Ping et al. demonstrate that mitochondria-to-plasma membrane anchoring is mediated by Num1 directly interacting with both organelles through two distinct and spatially separated lipid-specific binding domains. These findings suggest a general mechanism for interorganelle tethering.
Abstract
The mitochondria–ER cortex anchor (MECA) is required for proper mitochondrial distribution and functions by tethering mitochondria to the plasma membrane. The core component of MECA is the multidomain protein Num1, which assembles into clusters at the cell cortex. We show Num1 adopts an extended, polarized conformation. Its N-terminal coiled-coil domain (Num1CC) is proximal to mitochondria, and the C-terminal pleckstrin homology domain is associated with the plasma membrane. We find that Num1CC interacts directly with phospholipid membranes and displays a strong preference for the mitochondria-specific phospholipid cardiolipin. This direct membrane interaction is critical for MECA function. Thus, mitochondrial anchoring is mediated by a protein that interacts directly with two different membranes through lipid-specific binding domains, suggesting a general mechanism for interorganelle tethering.
Introduction
The shape and position of mitochondria are intimately connected to both mitochondrial and cellular function (Labbé et al., 2014; Lackner, 2014; Mishra and Chan, 2014). Central to mitochondrial positioning are molecular tethers, which allow spatial, temporal, and contextual control over the cellular position of the organelle. In neurons and immune cells, populations of mitochondria are tethered in synapses to meet the high energy demands and calcium buffering needs of these active cellular regions (Hollenbeck and Saxton, 2005; Quintana et al., 2007; Cai and Sheng, 2009). In budding yeast, mitochondrial tethering is critical to ensure mother cells retain and daughter cells inherit the essential mitochondrial compartment. In addition, mitochondrial tethering has been linked to aging in yeast, specifically in the maintenance of mother–daughter age asymmetry (McFaline-Figueroa et al., 2011). Tether-mediated interactions have also been proposed to facilitate the exchange of lipids, calcium, and other small molecules between mitochondria and other membrane compartments (Toulmay and Prinz, 2011; Helle et al., 2013). Although tethering plays a critical role in mitochondrial positioning and function in cells from yeast to neurons, the molecular mechanisms are poorly understood.
In yeast, the proper distribution of mitochondria between mother and daughter cells requires the activities of two distinct cortical tethers: the mitochondria–ER cortex anchor (MECA) and Mmr1. MECA is localized to mother cells and larger buds and functions in the distribution and mother-specific retention of mitochondria (Farkasovsky and Küntzel, 1995; Heil-Chapdelaine et al., 2000; Cerveny et al., 2007; Klecker et al., 2013; Lackner et al., 2013). At least two proteins (Num1 and Mdm36) and three organelles (mitochondria, the ER, and the plasma membrane) comprise the multisubunit MECA structure (Hammermeister et al., 2010; Lackner et al., 2013). In contrast to MECA, Mmr1 is predominately restricted to buds, where the protein functions in Myo2-driven bud-directed transport of mitochondria as well as mitochondrial retention by tethering mitochondria to cortical ER sheets at the bud tip (Itoh et al., 2004; Swayne et al., 2011). Thus, MECA and Mmr1 generate opposing mitochondrial tethering activities at spatially distinct cellular locations (Klecker et al., 2013; Lackner et al., 2013).
The core component of MECA is the cortical, multidomain protein Num1, which assembles into clusters at the cell cortex that function to tether mitochondria to the plasma membrane. Num1 also serves as a cortical anchor for dynein, and, as such, functions to harness dynein-mediated microtubule sliding along the cell cortex for nuclear migration into buds (Heil-Chapdelaine et al., 2000; Farkasovsky and Küntzel, 2001). However, the mitochondrial tethering function of Num1 is independent of its role in nuclear migration (Cerveny et al., 2007; Tang et al., 2012; Lackner et al., 2013). Num1 contains an N-terminal coiled coil (CC), an EF hand-like motif, a region of twelve 64-aa repeats, and a C-terminal pleckstrin homology (PH) domain (Fig. 1 A; Kormanec et al., 1991). The CC and PH domains of Num1 are required for association with mitochondria and the plasma membrane, respectively (Yu et al., 2004; Tang et al., 2009, 2012; Lackner et al., 2013). Although the PH domain of Num1 binds with high specificity to phosphatidylinositol 4,5-bisphosphate (PI4,5P2) in the plasma membrane (Yu et al., 2004; Tang et al., 2009), the exact nature of the Num1–mitochondria interaction is undefined. Here, we report that Num1 forms an extended structure in cells that interacts directly with the mitochondrial outer membrane via a novel membrane binding domain to mediate anchoring of mitochondria to the plasma membrane.
Figure 1.
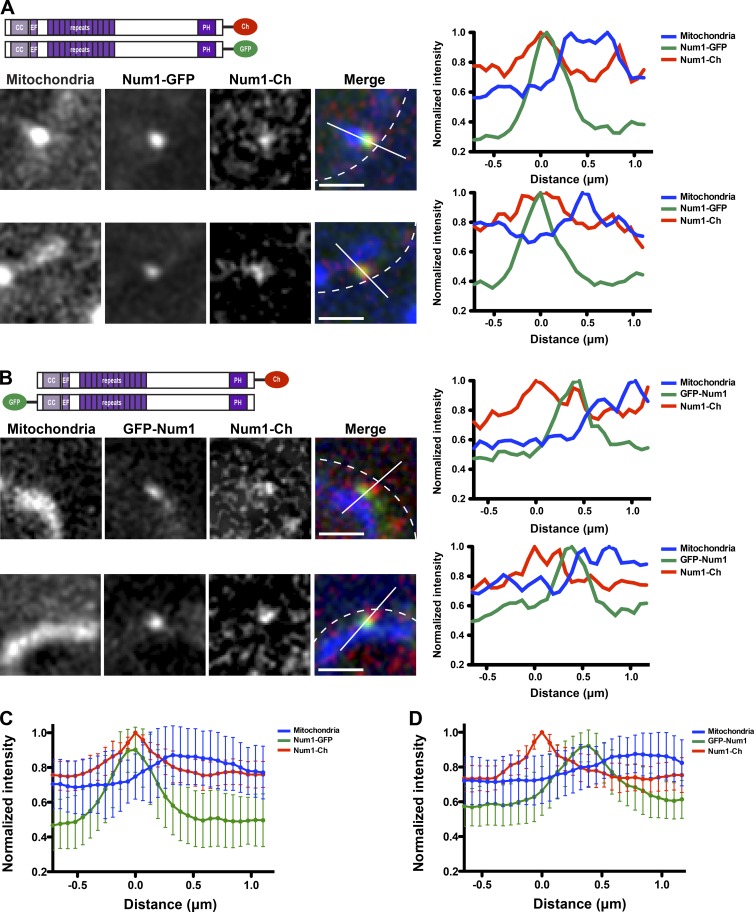
Num1 forms an extended, polarized structure. (A and B) Diploid cells expressing Num1-Cherry (Num1-Ch) and Num1-GFP (A) or Num1-Cherry and GFP-Num1 (B), as depicted in the schematics, and mito-BFP were visualized by fluorescence microscopy. Single focal planes are shown. The cell cortex is outlined with a dashed white line. Bars, 1 µm. The graphs show normalized fluorescence intensities of Num1-Cherry (red), Num1-GFP (A), or GFP-Num1 (B; green) and mitochondria (blue) along a line drawn through the center of Num1 foci as indicated in the corresponding image (white line). The position of peak Num1-Cherry intensity was set at 0 µm. Ch, Cherry; EF, EF-like hand motif. (C and D) Graphs of normalized fluorescence intensity profiles of lines analyzed as described in A and B. The mean intensities ± SD are plotted; n ≥ 30.
Results and discussion
MECA is an extended, polarized structure
The CC and PH domains of Num1 (Fig. 1 A) mediate interactions with mitochondria and the plasma membrane, respectively (Yu et al., 2004; Tang et al., 2009, 2012; Lackner et al., 2013). In addition to being required for an interaction with mitochondria, Num1CC is both necessary and sufficient for an interaction with Mdm36 (Lackner et al., 2013). Mdm36 is more proximal to mitochondria than the C terminus of Num1 (Lackner et al., 2013), suggesting that Num1 is an extended protein with the N terminus associated with mitochondria and the C terminus associated with the plasma membrane. To directly test this idea, we simultaneously coexpressed functional, differentially tagged versions of Num1 in diploid cells and analyzed their localization relative to one another and to mitochondria using matrix-targeted TagBFP (mito-BFP). One genomic copy of NUM1 was expressed with yEmCherry fused at its C terminus (Num1-Cherry), and the other genomic copy of NUM1 was expressed with yEGFP fused at either its N or C terminus (GFP-Num1 or Num1-GFP, respectively; Fig. 1, A and B).
In cells coexpressing Num1-GFP and Num1-Cherry, the proteins colocalized in foci at the cell cortex with nearly overlapping fluorescent signals (Fig. 1 A). Linescan analyses through the center of Num1 foci indicated that the intensity profiles for Num1-GFP and Num1-Cherry versus distance were similar (Fig. 1, A and C). In contrast, in cells coexpressing GFP-Num1 and Num1-Cherry, we observed adjacent, spatially resolved cortical foci (Fig. 1 B). The peak intensities of Num1-Cherry and GFP-Num1 foci were more proximal to the cell wall and mitochondria, respectively (Fig. 1, B and D). These results suggest that Num1 adopts an extended, polarized conformation in cells with its CC domain proximal to mitochondria and PH domain proximal to the plasma membrane.
