Abstract
Tissue clearing technology is currently one of the fastest growing fields in biomedical sciences. Tissue clearing techniques have become a powerful approach to understand further the structural information of intact biological tissues. Moreover, technological improvements in tissue clearing and optics allowed the visualization of neural network in the whole brain tissue with subcellular resolution. Here, we described an overview of various tissue-clearing techniques, with focus on the tissue-hydrogel mediated clearing methods, and discussed the main advantages and limitations of transparent tissue for clinical diagnosis.
Keywords: Optical Tissue Clearing; Imaging, Three-Dimensional; Immunolabeling; Neuroscience; Molecular Diagnosis
INTRODUCTION
We live in a 3-dimensional (3D) space and it is natural for us to perceive objects as 3D. However, most of the biomedical techniques are based on 2-dimensional (2D) cross-sectional images, and the comprehensive understanding of biological system may require a detailed 3D description of tissue and organ structures. Computed tomography, magnetic resonance imaging, and positron emission tomography have been developed to transform 2D information into 3D, for diagnosis [1-3]. Although these methods are powerful tools to obtain 3D information of an entire organism, they still lack the sensitivity to resolve the individual cell. Therefore, most histological or pathological observations are based on the physical characteristics of biological samples. In cases where 3D information is ultimately necessary, serial sections of the samples are imaged individually, aligned, and reconstructed digitally [4]. This is a highly time-consuming process, often causing image distortion or loss of information. Moreover, the entire structures of large and irregular objects are difficult to reconstruct using the known methods. For instance, neurons have highly branched axons and dendrites, and the structural complexity increases as their axons and dendrites travel long distances with widespread innervations. Hence, it is virtually impossible to obtain the whole morphology of neurons in intact biological tissues using traditional histological methods.
In the past decades, many tissue-clearing techniques were developed for 3D visualization of macromolecules without tissue sectioning (Table 1). Given that cellular-resolution imaging is primarily performed by light microscopy, optical clearing of tissue will greatly enhance the penetration of light deep into the tissue, allowing the monitoring of deep signals without discontinuation. The feasibility of understanding the anatomy and structure of biological tissue has increased because of the technical improvements in recent tissue clearing methods and light microscopy. Photons and light are dispersed when passing through the biological tissue of sufficient thickness. Therefore, light cannot penetrate deep into the tissues, with the maximal depth penetration by far-red light to be less than 100 μm [5]. Most of the incoming light is scattered between the lipophilic membrane and aqueous environment of the tissue because each component has a different refractive index (RI) in the intact tissue. The tissue contains a little luminous source, each sending light in every direction and allowing us to observe a thin layer of tissue.
Table 1.
Comparison of published tissue clearing methods
| Methods | Main chemicals | Clearing capability | Entire processing time | Tissue morphological change | Molecular integrity | Immunolabeling after tissue clearance | Fluorescence signal preservation | Long-term storage |
|---|---|---|---|---|---|---|---|---|
| BABB | Benzyl alcohol/benzyl benzoate | Strong | 3 Days | Shrinkage | Fluorescent quenching | No | No | Difficult |
| ClearT | Formamide | Medium | 3 Days (embryo) | No tissue expansion | No fluorescent protein quenching | Yes | Yes | Difficult |
| Sca/e | Urea/glycerol TritonX100 | Medium | 3 Weeks to 6 months | Large expansion in tissue volume/tissue becomes fragile | Partial denaturation and protein loss | No | No | Possible |
| 3DISCO | Dibenzyl ether and tetrahydrofuran | Strong | 5–7 Days | No tissue expansion | No fluorescent protein quenching within 1 day | No | No | Difficult |
| CLARITY | SDS, Boric acid/focusclear | Strong | 2 Weeks~ | Tissue expansion during the process | No fluorophore quenching | Yes | Yes (many rounds) | Possible |
| SeeDB | Fructose α-thioglycerol | Weak | 3 Days (brain slice) | No tissue expansion | No fluorescent protein quenching | No | No | Possible |
| CUBIC | Urea/aminoalcohol sucrose/nitrilotriethanol | Strong | 2 Weeks~ | Tissue expansion during the process | No fluorescent protein quenching | Yes | Partially yes | Difficult |
| PACT/PARS | SDS, Boric acid/histodenz | Strong | 2 Weeks~ | Tissue expansion during the process | No fluorescent quenching | Yes | Yes | Possible |
| iDISCO | Benzyl ether | Strong | 14–17 Days (whole brain) | Shrinkage | No fluorescent protein quenching within 2–3 days | No | Partially yes | Difficult |
| Sca/eS | Urea/sorbitol | Medium | 1 Week | No tissue expansion | No fluorescent quenching | Yes | Yes | Possible |
| SWITCH | SDS | Weak | 2 Weeks~ | Large expansion in tissue volume | No fluorescent quenching | Yes | Yes (many rounds) | Possible |
| ACT-PRESTO | SDS, boric acid/sucrose, urea | Strong | 2–3 Days | Tissue expansion during the process | No fluorescent quenching | Yes | Yes | Possible |
| EDC-CLARITY | SDS, boric acid | Strong | ~3 Weeks (brain slice) | Tissue expansion during the process | No fluorescent quenching | Yes | Yes | Possible |
The processing time indicates the time needed for the clearing of the whole brain or hemisphere of adult mouse.
