Aix et al. find that telomere dysfunction is a crucial signal for cardiomyocyte cell-cycle arrest during the postnatal period. After birth, mouse cardiomyocytes undergo telomere shortening. This leads to the appearance of dysfunctional telomeres that activate p21, causing cardiomyocyte cell-cycle withdrawal.
Abstract
The molecular mechanisms that drive mammalian cardiomyocytes out of the cell cycle soon after birth remain largely unknown. Here, we identify telomere dysfunction as a critical physiological signal for cardiomyocyte cell-cycle arrest. We show that telomerase activity and cardiomyocyte telomere length decrease sharply in wild-type mouse hearts after birth, resulting in cardiomyocytes with dysfunctional telomeres and anaphase bridges and positive for the cell-cycle arrest protein p21. We further show that premature telomere dysfunction pushes cardiomyocytes out of the cell cycle. Cardiomyocytes from telomerase-deficient mice with dysfunctional telomeres (G3 Terc−/−) show precocious development of anaphase-bridge formation, p21 up-regulation, and binucleation. In line with these findings, the cardiomyocyte proliferative response after cardiac injury was lost in G3 Terc−/− newborns but rescued in G3 Terc−/−/p21−/− mice. These results reveal telomere dysfunction as a crucial signal for cardiomyocyte cell-cycle arrest after birth and suggest interventions to augment the regeneration capacity of mammalian hearts.
Introduction
Heart failure is a leading cause of human death worldwide. This is in large part because of the extremely limited cardiomyocyte (CM) turnover of the adult human heart (Bergmann et al., 2009; Mollova et al., 2013), which is insufficient for substantial regeneration of the damaged myocardium after a heart attack. In marked contrast, mammalian CMs actively proliferate during embryonic development and the first days of postnatal life (Porrello and Olson, 2014; Senyo et al., 2014). During embryonic development (Soonpaa et al., 1994; Drenckhahn et al., 2008) and on postnatal day 1 (P1; Porrello et al., 2011), CMs in the mouse heart are able to proliferate, showing significant regeneration capacity. However, during the first two postnatal weeks, most mouse CMs become binucleated and withdraw from the cell cycle (Soonpaa et al., 1996; Ikenishi et al., 2012). As a consequence, the capacity for complete cardiac regeneration after injury is lost around P7 (Porrello et al., 2011, 2013). From P14 onward, heart injury provokes a pathological cardiac remodeling response similar to that seen in adult mice (Porrello et al., 2013).
Unlike mammals, zebrafish and other lower vertebrates retain CM proliferative ability in adult life, being able to efficiently regenerate lost myocardium after injury (Kikuchi and Poss, 2012). Why lower vertebrates maintain proliferation capacity throughout life whereas mammals lose it soon after birth is largely unknown. Although regulators of CM cell-cycle arrest have been identified in recent years, the molecular mechanisms driving permanent cell-cycle arrest of CMs remain poorly understood (Muralidhar et al., 2013; Porrello and Olson, 2014; Takeuchi, 2014; Quaife-Ryan et al., 2016). Identifying these mechanisms is crucial for the development of new therapies for the treatment of myocardial infarction and heart failure.
Telomeres are the physical ends of chromosomes and are essential for the maintenance of genome integrity (Muller, 1938; McClintock, 1939). Telomeres ensure proper segregation of chromosomes during cell division, which is essential for preventing chromosome fusions (McClintock, 1941). Telomeres consist of tandem DNA repeated sequences (TTAGGG) and a multiprotein complex (Liu et al., 2004; de Lange, 2005). Because of the end-replication problem and other degradative activities, telomeres shorten with each cell division (Watson, 1972; Olovnikov, 1973; Harley, 1991; Saretzki and Von Zglinicki, 2002). A minimum telomere length is essential to allow chromosome ends to be distinguished from damaged DNA (double-strand breaks; Chan and Blackburn, 2002; de Lange, 2002). Critically short telomeres activate the DNA damage response pathway, leading to apoptosis, senescence, or cell-cycle arrest (Artandi and Attardi, 2005). Telomeres can be elongated through the action of the enzyme telomerase, which is composed of a catalytic subunit (Tert) and an RNA template (Terc; Greider and Blackburn, 1985, 1987, 1989). Telomerase activity is clearly detected during embryonic development but is present only in few cells in most somatic adult tissues, including the heart (Borges and Liew, 1997; Ramirez et al., 1997; Oh et al., 2001; Harrington, 2004; Lansdorp, 2005; Richardson et al., 2012). Previous work indicates that telomerase and telomeres influence the proliferation and apoptosis rate in mature CMs (Leri et al., 2001, 2003; Oh et al., 2001; Boon et al., 2013; Bär et al., 2014), but their role in CMs during postnatal maturation and cardiac regeneration remains unknown.
Here, we show that ample telomere reserves are essential for CM proliferation and cardiac regeneration in newborn mice. Our results reveal that during postnatal cardiac maturation, telomerase is rapidly inactivated and CMs undergo telomere shortening, leading to the appearance of dysfunctional telomeres. We further demonstrate that dysfunctional telomeres activate a DNA damage response, trigger the formation of anaphase bridges, and up-regulate the cell-cycle inhibitor p21, leading to cell-cycle arrest of postnatal CMs.
Results
Cardiac telomerase activity and CM telomere length decline rapidly after birth
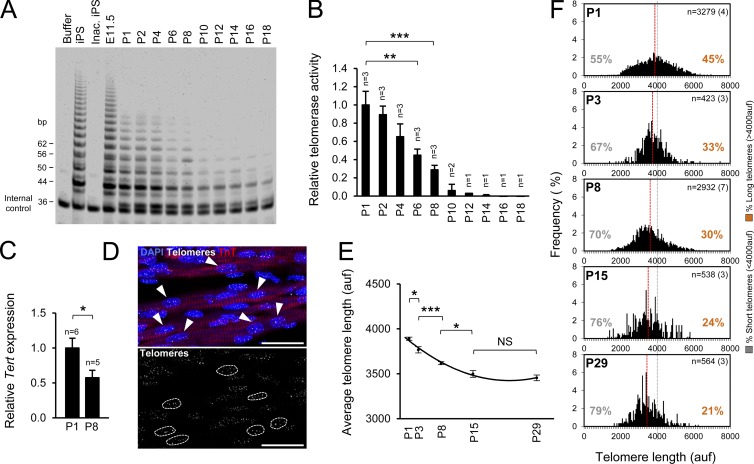
Telomerase is active during embryonic development of the heart but is present only in few cells in the adult heart (Borges and Liew, 1997; Oh et al., 2001; Richardson et al., 2012). We sought to investigate the temporal relationships among telomerase inactivation, CM cell-cycle arrest, and lack of a regenerative response after heart injury. We first examined the levels of telomerase activity in the heart during the embryonic and neonatal period using a fluorescence telomeric repeat amplification protocol (TRAP) assay. Consistent with previous studies, we found high levels of telomerase activity in embryonic hearts (embryonic day 11.5), but telomerase activity rapidly declined after birth, becoming almost undetectable from P10 onwards (Fig. 1, A and B). Parallel with the reduction in telomerase activity, we found a significant decrease in the gene expression of telomerase catalytic subunit (Tert) between P1 and P8 (Fig. 1 C), pointing to transcriptional repression as a likely mechanism underlying telomerase inactivation.
Figure 1.
Rapid decrease in cardiac telomerase activity and CM telomere length after birth. (A) Telomerase activity in the postnatal heart measured by fluorescent TRAP. iPS cells and embryonic heart (embryonic day 11.5 [E11.5]) are positive controls; lysis buffer and inactivated iPS cells (Inac. iPS) are negative controls. In total, three litters were analyzed. The gel shows representative results from one litter. (B) Relative telomerase activity quantification. n indicates the number of animals analyzed. (C) Relative Tert gene expression levels in P1 and P8 hearts measured by quantitative PCR. n indicates the number of animals analyzed. (D) Detail of a confocal maximum projection image of telomere Q-FISH and TnT immunofluorescence. CM nuclei were manually selected using the TnT immunofluorescence image. Only unambiguously identified CMs were considered for the analysis. Arrowheads indicate CMs. Dashed lines indicate telomere signal in those CMs. Bars, 25 µm. (E) Mean CM telomere length during postnatal maturation. (F) CM telomere length distribution and percentage of relatively short (<4,000 auf) and long telomeres (>4,000 auf). Gray lines at 4,000 auf facilitate comparisons of telomere size distribution between conditions. Red lines indicate mean telomere length. n indicates the total number of CMs analyzed per group. The number of animals is shown in brackets. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, nonsignificant.
We also measured CM telomere length by combining quantitative FISH (Q-FISH) for telomeres with immunofluorescence for the CM marker troponin T (TnT; Fig. 1 D). Consistent with the decrease in telomerase activity, CM telomere length dropped rapidly during the first 15 d after birth (Fig. 1, E and F). Remarkably, we detected no further significant changes from P15 to P29 (Fig. 1, E and F), indicating that after CMs have exited the cell cycle, the remaining telomere reserves in young animals are preserved. These results show that the proliferative capacity of CMs and the regenerative potential of the heart are associated with the presence of telomerase activity and sufficient telomere reserves.
