Abstract
Metastatic disease is responsible for the majority of prostate cancer deaths. The standard treatment for metastatic disease is surgical or chemical castration in the form of androgen deprivation therapy. Despite initial success and disease regression, resistance to therapy ultimately develops and the disease transitions to castration resistant prostate cancer, which is uniformly fatal. Thus, developing an understanding of genetic evolution in metastasis and in response to therapy has been a focus of recent studies. Large-scale sequencing studies have provided an expansive catalog of the mutation events that occur in the prostate cancer genome at various stages of disease progression. Smaller-scale studies have interrogated the genomic composition of multiple metastatic sites within individual patients, or have tracked clonal evolution longitudinally in tissues, circulating tumor cells, or circulating tumor DNA. Collectively, these efforts have provided a new conceptual framework for understanding the origin of prostate cancer, as well as the origin and evolution of metastatic disease. In this review, we will highlight these recent insights into the spatiotemporal landscape of genetic evolution of prostate cancer.
Keywords: metastasis, prostate, endocrine therapy resistance, cell lineage/genetics, clonal
Introduction
Prostate cancer is the most frequently diagnosed cancer in men and accounts for an estimated 142,000 deaths in developed countries each year (Torre, et al. 2015). Five year survival rates for localized disease are high (>90%) because localized cancers can be treated successfully with surgery and radiation. In normal prostate cells, the androgen receptor (AR) functions as a master transcriptional regulator activated by the androgens testosterone and dihydrotestosterone. Accordingly, prostate cancer presents as an androgen and AR- dependent disease (Dehm and Tindall 2011). For patients where surgery and radiation are not curative, the standard systemic treatment is functional suppression of AR transcriptional activity through androgen deprivation therapy (ADT). ADT includes surgical castration, pharmacologic castration, and antiandrogen therapy (Dehm and Tindall 2011). ADT is initially successful in most patients, but over time, castration resistant disease develops. Therefore, development of therapy resistance and transition to castration-resistant prostate cancer (CRPC) is a major clinical challenge. CRPC is characterized by rising PSA levels (signifying reactivation of AR transcriptional activity), an increase in tumor size, and metastatic spread (Knudsen and Scher 2009). Second-line ADT drugs that provide a more effective blockade of systemic androgen synthesis (abiraterone acetate) or act as higher-affinity antagonists of AR (enzalutamide) improve overall survival (Beer, et al. 2014; Ryan, et al. 2013). However, in many patients, second-line ADT fails to achieve control of tumor growth, which indicates frequent occurrence of primary resistance. Additionally, secondary resistance to these therapies inevitably develops in a relatively short time-frame and CRPC thus remains a uniformly fatal disease.
Clonal populations of cancer cells are in persistent evolution in response to environmental conditions. The concept of tumor evolution was proposed by Nowell in 1976 based on cytogenetic data (Nowell 1976). In this early model, a cell of origin acquires genetic alterations that promote neoplasia. Further genetic instability fuels clonal expansion of “fit” clones that ultimately leads to advanced malignancy, metastasis, and emergence of therapy-resistance (Nowell 1976). Large-scale genome sequencing studies in recent years have provided expansive catalogs of the genomic aberrations in primary and advanced prostate cancer and have offered insight into deregulated molecular pathways at each stage of disease for the development of novel targeted therapies (Abeshouse, et al. 2015; Robinson, et al. 2015). However, these large-scale genome sequencing studies have focused on single samples of bulk tumors collected at a single time point from individual patients, which does not enable a complete understanding of tumor heterogeneity. An additional limitation with this single-site/single-time sampling approach is that it does not reveal the subclonal changes that occur within tumor populations spanning disease progression and recurrence after therapeutic intervention.
More recently, smaller-scale studies focused on sampling of multiple metastatic sites or longitudinal sample collection combined with computational reconstructions of clonal evolution have confirmed that primary tumors and adjacent normal tissue consist of multiple clonal populations. These studies have further revealed that metastatic spread can occur through monoclonal or polyclonal seeding between metastases or in waves originating from the primary tumor. Moreover, genomic alterations associated with resistance to ADT have been identified, including AR and AR pathway components (further expanded upon and reviewed in (Watson, et al. 2015)), highlighting an opportunity for development of biomarkers representative of resistant subclones vis-a-vis personalized medicine approaches. In this review, we will discuss recent efforts to understand clonal evolution and map the clonal spread and expansion of metastatic prostate cancer. Collectively, these studies have illuminated several intricate mechanisms by which discrete clones can undergo selection and spread in individual patients.
Genomic approaches to interrogate clonal framework in tumor tissues
The multifocal and heterogeneous nature of prostate cancer can hinder efforts to understand tumor cell clonality, particularly in metastatic disease. Until recently, the tools available to assess clonality within multifocal and heterogeneous tumor samples relied upon histological assessment, cytogenetic approaches such as FISH, and molecular approaches such as PCR. Many malignancies exhibit multiclonality including AML, breast, melanoma, esophageal, and non-small cell lung cancer (Bolli, et al. 2014; Ding, et al. 2012; McFadden, et al. 2014; Merlo, et al. 2010; Nik-Zainal, et al. 2012; Rashid, et al. 2014). By applying the population genetics concept of the most-recent common ancestor it has been possible to quantify genomic aberrations and thereby define clonal and subclonal populations of cells responsible for metastatic seeding and evasion of therapy (Campbell, et al. 2008; Nik-Zainal et al. 2012). The first step in this strategy is to determine tumor purity, which is critical because resected tumors and biopsied tumor tissues harbor stromal cell infiltrates. This can be accomplished by identifying the fraction of tumor cells carrying clusters of mutations relative to normal tissue. (Van Loo, et al. 2010). To define clonal and subclonal populations of cells within metastases at different body sites, mutant allelic fraction in multiple tumor sections relative to normal tissues can be calculated, taking into account tumor purity and copy number (Campbell et al. 2008; Nik-Zainal et al. 2012). Any mutations present at a smaller proportion in comparison with the clonal population would be indicative of sub-clonal events.
