Mitochondria exert critical functions in lipid metabolism and promote the synthesis of major constituents of cellular membranes, such as phosphatidylethanolamine (PE). Here, Aaltonen et al. demonstrate that two pathways mediate PE synthesis: Ups2–Mdm35–dependent lipid transfer and MICOS-dependent membrane apposition.
Abstract
Mitochondria exert critical functions in cellular lipid metabolism and promote the synthesis of major constituents of cellular membranes, such as phosphatidylethanolamine (PE) and phosphatidylcholine. Here, we demonstrate that the phosphatidylserine decarboxylase Psd1, located in the inner mitochondrial membrane, promotes mitochondrial PE synthesis via two pathways. First, Ups2–Mdm35 complexes (SLMO2–TRIAP1 in humans) serve as phosphatidylserine (PS)-specific lipid transfer proteins in the mitochondrial intermembrane space, allowing formation of PE by Psd1 in the inner membrane. Second, Psd1 decarboxylates PS in the outer membrane in trans, independently of PS transfer by Ups2–Mdm35. This latter pathway requires close apposition between both mitochondrial membranes and the mitochondrial contact site and cristae organizing system (MICOS). In MICOS-deficient cells, limiting PS transfer by Ups2–Mdm35 and reducing mitochondrial PE accumulation preserves mitochondrial respiration and cristae formation. These results link mitochondrial PE metabolism to MICOS, combining functions in protein and lipid homeostasis to preserve mitochondrial structure and function.
Introduction
A defined lipid composition is crucial for the functional integrity of cellular membranes and depends on extensive lipid trafficking between cellular membranes (van Meer et al., 2008). Nonvesicular lipid transport is mediated by lipid transfer proteins, which comprise various conserved protein families with variable degrees of lipid specificity (Lev, 2010). We have identified heterodimeric complexes of yeast Ups1 (human PRELID1) and Mdm35 (human TRIAP1) as novel lipid transfer proteins in mitochondria (Connerth et al., 2012; Potting et al., 2013). These complexes shuttle phosphatidic acid (PA) imported from the ER across the mitochondrial intermembrane space (IMS) to the inner membrane (IM), where it is enzymatically converted to cardiolipin (CL). Impaired transport of PA and reduced CL accumulation compromises mitochondrial function and morphology and renders cells susceptible for apoptosis, illustrating the critical role of mitochondrial lipid trafficking for membrane homeostasis and cell survival (Potting et al., 2013; Tatsuta et al., 2014).
Ups1 and PRELID1 are members of a conserved protein family with two additional homologous proteins expressed in various organisms (Ups2 and Ups3 in yeast; SLMO1 and SLMO2 in humans; Dee and Moffat, 2005; Osman et al., 2009; Tamura et al., 2009). Although they lack sequence similarity with other classes of lipid transfer proteins, the crystal structures of Ups1–Mdm35 and SLMO1–TRIAP1 complexes revealed striking structural similarities with phosphatidylinositol transfer proteins and suggested similar transfer mechanisms (Miliara et al., 2015; Watanabe et al., 2015; Yu et al., 2015). However, the function of other members of the Ups1/PRELID1 family remained enigmatic.
Ups2 assembles with Mdm35 in the IMS and is required to maintain normal levels of PE in mitochondrial membranes, suggesting a role in PE homeostasis (Osman et al., 2009; Tamura et al., 2009). PE can be synthesized within mitochondria through decarboxylation of ER-derived PS by Psd1, which is located in the IM and exposes its catalytic domain to the IMS (Choi et al., 2005; Horvath et al., 2012). A fraction of PE is exported from mitochondria, transferred to the ER, and converted to PC, an abundant phospholipid in cellular membranes (Simbeni et al., 1990; Birner et al., 2001). Thus, mitochondrial PE synthesis is of pivotal importance for the lipid homeostasis of mitochondrial and other cellular membranes. However, it remained unclear how Ups2–Mdm35 complexes affected the accumulation of mitochondrial PE, as the activity of Psd1 and the synthesis of PC were not affected in ups2Δ cells (Osman et al., 2009; Tamura et al., 2012a) and the requirement of intermembrane space proteins for PS trafficking was questioned (Simbeni et al., 1990; Tamura et al., 2012b). Here, we identify Ups2–Mdm35 as PS-specific lipid transfer proteins and unravel an unexpected role of mitochondrial contact sites stabilized by MICOS for mitochondrial PE synthesis.
Results and discussion
Ups2–Mdm35 functions as a PS transfer protein complex
For functional characterization of Ups2–Mdm35, we established an in vitro assay system using the purified protein complex. To facilitate heterologous expression, we designed a Ups2 variant (Ups2*) that lacked cysteine residues but remained functionally active and restored the growth defect caused by the loss of Ups2 in Saccharomyces cerevisiae (Fig. S1 A). His-tagged Ups2* and Mdm35 were coexpressed in Escherichia coli, and the heterooligomeric complex was purified to homogeneity (Fig. S1 B).
