google Scholar]
- 4.Nyambat B, Meng CY, Vansith K, Vuthy U, Rin E, Kirkwood C, et al. Hospital-based surveillance for rotavirus diarrhoea in Phnom Penh, Cambodia, March 2005 through February 2007. Vaccine. 2009;27 Suppl 5:F81–4. doi: 10.1016/j.vaccine.2009.08.085. [DOI] [PubMed] [Google Scholar]
- 5.Lorrot M, Vasseur M. Physiopathologie de la diarrhée à rotavirus. Archives de Pédiatrie. 2007;14:S145–51. doi: 10.1016/s0929-693x(07)80018-2. [DOI] [PubMed] [Google Scholar]
- 6.Lundgren O, Svensson L. Pathogenesis of Rotavirus diarrhea. Microbes and Infection. 2001;3(13):1145–56. doi: 10.1016/s1286-4579(01)01475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephen J. Pathogenesis of infectious diarrhea. Can J Gastroenterol Hepatol. 2001;15(10):669–83. doi: 10.1155/2001/264096. [DOI] [PubMed] [Google Scholar]
- 8.Mai V, Draganov PV. Recent advances and remaining gaps in our knowledge of associations between gut microbiota and human health. World J Gastroenterol. 2009;15(1):81–5. doi: 10.3748/wjg.15.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young VB. The intestinal microbiota in health and disease. Curr Opin Gastroenterol. 2012;28(1):63–9. doi: 10.1097/MOG.0b013e32834d61e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez C, Antolin M, Santos J, Torrejon A, Casellas F, Borruel N, et al. Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am J Gastroenterol. 2008;103(3):643–8. doi: 10.1111/j.1572-0241.2007.01592.x. [DOI] [PubMed] [Google Scholar]
- 11.Thompson-Chagoyan OC, Vieites JM, Maldonado J, Edwards C, Gil A. Changes in faecal microbiota of infants with cow's milk protein allergy- A Spanish prospective case-control 6-month follow-up study. Pediatr Allergy Immunol. 2010;21(2 Pt 2):e394–400. doi: 10.1111/j.1399-3038.2009.00961.x. [DOI] [PubMed] [Google Scholar]
- 12.Nistal E, Caminero A, Vivas S, Ruiz de Morales JM, Saenz de Miera LE, Rodriguez-Aparicio LB, et al. Differences in faecal bacteria populations and faecal bacteria metabolism in healthy adults and celiac disease patients. Biochimie. 2012;94(8):1724–9. doi: 10.1016/j.biochi.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 13.Sha S, Xu B, Wang X, Zhang Y, Wang H, Kong X, et al. The biodiversity and composition of the dominant fecal microbiota in patients with inflammatory bowel disease. Diagn Microbiol Infect Dis. 2013;75(3):245–51. doi: 10.1016/j.diagmicrobio.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 14.Ma C, Wu X, Nawaz M, Li J, Yu P, Moore JE, et al. Molecular characterization of fecal microbiota in patients with viral diarrhea. Curr Microbiol. 2011;63(3):259–66. doi: 10.1007/s00284-011-9972-7. [DOI] [PubMed] [Google Scholar]
- 15.Yan H, Nguyen TA, Phan TG, Okitsu S, Li Y, Ushijima H. Development of rt-multiplex pcr assay for detection of adenovirus and group a and c rotaviruses in diarrheal fecal specimens from children in china. J Jpn Assoc Infect Dis. 2004;78(8):699–709. doi: 10.11150/kansenshogakuzasshi1970.78.699. [DOI] [PubMed] [Google Scholar]
- 16.Satokari RM, Vaughan EE, Akkermans AD, Saarela M, de Vos WM. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67(2):504–13. doi: 10.1128/AEM.67.2.504-513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez I, Ruiz-Larrea F, Cocolin L, Orr E, Phister T, Marshall M, et al. Design and evaluation of PCR primers for analysis of bacterial populations in wine by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2003;69(11):6801–7. doi: 10.1128/AEM.69.11.6801-6807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hovda MB, Sivertsvik M, Lunestad BT, Lorentzen G, Rosnes JT. Characterisation of the dominant bacterial population in modified atmosphere packaged farmed halibut (Hippoglossus hippoglossus) based on 16S rDNA-DGGE. Food Microbiol. 2007;24(4):362–71. doi: 10.1016/j.fm.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 19.van der Gast CJ, Whiteley AS, Lilley AK, Knowles CJ, Thompson IP. Bacterial community structure and function in a metal-working fluid. Environ Microbiol. 2003;5(6):453–61. doi: 10.1046/j.1462-2920.2003.00428.x. [DOI] [PubMed] [Google Scholar]
