Abstract
Background
Aspergillus cristatus is the dominant fungus involved in the fermentation of Chinese Fuzhuan brick tea. Aspergillus cristatus is a homothallic fungus that undergoes a sexual stage without asexual conidiation when cultured in hypotonic medium. The asexual stage is induced by a high salt concentration, which completely inhibits sexual development. The taxon is therefore appropriate for investigating the mechanisms of asexual and sexual reproduction in fungi. In this study, de novo genome sequencing and analysis of transcriptomes during culture under high- and low-osmolarity conditions were performed. These analyses facilitated investigation of the evolution of mating-type genes, which determine the mode of sexual reproduction, in A. cristatus, the response of the high-osmolarity glycerol (HOG) pathway to osmotic stimulation, and the detection of mycotoxins and evaluation of the relationship with the location of the encoding genes.
Results
The A. cristatus genome comprised 27.9 Mb and included 68 scaffolds, from which 10,136 protein-coding gene models were predicted. A phylogenetic analysis suggested a considerable phylogenetic distance between A. cristatus and A. nidulans. Comparison of the mating-type gene loci among Aspergillus species indicated that the mode in A. cristatus differs from those in other Aspergillus species. The components of the HOG pathway were conserved in the genome of A. cristatus. Differential gene expression analysis in A. cristatus using RNA-Seq demonstrated that the expression of most genes in the HOG pathway was unaffected by osmotic pressure. No gene clusters associated with the production of carcinogens were detected.
Conclusions
A model of the mating-type locus in A. cristatus is reported for the first time. Aspergillus cristatus has evolved various mechanisms to cope with high osmotic stress. As a fungus associated with Fuzhuan tea, it is considered to be safe under low- and high-osmolarity conditions.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2637-y) contains supplementary material, which is available to authorized users.
Keywords: Aspergillus cristatus, Genome, Mating-typing gene loci, HOG pathway, Mycotoxin
Background
Chinese commercial tea is classified as green, oolong, black, white, yellow and dark teas, according to the manufacturing process used. Fuzhuan brick tea is a type of dark tea that has been produced for more than 400 years [1]. The production of Fuzhuan brick tea exceeded 10 billion yuan in 2014, and it is very popular in China and north-eastern Asia.
Fuzhuan brick tea involves microbial fermentation, which exerts a major effect on its organoleptic qualities and health properties [2, 3]. Various fungal taxa are important during the production of Fuzhuan brick tea, which is produced under controlled temperature and moisture conditions [4]. Aspergillus, Eurotium and Penicillium species are the main fungal taxa isolated during fermentation. Aspergillus cristatus is the dominant taxon, termed the “Golden Flower Fungus” because of its yellow cleistothecium colour (Fig. 1) [5].
Fig. 1.
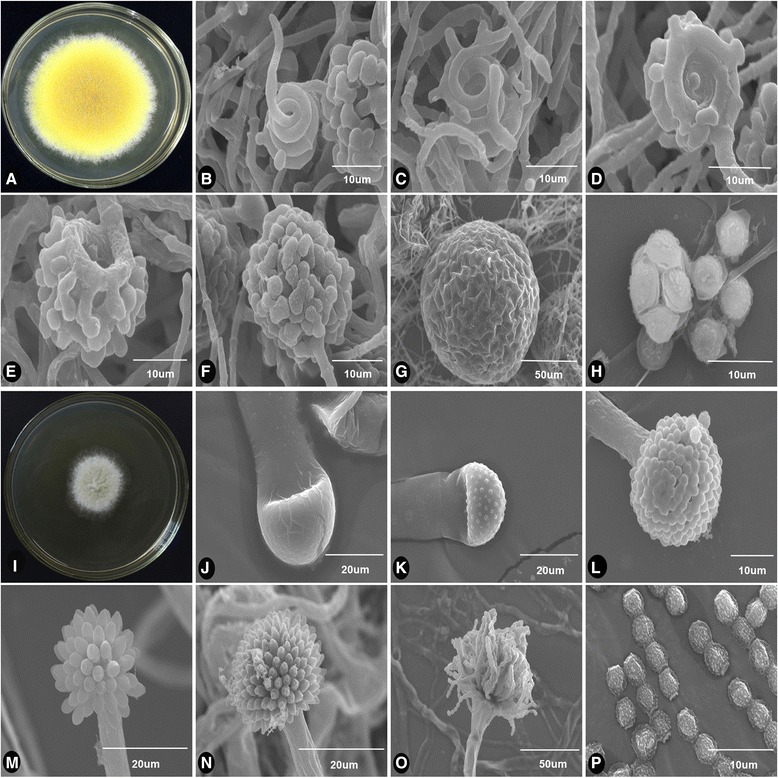
Sexual and asexual morphs of Aspergillus cristatus. a, i Dissecting microscopy; b-h, j-p Scanning electron microscopy. Sexual morph: colony phenotype in 0.5 M NaCl (a), ascogonium during the early and late stages (b-e), cleistothecium (f-g), ascus and ascospore (h). Asexual morph: colony phenotype in 3 M NaCl (i), conidiophores during the early and late stages (j-o), conidia (p)
Aspergilli have served as model organisms in genetic studies because of their multifaceted life cycle [6]. The majority of Aspergillus species (approximately two-thirds) reproduce only asexually, whilst those that exhibit sexual cycles are overwhelmingly homothallic in nature, there are few heterothallic species [7, 8]. Aspergilli are considered good candidates for genetic studies of reproduction in fungi [6, 9]. In the classical era of genetics, the optimal method of identifying gene function was screening for mutants that exhibit defective phenotypes related to their function. However, isolation of sexual reproduction mutants was problematic because of the priority production of conidia [10]. A range of genes involved in sexual reproduction have been identified in several Aspergillus species [9].
Aspergillus can develop asexual or sexual spores depending on growth conditions. High concentrations of salts, such as sodium chloride or potassium chloride, induce asexual reproduction but inhibit sexual spore formation in Aspergilli [11, 12]. Osmotic pressure also plays a key role in A. cristatus sporogenesis [13]. Aspergillus cristatus reproduces sexually only when grown in tea bricks and during culture in hypo-osmolar medium (Fig. 1a–h). Increased osmotic pressure results in greater numbers of asexual spores and fewer sexual spores. In ≥3 M NaCl conditions, only asexual conidia are produced (Fig. 1 i–p), and sexual reproduction is completely inhibited. Aspergillus cristatus employs different sexual and asexual reproductive strategies under different salt stress conditions, providing a robust genetic system for the study of eukaryotic sex development and cell biology.
Fuzhuan brick-tea is produced only in China [14]. As it is popular with certain ethnic groups in China and northeastern Asia, it is important to establish that A. cristatus does not produce carcinogenic mycotoxins, because this fungus dominates the fermentation process during brick-tea production. Many Aspergillus species produce mycotoxins by means of the mycotoxin pathway gene cluster, which comprises several genes [15–20]. Most of the clusters contain one or several central biosynthesis genes encoding extremely large, multidomain, multimodular enzymes belonging to the polyketide synthases (PKSs) or non-ribosomal peptide synthetases (NRPSs) [21]. Consequently, in this study, we focused on analysing the relationships between mycotoxin gene clusters and end products.