CM telomere length correlates positively with proliferation
To examine the relationship between CM telomere length and proliferative capacity, we used the CM marker TnT and the mitosis marker phospho–histone H3 Ser10 (pH3) to analyze telomere length by single-cell profiling in proliferative and nonproliferative CMs (Fig. 2 A). Proliferative (pH3+) CMs had a higher mean telomere length than nonproliferative (pH3−) CMs throughout the postnatal maturation period (Fig. 2, B and C). Interestingly, mean telomere-length differences (and the statistical significance) between proliferating and nonproliferating CMs gradually decrease at later time points (P15 and P29) compared with early time points (P1, P3, and P8), suggesting a reduction in the proliferative potential of proliferating (pH3+) CMs after birth. Furthermore, the proliferative CM subset included a higher percentage of CMs with long telomeres (Fig. 2 C). The correlation of long telomeres with CM proliferation suggested that the presence of long telomeres may be important for CM proliferation ability. In addition, the high levels of short telomeres in nonproliferative CMs may influence their inability to proliferate.
Figure 2.
Proliferative CMs possess long telomeres. (A) Detail of a maximum-intensity projection confocal image of telomere Q-FISH and TnT/pH3 immunofluorescence. Proliferative (pH3+) and nonproliferative (pH3−) CMs were manually selected using the pH3 and TnT immunofluorescence images. The white arrowhead indicates a pH3− CM; the yellow arrowhead indicates a pH3+ CM. Bar, 25 µm. (B) Mean telomere length in pH3+ and pH3− CMs. (C) Telomere-length distribution and percentage of relatively short (<4,000 auf) and long telomeres (>4,000 auf) in proliferative and nonproliferative CMs. Gray lines at 4,000 auf facilitate comparisons of telomere size distribution between conditions. Red lines indicate the mean telomere length. n indicates the number of CMs analyzed per group. The number of animals is shown in brackets. Data are mean ± SEM. *, P < 0.05; ***, P < 0.001; NS, nonsignificant.
Postnatal CMs activate the DNA damage response at dysfunctional telomeres and present anaphase bridges
Extensive telomere shortening could lead to the appearance of telomere-dysfunction–induced foci (TIFs), formed when telomeres reach critically short lengths and activate the DNA damage response (DDR). TIFs can be detected by the presence of DDR markers at the telomeres (d’Adda di Fagagna et al., 2003). Double immunofluorescence for the DDR marker phopho–histone H2AX Ser139 (γH2AX) and the CM marker troponin I (TnI), in combination with Q-FISH for telomeres, showed that the number of CMs with TIFs increases significantly from P1 to P8 (Fig. 3, A and B). Another consequence of telomere shortening is the generation of end-to-end-fusions that eventually cause DNA bridges when the chromosomes segregate to the mitotic poles during cell division (Davoli and de Lange, 2011; Murnane, 2012; Pampalona et al., 2012). To evaluate whether telomere shortening was producing DNA bridges, we examined CMs in anaphase-telophase. No DNA bridges were detected in CMs from P1 mice (Fig. 3 D). In contrast, the presence of CMs with shorter telomeres in the P8 neonates correlated with the presence of anaphase-telophase bridges (Fig. 3, C and D). When DNA bridges are formed, they normally break and often provoke the generation of micronuclei (Hoffelder et al., 2004). We also analyzed CMs from P1 and P8 mice and found CMs with a micronucleus only in P8 CMs (Fig. 3, E and F). The cell-cycle arrest of CMs at P8 thus correlates with an increase in telomere shortening, activation of the DDR at the telomeres, and the formation of anaphase bridges and micronuclei.
Figure 3.
Postnatal CMs with telomere shortening activate the DDR and form anaphase bridges. (A) Detail of telomere (Tel) Q-FISH and γH2AX/TnI immunofluorescence. Arrowheads indicate foci of the DNA-damage marker protein γH2AX at telomeres in a CM. Bars, 10 µm. (B) Quantification of the proportion of CMs with colocalized γH2AX and telomeres. n indicates the number of animals analyzed. (C) Confocal (left) and superresolution (right) images of DNA bridges in a dividing P8 CM. Arrowheads indicate a DNA bridge. Bars, 10 µm. (D) Proportion of anaphases-telophases with DNA bridges in CMs at P1 and P8. n indicates the number of animals analyzed per group. (E) P8 CM with a micronucleus. Bar, 25 µm. The arrowhead indicates the micronucleus. (F) Quantification of the proportion of CMs with a micronucleus at P1 and P8. n indicates the number of animals analyzed. Data are mean ± SEM. *, P < 0.05; **, P < 0.01.
Premature telomere shortening inhibits CM proliferation
To assess whether the rapid telomere shortening observed within the first 2 wk after birth contributes to the exit of CMs from the cell cycle, we generated mice with premature telomere shortening. This was achieved in the third generation (G3) of the telomerase RNA template knockout (Terc−/−; Blasco et al., 1997), generated by consecutive intercrossing of first-generation (G1) knockout mice. Telomere shortening was more patent in G3 Terc−/− mice than in G1 Terc−/− mice (Fig. S1 A), with mean telomere length and percentage of short telomeres at P1 equivalent to wild-type (WT) mice at P15 (Fig. S1, B and C).
Histological analysis of heart sections from P1 WT, and G3 Terc−/− mice did not reveal any phenotypic difference in heart morphology (Fig. 4 A). Although G3 Terc−/− P1 hearts were smaller than the WTs (Fig. 4 B), there was no effect on heart weight to body weight ratio (Fig. 4 C). CM size measured by wheat germ agglutinin (WGA) staining was also unchanged (Fig. 4, D and E). However, analysis of CM mitosis by double immunofluorescence for pH3 and TnT revealed significantly lower numbers of mitotic CMs in the G3 Terc−/− heart at P1 (Fig. 4, F and G). Moreover, immunofluorescence analysis of TnT and Aurora B kinase revealed significantly lower levels of CM cytokinesis in the P1 G3 Terc−/− neonates (Fig. 4, H and I). These data indicate that telomere shortening impedes CM proliferation.
Figure 4.
CM proliferation is diminished in G3 Terc−/− P1 neonates. (A) Representative Masson’s trichrome staining on WT and G3 Terc−/− heart sections at P1. n indicates the number of animals analyzed. RV, right ventricle; LV, left ventricle. Bars, 500 µm. (B) WT and G3 Terc−/− heart weight at P1. n indicates the number of animals analyzed. (C) Heart weight (HW) to body weight (BW) ratio in WT and G3 Terc−/− P1 hearts. n indicates the number of animals analyzed. (D) WGA staining in WT and G3 Terc−/− P1 hearts. Bars, 25 µm. (E) CM size quantification in P1 heart transverse sections; 100 CMs were analyzed per animal. n indicates the number of animals analyzed. (F) Detail of pH3 and TnT immunofluorescence at P1. Arrowheads indicate CMs in mitosis. Bars, 100 µm. (G) Quantification of mitotic CMs at P1. n indicates the number of animals analyzed. (H) Detail of Aurora B kinase and TnT immunofluorescence. The arrowhead indicates the Aurora B signal in the cleavage furrow. Bar, 10 µm. (I) Quantification of CM cytokinesis at P1. n indicates the number of animals analyzed. (J) Quantification of P1 WT and G3 Terc−/− CMs stained with the DDR protein γH2AX at telomeres. n indicates the number of animals analyzed. (K) Representative image of DNA bridges in P1 G3 Terc−/− CMs. Arrowheads indicate DNA bridges in dividing CMs. Bars, 10 µm. (L) Percentage of anaphases-telophases with DNA bridges in P1 WT and G3 Terc−/− CMs. n indicates the number of animals analyzed per group. (M) Detail of mono- and binucleated CMs. Bars, 25 µm. (N) Percentage of mono- and binucleated CMs at P1. n indicates the number of animals analyzed. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, nonsignificant.
Premature telomere shortening increases telomere damage, provokes the formation of anaphase bridges, and increases the proportion of binucleated CMs
The proportion of CMs with telomere damage in G3 Terc−/− neonates was already above normal at P1 (Fig. 4 J), consistent with an association between the weak CM proliferation in these mice and activation of the DDR at the telomeres. CMs of P1 G3 Terc−/− neonates also contained anaphase bridges (Fig. 4, K and L), and the percentage of binucleated CMs was significantly higher (Fig. 4, M and N). These results indicate that telomere shortening might contribute to CM binucleation by affecting the correct process of division.
Premature telomere shortening blocks the CM proliferative response and increases CM telomere damage after injury in P1 neonates
To further assess the effect of telomere shortening on CM proliferation, we evaluated injury-induced CM proliferation. The P1 neonatal mouse has the remarkable ability to regenerate damaged myocardium by increasing CM proliferation (Porrello et al., 2011, 2013). To study the proliferative response during the regeneration process in WT and G3 Terc−/− mice, we damaged the hearts of P1 mice by cryoinjury. Histological analysis confirmed the deposition of fibrotic tissue 7 d postinjury (dpi) in both genotypes (Fig. 5 A). In WT animals, the fibrotic area was significantly diminished at 28 dpi, whereas the fibrotic area in G3 Terc−/− mice was not significantly reduced (Fig. 5, A and B). Furthermore, persistent fibrotic tissue at 28 dpi in WT mice contained a high proportion of intermingled CMs, whereas significantly fewer CMs were detected in the scars in G3 Terc−/− hearts (Fig. 5, C and D). These results indicate that in P1 G3 Terc−/− hearts the regenerative response after cryoinjury is impaired.
Figure 5.