Using these methods, clonal evolution can be tracked across various disease states for which there is tissue available, and can be used to reconstruct a spatiotemporal mutational landscape of disease progression. For example, this approach was used to retrospectively construct a timeline of clonal evolution and metastatic spread in a rapid autopsy study of 10 subjects that died from prostate cancer (Gundem, et al. 2015). Alternatively, these methods enable monitoring of clonal evolution in real time. For example, one study collected longitudinal samples of blood and tumor biopsies from four patients with advanced prostate cancer as their disease progressed from localized prostate cancer, to biochemical recurrence, and ultimately to CRPC (Hong, et al. 2015). A separate study examined clonal evolution in response to selective pressures of therapy by targeted sequencing of plasma DNA and targeted deep-sequencing of tumor biopsies obtained from patients with ERG positive cancers (Carreira, et al. 2014). Together these studies build upon an existing catalog of known mutations, structural alterations, and altered pathways to inform our understanding the clonal evolution and spread of prostate cancer, particularly in response to AR targeted therapies.
Primary prostate cancer is multifocal and heterogeneous
Prostate cancer often presents as discrete foci within the prostate capsule (Ruijter, et al. 1999). Whether these discrete foci represent independent clones or geographically separated, yet related, clonal populations has been a subject of debate (Barry, et al. 2007; Boutros, et al. 2015; Boyd, et al. 2012; Cheng, et al. 1998; Cooper, et al. 2015; Kobayashi, et al. 2008; Lindberg, et al. 2013). For example, in a study of 254 prostatectomy specimens, nearly half harbored multiple individual tumor foci separated by at least 3mm within the resected gland (Villers, et al. 1992). In these cases, it is possible that two distinct tumor cell populations arose in the prostate. Alternatively, independent foci may represent related clonal populations. Similarly, a study of 17 radical prostatectomy specimens indicated the presence of multiple tumor foci in several samples. Analysis of discrete foci revealed concordant as well as discordant allelic imbalances, suggesting variability in clonal origin of tumor foci within an individual prostate gland (Ruijter et al. 1999). In a separate study of 47 prostatectomy specimens, ~20% were multifocal and exhibited varying grades of disease. Loss of heterozygosity (LOH) was observed within tumors, but intertumoral LOH was not predictable from intratumoral patterns of allelic loss (Hugel and Wernert 1999).
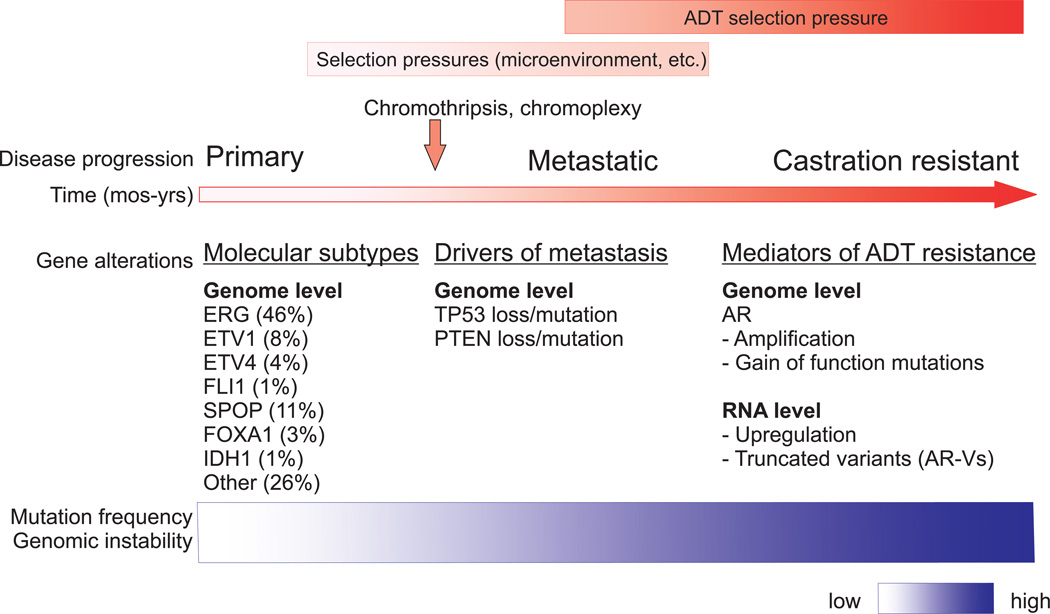
Primary prostate cancer can be sub-classified according to a recurrent set of mutually exclusive genomic alterations that occur early in disease development (Abeshouse et al. 2015). Specifically, analysis of 333 primary prostate carcinomas in The Cancer Genome Atlas (TCGA) project identified seven mutually exclusive genetic subtypes including ERG, ETV1, ETV4, FLI1, SPOP, FOXA1, and IDH1 (Abeshouse et al. 2015). Roughly 75% of prostate cancers fell into one of these genetic subtypes, however; ~25% remained uncharacterized (Figure 1). Identification of additional genetic alterations has provided evidence that primary tumors exhibit multiclonality coincident with multifocal disease. For example, whole genome sequencing (WGS) in a cohort of 5 patients (Gleason Scores 7–8) who underwent radical prostatectomies, provided the first WGS study demonstrating extensive genomic heterogeneity within prostate tumors (Boutros et al. 2015). One example of multiclonality within the resected prostate was a patient whose tumor was dissected into four regions, each of which harbored disease foci. The index lesion shared SPOP mutations and chromosomal deletions (on chr8, 16) with one focus, but not the remaining two dissected foci, which shared a deletion on chr19 (Boutros et al. 2015). Importantly, this study also discovered a previously unidentified recurrent amplification of MYCL associated with TP53 loss (Boutros et al. 2015). Similarly, morphologically normal tissues have been found to exhibit high levels of mutations and distinct ERG fusions that are present in malignant tissues (Cooper et al. 2015). In many cases, discrete tumor foci within the prostate are comprised of distinct clones representing different genetic subtypes or harboring different driver mutations, indicating independent clonal origins. For example, interrogation of two independent foci present in a prostatectomy specimen demonstrated they were distinct in clonal origin: one was ERG positive, while the other exhibited SPOP mutation (Barbieri, et al. 2012). It is important to note, however, that in this cohort (112 patients) the authors reported only this single case of multifocal disease. In a smaller study of four patients, three discrete prostate cancer foci had no common somatic ancestor as assessed by high frequency single nucleotide variants (SNVs) and copy number alteration profiling (Lindberg et al. 2013). In a study of 48 patients with ERG positive prostate cancer, areas of high grade prostatic intraepithelial neoplasia (PIN) and intraductal carcinoma proximal to areas of invasive carcinoma were found to be ERG positive (Haffner, et al. 2015). Higher-resolution studies of tissues from 7 of those patients demonstrated that subclonal PTEN loss present in the ERG positive invasive adenocarcinoma was also present in adjacent PIN lesions, indicating retrograde spreading to benign structures (Haffner et al. 2015).