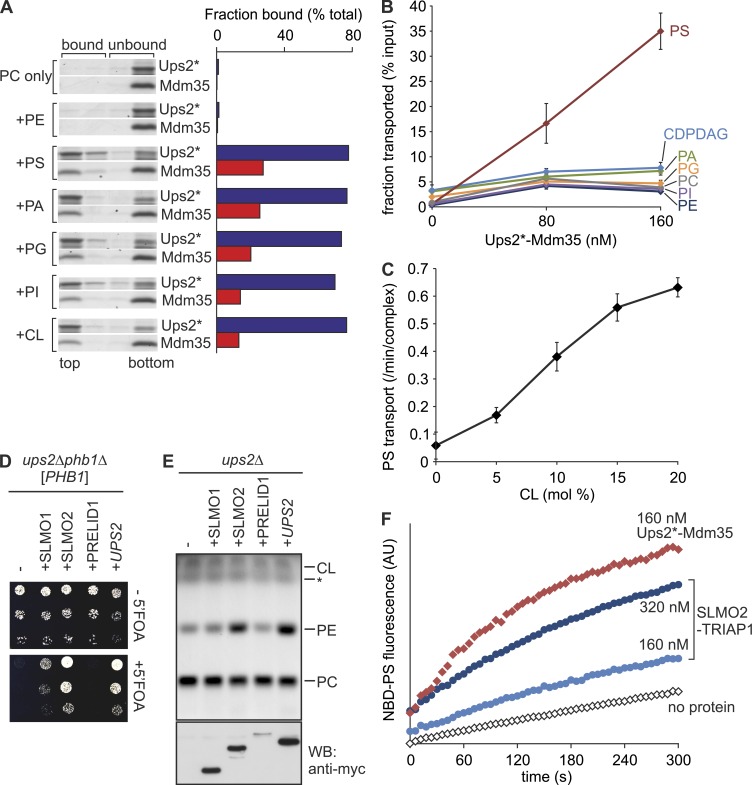
We first assessed whether Ups2*–Mdm35 binds phospholipids and performed flotation experiments using liposomes (Fig. 1 A). Ups2* bound to negatively charged phospholipids including PS and CL but not to neutral phospholipids (PC or PE; Fig. 1 A). A large proportion of Mdm35 dissociated from Ups2* upon binding to the membrane, as described previously for Ups1–Mdm35 (Connerth et al., 2012; Watanabe et al., 2015). To investigate possible phospholipid transfer activity, Ups2*–Mdm35 was incubated with donor liposomes containing phospholipids of all major classes and with acceptor liposomes devoid of these phospholipids (Fig. 1 B). After incubation, acceptor liposomes were isolated by density gradient centrifugation and analyzed by quantitative mass spectrometry (qMS). We observed selective and concentration-dependent transfer of PS to acceptor liposomes by Ups2*–Mdm35 (Fig. 1 B). Other phospholipids such as PA or cytidine diphosphate (CDP)–DAG were not transported or only with very low efficiency (Fig. 1 B). PS transfer by Ups2*–Mdm35 was accelerated by the presence of CL or other negatively charged phospholipids (PI or PG) in membranes, as revealed using a fluorescent-based dequenching assay (Fig. 1 C and Fig. S1, C and D). Thus, Ups2*–Mdm35 facilitates the selective transfer of PS, a lipid specificity that is strikingly different from that of Ups1–Mdm35, which is selective for PA (Connerth et al., 2012).
Figure 1.
Ups2–Mdm35 is a PS transfer protein complex. (A) Phospholipid binding. His-tagged Ups2C96S/C101S/C142S/C153S (Ups2*)–Mdm35 (5 µM) was incubated with liposomes composed of dioleoyl-PC (DOPC) and 20 mol% of the indicated dioleoyl-phospholipid (total lipid concentration 2 mM) for 10 min at 20°C. Upon flotation, four fractions were collected and analyzed by SDS-PAGE. Protein bands were analyzed densitometrically, and the sum of band intensities in all four lanes was set to 100%. The top two fractions represent bound material. (B) Phospholipid transfer. Ups2*–Mdm35 was incubated with donor liposomes (0.25 mM; DOPC/tetraoleoyl-CL/Lac-PE/17:0 PC/17:0 PE/DOPA/DOPS/DOPI/DOPG/DO-CDPDAG/DOPE/NBD-PE = 35/15/10/5/5/5/5/5/5/5/4.5/0.5 mol%, filled with 12.5% sucrose) and acceptor liposomes (1 mM, DOPC/DOPE/tetraoleoyl-CL/Lac-PE/rhodamine-PE = 50/24.95/15/10/0.05 mol%) for 5 min at 16°C. After isolation of acceptor liposomes by flotation, lipids were extracted and quantified by qMS. Error bars represent SEM. n = 4. (C) CL dependence of PS transfer activity by Ups2*–Mdm35. PS transfer was monitored by fluorescence dequenching (Fig. S1 C). Error bars represent SEM. n = 3. (D) Expression of human SLMO2 suppresses the lethality of ups2Δphb1Δ cells. Serial dilutions of ups2Δphb1Δ (PHB1) cells expressing C-terminally myc-tagged SLMO1, SLMO2, or PRELID1 were spotted on glucose-containing media with or without 5-fluoroorotic acid (5′FOA). (E) SLMO2 expression restores PE levels in Δups2 mitochondria. TLC of mitochondrial lipids. *, Unidentified lipid species. Same samples were subjected to SDS-PAGE and immunoblot analysis using anti-myc antibodies (WB). (F) SLMO2–TRIAP1 complex facilitates intermembrane transfer of NBD-PS. Transfer of NBD-PS was monitored by fluorescence dequenching. AU, arbitrary units.