G3 Terc−/− P1 neonates do not increase CM proliferation in response to cardiac cryoinjury. (A) Representative Masson’s trichrome staining in WT and G3 Terc−/− hearts after cryoinjury at P1. Bars, 1 mm. (B) Quantification of the fibrotic area. n indicates the number of animals analyzed. (C) Detail of Masson’s trichrome staining at 28 dpi. Bars, 200 µm. (D) Quantification of the area covered by CMs within the remaining fibrotic tissue at 28 dpi. n indicates the number of animals analyzed. (E) pH3 and TnT immunofluorescence detail (7 dpi). Arrowheads indicate CMs in mitosis. Controls are age-matched uninjured animals. Cryo, cryoinjury. Bars, 100 µm. (F) Quantification of CM mitosis in WT and G3 Terc−/− mice. n indicates the number of animals analyzed. (G) Detail of Aurora B kinase and TnT immunofluorescence at 7 dpi. The arrowhead indicates the Aurora B signal in the cleavage furrow. Bar, 10 µm. (H) Quantification of CM cytokinesis (7 dpi). n indicates the number of animals analyzed. (I) Detail of WGA staining at 28 dpi. Bars, 50 µm. (J) Quantification of CM area in transverse sections at 28 dpi; 100 CMs from the injury vicinity were analyzed per animal. n indicates the number of animals analyzed. (K) Quantification of CMs stained with the γH2AX DNA damage protein at telomeres in control (P8) and cryoinjured (7 dpi at P1) animals. n indicates the number of animals analyzed. (L) Detail of telomere (Tel) Q-FISH and 53BP1/TnT immunofluorescence at 7 dpi. Arrowheads indicate colocalization of telomere and 53BP1 signals. Bars, 10 µm. (M) Quantification of CMs with 53BP1 at telomeres in control (P8) and cryoinjured (7 dpi at P1) animals. n indicates the number of animals analyzed. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, nonsignificant.
In WT animals, cryoinjury caused a significant increase in CM mitosis (Fig. 5, E and F) and cytokinesis (Fig. 5, G and H), as measured by pH3 and Aurora B kinase immunofluorescence, respectively. The increase in proliferation was not restricted to the vicinity of the injury site but occurred throughout the section (Fig. 5, E, F, and H). Cryoinjured G3 Terc−/− hearts were unable to increase the number of proliferating CMs (Fig. 5, E, F, and H). The absence of a proliferative response in G3 Terc−/− mice was accompanied by a significant injury-induced hypertrophic increase in CM area in G3 Terc−/− hearts (Fig. 5, I and J). Hypertrophic growth of CMs is characterized by an increase in mitochondrial content to maintain energy balance (Rabinowitz and Zak, 1975; Goffart et al., 2004). As expected, G3 Terc−/− hearts showed a markedly higher expression of mitochondria-related genes (Fig. S2). These results indicate that, instead of proliferating, G3 Terc−/− CMs undergo hypertrophy in response to injury.
Examination of the DNA damage marker γH2AX revealed a significant cryoinjury-induced increase in the number of CMs with telomere damage in P1 G3 Terc−/− mice (Fig. 5 K). This result was corroborated by examination of 53BP1, another marker of activation of the DDR (Fig. 5, L and M). The absence of a proliferative response in G3 Terc−/− mice after injury is thus associated with the activation of the DDR at telomeres.
To evaluate the relative contributions of telomere shortening and the lack of telomerase activity to the inhibition of postinjury proliferation, we examined the effect of cryoinjury in P1 G1 Terc−/− mice. G1 Terc−/− and G3 Terc−/− mice both lack telomerase activity, but telomeres at P1 were longer in G1 Terc−/− CMs than in G3 Terc−/− counterparts and, in both cases, shorter than in WT CMs (Fig. S1 A). The fibrotic area in G1 Terc−/− hearts did not reduce significantly in the postinjury period (Fig. S3, A and B). However, G1 Terc−/− mice increased CM proliferation in response to injury (7 dpi; Fig. S3 C). The different CM proliferative responses in G1 Terc−/− and G3 Terc−/− mice confirmed that CM proliferation after injury requires sufficient telomere reserves. However, the results obtained comparing WT and G1 Terc−/− mice did not rule out the possibility that telomerase deficiency itself might contribute to CM proliferation and heart regeneration directly, independently of its role on telomere elongation.
Activation of the DDR by short telomeres blocks CM proliferation through p21 activation
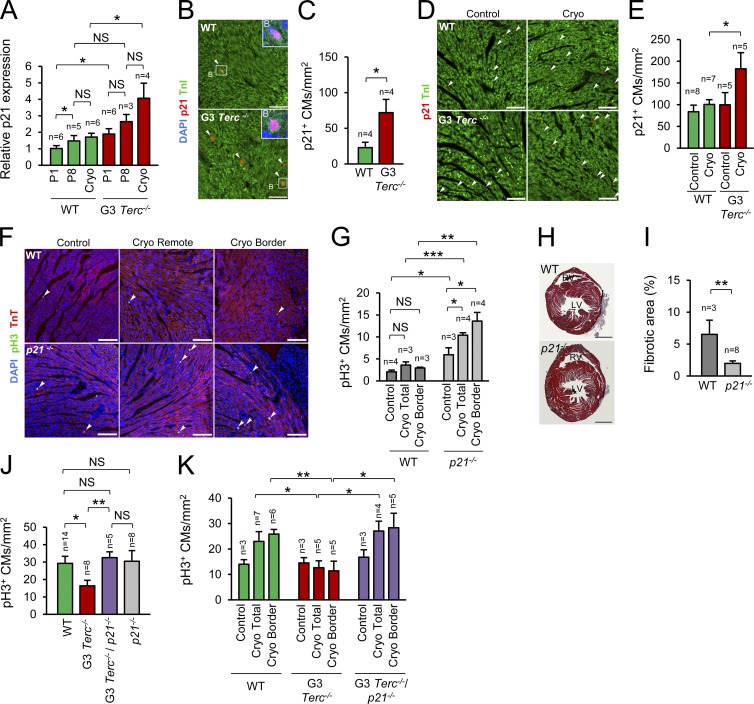
Once the DDR is activated, telomere shortening can result in apoptosis, senescence, or cell-cycle arrest (Artandi and Attardi, 2005). TUNEL staining and TnI immunofluorescence showed that G3 Terc−/− mice did not have a higher proportion of TUNEL-positive CMs after injury, indicating that telomere shortening did not lead to CM apoptosis (Fig. S4, A and B). Analysis of the expression of cyclin-dependent kinase inhibitors (CDKIs) in WT and G3 Terc−/− hearts at P1 and after cryoinjury did not detect significant differences in the expression of the inhibitors p27, p57, p15, p16, p18, or p19 (Fig. S4, C and D). However, P1 G3 Terc−/− hearts expressed significantly higher levels of the cell-cycle inhibitor p21 than P1 WT hearts, and p21 expression in G3 Terc−/− hearts increased further after injury (Fig. S4 E and Fig. 6 A). Immunofluorescence for p21 confirmed the specific increase in p21 in G3 Terc−/− CMs at P1 (Fig. 6, B and C) and after injury (Fig. 6, D and E).
Figure 6.
CM telomere dysfunction induces cell-cycle arrest through p21 activation. (A) Relative p21 gene expression measured by quantitative PCR. n indicates the number of animals analyzed. Cryo, cryoinjury at P1 analyzed at 7 dpi (at P8). (B) Detail of p21 and TnI immunofluorescence at P1. Arrowheads indicate p21-positive CMs. Bars, 50 µm. (C) Quantification of p21-positive CMs at P1. n indicates the number of animals analyzed. (D) Detail of p21 and TnI immunofluorescence analyzed 7 dpi at P1. Controls are age-matched uninjured animals. Arrowheads indicate p21-positive CMs. Bars, 50 µm. (E) Quantification of p21-positive CMs 7 dpi at P1. n indicates the number of animals analyzed. (F) Detail of pH3 and TnT immunofluorescence in WT and p21−/− mice analyzed 7 dpi at P7. Arrowheads indicate mitotic CMs. Bars, 100 µm. (G) Quantification of the number of CMs in mitosis in WT and p21−/− mice analyzed 7 dpi at P7. n indicates the number of animals analyzed. (H) Representative Masson’s trichrome staining in WT and p21−/− hearts cryoinjured at P7 and analyzed 28 dpi. Bars, 1 mm. (I) Quantification of fibrotic area in WT and p21−/− hearts cryoinjured at P7 and analyzed 28 dpi. n indicates the number of animals analyzed. (J) Number of CMs in mitosis in WT, G3 Terc−/−, G3 Terc−/−/p21−/−, and p21−/− mice at P1. n indicates the number of animals analyzed. (K) Number of CMs in mitosis in WT, G3 Terc−/−, and G3 Terc−/−/p21−/− mice cryoinjured at P1 and analyzed at 7 dpi. n indicates the number of animals analyzed. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, non-significant.
Interestingly, p21 transcript expression increased transiently in WT postnatal hearts, peaking around P8, when most CMs are undergoing cell-cycle arrest (Fig. S4 F). To confirm whether p21 is a regulator of CM cell-cycle arrest, we first compared WT and p21 knockout (p21−/−) hearts. Under homeostatic conditions CM proliferation was higher in p21−/− uninjured hearts than in WTs (Fig. 6, F and G). When injured at P7, WT mice showed no proliferative response. In contrast, p21−/− mice showed a significant injury-induced increase in CM proliferation at 7 dpi (Fig. 6, F and G). Accordingly, the postinjury fibrotic area at 28 dpi was significantly smaller in p21−/− hearts than in WT hearts (Fig. 6, H and I). The strong proliferative response after injury at P7 in p21−/− hearts further indicates that under physiological conditions the cell-cycle inhibitor p21 limits the extension of the postnatal window of CM proliferation.