BABB, benzyl alcohol and benzyl benzoate; 3DISCO, 3-dimensional imaging of solvent-cleared organs; CLARITY, clear lipid-exchanged acrylamide-hybridized rigid imaging/immunostaining/in situ hybridization-compatible tissue-hydrogel; SeeDB, see deep brain; CUBIC, clear unobstructed brain imaging cocktails and computational analysis; PACT, passive clear lipidexchanged acrylamide-hybridized rigid imaging/immunostaining/in situ hybridization-compatible tissue-hydrogel; PARS, perfusion-assisted agent release in situ; iDISCO, immunolabeling-enabled 3-dimensional imaging of solvent-cleared organs; SWITCH, system-wide control of interaction time and kinetics of chemicals; ACT, active clarity technique; PRESTO, pressure related efficient and stable transfer of macromolecules into the organs; EDC-CLARITY, 1-ethyl-3-3-dimethyl-aminopropyl carbodiimide-CLARITY; SDS, sodium dodecyl sulfate.
Many methods that optically clear intact tissue without sectioning have been developed. There are two major directions of tissue clearing. First, RI matching of the biological tissues is a unique method to improve tissue transparency. Biological tissues have intrinsic heterogeneities of RI (1.36–1.55) [5], and light is shattered multiple times resulting in an opaque tissue. Therefore, tissue transparency of biological samples submerged in a RI matching solutions will efficiently increase, especially in relatively small and soft tissues such as embryos. However, RI matching is not applicable to thick or hard tissue specimens; thus, researchers utilized a new method to resolve this problem. Second, active delipidation improves transparency because lipids are the main light-scattering factors in biological tissues. Combination of RI matching and delipidation substantially improves tissue transparency (Fig. 1).
Fig. 1.
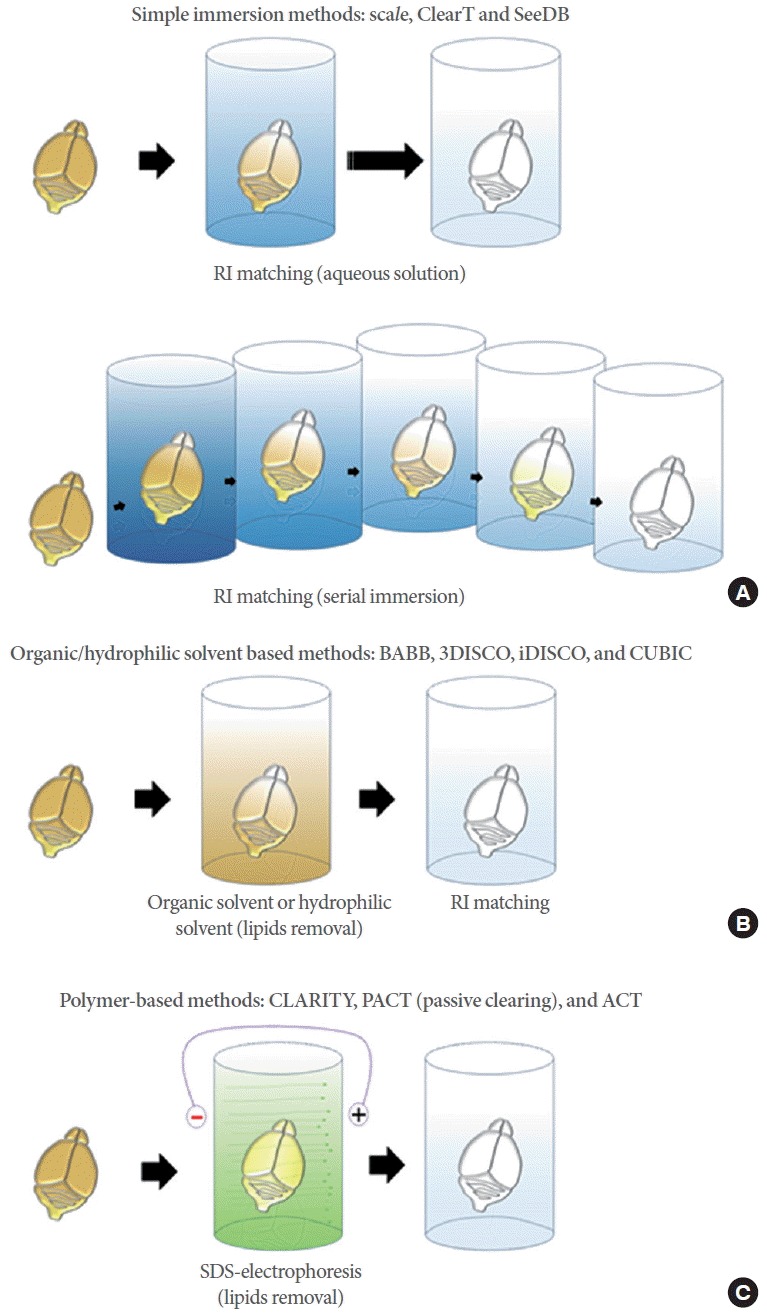
Tissue clearing technology. (A) Simple immersion methods. The tissue is placed in an aqueous clearing solution for days to months, and the solution is exchanged during the process (top). In other methods, samples are incubated in a series of immersion solutions with different concentrations. Finally, tissues greatly improve transparency (bottom). (B) Organic/hydrophilic solvent-based methods. Solvent-based clearing consists of 2 steps: tissue dehydration and lipid removal, and refractive index (RI) matching. (C) Polymer-based clearing methods. The lipid is removed in SDS-containing solution by electrophoresis or