Figure 1.
Gradual acquisition of genomic alterations is thought to be the primary driver of tumorigenesis. However, accelerated mechanisms of genomic alteration, such as chromothripsis, a massive and catastrophic reshuffling of entire chromosomes or regions of chromosomes, and chromoplexy, complex rearrangements that may arise from multiple rounds of DNA repair, could account for shortened time-frames of tumorigenesis and metastasis (Baca, et al. 2013; Stephens, et al. 2011). Indeed, chromoplexy has been shown to capture multiple genetic hits in one structural rearrangement event, which would be predicted to accelerate tumorigenesis and clonal evolution. For example, in one case of prostate cancer, the coding regions of several tumor suppressor genes including ETV6, ETV3, and CDKN1B, were disrupted by a single chromoplexy event involving 25 rearrangements spanning 3 chromosomes (Baca et al. 2013). In another case of prostate cancer, 27 rearrangements occurred in a single chromoplexy event to yield a SMAD rearrangement as well as an oncogenic TMPRSS2:ERG fusion (Baca et al. 2013).
The most common lesions in primary tumors, affecting ~50% of cancers, are ETS fusions, wherein AR-regulated or otherwise highly active promoters and/or enhancers such as TMPRSS2 are fused to the coding regions of ERG, ETV1, ETV4, or FLI1 (Abeshouse et al. 2015; Adamo and Ladomery 2015; Barbieri, et al. 2013; Taylor, et al. 2010). Interestingly, these gene fusion events have been shown to be promoted by androgen/AR-regulated changes in intra-nuclear chromatin organization (Lin, et al. 2009; Mani, et al. 2009; Weischenfeldt, et al. 2013). For example, AR binding sites have been shown to exist near TMPRSS2, ERG, and ETV1 fusion breakpoints, and androgen-induced binding of AR to these sites has been shown to bring 5’ and 3’ gene fusion partners into close proximity (Lin et al. 2009). Genotoxic stress has been shown to cooperate with the androgen-mediated proximity effect to induce TMPRSS2:ETS or TMPRSS2:ETV1 gene fusion events identical to those observed in clinical prostate cancer (Lin et al. 2009; Mani et al. 2009). Interestingly, this androgen-mediated proximity effect may promote the genesis of additional rearrangement events in the prostate cancer genome, as AR binding sites are frequently found adjacent to rearrangement breakpoints (Weischenfeldt et al. 2013).
Clonal Evolution and Spread of Metastatic Prostate Cancer
Relative to metastatic disease, primary tumors exhibit low mutation frequencies and genome instability (Abeshouse et al. 2015; Robinson et al. 2015). As the disease progresses to the metastatic and castration resistant phases, extensive mutations, structural rearrangements, and genome instability are observed. These genomic alterations are indicative of clonal evolution and subclonal selection to survive environmental pressures. Importantly, altered AR signaling after metastatic spread spans subtypes, occurs in greater than 50% of metastases, and remains a key target for therapies (Robinson et al. 2015). Commonly identified genomic alterations in metastatic CRPC are TP53 mutation or loss, PTEN loss and PI3K pathway defects, DNA repair pathway deficiencies, and amplification or mutation of AR (Figure 1) (Grasso, et al. 2012; Hong et al. 2015; Kim, et al. 2007; Lohr, et al. 2014; Robinson et al. 2015). It has been challenging to differentiate alterations that promote metastasis vs. alterations that promote therapy resistance because, historically, studies of metastatic prostate cancer have been performed using tissues from heavily-treated CRPC patients. Exome sequencing of CRPC metastases obtained at rapid autopsy from a cohort of 50 patients identified frequent mutations in TP53, AR, FOXA1, and PTEN, among others (Grasso et al. 2012). Indeed, the finding that mutation or amplification of AR do not occur in primary prostate cancer but are common genomic alterations in CRPC metastases appear to implicate AR as a driver of metastasis (Abeshouse et al. 2015). However, more detailed whole-genome sequencing studies have indicated AR as a driver of therapy resistance, rather than a driver of metastasis (Figure 1) (Carreira et al. 2014; Gundem et al. 2015; Hong et al. 2015; Romanel, et al. 2015). For instance, interrogation of subclonal structure within CRPC metastases demonstrated that different AR gene alterations can occur in different metastatic foci within a single patient, indicating these alterations occurred in response to therapy, after metastatic spread (Gundem et al. 2015). On the other hand, this same analysis indicated that TP53 loss, DNA repair pathway alterations, and PTEN loss occurred before metastatic spread (Gundem et al. 2015). Further, the frequencies of TP53 and PTEN inactivation were higher in metastatic CRPC when compared to primary prostate cancer, indicating driver roles in metastasis (Abeshouse et al. 2015).