To directly evaluate whether p21 up-regulation leads to cell-cycle arrest in CMs with dysfunctional telomeres, we analyzed CM proliferation at P1 and after injury in G3 Terc−/−/p21−/− double-knockout neonates. The deletion of p21 was sufficient to rescue the proliferative impairment of G3 Terc−/− mice (Fig. 6, J and K), confirming the role of p21 in arresting the cell cycle in CMs with dysfunctional telomeres.
Discussion
In this study, we demonstrate that the ability of CMs to proliferate postnatally depends on the presence of ample telomere reserves. We show that proliferation-competent CMs possess longer telomeres, indicating that in CMs telomere length is a marker of proliferation potential. In the absence of sufficient telomere reserves, CMs activate the DDR and present anaphase bridges, leading to the induction of cell-cycle arrest through the activation of p21 (Fig. 7). Telomeres are shortened during S phase because of the inability of the DNA replication machinery to duplicate the termini of linear DNA molecules (end-replication problem; Watson, 1972; Olovnikov, 1973; Harley, 1991). Therefore, the absence shortly after birth of the counteracting mechanism of telomerase activity, as reported here, predicts a loss of CM telomere reserves during their postnatal DNA division period. More surprising was the high pace of telomere loss starting at P1, leading to a significant increase in telomere DNA damage just one week later. Given this high rate of telomere erosion, we cannot discard a possible contribution to the loss of telomere reserves from other degradative activities. Interestingly, the level of reactive oxygen species, a telomere-shortening agent, increases significantly in mouse hearts during the first postnatal week (von Zglinicki, 2002; Puente et al., 2014).
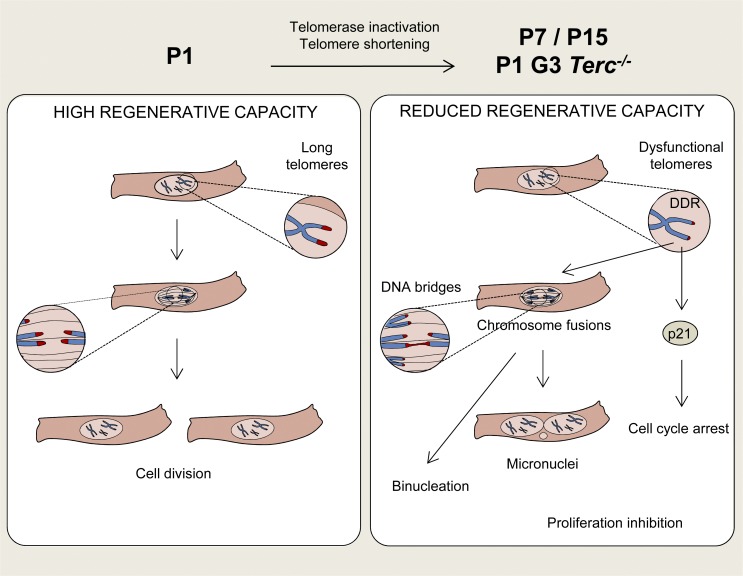
Figure 7.
Proposed model. At P1, a proportion of CMs presents long telomeres, giving them potential to proliferate during postnatal development and in response to cardiac injury. However, during the first two postnatal weeks, most CMs inactivate telomerase and shorten their telomeres. Telomere shortening leads to the appearance of dysfunctional damaged telomeres, chromosome fusions, micronuclei, and binucleation and at the same time activates p21, ultimately leading to CM cell-cycle arrest. CMs with premature telomere shortening (P1 G3 Terc−/− CMs) precociously activate the DNA damage response at telomeres, form anaphase bridges, up-regulate p21, and binucleate, outcomes that reinforce the role of telomere shortening in the withdrawal of CMs from the cell cycle.
A mechanism contributing to cell-cycle arrest in postnatal CMs may be related to the well-established role of telomere shortening in the origin of the fusion bridge–breakage cycle, a process that leads to tetraploidization, binucleation and proliferation-inhibition in cells with an intact p53 pathway (Margolis et al., 2003; Davoli and de Lange, 2011; Pampalona et al., 2012). In the fusion bridge–breakage cycle, chromosomes with dysfunctional telomeres fuse with each other, forming bridges during mitosis. These chromosome bridges can eventually break under forces emanating from the anaphase poles, but further proliferation is inhibited. In WT CMs, we found that telomere shortening was linked to the appearance of chromosome bridges between daughter nuclei: 8 d after birth, CMs showed significant telomere erosion and the presence of chromosome bridges. The percentage of chromosome bridges detected in our study was relatively small, but is likely an underestimate because (1) the analysis was performed on tissue sections, in which not all anaphases will have an orientation allowing bridge detection; (2) chromosome bridges might be hidden in polyploid nuclei; and (3) most chromosome bridges eventually break (Shimizu et al., 2005; Pampalona et al., 2012), rendering them undetectable. The presence of CMs with micronucleus, an outcome of DNA fragmentation, at P8 supports the idea that DNA bridges eventually break. Interestingly, chromosome bridging has also been detected in cultures of rat postnatal CMs stimulated to binucleate with serum (Engel et al., 2006). The premature anaphase bridging and binucleation in G3 Terc−/− CMs strongly suggests that telomere shortening contributes to the CM binucleation process. Potential genomic imbalance caused by breakage of chromosome bridges in binucleated CMs could constitute a barrier to proliferation. Our results do not exclude a contribution from other reported genetic or epigenetic factors to CM cell-cycle arrest after birth (Muralidhar et al., 2013; Porrello and Olson, 2014; Takeuchi, 2014; Quaife-Ryan et al., 2016).
Telomere shortening has also been implicated in the up-regulation of the cell-cycle inhibitor and tumor suppressor p21 (Ju et al., 2007). Our present results confirm at the transcriptional level previous observations indicating that p21 levels increase during the first postnatal week and decrease thereafter (Tane et al., 2014). Interestingly, the increase in p21 coincided with the appearance of telomeric damage, and p21 levels were precociously elevated in CMs that presented premature telomeric shortening, indicating a causal role of telomere shortening in p21 induction. Moreover, exacerbation of telomere shortening after cryoinjury in G3 Terc−/− hearts triggered a further increase in p21 levels and in the number of CMs with telomere damage. These findings strongly suggest that telomere shortening activates a DDR that up-regulates p21 in CMs during postnatal heart maturation and after injury, leading to cell-cycle arrest. The robust proliferation response of p21−/− CMs from 7-d-old hearts, an age by which WT CMs have lost their capacity to extensively proliferate after injury, further underlines the participation of p21 in stopping the cell cycle and inhibiting the regeneration response after injury in the early postnatal period. A similar situation is revealed when p21 is deleted in the context of chronic dysfunctional telomeres: late-generation telomerase-deficiency mice, in the absence of p21, show improved tissue function and extended lifespan without an increase in cancer risk (Choudhury et al., 2007). Loss of p21 in another progeria model, a mouse hypomorphic for the mitotic checkpoint protein BubR1, also delays senescence, although in a tissue-selective manner (Baker et al., 2013). Interestingly, deletion of p53, the upstream regulator of p21, does not improve tissue function but instead leads to accelerated atrophy of intestinal epithelium in mice harboring dysfunctional telomeres (Begus-Nahrmann et al., 2009) and to lens deterioration in mice with low BubR1 levels (Baker et al., 2013). The divergent effects of p21 and p53 deletion in progeria models might be explained by their different roles in cell-cycle arrest, cellular senescence, and apoptosis (Vousden and Prives, 2009).
Based on the results reported here, we propose that postnatal CM maturation involves the following set of events: telomerase inhibition, telomere shortening, telomere DNA damage, increased p21 levels, formation of anaphase bridges, binucleation, and cell-cycle arrest (Fig. 7).
The premature drop in telomere reserves in G3 Terc−/− hearts was linked to a high proportion of CMs with telomeric DNA damage, anaphase bridges, binucleation, and expression of p21. Unsurprisingly therefore, the number of proliferating CMs in G3 Terc−/− hearts was significantly lower than in WTs at P1, and P1 G3 Terc−/− hearts responded to injury not with CM proliferation but by mounting a hypertrophic response. The recovery of CM proliferation in P1 G3 Terc−/− neonates by deleting p21 confirmed that telomere dysfunction provokes CM cell-cycle arrest through p21 activation. Indeed, p21 deletion in late-generation Terc−/− mice is sufficient to rescue the proliferative defect in cells with high turnover, although without preventing the presence of short telomeres (Choudhury et al., 2007).
A corollary of these findings is that mechanisms that counteract acute telomere shortening would beneficially prolong CM proliferation after birth and in response to injury. Pioneering work by Oh et al. (2001) has shown that overexpression of human telomerase under a CM promoter elongates mouse heart telomeres and prolongs mouse CM proliferation in vivo. More recently, Puente et al. (2014) showed that scavenging of reactive oxygen species, a DNA-damage and telomere-shortening agent, extends the postnatal CM proliferation window. In addition, we have recently shown that the adult zebrafish mounts a full heart-regeneration response to injury when telomere reserves are preserved but fail to regenerate the injured heart when telomere reserves are lost (Bednarek et al., 2015).