passive clearing system. The tissue is then placed in RI matching solution. BABB, benzyl alcohol and benzyl benzoate; 3DISCO, 3-dimensional imaging of solvent-cleared organs; iDISCO, immunolabeling-enabled 3-dimensional imaging of solvent-cleared organs; CUBIC, clear unobstructed brain imaging cocktails and computational analysis; CLARITY, clear lipid-exchanged acrylamide-hybridized rigid imaging/immunostaining/in situ hybridization-compatible tissue-hydrogel; PACT, passive clear lipid-exchanged acrylamide-hybridized rigid imaging/immunostaining/in situ hybridization-compatible tissue-hydrogel; ACT, active clarity technique; SDS, sodium dodecyl sulfate.
In this review, we discuss the current variations of tissue clearing techniques and their future applications in 3D diagnostics.
UREA AND OPTICAL CLEARING REAGENT-BASED METHODS: scale, ClearT, AND SeeDB
Many researchers have developed techniques to increase the transparency via RI matching. RI matching methods are very simple and cost-effective; they use passive diffusion and require no special equipment. Easily available RI matching reagents, such as sugar, fructose, and formamide [6-8], have low molecular weight and RIs between 1.40 and 1.45. SeeDB (see deep brain) [6] and ClearT [7] are examples of simple immersion tissue clearing techniques (Fig. 1A). Biological samples are incubated in a series of immersion solutions with different percentages of fructose and formamide. The main advantage of the RI matching method is the maintenance of the structural information and fluorescence in biological tissue. However, they rely on the free diffusion of immersion solutions; thus, the reaction time is very slow and obtaining a completely transparent tissue is difficult. To improve the transparency, urea is often included in the RI matching solution because of its high RI, water solubility, and hydration in biological tissues [9,10]. The urea-based clearing method was named scale (Fig. 1A), and various versions of it have been reported, such as scaleA2 and scaleU2 [10]. Although transparency of tissues was markedly enhanced and the fluorescence of the biomarker was maintained, the samples swelled and softened during the clearing process with rapid decreased in fluorescence signal. These limitations were partially overcome by modification in the recipe of the solution, termed scaleS (sorbitol-based optical clearing method) [11], which allows minimal volume changes and fluorescence quenching.