In this paper, we investigated the relationship between the HOG pathway and osmotic pressure, verified the safety of A. cristatus under low- and high-osmotic pressure conditions, and established the evolutionary patterns of the mating-type genes through genome sequencing and RNA-Seq transcriptomic data from A. cristatus at two developmental stages. This is the first high-quality genome sequence of A. cristatus to be published and the first report of its safety using genomic data mining. This study may serve as a model for further investigations of the relationship between osmotic pressure and reproduction mode.
Results and discussion
Genome sequencing and assembly
The genome of A. cristatus was sequenced using a whole-genome shotgun approach. A total of 3,489 Mb raw sequence data were generated from the Illumina Hiseq 2000 platform at BGI-ShenZhen. After filtering, the total assembly size of the genome of A. cristatus was 27.9 Mb, which was assembled into 168 contigs and 68 scaffolds, with an N50 length of 2.3 Mb (Fig. 2, Table 1). With the exception of A. clavatus, A. rambellii and A. ruber, the genome of A. cristatus is smaller than other sequenced Aspergillus genomes [22] (Additional file 1: Table S1).
Fig. 2.
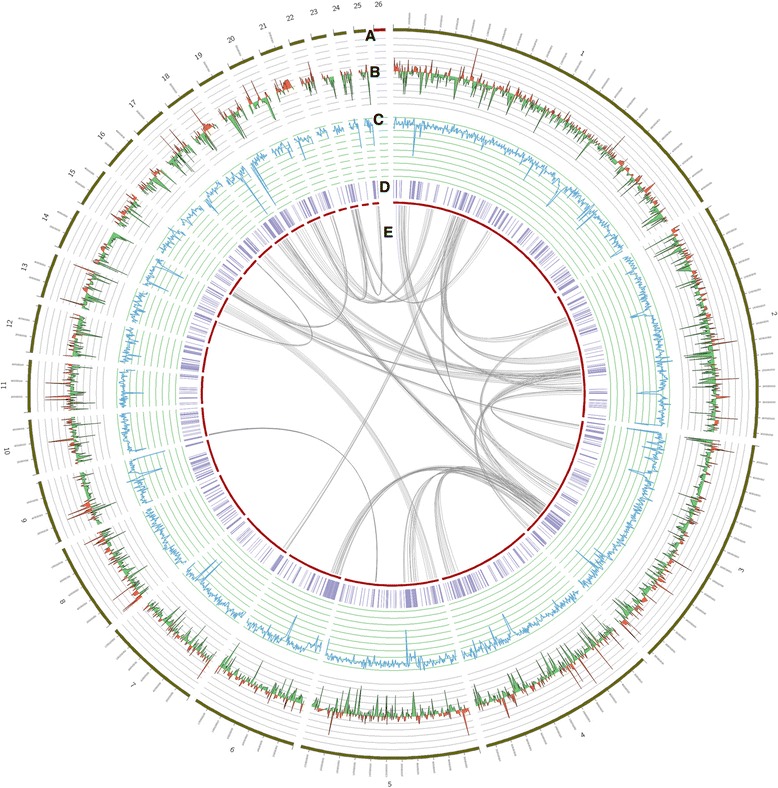
Circular representation of the Aspergillus cristatus genome. The following data are shown (from the outside, in): a Size of scaffolds > 15 kb. b Differential expression as the log2 ratio of expression at high salinity (3 M NaCl), with increased expression in red and decreased expression in green. c GC content of each scaffold. d Locations of transposable elements. e Gene duplications and links. Linked locations were determined by aligning the predicted proteins to the genome using Exonerate (cut-off: 1e-10) [79]
Table 1.
General feature of Aspergillus cristatus genome assembly
| Genome | Value |
|---|---|
| Nuclear genome | |
| General information | |
| Size (Mb) | 27.9 |
| Number of scaffolds | 68 |
| N50 (bp) | 2308221 |
| G + C content (%) | 49.68 |
| Coding (%) | 51.92 |
| Protein coding genes | 10136 |
| Mean gene length (bp) | 1573.31 |
| Genes with intron (%) | 9.15 |
| Exons | |
| Total exon number | 30219 |
| Total exon length | 14486784 |
| Mean number per gene | 2.98 |
| Mean length (bp) | 479.39 |
| G + C content (%) | 53.72 |
| Introns | |
| Total intron number | 20083 |
| Total intron length (bp) | 1460267 |
| Mean number per gene | 1.98 |
| Mean length (bp) | 72.71 |
| G + C content (%) | 44.54 |
Gene prediction and annotation
A total of 10,136 genes were predicted via ab initio and homology-based analyses. The gene density was 2.98 kb per gene, which is higher than those of other sequenced Aspergillus species (Additional file 1: Table S1). In A. cristatus, the annotated coding regions accounted for 51.92 % of the genome, with an average coding length of 1,573 bp and 2.98 exons per gene; the average exon length was 479 bp. The overall GC content was 49.68 %, while the average GC content of the open reading frames was 53.72 % (Table 1).
Gene ontology analysis categorised the gene set into 443 functional groups. Subsets of these functional groups were annotated within the “mating projection” category. Kyoto Encyclopaedia of Genes and Genomes (KEGG) analysis was used to assign 5,159 genes to 311 pathways [23]. Thirteen genes were predicted to be involved in the HOG signalling pathway, with the exception of Sln1, which controls adaptation to different osmolarities [24] (Table 2).
Table 2.
HOG MAP-kinase pathway genes in Aspergillus cristatus and the expression under different osmolarities
| S. cerevisiae | A. nidulans | A. cristatus | Conidia/Asexual (log2 FPKM) |
Ascospore/Sexual (log2 FPKM) |
P-value |
|---|---|---|---|---|---|
| Ctt1 | catA | SI65_01337 | −1.302475 | −2.95113888 | 1.03E-07 |
| Msn2 | AN4013 | SI65_01521 | 3.5055734 | 3.439995398 | 5.86E-26 |
| ssk2 | sskB | SI65_02844 | 2.8020422 | 2.834553469 | 1.26E-31 |
| Msn4 | brlA | SI65_02778 | 3.9057937 | 6.439433647 | 6.09E-32 |
| Ste11 | steC | SI65_03137 | 2.8353663 | 5.874860888 | 9.51E-185 |
| ssk1 | sskA | SI65_09147 | 4.1617507 | 4.469430179 | 0.000218894 |
| Pbs2 | PbsB | SI65_06242 | 5.001496 | 5.74608968 | 1.16E-27 |
| Sho1 | shoA | SI65_09148 | 8.595541 | 8.395251352 | 6.76E-32 |
| Mcm1 | mcmA | SI65_10214 | 5.9391184 | 7.177598929 | 0.01221206 |
| Ste20 | AN2067 | SI65_07597 | 5.1717752 | 6.7099798 | 0.000886426 |
| ypd1 | YpdA | SI65_00763 | 6.5681294 | 5.056717908 | 0 |
| Glo1 | AN4174 | SI65_07143 | 6.7967535 | 6.2913457 | 7.64E-12 |
| Hog1 | HogA | SI65_07698 | 7.2766828 | 7.036393435 | 1.36E-63 |
Phylogenetic relationships
The phylogenetic relationships between filamentous fungi have been established based on ribosomal DNA sequences or single-gene families [25]. We selected a set of 1,034 single-copy predicted orthologous genes from A. cristatus and 22 sequenced genes from filamentous fungi. These orthologous genes were used to construct a maximum-likelihood tree (Additional file 1: Table S2). Penicillium marneffei was used as an outgroup taxon to root the tree. The topology (all internal branches had 100 % bootstrap support) of this tree was consistent with previous reports [22]. Aspergillus cristatus, A. glaucus and A. ruber clustered within the same clade, indicating closer genetic relationships than that with A. nidulans (Fig. 3). The teleomorph of three species are Eurotium spp., and they are homothallic. These microorganisms are widely distributed in the natural environment, can adapt to high concentrations of sodium chloride and other ions, and thus are a valuable resource for investigation of stress resistance mechanisms [26, 27].