In humans, CM division rate peaks in infants and declines thereafter (Mollova et al., 2013; Bergmann et al., 2015), which is associated with rapid telomerase inhibition in low-turnover tissues and gradual telomere shortening with aging (Wright et al., 1996; Daniali et al., 2013). Interestingly, Haubner et al. (2016) recently reported a case of functional recovery from severe myocardial infarction in a newborn child, at the time when telomeres are at their longest. In contrast, some individuals from families with premature telomere shortening suffer from dilated cardiomyopathy, severe cardiac fibrosis, and myocardial infarction at young ages (Vulliamy et al., 2004; Basel-Vanagaite et al., 2008), and humans with genetic variants affecting telomere length are at above-average risk of developing coronary heart disease (Maubaret et al., 2013; Ding et al., 2014). These observations, together with the other findings presented in this study, highlight the value of studying the role of telomere-length regulators in modulating the homeostasis and regenerative capacity of the mammalian heart.
Materials and methods
Experimental animals
Non-WT mouse strains used were Terc−/− (Blasco et al., 1997) and p21−/− (Brugarolas et al., 1995; C57BL/6). Animal experiments were performed according to Institutional and European Union guidelines.
Heart weight to body weight ratio
After weighing the neonates, the hearts were extracted, washed with PBS, carefully drained, and weighed.
Neonatal cryoinjury
P1 and P7 neonatal mice were anesthetized by hypothermia. A left ventrolateral thoracotomy was performed and a 0.8-mm-diameter metal probe cooled in liquid nitrogen was applied to the left ventricle anterior wall for ∼5 s. The cryoinjured area was identified as a white region. The intercostal space and the skin were sutured with a 6–0-gauge black-braided, nonabsorbable silk suture, and the animals were rewarmed until recovery and returned to the mother.
Determination of fibrotic area and the percentage of CMs inside the fibrotic region
Hearts were fixed overnight in 4% PFA and embedded in paraffin. To determine the fibrotic area, the whole heart was transversely sectioned (7 µm), and sections were selected for analysis every 70 to 140 µm. Masson’s trichrome staining was performed according to standard procedures. Images were acquired with a BX51 microscope (Olympus), a DP71 camera (Nikon), and cellSens software. The fibrotic area was measured in the section with the largest amount of fibrotic tissue as a percentage of the total section. Likewise, the percentage of the fibrotic region occupied by CMs was quantified in the same section. The calculations were done with Metamorph (Molecular Devices).
Immunofluorescence
Hearts were fixed overnight in 4% PFA and processed for paraffin sectioning (7 µm). Antigens were retrieved by boiling in 10 mM, pH 6, citrate buffer for 15 min. Slides were permeabilized with PBS 0.3% Triton for 20 min, and nonspecific binding sites were blocked by incubation for 2 h in blocking solution containing 3% BSA, 20 mM MgCl2, 0.3% Tween-20, and 5% donkey serum (for p21 immunofluorescence) or 5% goat serum (for other immunostainings). For staining with biotinylated antibodies, endogenous biotin was blocked with the avidin-biotin blocking kit (Vector Laboratories). Primary antibody incubations were conducted at the indicated dilution overnight at 4°C, and secondary antibody incubations at 1:200 for 1 h at room temperature.
Primary antibodies for pH3 and TnT immunofluorescence were rabbit anti-pH3 mitosis marker (1:100; EMD Millipore) and mouse anti-TnT (1:50; Developmental Studies Hybridoma Bank), and the secondary antibodies were biotin goat anti–rabbit (Jackson ImmunoResearch Laboratories, Inc.), streptavidin–Alexa Fluor 488 (Invitrogen), and goat anti–mouse–Alexa Fluor 647 or Cy3 (Jackson ImmunoResearch Laboratories, Inc.). For Aurora B and TnT immunofluorescence, endogenous peroxidases were blocked with 1% H2O2 in methanol before permeabilization. The slides were incubated with rabbit anti–Aurora B (1:50; Sigma-Aldrich) and then with biotin goat anti–rabbit secondary antibody. The slides were then incubated with the Vectastain Elite ABC kit (Vector Laboratories) and the TSA Plus Tetramethylrhodamine System (1:50; PerkinElmer). After this, the slides were incubated with mouse anti-TnT followed by goat anti–mouse Alexa Fluor 488 (Invitrogen). For 53BP1 and TnT immunofluorescence, the primary antibodies were rabbit anti-53BP1 (1:500; Abcam) and mouse anti-TnT, and the secondary antibodies were goat anti–rabbit Alexa Fluor 488 (Invitrogen) and goat anti–mouse Alexa Fluor 647. For γH2AX and TnI, the primary antibodies were mouse anti-γH2A.X (Ser139, 1:300: EMD Millipore) and rabbit anti-TnI (1:100; Abcam), and the secondary antibodies were goat biotin anti-mouse (Jackson ImmunoResearch Laboratories, Inc.), streptavidin–Alexa Fluor 488, and goat anti–rabbit Alexa Fluor 647 (Jackson ImmunoResearch Laboratories, Inc.). For p21 and TnI, the endogenous peroxidases were blocked as described in the previous paragraph and the slides were incubated with goat anti-p21 (1:100; Santa Cruz Biotechnology, Inc.) followed by biotin donkey anti–goat (Jackson ImmunoResearch Laboratories, Inc.). Slides were then incubated with the Vectastain Elite ABC kit and the TSA Plus Cyanine 5 System (1:50; PerkinElmer). After this, the slides were incubated with rabbit anti-TnI followed by donkey anti–rabbit Alexa Fluor 488 (Invitrogen).
In all cases, slides were stained with DAPI (1:1,000; Merck Millipore) and mounted with Vectashield. Images were acquired with an SP5 confocal microscope (Leica) with LAS AF and Matrix Screener software or with an A1R confocal microscope (Nikon) with NIS Elements software. Immunofluorescence signals were quantified manually in images of whole transverse sections (2–10 per heart) with NIS Elements software. For Aurora B, quantifications were performed in maximum-intensity projection images, and the putative CMs with signal in the cleavage furrow were confirmed as positive in tridimensional images. Telomere colocalization of 53BP1 or γH2AX was quantified in z stacks. All other immunofluorescence signals were quantified in maximum-intensity projection images.
Telomere-length analysis
Telomere length was measured by Q-FISH. Hearts were fixed in 4% PFA, and paraffin-embedded tissue sections of the same thickness (7 µm) were first immunostained for pH3 and TnT, 53BP1 and TnT, or γH2AX and TnI (see Materials and methods). Slides were then fixed in 4% formaldehyde for 20 min and hybridized with a Cy3-labeled peptide nucleic acid probe targeting the telomere repeat sequence (Zijlmans et al., 1997; Herrera et al., 1999; González-Suárez et al., 2000; Flores et al., 2008). The intensity of the fluorescent signal for a given telomere is directly proportional to telomere length, providing a quantitative measure of telomere length (Lansdorp et al., 1996).
For telomere-length analysis, DAPI, Cy3 (telomeres), and immunofluorescence signals were acquired sequentially in separate channels of a confocal microscope (SP5) fitted with a 40× objective (1.5× zoom) and linked to LasAF and MatrixScreener software. All slides were stained at the same time, and all images were acquired at the same laser intensity. For image quantification, Nikon NIS Elements software was used to subtract Cy3 channel background and generate maximum-intensity projections from 16-bit image stacks (15 sections at steps of 1.0 µm). Quantitative image analysis was performed using the Metamorph platform as described previously (Flores et al., 2008). The DAPI images were signal-intensity thresholded. After conversion to a 1-bit binary mask, the DAPI image was used to define the nuclear area and the Cy3 image to quantify telomere fluorescence. The binary DAPI mask was applied to the Cy3 image to obtain a combined image with the telomere fluorescence information for each nucleus. Cy3 fluorescence intensity was measured as the mean gray value in each nucleus in arbitrary units of fluorescence (auf). Telomere intensity values were exported to Excel. For the determination of total, pH3-positive, and pH3-negative CM telomere length, CMs nuclei were manually selected with Metamorph using the pH3 and TnT immunofluorescence images. Only cells clearly identifiable as CMs where considered for the analysis. Percentages of cells with short or long telomeres were calculated stablishing an arbitrary limit of 4,000 auf.
Telomerase activity
Telomerase activity was measured by fluorescent TRAP (Herbert et al., 2006), with some modifications. Mice from the same litter were sacrificed at different ages, and the hearts were rapidly homogenized for protein extraction with a motorized disposable pestle (Sigma-Aldrich) in CHAPS lysis buffer. The experiment was repeated with three litters. Induced pluripotent stem (iPS) cells and embryonic day 11.5 heart (pool of four hearts) were used as positive controls, and lysis buffer and inactivated iPS cells (85°C, 30 min) were used as negative controls. The samples were incubated for 30 min at 4°C and centrifuged at 14,000 rpm for 20 min, and the supernatants were collected and stored at −80°C. Protein concentrations were determined by the Bradford method (DC Protein Assay; Bio-Rad Laboratories). Protein samples (0.01 µg of iPS cells, 0.1 µg of embryonic day 11.5 hearts, and 0.5 µg of postnatal hearts) in a total volume of 1 µl were incubated at 30°C for 30 min with 47.6 µl TRAP reaction mix (20 mM Tris-HCl, pH 8.3, 1.5 mM MgCl2, 63 mM KCl, 0.05% Tween-20, 1 mM EGTA, 0.05 mM dNTP, and diethylpyrocarbonate-treated water; Sigma-Aldrich) and 2 ng/µl Cy5-TS primer (5′-AATCCGTCGAGCAGAGTT-3′). A final 50-µl reaction volume was prepared by adding 2 U Taq polymerase (5 U/µl) and a specific TRAP primer mix consisting of 100 ng/µl Ancor primer (5′-GCGCGGCTAACCCTAACCCTAACC-3′), 100 ng/µl OG primer (5′-ATGGCATCACCGGCTTAT-3′), and 0.01 attamol per microliter of TSOG primer (5′-AATCCGTCGAGCAGAGTTATAAGCCGGTGATGCCAT-3′). The samples were heated at 95°C for 5 min to inactivate telomerase and then subjected to 30 amplification cycles (95°C for 30 s, 55°C for 30 s, and 72°C for 30 s), followed by a single extension cycle at 72°C for 7 min. The PCR products were separated on 10% nondenaturing polyacrylamide gels and visualized with Typhoon 9400. Relative telomerase activity was calculated as the ratio of the intensity of the telomerase product ladder to that of the internal control band and was normalized to the protein concentration in each sample. Quantification was done using Quantity One and is expressed as fold change with respect to P1 heart.