Clonal spread between the primary tumor and distant metastatic sites is multidirectional
Metastases are thought to be monoclonal in origin with respect to the primary tumor, with clones sharing a number of common mutations but exhibiting divergent somatic mutations between different metastatic sites (Gundem et al. 2015; Hong et al. 2015). Early rapid autopsy investigations observed that PSA immunostaining varied across and within metastatic foci, suggesting that multiple subclonal populations existed within metastases (Roudier, et al. 2003; Shah, et al. 2004). Genome-wide copy number analysis and targeted sequencing studies revealed that metastases within a patient shared common mutations and therefore shared clonal origins (Liu, et al. 2009; Robbins, et al. 2011). However, these same studies also demonstrated these tumors harbored sets of divergent mutations, indicating that subclonality existed between individual tumor foci. For example, analysis of three spatially distinct metastases from a single autopsy subject revealed shared amplifications at chromosomes 5p and 14q as well as a set of shared somatic mutations (Robbins et al. 2011). The presence of common copy number alterations and somatic mutations across multiple metastases indicated a clonal progenitor cell, presumably from the primary tumor. However, in this study primary tumor tissue was not available for this patient (Robbins et al. 2011). Similarly, copy number analysis and targeted resequencing of 80 anatomically distinct metastatic foci isolated from 24 patients with CRPC indicated that metastases shared a clonal origin in most patients (Liu et al. 2009). Importantly, metastases shared clonal copy number variations with the primary tumor in five patients with available tissues (Liu et al. 2009). These observations indicate that prostate cancer metastases originate from a common clonal progenitor in the prostate. However, as discussed below, alternate explanations may also be possible in light of recent data demonstrating that cells from prostate cancer metastases may be able to re-seed the surgical bed where the original primary tumor existed (Hong et al. 2015; Liu et al. 2009).
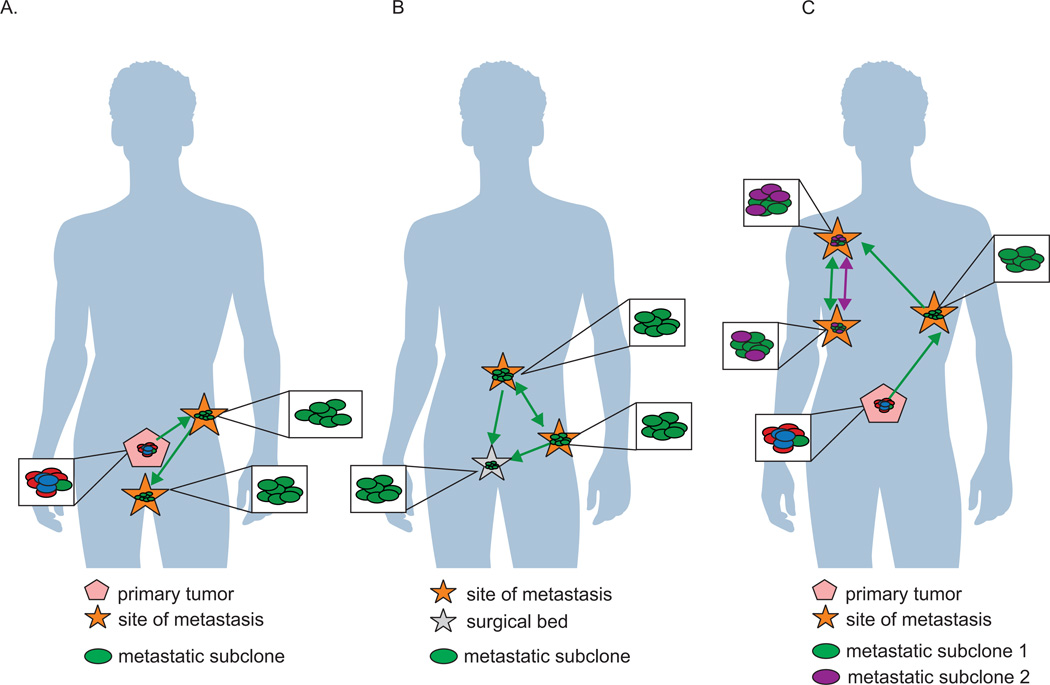
A commonly accepted model of metastasis depicts subclonal populations colonizing metastatic sites in waves originating from the primary tumor (Wan, et al. 2013). In this model, subclonal populations within the primary tumor compete for dominance and are selected for survival and growth by environmental selection pressures (Greaves and Maley 2012). In support of this model, a longitudinal study of four patients demonstrated that metastases were seeded in temporally separated waves originating from the primary tumor (Hong et al. 2015). Presumably, as the primary tumor acquired new mutations, structural variations, and copy number alterations, new metastatic subclonal populations were released to seed and re-seed organ sites (Hong et al. 2015) (Figure 2A). Similarly, in a single case study, anatomically distinct metastatic sites from a patient with CRPC were found to share many genetic alterations including high-level amplification of the AR locus, PTEN loss, TP53 loss, and mutation of SPOP. As a result of these shared genomic alterations, these clones were interpreted to be monoclonal in origin (Haffner, et al. 2013). Interestingly, examination of the microdissected primary tumor revealed that a small 2.2×1.3 mm well-differentiated Gleason pattern 3 lesion exhibited PTEN negative immunohistological staining. DNA sequencing of this lesion revealed the same 4 base pair deletion in PTEN observed in the metastatic clones concurrent with TP53 mutation, suggesting that this lesion harbored the progenitor cell that seeded distant metastases (Haffner et al. 2013). Surprisingly, the surrounding higher grade Gleason pattern 4 tumor tissue did not harbor the same underlying mutations as the metastases. These data indicate that the primary index lesion does not always harbor the lethal clone that is ultimately responsible for seeding distant metastasis in the patient.
Figure 2.
Another common mechanism of metastasis in advanced prostate cancer is via seeding of de novo metastatic sites by cells originating from distant metastasis (Gundem et al. 2015) (Figure 2B). Furthermore, “local recurrences” after surgery may also arise via a similar mechanism of cells from distant metastases seeding the prostatic cavity after radical prostatectomy (Hong et al. 2015) (Figure 2B). The mechanistic underpinnings of metastasis-to-metastasis and metastasis-to-primary site seeding remain to be clarified.