Human mitochondria harbor three Ups/PRELI family proteins, termed SLMO1, SLMO2, and PRELID1 (Dee and Moffat, 2005). Whereas PRELID1–TRIAP1 mediates PA transfer (Potting et al., 2013), the function of SLMO1 and SLMO2 remained enigmatic. To identify the functional ortholog of Ups2, we expressed human Ups/PRELI family members in yeast cells lacking Ups2. Deletion of UPS2 is lethal in yeast cells lacking Phb1, a subunit of prohibitin membrane scaffolds (Osman et al., 2009). Expression of SLMO2 and, to a lesser extent, SLMO1 allowed growth of ups2Δphb1Δ cells (Fig. 1 D). We observed normal PE accumulation in ups2Δ mitochondria harboring SLMO2, whereas expression of SLMO1 or PRELID1 did not significantly affect mitochondrial PE (Fig. 1 E). Consistent with these in vivo results, purified SLMO2–TRIAP1 facilitated intermembrane transfer of NBD-PS but not NBD-PA (Figs. 1 F and S1 E) in vitro. We conclude from these experiments that Ups2–Mdm35 and SLMO2–TRIAP1 are conserved PS transfer proteins.
Mitochondrial PE synthesis can proceed independently of Ups2 in vivo
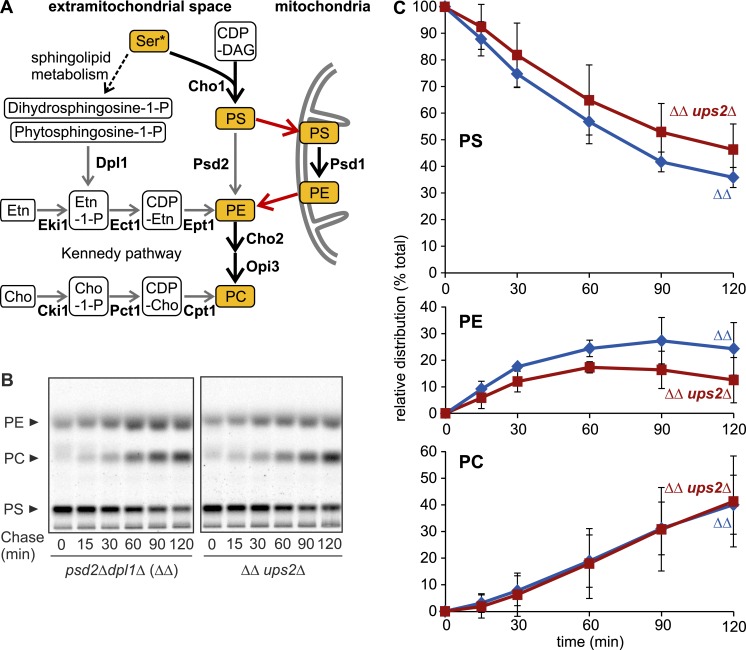
Our in vitro data suggest that Ups2–Mdm35 transports PS across the IMS for Psd1-dependent PE synthesis. We therefore performed pulse-labeling experiments in Ups2-deficient cells using [14C]serine, which is incorporated into PS. PE can be synthesized using different cellular metabolic pathways (Birner et al., 2001; Fig. 2 A). Thus we used yeast cells, which lack Psd2 and Dpl1 (psd2Δdpl1Δ), key enzymes in nonmitochondrial PE synthesis. The formation of PE depended solely on Psd1 in psd2Δdpl1Δ cells in the absence of ethanolamine (Birner et al., 2001; Fig. 2 A). We monitored Psd1-dependent decarboxylation of radiolabeled PS to PE and its subsequent conversion to PC by PE methylases in the ER (Fig. 2 B). Loss of Ups2 in psd2Δdpl1Δ cells (referred to as ΔΔ in the figures) moderately impaired the accumulation of PE but did not affect its conversion to PC (Fig. 2, B and C). These results are consistent with decreased PE levels in ups2Δ mitochondria (Osman et al., 2009; Tamura et al., 2009) and the function of Ups2–Mdm35 as a lipid transfer protein complex in vivo. However, these observations indicate that Psd1-dependent PE synthesis can proceed independently of PS transfer by Ups2–Mdm35 in vivo.
Figure 2.
PE biogenesis can proceed in the absence of Ups2. (A) PE/PC biogenesis in S. cerevisiae. Thick arrows represent the reactions proceeding in the experiments in B and in Fig. 4 A, where nonmitochondrial PE/PC synthesis was shut down by deletion of DPL1 and PSD2 and the lack of ethanolamine (Etn) or choline (Cho) in the medium. Red arrows represent lipid exchange. (B) Assessment of PE/PC synthesis in vivo. After pulse labeling with [14C]serine and further incubation in medium with unlabeled serine (chase), lipids were extracted and analyzed by TLC and autoradiography. ΔΔ, dpl1Δpsd2Δ. (C) Levels of PS, PE, and PC were quantified and are shown as a fraction of total lipids. Error bars represent SD. n = 3.