Num1CC interacts directly with Mdm36 and phospholipid membranes
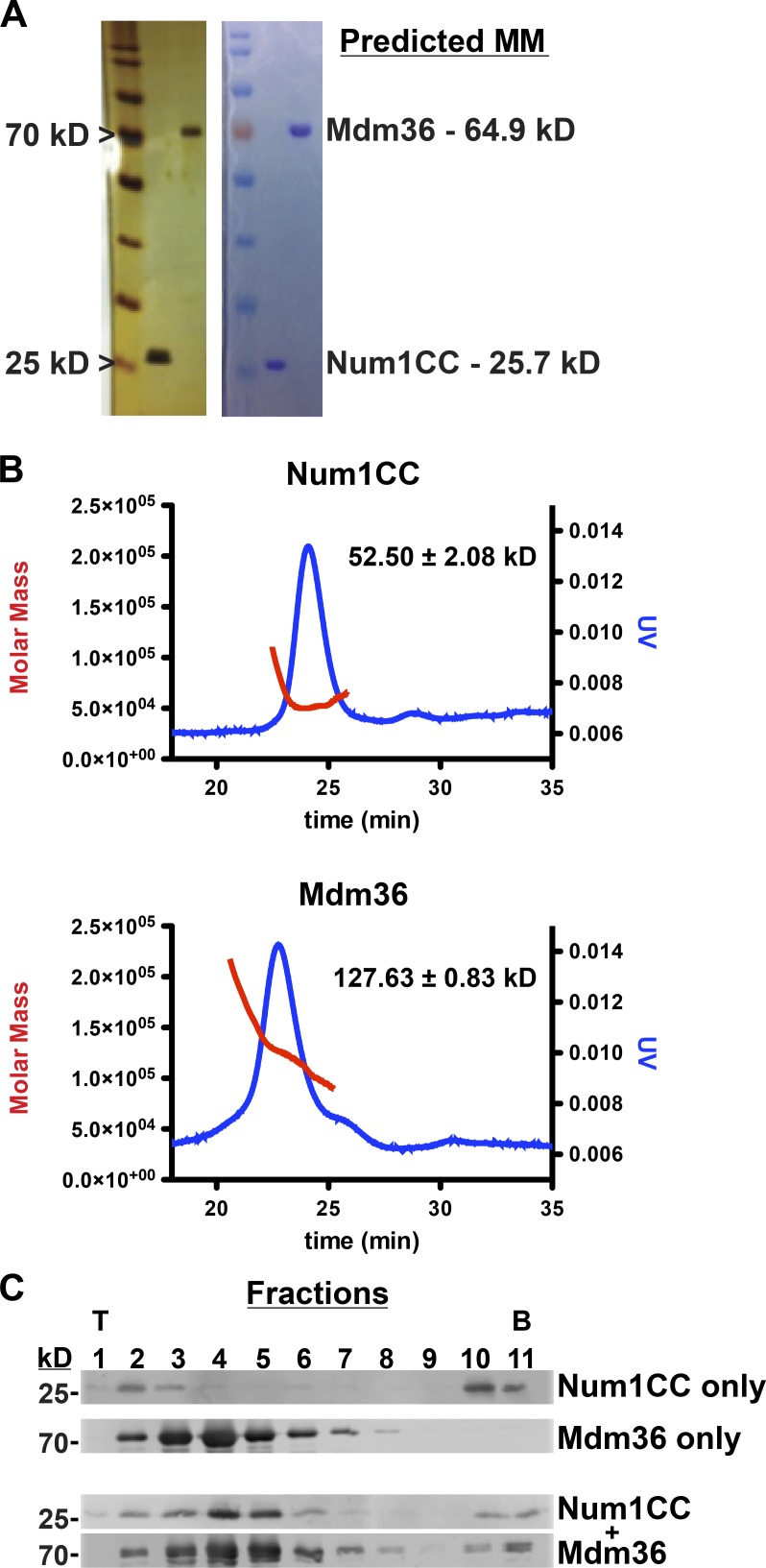
Our imaging analyses place Num1CC and Mdm36 near the mitochondrial membrane (Fig. 1; Lackner et al., 2013). To examine the mechanistic contributions of Num1CC and Mdm36 to MECA’s mitochondrial interaction, we examined the assembly and membrane binding properties of Num1CC and Mdm36 in vitro. Recombinant Num1CC (amino acids 97–294) and Mdm36 were purified from Escherichia coli (Fig. 2 A). Size exclusion chromatography with inline multiangle light scattering (SEC-MALS) revealed that both proteins exist as dimers (Fig. 2 B). The dimerization of Num1CC is consistent with previous studies (Tang et al., 2012). To determine if Num1CC and Mdm36 form a complex under physiological conditions, Num1CC and Mdm36 alone, and in combination, were subjected to sucrose density gradient centrifugation. Analysis of the sucrose gradient fractions revealed that Num1CC and Mdm36 migrated farther into the gradient after incubation of the two proteins in comparison to either protein alone, indicative of complex formation (Fig. 2 C). These results demonstrate that Num1CC interacts directly with Mdm36.
Figure 2.
Num1CC and Mdm36 form a complex. (A) Silver (left) and Coomassie-stained (right) SDS-PAGE gels showing purified Num1CC and Mdm36. The 25- and 70-kD markers are indicated on the left, and the predicted molecular masses (MMs) of each protein are shown on the right. (B) SEC-MALS analyses of Num1CC and Mdm36. The graphs show the elution profile as assessed by absorbance at 280 nm (blue) and determined molecular mass (red) of each protein. Masses were determined to be 52.50 ± 2.08 kD and 127.63 ± 0.83 kD for Num1CC and Mdm36, respectively (n = 3, mean ± SD). (C) Purified Num1CC (10 µM) and Mdm36 (10 µM) alone and in combination were subjected to sucrose density gradient analysis. Gradients were manually fractionated from top (T) to bottom (B) and analyzed by SDS-PAGE and Western blot analysis.
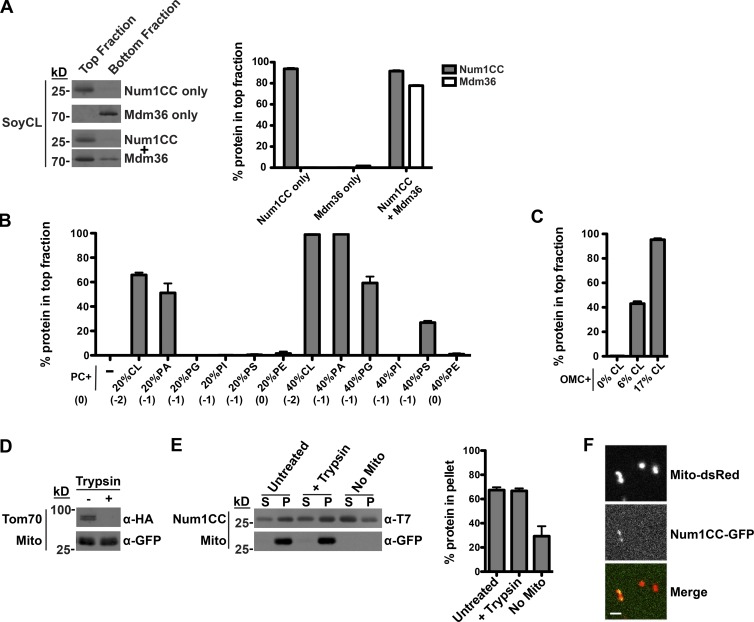
We next asked whether Num1CC, Mdm36, or the Num1CC–Mdm36 complex were able to directly interact with phospholipid membranes. We measured membrane association by liposome floatation using liposomes made from soybean polar lipid extract spiked with 6% cardiolipin (SoyCL), which closely approximates the phospholipid composition of the mitochondrial outer membrane (MOM; Zinser and Daum, 1995). Num1CC readily associated with the liposomes, whereas Mdm36 alone did not (Fig. 3 A). However, Mdm36 was recruited to liposomes in the presence of Num1CC (Fig. 3 A), indicating that Num1CC simultaneously interacts with phospholipid membranes and Mdm36.
Figure 3.