WGA staining and measurement of CM size
WGA staining was performed on paraffin sections in combination with TnI immunofluorescence. Slides were incubated for 1 h with 10 µg/ml WGA–Alexa Fluor 647 conjugate (Invitrogen), and TnI immunofluorescence was performed as above, using goat anti–rabbit-Cy3 (Jackson ImmunoResearch Laboratories, Inc.) as secondary antibody. The images were acquired with an SP5 confocal microscope and LAS AF and Matrix Screener software. For CM size determination, the borders of the CMs were manually defined using NIS Elements software.
TUNEL
TUNEL staining was performed on paraffin sections using terminal deoxynucleotidyl transferase and biotin-16-29-deoxyuridine-59-triphosphate (both from Roche) in combination with TnI immunofluorescence (performed as above). The secondary antibodies used were Streptavidin-488 (1:200; Molecular Probes, Invitrogen) and goat anti–rabbit Alexa Fluor 647 (1:200; Jackson ImmunoResearch Laboratories, Inc.). Images were acquired with a Nikon A1R confocal microscope. TUNEL-positive CMs were counted manually in whole transverse sections using NIS Elements software.
Detection of anaphase bridges
Paraffin sections were incubated with DAPI (1:1,000; EMD Millipore), and images were acquired with an SP5 confocal microscope and LAS AF software. The presence of anaphase bridges was confirmed by staining paraffin sections with Sytox Green (1:200; Molecular Probes) and image acquisition with an SP8-STED confocal superresolution microscope (Leica).
CM isolation and binucleation analysis
CMs were isolated from P1 neonatal hearts as described previously (Mollova et al., 2013). Fresh hearts were cut in small pieces, fixed in 4% PFA for 2 h, and digested with collagenase B (1.8 mg/ml) and D (2.4 mg/ml) at 37°C overnight. For nucleation counts, the CMs were incubated with blocking solution for 1 h, rabbit anti-TnI antibody (1:100; Abcam) for 1 h, and goat anti–rabbit-Cy3 (1:200; Jackson ImmunoResearch Laboratories, Inc.) and DAPI (1:1,000; EMD Millipore) for 1 h at room temperature. Images were acquired with an A1R confocal microscope. Approximately 500 CMs per animal were analyzed.
Detection of micronuclei
CMs were isolated (as above) and incubated with blocking solution for 1 h, mouse anti-TnT antibody for 1 h (1:50; Developmental Studies Hybridoma Bank), and goat anti–mouse-Cy3 (1:200; Jackson InmunoResearch Laboratories, Inc.) and DAPI (1:1,000) for 1 h. Images were acquired with an SP5 confocal microscope.
RNA extraction
Total RNA was extracted using the QIAGEN RNeasy Mini kit or the Zymo DIRECT-ZOL RNA miniprep kit, with an additional DNase (QIAGEN) treatment. RNA quantity and integrity were determined using Nanodrop, agarose gels, and the Agilent 2100 Bioanalyzer.
RNA-sequencing library production
One microgram total RNA was used with the TruSeq RNA Sample Preparation v2 kit (Illumina) to construct index-tagged cDNA libraries. Quality, quantity, and size distribution of the Illumina libraries were determined using the DNA-1000 kit (Agilent Bioanalyzer; Agilent Technologies). Libraries were sequenced (single-end mode and length 75 bp) on the Genome Analyzer IIx System, using the standard RNA-sequencing protocol in the TruSeq SBS kit v5. Fastq files containing reads for each library were extracted and demultiplexed using the Casava v1.8.2 pipeline.
RNA-sequencing analysis
Sequencing adaptor contaminations were removed from reads using cutadapt software (Martin, 2011), and the resulting reads were mapped and quantified on the transcriptome (Ensembl gene-build 66) using RSEM v1.2.3. Only genes with at least two counts per million in at least five samples were considered for statistical analysis. Data were then normalized and deferential expression was tested using the bioconductor package EdgeR (Robinson et al., 2010). We considered as differentially expressed those genes with a Benjamini-Hochberg adjusted p-value <0.05.
Quantitative RT-PCR
cDNA was synthesized with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) using 0.5 µg total RNA per reaction. Quantitative PCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems) and the following primers: Tert (forward, 5′-GCCTCACCTTCCAGAGTGTCTTC-3′; reverse, 5′-CTCAGAGGTCCCAGGAGACATTTG-3′); p15 (forward, 5′-CAGTTGGGTTCTGCTCCGT-3′; reverse, 5′-AGATCCCAACGCCCTGAAC-3′); p16 (forward, 5′-GGGTTTCGCCCAACGCCCCGA-3′; reverse, 5′-TGCAGCACCACCAGCGTGTCC-3′); p18 (forward, 5′-CTCCGGATTTCCAAGTTTCA-3′; reverse, 5′-GGGGGACCTAGAGCAACTTAC-3′); p19 (forward, 5′-GTCCTGGACATTGGGGCT-3′; reverse, 5′-AACCGCTTCGGCAAGAC-3′); p21 (forward, 5′-ATCACCAGGATTGGACATGG-3′; reverse, 5′-CGGTGTCAGAGTCTAGGGGA-3′); p27 (forward, 5′-GGGGAACCGTCTGAAACATT-3′; reverse, 5′-AGTGTCCAGGGATGAGGAAG-3′); p57 (forward, 5′-TTCTCCTGCGCAGTTCTCTT-3′; reverse, CTGAAGGACCAGCCTCTCTC-3′); GusB (forward, 5′-ACTCCTCACTGAACATGCGA-3′; reverse, 5′-ATAAGACGCATCAGAAGCCG-3′); and 36B4 (forward, 5′-GCACATCACTCAGAATTTCAATGG-3′; reverse, 5′-GGACCCGAGAAGACCTCCTT-3′).
Statistical analysis
Data are presented as mean ± SEM. A Wilcoxon’s ram sum test was used to calculate the statistical significance of differences between telomere-length distributions. Other comparisons were made by two-tailed unpaired t test. Calculations were made in Microsoft Excel and Prism. Differences were considered significant at P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***).
Online supplemental material
Fig. S1 compares CM telomere length in WT, G1 Terc−/−, and G3 Terc−/− neonatal mice, demonstrating that CMs in G3 Terc−/− neonates undergo premature telomere shortening. Fig. S2 shows increased expression of oxidative phosphorylation genes in G3 Terc−/− hearts after cryoinjury, corroborating the hypertrophic response to cardiac injury. Fig. S3 shows the cardiac response to cryoinjury in G1 Terc−/− neonates. Fig. S4 shows analysis of TUNEL and CDKI expression in WT and G3 Terc−/− neonates. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201510091/DC1.
Supplementary Material
Acknowledgments
We thank the Centro Nacional de Investigaciones Cardiovasculares (CNIC) Genomics Unit for RNA sequencing; the CNIC Bioinformatics Unit for help in RNA-sequencing analysis; the CNIC Histology Unit for Masson’s trichrome staining; F. Sánchez-Cabo for help with statistics; M.J. Gómez for help with RNA-sequencing analysis; the CNIC Animal Facility for mouse care; the CNIC Microscopy Unit for support with image acquisition; I. de Diego (CNIC) for help with genotyping; M. Torres, J.L de la Pompa, N. Mercader, V. Andrés, and E. Lara-Pezzi (CNIC) for critical reading of the manuscript; and S. Bartlett (CNIC) for text editing.