Fig. 3.
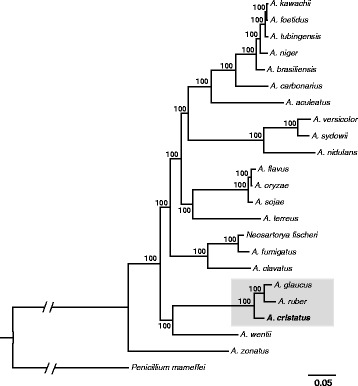
Phylogenetic relationships among Aspergillus genomes. The grey rectangle represents the clade in which Aspergillus cristatus is located. A total of 1,034 single copy orthologous proteins were concatenated and a phylogenetic tree constructed using the RaxML 7.2.8 software [69], with the best model, which was simulated with ProtTest 1.4 [70], 1000 bootstrap replicates were used
Sex and evolution of mating-type loci
Sexual reproduction in ascomycetes is governed by two mating-type genes: one gene encodes a protein with an alpha-box domain (mating-type gene MAT1-1-1), and the other encodes a protein with a high mobility group (HMG) domain (mating-type gene MAT1-2-1) [28]. The sexual development of the genus Aspergillus is mainly homothallic, and the two mating-type genes are located on different chromosomes [6]. Heterothallic fungi have only one mating-type gene and require a partner with a different mating-type gene. The two mating-type genes of heterothallic fungi typically occupy the same chromosomal location in different haploid genomes but are not obviously related in terms of structure or common descent; these genes are termed idiomorphs [29].
Two models for the evolution of the MAT locus in Aspergilli have been proposed (Fig. 5a, b). The first model is evolution of heterothallism from homothallism [30]. The second model is evolution of homothallism from heterothallism [31]. In the first model, the shared homothallic ancestor included two adjacent genes (MAT1-1-1 and MAT1-2-1), which are flanked by the SLA2 and APN1 genes. The alpha box and HMG domain genes in one lineage were located on different chromosomes flanked by either the SLA2 or APN1 gene through chromosomal breakage, resulting in heterothallic species. In addition, the ancestor evolved into heterothallic species with an alpha box or HMG domain gene at the same locus due to chromosomal segregation and gene loss. In the second model, the shared heterothallic ancestor contained either the alpha box or the HMG domain gene at the same locus, flanked by the SLA2 and APN1 genes. The alpha box and HMG genes of the ancestor separated to different chromosomes flanked by the SLA2 and APN1 genes, respectively, due to chromosomal breakage, translocation and rearrangement, resulting in the evolution of homothallic species. Moreover, when the ancestor underwent gene duplication and chromosomal translocation, it evolved into other homothallic species (Neosartorya fischeri), which contained the alpha box domain gene at the original loci flanked by SLA2 and APN1, while the HMG gene was located at a separate locus flanked by dSLA2 and dAPN1, which are pseudogenes of SLA2 and APN1, respectively [31]. The sequence upstream of the N. fischeri MAT2 locus contains numerous regions with sequence similarity to transposase genes from other fungi.
Fig. 5.
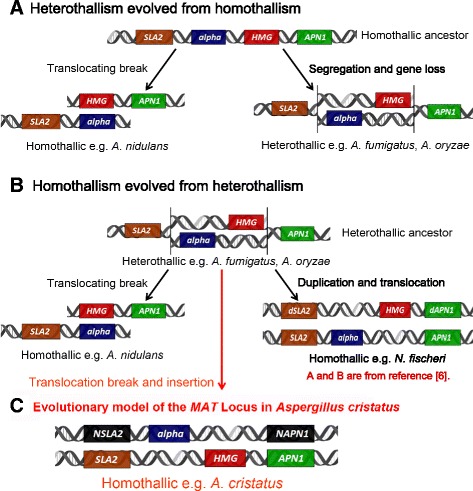
Evolutionary model of the MAT locus in Aspergillus cristatus. a The mode of evolution of MAT loci from homothallic to heterothallic species within the genus Aspergillus; b The mode of evolution of MAT loci from heterothallic to homothallic species within the genus Aspergillus; c evolutionary model of MAT loci in A. cristatus, the HMG (MAT2) locus is flanked by SLA2 and APN1, and the alpha (MAT1) locus is flanked by NSLA2 and NAPN1, NAPN1 and NSLA2 indicated that the protein flanking the MAT1 locus has no homology with SLA2 and APN1. a and b are from reference [6]. c Model of the evolution of MAT loci in A. cristatus
The annotation results of the A. cristatus genome showed that MAT1-1-1 (SI65_05562) and MAT1-2-1 (SI65_06277) are distributed in scaffolds 5 and 6, respectively. The BLASTp analysis indicated that these genes are present as a single copy in the A. cristatus genome. Fgenesh + analysis suggested that MAT1-1-1 contains two exons and an alpha-box sequence and encodes 384 amino acid residues, while MAT1-2-1 contains three exons and one HMG-box sequence and encodes 357 amino acid residues. The conserved amino acid sequences of the two genes were subject to a BLAST search against those of other Aspergillus species using Jalview version 2.0 [32] (Additional file 2: Figure S1). The results demonstrated that MAT genes are conserved in A. cristatus.