Num1CC interacts directly with phospholipid membranes. (A) A total of 3 µM Num1CC and 1.5 µM Mdm36 were incubated with soybean liposomes spiked with 6% CL (SoyCL), which approximates the phospholipid composition of the MOM. The association of the protein with liposomes was assessed by its ability to float with liposomes, as indicated by the amount of protein in the top fraction of the gradient. Equivalent amounts of the top and bottom fractions of the floatation gradients were subjected to SDS-PAGE and Western blot analysis. The percentage of protein found in the top fraction is shown as the mean ± SEM; n = 3 independent experiments. (B) A total of 3 µM Num1CC was incubated with liposomes composed of PC and the indicated mol% of a second phospholipid, and the reactions were subjected to liposome floatation and analysis as described in A. Data are shown as the mean ± SEM; n = 3 independent experiments. The net charge of the phospholipid headgroups is indicated below the graph in parentheses. (C) A total of 3 µM Num1CC was incubated with OMC + 0%, 6%, or 17% CL, as indicated, and the reactions were subjected to liposome floatation and analysis as described in A. Data are shown as the mean ± SEM; n = 3 independent experiments. (D) Mitochondria were isolated from Δnum1 cells expressing matrix-targeted GFP and Tom70-HA. After isolation, mitochondria were left untreated or treated with trypsin and subjected to SDS-PAGE and Western blot analysis. Tom70-HA served as a control for efficient trypsin treatment and GFP served as a marker for mitochondria. (E) A total of 1 µM Num1CC was incubated with isolated mitochondria (described in D) that were left untreated or treated with trypsin, as indicated. The association of the protein with mitochondria was assessed by its ability to pellet with mitochondria. Equivalent amounts of supernatant (S) and pellet (P) fractions were subjected to SDS-PAGE and Western blot analysis. The percentage of protein found in the pellet fraction is shown as the mean ± SEM; n = 3 independent experiments. (F) Num1CC-GFP was incubated with trypsin-treated mitochondria isolated from Δnum1 cells expressing matrix-targeted dsRED. The association of the protein with mitochondria was visualized by fluorescence microscopy. A single focal plan is shown. Bar, 2 µm.
Num1CC interacts preferentially with phospholipid membranes containing cardiolipin
We next examined the nature of the Num1CC-phospholipid interaction. We found that Num1CC robustly associated with liposomes in an electrostatic-dependent manner (Fig. S1 A). To further define the specificity of the Num1CC-phospholipid interaction, we assessed the ability of Num1CC to bind to liposomes composed of the neutral phospholipid phosphatidylcholine (PC; 80 mol%) and an additional phospholipid (20 mol%): cardiolipin (CL), phosphatidic acid (PA), phosphatidylglycerol (PG), phosphatidylinositol (PI), or phosphatidylserine (PS), which are negatively charged phospholipids, or phosphatidylethanolamine (PE), a neutral phospholipid (Fig. 3 B). Num1CC preferentially associated with liposomes that contained CL and PA. Interestingly, Num1CC did not interact with PG, PI, and PS liposomes, indicating that it is not the negative charge of the head group alone that is required for the liposome interaction. When present at 40%, PG and PS were able to support the Num1CC–phospholipid interaction, albeit not as effectively as CL and PA (Fig. 3 B). Interestingly, CL and PA, as well as PE, belong to a class of phospholipids referred to as nonbilayer or cone-shaped lipids, which induce membrane curvature or create distinct microenvironments in a planar bilayer. These data indicate that Num1CC preferentially binds to negatively charged, cone-shaped phospholipids.
The 20% and 40% PA, PG, and PS used in the aforementioned assays far exceeds their concentrations in the MOM, which have been reported to be ∼1–4% (Simbeni et al., 1991; Zinser and Daum, 1995; Connerth et al., 2012). In contrast, CL is present at 6% in the MOM and 17% at contact sites between the mitochondrial outer and inner membranes (Simbeni et al., 1991), which are concentrations closer to the 20% used in the assay. To test whether Num1CC associates with a physiological phospholipid composition, we used individual phospholipids to make chemically defined liposomes that mimic the composition of the MOM (outer membrane composition [OMC]) and varied the concentration of CL present in these liposomes (0%, 6%, and 17%). In floatation assays, Num1CC bound to OMC liposomes containing 6% and 17% CL, but not to OMC liposomes that lack CL (Fig. 3 C). Therefore, Num1CC can directly interact with phospholipid membranes and displays a strong preference for the mitochondria-specific phospholipid CL.
We next tested if Num1CC directly associates with mitochondrial membranes. Mitochondria were purified from Δnum1 cells and protease treated to remove proteins from the mitochondrial surface. Tom70, a MOM-anchored protein, was used as a control to assess the efficiency of the protease treatment (Fig. 3 D). In mitochondrial pelleting assays, Num1CC pelleted with untreated and protease-treated mitochondria (Fig. 3 E), indicating that the Num1CC–mitochondria interaction is protein independent. We also visualized the association of purified, recombinant Num1CC-GFP with protease-treated mitochondria using fluorescence microscopy. We observed Num1CC-GFP colocalizing with mitochondria labeled with matrix-targeted dsRED (mito-RED; Fig. 3 F). These data indicate that Num1CC can directly interact with the native mitochondrial membrane.
The CC region is necessary and sufficient for an interaction with mitochondria
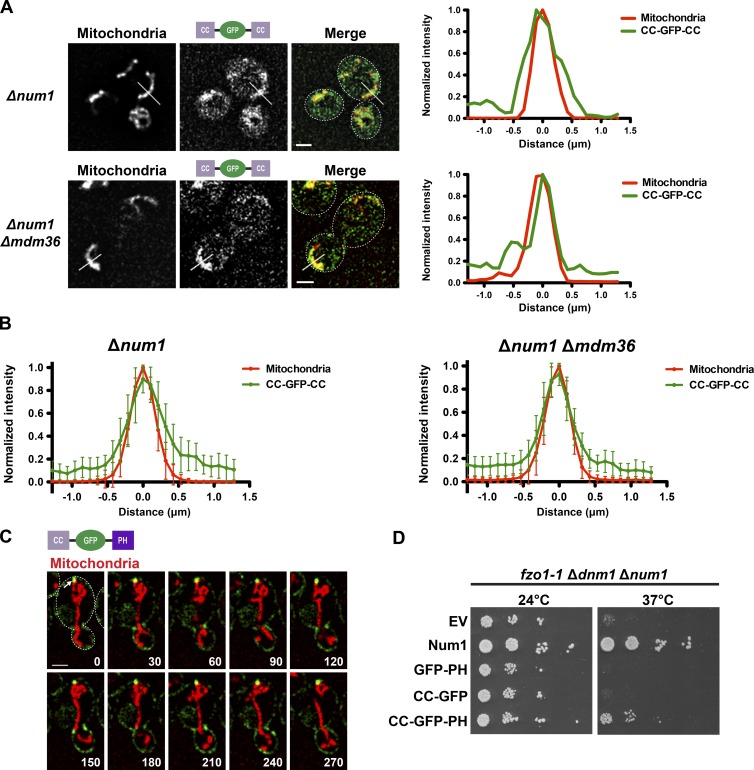
To test whether Num1CC interacts with mitochondria in cells, we fused Num1CC to the N terminus of GFP (CC-GFP) or both termini of GFP (CC-GFP-CC), to increase the binding affinity of the GFP fusion for its target membrane. We expressed CC-GFP or CC-GFP-CC along with mito-RED in Δnum1 cells. We observed an enrichment of CC-GFP on mitochondria in addition to a diffuse cytosolic pool of the protein (Fig. S2, A and B), and the CC-GFP-CC fusion protein exhibited a striking enrichment on mitochondria (Fig. 4, A and B). Mitochondrial-associated CC-GFP and CC-GFP-CC were also observed in the absence of Mdm36 (Fig. S2, A and B; and Fig. 4, A and B), consistent with our in vitro results demonstrating that Num1CC can interact with phospholipid membranes in the absence of Mdm36. These results indicate that Num1CC is sufficient for an interaction with mitochondria.
Figure 4.
Num1CC interacts with the mitochondrial membrane to mediate mitochondrial tethering. (A) Δnum1 (top) and Δnum1 Δmdm36 (bottom) cells expressing CC-GFP-CC (as seen in the schematic) and mito-RED were analyzed by fluorescence microscopy. Single focal planes are shown. The cell cortex is outlined with a dashed white line. Bars, 2 µm. The graphs show normalized fluorescence intensities of mito-RED (red) and CC-GFP-CC (green) along a line drawn perpendicular to a mitochondrial tubule as indicated in the corresponding image (white line). The position of peak mito-RED intensity was set at 0 µm. (B) Graphs of normalized fluorescence intensity profiles for lines analyzed as described in A. The mean intensities ± SD are plotted; n ≥ 20. (C) Δnum1 cells expressing CC-GFP-PH (as seen in the schematic) and mito-RED were analyzed by fluorescence microscopy. A single focal plane is shown. The cell cortex is outlined with a dashed white line. The arrow indicates a tether point. Time is shown in seconds. Bar, 2 µm. (D) Expression of CC-GFP-PH rescues the growth of fzo1-1 Δdnm1 Δnum1 cells. Serial dilutions of fzo1-1 Δdnm1 Δnum1 cells expressing Num1, GFP-PH, CC-GFP, or CC-GFP-PH were plated on SC-TRP + DEX medium and grown at 24 or 37°C, as indicated. EV, empty vector.
Our data, in combination with previous studies, suggests that Num1 contains two membrane binding domains, the CC and PH domains, which interact directly with the mitochondrial membrane and plasma membrane, respectively (Yu et al., 2004; Tang et al., 2009, 2012; Lackner et al., 2013). To test whether the membrane binding domains of Num1 alone are sufficient for mitochondria–plasma membrane tethering, we constructed a minimal Num1 construct by fusing Num1CC and Num1PH to the N and C termini of GFP, respectively (CC-GFP-PH). We expressed this construct along with mito-RED in Δnum1 cells. In addition to a diffuse cortical distribution, CC-GFP-PH localized to clusters at the cell cortex (Fig. 4 C). Strikingly, the CC-GFP-PH clusters were observed adjacent to regions of the mitochondrial network that were persistently localized to the cell cortex. These data suggest that the two membrane binding domains of Num1, Num1CC, and Num1PH are sufficient to tether mitochondria to the plasma membrane.