Psd1 can convert PS in juxtaposed membranes
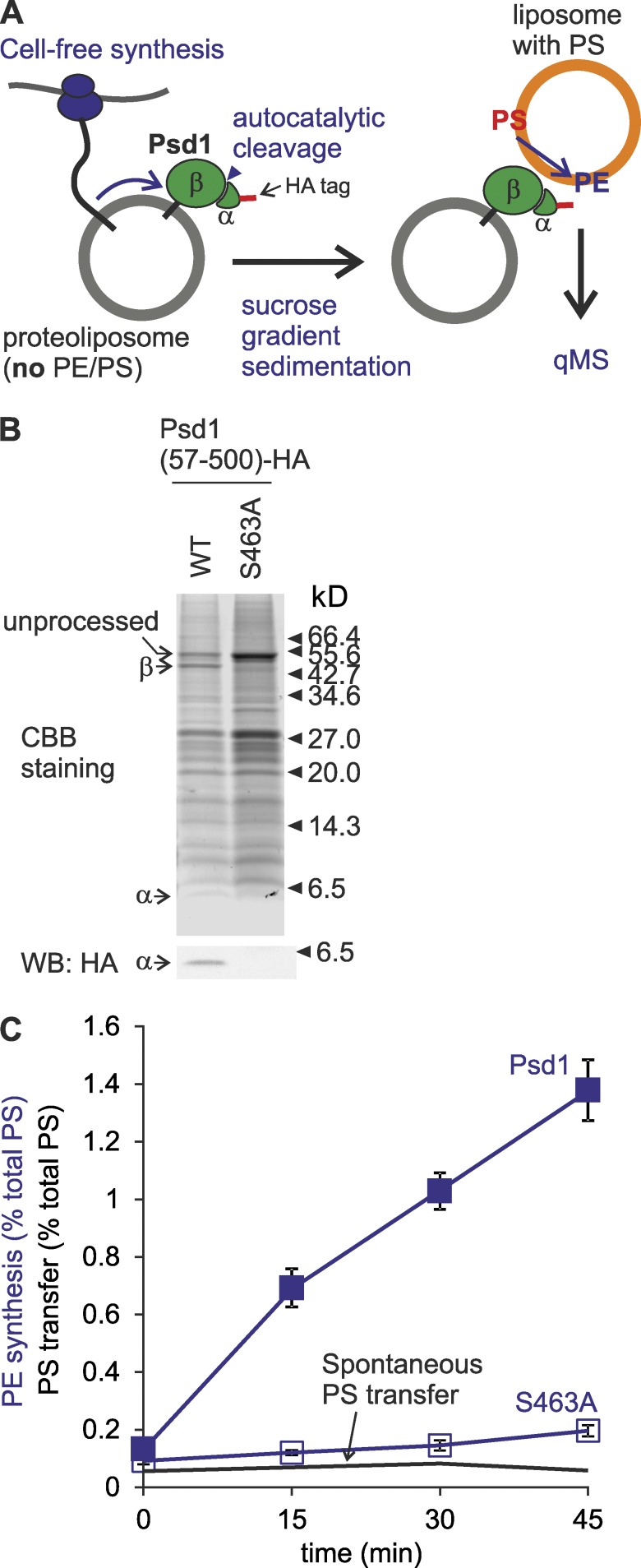
The Ups2-independent PE synthesis by Psd1 in vivo suggests that PS can reach Psd1 in the inner membrane by alternative routes that do not involve PS transfer by Ups2–Mdm35 across the IMS. Other members of the Ups/PRELI family of lipid transfer proteins may at least partially substitute for the loss of Ups2 and preserve PS transport. However, psd2Δdpl1Δ cells lacking all Ups-like proteins (Ups1–3) were not ethanolamine auxotrophs, demonstrating that Psd1-dependent PE synthesis can occur in the absence of these lipid transfer proteins (Fig. S2 A). An alternative scenario is that the IMS catalytic domains of Psd1 (Choi et al., 2005; Horvath et al., 2012) are capable of decarboxylating PS in the outer membrane (OM), that is, independently of PS transfer to the IM. To test this possibility, we synthesized Psd1 in vitro and reconstituted it into liposomes lacking PS and PE (Fig. 3 A). The decarboxylase activity of Psd1 depends on its autocatalytic processing, which is abolished upon mutating catalytically active S463 (Horvath et al., 2012; Onguka et al., 2015). We observed autocatalytic cleavage of a significant fraction of reconstituted Psd1 but not Psd1S463A, indicating that liposome-bound Psd1 reached the functional state (Fig. 3 B). Proteoliposomes harboring Psd1 or Psd1S463A were incubated with liposomes containing PS, and the formation of PE was assessed by qMS (Fig. 3 C). Psd1 but not Psd1S463A promoted PS decarboxylation and the formation of PE in liposomes in trans (Fig. 3 C). Notably, PE synthesis depended on Psd1 and did not involve PS transfer between liposomes, as significant spontaneous PS transfer did not occur (Fig. 3 C). Moreover, no liposome fusion was detected (Fig. S2 B). We conclude that Psd1 can catalyze PS decarboxylation and conversion to PE in a juxtaposed membrane, providing a possible rationale for the Psd1-dependent PE synthesis in the absence of PS transfer by Ups2–Mdm35 in vivo.
Figure 3.
Psd1 is able to convert PS to PE at juxtaposed membranes in trans. (A) Monitoring of trans-decarboxylation by reconstituted Psd1. (B) Autocatalytic processing of reconstituted Psd1. The Psd1-containing fraction after flotation was subjected to SDS-PAGE and immunoblot analysis using HA-specific antibodies. CBB, Coomassie brilliant blue. (C) Psd1 decarboxylates PS to PE at juxtaposed membrane. S463A, Psd1S463A. Spontaneous PS transfer under identical conditions was plotted for comparison. PE produced or PS transferred at given time points are shown as percentage of total PS input. Error bars represent SEM. n = 3.