Multidirectional spread can lead to polyclonal seeding of metastatic sites
Similar to primary prostate cancer, a common model to explain intratumoral heterogeneity at metastatic foci has been that cells acquiring new genetic alterations are under constant environmental and therapeutic selection pressure, leading to continuous expansion and contraction of tumor subclones. However, in addition to this monoclonal model of metastatic evolution and spread, a polyclonal pattern has also been described. For example, Gundem and colleagues assessed the clonal relationship between subclones occupying different metastatic sites. On a phylogenetic tree, a truncal mutation in a pair of samples would be apparent if the same mutation was present in 100% in the cancer cell fraction at two different metastatic sites. Conversely, non-truncal (or branch) mutations would be apparent if the same mutation was present in less than 100% of the cancer cell fraction at two different metastatic sites. Using this logic, it was found that 50% of the subjects in a rapid autopsy study exhibited polyclonal seeding of multiple metastatic sites (Gundem et al. 2015). Polyclonal seeding was defined in this study as multiple genetically distinct subclones colonizing a single metastatic site (Figure 2C). Interestingly, all instances of polyclonal seeding involved mutations in genes with known involvement in therapy resistance, indicating that polyclonal seeding and interclonal cooperation may be required to evade therapy.
Overall, these studies have illuminated details of the complex mechanisms underlying metastatic seeding. This process can be initiated by monoclonal or polyclonal populations originating in both the primary tumor and metastases in other sites (Gundem et al. 2015; Hong et al. 2015). Polyclonal populations involved in these seeding events were more similar to one another than to clones occupying other metastatic sites (Gundem et al. 2015). Whether that is due to proximity effects on subclonal evolution or, conversely, characteristics of the metastatic niche that impart habitability remains to be tested. Importantly, it has become clear from these collective data that multiple mechanisms of seeding give rise to metastases in different body sites, highlighting the need for personalized medicine via monitoring of genetic changes in tumor cells within patients over the course of their disease and stages of treatment.
Subclonal expansion to escape targeted therapy
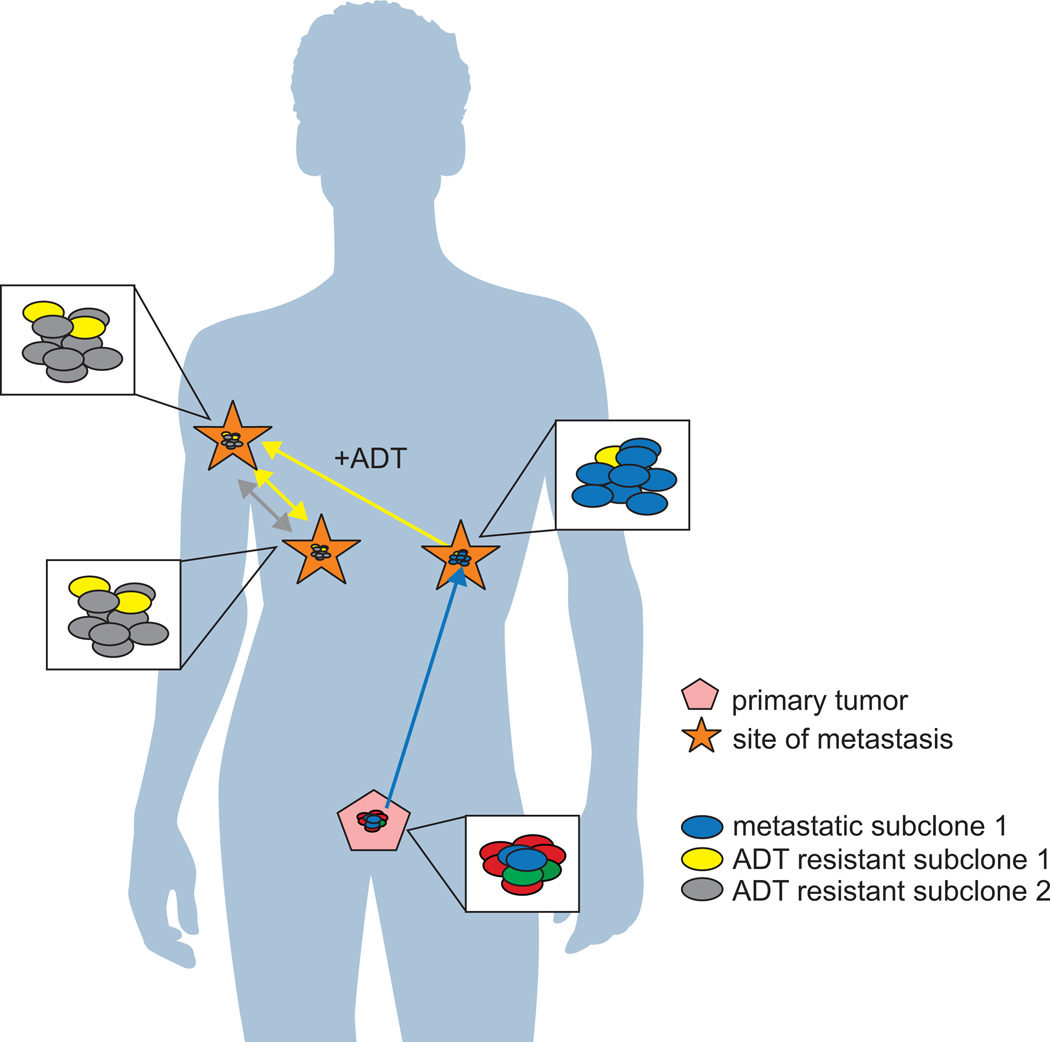
Treatment with AR-targeted ADT imparts a selection pressure upon tumor foci (Figure 3). Clones harboring gene alterations that promote ADT resistance, such as AR point mutations, alterations in AR pathway components, and alterations in MYC and CTNNB1, were shown to seed and reseed multiple sites (Gundem et al. 2015). Those rare sub-populations of cells within tumor foci that reactivate AR through acquisition of mutations, copy number alterations, or synthesis of constitutively active AR splice variants will evade ADT (Watson et al. 2015). This concept is supported by a recent targeted sequencing study wherein germline, plasma, and tumor samples from ERG positive patients were collected before, during, and after treatment with abiraterone acetate or enzalutamide. This longitudinal analysis identified AR amplification and the appearance of AR point mutations in response to therapy (Carreira et al. 2014). An additional mechanism of increased AR signaling in response to ADT is expression of truncated AR variants (Dehm, et al. 2008). Expression of AR-Vs has been detected in primary tumors, metastases, as well as circulating tumor cells (Abeshouse et al. 2015). AR-Vs function as constitutively active transcription factors and can enable CRPC cells to escape AR-directed therapies that require the intact AR ligand binding domain (Dehm et al. 2008; Guo, et al. 2009; Hu, et al. 2012; Li, et al. 2011; Sun, et al. 2010; Watson, et al. 2010). Intriguingly, RNA-seq studies have indicated that AR-V expression relative to full-length AR does not increase dramatically in metastatic CRPC tissues relative to primary tumor tissues (Abeshouse et al. 2015; Robinson et al. 2015). This finding suggests that subclonal expansion of AR-V-expressing cells does not occur in metastatic disease, and may instead represent a property of rare populations of tumor cells that are resistant to ADT. However, these findings from RNA-seq-based studies are in contrast to RT-PCR-based studies, which have found higher expression of AR-Vs in CRPC tissues relative to primary prostate cancer, as well as correlations between AR-V expression and with poor outcomes (Hornberg, et al. 2011; Hu, et al. 2009; Sun et al. 2010).