E. Aix was supported by a Formación de Profesorado Universitario predoctoral fellowship from the Ministerio de Educación. C. Sánchez-Ferrer is supported by a predoctoral fellowship from the Ministerio de Economía y Competitividad. T. Aguado is a recipient of a Fundación Científica Asociación Española Contra el Cáncer postdoctoral fellowship. This study was supported by grants from the Ministerio de Economía y Competitividad (SAF2012-38449 to I. Flores) and the Red Temática de Investigación Cooperativa en Enfermedades Cardiovasculares (RD12/0042/0045 to I. Flores). The CNIC is supported by the Ministerio de Economía y Competitividad and the Pro-CNIC Foundation and is a Severo Ochoa Center of Excellence (Ministerio de Economía y Competitividad award SEV-2015-0505).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- 53BP1
- p53 binding protein 1
- auf
- arbitrary units of fluorescence
- CDKI
- cyclin-dependent kinase inhibitor
- CM
- cardiomyocyte
- DDR
- DNA damage response
- dpi
- days postinjury
- G
- generation
- γH2AX
- phopho–histone H2AX Ser139
- iPS
- induced pluripotent stem
- P
- postnatal day
- pH3
- phospho–histone H3 Ser10
- Q-FISH
- quantitative FISH
- TIF
- telomere-dysfunction–induced focus
- Tn
- troponin
- TRAP
- telomeric repeat amplification protocol
- WT
- wild type
References
- Artandi S.E., and Attardi L.D.. 2005. Pathways connecting telomeres and p53 in senescence, apoptosis, and cancer. Biochem. Biophys. Res. Commun. 331:881–890. 10.1016/j.bbrc.2005.03.211 [DOI] [PubMed] [Google Scholar]
- Baker D.J., Weaver R.L., and van Deursen J.M.. 2013. p21 both attenuates and drives senescence and aging in BubR1 progeroid mice. Cell Reports. 3:1164–1174. 10.1016/j.celrep.2013.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bär C., Bernardes de Jesus B., Serrano R., Tejera A., Ayuso E., Jimenez V., Formentini I., Bobadilla M., Mizrahi J., de Martino A., et al. 2014. Telomerase expression confers cardioprotection in the adult mouse heart after acute myocardial infarction. Nat. Commun. 5:5863 10.1038/ncomms6863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L., Dokal I., Tamary H., Avigdor A., Garty B.Z., Volkov A., and Vulliamy T.. 2008. Expanding the clinical phenotype of autosomal dominant dyskeratosis congenita caused by TERT mutations. Haematologica. 93:943–944. 10.3324/haematol.12317 [DOI] [PubMed] [Google Scholar]
- Bednarek D., González-Rosa J.M., Guzmán-Martínez G., Gutiérrez-Gutiérrez Ó., Aguado T., Sánchez-Ferrer C., Marques I.J., Galardi-Castilla M., de Diego I., Gómez M.J., et al. 2015. Telomerase is essential for Zebrafish heart regeneration. Cell Reports. 12:1691–1703. 10.1016/j.celrep.2015.07.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begus-Nahrmann Y., Lechel A., Obenauf A.C., Nalapareddy K., Peit E., Hoffmann E., Schlaudraff F., Liss B., Schirmacher P., Kestler H., et al. 2009. p53 deletion impairs clearance of chromosomal-instable stem cells in aging telomere-dysfunctional mice. Nat. Genet. 41:1138–1143. 10.1038/ng.426 [DOI] [PubMed] [Google Scholar]
- Bergmann O., Bhardwaj R.D., Bernard S., Zdunek S., Barnabé-Heider F., Walsh S., Zupicich J., Alkass K., Buchholz B.A., Druid H., et al. 2009. Evidence for cardiomyocyte renewal in humans. Science. 324:98–102. 10.1126/science.1164680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O., Zdunek S., Felker A., Salehpour M., Alkass K., Bernard S., Sjostrom S.L., Szewczykowska M., Jackowska T., Dos Remedios C., et al. 2015. Dynamics of cell generation and turnover in the human heart. Cell. 161:1566–1575. 10.1016/j.cell.2015.05.026 [DOI] [PubMed] [Google Scholar]
- Blasco M.A., Lee H.W., Hande M.P., Samper E., Lansdorp P.M., DePinho R.A., and Greider C.W.. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 91:25–34. 10.1016/S0092-8674(01)80006-4 [DOI] [PubMed] [Google Scholar]
- Boon R.A., Iekushi K., Lechner S., Seeger T., Fischer A., Heydt S., Kaluza D., Tréguer K., Carmona G., Bonauer A., et al. 2013. MicroRNA-34a regulates cardiac ageing and function. Nature. 495:107–110. 10.1038/nature11919 [DOI] [PubMed] [Google Scholar]
- Borges A., and Liew C.C.. 1997. Telomerase activity during cardiac development. J. Mol. Cell. Cardiol. 29:2717–2724. 10.1006/jmcc.1997.0503 [DOI] [PubMed] [Google Scholar]
- Brugarolas J., Chandrasekaran C., Gordon J.I., Beach D., Jacks T., and Hannon G.J.. 1995. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 377:552–557. 10.1038/377552a0 [DOI] [PubMed] [Google Scholar]
- Chan S.W., and Blackburn E.H.. 2002. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene. 21:553–563. 10.1038/sj.onc.1205082 [DOI] [PubMed] [Google Scholar]
- Choudhury A.R., Ju Z., Djojosubroto M.W., Schienke A., Lechel A., Schaetzlein S., Jiang H., Stepczynska A., Wang C., Buer J., et al. 2007. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat. Genet. 39:99–105. 10.1038/ng1937 [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., and Jackson S.P.. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 426:194–198. 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- Daniali L., Benetos A., Susser E., Kark J.D., Labat C., Kimura M., Desai K., Granick M., and Aviv A.. 2013. Telomeres shorten at equivalent rates in somatic tissues of adults. Nat. Commun. 4:1597 10.1038/ncomms2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T., and de Lange T.. 2011. The causes and consequences of polyploidy in normal development and cancer. Annu. Rev. Cell Dev. Biol. 27:585–610. 10.1146/annurev-cellbio-092910-154234 [DOI] [PubMed] [Google Scholar]
- de Lange T. 2002. Protection of mammalian telomeres. Oncogene. 21:532–540. 10.1038/sj.onc.1205080 [DOI] [PubMed] [Google Scholar]
- de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19:2100–2110. 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- Ding H., Yan F., Zhou L.L., Ji X.H., Gu X.N., Tang Z.W., and Chen R.H.. 2014. Association between previously identified loci affecting telomere length and coronary heart disease (CHD) in Han Chinese population. Clin. Interv. Aging. 9:857–861. 10.2147/CIA.S60760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenckhahn J.D., Schwarz Q.P., Gray S., Laskowski A., Kiriazis H., Ming Z., Harvey R.P., Du X.J., Thorburn D.R., and Cox T.C.. 2008. Compensatory growth of healthy cardiac cells in the presence of diseased cells restores tissue homeostasis during heart development. Dev. Cell. 15:521–533. 10.1016/j.devcel.2008.09.005 [DOI] [PubMed] [Google Scholar]
- Engel F.B., Schebesta M., and Keating M.T.. 2006. Anillin localization defect in cardiomyocyte binucleation. J. Mol. Cell. Cardiol. 41:601–612. 10.1016/j.yjmcc.2006.06.012 [DOI] [PubMed] [Google Scholar]
- Flores I., Canela A., Vera E., Tejera A., Cotsarelis G., and Blasco M.A.. 2008. The longest telomeres: a general signature of adult stem cell compartments. Genes Dev. 22:654–667. 10.1101/gad.451008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffart S., von Kleist-Retzow J.C., and Wiesner R.J.. 2004. Regulation of mitochondrial proliferation in the heart: power-plant failure contributes to cardiac failure in hypertrophy. Cardiovasc. Res. 64:198–207. 10.1016/j.cardiores.2004.06.030 [DOI] [PubMed] [Google Scholar]
- González-Suárez E., Samper E., Flores J.M., and Blasco M.A.. 2000. Telomerase-deficient mice with short telomeres are resistant to skin tumorigenesis. Nat. Genet. 26:114–117. 10.1038/79089 [DOI] [PubMed] [Google Scholar]
- Greider C.W., and Blackburn E.H.. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 43:405–413. 10.1016/0092-8674(85)90170-9 [DOI] [PubMed] [Google Scholar]
- Greider C.W., and Blackburn E.H.. 1987. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 51:887–898. 10.1016/0092-8674(87)90576-9 [DOI] [PubMed] [Google Scholar]
- Greider C.W., and Blackburn E.H.. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 337:331–337. 10.1038/337331a0 [DOI] [PubMed] [Google Scholar]
- Harley C.B. 1991. Telomere loss: mitotic clock or genetic time bomb? Mutat. Res. 256:271–282. 10.1016/0921-8734(91)90018-7 [DOI] [PubMed] [Google Scholar]
- Harrington L. 2004. Does the reservoir for self-renewal stem from the ends? Oncogene. 23:7283–7289. 10.1038/sj.onc.1207948 [DOI] [PubMed] [Google Scholar]
- Haubner B.J., Schneider J., Schweigmann U., Schuetz T., Dichtl W., Velik-Salchner C., Stein J.-I., and Penninger J.M.. 2016. Functional recovery of a human neonatal heart after severe myocardial infarction. Circ. Res. 118:216–221. 10.1161/CIRCRESAHA.115.307017 [DOI] [PubMed] [Google Scholar]
- Herbert B.S., Hochreiter A.E., Wright W.E., and Shay J.W.. 2006. Nonradioactive detection of telomerase activity using the telomeric repeat amplification protocol. Nat. Protoc. 1:1583–1590. 10.1038/nprot.2006.239 [DOI] [PubMed] [Google Scholar]
- Herrera E., Samper E., Martín-Caballero J., Flores J.M., Lee H.W., and Blasco M.A.. 1999. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 18:2950–2960. 10.1093/emboj/18.11.2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffelder D.R., Luo L., Burke N.A., Watkins S.C., Gollin S.M., and Saunders W.S.. 2004. Resolution of anaphase bridges in cancer cells. Chromosoma. 112:389–397. 10.1007/s00412-004-0284-6 [DOI] [PubMed] [Google Scholar]