MICOS coordinates mitochondrial PE metabolism
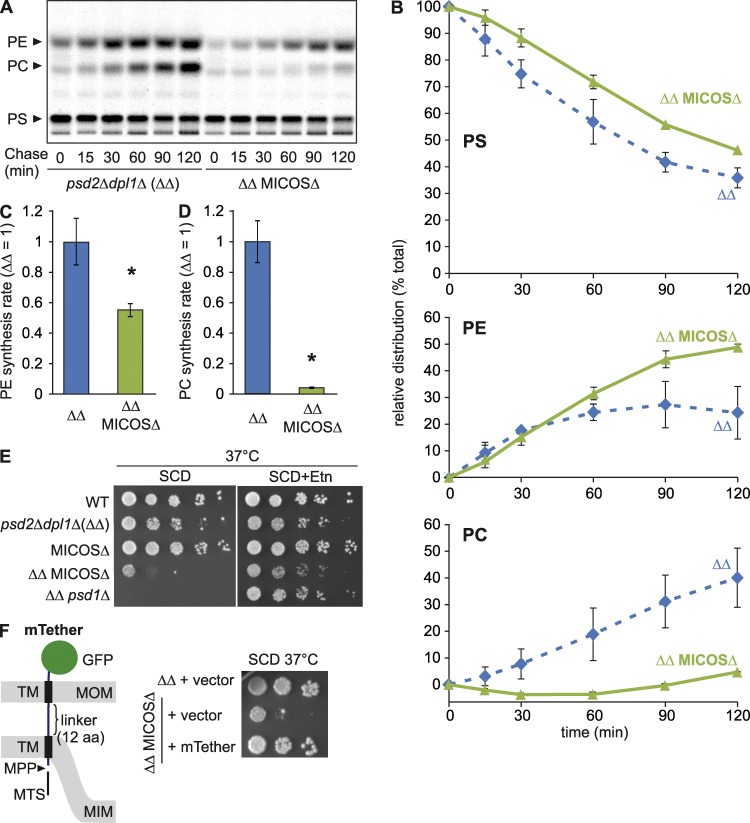
These results imply that mitochondrial PE synthesis may be modulated by the spatial positioning of OM and IM relative to each other. The mitochondrial contact site and cristae organizing system (MICOS) has been identified as a master regulator of mitochondrial shape and organization (Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Alkhaja et al., 2012). Heterooligomeric MICOS complexes in the IM are associated with the maintenance and formation of mitochondrial cristae and membrane contacts to the OM, which support coordinated protein translocation across mitochondrial membranes (Friedman et al., 2015; Horvath et al., 2015). Interestingly, MICOS subunits show strong negative genetic interactions with the cardiolipin synthesis pathway (Hoppins et al., 2011), reminiscent of components involved in the metabolism of PE (Gohil et al., 2005). We therefore pursued the possibility that MICOS may ensure efficient PE synthesis by Psd1. We deleted PSD2 and DPL1 in yeast cells lacking all six MICOS subunits (MICOSΔ; Friedman et al., 2015) to abolish nonmitochondrial PE synthesis (Fig. 2 A) and performed pulse-labeling experiments using [14C]serine in vivo. Loss of MICOS significantly reduced the rate of Psd1-dependent PE synthesis and more severely affected the rate of methylation of PE to PC (Figs. 4, A–D). Determination of the proportion of accumulated PE and PC relative to total PS, PC, and PE revealed a drastic decrease in PC synthesis in MICOS-deficient cells, whereas the relative fraction of PE was increased (Fig. 4 B). This is in contrast to ups2Δ cells (Fig. 2 C), although loss of MICOS complexes or Ups2 caused a similar reduction in the absolute amounts of PE accumulating in the cells (Fig. S2 C).
Figure 4.
MICOS assists mitochondrial synthesis of PE and its conversion to PC in the ER. (A) Synthesis of PS, PE, and PC in cells labeled with [14C]serine. ΔΔ, dpl1Δpsd2Δ. (B) Relative distributions of radioactivity in PS, PE, or PC bands are represented as in Fig. 2 C. Error bars represent SD. n = 3. (C and D) Psd1-dependent formation of PE and PC from [14C]PS. Synthesis rates for PE (C) and PC (D) in cells lacking MICOS are shown. Synthesis rates were obtained by plotting kinetic changes in band intensities of the sum of PE and PC (C) or PC (D) and determining the slope of the fitted curve (between t = 15 min and t = 90 min). Mean values of ΔΔ were set to 1. Error bars represent SEM. n = 3. *, P < 0.05. (E) Loss of nonmitochondrial PE synthesis impairs growth of MICOSΔ cells. Cell growth was assessed at 37°C on glucose-containing SC medium (SCD) supplemented with ethanolamine (Etn; 10 mM) where indicated. WT, wild-type. (F) Expression of an artificial tether between two mitochondrial membranes (mTether) suppresses ethanolamine auxotrophy of cells lacking MICOS and nonmitochondrial PE synthesis. Left, schematic representation of mTether; right, cell growth on SCD at 37°C.
In agreement with a role of MICOS complexes in mitochondrial PE metabolism, inhibition of nonmitochondrial PE synthesis impaired the growth of MICOS-deficient cells on the fermentable carbon source dextrose (Fig. 4 E). Similarly, Psd1 showed a negative genetic interaction in cells lacking Psd2 and Dpl1 (Fig. 4 E). In both cases, growth was supported by supplementation of the medium with ethanolamine, which restores nonmitochondrial PE synthesis, demonstrating that a deficiency in PE biosynthesis causes the observed growth defect (Fig. 4 E). Psd1 activity was not affected by loss of MICOS subunits (Fig. S2 D), suggesting that PS delivery to Psd1 is rate-limiting for mitochondrial PE synthesis in these cells. Together, our data indicate that MICOS complexes modulate mitochondrial PE synthesis and promote the formation of PC in the ER from mitochondrial-derived PE. Thus, we propose that MICOS ensures close apposition of both mitochondrial membranes, allowing decarboxylation of PS in the OM by Psd1.