HYDROPHILIC SOLVENT-BASED METHOD: CUBIC
The aqueous component-based simple immersion clearing exhibits tissue thickness-related limitation as discussed earlier. However, addition of hydrophilic solvent in the RI matchin solution can greatly improve tissue transparency. Clear unobstructed brain imaging cocktails and computational analysis (CUBIC) includes both hydrophilic solvent-based lipid elimination and RI adjustment steps (Fig. 1B) [12]. Advantages of this method include consistent tissue size, high transparency rate, and relatively easy procedure. Therefore, this procedure can be modified for whole-body clearing with cardiac perfusion of clearing solution [13]. It also maintains signals from green fluorescent protein (GFP) variants, and the antibody can penetrate the processed tissue. However, white matter or other lipid-rich regions remain translucent, and the tissue becomes very soft after prolonged lipid removal. Moreover, the overall processing time is relatively long (15–19 days for whole mouse brain clearing).
ORGANIC SOLVENT-BASED METHODS: BABB, 3DISCO, AND iDISCO
Organic solvents can effectively remove lipids of biological tissue by a strong dehydration effect, resulting in a hard and crystal-clear tissue. Methyl salicylate, benzyl alcohol, benzyl benzoate, dichloromethane, and dibenzylether have been used as organic solvents [14-17]. Benzyl alcohol and benzyl benzoate (BABB) was the first established organic solvents for tissue clearing and has been used for many different types of biological tissues in the past decades [14,18,19]. Although high tissue transparency is achieved, fluorescence signal is rapidly reduced during the clearing process. 3D imaging of solvent-cleared organs (3DISCO) [16] shows better fluorescence preservation compared with BABB [14]. Immunolabeling-enabled 3D imaging of solvent-cleared organs (iDISCO) [17] is a recently reported, modified version of 3DISCO (Fig. 1B). These 3 methods for tissue clearing are composed of a 2-step process: delipidation of biological samples, followed by RI matching. Samples become dramatically transparent in organic solvent within several hours [20]. Fixatives such as methanol or acetone do not generate covalent bonding of macromolecules but precipitate by dehydration; thus, proteins are not stably immobilized. This cause structural changes in proteins and rapid quenching of reporter fluorophores such as GFPs [12]. In addition, dehydration and delipidation cause remarkable shrinkage of the samples, which is not desirable for accurate quantifications. However, shrinkage of samples is beneficial in some cases because it reduces the burden for image acquisition and postprocessing load. Another concern on these methods is the use of a highly toxic organic solvent that requires special care.
POLYMER-BASED METHODS: CLARITY, PACT, and ACT
CLARITY (clear lipid-exchanged acrylamide-hybridized rigid imaging/immunostaining/in situ hybridization-compatible tissue-hydrogel) is a new tissue clearing method has been recently developed [21]. Unlike other technologies, lipids are selectively removed by electrophoresis, whereas DNA and proteins are retained in the biological tissue (Fig. 1C). To keep the important substances intact, acrylamide copolymerization is applied after molecular cross-linking by fixation. Addition of polymer into the tissue can support the integrity of the sample, especially after lipid removal. Complete removal of membranous components markedly increases the transparency of the tissues. The porous nature of hydrogel also allows fast penetration of reagents into thick tissues. The fluorescent probes are maintained, and immunolabeling can be performed repeatedly in the same sample. In CLARITY, tissues are incubated in a 4% paraformaldehyde (PFA) and hydrogel monomer solution (4% acrylamide and 0.05% bisacrylamide solution), which induces cross-linking between tissue and hydrogel monomers. A fine hydrogel mesh structure is created in intact biological tissue with higher concentration of hydrogel monomers in immersion solution [21]. As a result, tissue clearing takes a very long time if lipids are to be removed by simple diffusion of detergents. Therefore, the extraction of all non–cross-linked lipids is achieved by active electrophoresis with sodium dodecyl sulfate (SDS)-containing tissue clearing buffer (Fig. 2A) [21-23]. CLARITY technique requires apparatus for electrophoretic tissue clearing (ETC), and the efficacy of the clearing is highly dependent on the ETC conditions, such as temperature, pH, and currents.
Fig. 2.