- Ikenishi A., Okayama H., Iwamoto N., Yoshitome S., Tane S., Nakamura K., Obayashi T., Hayashi T., and Takeuchi T.. 2012. Cell cycle regulation in mouse heart during embryonic and postnatal stages. Dev. Growth Differ. 54:731–738. 10.1111/j.1440-169X.2012.01373.x [DOI] [PubMed] [Google Scholar]
- Ju Z., Choudhury A.R., and Rudolph K.L.. 2007. A dual role of p21 in stem cell aging. Ann. N. Y. Acad. Sci. 1100:333–344. 10.1196/annals.1395.036 [DOI] [PubMed] [Google Scholar]
- Kikuchi K., and Poss K.D.. 2012. Cardiac regenerative capacity and mechanisms. Annu. Rev. Cell Dev. Biol. 28:719–741. 10.1146/annurev-cellbio-101011-155739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdorp P.M. 2005. Role of telomerase in hematopoietic stem cells. Ann. N. Y. Acad. Sci. 1044:220–227. 10.1196/annals.1349.027 [DOI] [PubMed] [Google Scholar]
- Lansdorp P.M., Verwoerd N.P., van de Rijke F.M., Dragowska V., Little M.T., Dirks R.W., Raap A.K., and Tanke H.J.. 1996. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 5:685–691. 10.1093/hmg/5.5.685 [DOI] [PubMed] [Google Scholar]
- Leri A., Barlucchi L., Limana F., Deptala A., Darzynkiewicz Z., Hintze T.H., Kajstura J., Nadal-Ginard B., and Anversa P.. 2001. Telomerase expression and activity are coupled with myocyte proliferation and preservation of telomeric length in the failing heart. Proc. Natl. Acad. Sci. USA. 98:8626–8631. 10.1073/pnas.151013298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leri A., Franco S., Zacheo A., Barlucchi L., Chimenti S., Limana F., Nadal-Ginard B., Kajstura J., Anversa P., and Blasco M.A.. 2003. Ablation of telomerase and telomere loss leads to cardiac dilatation and heart failure associated with p53 upregulation. EMBO J. 22:131–139. 10.1093/emboj/cdg013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., O’Connor M.S., Qin J., and Songyang Z.. 2004. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 279:51338–51342. 10.1074/jbc.M409293200 [DOI] [PubMed] [Google Scholar]
- Margolis R.L., Lohez O.D., and Andreassen P.R.. 2003. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J. Cell. Biochem. 88:673–683. 10.1002/jcb.10411 [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 17:10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Maubaret C.G., Salpea K.D., Romanoski C.E., Folkersen L., Cooper J.A., Stephanou C., Li K.W., Palmen J., Hamsten A., Neil A., et al. EARSII consortium . 2013. Association of TERC and OBFC1 haplotypes with mean leukocyte telomere length and risk for coronary heart disease. PLoS One. 8:e83122 10.1371/journal.pone.0083122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1939. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl. Acad. Sci. USA. 25:405–416. 10.1073/pnas.25.8.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1941. The stability of broken ends of chromosomes in Zea mays. Genetics. 26:234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollova M., Bersell K., Walsh S., Savla J., Das L.T., Park S.Y., Silberstein L.E., Dos Remedios C.G., Graham D., Colan S., and Kühn B.. 2013. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc. Natl. Acad. Sci. USA. 110:1446–1451. 10.1073/pnas.1214608110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H.J. 1938. The remaking of chromosomes. Collecting Net. 13:181–198. [Google Scholar]
- Muralidhar S.A., Mahmoud A.I., Canseco D., Xiao F., and Sadek H.A.. 2013. Harnessing the power of dividing cardiomyocytes. Glob. Cardiol. Sci. Pract. 2013:212–221. 10.5339/gcsp.2013.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane J.P. 2012. Telomere dysfunction and chromosome instability. Mutat. Res. 730:28–36. 10.1016/j.mrfmmm.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Taffet G.E., Youker K.A., Entman M.L., Overbeek P.A., Michael L.H., and Schneider M.D.. 2001. Telomerase reverse transcriptase promotes cardiac muscle cell proliferation, hypertrophy, and survival. Proc. Natl. Acad. Sci. USA. 98:10308–10313. 10.1073/pnas.191169098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov A.M. 1973. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41:181–190. 10.1016/0022-5193(73)90198-7 [DOI] [PubMed] [Google Scholar]
- Pampalona J., Frías C., Genescà A., and Tusell L.. 2012. Progressive telomere dysfunction causes cytokinesis failure and leads to the accumulation of polyploid cells. PLoS Genet. 8:e1002679 10.1371/journal.pgen.1002679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello E.R., and Olson E.N.. 2014. A neonatal blueprint for cardiac regeneration. Stem Cell Res. (Amst.). 13:556–570. 10.1016/j.scr.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello E.R., Mahmoud A.I., Simpson E., Hill J.A., Richardson J.A., Olson E.N., and Sadek H.A.. 2011. Transient regenerative potential of the neonatal mouse heart. Science. 331:1078–1080. 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello E.R., Mahmoud A.I., Simpson E., Johnson B.A., Grinsfelder D., Canseco D., Mammen P.P., Rothermel B.A., Olson E.N., and Sadek H.A.. 2013. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl. Acad. Sci. USA. 110:187–192. 10.1073/pnas.1208863110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente B.N., Kimura W., Muralidhar S.A., Moon J., Amatruda J.F., Phelps K.L., Grinsfelder D., Rothermel B.A., Chen R., Garcia J.A., et al. 2014. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 157:565–579. 10.1016/j.cell.2014.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaife-Ryan G.A., Sim C.B., Porrello E.R., and Hudson J.E.. 2016. Resetting the epigenome for heart regeneration. Semin. Cell Dev. Biol. 10.1016/j.semcdb.2015.12.021 [DOI] [PubMed] [Google Scholar]
- Rabinowitz M., and Zak R.. 1975. Mitochondria and cardiac hypertrophy. Circ. Res. 36:367–376. 10.1161/01.RES.36.3.367 [DOI] [PubMed] [Google Scholar]
- Ramirez R.D., Wright W.E., Shay J.W., and Taylor R.S.. 1997. Telomerase activity concentrates in the mitotically active segments of human hair follicles. J. Invest. Dermatol. 108:113–117. 10.1111/1523-1747.ep12285654 [DOI] [PubMed] [Google Scholar]
- Richardson G.D., Breault D., Horrocks G., Cormack S., Hole N., and Owens W.A.. 2012. Telomerase expression in the mammalian heart. FASEB J. 26:4832–4840. 10.1096/fj.12-208843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saretzki G., and Von Zglinicki T.. 2002. Replicative aging, telomeres, and oxidative stress. Ann. N. Y. Acad. Sci. 959:24–29. 10.1111/j.1749-6632.2002.tb02079.x [DOI] [PubMed] [Google Scholar]
- Senyo S.E., Lee R.T., and Kühn B.. 2014. Cardiac regeneration based on mechanisms of cardiomyocyte proliferation and differentiation. Stem Cell Res. (Amst.). 13:532–541. 10.1016/j.scr.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N., Shingaki K., Kaneko-Sasaguri Y., Hashizume T., and Kanda T.. 2005. When, where and how the bridge breaks: anaphase bridge breakage plays a crucial role in gene amplification and HSR generation. Exp. Cell Res. 302:233–243. 10.1016/j.yexcr.2004.09.001 [DOI] [PubMed] [Google Scholar]
- Soonpaa M.H., Koh G.Y., Klug M.G., and Field L.J.. 1994. Formation of nascent intercalated disks between grafted fetal cardiomyocytes and host myocardium. Science. 264:98–101. 10.1126/science.8140423 [DOI] [PubMed] [Google Scholar]
- Soonpaa M.H., Kim K.K., Pajak L., Franklin M., and Field L.J.. 1996. Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiol. 271:H2183–H2189. [DOI] [PubMed] [Google Scholar]
- Takeuchi T. 2014. Regulation of cardiomyocyte proliferation during development and regeneration. Dev. Growth Differ. 56:402–409. 10.1111/dgd.12134 [DOI] [PubMed] [Google Scholar]
- Tane S., Ikenishi A., Okayama H., Iwamoto N., Nakayama K.I., and Takeuchi T.. 2014. CDK inhibitors, p21(Cip1) and p27(Kip1), participate in cell cycle exit of mammalian cardiomyocytes. Biochem. Biophys. Res. Commun. 443:1105–1109. 10.1016/j.bbrc.2013.12.109 [DOI] [PubMed] [Google Scholar]
- von Zglinicki T. 2002. Oxidative stress shortens telomeres. Trends Biochem. Sci. 27:339–344. 10.1016/S0968-0004(02)02110-2 [DOI] [PubMed] [Google Scholar]
- Vousden K.H., and Prives C.. 2009. Blinded by the light: The growing complexity of p53. Cell. 137:413–431. 10.1016/j.cell.2009.04.037 [DOI] [PubMed] [Google Scholar]
- Vulliamy T., Marrone A., Szydlo R., Walne A., Mason P.J., and Dokal I.. 2004. Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat. Genet. 36:447–449. 10.1038/ng1346 [DOI] [PubMed] [Google Scholar]
- Watson J.D. 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239:197–201. 10.1038/newbio239197a0 [DOI] [PubMed] [Google Scholar]
- Wright W.E., Piatyszek M.A., Rainey W.E., Byrd W., and Shay J.W.. 1996. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 18:173–179. [DOI] [PubMed] [Google Scholar]
- Zijlmans J.M., Martens U.M., Poon S.S., Raap A.K., Tanke H.J., Ward R.K., and Lansdorp P.M.. 1997. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. USA. 94:7423–7428. 10.1073/pnas.94.14.7423 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.