To assess whether MICOS functions in mitochondrial PE synthesis via tethering of the outer and inner membranes, we generated a chimeric protein containing the mitochondrial targeting signal and transmembrane domain of the IM protein Yme2 (aa 279–308) and the transmembrane domain of the OM protein Tom70 (aa 10–30) fused to GFP (Fig. 4 F). Transmembrane domains were joined by a linker of 12 aa. We expressed the chimeric protein from a centromeric plasmid in MICOS-deficient cells lacking Psd2 and Dpl1, whose growth strictly depends on mitochondrial PE synthesis by Psd1 or supplementation with exogenous ethanolamine (Fig. 4 E). The artificial tether protein accumulated in mitochondria in its mature form and exposed the GFP domain to the outside (Fig. S2, E and F). Expression of the tether protein allowed growth of MICOS-deficient cells in the absence of ethanolamine (Fig. 4 F). Thus, an artificial tether can at least partially replace MICOS in PE metabolism, consistent with a requirement for a close apposition of mitochondrial membranes in PE synthesis by Psd1. However, [14C]serine labeling experiments revealed only moderately increased PE synthesis in these cells, and respiratory growth of MICOS-deficient cells expressing the tether protein was not restored (Fig. S2 G and not depicted), demonstrating that the artificial membrane tether substitutes for only some functions of MICOS as a membrane organizing complex.
Deletion of UPS2 preserves respiratory growth and cristae morphology in the absence of MICOS subunits
Disturbances in PE or CL synthesis impair cristae morphogenesis in various organisms (Signorell et al., 2009; Connerth et al., 2012), highlighting the importance of the phospholipid environment for mitochondrial ultrastructure. Cristae morphology is disturbed upon inhibition of CL synthesis but restored when PA transfer to the IM by Ups1–Mdm35 is impaired (Connerth et al., 2012), indicating that moderate alterations in lipid composition can have dramatic effects on cristae morphogenesis. We therefore examined whether alterations in the mitochondrial PE metabolism contribute to cristae morphology or ultrastructure defects in MICOS-deficient cells.
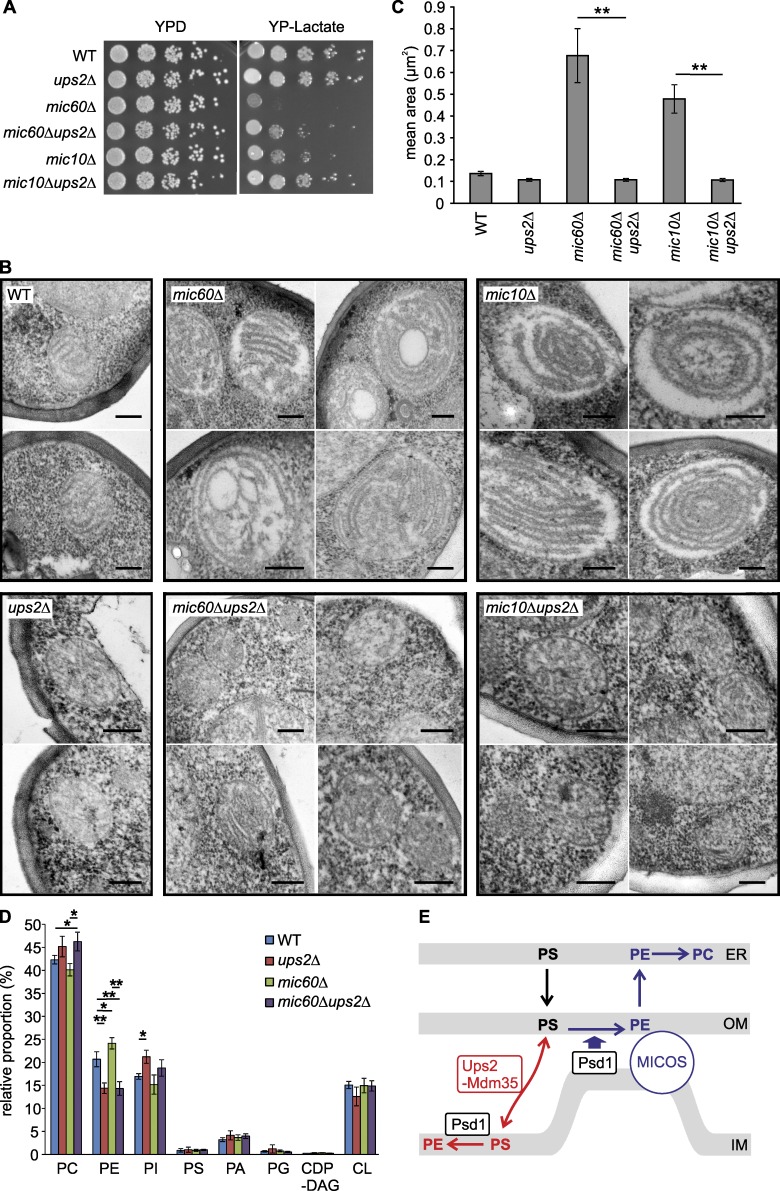
We deleted UPS2 in cells lacking the core MICOS subunits Mic10 or Mic60 and assessed cell growth (Fig. 5 A). The growth of mic10Δ and mic60Δ cells on nonfermentable carbon sources was significantly improved in the absence of Ups2 (Fig. 5 A). Similarly, deletion of UPS2 promoted respiratory growth of MICOSΔ cells (Fig. S3 A) as well as respiratory growth of mic10Δ or mic60Δ cells devoid of extramitochondrial PE synthesis (Fig. S3 B). Electron microscopy of cells lacking MICOS subunits showed enlarged mitochondria with stacked, multilamellar cristae (Rabl et al., 2009; Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Alkhaja et al., 2012; Friedman et al., 2015), whereas the absence of Ups2 did not affect the overall mitochondrial architecture (Fig. 5 B). Deletion of UPS2 grossly restored the mean size of mitochondria and cristae morphogenesis in mic60Δ or mic10Δ cells (Fig. 5, B and C), whereas fluorescence microscopy revealed that tubular mitochondria were not restored in mic60Δ cells (Fig. S3 C). These results demonstrate that cristae morphology and respiratory defects in MICOS-deficient cells can be at least partially bypassed by inhibiting Ups2-mediated PS transfer to the inner membrane.