Num1-mediated mitochondria–plasma membrane tethering is essential in the absence of the mitochondrial division and fusion proteins, Dnm1 and Fzo1, respectively (Lackner et al., 2013). To further test the mitochondria–plasma membrane tethering activity of CC-GFP-PH, we examined the ability of the minimal tethering construct to rescue the lethality of fzo1 dnm1 num1 cells. We expressed CC-GFP-PH or constructs lacking the CC (GFP-PH) or PH (CC-GFP) domains in fzo1-1 Δdnm1 Δnum1 cells, which contain a temperature-sensitive allele of FZO1, fzo1-1 (Hermann et al., 1998). In contrast to GFP-PH and CC-GFP, expression of CC-GFP-PH suppressed the growth defect of fzo1-1 Δdnm1 Δnum1 cells grown at the nonpermissive temperature (Fig. 4 D), consistent with the ability of CC-GFP-PH to tether mitochondria to the plasma membrane.
Defects in Num1CC phospholipid binding disrupt MECA-mediated mitochondrial tethering
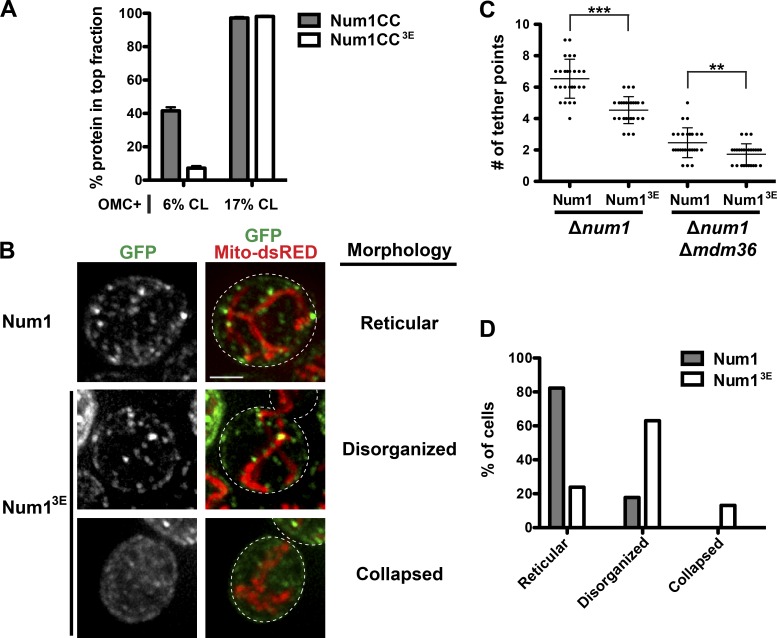
We next sought to identify a mutant that disrupts the Num1CC-phospholipid interaction in vitro and examine its mitochondrial tethering ability in cells. However, we did not want to disrupt the interaction between Num1CC and Mdm36, as this interaction is predicted to affect the robustness of MECA-mediated mitochondrial tethering (Tang et al., 2012; Lackner et al., 2013). Given the affinity of Num1CC for negatively charged lipids, we constructed a series of Num1CC mutants in which the charge of several basic amino acids was reversed. We identified one mutant, Num1CC3E (K121E + R262E + R265E), which fit our criteria. Num1CC3E interacted with Mdm36 as assessed by sucrose density gradient and yeast two-hybrid analyses (Fig. S3, A and B). However, the interaction of Num1CC3E with OMC + 6% CL liposomes was dramatically reduced in comparison to that of Num1CC (Fig. 5 A). Increasing the concentration of CL to 17% in OMC liposomes rescued the binding of Num1CC3E to near wild-type Num1CC levels, indicating that phospholipid binding is impaired but not fully disrupted.
Figure 5.
A direct interaction between Num1 and the mitochondrial outer membrane is critical for MECA function. (A) Num1CC and Num1CC3E were incubated with OMC + 6% or 17% CL, as indicated, and subjected to liposome floatation and analysis as described in Fig. 3 A. Data are shown as the mean ± SEM; n = 3 independent experiments. (B) Δnum1 cells expressing full length GFP-Num1 or GFP-Num13E and mito-RED were analyzed by fluorescence microscopy. Whole-cell projections are shown. The cell cortex is outlined with a dashed white line. Bar, 2 µm. (C) Quantification of the number of mitochondrial tether points in Δnum1 and Δnum1 Δmdm36 cells expressing GFP-Num1 or GFP-Num13E. Tether points in ≥20 cells were counted, and the data are shown as the mean ± SD. To be considered a tether point, mitochondria had to remain associated with a Num1 cluster for ≥90 s. ***, P < 0.0001; **, P < 0.01. (D) Characterization and quantification of the mitochondrial morphology observed in Δnum1 cells expressing GFP-Num1 or GFP-Num13E and mito-RED; n ≥ 60 cells. Examples of each mitochondrial morphology classification are shown in B.
Introduction of the 3E mutations into full-length GFP-tagged Num1 (GFP-Num13E) resulted in a significant reduction in the number of mitochondrial tether points in cells (Fig. 5, B and C). To be considered a tether point, mitochondria had to remain associated with a Num1 cluster for ≥90 s. Also, in a majority of cells expressing GFP-Num13E, the mitochondrial network appeared disorganized, in contrast to the wild-type reticular network observed in cells expressing GFP-Num1 (Fig. 5, B and D). Similar to wild-type Num1, the number of GFP-Num13E mitochondrial tether points was further reduced in cells lacking Mdm36 (Fig. 5 C), which is consistent with our data that the 3E mutations do not disrupt the Num1CC-Mdm36 interaction (Fig. S3). Together, these data indicate that the reduced ability of Num1CC to bind phospholipid membranes disrupts mitochondrial tethering and distribution and, therefore, a direct interaction between Num1 and the MOM is critical for MECA function.
Here, we identify a novel membrane binding domain within the CC of Num1 that interacts directly with the mitochondrial membrane. Thus, Num1 contains two membrane binding domains, a CC domain and PH domain, that preferentially interact with organelle-specific lipids: CL in mitochondria and PI4,5P2 in the plasma membrane (Yu et al., 2004). The two membrane binding domains of Num1 are at opposite ends of an extended structure. This extended structure may be required to accommodate yet a third membrane, cortical ER, at sites of mitochondria–plasma membrane tethering (Lackner et al., 2013). The direct membrane interactions mediated by the two lipid binding domains of Num1 are critical for MECA-mediated tethering, and our data suggest integral membrane domains are not required. This is in contrast to many other interorganelle tethers, such as the ER–mitochondria encounter structure, in which one or more protein components are integral membrane proteins (Kornmann et al., 2009; Prinz, 2014). Thus, our data suggest a mechanism for interorganelle tethering in which membrane binding domains with different lipid specificities mediate direct contact of a protein, or protein complex, with two distinct organelles.
Num1CC has been suggested to be a dimeric BAR (Bin/Amphiphysin/Rvs-like) domain (Tang et al., 2012). BAR domains are membrane-interacting domains that sense, generate, and/or stabilize specific membrane structures (Frost et al., 2009). The direct Num1CC–phospholipid membrane interaction we observe is consistent with a BAR domain–like function. Num1CC has been proposed to associate with or generate specific structures on the plasma membrane (Tang et al., 2012; Klecker et al., 2013). We find that Num1CC does not associate with PI4,5P2 in vitro (Fig. S1 B) or with the plasma membrane in cells. Our data indicate that Num1CC associates directly with the MOM. Num1CC preferentially associates with CL and PA, which are negatively charged, cone-shaped phospholipids. This phospholipid preference suggests that, rather than interacting with a specific phospholipid, Num1CC recognizes a specific membrane structure on the mitochondrial surface. This membrane structure is likely formed and/or maintained by CL and additional factors, such as proteins and other cone-shaped phospholipids that reside in the MOM or mitochondrial outer–inner membrane contact sites (MMCSs). CL is not essential for MECA-mediated mitochondrial tethering, as cells lacking CL exhibit normal mitochondrial distribution (Chen et al., 2010). Therefore, additional factors likely compensate for the lack of CL when CL synthesis is disrupted. For example, PE, a cone-shaped but neutral lipid, has overlapping functions with CL. Cells can survive in the absence of either CL or PE, but the absence of both is lethal (Gohil et al., 2005). The idea that other factors can compensate for CL is further supported by the finding that mitochondria in cells lacking Ups1, a mitochondrial lipid transfer protein, have decreased levels of CL but increased levels of PA, most notably at MMCSs (Connerth et al., 2012). The membrane structures Num1CC associates with are, at this point, unknown. However, given that negatively charged, cone shaped lipids are enriched at the MMCSs (Simbeni et al., 1991; Connerth et al., 2012), Num1 could specifically bind to these sites.
CL plays critical structural and regulatory roles within mitochondria (Claypool and Koehler, 2012), and MMCSs serve as functional hubs for protein import and phospholipid transport, processes critical for mitochondrial function (Tatsuta et al., 2014; Horvath et al., 2015). Similarly, PI4,5P2 serves both structural and signaling roles at the plasma membrane and is a key regulator of many cellular processes (Strahl and Thorner, 2007). Thus, Num1 interacts with phospholipids and/or membrane structures in the MOM and plasma membrane that serve as hubs for key cellular functions. In this context, we speculate that direct interaction with these functionally relevant phospholipids serves to provide spatial and temporal regulation to MECA function and a mechanism to integrate mitochondria–plasma membrane anchoring with mitochondrial and cellular function.