BLAST analysis of the flanking regions of A. cristatus MAT 1-2-1 revealed the presence of putative cytoskeleton assembly control (SLA2) and DNA lyase (APN1) genes upstream and downstream of MAT 1-2-1, respectively. The upstream and downstream regions of the A. cristatus MAT 2 locus exhibit collinearity with the upstream and downstream regions of the MAT 1 locus of N. fischeri (Fig. 4a). However, analysis of the flanking genes of the A. cristatus MAT1 locus showed that these genes have no homology to SLA2 and APN1 (Fig. 4b). The sequences flanking the A. cristatus MAT1 locus encode proteins that have no homology with SLA2 and APN1; therefore, these loci were termed NSLA2 and NAPN1 to distinguish them from SLA2 and APN1 (Fig. 5c). In addition, the sequences upstream and downstream of the A. cristatus MAT1 locus did not encode transposase genes with similarity to those of other fungi. These results indicated that the evolutionary model of the MAT loci of A. cristatus is different from those of other Aspergillus species, possibly due to gene translocation breakage and insertion (Fig. 5c). This model supports the evolution of homothallism from heterothallism.
Fig. 4.
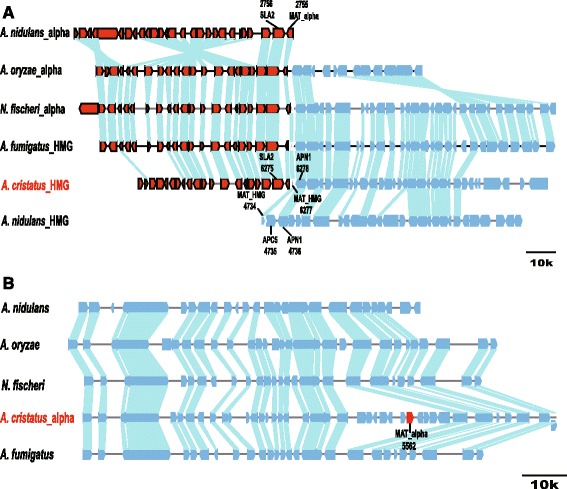
Comparison of Aspergillus MAT loci. Coding regions are indicated by arrows, with the direction of the arrow denoting the direction of transcription. a The Aspergillus cristatus MAT-HMG locus is co-linear with other Aspergillus MAT loci. Red genes show orthologs from the left flank (as drawn) of the A. nidulans alpha locus with the left flanks of A. oryzae, N. fischeri and A. cristatus loci. Blue genes indicate orthologs to the right flank of the A. nidulans HMG locus. Genes labelled and outlined in black are associated with MAT loci in other fungi. b The A. cristatus MAT-alpha locus has no collinearity with any other Aspergillus species. The red gene indicates the MAT-alpha gene of A. cristatus
MAPK signalling transduction
The cell surface and nucleus of eukaryotic cells receive and respond to environmental signals via the MAPK pathway, which is a highly conserved eukaryotic signal transduction system [33]. This pathway orchestrates cell growth, morphogenesis and cell division in response to hormones, stress and other abiotic signals [25]. In the model yeast Saccharomyces cerevisiae, four signalling pathways are associated with the MAPK pathway: the pheromone pathway, the hypotonic pathway, the high-osmolarity pathway and the starvation pathway. Among these, the HOG pathway is involved in responses to osmotic pressure, and its activation depends on the induction of several genes in response to increased osmotic pressure [34].
In S. cerevisiae, the HOG pathway includes two branches: the Sln1-branch and the Sho1-branch. Regulation of these branches converges on the MAPK kinase (MAPKK) Pbs2 [35]. Under normal conditions, Sln1, a sensor histidine kinase, is constitutively activated by autophosphorylation and subsequently phosphorylates the phosphotransfer protein Ypd1, which, in turn, transfers phosphate to the Ssk1 response regulator. Ssk1 is phosphorylated and inactive under low-osmolarity conditions, which blocks activation of the Pbs2 MAPKK-Hog1 MAPK system. In response to stress, the two-component phosphorelay system is rapidly repressed, resulting in the activation of Ssk2 and Ssk22, which activate Pbs2 and Hog1 [36]. Under high-osmolarity conditions, Sho1, which contains four transmembrane domains and a carboxy-terminal SH3 domain, utilises Ste20 and Ste50 to activate the MAPKK kinase Ste11, which then activates Pbs2 [37, 38]. Pbs2 then phosphorylates the MAPK Hog1, resulting in the translocation of Hog1 into the nucleus and induction of the transcription of a large number of genes, some of which are responsible for glycerol production [39].
As in S. cerevisiae, the HogA (SakA) pathway of A. nidulans is activated in an osmotic and oxidative manner [40, 41]. Interestingly, a sakA-null mutant showed only slight sensitivity to high osmolarity stress, and PbsB (a homolog to S. cerevisiae Pbs2) in A. nidulans lacks the Pro-rich motif necessary for binding to Sho1p. This indicates that osmo-regulation in A. nidulans differs from that in yeast [42].
A. cristatus can grow and develop under both low- and high-osmolarity conditions; moreover, most HOG pathway genes in S. cerevisiae have homologs in the A. cristatus genome, with the exception of Sln1. Therefore, we hypothesised that the expression of components of the HOG MAPK cascade pathway would respond to a change in osmotic pressure. To test this hypothesis, we used RNA-Seq to compare gene expression levels in A. cristatus cultured in 0.5 M NaCl (sexual stage developed) and 3 M NaCl (asexual stage developed). Interestingly, there was no significant change in the level of expression of most key genes in the HOG pathway—such as sho1, hog1, and ste20—between high and low osmotic pressure conditions (Fig. 6), indicating that the HOG MAPK cascade pathway is not involved in the response to changes in the osmotic pressure of the medium from hypotonic (0.5 M NaCl) to hypertonic (3 M NaCl). Thus, other mechanisms, such as protein modifications, might be involved in the response to changes in osmotic pressure [24].
Fig. 6.
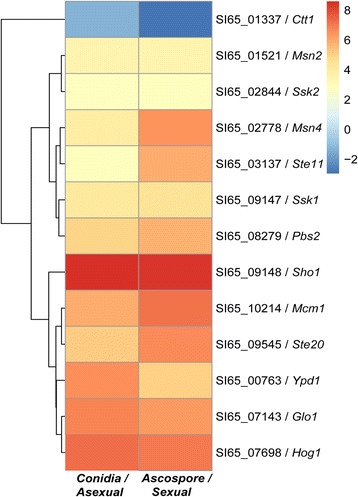
Expression of HOG MAPK pathway genes according to osmolarity. Thirteen genes of the HOG pathway are shown, and background colour changes from blue to red indicate changes in log2 FPKM values (−2, 0, 2, 4, 6, 8; upper right) of gene expression levels under low and high osmolarity conditions. Numbers at right, such as SI65_01337/Ctt1, are Gene IDs or names. Treatment conditions are detailed beneath the images
Mycotoxin gene cluster analysis and mycotoxin detection
Mycotoxins are important because they can affect human health [25, 43]. Mycotoxins are produced by various filamentous ascomycetes, especially members of the genera Aspergillus and Fusarium, through well-defined biosynthetic pathways. This issue is relevant to A. cristatus because of its ‘generally recognised as safe’ status and its use in the production of Fuzhuan brick tea. The biosynthetic genes and pathways for six mycotoxins (aflatoxin, fumonisin, gliotoxin, ochratoxin, sterigmatocystin and zearalenone) were searched in the genome [15–20]. Aspergillus cristatus contains 39 secondary metabolite gene clusters (Table 3). Several backbone enzymes of the mycotoxin gene clusters, which were predicted by antiSMASH [44], are orthologs of the backbone enzymes of some mycotoxin gene clusters (Table 3). The genome contains 14 NRPS- and 14 PKS-encoding genes, most of which are located in clusters. As shown in Additional file 1: Table S3, 85 potential homologs of mycotoxin biosynthetic genes were found in the A. cristatus genome using BLASTp and InParanoid 7.0 [45]. It should be noted that in all cases, the homologs of mycotoxin biosynthetic genes are not located in mycotoxin gene clusters (Additional file 1: Table S3). None of the predicted gene clusters showed collinearity with known mycotoxin gene clusters. Thus, there are no known mycotoxin biosynthetic gene clusters in the A. cristatus genome.
