Concentrating factors in nuclear bodies is thought to promote efficient gene expression. Tatomer et al. show that the histone locus body (HLB) concentrates pre-mRNA processing factors at replication-dependent histone genes, resulting in optimal 3′ end formation of histone mRNAs coupled with transcription termination.
Abstract
The histone locus body (HLB) assembles at replication-dependent histone genes and concentrates factors required for histone messenger RNA (mRNA) biosynthesis. FLASH (Flice-associated huge protein) and U7 small nuclear RNP (snRNP) are HLB components that participate in 3′ processing of the nonpolyadenylated histone mRNAs by recruiting the endonuclease CPSF-73 to histone pre-mRNA. Using transgenes to complement a FLASH mutant, we show that distinct domains of FLASH involved in U7 snRNP binding, histone pre-mRNA cleavage, and HLB localization are all required for proper FLASH function in vivo. By genetically manipulating HLB composition using mutations in FLASH, mutations in the HLB assembly factor Mxc, or depletion of the variant histone H2aV, we find that failure to concentrate FLASH and/or U7 snRNP in the HLB impairs histone pre-mRNA processing. This failure results in accumulation of small amounts of polyadenylated histone mRNA and nascent read-through transcripts at the histone locus. Thus, the HLB concentrates FLASH and U7 snRNP, promoting efficient histone mRNA biosynthesis and coupling 3′ end processing with transcription termination.
Introduction
Structures termed nuclear bodies (NBs) create microenvironments within the nucleus that promote the organized regulation of genome function. Defined microscopically as a concentration and colocalization of a set of specific RNPs and/or proteins, each NB contains a suite of factors associated with particular nuclear processes including RNA biosynthesis, DNA replication, and DNA repair (Gall, 2000; Matera et al., 2009; Nizami et al., 2010; Morimoto and Boerkoel, 2013). NBs assemble through protein-nucleic acid and protein-protein interactions. For example, the nucleolus is seeded by rRNA (Falahati et al., 2016) paraspeckles are seeded by Men ε/β long noncoding RNAs (Mao et al., 2011a), and the protein Coilin organizes the Cajal body (CB; Mao et al., 2011b). NBs are not membrane bound, and their components can freely exchange with the nucleoplasm (Dundr et al., 2004). By concentrating reactants and substrates, NBs are postulated to provide discrete microenvironments that increase rates of nuclear processes (Matera et al., 2009; Dundr, 2011; Mao et al., 2011b). Although there is some evidence for this hypothesis (Klingauf et al., 2006; Stanek et al., 2008; Strzelecka et al., 2010; Novotný et al., 2011), disruption of NBs does not always obviously impact associated reactions (Deryusheva and Gall, 2009; Liu et al., 2009). Thus, determining the specific contribution that a NB makes to an in vivo reaction has proven challenging. We have been addressing this question by studying the histone locus body (HLB).
Joe Gall et al. identified the HLB in Drosophila melanogaster in 2006 (Liu et al., 2006). It then became apparent that an NB initially described as a specialized CB in which U7 small nuclear RNP (snRNP; Frey and Matera, 1995) and the protein NPAT (Ma et al., 2000; Zhao et al., 2000) are concentrated was the mammalian HLB. The HLB is involved in replication-dependent (RD) histone mRNA synthesis (Ye et al., 2003; White et al., 2011), which occurs at high levels only during S phase of the cell cycle to package newly replicated DNA (Marzluff et al., 2008). RD histone genes encode the only known eukaryotic mRNAs that are not polyadenylated, ending instead in a stem loop at the 3′ end (Marzluff et al., 2008). Two factors required for formation of the 3′ end of histone mRNAs, FLASH and U7 snRNP, are concentrated in and continuously present in the HLB (Yang et al., 2009). In addition to FLASH and U7 snRNP, formation of the 3′ end of histone mRNAs requires a complex of polyadenylation factors termed the histone cleavage complex (HCC; Yang et al., 2013). Association of the HCC with U7 snRNP requires a direct interaction between FLASH and the U7 snRNP component Lsm11 (Sabath et al., 2013; Yang et al., 2013).
The HLB is present throughout interphase of the cell cycle (Ma et al., 2000; Zhao et al., 2000; Liu et al., 2006; White et al., 2007, 2011; Ghule et al., 2008; Tatomer et al., 2014), and expression of the histone genes is activated by cyclin E/Cdk2, which phosphorylates NPAT (Mxc in Drosophila; Ma et al., 2000; Zhao et al., 2000; Ye et al., 2003; White et al., 2007, 2011). A specific DNA element in the Drosophila histone gene cluster nucleates the HLB, suggesting direct association between the HLB and chromatin at the histone locus (Salzler et al., 2013). The HLB begins to assemble in the Drosophila embryo before the onset of zygotic histone mRNA expression (White et al., 2011; Salzler et al., 2013). The recruitment of FLASH and U7snRNP to the HLB does not require the cis elements in the histone pre-mRNA with which they associate (Salzler et al., 2013). These observations indicate that the HLB is not merely a reflection of high, localized gene expression and instead suggest that the HLB has evolved as a feature of the metazoan nucleus that optimizes cell cycle–regulated biosynthesis of histone mRNAs.
An ideal system to understand HLB function requires the ability to manipulate the localization and activity of HLB components without destroying the HLB, as well as the ability to directly assay the molecular and biological effects of such manipulations. Here, we describe such a system for the Drosophila HLB. By using both a transgenic complementation system and genetic alteration of HLB composition, we find that when FLASH and/or U7 snRNP are present at normal levels in the nucleus, but not concentrated in the HLB, histone pre-mRNA processing is slowed. This results in transcriptional read-through and the accumulation of unprocessed histone transcripts at the histone locus, as well as small amounts of misprocessed, polyadenylated mRNA. These data indicate that concentrating factors in the HLB increases the rate of histone pre-mRNA processing coupling 3′ end formation with transcription termination.
Results
FLASH is essential for histone mRNA 3′ end formation in Drosophila
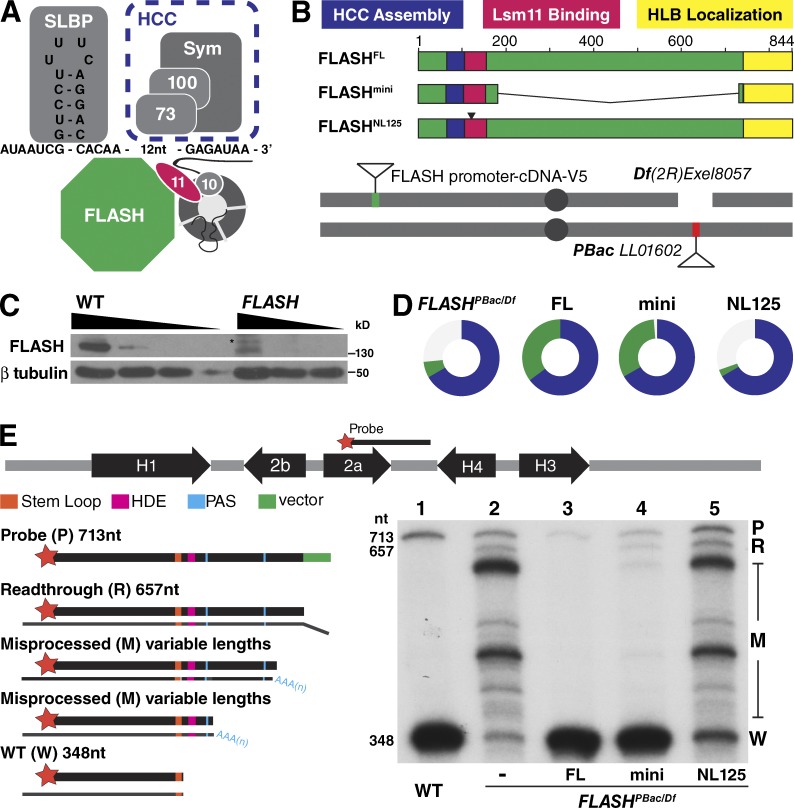
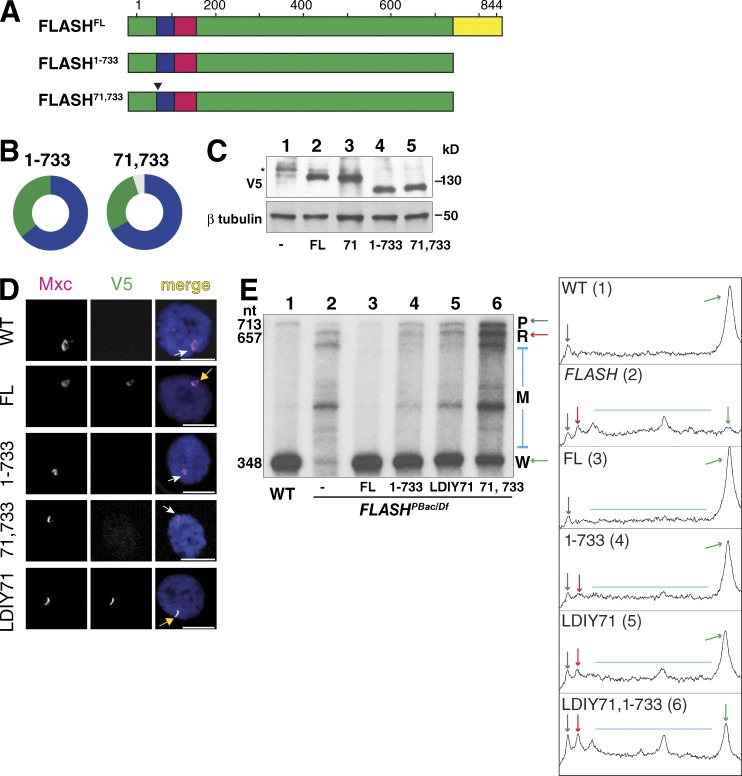
FLASH plays a critical biochemical role in histone pre-mRNA processing as a bridge between the U7 snRNP and the HCC (Sabath et al., 2013; Yang et al., 2013; Fig. 1 A). To determine the contribution that concentrating FLASH in the HLB makes to RD histone pre-mRNA processing, we first established a transgenic complementation system to identify the protein domains necessary for FLASH function in vivo (Fig. 1 B). A piggyback transposon insertion in the 5′ UTR of the FLASH gene (Figs. S1 A and 1 B) results in a large reduction of FLASH protein (Fig. 1 C; and see also Fig. 5). This hypomorphic allele (FLASHPBac) in trans to a deficiency of FLASH (FLASHDf) reduces viability (20% of expected), and the adult flies that eclose are female sterile but male fertile (Fig. 1 D). Expression of C-terminally V5-tagged full-length (FL) FLASH from a single-copy transgene using the endogenous FLASH promoter completely rescued the reduced viability and sterility caused by FLASHPBac/Df (Fig. 1 D and Table 1). This system provides a means for testing the ability of transgenes expressing different alleles of FLASH to rescue FLASH mutant phenotypes.
Figure 1.