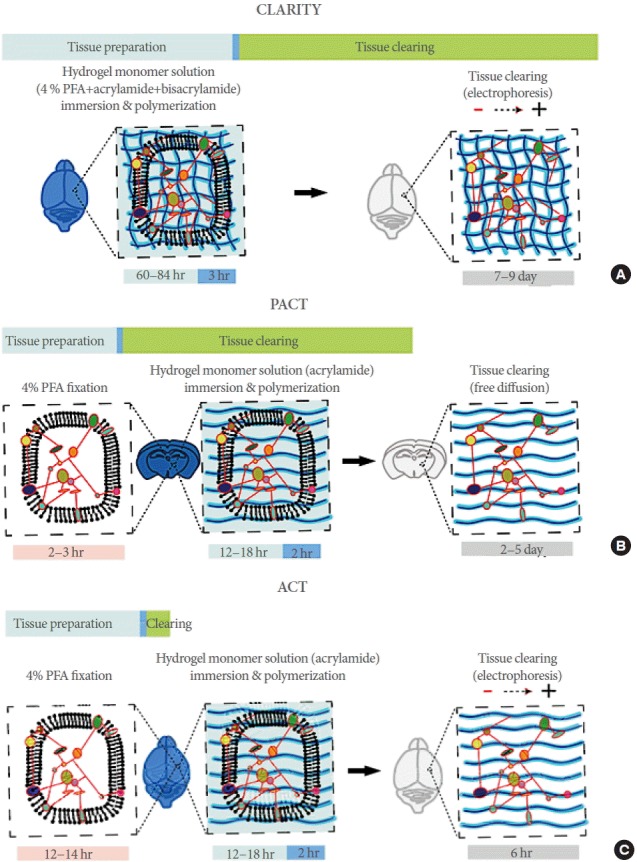
Comparison of polymer-based methods. (A) CLARITY tissue-polymer system and processing time. Hydrogel monomer mixture contains 4% PFA, 4% acrylamide, and 0.05% bis-acrylamide. After polymerization, cross-linking between tissue and hydrogel monomers occurs. The entire processing time to clarify the whole brain of adult mouse is over 2 weeks. (B) PACT tissue-polymer system and processing time. This method uses a thick-sliced, fixed sample and 4% acrylamide solution (hydrogel monomer solution), and the lipid is passively diffused out from the tissue. The entire processing time to clarify adult mouse brain slices (2-to 3-mm thickness) is over 1 week. (C) ACT tissue-polymer system and processing time. Fixed whole brain is incubated in 4% acrylamide solution. After polymerization, the tissue has a less complex tissue-hybrid network formation than CLARITY, and lipid is removed by electrophoretic tissue clearing within 6 hours. CLARITY, clear lipid-exchanged acrylamide-hybridized rigid imaging/immunostaining/in situ hybridization-compatible tissue-hydrogel; PFA paraformaldehyde; PACT, passive clear lipid-exchanged acrylamide-hybridized rigid imaging/immunostaining/in situ hybridization-compatible tissue-hydrogel; ACT, active clarity technique; SDS, sodium dodecyl sulfate .
PACT (passive clear lipid-exchanged acrylamide-hybridized rigid imaging/immunostaining/in situ hybridization-compatible tissue-hydrogel) is a modified version of CLARITY wherein tissue fixation and hydrogel infusion steps are separated. Because fixation-based protein and polymer networks have less interactions, resulting tissues are more porous and extraction of lipids can be achieved by simple diffusion without electrophoresis (Fig. 2B) [24]. Although this method allows 3D visualization of macromolecules in intact tissue, the entire process for clarifying the whole mouse brain is time-consuming, requiring several weeks.
Rapid clearing of tissues is essential for the application of this method to routine basic or clinical laboratory methodology for life science research or volume diagnosis. To overcome the limitations of CLARITY technique, active clarity technique (ACT) was recently developed. It includes optimized ETC systems and tissue hybridization condition for rapid tissue clearing [25]. With this technique, it is possible to increase the speed of tissue transparency by 30–100 times than CLARITY or other tissue transparent techniques. It takes about 6–24 hours to clarify the whole organs of the mouse. Because the tissue-hydrogel formed in ACT procedure is highly porous as PACT, penetration of small compounds and macromolecules such as antibodies is more efficient (Fig. 2C). ACT requires several hours for the completion of tissue clearing; thus, 3–7 days are required for the entire process of 3D immunolabeling with thick sections or organs, from sampling to image acquisition [25]. Rapid processing time of ACT is by far the most suitable for routine clinical diagnostics. The overall times for 3D imaging with ACT processing and 2D imaging with conventional paraffin embedding are similar (<1 week).