Table 3.
Secondary metabolism gene clusters in Aspergillus cristatus
| Cluster | Scafflod | Backbone enzymes | Genes | Predicted products |
|---|---|---|---|---|
| Cluster1 | 1 | PKS | SI65_01032-01050 | stigmatellin |
| Cluster2 | 1 | Lanthionine | SI65_01066-01075 | |
| Cluster3 | 1 | Siderophore | SI65_01345-01350 | |
| Cluster4 | 1 | Terpene | SI65_01890-01900 | |
| Cluster5 | 1 | Terpene | SI65_01982-01992 | |
| Cluster6 | 2 | PKS | SI65_02155-02174 | hypothemycin |
| Cluster7 | 2 | PKS | SI65_02372-02384 | |
| Cluster8 | 2 | NRPS | SI65_02625-02642 | |
| Cluster9 | 2 | PKS | SI65_02638-02656 | |
| Cluster10 | 2 | PKS | SI65_02843-02864 | |
| Cluster11 | 3 | PKS | SI65_03265-03285 | |
| Cluster12 | 3 | PKS | SI65_03715-03729 | |
| Cluster13 | 3 | NRPS | SI65_03775-03795 | |
| Cluster14 | 3 | PKS | SI65_03940-03953 | |
| Cluster15 | 4 | NRPS | SI65_04496-04511 | |
| Cluster16 | 4 | NRPS | SI65_05048-05065 | |
| Cluster17 | 5 | NRPS | SI65_05225-05238 | |
| Cluster18 | 5 | NRPS | SI65_05406-05426 | |
| Cluster19 | 5 | PKS | SI65_05759-05776 | |
| Cluster20 | 6 | NRPS | SI65_06121-06138 | |
| Cluster21 | 7 | PKS | SI65_06667-06682 | |
| Cluster22 | 8 | NRPS | SI65_07091-07104 | |
| Cluster23 | 8 | Terpene | SI65_07245-07259 | |
| Cluster24 | 9 | PKS | SI65_07348-07363 | |
| Cluster25 | 9 | Lanthionine | SI65_07571-07579 | |
| Cluster26 | 10 | Terpene | SI65_07672-07681 | |
| Cluster27 | 11 | Siderophore | SI65_07931-07938 | |
| Cluster28 | 12 | NRPS | SI65_08199-08214 | |
| Cluster29 | 12 | NRPS | SI65_08311-08325 | |
| Cluster30 | 13 | NRPS | SI65_08399-08408 | |
| Cluster31 | 13 | NRPS | SI65_08558-08578 | |
| Cluster32 | 13 | PKS | SI65_08470-08482 | |
| Cluster33 | 14 | Terpene | SI65_08621-08631 | |
| Cluster34 | 21 | PKS | SI65_09731-09746 | |
| Cluster35 | 21 | Terpene | SI65_09793-09801 | |
| Cluster36 | 22 | PKS | SI65_09879-09899 | |
| Cluster37 | 24 | Terpene | SI65_10001-10023 | |
| Cluster38 | 25 | NRPS | SI65_10088-10103 | |
| Cluster39 | 26 | NRPS | SI65_10253-10268 |
In general, the biosynthesis genes for fungal secondary metabolites are located in clusters [21]; however, the mycotoxin genes in A. crastatus were not found located in clusters. Moreover, transcription data showed that most of the mycotoxin genes are expressed at low and high osmotic pressure (Additional file 1: Table S3). This result suggested that A. crastatus might produce mycotoxins during culture under low and high osmolarity conditions. To confirm this, we assayed six mycotoxins by High Performance Liquid Chromatography (HPLC). Only fumonisin B1 was detected at 0.17 and 0.15 ppm at low and high osmolarities, respectively. However, this is lower than the standard (2 ppm) set by the US Food and Drug Administration (FDA) [46] (Additional file 2: Figure S2). These data suggest that the strain is safe under low- and high-osmolarity conditions, and the locations of the mycotoxin genes did not provide information regarding mycotoxin production by the fungus.
Conclusions
Comparison of the MAT loci of A. cristatus with those of other Aspergillus species revealed that the evolution of the A. cristatus MAT locus differs from those of other Aspergillus species. The findings regarding the Aspergillus MAT loci supported the evolution of homothallism from heterothallism. The majority of sex-related components identified in other ascomycetes are also present in A. cristatus. The initial analysis of the evolution of the MAT loci, associated with sex-related components, provides information for further investigation of sexual development in A. cristatus.
The components of the HOG pathway were conserved in A. cristatus. Gene expression analysis demonstrated that the HOG pathway of A. cristatus was not involved in the response to high osmotic pressure. Thus, A. cristatus may respond to high osmolarity stress via mechanisms other than the HOG pathway.
A mycotoxin gene cluster collinearity analysis indicated that the mycotoxin biosynthetic gene clusters responsible for production of six toxins were not present in the A. cristatus genome. An HPLC assay indicated that the strain is safe under low- and high-osmolarity conditions; moreover, the locations of the mycotoxin genes did not provide information regarding mycotoxin production by this fungus.
Methods
Strains, growth conditions, and genomic DNA and RNA extraction
A. cristatus E4 (CGMCC 7.193) was isolated from Fuzhuan brick tea in a Yiyang Tea Factory in Yi Yang City, Hunan Province, China. The fungus was cultured in liquid medium (malt extract 20 g, yeast extract powder 20 g, sucrose 30 g, and water 1000 mL) with shaking at 180 rpm at 28 °C for 5 days. Mycelia were collected by filtration on Waterman paper and placed in a mortar, liquid nitrogen was added, and the samples were crushed using a pestle. Genomic DNA from fungal mycelia was extracted using the CTAB method [47]. The DNA pellet was dissolved in sterile water and adjusted to a concentration of 500 μg/mL. The fungus was cultured on a cellulose membrane on MYA (malt extract 20 g, yeast extract powder 20 g, sucrose 30 g, agar powder 18 g, and water 1000 mL) plates in the dark at 28 °C for 7 days. Mycelia were collected after sporulation. Total RNA was extracted from fresh mycelia using TRIzol reagent according to the manufacturer’s instructions (Life Technologies Co. Ltd., Carlsbad, CA, USA), and the RNA pellet was dissolved in sterile water containing diethylpyrocarbonate diluted 1:1000 in sterile water. DNA and RNA were quantified using a Nano Drop 2000 UV–vis spectrophotometer (Thermo Fisher Scientific Inc., Boston, MA, USA) based on the absorbance at 260 and 280 nm, respectively. DNA and RNA samples were subjected to genome and transcriptome sequencing by BGI-Shenzhen (Shenzhen, China).