Histone pre-mRNA processing is essential in vivo FLASH function. (A) The histone pre-mRNA processing machinery. Slbp binds the stem loop on the pre-mRNA. The U7 snRNP interacts with the HDE in histone pre-mRNA. FLASH binds the U7 snRNP protein Lsm11 (11), and the FLASH/U7 snRNP complex recruits the HCC containing the Symplekin scaffold (Sym) and the CPSF100/CPSF73 endonuclease (100/73). 10, Lsm10. (B) Schematic of functional domains of FLASH and the transgene rescue system. Transgenes were inserted at chromosomal location 25C6 and recombined with a deletion of the FLASH locus (Df(2R)Exel8057). All FLASHPBac/Df experimental flies contained a maternal Df chromosome in trans to a paternal PBac LL01602 chromosome (see Fig. S1). NL125 is a 2-aa mutation in the Lsm11 binding region (see Fig. 2 A). (C) Anti-FLASH Western analysis of WT and FLASHPBac/Df wandering third-instar larvae using threefold serial dilutions. Asterisk, cross-reacting protein. (D) Visual representation of transgenic rescue of FLASHPBac/Df. Circles indicate the proportion of FLASHPBac/Df mutant with the indicated transgene (green) and control heterozygous sibling (blue) adult flies obtained. The expected fraction of control siblings is two thirds. Thus, one third green indicates full viability, and the absence of green indicates lethality. Table 1 shows the data for this analysis. (E) S1 nuclease protection assay. The map of a Drosophila histone gene repeat is shown. Equal amounts (5 µg) of total RNA from wandering third-instar larvae of WT, the FLASHPBac/Df mutant, or the mutant complemented with the indicated transgenes were analyzed using a 3′-labeled DNA probe complementary to H2a mRNA that extends to the H4 HDE followed by 56 nt vector sequence (green). The hybrids were incubated with S1 nuclease, and the protected fragments were resolved on a 6% acrylamide-7M urea gel and visualized by autoradiography. Properly processed H2a mRNA protects a 348-nt fragment from S1 digestion (W). The H2a gene contains multiple downstream cryptic polyadenylation signals (PAS; blue) that protect a heterogeneous set of longer fragments (M). Transcripts that extend beyond the complementary region result in a single protected read-through fragment (R) that is distinct from undigested probe (P). The sizes (in nucleotides) of the processed mRNA, R RNA, and undigested probe are indicated.
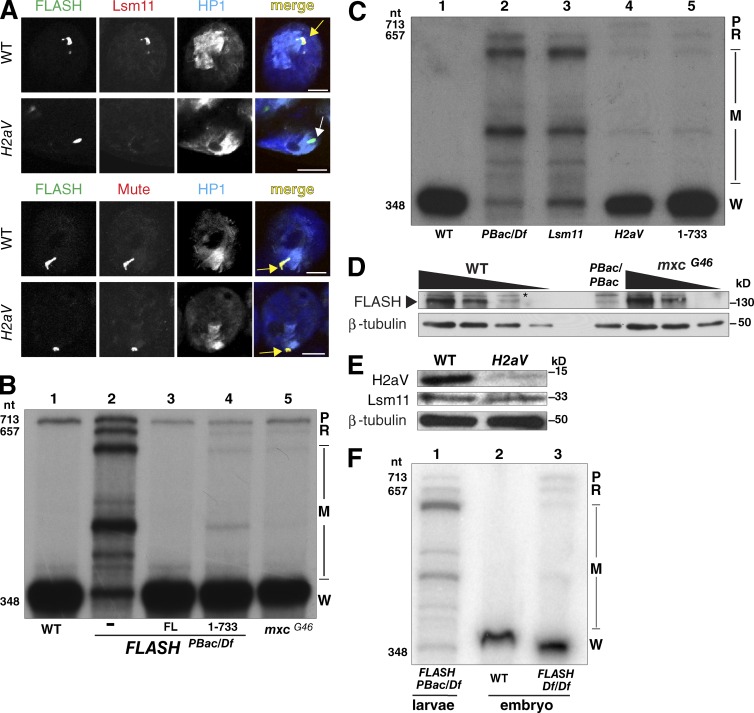
Figure 5.
Failure to concentrate FLASH and/or U7snRNP in the HLB results in inefficient histone pre-mRNA processing. (A) Larval salivary glands from H2aV810 mutants were stained for HLB markers and HP1. Arrows as in Figs. 2 and 3. Bars, 10 µm. (B) Equal amounts of RNA from ovaries of WT (lane 1) or FLASHPbac/Df (lane 2), FLASHFL (lane 3), FLASH1–733 (lane 4), and mxcG46 mutants were analyzed by S1 nuclease protection assay as in Fig. 1 E. (C) Equal amounts of RNA from WT (lane 1), FLASHPbac/Df (lane 2), Lsm11-null mutant (Godfrey et al., 2009; lane 3), H2aV mutant (lane 4), and FLASH1–733 (lane 5) third-instar larvae were analyzed by S1 nuclease protection assay as in Fig. 1 E. (D) Western analysis of threefold serial dilutions of extracts from WT, FLASHPBac/PBac, and mxcG46 ovaries using anti-FLASH antibody and tubulin as a loading control. Asterisk, cross-reacting protein. (E) Anti-H2aV or anti-Lsm11 Western analysis of proteins from H2aV810 mutant larvae, with tubulin as a loading control. See also Fig. S5. (F) Equal amounts of RNA from 14–18 h WT embryos (lane 2) or FLASHDf/Df embryos (lane 3) were analyzed by S1 nuclease mapping as in E. Lane 1 is analysis of RNA from FLASHPbac/Df larvae.
Table 1. Phenotypes of each of the transgenic FLASH constructs in the FLASHPBac/Df mutant background.
| Flies counted | Mendelian? (χ2 P = 0.05) | Student’s t test | Fertility | HLB | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Allele | Control sibling | Experimental | Total | Significantly different from Dfmat/pBacpat mutant? | Female | Male | ||||
| Df(maternal) × pBac(paternal) | 610 | 62 | 672 | N | N | Y | ||||
| pBac(maternal) × Df(paternal) | 509 | 213 | 722 | N | Y, P < 0.001a | N | Y | |||
| pBac × pBac | 450 | 84 | 534 | N | Y, P < 0.05 | N | Y | |||
| FL | 606 | 329 | 935 | Y | Y, P < 0.001a | Y | Y | Y | ||
| Mini | 443 | 213 | 656 | Y | Y, P < 0.001a | Y | Y | Y | ||
| NL 125 | 607 | 27 | 634 | N | Y, P < 0.05b | N | n/d | Y | ||
| 78–844 | 608 | 0 | 608 | N | Y, P < 0.001b | n/a | n/a | Y | ||
| 65–844 | 596 | 294 | 890 | Y | Y, P < 0.001a | Y | Y | Y | ||
| LDIY 71 | 387 | 164 | 551 | Y | Y, P < 0.001a | Y | Y | Y | ||
| LDIY 45 | 381 | 206 | 587 | Y | Y, P < 0.001a | Y | Y | Y | ||
| LDIY 45,71 | 621 | 7 | 628 | N | Y, P < 0.001b | N | n/d | Y | ||
| 1–733 | 338 | 189 | 527 | Y | Y, P < 0.001a | Y | Y | N | ||
| LDIY 71, 1–733 | 589 | 255 | 844 | Y | Y, P < 0.001a | N | Y | N | ||
| 1–733, 101 | 467 | 221 | 688 | Y | Y, P < 0.001a | N | n/d | Y | ||
| LDIY71,1–733,101 | 479 | 195 | 674 | Y | Y, P < 0.001a | N | n/d | Y | ||
The data for each of the genetic rescue experiments presented as circle charts in Figs. 1, 2, and 3 is shown. For the transgenic rescue crosses, a recombinant chromosome containing FLASHDf and a FLASH transgene was provided maternally. For each experiment, a χ2 analysis was performed under the null hypothesis that the phenotypic classes of adult flies were present in expected Mendelian ratios (two thirds CyO/mutant control siblings and one third homozygous mutant genotype). A Student’s t test was used to determine whether the number of flies in the experiment class was statistically different from when no transgene was present in the FLASHPBac/Df background. Note that three of the transgenes, FLASHNL125, FLASH 78–844, and FLASHLDIY45,71, behaved as dominant negatives, consistent with them being incorporated into the HLB and competing with the small amount of endogenous FLASH. The FLASHPBac/PBac phenotype suggests the presence of a second site mutation on the PBac chromosome. Assessment of fertility for each genotype is also noted. The last column indicates transgenic FLASH proteins capable of localizing to the HLB. N, no; n/a, not applicable; n/d, not determined; Y, yes.
A difference caused by transgene rescue of partial lethality.
A difference caused by a more severe mutant phenotype resulting from transgene expression.
We determined whether the developmental phenotypes of the Drosophila FLASHPBac/Df mutant were caused by histone pre-mRNA processing defects or loss of another FLASH function, as FLASH was originally implicated in Fas-mediated apoptosis in mammalian cells (Imai et al., 1999). The 844-aa FLASH protein has three known biochemical functions (Fig. 1 B). The N-terminal 154 aa of FLASH is essential for histone pre-mRNA processing in vitro, and aa 105–150 in this region bind the N terminus of the U7 snRNP protein Lsm11 (Burch et al., 2011). The Lsm11–FLASH N-terminal interaction creates an interface that recruits the HCC (Sabath et al., 2013; Yang et al., 2013), which contains the endonuclease CPSF-73 (Dominski et al., 2005). The C-terminal 111 aa of FLASH are necessary for localization to the HLB (Burch et al., 2011). To test if these three activities are sufficient for FLASH function in vivo, we expressed a transgene encoding a 547-aa internal deletion of the 844-aa FLASH protein, fusing the HCC assembly and Lsm11 binding domains to the HLB localization domain (FLASHmini; Fig. 1 B). This transgene rescued the reduced viability and sterility of the FLASHPBac/Df mutant (Fig. 1 D and Table 1), suggesting that the essential function of FLASH in Drosophila is histone mRNA 3′ end formation as a component of the HLB (see Fig. 4).
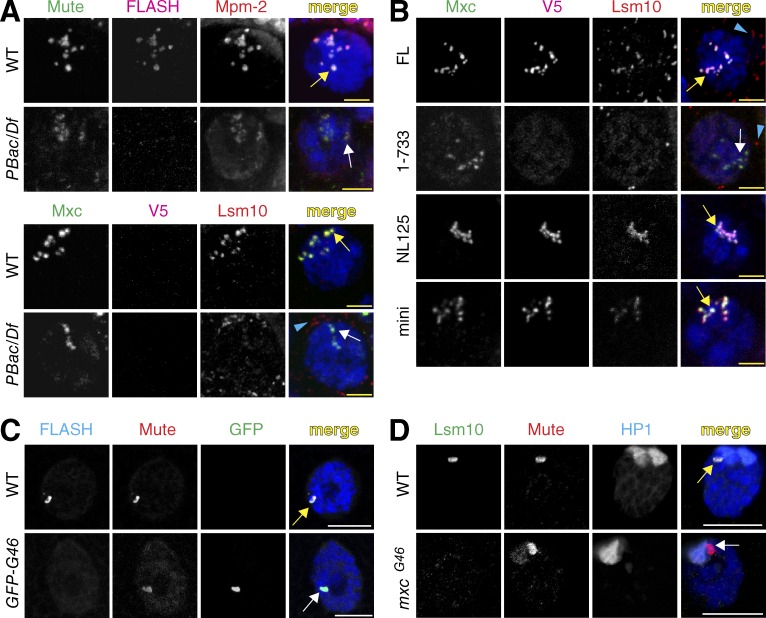
Figure 4.