Materials and methods
Strains and plasmids
Strains W303 (ade2–1; leu2–3; his3–11, 15; trp1–1; ura3–1; can1–100; Naylor et al., 2006) and W303 NUM1-yEGFP::HIS, W303 NUM1-yEmCherry::HIS, and W303 fzo1-1 Δdnm1::HIS Δnum1::KAN (Lackner et al., 2013) were described previously. Strain WDH Δpep4 (ura3-52; trp1; leu2-delta1; his3-delta200; pep4::HIS3; prb1-delta1.6R; can1 GAL; Δpep4) was a gift from W.D. Heyer (University of California, Davis, Davis, CA). The yeast two-hybrid strains Y187 and Y2HGold were purchased from Takara Bio Inc.
The plasmids pVT100-GFP (matrix-targeted GFP; Westermann and Neupert, 2000), pXY142-mito-dsRED (mito-RED; Friedman et al., 2011), pAGL (agift from D.E. Gottschling, Fred Hutchinson Cancer Research Center, Seattle, WA; Veatch et al., 2009), pLY23 (a gift from E.L. Weiss, Northwestern University, Evanston, IL; Louvion et al., 1993), p414GalL and p414MET25 (Mumberg et al., 1994), pET22b (EMD Millipore), pGBKT7 and pGADT7 (Takara Bio Inc.), pYES-TagBFP (Murley et al., 2013), p426-MET25::STII-T7 (Lackner et al., 2009), pGADT7-BamHI, pGBKT7 BamHI-Mdm36, pGADT7 BamHI-Num1CC, p414MET25::yEGFP, p416MET25::yEGFP-Num1, p414MET25::yEGFP-Num1, and p416GPD::Tom70-GFP, p416GPD::GFP-PH and p416GPD::Tom70mito-GFP-Num1PH (Lackner et al., 2013) were described previously.
The following W303 gene deletion strains were obtained by replacing the complete ORF of the genes by the indicated cassette using PCR-based targeted homologous recombination: Δnum1::HIS3MX6, Δmdm36::HISMX6, and Δmdm36::NATNT2 (Longtine et al., 1998). The functional C-terminally tagged strains NUM1-yEGFP::KAN (Num1-GFP), NUM1-yEGFP::HIS (Num1-GFP), NUM1-yEmCherry::HISMX6 (Num1-Ch) were constructed by PCR-based targeted homologous recombination using yEGFP::KAN (Sheff and Thorn, 2004), yEGFP::SpHIS5MX6 (Sheff and Thorn, 2004), and yEmCherry-SpHIS5MX6 (Lackner et al., 2013). The functional N-terminally tagged strain GalL::yEGFP-NUM1::NATNT2 (GFP-Num1) was constructed by PCR-based targeted homologous recombination using pYM-N29 (Janke et al., 2004). Haploid double-mutant/tagged strains were generated by crossing, followed by sporulation or by PCR-based targeted homologous recombination. Diploid strains were generated by mating haploid strains.
Strains expressing GAL4-EstrogenBD-VP16::NATMX6 or GAL4-EstrogenBD-VP16::URA3, which are used for estradiol control of GAL promoters, were constructed by transforming Mss1-linearized pAGL (Veatch et al., 2009) or Bsp1191-linearized pYL22 (Louvion et al., 1993), respectively, into W303 Δnum1::HIS, Δnum1::HIS Δmdm36::HIS, Δnum1::HIS Δmdm36::NAT.
p412ADH1::mito-tagBFP (mito-BFP) was constructed by subcloning mito-TagBFP from pYES-TagBFP into p412ADH1 using SpeI–XhoI sites. p414-GalL::Num1CC-GFP-Num1PH (CC-GFP-PH) was constructed in multiple steps. First, p414MET25::Tom70mito-GFP-Num1PH was constructed by subcloning Tom70mito-GFP-Num1PH from p416GPD::Tom70mito-GFP-Num1PH into p414MET25 using BamHI–XhoI sites. Num1CC (amino acids 94–294) was PCR amplified and inserted into p414MET25::Tom70mito-GFP-Num1PH using BamHI–SalI sites to construct p414MET25::Num1CC-GFP-Num1PH. Num1CC-GFP-Num1PH was then subcloned from p414MET25::Num1CC-GFP-Num1PH into p414GalL using BamHI–XhoI sites. p414GalL::Num1CC-GFP-Num1PH was transformed into a W303 Δnum1::HIS GAL4-EstrogenBD-VP16::NATMX6 strain along with pXY142-mito-dsRED.
p414GalL::Num1CC-GFP (CC-GFP) was constructed in multiple steps. First, p414GPD::Tom70-GFP was constructed by subcloning Tom70-GFP from p416GPD::Tom70-GFP into p414GPD using BamHI–XhoI sites. GFP was then subcloned from p414-GPD::Tom70-GFP and inserted into p414-GalL::Num1CC-GFP-PH using PacI–XhoI sites. To construct p414-GalL::Num1CC-GFP-Num1CC (CC-GFP-CC), Num1CC was subcloned from pGADT7 BamHI-Num1CC using BamHI–XhoI sites and inserted into p414-GalL::Num1CC-GFP-Num1PH digested with BglII–XhoI. p414GalL::Num1CC-GFP and p414-GalL::Num1CC-GFP-Num1CC were transformed into W303 Δnum1::HIS GAL4-EstrogenBD-VP16::NATMX6 and W303 Δnum1::HIS Δmdm36::NAT GAL4-EstrogenBD-VP16::URA3 strain along with pXY142-mito-dsRED.
p416GalL::yEGFP-Num13E (GFP-Num13E), which harbors the mutations K121E+R262E+R265E, was constructed in multiple steps. First, the wild-type Num1 sequence in p416MET25::yEGFP-Num1 was replaced with the Num13E sequence by gap repair in a Δnum1::HIS GAL4-EstrogenBD-VP16::NATMX6 strain (Orr-Weaver and Szostak, 1983; Oldenburg et al., 1997; Andersen, 2011). Num1CC3E was ordered as a gBlock gene fragment from Integrated DNA Technologies and amplified by PCR with additional regions of sequence homology to the template plasmid. After gap repair transformation, plasmids were sequenced to verify the 3E mutations were present. Gap repair was then used to replace the MET25 promoter with the GalL promoter.
To construct pGADT7 BamHI::Num1CC3E, Num1CC3E was amplified from the gBlock containing the Num1CC3E sequence by PCR and inserted into pGADT7 BamHI using BamHI–XhoI sites.
p414GalL::yEGFP-Num1 (GFP-Num1) was constructed in multiple steps. yEGFP from p414MET25::yEGFP was subcloned into p414GalL using SpeI–BamHI. Num1 was then subcloned from p416MET25::yEGFP-Num1 and inserted into p414GalL::yEGFP using BamHI–XhoI sites.
pET22b His6-T7 was used to overexpress proteins in E. coli. To construct pET22b His6-T7, a fragment encoding the 6xHis-T7tag (MGSSHHHHHHGMASMTGGQQMGGS), which was generated by annealing oligos, was cloned into pET22b using NdeI–BamHI sites. To construct pET22b His6-T7-Num1CC, Num1CC was subcloned from pGADT7 BamHI-Num1CC using BamHI–XhoI sites. To construct pET22b His6-T7-Mdm36, Mdm36 was amplified via PCR from genomic DNA, digested with BglII–XhoI, and inserted into pET22b His6-T7, which was digested with BamHI–XhoI. To construct pET22b His6-T7-Num1CC3E, the mutant sequence was amplified via PCR from a designed gBlock and inserted into pET22b H6-T7 using BamHI–XhoI sites.
To construct p426-MET25::STII-T7-Num1CC-GFP, Num1CC-GFP was amplified via PCR from p414GalL::Num1CC-GFP and inserted into p426-MET25::STII-T7 using gap repair. STII-T7 designates the StrepII tag affinity tag and T7 epitope tag.
Imaging and image analysis
To visualize the differentially tagged Num1 fusions, W303 NUM1-yEmCherry/NUM1-yEGFP and W303 NUM1-yEmCherry/GalL::yEGFP-NUM1 diploid strains harboring mito-BFP were grown in SC-URA-LEU + DEX, 2× Ade, and 1 nM estradiol. The concentration of estradiol used resulted in steady-state levels of GFP-Num1 that were comparable to Num1-GFP expressed from the endogenous Num1 promoter as assessed by Western blot analysis of whole-cell extracts using rabbit anti-GFP (A11122; Thermo Fisher Scientific) as the primary antibody and goat anti–rabbit IgG DyLight 800 (Thermo Fisher Scientific) as the secondary antibody. The immunoreactive bands were detected with the Odyssey Infrared Imaging System (LI-COR Biosciences) and quantified using the accompanying software.
To visualize CC-GFP, CC-GFP-CC, and CC-GFP-PH, W303 GAL4-EstrogenBD-VP16 Δnum1::HIS and Δnum1::HIS Δmdm36::NAT strains harboring p414-GalL::Num1CC-GFP, p414-GalL::Num1CC-GFP-Num1CC, or p414-GalL::Num1CC-GFP-Num1PH, respectively, and pXY142-mito-dsRED were grown in SC-LEU-TRP + DEX media with 2× Ade and 0.5 nM estradiol.