- 20.Sanz Y, Sanchez E, Marzotto M, Calabuig M, Torriani S, Dellaglio F. Differences in faecal bacterial communities in coeliac and healthy children as detected by PCR and denaturing gradient gel electrophoresis. FEMS Immunol Med Microbiol. 2007;51(3):562–8. doi: 10.1111/j.1574-695X.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- 21.Scanlan PD, Shanahan F, Marchesi JR. Human methanogen diversity and incidence in healthy and diseased colonic groups using mcrA gene analysis. BMC Microbiology. 2008;8(1):79. doi: 10.1186/1471-2180-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makos RP, Dikii BN, Zasukha GP. Dysbacteriosis in patients with viral hepatitis. Vrach Delo. 1988;(7):114–6. [PubMed] [Google Scholar]
- 23.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107(33):14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan W, Huo G, Li X, Yang L, Duan C, Wang T, et al. Diversity of the intestinal microbiota in different patterns of feeding infants by Illumina high-throughput sequencing. World J Microbiol Biotechnol. 2013;29(12):2365–72. doi: 10.1007/s11274-013-1404-3. [DOI] [PubMed] [Google Scholar]
- 25.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20(4):593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reuter G. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr Issues Intest Microbiol. 2001;2(2):43–53. [PubMed] [Google Scholar]
- 27.Boesten RJ, de Vos WM. Interactomics in the human intestine: Lactobacilli and Bifidobacteria make a difference. J Clin Gastroenterol. 2008;42 Suppl 3 Pt 2:S163–7. doi: 10.1097/MCG.0b013e31817dbd62. [DOI] [PubMed] [Google Scholar]
- 28.Macho Fernandez E, Valenti V, Rockel C, Hermann C, Pot B, Boneca IG, et al. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut. 2011;60(8):1050–9. doi: 10.1136/gut.2010.232918. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto S, Watanabe N, Imaoka A, Okabe Y. Preventive effects of Bifidobacterium- and Lactobacillus-fermented milk on the development of inflammatory bowel disease in senescence-accelerated mouse P1/Yit strain mice. Digestion. 2001;64(2):92–9. doi: 10.1159/000048846. [DOI] [PubMed] [Google Scholar]
- 30.Kimura K, McCartney AL, McConnell MA, Tannock GW. Analysis of fecal populations of bifidobacteria and lactobacilli and investigation of the immunological responses of their human hosts to the predominant strains. Appl Environ Microbiol. 1997;63(9):3394–8. doi: 10.1128/aem.63.9.3394-3398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Azevedo MS, Wen K, Gonzalez A, Saif LJ, Li G, et al. Probiotic Lactobacillus acidophilus enhances the immunogenicity of an oral rotavirus vaccine in gnotobiotic pigs. Vaccine. 2008;26(29-30):3655–61. doi: 10.1016/j.vaccine.2008.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuzon G, Naas T, Fortineau N, Nordmann P. Novel chromogenic medium for detection of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. J Clin Microbiol. 2008;46(7):2442–4. doi: 10.1128/JCM.00492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rolhion N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13(10):1277–83. doi: 10.1002/ibd.20176. [DOI] [PubMed] [Google Scholar]
- 34.Elseviers MM, Van Camp Y, Nayaert S, Dure K, Annemans L, Tanghe A, et al. Prevalence and management of antibiotic associated diarrhea in general hospitals. BMC Infect Dis. 2015;15:129. doi: 10.1186/s12879-015-0869-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek. 1998;73(1):127–41. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 36.Neilson JW, Jordan FL, Maier RM. Analysis of artifacts suggests DGGE should not be used for quantitative diversity analysis. J Microbiol Methods. 2013;92(3):256–63. doi: 10.1016/j.mimet.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]