Analysis of the HLB in FLASH mutants. (A and B) The presence of four HLB components was assessed in nurse cells of the indicated genotypes at stage 5 of oogenesis with antibodies against FLASH; Lsm10; Mute, a constitutive HLB factor of unknown function (Bulchand et al., 2010); Mxc; and MPM-2 monoclonal antibodies, which detect phosphorylated Mxc (White et al., 2011). Note that at these stages of oogenesis, the chromatids of the polyploid nurse cells are dispersed, resulting in many HLB foci in close proximity. Yellow arrows indicate proper localization, and white arrows indicate mislocalization. Blue arrowheads indicate signal from the Lsm10 antibody that is not present in the nucleus; the source of this signal is unknown. Bars, 5 µm. (C and D) HLB assembly in polyploid salivary gland nuclei of mxcG46 mutants (D) or mxcG48-null mutants expressing transgenic GFP-MxcG46 protein (GFP-G46; C) detected with anti-GFP antibodies. Note that salivary gland chromosomes are polytene, resulting in a single, large HLB. HP1 staining visualizes the chromocenter located near the histone locus. Bars, 15 µm.
To understand the molecular basis for the FLASH mutant phenotype, we examined the histone mRNA species produced in various FLASH mutant genotypes (Fig. 1 E). When histone pre-mRNA processing is compromised in Drosophila, cryptic polyadenylation signals located downstream of the normal processing site are used, resulting in the cytoplasmic accumulation of longer, polyadenylated histone mRNA (Sullivan et al., 2001; Lanzotti et al., 2002; Godfrey et al., 2006). We also detect longer nascent transcripts at the histone locus by in situ hybridization (Lanzotti et al., 2002). To detect misprocessed histone RNA species, we use an S1 nuclease protection assay that provides a quantitative measurement of the amounts of properly processed histone mRNA (Fig. 1 E, “W”) compared with misprocessed, polyadenylated histone mRNA (Fig. 1 E, “M”). The assay also detects “read-through” transcripts that represent the unprocessed nascent pre-mRNA (Fig. 1 E, “R”; Sullivan et al., 2009).
FLASHPBac/Df mutant third-instar larvae express large amounts of misprocessed, polyadenylated histone mRNA (Fig. 1 E, lane 2; and Fig. S1 C), confirming that a major function of FLASH is histone mRNA 3′ end formation. We additionally detected small amounts of properly processed mRNA, consistent with this being a hypomorphic allele, as well as small amounts of unprocessed read-through RNA (Fig. 1 E, lane 2). Expression of FLASHFL protein completely rescued this mutant RNA phenotype (Fig. 1 E, lane 3). Expression of FLASHmini also rescued the mutant RNA phenotype, although small amounts of both misprocessed and read through histone mRNA were detected in FLASHmini larvae (Fig. 1 E, lane 4). FLASHmini is almost fully proficient in processing in vivo, consistent with it rescuing viability and fertility of the FLASHPBac/Df mutant (Fig. 1 D). The developmental phenotypes observed with FLASHPBac/Df are consistent with our previous in vivo analyses of processing factors stem-loop binding protein (Slbp) and U7 snRNP (Sullivan et al., 2001; Godfrey et al., 2009), supporting the idea that the primary role for FLASH is histone mRNA 3′ end formation.
To directly ask if histone mRNA processing is an essential FLASH function, we changed to alanines the asparagine-leucine residues 125–126 in FLASH that are required for in vitro binding of the N terminus of FLASH with Lsm11 (Burch et al., 2011; Figs. 1 B and 2 A). FLASHNL125 did not rescue FLASHPBac/Df reduced viability (Fig. 1 D) or the misprocessed histone mRNA phenotype (Fig. 1 E, lane 5). FLASHNL125 protein and U7 snRNP were present in the HLB (see Fig. 4 B), indicating there is an intact HLB in this mutant. We previously showed that a 2-aa mutation in Lsm11 that prevents FLASH binding also results in misprocessed histone mRNA and lethality (Burch et al., 2011). Thus, the interaction between FLASH and Lsm11 is essential for proper histone mRNA 3′ end formation and normal development, but not for localization of either component to the HLB.
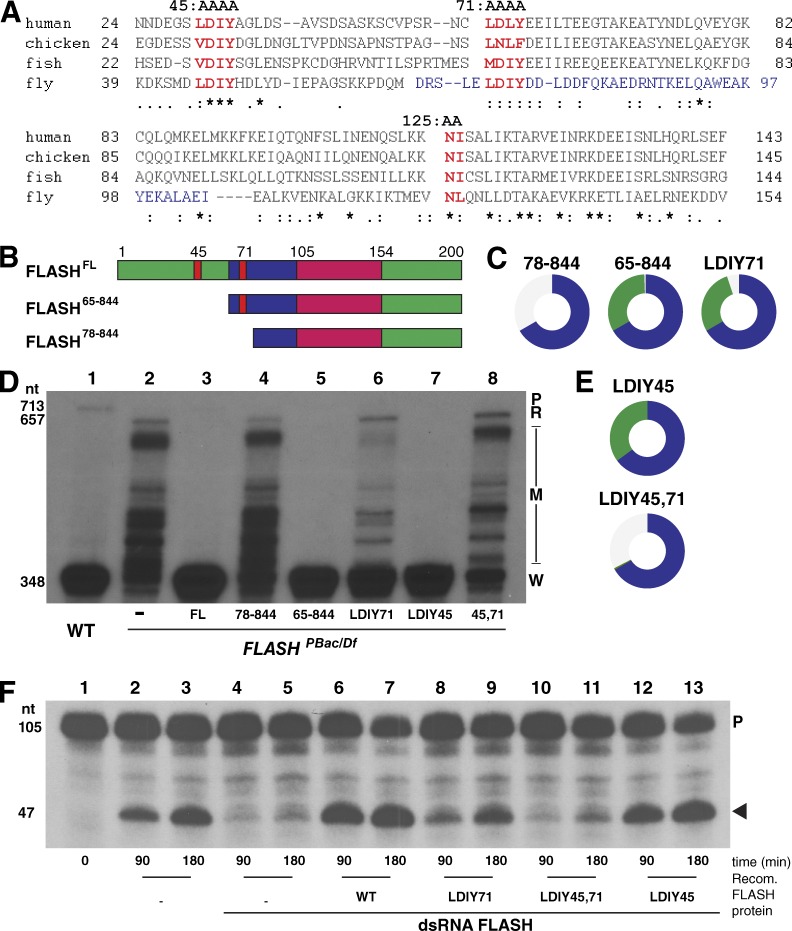
Figure 2.
Function of FLASH N-terminal mutants in histone pre-mRNA processing. (A) Alignment of FLASH N-terminal sequences indicating the region required for HCC binding (blue), the conserved LDIY motifs (red; mutated to AAAA in FLASHLDIY45 and FLASHLDIY71), and the NL residues (red) of the Lsm11 binding region mutated to AA in FLASHNL125. (B) Diagram of the first 200 aa of the N-terminal FLASH deletion mutants. (C) Visualization of rescue of FLASHPBac/Df with transgenic FLASH N-terminal mutants as in Fig. 1 D. (D) Equal amounts total RNA from WT and mutant larvae complemented with the indicated FLASH transgenes were analyzed by S1 nuclease protection as in Fig. 1 E. A PhosphorImager analysis of the same gel is in Fig. S1. (E) Visualization of rescue of FLASHPBac/Df with transgenic FLASH LDIY mutants. (F) Biochemical activity of FLASH LDIY mutant proteins was assessed using an in vitro histone pre-mRNA processing assay (Sabath et al., 2013). FLASH-depleted nuclear extracts were complemented with 100 ng of the indicated recombinant (Recomb.) N-terminal FLASH-GST fusion protein (aa 1–178). 32P-labeled synthetic H3 pre-mRNA was incubated with the extract for 90 or 180 min, and the resulting undigested probe (P) and cleaved probe (arrowhead) were resolved on an 8% polyacrylamide-7M urea gel. See also Fig. S1.
The FLASH N terminus is necessary for histone mRNA 3′ end formation in vivo
We have defined a domain N terminal of the Lsm11 binding domain that is essential for histone pre-mRNA processing in vitro (Sabath et al., 2013; Yang et al., 2013). In both mammalian and Drosophila nuclear extracts, this domain is necessary for recruitment of the HCC to the FLASH–Lsm11 complex. Deletion of residues 1–77 from Drosophila FLASH or mutation to AAAA of a highly conserved LDIY motif (aa 71–74; Fig. 2 A) inhibited processing in vitro and blocked recruitment of the HCC in nuclear extracts (Sabath et al., 2013).
To determine the in vivo requirement for this region of FLASH, we tested the ability of an N-terminal truncation of FLASH containing aa 71–74 (FLASH65–844), which functions normally in vitro, as well as FLASH78–844 and FLASHLDIY71 (with aa 71–74 mutated to AAAA), which are defective in processing in vitro, to rescue FLASHPBac/Df mutant phenotypes (Fig. 2, B–D; and Fig. S2). Expression of FLASH65–844 fully rescued FLASHPBac/Df-reduced viability and sterility, whereas the mutant lacking 13 additional aa (FLASH78–844) did not (Figs. 1 D and 2 C and Table 1). Surprisingly FLASHLDIY71, which has reduced processing activity in vitro (Fig. 2 F, lanes 8 and 9; Sabath et al., 2013), rescued FLASHPBac/Df-reduced viability and sterility (Fig. 2 C). Consistent with this finding, FLASHLDIY71 mutants contained large amounts of properly processed histone mRNA with a normal 3′ end, although we could detect small amounts of misprocessed and read-through histone mRNA species (Fig. 2 D, lane 6; and Fig. S2). We conclude that the FLASHLDIY71 protein supports histone pre-mRNA processing in vivo, in contrast to its low activity in vitro.
There is a second LDIY motif at aa 45–48 in Drosophila FLASH that is also conserved in vertebrates (Fig. 2 A). This motif is not essential, because the FLASH65–844 transgene completely rescues viability and histone pre-mRNA processing. To test whether it might play any role in processing, we mutated these four amino acids to alanine and generated transgenes expressing FLASH proteins with this mutation (FLASHLDIY45) or with mutation of both LDIY motifs (FLASHLDIY45,71). The FLASHLDIY45 protein rescued FLASHPBac/Df reduced viability and sterility and completely restored histone pre-mRNA processing (Fig. 2, D [lane 7] and E). In contrast, FLASHLDIY45,71 failed to rescue FLASHPBac/Df-reduced viability (Fig. 2 E) and had a more severe histone mRNA processing defect than the FLASHLDIY71 mutant (Fig. 2 D, lanes 6–8). In addition, fewer FLASHLDIY45,71 and FLASH78–844 flies eclosed than FLASHPBac/Df flies (Fig. 1 D; Fig. 2, D and E; and Table 1). These mutant FLASH proteins can bind Lsm11 normally, suggesting a dominant-negative effect in the presence of the small amount of wild-type (WT) FLASH present in FLASHPBac/Df animals.