To visualize wild-type Num1 and Num13E tether points, W303 GAL4-EstrogenBD-VP16 Δnum1::HIS and Δnum1::HIS Δmdm36::NAT or Δnum1::HIS Δmdm36::HIS, respectively, strains harboring p414GalL::yEGFP-Num1 or p416GalL::yEGFP-Num13E and pXY142-mito-dsRED were grown in SC-LEU-TRP + DEX or SC-LEU-URA + DEX, respectively, and 1 nM estradiol.
For all imaging, cells were grown to mid-log phase, concentrated by centrifugation, and mounted on a 4% wt/vol agarose pad. All imaging was performed at 24°C. To image the differentially tagged Num1 fusions and the localization of GFP-Num1 and GFP-Num13E in Δnum1 cells, z-series of cells were imaged at a single time point using a DeltaVision Core microscope fit with a U PLAN S APO 100×, 1.4 NA objective (Olympus) and CoolSnapHQ2 Camera (Photometrics). A step size of 0.2 µm was used. The images were captured and deconvolved using softWoRx’s (Applied Precision Ltd.) iterative, constrained three-dimensional deconvolution method. ImageJ and Photoshop (Adobe) were used to make linear adjustments to brightness and contrast. Deconvolved, smoothed images are shown.
To image CC-GFP, CC-GFP-CC, and CC-GFP-PH and to quantify tether points in Δnum1 cells expressing GFP-Num1 and GFP-Num13E, z-series of cells were imaged at a single time point or over time using a Spinning Disk Confocal System (Leica) fit with a CSU-X1 spinning disk head (Yokogawa Electric Corporation), a PLAN APO 100×, 1.44 NA objective (Leica), and an Evolve 512 Delta EMCCD camera (Photometrics). A step size of 0.4 µm was used. Image capture was done using Metamorph (Molecular Devices). The images were deconvolved using AutoQuant X3’s (Media Cybernetics) iterative, constrained three-dimensional deconvolution method. FIJI and Photoshop (Adobe) were used to make linear adjustments to brightness and contrast. Deconvolved images are shown. FIJI was used to measure the fluorescent intensity of cells expressing GFP-Num1 and GFP-Num13E, and cells with similar fluorescence intensities were used to quantify tether points and mitochondrial morphology.
For linescan analyses, lines were drawn through a region of interest in ImageJ/FIJI. Intensity profiles across the line were generated for all channels imaged and were exported to Excel (Microsoft). The data were normalized so that the peak intensity for each channel was set to 1. The position of peak Num1-Cherry or mito-RED intensity was set at 0 µm for the data shown in Figs. 1, 4, and S2. The mean intensity and standard deviation were generated for all lines and plotted using GraphPad (Prism).
Protein purification
Num1CC, Num13E, and Mdm36 were purified from E. coli as follows: Starter cultures of BL21(λDE3)/RIPL cells harboring plasmids pET22b His6-T7-Num1CC, pET22b His6-T7-Num1CC3E, or pET22b His6-T7-Mdm36, from which expression of the genes is driven by the T7 promoter, were grown overnight in Luria–Bertani medium with 150 µg/ml ampicillin, 25 µg/ml chloramphenicol, and 0.04% glucose. The starter cultures were used to inoculate 2 l Luria–Bertani medium with 150 µg/ml ampicillin, 25 µg/ml chloramphenicol, and 0.04% glucose. The cells were grown at 37°C until an OD600 of 0.5 was reached. To induce protein expression, IPTG was added to 250 µM and growth was continued for 16 h at 18°C. The cells were then harvested by velocity centrifugation at 3,500 g for 15 min. The resulting pellet was resuspended in 1/200 volume of resuspension buffer (RB; 20 mM Tris, pH 8.0, 500 mM NaCl, and 1.89 mM 2-mercaptoethanol), quickly frozen in liquid N2, and stored at −80°C. The cell suspension was quickly thawed in a room-temperature water bath, PMSF was added to 0.5 mM, and the thawed cell suspension was subjected to two more freeze–thaw cycles. The homogenate was sonicated briefly to further lyse cells and clarified by centrifugation at 17,000 g for 45 min at 4°C. Num1CC, Num1CC3E, or Mdm36 was purified from the supernatant using HisPur Ni-NTA resin (Thermo Fisher Scientific). The supernatant was incubated with resin at 1/7 volume for 1 h at 4°C, and the resin was then pelleted at 3,000 g for 3 min. The resin was washed three times with RB and three times with wash buffer (RB + 30 mM imidazole) and loaded into a chromatography column. Protein was eluted from the column using a step gradient of RB + 60–300 mM imidazole. Four 500-µl elutions were collected for each imidazole step: 60, 90, 120, 150, and 300 mM imidazole. Then, 5 µl of each elution was mixed with sample buffer, run on a SDS-PAGE gel, and Coomassie stained. Num1CC, Num1CC3E, or Mdm36 containing elutions were pooled and dialyzed overnight in 20 mM Tris, pH 8.0, 500 mM NaCl. Glycerol was added to 10%, and the protein was aliquoted, frozen in liquid N2, and stored at −80°C. The concentration of the purified proteins was determined using a BCA protein assay kit (Thermo Fisher Scientific).
STII-T7-Num1CC-GFP (Num1CC-GFP) was purified from yeast as follows: Starter cultures of WDH Δpep4 harboring plasmids to overexpress Num1CC-GFP (p426-MET25::STII-T7-Num1CC-GFP) were grown overnight in SC-URA + MET. Each starter culture was used to inoculate 4 liters of SC-URA-MET at an OD600 of 0.0075, and the cells were grown at 30°C until an OD600 of 1.5–3.0 was reached. The cells were then harvested by centrifugation at 3,000 g for 5 min and washed with sterile water. The resulting cell pellet was resuspended in an equal volume of lysis buffer (50 mM Hepes, pH 8.0, and 300 mM NaCl) and 1× Protease Inhibitor Cocktail (Invitrogen) was added. The cell-buffer suspension was added dropwise to liquid N2 to create yeast cell-buffer pellets, and the pellets were stored at −80°C. The frozen cells were lysed using a cryogenic freezer mill (Mixer Mill MM400; Retsch), and the resulting cell lysate powder was thawed in a room-temperature water bath. 1× Protease Inhibitor Cocktail (Invitrogen) was added to the lysate, and large cellular debris was removed with a low-speed spin (3,000 g) for 10 min at 4°C. The resulting supernatant was subjected to a high-speed spin (17,000 g) for 45 min at 4°C. The resulting supernatant was incubated on ice for 10 min with 400 µl BioLock, Biotin Blocking Solution (IBA), and 2 M NaOH was added to adjust pH to 7.0–8.0. The supernatant was then passed over a 0.5-ml column bed of Strep-Tactin Superflow Agarose (IBA) that had been preequilibrated in lysis buffer. The affinity matrix was washed with 5 ml lysis buffer, and Num1CC-GFP was eluted using lysis buffer + 2.5 mM desthiobiotin (Sigma-Aldrich). Glycerol was added to 20%, and the protein was aliquoted, frozen in liquid N2, and stored at −80°C. The concentration of the purified protein was determined using the BCA Protein Assay (Thermo Fisher Scientific).
SEC-MALS
SEC experiments were performed using a Superdex 200 HR column attached to an inline Wyatt Dawn Heleos II MALS detector and an Optilab T-rEX differential refractometer. Samples were prepared from protein dialyzed overnight in 20 mM Hepes, pH 7.0, and 450 mM NaCl using Thermo Fisher Scientific Slide-A-Lyzer Dialysis Cassettes. Dialyzed protein was centrifuged for 4 min at 3,000 g at 4°C to remove debris. Samples containing ∼300 µg of protein were injected into the instrument. The light scattering and refractive index profiles of the various molecular species were recorded. The experiments were conducted in a degassed 20 mM Hepes, pH 7.0, and 450 mM NaCl buffer. The refractive index increment (dn/dc) of the solute/solvent system was calibrated with known standards before analysis. The molar mass of each resolved species was quantified using the ASTRA software (Wyatt Technology). The data were plotted using GraphPad Prism.
Sucrose gradient sedimentation assays
Purified proteins were added to a continuous sucrose gradient. Continuous sucrose gradients were prepared by overlaying 2.5 ml of 17.5% sucrose in gradient reaction buffer (GRB; 20 mM Hepes, pH 7.0, and 150 mM NaCl) with 2.5 ml of 2.5% sucrose in GRB in a 13 × 51 mm polyallomer centrifuge tube (Beckman Coulter). The centrifuge tube was sealed with parafilm, set horizontally for 2 h at room temperature, then set vertically for 1 h at 4°C. A molecular weight marker mix (HMW Native Marker kit; GE Healthcare) was added to a separate continuous sucrose gradient. Sucrose gradients were centrifuged in an SW55 rotor (Beckman Coulter) at 100,000 g at 4°C for 16 h. Then, 500-µl fractions were pipetted from the top and the pellet was resuspended in 500 µl of 17.5% sucrose in GRB, resulting in a total of 11 fractions. Equal volumes of each fraction from the molecular weight marker gradients were analyzed by SDS-PAGE followed by silver stain analysis. Equal volumes of each fraction from the purified protein gradients were analyzed by SDS-PAGE followed by Western blot analysis using mouse monoclonal anti-T7 (EMD Millipore) for the primary antibody and goat anti–mouse IgG Dylight 680 (Thermo Fisher Scientific) for the secondary antibody. The immunoreactive bands were detected with the Odyssey Infrared Imaging System (LI-COR Biosciences).