Figure 3.
One provocative observation in recent studies has been that the acquisition of various AR mutation and amplification events at metastatic sites can occur through polyclonal seeding mechanisms (Figure 3) (Gundem et al. 2015; Hong et al. 2015). For example, in one patient, two subclones involved in polyclonal seeding harbored two different alterations that promote ADT resistance, including MYC amplification and a pathogenic AR substitution (Gundem et al. 2015). Similarly, in a longitudinal biopsy study, a metastatic site in the iliac crest displayed profound clonal evolution in response to ADT, ultimately becoming enriched for a sub-population of cells that had originated in a sacral metastasis (Hong et al. 2015). In patients that exhibited polyclonal seeding, several subclones across metastases were shared and were therefore more similar to each other than to the primary tumor (Gundem et al. 2015). Together these findings suggest that interclonal cooperation may occur, and that multiple independent ADT resistant subclones may arise and ultimately cooperate to drive subclonal expansion in response to ADT. There is precedent for polyclonal seeding in murine models of mammary tumors, but until recently this has not been observed in human tissues (Cleary, et al. 2014; Marusyk, et al. 2014). For example, in Wnt driven mammary tumors, Hrasmt Wntlow basal and Hraswt Wnthigh luminal subclones cooperate interclonally to propogate tumor growth and response to Wnt targeted therapy (Cleary et al. 2014). In these cases, acquisition of multiple mutations across subclonal populations appears to promote and may be necessary to drive tumor growth and clonal expansion under treatment conditions.
Clones representative of multiple stages of disease persist in blood
Tumor cells representing various stages and sites of disease are detectable in blood. Thus, circulating tumor cells and cell-free circulating tumor DNA provide a window into the totality of disease within a patient. Recently, a number of studies have demonstrated detection of biomarkers of advanced disease and therapy resistance in patients with advanced prostate cancer. For example, a clone originating from a primary tumor was detected by targeted sequencing of genomic DNA isolated from blood three years after resection of the prostate (Hong et al. 2015). In the same patient, a metastatic clone that originated in the primary tumor but seeded a metastatic site in the seminal vesicle was detectable in blood (Hong et al. 2015). Targeted sequencing of DNA isolated from plasma revealed the emergence of T878A and L702H mutations in the AR during progression on abiraterone (Romanel et al. 2015). In the same study, patients with AR amplification or AR mutation were less likely to experience a PSA decline after initiation of therapy with abiraterone acetate. Importantly, the presence of mutant AR alleles was more common in patients that did not display evidence for AR gene amplification, indicating that clones exhibiting single alterations in AR may be sufficient to impart therapy resistance (Romanel et al. 2015). Accordingly, AR amplification was detected in circulating cell-free DNA and was shown to be associated with enzalutamide and abiraterone treatment resistance in a cohort of 62 CRPC patients, albeit at a higher frequency in those treated with enzalutamide versus those treated with abiraterone acetate (Azad, et al. 2015).
AR variants that are detectable at the mRNA level in circulating tumor cells are predictive of poor response to treatment with abiraterone acetate or enzalutamide (Antonarakis, et al. 2015; Antonarakis, et al. 2014). In a cohort of patients with castration resistant disease, more than 50% of patients had detectable expression of at least one AR-V mRNA in single circulating tumor cells isolated from blood, and ~17% expressed more than one AR-V in an individual circulating tumor cell (Miyamoto, et al. 2015). In the same study, the authors observed alterations in GR and Wnt signaling in different subsets of circulating tumor cells. Variations in subclonal abundance were observed in patients with CRPC before, during, and after treatment with abiraterone (Carreira et al. 2014). In one patient, biopsy of a liver metastasis revealed a W742C mutation in AR, whereas circulating DNA revealed AR amplification. Treatment with docetaxel resolved the liver metastasis, but the patient exhibited no response to abiraterone acetate, as defined by a 50% decline from baseline in serum PSA levels. This was likely due to the occult subclonal lesions harboring AR amplification, which were detectable in plasma DNA (Carreira et al. 2014). Together, these studies indicate the presence of multiple subclonal populations within the circulation of a single patient. Overall, capture of circulating tumor cells or cell-free DNA has provided a useful approach to gain a snapshot of subclonal populations and their evolution during disease progression. As such, these studies have generated enthusiasm around the ultimate use of circulating tumor cell or cell-free DNA analysis to help inform treatment decisions for individual patients.
Summary
Retrospective and real-time computational renderings of the clonal architecture within patient primary tumors and metastases have provided an unprecedented window into clonal evolution across progressive disease. A tremendous amount of genetic information is available describing the mutations, structural alterations, and altered pathways in both primary and metastatic prostate cancer, yet they are limited in that they can only provide genetic alterations across bulk tissue samples (Abeshouse et al. 2015; Grasso et al. 2012; Robinson et al. 2015). Here, we have reviewed studies that confirm the clonal heterogeneity of primary and metastatic prostate cancer, and provide early evidence that metastatic spread can occur via several mechanisms and not one distinct linear pathway. Furthermore, the studies discussed here have provided the first insights into the temporal acquisition of gene alterations during metastasis and therapy resistance of prostate cancer.