We compared the biochemical activity in processing of the FLASHLDIY45, FLASHLDIY45,71, and FLASHLDIY71 proteins using an in vitro processing assay (Fig. 2 F, lanes 1–3). Extracts from S2 cells depleted of FLASH by double-stranded RNA are deficient in processing synthetic histone pre-mRNA substrates (Fig. 2 F, lanes 4 and 5), and this deficiency can be biochemically complemented by addition of Escherichia coli–derived proteins containing the N-terminal 178 residues of FLASH (Fig. 2 F, lanes 6 and 7; Sabath et al., 2013). In this assay, the FLASHLDIY45 N-terminal fragment was active (Fig. 2 F, lanes 12 and 13), FLASHLDIY71 had reduced activity relative to the WT protein (Fig. 2 F, compare lanes 6 and 7 to 8 and 9), and FLASHLDIY45,71 was inactive (Fig. 2 F, lanes 10 and 11). Together, these genetic and biochemical data indicate that both the LDIY71 motif and the LDIY45 motif contribute to processing, with the LDIY45 being critical only when the LDIY71 is mutated.
Concentrating FLASH in the HLB facilitates histone mRNA 3′ end formation
The FLASHLDIY71 mutant functioned in vivo better than expected from its reduced activity in vitro, suggesting that some aspect of the FLASHLDIY71 processing defect is compensated for in an intact cell. We tested whether concentrating FLASHLDIY71 in the HLB contributed to histone 3′ end formation in vivo. FLASH1–733 (Fig. 3 A) lacks the C-terminal 111 aa and does not concentrate in the HLB in cultured S2 cells (Burch et al., 2011) or transgenic animals (Figs. 3 D and S3), although it rescued FLASHPBac/Df-reduced viability and sterility (Fig. 3 B and Table 1). FLASH1–733 accumulates to similar levels as FLASHFL as assayed by Western blotting (Figs. 3 C and S4), but the low levels of FLASH throughout the nucleoplasm prevent its detection by immunofluorescence. We detected both misprocessed and read through transcripts in FLASH1–733 animals, suggesting that concentrating FLASH in the HLB may be required for full FLASH function in histone pre-mRNA processing in vivo (Fig. 3 E, lane 4; and Fig. S4). The read-through transcripts in the FLASH1–733 mutant were present in similar amounts as the misprocessed RNAs (Fig. 3 E, compare R to M in lanes 2 and 4). The histone mRNA species in FLASH1–733 animals are qualitatively like those in FLASHLDIY71 animals, which also had similar amounts of read-through transcripts and misprocessed RNAs (Fig. 3 E, lanes 4 and 5). In contrast, when FLASH is severely depleted (e.g., FLASHPBac/Df) or inactive in processing (e.g., FLASHLDIY45,71 and FLASHNL125), the misprocessed mRNAs are predominant, as they also are in Slbp and U7 mutants (Lanzotti et al., 2002; Godfrey et al., 2006). Thus, these two FLASH mutants (FLASH1–733 and FLASHLDIY71) with very different biochemical defects resulted in a similar alteration in histone mRNA biosynthesis in vivo.
Figure 3.
Concentrating FLASH in the HLB promotes histone 3′ end formation. (A) Diagram of C-terminal FLASH deletion mutants. (B) Visualization of rescue of FLASHPBac/Df with transgenic FLASH C-terminal mutants as in Fig. 1 D. (C) Anti-V5 Western analysis of total protein from larvae expressing FLASHFL, FLASHLDIY71, FLASH1–733, and FLASHLDIY71, 1–733 rescue transgenes. Asterisk, cross-reacting protein. (D) Localization of FLASH transgenic proteins (V5) in larval polyploid salivary gland nuclei stained for an HLB marker (Mxc). WT transgenic FLASH protein localizes to the HLB (yellow arrow), but the C-terminal truncation mutants do not (white arrows). Bars, 10 µm. (E) Total RNA from WT third-instar larvae, the FLASHPbac/Df mutant and the mutant rescued with the indicated FLASH transgenes was analyzed by S1 nuclease mapping as in Fig. 1 E. An autoradiograph is shown and the band intensities of a PhosphorImage of each lane were plotted using the ImageJ gel-analysis function.
These results suggest that both concentrating FLASH in the HLB and FLASH’s intrinsic processing activity may independently contribute to efficient pre-mRNA processing in vivo. If this is true, mislocalizing the partially functional FLASHLDIY71 protein by deleting the C-terminal 111 aa (Fig. 3 A, FLASH71,1–733) should exacerbate its mutant phenotype. FLASH71,1–733 did not concentrate in the HLB (Fig. 3 D) but accumulated to similar levels as FLASHFL, FLASHLDIY71, and FLASH1–733 (Fig. 3 C). Although FLASH71,1–733 rescued FLASHPac/Df-reduced viability (Fig. 3 B), it did not restore fertility (Table 1). Consistent with this more severe developmental phenotype, the histone pre-mRNA processing defect is exacerbated in FLASHLDIY71,1–733 relative to either single mutant (Fig. 3 E, compare lanes 4 and 5 with lane 6). Thus, the FLASHLDIY71 protein is more effective in promoting histone pre-mRNA processing when it is concentrated in the HLB. The synergistic effect of disrupting both processing activity and localization is consistent with independent contributions from each domain in 3′ end formation in vivo.
A second possibility to explain the FLASH1–733 RNA phenotype is that a region in the C terminus of FLASH also contributes to processing, independent of its role in localizing FLASH to the HLB. This does not seem likely, because aa 1–178 of FLASH are fully active in vitro, and we have not detected any interaction of the C terminus of Drosophila FLASH with Lsm11 using in vitro binding assays (unpublished data). However, to address this possibility, we examined a situation in which FL FLASH is mislocalized by using a mutant allele of the multi sex combs (mxc) gene. mxc encodes a 1,837-aa protein orthologous to human NPAT that likely acts as a scaffold for HLB assembly (White et al., 2011; Terzo et al., 2015). mxcG46 encodes a C-terminal truncation of Mxc protein lacking aa 1643–1837, and mxcG46 mutants are viable but female sterile. In the mxcG46 mutant, FLASH is also mislocalized (Fig. 4 C). Importantly, the total amount of FLASH protein in mxcG46 mutant ovaries was similar to the normal levels of WT FLASH (Fig. 5 D). Thus the C-terminal region of Mxc is required for localization of FLASH to the HLB (Fig. 4 C). Because our C-terminal anti-Mxc antibody does not detect MxcG46 protein (White et al., 2011; Fig. 4 C), we stained salivary glands of transgenic GFP-MxcG46–rescued animals with anti-GFP antibodies to confirm that MxcG46 protein supports HLB assembly (Fig. 4 C; Terzo et al., 2015).
In mxcG46 mutants, we detected small amounts of both misprocessed H2a RNA and read-through transcripts, although the majority of the H2a mRNA was properly processed (Fig. 5 B). These results are similar to the FLASH1–733 and FLASHLDIY71 mutants (Fig. 2 D). We also observed read-through transcripts by in situ hybridization in both the FLASH1–733 and MxcG46 nurse cells (Fig. 6, yellow arrows). Thus, mislocalizing WT, FL FLASH by truncating Mxc resulted in essentially identical effects on histone mRNA as removal of the FLASH C terminus. Although we cannot rigorously exclude another contribution to processing from the FLASH C terminus, these results are consistent with the effect being caused by mislocalizing FLASH.
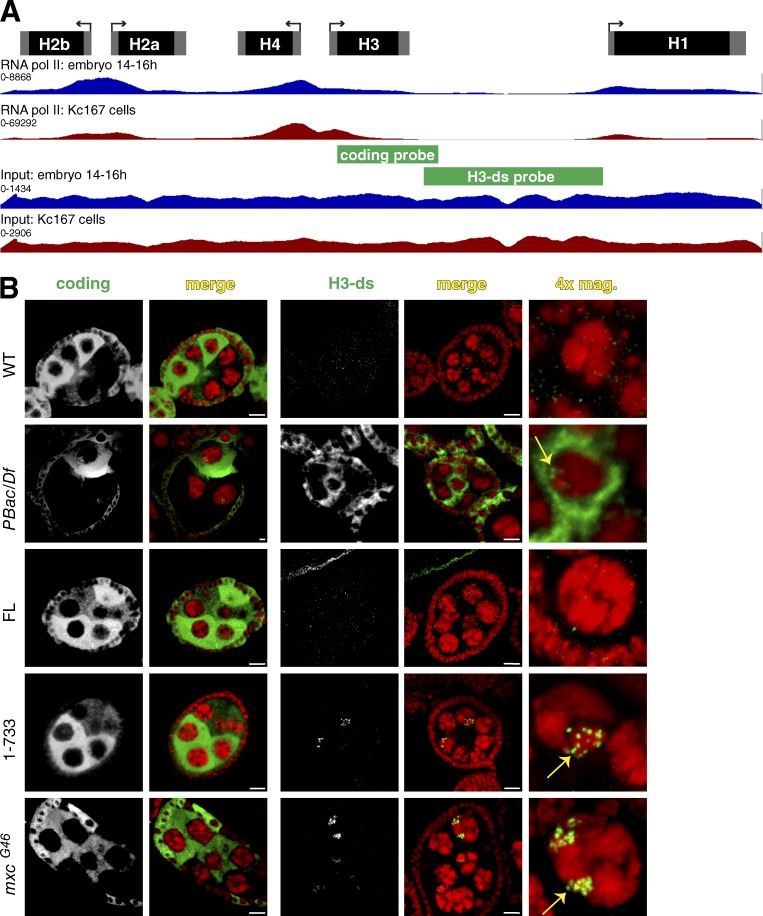
Figure 6.
Histone mRNA expression analyzed by in situ hybridization. (A) RNA pol II occupancy data for 14–16 h embryos and Kc167 cells (Kharchenko et al., 2011) visualized on a single histone repeat. The location of in situ hybridization probes used in B is indicated. (B) Fluorescent in situ hybridization of ovaries of the indicated genotypes using an H3 coding probe or a probe (H3-ds) that only detects misprocessed or read-through transcripts. Note that histone H3 mRNA accumulates only during S phase and that the nurse cell endocycles are asynchronous. The cytoplasmic H3-ds signal in FLASHPBac/Df is from misprocessed H3 transcripts that were polyadenylated and exported. The small amounts of misprocessed RNA in FLASH1−733 and mxcG46 (see Fig. 5 B) mRNAs are below our threshold of detection. The nuclear foci in FLASH1-733 and mxcG46 mutants detected with the H3-ds probe (yellow arrows) are nascent transcripts at the histone locus. Bars, 10 µm.