Sequencing and assembly
From the genomic DNA of A. cristatus, 500 bp and 6 kb DNA sequencing libraries were constructed using 5 μg and 20 μg DNA [48], respectively. A total of 2,364 and 1,125 Mb reads were generated by an Illumina Hiseq™ 2000 at BGI-Shenzhen (Shenzhen, China). To ensure the accuracy of the assembly, reads with 36 low-quality (≤ Q2) bases, 9 % Ns, or 3-bp overlaps between the adapter and duplications were filtered. The short reads from the two libraries were assembled using SOAPdenovo 1.04 [49, 50], with optimal assembly acquired using the key parameter K = 55.
Gene prediction and annotation
Gene models were predicted independently using a set of gene finders including Augustus [51], GeneMark-ES [52], GeneId 1.2 [53] and Fgenesh + [54]. Augustus parameters were trained on gene models in Aspergillus (A. fumigatus, A. nidulans and A. oryzae) using the transcriptomic data as hints. GeneMark-ES functions in a self-training manner. The available fungal genome sequence of A. nidulans was used for the GeneId gene predictor. The predicted gene models were then combined into consensus gene structure annotations using EvidenceModeler [55].
BLASTp searches against the UniProt/SwissProt, KEGG and COG databases were performed to assign general protein function profiles [56–58]. Pfam was used to scan for significant domains using HMMER [59, 60]. Blast2go was used for Gene Ontology (GO) and InterPro annotation [61–63]. Predicted proteins were classified as proteases by querying the MEROPS database using BLASTp (E-value cut-off of 1e-10) [64]. Potential secondary metabolite key enzyme genes were identified using antiSMASH 1.2.2 [44]. Transposons and retrotransposons encoding transposases and retrotransposases were classified by BLASTp analysis against the Repbase database [65].
Orthology and phylogenetic analysis
Orthologous groups were clustered using the OrthoMCL version 2.0 software with an E-value cut-off of 1e-5 and percentage match cut-off of 50 [66, 67]. In total, 1,034 single-copy orthologous proteins were acquired and aligned using MAFFT 7.221 [68]. A maximum-likelihood phylogenetic tree was created using the concatenated amino acid sequences and the RaxML 7.2.8 software [69], with the best model, which was simulated using the ProtTest 1.4 software [70].
Transcriptome analysis
Raw data generated by the sequencer were converted to raw nucleotide reads by Illumina GAPipeline 1.6. Clean reads were acquired by removing the adaptor and the low-quality reads (Q ≤ 5), and were mapped to the genome using Tophat [71]. Up to two base mismatches were allowed. The abundance of each clean read was converted to transcripts per million for quantitative comparison among samples. We used the false discovery rate (FDR ≤0.001) to estimate the level of differential gene expression among samples under different induction conditions [72]. Genes with FDR values less than 0.001 and log2-fold changes greater than 2.0 or lower than −2.0 were considered to be differentially expressed.
Mycotoxin gene clusters analysis and mycotoxin detection
The mycotoxin gene clusters in this study were obtained from GenBank (Additional file 1: Table S3), and their sequences applied as BLAST queries against the protein sequences of A. cristatus (E-value < 1e-5). A homology analysis was performed using InParanoid 7.0 (E-value < 0.01, score >50) [45]. To detect mycotoxins in the end products, mycelia were incubated in the dark at 28 °C for 9 days with 0.5 M or 3.0 M NaCl and collected on cellulose membranes after sporulation. Mycelia were processed using various methods [73–78] and then analysed by HPLC (Hitachi L-2000).
Availability of data
All data contributing to this genome initiative has been deposited at the NCBI under BioProject PRJNA271918; the genome accession number is [JXNT00000000]. The genome version described in this paper is the first version and the accession number is [JXNT01000000]. The RNA-seq expression dataset has been deposited at the NCBI’s Gene Expression Omnibus under the accession code GSE65662. The phylogenetic analysis results are deposited in TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S19105).
Ethics statement
No specific permissions were required for these locations/activities. The field studies did not involve endangered or protected species and were conducted in accordance with local legislation.
Acknowledgments
We thank Bioinformatics-platform of state key laboratory of mycology for helping with the data analysis. We would like to express our gratitude to Professor Xingzhong Liu (State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences) and Professor Chengshu Wang (Institute of Plant Physiology & Ecology, Shanghai Institute for Biology Science, Chinese Academy of Sciences) for discussions and critical reviewing of the manuscript.
Funding
This work was supported by special grants from Guizhou Academy of Agriculture Science (GAAS-SP-2011/037) and Department of Science and Technology of Guizhou Province (GSTD-LH-[2015]-7683).
Abbreviations
- FDA
Food and drug administration
- FPKM
Fragments per kilobase of exon per million fragments mapped
- HMG
High-mobility group
- HOG
The high-osmolarity glycerol
- HPLC
High performance liquid chromatography
- MAT
Mating-typing gene
- MAPK
Mitogen-activated oprotein kinase
- MAPKK
MAPK kinase
- MAPKKK
MAPKK kinase
Additional files
Aspergillus cristatus genome statistics compared to that of other sequenced Aspergillus fungi. Table S2. Genes used for Phylogenetic Analysis. Table S3. Homology Genes of Mycotoxin Biosynthesis in Aspergillus cristatus Genome. (PDF 808 kb)
Results of MAT1-1-1 and MAT1-2-1 amino acid sequence analysis of members of the genus Aspergillus using Jalview version 2.0 [32]. Figure S2. Detection of six mycotoxins by HPLC. (PDF 401 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ZYL designed and coordinated the study. YYG prepared the genomic DNA and RNA sample of A. cristatus for genome and transcriptome sequencing, participated in the interpretation of the results and writing the manuscript. YCW conducted data analysis, participated in bioinformatics analyses, in the interpretation of the results and drafted the manuscript. YXL isolated the fungus from tea and participated in the coordinated of the study. YMT, XXR and KH participated in writing the manuscript. XYZ performed bioinformatics analyses, participated in the interpretation of the results. YFL assisted the bioinformatics analyses. All authors read, corrected and approved the final manuscript.