Figure 5.
Deletion of UPS2 restores respiratory growth and mitochondrial architecture in cells lacking MICOS. (A) Cell growth on fermentable and nonfermentable carbon sources. Serial dilutions of the indicated strains were analyzed on YPD or YP-lactate plates at 30°C. WT, wild-type. (B) EM analysis of cells lacking Ups2, Mic60, and/or Mic10. Representative micrographs of cells grown on YP-lactate are shown. Bars, 200 nm. (C) Mean area of mitochondria in electron micrographs from indicated strains. Error bars represent SEM. n > 15. **, P < 0.01. (D) qMS of the mitochondrial phospholipidome in cells grown in lactate medium. Error bars represent SEM. n = 3. *, P < 0.05; **, P < 0.01. (E) Model for the roles of Ups2–Mdm35 and MICOS for Psd1-dependent PE synthesis.
To corroborate these findings, we determined the mitochondrial phospholipid profile in mic60Δ, ups2Δ, and mic60Δups2Δ cells by qMS (Figs. 5 D and S3 D). The loss of Mic60 impaired mitochondrial architecture (Fig. 5 B) but did not significantly affect the phospholipid composition in mitochondrial membranes or the acyl-chain distribution in various PE species (Figs. 5 D and S3 D). Deletion of UPS2 resulted in decreased PE levels in mitochondria (Osman et al., 2009; Tamura et al., 2009), which was also evident in the absence of Mic60 or all MICOS subunits (Figs. 5 D and S3 E). We observed an altered acyl-chain composition of PE in ups2Δ and ups2Δmic60Δ mitochondria (Fig. S3 D). Thus, loss of Ups2 leads to significant alterations in PE levels in mitochondrial membranes of mic60Δ cells and grossly restores mitochondrial ultrastructure. The beneficial effect of decreased PE levels for mitochondrial function in MICOS-deficient cells was further substantiated by deleting PSD1 (Fig. S3 F). Deletion of PSD1 in wild-type cells drastically reduced PE accumulation in mitochondria and impaired respiratory growth (Birner et al., 2001). However, loss of Psd1 in MICOS-deficient cells significantly improved respiratory growth (Fig. S3 F). We therefore conclude that cristae morphogenesis and mitochondrial respiration can be maintained in MICOS-deficient cells by reducing PS transfer to the IM, which limits the accumulation of PE, demonstrating that the membrane lipid environment and mitochondrial structure and function are intimately linked.
Together, our results demonstrate an intimate cooperation of Ups2–Mdm35 lipid transfer proteins with MICOS in mitochondrial PE synthesis, highlighting the importance of membrane contacts for mitochondrial phospholipid homeostasis. We propose that PE is synthesized within mitochondria along two Psd1-dependent pathways (Fig. 5 E): first, Ups2–Mdm35–dependent PS transfer to the IM results in its decarboxylation by Psd1 and the accumulation of PE in the IM; second, MICOS ensures close apposition of mitochondrial membranes, allowing the Psd1-dependent but Ups2–Mdm35–independent formation of PE in the OM. This PE pool can be released from mitochondria and converted to PC in the ER.
Several lines of evidence suggest that lipid exchange between the OM and IM occurs at contact sites (Simbeni et al., 1990). CL binding may recruit Ups2–Mdm35 to these sites, which are enriched in CL (Simbeni et al., 1991; Connerth et al., 2012). MICOS likely facilitates intramitochondrial PS transfer by Ups2–Mdm35 at these sites, supporting a close membrane apposition. However, our results reveal an additional function of MICOS in PE metabolism: the close spatial proximity of both mitochondrial membranes allows Psd1 to decarboxylate PS in the OM. Therefore, Psd1-dependent synthesis of PE and its conversion in PC can be maintained in the absence of Ups2–Mdm35. It will be of interest to examine whether MICOS also enables other IM proteins to act on substrates in the OM, such as the i-AAA protease Yme1 that cleaves Atg32 in the OM of yeast mitochondria (Wang et al., 2013). Moreover, our studies are reminiscent of the role of tethering complexes at plasma membrane–ER contact sites in PC synthesis (Tavassoli et al., 2013). Assisting trans-activity of lipid-modifying enzymes is therefore emerging as a general principle of how membrane contacts contribute to cellular lipid homeostasis.