High concentrations of FLASH are necessary for U7 snRNP accumulation in the HLB
To determine whether some of the effects of mutating FLASH might be mediated through changes in HLB composition, we visualized HLBs in different FLASH mutant animals with antibodies against a panel of HLB markers (Figs. 4 and S3). We analyzed ovaries of the small number of FLASHPBac/Df mutant females that survive to adulthood. Although these females are sterile and we do not observe fully developed eggs, oogenesis consistently proceeded to stage 8, allowing us to examine stage 5 polyploid nurse cells, which provide excellent cytology for HLBs (Liu et al., 2006). We did not detect FLASH in the HLBs of FLASHPBac/Df mutant ovaries, although Mxc and Mute were present (Fig. 4 A). We also did not detect the U7 snRNP-specific protein Lsm10 in these HLBs (Fig. 4 A). Expressing FLASHFL protein in the FLASHPBac/Df mutant restored Lsm10 localization to the HLB, as did expressing FLASHmini and FLASHNL125 (Fig. 4 B). In contrast, expressing the FLASH1–733 mutant protein resulted in accumulation of low levels of Lsm10 in the HLB (Fig. 4 B), which was only detectable in some experiments and is possibly because of a small amount of FL FLASH produced by the FLASHPBac/Df genotype. These data suggest that high concentrations of FLASH in the HLB are needed for U7 snRNP accumulation in the HLB. In addition, the FLASHNL125 results demonstrate that the interaction of the N terminus of FLASH and Lsm11 is not necessary for concentration of U7 snRNP in the HLB, even though this interaction is required for pre-mRNA processing. Lsm10 also does not accumulate in the HLBs of the mxcG46 mutant (Fig. 4 D), consistent with the results with the mislocalized FLASH1–733 protein. Collectively, our data suggest that two distinct regions of FLASH function within the HLB to (a) localize FLASH and U7 snRNP to the HLB (C terminus of FLASH) and (b) bind the Lsm11 protein of U7 snRNP and recruit the HCC to the U7 snRNP to activate histone pre-mRNA processing.
Factors in addition to FLASH are required to concentrate U7 snRNP in the HLB
The Drosophila H2aV gene produces a polyadenylated mRNA that encodes a variant H2a histone protein with the combined roles of mammalian H2AX and H2AZ, functioning in transcriptional regulation and the DNA damage response (Redon et al., 2002). The proper ratio of H2aV to canonical H2a histone protein is critical for viability (Li et al., 2014). We have previously shown that U7 snRNP was not localized to the HLB in H2aV mutant animals or H2aV-depleted cells (Wagner et al., 2007). Because these studies were done before our discovery of FLASH, we revisited H2aV mutants to determine whether loss of H2aV also affects FLASH localization. FLASH and two other HLB markers, Mxc and Mute, colocalize in a prominent focus in H2aV810 mutant salivary glands (Fig. 5 A). Lsm11 was absent from these HLBs (Fig. 5 A).
In H2aV810 mutants we detected both misprocessed H2a RNA and read-through transcripts, although the majority of the H2a mRNA was properly processed (Fig. 5, B and C). These results are essentially indistinguishable from the RNA phenotype of the mxcG46 and FLASH1–733 mutants (Fig. 5, B and C), supporting the idea that localization of FLASH and U7 snRNP is necessary for efficient processing of histone mRNA. Importantly, the levels of Lsm11 were similar to WT in the H2aV mutant larvae (Fig. 5 E), and we previously showed that nuclear extracts prepared from H2aV depleted cells are as active as extracts from control cells in histone pre-mRNA processing (Wagner et al., 2007). As with FLASH, these results indicate that high local concentrations of processing-competent U7 snRNP in the HLB, and not its overall availability in the nucleus, are necessary for efficient histone pre-mRNA processing in vivo.
The HLB promotes rapid cotranscriptional histone pre-mRNA processing
Our transgenic analysis found that mutants with reduced local FLASH activity, either as a result of mislocalizing FLASH (FLASH1–733, mxcG46) or U7 snRNP (H2aV810), or reducing FLASH activity in the HLB (FLASHLDLY71), produce read-through and misprocessed RNA in similar amounts (as well as large amounts of WT mRNA). The production of read-through and misprocessed histone mRNAs indicates that RNA polymerase II (pol II) must have transcribed well past the normal cleavage site. Analysis of pol II occupancy by ChIP sequencing in WT embryos and cultured Kc cells from the modENCODE project revealed that there is little or no RNA pol II present in the intergenic regions between histone genes (Fig. 6 A). This profile suggests that normally there is rapid cleavage followed by efficient transcription termination.
Failure to properly process histone pre-mRNA and terminate transcription results in longer transcripts that are detected at the site of transcription using a probe derived from the region downstream of the H3 gene (H3-ds; Fig. 6 A; Lanzotti et al., 2002; Godfrey et al., 2006, 2009). We tested whether similar transcripts can be detected in mutants with mislocalized FLASH. The fluorescent H3-ds probe does not detect any transcripts in WT cells but detected abundant misprocessed, polyadenylated histone mRNA in the nurse cell cytoplasm of the FLASHPbac/Df mutant (Fig. 6 B). This phenotype was rescued by FLASHFL (Fig. 6 B). FLASH1–733 and mxcG46 nurse cells contained predominantly cytoplasmic WT H3 mRNA that was detected with a coding probe, but not the H3-ds probe (Fig. 6 B). With the H3-ds probe, we detected foci of nascent transcripts in FLASH1–733 and mxcG46 nurse cells, but no cytoplasmic misprocessed mRNA (Fig. 6 B). Because the steady-state level of normally processed histone mRNAs is similar in the FLASH1–733, mxcG46 and WT animals, many of these longer, nascent transcripts are likely ultimately cleaved at the normal site. The increase in steady-state amount of unprocessed, nascent histone H3 pre-mRNA suggests that when FLASH or U7 snRNP is not concentrated in the HLB, the rate of histone pre-mRNA processing slows.
To further support the possibility that the efficiency of 3′ end formation is sensitive to the amount of FLASH activity in the HLB, we examined the expression of histone mRNA in a FLASH-null mutant during embryogenesis. Maternally deposited WT FLASH protein accumulates in the HLB before activation of zygotic histone transcription (White et al., 2011). Therefore, zygotic FLASH mutant phenotypes will not be apparent until multiple rounds of cell division deplete the maternal FLASH protein. This “maternal run out” situation provides an opportunity to examine embryos with reduced concentrations of FLASH in the HLB but before FLASH is completely depleted. Stage 17 (14–18 h) FLASHDf/Df mutant embryos had both misprocessed and read-through histone mRNAs in similar amounts (Fig. 5 F, lane 3), a phenotype qualitatively different from the effect of severely depleting FLASH that occurs by larvae stages. Thus, the initial phenotype as maternal FLASH is depleted in the FLASH mutant is similar to the effect of mislocalizing normal amounts of FLASH, as seen in the mxcG46 and FLASH1–733 mutants. This effect is likely a result of reducing the local concentration of FLASH in the HLB, supporting the conclusion that concentrating factors in the HLB is important for efficient histone mRNA 3′ end formation.
Discussion
Our systematic analysis of Drosophila FLASH mutants and mutants in other HLB components has allowed us to attribute a function in histone mRNA metabolism to the HLB: promoting rapid histone pre-mRNA processing, resulting in efficient coupling of transcription termination with 3′ end formation.
The HLB contributes to multiple steps in histone mRNA biosynthesis
HLB assembly initiates with formation of a complex containing FLASH and Mxc, followed by incorporation of other components including U7 snRNP (White et al., 2011). How Mxc and FLASH are specifically targeted to the histone locus is not known, but this process does not require the 3′ processing signals in the histone pre-mRNA and is coordinated by the Drosophila H3-H4 promoter (Salzler et al., 2013). An emerging property of HLB components is the presence of independent biochemical and HLB targeting domains (Yang et al., 2014; Terzo et al., 2015). For instance, the FLASH C terminus is required for localization to the HLB, whereas the N-terminal region required for histone pre-mRNA processing interacts with U7 snRNP. Similarly, U7 snRNP recruitment to the HLB requires the C termini of FLASH and Mxc but does not require the interaction between the N termini of FLASH and Lsm11 that is required for histone pre-mRNA processing (Burch et al., 2011).
Once the HLB assembles in early development, Mxc, FLASH, and U7 snRNP are concentrated in the HLB throughout interphase, independent of whether the histone genes are being expressed (White et al., 2011). Thus, activation of histone mRNA biosynthesis is distinct from recruitment of HLB components to the histone locus. The constitutive presence of FLASH and U7 snRNP contrasts with cell cycle–dependent enrichment of the HCC component Symplekin in the HLB (Tatomer et al., 2014). The uncoupled process for HLB localization of and interaction between U7 snRNP and FLASH suggests that changes within the HLB during S phase may facilitate HCC recruitment to and subsequent cotranscriptional cleavage of histone pre-mRNA.
Concentrating factors within the HLB ensures efficient histone mRNA synthesis
Here, we have analyzed three different situations to determine that high concentrations of FLASH and U7 snRNP in the HLB are necessary for efficient histone mRNA synthesis: (1) FLASH mutants with impaired localization, (2) mutants in Mxc and H2aV that result in mislocalization of WT FLASH and/or U7 snRNP, and (3) diminished WT FLASH levels as the maternal supply of FLASH is reduced during development of FLASH mutant embryos. A direct way to do this would be to retarget FLASH1–733 to the HLB by replacing the C terminus of FLASH with another HLB localization signal. We attempted to directly test the possibility that localization of FLASH is critical for histone pre-mRNA processing by retargeting FLASH1–733 to the HLB with the N-terminal 101 aa of Mxc, which can target GFP to the HLB and is essential to target Mxc to the HLB (Terzo et al., 2015). Although this signal targeted FLASH1–733 to the HLB in cultured cells and partially rescued the FLASH mutant phenotype (Fig. S4), only a small amount of the fusion protein accumulates in transgenic animals in some tissues (Fig. S5), precluding us from making any firm conclusions regarding additional functions of the FLASH C terminus.