Liposome floatation assays
Phospholipids were supplied in chloroform from Avanti Polar Lipids, Inc. For soybean + 6% CL liposomes (SoyCL), soybean lipid polar extract was mixed with CL to achieve a final mol% of 6% CL. For OMC liposomes, palmitoyl-oleoyl phosphatidylcholine, palmitoyl-oleoyl PE, soybean PI, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (PA), 1-palmitoyl-2-oleoyl-sn-glycero-3-[phosphor-l-serine] (PS), tetraoleoyl-cardiolipin (CL), all at 10 mg/ml in chloroform, were mixed to achieve a composition that mimics the mitochondrial outer membrane: 46% PC, 33% PE, 10% PI, 4% PA, 1% PS, 6% CL (Zinser and Daum, 1995). For OMC + 0% CL and OMC + 17% CL, compensatory changes were made in the percentage of PC present in the lipid mixture. Headgroup-labeled lissamine rhodamine B phosphatidylethanolamine was added to all liposome mixtures in trace amounts.
Lipid mixtures were placed in a vacuum chamber overnight. The lipid films were rehydrated with 20 mM Hepes, pH 7.0, to a final lipid concentration of 2 mg/ml at room temperature for 1 h. Lipid mixtures were pipetted up and down to create a heterogeneous population of liposomes. Purified proteins and liposomes, as indicated, were added to GRB for a total volume of 100 µl. This reaction was left at room temperature for 20 min, and 400 µl of 50% sucrose in GRB was added to the reaction mixture and then added to the bottom of a 13 × 51–mm polycarbonate centrifuge tube (Beckman). The reaction plus sucrose mixture was overlaid with 1 ml 30% sucrose in GRB, 500 µl 10% sucrose in GRB, and 250 µl 0% sucrose in GRB, for a total volume of 2.5 ml. Sucrose gradients were subjected to centrifugation in a Beckman SW55 rotor at 200,000 g at 4°C for 2 h. Two 1.25-ml fractions were pipetted from the top, resulting in a top fraction and bottom fraction. To monitor the efficiency of the liposome floats, the rhodamine fluorescence of each fraction was quantified using a SpectraMax M5 plate reader (Molecular Devices) with the excitation and emission monochromators set at 550 nm and 590 nm, respectively. In all cases, >85% of liposomes were observed in the top fraction. To quantify the fraction of protein that floated with the liposomes, equal volumes of top and bottom fractions were analyzed by SDS-PAGE followed by Western analysis using mouse monoclonal anti-T7 (EMD Millipore) for the primary antibody and goat anti–mouse IgG Dylight 680 (Thermo Fisher Scientific) for the secondary antibody. The immunoreactive bands were detected with the Odyssey Infrared Imaging System (LI-COR Biosciences) and quantified using the accompanying software. For the high-salt floats, the 150 mM NaCl in GRB was replaced with 450 mM NaCl.
Mitochondrial pelleting assays and fluorescence microscopy
Mitochondria were isolated as previously described (Meeusen and Nunnari, 2007). In brief, 2 d before isolation, a 50 ml starter culture of W303 Δnum1::KAN TOM70-HA::HIS cells harboring plasmids expressing matrix-targeted GFP or mito-RED was inoculated into synthetic minimal complete media. The next day, a 2-l Erlenmeyer flask containing rich yeast extract/peptone/glycerol/ethanol medium was inoculated to an OD600 of 0.1 and placed on a rotary shaker at 200 rpm until an OD600 of 0.8–1.0 was reached. Cells were harvested by centrifugation at 3,000 g for 5 min and washed once in distilled H2O. Cells were then resuspended at 20 OD600/ml in Tris-β-me buffer and shaken at ∼30 rpm for 30 min at room temperature. Cells were pelleted at 1,900 g for 5 min and washed in 35 ml 1.2 M sorbitol. Cells were then resuspended at 50 OD600/ml in 1.2 M sorbitol containing 4 mg/ml zymolyase and shaken at ∼30 rpm for 1 h. Generated spheroplasts were then pelleted at 1,900 g for 5 min and washed with 50 ml 1.2 M sorbitol and repelleted. The spheroplast pellet was resuspended in 2 ml cold NMIB buffer (NMIB; 20 mM Hepes, pH 7.4, 100 mM KOAc, 50 mM KCl, 5 mM MgCl2, and 0.6 M sorbitol) and immediately transferred to chilled tight dounces that were prewashed with NMIB. The resuspension was dounced 100 times, transferred to a 50-ml conical vial, and subjected to a 2,400-g spin for 5 min. The resulting supernatant was transferred to a new vial and pelleted at 13,200 g for 15 min to yield a mitochondrial-enriched pellet. The mitochondrial pellet was resuspended in NMIB to a concentration of 1.5 mg/ml. Protein concentration was determined using a Bradford assay.
For protease treatment, trypsin was added to a final concentration of 50 µg/ml, and the reaction was placed on ice for 20 min. Trypsin was inhibited by adding soybean trypsin inhibitor to a final concentration of 0.5 mg/ml followed by a 15-min incubation on ice. Mitochondria were washed two times with NMIB and resuspended in NMIB at a concentration of 1.5 mg/ml. The absence of Tom70-HA in the treated sample, as analyzed by Western analysis using mouse monoclonal anti-HA (a gift from R.A. Lamb, Northwestern University, Evanston, IL) and goat anti–mouse IgG Dylight 680, was used to monitor the efficiency of the protease treatment.
For the mitochondrial pelleting assay, 1 µM Num1CC (final) was incubated with untreated or trypsin-treated mitochondria labeled with 0.75 mg/ml matrix-targeted GFP (final) in NMIB for 20 min at room temperature. The reactions were then subjected to a 10,000-g spin for 1 min. The supernatant was removed and the pellet was resuspended in an equivalent volume of NMIB. To quantify the fraction of protein that pelleted with mitochondria, equal volumes of supernatant and pellet fractions were analyzed by SDS-PAGE followed by Western analysis using mouse monoclonal anti-T7 (EMD Millipore) and rabbit polyclonal anti-GFP (Thermo Fisher Scientific) for primary antibodies and goat anti–mouse IgG Dylight 680 and goat anti-rabbit IgG Dylight 880 (Thermo Fisher Scientific) for secondary antibodies, respectively. The immunoreactive bands were detected with the Odyssey Infrared Imaging System (LI-COR Biosciences) and quantified using the accompanying software.
To visualize the association of CC-GFP with trypsin-treated mitochondria labeled with matrix-targeted ds-RED, the reactions were set up as described for the pelleting assay. After the 20-min incubation, 4 µl of the reaction was placed on a glass slide and imaged using the Spinning Disk Confocal System described in the Imaging and image analysis section.
Growth assay
For analysis of growth by serial dilution, cells were grown overnight in synthetic complete medium-tryptophan (SC-TRP) + 2% (wt/vol) dextrose (DEX), pelleted, and resuspended in water at a concentration of 0.2 OD600/ml, and tenfold serial dilutions were performed. Cells were spotted onto SC-TRP + DEX medium and were grown at the indicated temperature.
Yeast two-hybrid analysis
Y187 and Y2HGold were transformed with the indicated Gal4AD and Gal4BD fusions, respectively. Y187 cells harboring the indicated Gal4AD fusions then were mated with Y2HGold cells harboring the indicated Gal4BD fusions. Diploids were selected by plating on SC-LEU-TRP + DEX medium, and protein–protein interactions were assessed by growth on SC-LEU-TRP-HIS + DEX medium. The expression of all Gal4AD and Gal4BD fusion proteins was confirmed by Western blot analysis.
Online supplemental material
Fig. S1 provides evidence that Num1CC interacts with liposomes in an electrostatic-dependent manner and does not associate with PI4,5P2. Fig. S2 provides additional evidence that Num1CC associates with mitochondria in cells. Fig. S3 provides evidence that the Num13E mutant interacts with Mdm36. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201511021/DC1.
Supplementary Material
Acknowledgments
We thank Suzanne Hoppins, Jason Brickner, Jodi Nunnari, and members of the Weiss Laboratory for suggestions and critical scientific discussions. We thank staff and instrumentation support from the Biological Imaging and Keck Biophysics Facilities at Northwestern University.