A consensus finding from these studies is that loss or mutation of TP53 and PTEN loss occur prior to or early in metastasis, indicating that they drive metastatic spread. The recently identified mutually exclusive molecular subtypes of primary prostate cancer including ETS fusions, mutations in FOXA1, FLI1, SPOP, and IDH1 represent early drivers of tumorigenesis, and at least some molecular basis for inter-patient heterogeneity. However, acquisition of mutations in drivers of metastasis does not appear to occur preferentially in any one of these molecular subtypes, indicating that the transition to metastatic disease may follow a somewhat unified biological mechanism. Another key finding in these studies is that AR, which is altered in >60% of metastatic prostate cancer and known to be a mediator of ADT resistance (reviewed in (Watson et al. 2015)), undergoes alterations after metastasis has occurred. This novel finding indicates that metastasis and therapy resistance are temporally separate processes. However, it remains unclear whether rare subclones originating in the primary tumor or early metastases harbor AR alterations that later promote ADT resistance or, alternatively, whether such alterations arise after metastasis and initial treatment with ADT. Further analysis of circulating tumor cells from patients at various stages of disease progression may help to understand if resistant tumor clones are present prior to ADT.
In addition to providing insight into the temporal landscape of genetic alterations across prostate cancer development, the studies reviewed here have provided views of the mechanisms of metastatic spread. Foremost, these studies have established that metastatic spread can occur through monoclonal or polyclonal seeding between metastases or in waves originating from the primary tumor. Furthermore, they have demonstrated that clonal spread is not unidirectional, as metastatic clones can reseed the surgical bed of the resected prostate. The mechanistic underpinnings of metastasis-to-metastasis seeding remain to be understood. For example, the properties of the metastatic niche that are required for either monoclonal or polyclonal seeding or re-seeding by metastatic clonal populations are largely unknown. Nevertheless, these studies have highlighted the complexity of patterns of clonal spread and subclonal evolution in response to environmental and therapeutic pressures such as ADT. Monitoring circulating tumor cells and circulating cell-free DNA provides an opportunity to track the totality of these complex disease processes within an individual patient, and possibly identify subclones harboring clinically actionable mutations or early indicators of therapeutic resistance.
Acknowledgments
Funding
SMD is currently funded by a Movember/Prostate Cancer Foundation Challenge Award, American Cancer Society Research Scholar Grant RSG-12-031-01-TBE, NIH grant R01CA174777, US Department of Defense Prostate Cancer Research Program grants W81XWH-12-2-0093, W81XWH-13-1-0518, W81XWH-15-1-0633, and W81XWH-15-1-0501, and a grant from the Minnesota Partnership for Biotechnology and Medical Genomics. JLV is supported by the Cancer Biology Training Grant T32 CA009138.
Footnotes
Declaration of potential conflict of interest
SMD has served as a paid consultant/advisor for Astellas/Medivation.
References
- Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry Christopher D, Annala M, Aprikian A, Armenia J, Arora A, et al. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo P, Ladomery MR. The oncogene ERG: a key factor in prostate cancer. Oncogene. 2015 doi: 10.1038/onc.2015.109. [DOI] [PubMed] [Google Scholar]
- Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Nakazawa M, Nadal R, Paller CJ, Denmeade SR, Carducci MA, et al. Androgen Receptor Splice Variant 7 and Efficacy of Taxane Chemotherapy in Patients With Metastatic Castration-Resistant Prostate Cancer. JAMA Oncol. 2015;1:582–591. doi: 10.1001/jamaoncol.2015.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad AA, Volik SV, Wyatt AW, Haegert A, Le Bihan S, Bell RH, Anderson SA, McConeghy B, Shukin R, Bazov J, et al. Androgen Receptor Gene Aberrations in Circulating Cell-Free DNA: Biomarkers of Therapeutic Resistance in Castration-Resistant Prostate Cancer. Clin Cancer Res. 2015;21:2315–2324. doi: 10.1158/1078-0432.CCR-14-2666. [DOI] [PubMed] [Google Scholar]
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Bangma CH, Bjartell A, Catto JW, Culig Z, Gronberg H, Luo J, Visakorpi T, Rubin MA. The mutational landscape of prostate cancer. Eur Urol. 2013;64:567–576. doi: 10.1016/j.eururo.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry M, Perner S, Demichelis F, Rubin MA. TMPRSS2-ERG fusion heterogeneity in multifocal prostate cancer: clinical and biologic implications. Urology. 2007;70:630–633. doi: 10.1016/j.urology.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, Iversen P, Bhattacharya S, Carles J, Chowdhury S, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371:424–433. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, Dawson KJ, Iorio F, Nik-Zainal S, Bignell GR, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, Meng A, Hennings-Yeomans PH, McPherson A, Sabelnykova VY, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47:736–745. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- Boyd LK, Mao X, Xue L, Lin D, Chaplin T, Kudahetti SC, Stankiewicz E, Yu Y, Beltran L, Shaw G, et al. High-resolution genome-wide copy-number analysis suggests a monoclonal origin of multifocal prostate cancer. Genes Chromosomes Cancer. 2012;51:579–589. doi: 10.1002/gcc.21944. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Goodhead I, Follows GA, Green AR, Futreal PA, Stratton MR. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci U S A. 2008;105:13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Romanel A, Goodall J, Grist E, Ferraldeschi R, Miranda S, Prandi D, Lorente D, Frenel JS, Pezaro C, et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med. 2014;6:254ra125. doi: 10.1126/scitranslmed.3009448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Song SY, Pretlow TG, Abdul-Karim FW, Kung HJ, Dawson DV, Park WS, Moon YW, Tsai ML, Linehan WM, et al. Evidence of independent origin of multiple tumors from patients with prostate cancer. J Natl Cancer Inst. 1998;90:233–237. doi: 10.1093/jnci/90.3.233. [DOI] [PubMed] [Google Scholar]