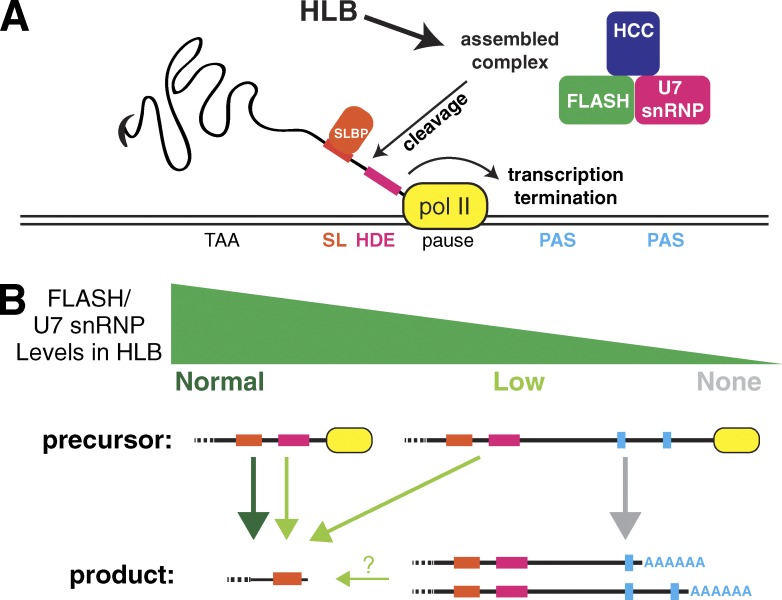
The processing of histone pre-mRNAs in Drosophila is normally very efficient. There is a strong pause site just downstream of the U7 binding site (the histone downstream element [HDE]; Adamson and Price, 2003), and no transcripts are detected extending 3′ of the processed RNA in WT animals (Lanzotti et al., 2002). In addition, RNA pol II is not present downstream of the HDE in both Drosophila (Fig. 6 A) and mammalian cells (Anamika et al., 2012; Cheng et al., 2012). Thus, termination of transcripts from the histone genes is normally tightly coupled with histone pre-mRNA processing. Mutations in Drosophila FLASH, Slbp, or U7 snRNP that prevent histone pre-mRNA processing allow transcription to proceed 3′ of the histone genes, resulting in formation of polyadenylated histone mRNAs via cryptic, downstream signals (Lanzotti et al., 2002; Godfrey et al., 2006). Here, we showed that subtler perturbations that affect the localization of FLASH and/or U7 snRNP to the HLB, or that partially impair FLASH activity within the HLB, also result in read-through transcripts at the histone locus, although large amounts of properly processed histone mRNA still accumulate. The appearance of these longer, nascent transcripts suggests that normal processing is delayed, resulting in loss of coupling between processing and transcription termination.
What is the basis for the mutant RNA phenotype in cases where the activity of processing factors is reduced rather than abolished? Mechanistic differences between processing 3′ ends of polyadenylated mRNAs and histone mRNAs suggest an answer. The interaction of polyadenylation factors with the C-terminal domain of RNA pol II couples polyadenylation with transcription (McCracken et al., 1997; Adamson et al., 2005) and the C-terminal domain may participate directly in promoting cleavage or polyadenylation (Hirose and Manley, 1998). In contrast, there are no known interactions between RNA pol II and components specific to the histone-processing complex. Unlike polyadenylation, cotranscriptional processing does not increase the rate of histone mRNA 3′ end formation in vitro (Adamson and Price, 2003). These results suggest that processing histone pre-mRNA at the normal location can occur after the polymerase has transcribed well past the HDE (Fig. 7).
Figure 7.
Model of HLB participation in histone pre-mRNA processing. (A) RNA pol II (yellow) pauses 3′ of the HDE, allowing Slbp (orange) to bind the stem loop (SL) in the pre-mRNA downstream of the stop codon and U7 snRNP (pink), FLASH (green), and the HCC (blue) to assemble into a complex that associates with the pre-mRNA at the HDE. Processing occurs followed by transcription termination. (B) High levels of active FLASH and U7 snRNP (dark green arrow) in the HLB result in rapid cleavage while pol II is paused leading to transcription termination, preventing RNA pol II from encountering downstream polyadenylation signals (PASs). Low levels of FLASH (light green arrows) slow normal processing resulting in release of some pol II from the pause site. This leads to production of longer pre-mRNAs, which are detected at the site of histone gene transcription by in situ hybridization. Most of these transcripts are ultimately processed at the normal site, and a small proportion of these are polyadenylated (thin, green arrow). In this situation, there are multiple possible precursor RNAs that could give rise to properly processed histone mRNA. Some will be processed while the RNA pol II is paused, others will be processed after the pol II releases from the pause site, and it is possible (question marks) that some of the polyadenylated transcripts could be “reprocessed” as described in the text. Absence or severe reduction in any histone pre-mRNA processing factor, whether or not it is an HLB component, results in production of a long precursor pre-mRNA that is processed by the polyadenylation machinery (gray arrow).
A second possible way to accumulate properly processed histone mRNA from longer transcripts is to “reprocess” a transcript that has already been cleaved and polyadenylated. Our previous study suggested in mammalian cells that utilization of a processing site downstream of the stem loop or HDE might stabilize a longer transcript, providing time for partially defective histone processing machinery to generate a normal histone mRNA 3′ end (Liu et al., 1989). Indeed, in Caenorhabditis elegans, which lacks FLASH and U7 snRNP but contains Slbp, there is evidence that the normal pathway of histone mRNA processing is polyadenylation followed by cleavage after the stem loop by an siRNA-like mechanism (Mangone et al., 2010; Avgousti et al., 2012).
Cellular microenvironments that enhance biological processes
Our results support the idea that NBs facilitate reactions by concentrating factors (Matera et al., 2009; Mao et al., 2011b). The exchange of NB components with the adjacent environment is slower than expected by diffusion, promoting concentration in NBs (Dundr et al., 2004). Such concentration of components creates discrete microdomains with different physical chemical properties than the surrounding nucleoplasm (Brangwynne et al., 2011; Han et al., 2012; Kato et al., 2012). A role for CBs in facilitating snRNP assembly by concentrating factors has been previously suggested (Klingauf et al., 2006; Stanek et al., 2008; Novotný et al., 2011). In addition, a reduced rate of tri-snRNP assembly was postulated to cause the splicing defects observed in zebrafish lacking CBs because of Coilin depletion (Strzelecka et al., 2010). In the case of the HLB, our data provide evidence for two roles: enhancing the rate of pre-mRNA processing and promoting coupling of processing with transcription termination. Identification and characterization of additional HLB components in the future will facilitate our ability to further test this model of NB function.
Materials and methods
Drosophila strains
PBac[PB]FLASHLL01602 was a gift from G. Matera (University of North Carolina, Chapel Hill, NC), who obtained it from the Drosophila Genomics Resources Center. The site of insertion was determined by PCR and sequencing (Fig. S1). FLASH constructs previously generated in pIZ/V5 (Thermo Fisher Scientific) were inserted into pATTB (Bischof et al., 2007) with KpnI and XhoI for φC31-mediated transgenesis (Best Gene) and integration into the attP40 landing site. Transgenic GFP-MxcG46 integrated at VK00033 was expressed with the ubiquitin promoter of pUGW in the mxcG48 null background (White et al., 2011). Additional strains are summarized in Table 2. The features of the FLASH transgenes are summarized in Table 3.
Table 2. Drosophila strains.
| Strain | Source | Reference |
|---|---|---|
| PBac LL01602 | Drosophila Genomics Resource Center: 140418 | — |
| Df (2R) 8057 | Bloomington: 7871 | — |
| PBac c02047 (Lsm11) | Exelixis Harvard: c02047 | Godfrey et al., 2009 |
| His2aV810 | Bloomington: 9264 | van Daal and Elgin, 1992 |
| MxcG46 | Bloomington: 32114 | White et al., 2011 |
| MxcG48 | Bloomington: 7141 | White et al., 2011 |
Table 3. Construct features.
| FLASH transgene feature | Sequence |
|---|---|
| attB–KpnI–promoter | 5′-GATCTCTAGAGGTACCcgagattattgtttgtattgtgatgtttttttttaacgaatttaa-3′ |
| V5–XhoI–AttB | 5′-GGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGTAACTCGAGCCGCGGCCGCAGAT-3′ |
| 5′ UTR–EcoRI–coding | 5′-GAAGAAACGTAAGCGATTGGTGAATTCCAAAATG-3′ |
| EcoRI–FL | 5′-GAATTCCAAAATGGAAACGCCTGCATATGCCAC-3′ |
| EcoRI–aa 65 | 5′-GAATTCCAAAATGGACAGATCCCTTGAACTGGAC-3′ |
| EcoRI–aa 78 | 5′-GAATTCCAAAATGGACGACTTTCAGAAGGCCGA-3′ |
| aa 844–SacII–V5 | 5′-CTGCTGCgGACAAACCAATTCCGCGGTTCGAA-3′ GGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACG-3′ |
| aa 733–SacII–V5 | 5′-CTCACCCAGACTCCAAAACAGGCTCCGCGGTTCGGA-3′ GGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACG-3′ |
| FLASHmini junction | 5′-CGAAAAGGACGATGT/ACCCAGACTCCAAAA-3′ |
Engineered restriction sites are underlined, start and stop codons are bold, the FLASH promoter sequence is lowercase, the V5 tag is italicized, and “/” indicates the unique junction in the FLASHmini open reading frame.
Genomic PCR
Genomic DNA was prepared by incubating a squashed fly in 50 µl of squash buffer (10 mM Tris, pH 8.2, 25 mM NaCl, 1 mM EDTA, and 200 µg/ml Proteinase K; New England Biolabs, Inc.) at 37°C for 30 min. After 10 min at 85°C, 1 µl per reaction was used to amplify each product with TAQ DNA polymerase (Thermo Fisher Scientific). PCR products were purified (Thermo Fisher Scientific) and subjected to Sanger sequencing. The primers used in each PCR reaction are summarized in Table 4.
Table 4. Sequences for primers used to identify the PBac LL01602.
| Primer name | Sequence |
|---|---|
| FLASH sequencing F | 5′-GAACGGTACCCGAGATTATTGTTTGTATTGTGATGT-3′ |
| FLASH sequencing R | 5′-GTCTTCCAAATCCATCAAGTCCTC-3′ |
| 5′ Pbac | 5′-CGCGATAAATCTTTCTCTCTCG-3′ |
| 3′ Pbac | 5′-CCGATAAAACACATGCGTCA-3′ |
Histone mRNA analysis
A total of 5 µg total cellular RNA extracted with Trizol reagent (Invitrogen) was used for each S1 nuclease protection reaction. The probe was created by 5′ end labeling BspEII cut H2a DNA with α-32P-dCTP using the Klenow fragment of DNA polymerase I (New England Biolabs, Inc.). The probe was gel purified and hybridized to either total larval RNA or control yeast tRNA followed by digestion with S1 nuclease (Lanzotti et al., 2002). Protected fragments were resolved on a 6% polyacrylamide-7M urea gel and visualized by autoradiography. The band intensities for each lane were plotted by the ImageJ gel-analysis function.
Immunoblotting
Ovaries of the indicated genotypes were dissected in Grace’s media (Gibco) and lysed in RIPA buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris, pH 7.5, 0.1% SDS, 0.5% sodium-deoxycholate, and protease inhibitors; Thermo Fisher Scientific). Brains and imaginal discs were dissected from wandering third-instar larvae and lysed by boiling in sample buffer. All samples were passed through a 25-gauge needle 100 times before resolving through a 10% gel by SDS-PAGE and detection with ECL Prime (GE Healthcare) using the antibodies summarized in Table 6.