References
- 1.Schoch CL, Seifert KA. S Huhndorf, Robert V, Spouge JL, Levesque CA, Chen W, Bolchacova E, Voigt K, Crous PW. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci. 2012;109(16):6241–6. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X, Yan M, Zhu Y. Influence of fungal fermentation on the development of volatile compounds in the Puer tea manufacturing process. Engineering in Life Sciences. 2005;5(4):382–6. doi: 10.1002/elsc.200520083. [DOI] [Google Scholar]
- 3.Mo H, Xu X, Yan M, Zhu Y. Microbiological analysis and antibacterial effects of the indigenous fermented Puer tea. Agro food industry hi-tech. 2005;16(6):16–8. [Google Scholar]
- 4.Porter CH, Collins FH. Species-diagnostic differences in a ribosomal DNA internal transcribed spacer from the sibling species Anopheles freeborni and Anopheles hermsi (Diptera: Culicidae) Am J Trop Med Hyg. 1991;45(2):271–9. doi: 10.4269/ajtmh.1991.45.271. [DOI] [PubMed] [Google Scholar]
- 5.Mo H, Zhu Y, Chen Z. Microbial fermented tea–a potential source of natural food preservatives. Trends Food Sci Technol. 2008;19(3):124–30. doi: 10.1016/j.tifs.2007.10.001. [DOI] [Google Scholar]
- 6.Lee SC, Ni M, Li W, Shertz C, Heitman J. The evolution of sex: a perspective from the fungal kingdom. Microbiol Mol Biol Rev. 2010;74(2):298–340. doi: 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heitman J, Kronstad JW, Taylor JW, Casselton L. Sex in fungi: molecular determination and evolutionary implications. Washington: ASM Press; 2007.
- 8.Kwon-Chung KJ, Sugui JA. Sexual reproduction in Aspergillus species of medical or economical importance: why so fastidious? Trends Microbiol. 2009;17(11):481–7. doi: 10.1016/j.tim.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyer PS, O’Gorman CM. Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol Rev. 2012;36(1):165–92. doi: 10.1111/j.1574-6976.2011.00308.x. [DOI] [PubMed] [Google Scholar]
- 10.Han KH. Molecular genetics of Emericella nidulans sexual development. Mycobiology. 2009;37(3):171–82. doi: 10.4489/MYCO.2009.37.3.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han KH, Lee DB, Kim JH, Kim MS, Han KY, Kim WS, Park YS, Kim HB, Han DM. Environmental factors affecting development of Aspergillus nidulans. J Microbiol. 2003;41(1):34–40. [Google Scholar]
- 12.Song MH, Jy N, Han YS, Han DM, Chae KS. Promotion of conidial head formation in Aspergillus oryzae by a salt. Biotechnol Lett. 2001;23(9):689–91. doi: 10.1023/A:1010308601469. [DOI] [Google Scholar]
- 13.Liu ZY, Qin J. li NL. Study of conditions of Sporogenesis of Aspergillus chevalieri var.intermedius (synonym of Aspergillus cristatus) in FuZhuan Tea. Southwest China Journal of Agricultural Science. 1991;4(1):73–7. [Google Scholar]
- 14.Xu X, Mo H, Yan M, Zhu Y. Analysis of characteristic aroma of fungal fermented Fuzhuan brick-tea by gas chromatography/mass spectrophotometry. J Sci Food Agric. 2007;8(7):1502–4. doi: 10.1002/jsfa.2874. [DOI] [Google Scholar]
- 15.Yu JJ, Chang PK, Ehrlich KC, Cary JW, Bhatnagar D, Cleveland TE, Payne GA, Linz JE, Woloshuk CP, Bennett JW. Clustered Pathway Genes in Aflatoxin Biosynthesis. Appl Environ Microbiol. 2004;70(3):1253–62. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robert H, Proctor RH, Busman M, Seo JA. Yin Won Lee YW, Plattner RD. A fumonisin biosynthetic gene cluster in Fusarium oxysporum strain O-1890 and the genetic basis for B versus C fumonisin production. Fungal Genet Biol. 2008;45(6):1016–26. doi: 10.1016/j.fgb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Gardiner DM, Howlett BJ. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiology Letter. 2005;248(2):241–8. doi: 10.1016/j.femsle.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 18.Karolewiez J, Geisen R. Cloning a part of the ochratoxin A biosynthetic gene cluster of Penicillium nordicum and characterization of the ochratoxin polyketide synthase gene. Syst Appl Microbiol. 2005;28(7):588–95. doi: 10.1016/j.syapm.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Kim YT, Lee YR, Jin JM, Han KH, Kim H, Kim JC, Lee T, Yun SH, Lee YW. Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol Microbiol. 2005;58(4):1102–13. doi: 10.1111/j.1365-2958.2005.04884.x. [DOI] [PubMed] [Google Scholar]
- 20.Brown DW, Yu JH, Kelkar HS, Fernandes M, Nesbitt TC, Keller NP, Adams TH, Leonard TJ. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci. 1996;93(4):1418–22. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brakhage AA. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11(1):21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 22.Gibbons JG, Rokas A. The function and evolution of the Aspergillus genome. Trends Microbiol. 2013;21(1):14–22. doi: 10.1016/j.tim.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79(1):143–80. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 25.Pel HJ, De Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, Vries RP de, Albang R, Albermann K. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–31. [DOI] [PubMed]
- 26.Kis-Papo T, Weig AR, Riley R, Persoh D, Salamov A, Sun H, Lipzen A, Wasser SP, Rambold G, Igor V, Grigoriev IV, Nevo E. Genomic adaptations of the halophilic Dead Sea filamentous fungus Eurotium rubrum. Nat Commun. 2014;5:1–8. doi: 10.1038/ncomms4745. [DOI] [PubMed] [Google Scholar]
- 27.Liu XD, Xie LX, Wei Y, Zhou XY, Jia BL, Liu JL, Zhang SH. Abiotic Stress Resistance, a Novel Moonlighting Function of Ribosomal Protein RPL44 in the Halophilic Fungus Aspergillus glaucus. Appl Environ Microbiol. 2014;80(14):4294–300. doi: 10.1128/AEM.00292-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turgeon BG, Yoder OC. Proposed Nomenclature for Mating Type Genes of Filamentous Ascomycetes. Fungal Genet Biol. 2000;31(1):1–5. doi: 10.1006/fgbi.2000.1227. [DOI] [PubMed] [Google Scholar]
- 29.Metzenberg RL, Glass NL. Mating type and mating strategies in Neurospora. BioEssays. 1990;12(2):53–9. doi: 10.1002/bies.950120202. [DOI] [PubMed] [Google Scholar]
- 30.Galagan JE, Calve SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee S-I, Basturkmen M, Spevak C, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, Enfert CD, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, J. Yu, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, M. Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438(7071):1105–15. [DOI] [PubMed]
- 31.Rydholm C, Dyer PS, Lutzoni F. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryotic Cell. 2007;6(5):868–74. doi: 10.1128/EC.00319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2.0-a multiple sequence alignment editor and analysis workbench. Bioninformatics. 2009;25(9):1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayram Ö, Bayram ÖS, Ahmed YL, Maruyama J-i, Valerius O, Rizzoli SO, Ficner R, Irniger S, Braus GH. The Aspergillus nidulans MAPK Module AnSte11-Ste50-Ste7-Fus3 Controls Development and Secondary Metabolism. PLoS Genet. 2012;8(7):e1002816. doi: 10.1371/journal.pgen.1002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krantz M, Becit E, Hohmann S. Comparative genomics of the HOG-signalling system in fungi. Curr Genet. 2006;49:137–51. doi: 10.1007/s00294-005-0038-x. [DOI] [PubMed] [Google Scholar]