GLUTARALDEHYDE-BASED METHOD: SWITCH
SWITCH (system-wide control of interaction time and kinetics of chemicals) is the most recently developed method for proteomic imaging of intact tissues [26]. Aggressive cross-linking of proteins with glutaraldehyde makes tissues durable to the subsequent harsh processes such as SDS treatments and exposure to high temperature. Therefore, the authors obtained dozens of protein information from the same biological tissue and analyzed their significance. The advantages of this technology are expected to be used as a molecular diagnostic technique by multilabeling of same biological tissue. However, one limitation of this strategy is that glutaraldehyde is a very strong fixative, which should inactivate many epitope, and the robust screening of antibody markers may be required.
DETECTION OF PROTEIN, RNA, AND OTHER MACROMOLECULES AFTER TISSUE CLEARING
Most of the tissue clearing methods described here allow fluorescence imaging of proteins and mRNA in the biological tissues [21,25]. In order to obtain the distribution of proteins in transparent tissue, we should first consider the stability of antigenicity and penetration efficiency of antibodies. Among tissue clearing methods, iDISCO [17], ACT [25], and SWITCH [26] reported that tissue antigenicity is relatively well preserved throughout the clearing process. However, proteins are exposed to different solvents and temperatures, and, thus, the compatibility of each antibody should be empirically tested. Another important factor is the diffusion of macromolecules into thick tissues. The pore size of the processed tissue is a critical determinant for the extent of macromolecule diffusion. Therefore, highly porous structures are desirable in CUBIC, PACT, or ACT, whereas CLARITY or iDISCO has significant limitation of free diffusion. Although it is generally true that lipid removal increases the pore size of the tissues and penetrability, dense connective tissues may also block the diffusion of macromolecules. To overcome this limitation, many methods are designed to actively deliver macromolecules into deep tissues by altering electrical, physical, and chemical factors, such as stochastic electrotransport [27], PRESTO (pressure related efficient and stable transfer of macromolecules into the organs) [25], and SWITCH [26], respectively. These techniques could be applicable to the wide variety of tissue labeling techniques.
While achieving covalent linkages between amino acid residues in the process of tissue fixation, proteins form a network that improves their stability after tissue clearing. However, RNA and DNA do not have amino acid residues, and thus they are not stable in fixed biological tissue. There is a possibility that the position of RNA and DNA is changed by their electrical properties in techniques using electrophoresis for tissue clearing such as CLARITY [21] and ACT [25]. Therefore, it is necessary to use a polymer that has the ability to capture RNA and form a stable cross-linking within tissue-hydrogel polymer. Recently, EDC-CLARITY (1-ethyl-3-3-dimethyl-aminopropyl carbodiimide-CLARITY) [28] was developed to achieve high resolution imaging of diverse coding and non-coding RNAs within intact tissue. For fixation of targeting functional groups on the RNA molecules, researchers used EDC and PFA. This method enables volumetric RNA detection in transparent biological tissue [28]. However, EDC-CLARITY has limitations including complex experimental approach and specialized apparatus; thus, it still requires improvements in the procedure.
CONCLUSION
We are now able to see through the tissues using clearing technologies mentioned above. It is expected that tissue-clearing methods will soon be applied to 3D diagnosis. In order to do so, development of better imaging techniques is necessary. Rapid imaging is essential in clinical diagnosis; the whole process from tissue collection to image analysis should be finished within a few days. Currently, ACT can achieve the most rapid tissue clearing and immunolabeling within 5–7 days. Conventional confocal microscopy requires 24–50 hours for completion of 1 mm3 in general. Imaging speed and resolution are very important factors in diagnosis, and fast fluorescence imaging techniques such as single plane illumination microscopy [29] or a high resolution fine structure imaging technology using scattering of light such as optical coherence tomography [30] may improve the speed and reliability of 3D imaging for clinical diagnosis.
Footnotes
Fund Support
This work was supported by the Brain Research Program through the National Research Foundation (NRF) funded by the Korean Ministry of Science, ICT & Future Planning (NRF-2015M3C7A1028790).