Table 6. Summary of antibodies used for both immunofluorescence and Western blot analysis.
| Antibody | Raised in or recognizes | Source | Concentration | Incubation |
|---|---|---|---|---|
| Primary | ||||
| FLASH | Rabbit | Yang et al., 2009 | 1:2,000 | 4°C, overnight |
| V5 | Mouse | Invitrogen | 1:1,000 | 4°C, overnight |
| Mxc | Guinea pig | White et al., 2011 | 1:2,000 | 4°C, overnight |
| Mute | Guinea pig | Bulchand et al., 2010 | 1:5,000 | 4°C, overnight |
| Lsm10 | Rabbit | Liu et al., 2006 | 1:2,000 | 4°C, overnight |
| Lsm11 | Rabbit | Liu et al., 2006 | 1:2,000 | 4°C, overnight |
| HP1 | Mouse | Developmental Studies Hybridoma Bank | 1:1,000 | 4°C, overnight |
| H2aV | Rabbit | Leach et al., 2000 | 1:1,000 | 4°C, overnight |
| GFP | Chicken | EMD Millipore | 1:1,000 | 4°C, overnight |
| β-Tubulin | Rabbit | Abcam | 1:5,000 | Room temperature, 2 h |
| Secondary | ||||
| Alexa Fluor 488 | Rabbit | Invitrogen | 1:2,000 | Room temperature, 2 h |
| Alexa Fluor 488 | Mouse IgG2a | Invitrogen | 1:1,000 | Room temperature, 2 h |
| Cy3 | guinea pig | Jackson ImmunoResearch Laboratories, Inc. | 1:1,000 | Room temperature, 2 h |
| Alexa Fluor 555 | Mouse IgG1 | Invitrogen | 1:1,000 | Room temperature, 2 h |
| Cy5 | rabbit | Abcam | 1:2,000 | Room temperature, 2 h |
| Cy5 | rabbit | Jackson ImmunoResearch Laboratories, Inc. | 1:1,000 | Room temperature, 2 h |
| ECL donkey HRP | rabbit | GE Healthcare | 1:10,000 | Room temperature, 1 h |
| ECL prime donkey HRP | mouse | GE Healthcare | 1:10,000 | Room temperature, 1 h |
Immunofluorescence
For immunofluorescence, salivary glands were dissected in PBS + 0.1% Triton X-100 and ovaries were dissected in Grace’s medium (Gibco). Fixation and permeablization conditions are summarized in Table 5. Confocal images were taken at room temperature at a zoom of 1.0–3.0 with a 40× (numerical aperture 1.30) Plan Neofluor objective on a 510 laser scanning confocal microscope (ZEISS) using the LSM data acquisition software (ZEISS). Image false coloring and contrast was adjusted, and images were digitally zoomed using Photoshop (Adobe). Staining conditions and antibodies are presented in Table 6.
Table 5. Tissue preparation for antibody staining for immunofluorescence.
| Tissue | Fixation | Permeablization |
|---|---|---|
| Salivary gland (except in Figs. 4 C and 5 A) | 7% formaldehyde, 20 min | 0.2% Tween-80, 15 min |
| Salivary gland (Fig. 4 C) | 3.7% formaldehyde, 20 min | Not performed |
| Salivary gland (Fig. 5 A) | 3.7% formaldehyde, 15 min | 1% Triton X-100, 15 min |
| Ovary | 7% formaldehyde, 20 min | 0.2% Tween-80, 15 min |
Fluorescence in situ hybridization
Ovaries were dissected and fixed in 4% formaldehyde in PBS for 20 min and postfixed in 4% formaldehyde in PBS + 0.1% Tween-80 (PBTw). Samples were washed three times in PBTw for 2 min and then incubated in 3 µg/ml proteinase K in PBTw for 10 min at room temperature followed by 30 min on ice. Digestion was quenched by incubation in 2 µg/ml glycine/PBTw for 2 min twice. After two PBTw rinses, ovaries were postfixed for 20 min in 4% formaldehyde. Ovaries were washed five times in PBTw (Lécuyer et al., 2008). Samples were then hybridized to the indicated probe as previously described (Tomancak et al., 2002). Detection was performed with 1:200 peroxidase-conjugated anti-DIG (Roche) and 1:100 Cy5 tyramide reagent (PerkinElmer).
Fluorescent in situ probes were synthesized by in vitro transcription from vector template DNA that was linearized by digestion with ApaI for H3-coding and XhoI for H3-ds as previously described (Lanzotti et al., 2002). The H3 coding probe was transcribed with T3 RNA polymerase (Ambion), and the H3-ds probe was transcribed with T7 RNA polymerase (Ambion) using the DIG RNA labeling mix (Roche). RNA synthesis was verified by agarose gel electrophoresis. The sequence boundaries for each probe are summarized in Table 7.
Table 7. Boundaries for fluorescent in situ hybridization probe sequences.
| Probe | Sequence |
|---|---|
| H3-coding 5′ end of probe | 5′-ATGGCTCGTACCAAGCAAAC-3′ |
| H3-coding 3′ end of probe | 5′-GCTCAACATTTATTAAATTA-3′ |
| H3-ds 5′ end of probe | 5′-CGGTACTGGGTCTTAAATCA-3′ |
| H3-ds 3′ end of probe | 5′-AGTGCTCTCCTCCTCGATTC-5′ |
In vitro processing assay
Depletion of FLASH from S2 cells, preparation of nuclear extracts, and in vitro processing assays were performed as previously described (Sabath et al., 2013). The details for nuclear extract preparation are as follows. Cells were harvested by centrifugation (5 min, 3,000 rpm, 4°C) and washed once in PBS. The cell pellet was resuspended in three times the estimated pellet volume in buffer A (10 mM Hepes/KOH, pH 7.9, 1.5 mM MgCl2, 10mM KCl, and 0.5 mM DTT [add fresh]). Cells were transferred and lysed with 10–12 strokes in a 2-ml glass Dounce. Nuclei were pelleted by centrifugation (5 min, 5,000 rpm, 4°C). The volume of the nuclear pellet was estimated and resuspended in 1.5 vol of low-salt buffer C (20 mM Hepes/KOH, pH 7.9, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT [add fresh], and 0.02 M NaCl). The resuspended nuclei were transferred to a fresh tube with a micro–stir bar. High-salt buffer C (20 mM Hepes/KOH, pH 7.9, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT [add fresh], and 1.2 M NaCl) was added drop by drop to the nuclei. The total amount of high-salt buffer added brought the final concentration of Na+ to between 0.26 and 0.28 M. The amount was calculated with the following equation: high-salt buffer C = (volume resuspended nuclei × 0.25/0.95 + volume resuspended nuclei × 0.28/0.98)/2. After 1 h mixing on ice, the lysate was transferred to a new Eppendorf and cleared by centrifugation (10 min, 10,000 rpm, 4°C). The supernatant was transferred to a Slide-A Lyzer Dialysis cassette (3,500 molecular weight cut-off and 0.1–0.5 ml capacity; Thermo Fisher Scientific) with a syringe and 18-gauge needle. The nuclear extract was dialyzed buffer D (20 mM Hepes/KOH, pH 7.9, 20% glycerol, 0.1 M KCl, 0.2 mM EDTA, and 0.5 mM DTT [add fresh]) for 2 h at 4°C, and this step was repeated with fresh buffer D. The extract was removed with a syringe and cleared by centrifugation (10 min, 10,000 rpm, 4°C). The supernatant was aliquoted, frozen on dry ice/EtOH, and stored at −80°C.
RNA pol II localization
High-throughput sequencing reads were processed as previously described (McKay and Lieb, 2013). The following exceptions were made to unambiguously map reads to the histone locus. The exact number of histone gene repeats in the Drosophila genome is unknown, and the reference genome sequence is thus incomplete. To circumvent this problem, a custom reference genome was created by removing all canonical histone gene-repeat sequences and adding back a single 5-kb histone gene-repeat unit. An unlimited number of reads were then mapped to this custom genome with bowtie (Langmead et al., 2009) using the options “–nomaqround” and “–best.” Coverage values were then calculated for each base in the genome, and the data were visualized using Integrative Genomics Viewer (Robinson et al., 2011). Datasets modENCODE_5122 and modENCODE_5569 were downloaded from the modENCODE ftp site (Kharchenko et al., 2011).
Online supplemental material
Fig. S1 is a characterization of FLASHLL01602 PBac. Fig. S2 shows expression and localization of transgenic FLASH proteins. Fig. S3 shows RNA phenotypes in animals where FLASH concentration and/or biochemical activity of FLASH in the HLB is altered. Fig. S4 shows attempts to target FLASH1–733 to the HLB using the HLB localization site from Mxc. Fig. S5 shows HLB localization analysis of transgenic FLASH-mxc fusion constructs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201504043/DC1.
Supplementary Material
Acknowledgments
We thank B. Burch, X.-C. Yang, and S. Lyons for technical assistance, J. Gall for the Lsm10 antibody, S. Nowotarski and K. Boltz for critically reading the manuscript, and G. Matera for helpful discussions.