- 35.O’Rourke SM, Herskowitz I. A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch. Mol Cell Biol. 2002;22(13):4739–49. doi: 10.1128/MCB.22.13.4739-4749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bahn Y-S, Xue CY, Idnurm A, Rutherford JC, Heitman J, Cardenas ME. Sensing the environment: lessons from fungi. Nat Rev Microbiol. 2007;5(1):57–69. doi: 10.1038/nrmicro1578. [DOI] [PubMed] [Google Scholar]
- 37.O’Rourke SM, Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12(18):2874–86. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Drogen F, O’Rourke SM, Stucke VM, Jaquenoud M, Neiman AM, Peter M. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr Biol. 2000;10(11):630–9. doi: 10.1016/S0960-9822(00)00511-X. [DOI] [PubMed] [Google Scholar]
- 39.Reiser V, Ruis H, Ammerer G. Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10(4):1147–61. doi: 10.1091/mbc.10.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagiwara D, Asano Y, Marui J, Furukawa K, Kanamaru K, Kato M, Abe K, Kobayashi T, Yamashino T, Mizuno T. The SskA and SrrA response regulators are implicated in oxidative stress responses of hyphae and asexual spores in the phosphorelay signaling network of Aspergillus nidulans. Biosci Biotechnol Biochem. 2007;71(4):1003–14. doi: 10.1271/bbb.60665. [DOI] [PubMed] [Google Scholar]
- 41.Vargas-Pérez I, Sánchez O, Kawasaki L, Georgellis D, Aguirre J. Response regulators SrrA and SskA are central components of a phosphorelay system involved in stress signal transduction and asexual sporulation in Aspergillus nidulans. Eukaryotic Cell. 2007;6(9):1570–83. doi: 10.1128/EC.00085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furukawa K, Hoshi Y, Maeda T, Nakajima T, Abe K. Aspergillus nidulans HOG pathway is activated only by two-component signalling pathway in response to osmotic stress. Mol Microbiol. 2005;56(5):1246–61. doi: 10.1111/j.1365-2958.2005.04605.x. [DOI] [PubMed] [Google Scholar]
- 43.Keller NP, Turner G, Bennett JW. Fungal secondary metabolism-from biochemistry to genomics. Nat Rev Microbiol. 2005;3(12):937–47. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 44.Medema MH, Blin K, Cimermancic P, Jager V. de, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39(suppl 2):W339–46. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer ELL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2009;38:D196–203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.http://www.fda.gov/Food/GuidanceRegulation/GuidanceDocumentsRegulatoryInformation/ChemicalContaminantsMetalsNaturalToxinsPesticides/ucm109231.htm. Accessed 5 May.
- 47.Rogers SO, Bendich AJ. Extraction of total cellular DNA from plants, algae and fungi. Plant Mol Biol Manual. 1994;DI:1–8.
- 48.Liu DB, Gong J, Dai WK, Kang XC, Huang Z, Zhang HM, Liu W, Liu L, Ma JP, Xia ZL, Chen YX, Chen YW, Wang DP, Ni PX, Guo AY, Xiong XY. The Genome of Ganderma lucidum Provide Insights intoTriterpense Biosynthesis and Wood Degradation. PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0036146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–4. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 50.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2009;20:265–72. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanke M, Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33(suppl 2):W465–7. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18(12):1979–90. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blanco E, Parra G. GuigóR. Using geneid to identify genes. Current protocols in Bioinformatics. 2007;4(3):1–28. doi: 10.1002/0471250953.bi0403s18. [DOI] [PubMed] [Google Scholar]
- 54.Solovyev V, Kosarev P, Seledsov I, Vorobyev D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006;7(Suppl 1):1–12. doi: 10.1186/gb-2006-7-s1-s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9(1):R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.UniProt Consortium The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–8. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–60. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4(1):1–14. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J. The Pfam protein families database. Nucleic Acids Res. 2012;40(D1):D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Suppl 2):W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–35. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ashburner M. A. Ball C, A. Blake J, Botstein D, Butler H, Cherry JM, P. Davis A, Dolinski K, S. Dwight S, T. Eppig J, A. Harris M, P. HilL D, Issel-Tarver L, Kasarskis A, Lewis S, C. Matese J, E. Richardson J, Ringwald M, M. Rubin G and Sherlock G. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, McMenamin C, Mi HY, Mutowo-Muellenet P, Mulder N, Natale D, Orengo C, Pesseat S, Punta M, Quinn AF, Rivoire C, Sangrador-Vegas A, Selengut JD, Sigrist CJA, Scheremetjew M, Tate J, Thimmajanarthanan M, Thomas PD, Wu CH, Yeats C, Yong SY. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40(D1):D306–12. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42(1):D503–9. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Repbase database. [http://girinst.org/]. Accessed 5 May.
- 66.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen F, Mackey AJ, Stoeckert CJ, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34(suppl 1):D363–8. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 70.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–5. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 71.Trapnell C, Pachter L, Salzberg SL. Tophat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29(4):1165–88. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
- 73.Determination aflatoxins content in food-Clean up by immunoaffinity chromatogaphy and determination by high-performance liquid chromatigraphy and fluor-ometer (Standard NO: GB/T 18979–2003), Standardization Administration of the People’s Republic of China, 2003.
- 74.Inspection of grain and oils-Determination of ochratoxin A in grains by high performance liquid chromatography and fluorometer (Standard NO: GB/T 25220–2010), Standardization Administration of the People’s Republic of China, 2010.
- 75.Determination of zearalenone in food-High performance liquid chromatographic method with immunoaffinity column clean-up (Standard NO: GB/T 23504–2009), Standardization Administration of the People’s Republic of China, 2009.
- 76.Inspection of grain and oils-Determination of fumonisins in corn and its products by high liquid chromatography and fluorometer with immunoaffinity column cleanup (Standard NO: GB/T 25228–2010), Standardization Administration of the People’s Republic of China, 2010.
- 77.Pereyra CM, Alonso VA, Rosa CAR, Chiacchiera SM, Dalcero AM, Cavaglieri LR. Gliotoxin natural incidence and toxigenicity of Aspergillus fumigatus isolated from corn silage and ready dairy cattle feed. World Mycotoxin J. 2008;1(4):457–62. doi: 10.3920/WMJ2007.1012. [DOI] [Google Scholar]
- 78.Yuan J, Du J, Wang HF, Ju XR. Determination of Sterigmatocysin in Wheat by HPLC. Food Sci. 2011;32(12):174–7. [Google Scholar]
- 79.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6(1):31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]