Conflict of Interest
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Pelizzari CA, Chen GT, Spelbring DR, Weichselbaum RR, Chen CT. Accurate three-dimensional registration of CT, PET, and/or MR images of the brain. J Comput Assist Tomogr. 1989;13:20–6. doi: 10.1097/00004728-198901000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Clarke LP, Velthuizen RP, Camacho MA, Heine JJ, Vaidyanathan M, Hall LO, et al. MRI segmentation: methods and applications. Magn Reson Imaging. 1995;13:343–68. doi: 10.1016/0730-725x(94)00124-l. [DOI] [PubMed] [Google Scholar]
- 3.Judenhofer MS, Wehrl HF, Newport DF, Catana C, Siegel SB, Becker M, et al. Simultaneous PET-MRI: a new approach for functional and morphological imaging. Nat Med. 2008;14:459–65. doi: 10.1038/nm1700. [DOI] [PubMed] [Google Scholar]
- 4.Griffini P, Smorenburg SM, Verbeek FJ, van Noorden CJ. Three-dimensional reconstruction of colon carcinoma metastases in liver. J Microsc. 1997;187(Pt 1):12–21. doi: 10.1046/j.1365-2818.1997.2140770.x. [DOI] [PubMed] [Google Scholar]
- 5.Berke IM, Miola JP, David MA, Smith MK, Price C. Seeing through musculoskeletal tissues: improving in situ imaging of bone and the lacunar canalicular system through optical clearing. PLoS One. 2016;11: doi: 10.1371/journal.pone.0150268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ke MT, Fujimoto S, Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013;16:1154–61. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- 7.Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, Mason C. ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development. 2013;140:1364–8. doi: 10.1242/dev.091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai PS, Kaufhold JP, Blinder P, Friedman B, Drew PJ, Karten HJ, et al. Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels. J Neurosci. 2009;29:14553–70. doi: 10.1523/JNEUROSCI.3287-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stumpe MC, Grubmüller H. Aqueous urea solutions: structure, energetics, and urea aggregation. J Phys Chem B. 2007;111:6220–8. doi: 10.1021/jp066474n. [DOI] [PubMed] [Google Scholar]
- 10.Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, et al. scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–8. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 11.Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, et al. scaleS: an optical clearing palette for biological imaging. Nat Neurosci. 2015;18:1518–29. doi: 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
- 12.Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–39. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 13.Tainaka K, Kubota SI, Suyama TQ, Susaki EA, Perrin D, Ukai-Tadenuma M, et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell. 2014;159:911–24. doi: 10.1016/j.cell.2014.10.034. [DOI] [PubMed] [Google Scholar]
- 14.Dodt HU, Leischner U, Schierloh A, Jahrling N, Mauch CP, Deininger K, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods. 2007;4:331–6. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- 15.Becker K, Jährling N, Kramer ER, Schnorrer F, Dodt HU. Ultramicroscopy: 3D reconstruction of large microscopical specimens. J Biophotonics. 2008;1:36–42. doi: 10.1002/jbio.200710011. [DOI] [PubMed] [Google Scholar]
- 16.Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc. 2012;7:1983–95. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- 17.Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Keller PJ, Dodt HU. Light sheet microscopy of living or cleared specimens. Curr Opin Neurobiol. 2012;22:138–43. doi: 10.1016/j.conb.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Dent JA, Polson AG, Klymkowsky MW. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development. 1989;105:61–74. doi: 10.1242/dev.105.1.61. [DOI] [PubMed] [Google Scholar]
- 20.Azaripour A, Lagerweij T, Scharfbillig C, Jadczak AE, Willershausen B, Van Noorden CJ. A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Prog Histochem Cytochem. 2016 Apr 14; doi: 10.1016/j.proghi.2016.04.001. [Epub]. http://dx.doi.org/10.1016/j.proghi.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–7. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9:1682–97. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richardson DS, Lichtman JW. Clarifying tissue clearing. Cell. 2015;162:246–57. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–58. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee E, Choi J, Jo Y, Kim JY, Jang YJ, Lee HM, et al. ACT-PRESTO: rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Sci Rep. 2016;6:18631. doi: 10.1038/srep18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray E, Cho JH, Goodwin D, Ku T, Swaney J, Kim SY, et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell. 2015;163:1500–14. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SY, Cho JH, Murray E, Bakh N, Choi H, Ohn K, et al. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc Natl Acad Sci U S A. 2015;112:E6274–83. doi: 10.1073/pnas.1510133112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sylwestrak EL, Rajasethupathy P, Wright MA, Jaffe A, Deisseroth K. Multiplexed intact-tissue transcriptional analysis at cellular resolution. Cell. 2016;164:792–804. doi: 10.1016/j.cell.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–9. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 30.Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, Chang W, et al. Optical coherence tomography. Science. 1991;254:1178–81. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]