This work was supported by National Institutes of Health grants GM58921 (to W.F. Marzluff and R.J. Duronio) and F31GM106698 (to E. Terzo).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CB
- Cajal body
- FL
- full length
- FLASH
- Flice-associated huge protein
- HCC
- histone cleavage complex
- HDE
- histone downstream element
- mxc
- multi sex combs
- NBs
- nuclear bodies
- pol II
- polymerase II
- RD
- replication dependent
- Slbp
- stem-loop binding protein
- snRNP
- small nuclear RNP
- WT
- wild type
References
- Adamson T.E., and Price D.H.. 2003. Cotranscriptional processing of Drosophila histone mRNAs. Mol. Cell. Biol. 23:4046–4055. 10.1128/MCB.23.12.4046-4055.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson T.E., Shutt D.C., and Price D.H.. 2005. Functional coupling of cleavage and polyadenylation with transcription of mRNA. J. Biol. Chem. 280:32262–32271. 10.1074/jbc.M505532200 [DOI] [PubMed] [Google Scholar]
- Anamika K., Gyenis À., Poidevin L., Poch O., and Tora L.. 2012. RNA polymerase II pausing downstream of core histone genes is different from genes producing polyadenylated transcripts. PLoS One. 7:e38769 10.1371/journal.pone.0038769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgousti D.C., Palani S., Sherman Y., and Grishok A.. 2012. CSR-1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J. 31:3821–3832. 10.1038/emboj.2012.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R.K., Hediger M., Karch F., and Basler K.. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 104:3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Mitchison T.J., and Hyman A.A.. 2011. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA. 108:4334–4339. 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulchand S., Menon S.D., George S.E., and Chia W.. 2010. Muscle wasted: a novel component of the Drosophila histone locus body required for muscle integrity. J. Cell Sci. 123:2697–2707. 10.1242/jcs.063172 [DOI] [PubMed] [Google Scholar]
- Burch B.D., Godfrey A.C., Gasdaska P.Y., Salzler H.R., Duronio R.J., Marzluff W.F., and Dominski Z.. 2011. Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA. 17:1132–1147. 10.1261/rna.2566811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B., Li T., Rahl P.B., Adamson T.E., Loudas N.B., Guo J., Varzavand K., Cooper J.J., Hu X., Gnatt A., et al. 2012. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol. Cell. 45:38–50. 10.1016/j.molcel.2011.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryusheva S., and Gall J.G.. 2009. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol. Biol. Cell. 20:5250–5259. 10.1091/mbc.E09-09-0777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Yang X.C., and Marzluff W.F.. 2005. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 123:37–48. 10.1016/j.cell.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Dundr M. 2011. Seed and grow: a two-step model for nuclear body biogenesis. J. Cell Biol. 193:605–606. 10.1083/jcb.201104087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Hebert M.D., Karpova T.S., Stanek D., Xu H., Shpargel K.B., Meier U.T., Neugebauer K.M., Matera A.G., and Misteli T.. 2004. In vivo kinetics of Cajal body components. J. Cell Biol. 164:831–842. 10.1083/jcb.200311121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahati H., Pelham-Webb B., Blythe S., and Wieschaus E.. 2016. Nucleation by rRNA Dictates the Precision of Nucleolus Assembly. Curr. Biol. 26:277–285. 10.1016/j.cub.2015.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M.R., and Matera A.G.. 1995. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. USA. 92:5915–5919. 10.1073/pnas.92.13.5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J.G. 2000. Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16:273–300. 10.1146/annurev.cellbio.16.1.273 [DOI] [PubMed] [Google Scholar]
- Ghule P.N., Dominski Z., Yang X.C., Marzluff W.F., Becker K.A., Harper J.W., Lian J.B., Stein J.L., van Wijnen A.J., and Stein G.S.. 2008. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 105:16964–16969. 10.1073/pnas.0809273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey A.C., Kupsco J.M., Burch B.D., Zimmerman R.M., Dominski Z., Marzluff W.F., and Duronio R.J.. 2006. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA. 12:396–409. 10.1261/rna.2270406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey A.C., White A.E., Tatomer D.C., Marzluff W.F., and Duronio R.J.. 2009. The Drosophila U7 snRNP proteins Lsm10 and Lsm11 play an essential role in development independent of histone pre-mRNA processing. RNA. 15:1661–1672. 10.1261/rna.1518009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., and McKnight S.L.. 2012. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 149:768–779. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Hirose Y., and Manley J.L.. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 395:93–96. 10.1038/25786 [DOI] [PubMed] [Google Scholar]
- Imai Y., Kimura T., Murakami A., Yajima N., Sakamaki K., and Yonehara S.. 1999. The CED-4-homologous protein FLASH is involved in Fas-mediated activation of caspase-8 during apoptosis. Nature. 398:777–785. 10.1038/19709 [DOI] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 149:753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko P.V., Alekseyenko A.A., Schwartz Y.B., Minoda A., Riddle N.C., Ernst J., Sabo P.J., Larschan E., Gorchakov A.A., Gu T., et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 471:480–485. 10.1038/nature09725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingauf M., Stanek D., and Neugebauer K.M.. 2006. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol. Biol. Cell. 17:4972–4981. 10.1091/mbc.E06-06-0513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti D.J., Kaygun H., Yang X., Duronio R.J., and Marzluff W.F.. 2002. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ end processing in vivo. Mol. Cell. Biol. 22:2267–2282. 10.1128/MCB.22.7.2267-2282.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach T.J., Mazzeo M., Chotkowski H.L., Madigan J.P., Wotring M.G., and Glaser R.L.. 2000. Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. J. Biol. Chem. 275:23267–23272. 10.1074/jbc.M910206199 [DOI] [PubMed] [Google Scholar]
- Lécuyer E., Parthasarathy N., and Krause H.M.. 2008. Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods Mol. Biol. 420:289–302. 10.1007/978-1-59745-583-1_18 [DOI] [PubMed] [Google Scholar]
- Li Z., Johnson M.R., Ke Z., Chen L., and Welte M.A.. 2014. Drosophila lipid droplets buffer the H2Av supply to protect early embryonic development. Curr. Biol. 24:1485–1491. 10.1016/j.cub.2014.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.L., Murphy C., Buszczak M., Clatterbuck S., Goodman R., and Gall J.G.. 2006. The Drosophila melanogaster Cajal body. J. Cell Biol. 172:875–884. 10.1083/jcb.200511038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.L., Wu Z., Nizami Z., Deryusheva S., Rajendra T.K., Beumer K.J., Gao H., Matera A.G., Carroll D., and Gall J.G.. 2009. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol. Biol. Cell. 20:1661–1670. 10.1091/mbc.E08-05-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.-J., Levine B.J., Skoultchi A.I., and Marzluff W.F.. 1989. The efficiency of 3′-end formation contributes to the relative levels of different histone mRNAs. Mol. Cell. Biol. 9:3499–3508. 10.1128/MCB.9.8.3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Van Tine B.A., Wei Y., Garrett M.D., Nelson D., Adams P.D., Wang J., Qin J., Chow L.T., and Harper J.W.. 2000. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14:2298–2313. 10.1101/gad.829500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M., Manoharan A.P., Thierry-Mieg D., Thierry-Mieg J., Han T., Mackowiak S.D., Mis E., Zegar C., Gutwein M.R., Khivansara V., et al. 2010. The landscape of C. elegans 3'UTRs. Science. 329:432–435. 10.1126/science.1191244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.S., Sunwoo H., Zhang B., and Spector D.L.. 2011a Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 13:95–101. 10.1038/ncb2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.S., Zhang B., and Spector D.L.. 2011b Biogenesis and function of nuclear bodies. Trends Genet. 27:295–306. 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W.F., Wagner E.J., and Duronio R.J.. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9:843–854. 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G., Izaguire-Sierra M., Praveen K., and Rajendra T.K.. 2009. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev. Cell. 17:639–647. 10.1016/j.devcel.2009.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., and Bentley D.L.. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 385:357–361. 10.1038/385357a0 [DOI] [PubMed] [Google Scholar]
- McKay D.J., and Lieb J.D.. 2013. A common set of DNA regulatory elements shapes Drosophila appendages. Dev. Cell. 27:306–318. 10.1016/j.devcel.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto M., and Boerkoel C.F.. 2013. The role of nuclear bodies in gene expression and disease. Biology (Basel). 2:976–1033. 10.3390/biology2030976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizami Z., Deryusheva S., and Gall J.G.. 2010. The Cajal body and histone locus body. Cold Spring Harb. Perspect. Biol. 2:a000653 10.1101/cshperspect.a000653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotný I., Blažíková M., Staněk D., Herman P., and Malinsky J.. 2011. In vivo kinetics of U4/U6·U5 tri-snRNP formation in Cajal bodies. Mol. Biol. Cell. 22:513–523. 10.1091/mbc.E10-07-0560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon C., Pilch D., Rogakou E., Sedelnikova O., Newrock K., and Bonner W.. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162–169. 10.1016/S0959-437X(02)00282-4 [DOI] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., and Mesirov J.P.. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath I., Skrajna A., Yang X.C., Dadlez M., Marzluff W.F., and Dominski Z.. 2013. 3′-End processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors. RNA. 19:1726–1744. 10.1261/rna.040360.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzler H.R., Tatomer D.C., Malek P.Y., McDaniel S.L., Orlando A.N., Marzluff W.F., and Duronio R.J.. 2013. A sequence in the Drosophila H3-H4 Promoter triggers histone locus body assembly and biosynthesis of replication-coupled histone mRNAs. Dev. Cell. 24:623–634. 10.1016/j.devcel.2013.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek D., Pridalová-Hnilicová J., Novotný I., Huranová M., Blazíková M., Wen X., Sapra A.K., and Neugebauer K.M.. 2008. Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol. Biol. Cell. 19:2534–2543. 10.1091/mbc.E07-12-1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka M., Trowitzsch S., Weber G., Lührmann R., Oates A.C., and Neugebauer K.M.. 2010. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat. Struct. Mol. Biol. 17:403–409. 10.1038/nsmb.1783 [DOI] [PubMed] [Google Scholar]
- Sullivan E., Santiago C., Parker E.D., Dominski Z., Yang X., Lanzotti D.J., Ingledue T.C., Marzluff W.F., and Duronio R.J.. 2001. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 15:173–187. 10.1101/gad.862801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K.D., Steiniger M., and Marzluff W.F.. 2009. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol. Cell. 34:322–332. 10.1016/j.molcel.2009.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatomer D.C., Rizzardi L.F., Curry K.P., Witkowski A.M., Marzluff W.F., and Duronio R.J.. 2014. Drosophila Symplekin localizes dynamically to the histone locus body and tricellular junctions. Nucleus. 5:613–625. 10.4161/19491034.2014.990860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzo E.A., Lyons S.M., Poulton J.S., Temple B.R.S., Marzluff W.F., and Duronio R.J.. 2015. Distinct self-interaction domains promote Multi Sex Combs accumulation in and formation of the Drosophila histone locus body. Mol. Biol. Cell. 26:1559–1574. 10.1091/mbc.E14-10-1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P., Beaton A., Weiszmann R., Kwan E., Shu S., Lewis S.E., Richards S., Ashburner M., Hartenstein V., Celniker S.E., and Rubin G.M.. 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3:H0088 10.1186/gb-2002-3-12-research0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daal A., and Elgin S.C.. 1992. A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol. Biol. Cell. 3:593–602. 10.1091/mbc.3.6.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E.J., Burch B.D., Godfrey A.C., Salzler H.R., Duronio R.J., and Marzluff W.F.. 2007. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol. Cell. 28:692–699. 10.1016/j.molcel.2007.10.009 [DOI] [PubMed] [Google Scholar]
- White A.E., Leslie M.E., Calvi B.R., Marzluff W.F., and Duronio R.J.. 2007. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol. Biol. Cell. 18:2491–2502. 10.1091/mbc.E06-11-1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A.E., Burch B.D., Yang X.C., Gasdaska P.Y., Dominski Z., Marzluff W.F., and Duronio R.J.. 2011. Drosophila histone locus bodies form by hierarchical recruitment of components. J. Cell Biol. 193:677–694. 10.1083/jcb.201012077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.C., Burch B.D., Yan Y., Marzluff W.F., and Dominski Z.. 2009. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol. Cell. 36:267–278. 10.1016/j.molcel.2009.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.C., Sabath I., Dębski J., Kaus-Drobek M., Dadlez M., Marzluff W.F., and Dominski Z.. 2013. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Mol. Cell. Biol. 33:28–37. 10.1128/MCB.00653-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.C., Sabath I., Kunduru L., van Wijnen A.J., Marzluff W.F., and Dominski Z.. 2014. A conserved interaction that is essential for the biogenesis of histone locus bodies. J. Biol. Chem. 289:33767–33782. 10.1074/jbc.M114.616466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Wei Y., Nalepa G., and Harper J.W.. 2003. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol. Cell. Biol. 23:8586–8600. 10.1128/MCB.23.23.8586-8600.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Kennedy B.K., Lawrence B.D., Barbie D.A., Matera A.G., Fletcher J.A., and Harlow E.. 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14:2283–2297. 10.1101/gad.827700 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.