- Cleary AS, Leonard TL, Gestl SA, Gunther EJ. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature. 2014;508:113–117. doi: 10.1038/nature13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CS, Eeles R, Wedge DC, Van Loo P, Gundem G, Alexandrov LB, Kremeyer B, Butler A, Lynch AG, Camacho N, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47:367–372. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer. 2011;18:R183–R196. doi: 10.1530/ERC-11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, Brewer DS, Kallio HM, Hognas G, Annala M, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, Chen H, Kong X, Melamed J, Tepper CG, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, Mosbruger T, Esopi DM, Fedor H, Heaphy CM, Walker DA, Adejola N, Gurel M, Hicks J, Meeker AK, et al. Tracking the clonal origin of lethal prostate cancer. J Clin Invest. 2013;123:4918–4922. doi: 10.1172/JCI70354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, Weier C, Xu MM, Vaghasia A, Gurel B, Gumuskaya B, Esopi DM, Fedor H, Tan HL, Kulac I, et al. Molecular evidence that invasive adenocarcinoma can mimic prostatic intraepithelial neoplasia (PIN) and intraductal carcinoma through retrograde glandular colonization. J Pathol. 2015 doi: 10.1002/path.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong MK, Macintyre G, Wedge DC, Van Loo P, Patel K, Lunke S, Alexandrov LB, Sloggett C, Cmero M, Marass F, et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat Commun. 2015;6:6605. doi: 10.1038/ncomms7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberg E, Ylitalo EB, Crnalic S, Antti H, Stattin P, Widmark A, Bergh A, Wikstrom P. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One. 2011;6:e19059. doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, Han M, Partin AW, Vessella RL, Isaacs WB, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, Edwards J, Isaacs WB, Nelson PS, Bluemn E, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–3462. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugel A, Wernert N. Loss of heterozygosity (LOH), malignancy grade and clonality in microdissected prostate cancer. Br J Cancer. 1999;79:551–557. doi: 10.1038/sj.bjc.6690087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Dhanasekaran SM, Mehra R, Tomlins SA, Gu W, Yu J, Kumar-Sinha C, Cao X, Dash A, Wang L, et al. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res. 2007;67:8229–8239. doi: 10.1158/0008-5472.CAN-07-1297. [DOI] [PubMed] [Google Scholar]
- Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15:4792–4798. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Ishida H, Shindo T, Niwa S, Kino M, Kawamura K, Kamiya N, Imamoto T, Suzuki H, Hirokawa Y, et al. Molecular analysis of multifocal prostate cancer by comparative genomic hybridization. Prostate. 2008;68:1715–1724. doi: 10.1002/pros.20832. [DOI] [PubMed] [Google Scholar]
- Li Y, Alsagabi M, Fan D, Bova GS, Tewfik AH, Dehm SM. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res. 2011;71:2108–2117. doi: 10.1158/0008-5472.CAN-10-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg J, Klevebring D, Liu W, Neiman M, Xu J, Wiklund P, Wiklund F, Mills IG, Egevad L, Gronberg H. Exome sequencing of prostate cancer supports the hypothesis of independent tumour origins. Eur Urol. 2013;63:347–353. doi: 10.1016/j.eururo.2012.03.050. [DOI] [PubMed] [Google Scholar]
- Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, Chen L, Ewing CM, Eisenberger MA, Carducci MA, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514:54–58. doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, Stewart C, Carter SL, Cibulskis K, Bhutkar A, McKenna A, Dooley A, Vernon A, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 2014;156:1298–1311. doi: 10.1016/j.cell.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo LM, Shah NA, Li X, Blount PL, Vaughan TL, Reid BJ, Maley CC. A comprehensive survey of clonal diversity measures in Barrett's esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev Res (Phila) 2010;3:1388–1397. doi: 10.1158/1940-6207.CAPR-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Rashid NU, Sperling AS, Bolli N, Wedge DC, Van Loo P, Tai YT, Shammas MA, Fulciniti M, Samur MK, Richardson PG, et al. Differential and limited expression of mutant alleles in multiple myeloma. Blood. 2014;124:3110–3117. doi: 10.1182/blood-2014-04-569327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins CM, Tembe WA, Baker A, Sinari S, Moses TY, Beckstrom-Sternberg S, Beckstrom-Sternberg J, Barrett M, Long J, Chinnaiyan A, et al. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanel A, Tandefelt DG, Conteduca V, Jayaram A, Casiraghi N, Wetterskog D, Salvi S, Amadori D, Zafeiriou Z, Rescigno P, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med. 2015;7:312re310. doi: 10.1126/scitranslmed.aac9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier MP, True LD, Higano CS, Vesselle H, Ellis W, Lange P, Vessella RL. Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum Pathol. 2003;34:646–653. doi: 10.1016/s0046-8177(03)00190-4. [DOI] [PubMed] [Google Scholar]
- Ruijter ET, Miller GJ, van de Kaa CA, van Bokhoven A, Bussemakers MJ, Debruyne FM, Ruiter DJ, Schalken JA. Molecular analysis of multifocal prostate cancer lesions. J Pathol. 1999;188:271–277. doi: 10.1002/(SICI)1096-9896(199907)188:3<271::AID-PATH359>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, Fizazi K, Mainwaring P, Piulats JM, Ng S, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368:138–148. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, Macvicar GR, Varambally S, Harwood J, Bismar TA, et al. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 2004;64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, Page ST, Coleman IM, Nguyen HM, Sun H, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH, Sun W, Weigman VJ, Marynen P, Zetterberg A, Naume B, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villers A, McNeal JE, Freiha FS, Stamey TA. Multiple cancers in the prostate. Morphologic features of clinically recognized versus incidental tumors. Cancer. 1992;70:2313–2318. doi: 10.1002/1097-0142(19921101)70:9<2313::aid-cncr2820700917>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Wan L, Pantel K, Kang Y. Tumor metastasis: moving new biological insights into the clinic. Nat Med. 2013;19:1450–1464. doi: 10.1038/nm.3391. [DOI] [PubMed] [Google Scholar]
- Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, Kim K, Sawyers CL. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. 2010;107:16759–16765. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S, Wuttig D, Warnatz HJ, Stehr H, Rausch T, et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell. 2013;23:159–170. doi: 10.1016/j.ccr.2013.01.002. [DOI] [PubMed] [Google Scholar]