L.M. Kraft is supported by a National Institutes of Health National Institute of General Medical Sciences training grant (T32GM008061) and A.E. Nilles and W. Chen by grant T32GM008382. L.L. Lackner is supported by the Robert H. Lurie Comprehensive Cancer Center – The Lefkofsky Family Foundation/Liz and Eric Lefkofsky Innovation Research Award.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BAR
- Bin/Amphiphysin/Rvs-like
- CC
- coiled coil
- CL
- cardiolipin
- GRB
- gradient reaction buffer
- MECA
- mitochondria–ER cortex anchor
- MMCS
- mitochondrial outer-inner membrane contact site
- MOM
- mitochondrial outer membrane
- Num1CC
- N-terminal coiled-coil domain
- OMC
- outer membrane composition
- PA
- phosphatidic acid
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PH
- pleckstrin homology
- PI
- phosphatidylinositol
- PI4,5P2
- phosphatidylinositol 4,5-bisphosphate
- PS
- phosphatidylserine
- RB
- resuspension buffer
- SEC-MALS
- size exclusion chromatography with inline multiangle light scattering
References
- Andersen E.C. 2011. PCR-directed in vivo plasmid construction using homologous recombination in baker’s yeast. Methods Mol. Biol. 772:409–421. 10.1007/978-1-61779-228-1_24 [DOI] [PubMed] [Google Scholar]
- Cai Q., and Sheng Z.H.. 2009. Mitochondrial transport and docking in axons. Exp. Neurol. 218:257–267. 10.1016/j.expneurol.2009.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny K.L., Studer S.L., Jensen R.E., and Sesaki H.. 2007. Yeast mitochondrial division and distribution require the cortical num1 protein. Dev. Cell. 12:363–375. 10.1016/j.devcel.2007.01.017 [DOI] [PubMed] [Google Scholar]
- Chen S., Liu D., Finley R.L. Jr., and Greenberg M.L.. 2010. Loss of mitochondrial DNA in the yeast cardiolipin synthase crd1 mutant leads to up-regulation of the protein kinase Swe1p that regulates the G2/M transition. J. Biol. Chem. 285:10397–10407. 10.1074/jbc.M110.100784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool S.M., and Koehler C.M.. 2012. The complexity of cardiolipin in health and disease. Trends Biochem. Sci. 37:32–41. 10.1016/j.tibs.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerth M., Tatsuta T., Haag M., Klecker T., Westermann B., and Langer T.. 2012. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 338:815–818. 10.1126/science.1225625 [DOI] [PubMed] [Google Scholar]
- Farkasovsky M., and Küntzel H.. 1995. Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J. Cell Biol. 131:1003–1014. 10.1083/jcb.131.4.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky M., and Küntzel H.. 2001. Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J. Cell Biol. 152:251–262. 10.1083/jcb.152.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., and Voeltz G.K.. 2011. ER tubules mark sites of mitochondrial division. Science. 334:358–362. 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A., Unger V.M., and De Camilli P.. 2009. The BAR domain superfamily: membrane-molding macromolecules. Cell. 137:191–196. 10.1016/j.cell.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohil V.M., Thompson M.N., and Greenberg M.L.. 2005. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. J. Biol. Chem. 280:35410–35416. 10.1074/jbc.M505478200 [DOI] [PubMed] [Google Scholar]
- Hammermeister M, Schodel K, and Westermann B. 2010. Mdm36 is a mitochondrial fission-promoting protein in Saccharomyces cerevisiae. Mol. Biol. Cell. 21:2443–2452. 10.1091/mbc.E10-02-0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine R.A., Oberle J.R., and Cooper J.A.. 2000. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J. Cell Biol. 151:1337–1344. 10.1083/jcb.151.6.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle S.C., Kanfer G., Kolar K., Lang A., Michel A.H., and Kornmann B.. 2013. Organization and function of membrane contact sites. Biochim. Biophys. Acta. 1833:2526–2541. 10.1016/j.bbamcr.2013.01.028 [DOI] [PubMed] [Google Scholar]
- Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., and Shaw J.M.. 1998. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143:359–373. 10.1083/jcb.143.2.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck P.J., and Saxton W.M.. 2005. The axonal transport of mitochondria. J. Cell Sci. 118:5411–5419. 10.1242/jcs.02745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S.E., Rampelt H., Oeljeklaus S., Warscheid B., van der Laan M., and Pfanner N.. 2015. Role of membrane contact sites in protein import into mitochondria. Protein Sci. 24:277–297. 10.1002/pro.2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Toh-E A., and Matsui Y.. 2004. Mmr1p is a mitochondrial factor for Myo2p-dependent inheritance of mitochondria in the budding yeast. EMBO J. 23:2520–2530. 10.1038/sj.emboj.7600271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Klecker T., Scholz D., Förtsch J., and Westermann B.. 2013. The yeast cell cortical protein Num1 integrates mitochondrial dynamics into cellular architecture. J. Cell Sci. 126:2924–2930. 10.1242/jcs.126045 [DOI] [PubMed] [Google Scholar]
- Kormanec J., Schaaff-Gerstenschläger I., Zimmermann F.K., Perecko D., and Küntzel H.. 1991. Nuclear migration in Saccharomyces cerevisiae is controlled by the highly repetitive 313 kDa NUM1 protein. Mol. Gen. Genet. 230:277–287. 10.1007/BF00290678 [DOI] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481. 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé K., Murley A., and Nunnari J.. 2014. Determinants and functions of mitochondrial behavior. Annu. Rev. Cell Dev. Biol. 30:357–391. 10.1146/annurev-cellbio-101011-155756 [DOI] [PubMed] [Google Scholar]
- Lackner L.L. 2014. Shaping the dynamic mitochondrial network. BMC Biol. 12:35 10.1186/1741-7007-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner L.L., Horner J.S., and Nunnari J.. 2009. Mechanistic analysis of a dynamin effector. Science. 325:874–877. 10.1126/science.1176921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner L.L., Ping H., Graef M., Murley A., and Nunnari J.. 2013. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc. Natl. Acad. Sci. USA. 110:E458–E467. 10.1073/pnas.1215232110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Louvion J.F., Havaux-Copf B., and Picard D.. 1993. Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene. 131:129–134. 10.1016/0378-1119(93)90681-R [DOI] [PubMed] [Google Scholar]
- McFaline-Figueroa J.R., Vevea J., Swayne T.C., Zhou C., Liu C., Leung G., Boldogh I.R., and Pon L.A.. 2011. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell. 10:885–895. 10.1111/j.1474-9726.2011.00731.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S.L., and Nunnari J.. 2007. Mitochondrial fusion in vitro. Methods Mol. Biol. 372:461–466. 10.1007/978-1-59745-365-3_32 [DOI] [PubMed] [Google Scholar]
- Mishra P., and Chan D.C.. 2014. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 15:634–646. 10.1038/nrm3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Müller R., and Funk M.. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767–5768. 10.1093/nar/22.25.5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A., Lackner L.L., Osman C., West M., Voeltz G.K., Walter P., and Nunnari J.. 2013. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2:e00422 10.7554/eLife.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor K., Ingerman E., Okreglak V., Marino M., Hinshaw J.E., and Nunnari J.. 2006. Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J. Biol. Chem. 281:2177–2183. 10.1074/jbc.M507943200 [DOI] [PubMed] [Google Scholar]
- Oldenburg K.R., Vo K.T., Michaelis S., and Paddon C.. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25:451–452. 10.1093/nar/25.2.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T.L., and Szostak J.W.. 1983. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. USA. 80:4417–4421. 10.1073/pnas.80.14.4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz W.A. 2014. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol. 205:759–769. 10.1083/jcb.201401126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana A., Schwindling C., Wenning A.S., Becherer U., Rettig J., Schwarz E.C., and Hoth M.. 2007. T cell activation requires mitochondrial translocation to the immunological synapse. Proc. Natl. Acad. Sci. USA. 104:14418–14423. 10.1073/pnas.0703126104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff M.A., and Thorn K.S.. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670. 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Simbeni R., Pon L., Zinser E., Paltauf F., and Daum G.. 1991. Mitochondrial membrane contact sites of yeast. Characterization of lipid components and possible involvement in intramitochondrial translocation of phospholipids. J. Biol. Chem. 266:10047–10049. [PubMed] [Google Scholar]
- Strahl T., and Thorner J.. 2007. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae . Biochim. Biophys. Acta. 1771:353–404. 10.1016/j.bbalip.2007.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne T.C., Zhou C., Boldogh I.R., Charalel J.K., McFaline-Figueroa J.R., Thoms S., Yang C., Leung G., McInnes J., Erdmann R., and Pon L.A.. 2011. Role for cER and Mmr1p in anchorage of mitochondria at sites of polarized surface growth in budding yeast. Curr. Biol. 21:1994–1999. 10.1016/j.cub.2011.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Punch J.J., and Lee W.L.. 2009. A CAAX motif can compensate for the PH domain of Num1 for cortical dynein attachment. Cell Cycle. 8:3182–3190. 10.4161/cc.8.19.9731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Germain B.S., and Lee W.L.. 2012. A novel patch assembly domain in Num1 mediates dynein anchoring at the cortex during spindle positioning. J. Cell Biol. 196:743–756. 10.1083/jcb.201112017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T., Scharwey M., and Langer T.. 2014. Mitochondrial lipid trafficking. Trends Cell Biol. 24:44–52. 10.1016/j.tcb.2013.07.011 [DOI] [PubMed] [Google Scholar]
- Toulmay A., and Prinz W.A.. 2011. Lipid transfer and signaling at organelle contact sites: the tip of the iceberg. Curr. Opin. Cell Biol. 23:458–463. 10.1016/j.ceb.2011.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch J.R., McMurray M.A., Nelson Z.W., and Gottschling D.E.. 2009. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 137:1247–1258. 10.1016/j.cell.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B., and Neupert W.. 2000. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 16:1421–1427. [DOI] [PubMed] [Google Scholar]
- Yu J.W., Mendrola J.M., Audhya A., Singh S., Keleti D., DeWald D.B., Murray D., Emr S.D., and Lemmon M.A.. 2004. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 13:677–688. 10.1016/S1097-2765(04)00083-8 [DOI] [PubMed] [Google Scholar]
- Zinser E., and Daum G.. 1995. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast. 11:493–536. 10.1002/yea.320110602 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.