Lipid transfer activity has been assigned to some tethering complexes between cellular membranes (Reinisch and De Camilli, 2015). Accordingly, MICOS might mediate intramitochondrial lipid transfer directly, for instance facilitating PE export from the IM. This may explain why expression of an artificial membrane tether in MICOS-deficient cells can restore growth in the absence of ethanolamine but not other phenotypes associated with the loss of MICOS. Respiratory growth of MICOS-deficient cells is not restored upon expression of this membrane tether, consistent with additional functions of MICOS as a general membrane organizing system for cristae morphogenesis, protein translocation, and respiratory chain assembly (Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Friedman et al., 2015; Horvath et al., 2015). However, these functions appear to be intimately linked to mitochondrial PE metabolism, as limiting Ups2–Mdm35–dependent PS transport to the IM promoted respiratory growth and cristae morphogenesis in the absence of MICOS. Thus, moderately altered phospholipid composition in the IM, such as increased PE levels, has profound effects on mitochondrial ultrastructure. The exact mechanism of this suppressive effect remains to be determined. Similar to Ups2, respiratory complexes III and IV and ATP synthase are required to generate aberrant cristae in MICOS-deficient cells (Hoppins et al., 2011; Friedman et al., 2015). It therefore appears likely that MICOS ensures mitochondrial structure and function by fine-tuning mitochondrial membrane properties and phospholipid homeostasis with the activities of other proteinaceous membrane organizers.
Materials and methods
Yeast strains and growth conditions
Yeast strains used in this study are listed in Table S1. Genes were deleted by PCR-targeted homologous recombination (Wach et al., 1994). Yeast cells were cultivated in YP or synthetic complete (SC) medium supplemented with 2% glucose, 2% galactose, or 2% lactate or in lactate medium. To assess ethanolamine auxotrophy, SC media were supplemented with 10 mM ethanolamine.
Cloning, expression, and purification of Ups2–Mdm35 and SLMO2–TRIAP1 complexes
DNA fragments encoding Ups2 with an N-terminal hexahistidine tag and Mdm35 were cloned into a pET-Duet-1 vector. All four codons coding cysteine in UPS2 were replaced by codons coding serine by PCR-mediated site-directed mutagenesis. Proteins were expressed in E. coli Rosetta-gami 2 (DE3; Promega) or Suffle T7 (New England Biolabs, Inc.) cells. After incubation at 37°C for 2 h, the culture was shifted to 18°C for 1 h, and Ups2 and Mdm35 were expressed by adding IPTG (0.2 mM) for 14 h. The Ups2–Mdm35 complex was purified according to the purification protocol for Ups1–Mdm35 complex, with some modifications (Connerth et al., 2012). In brief, cells were lysed in buffer B (50 mM Tris/HCl, pH 8, 250 mM NaCl, 1× complete EDTA-free protease inhibitor cocktail [Roche], 1 mM PMSF, and 20 mM imidazole). The lysate was spun at 30,000 g for 20 min, and the supernatant was subjected to column chromatography using HisTrap and HiLoad 16/60 Superdex-75 pg column (GE Healthcare). NBD-PS transfer activities were determined in elution fractions. Fractions containing the highest PS transfer activity were combined, concentrated, dialyzed against buffer C (10 mM Tris/HCl, pH 7.4, and 150 mM NaCl), and stored at −80°C.
DNA fragments encoding human SLMO2 with N-terminal hexahistidine tag and TRIAP1 were amplified from cDNA by PCR and cloned into pET-Duet-1 vector. SLMO2 and TRIAP1 were expressed in cell-free lysate as described previously (Schwarz et al., 2007) in the absence of detergent and reducing agents. The lysate was spun at 16,100 g for 20 min, and the supernatant (60 µl) was mixed with 540 µl buffer B and 20 µl of a 50% slurry of Ni-Sepharose HP beads (GE Healthcare). After incubation for 1.5 h at 4°C, beads were washed with buffer B containing imidazole (40 mM), and bound protein complexes were eluted with buffer B containing 300 mM imidazole (100 µl).
Assessment of PE/PC synthesis in vivo
Yeast cells were incubated in YP medium supplemented with 2% galactose. Overnight cultures were diluted in SC medium without inositol supplemented with 2% galactose (SC-gal) at OD 0.4 and incubated at 30°C until OD 1–2. Logarithmically growing cells were pulse labeled by [14C]serine (5 µCi/ml) in SC-gal medium lacking inositol and serine for 15 min at 30°C. Cells were collected, washed, and resuspended in medium containing unlabeled serine. After further incubation at 30°C, samples were subjected to lipid extraction using chloroform/methanol (2:1 [vol/vol]), and lipids were analyzed by TLC (chloroform/methanol/25% ammonia at 65:35:5 [vol/vol/vol]). Autoradiographs of TLC plates were quantified.
Reconstitution of Psd1
A DNA fragment encoding Psd1 (amino acid positions 57–500) containing an N-terminal heptahistidine peptide followed by a TEV cleavage site and C-terminal HA-peptide was amplified by PCR and cloned into pET16b. The Psd1 variant was expressed in bacterial cell–free lysate essentially as described (Schwarz et al., 2007) in the presence of 5 mM liposome P (69.25% 1,2-dioleoyl-sn-glycero-3-phosphocholine [DOPC], 20% tetraoleoyl-CL, 10% dioleoyl-PG, 0.25% NBD-PE, and 0.5% rhodamine-PE, reconstituted in buffer LP [5 mM Tris-HCl, pH 8.5, and 10 mM potassium acetate]), but in the absence of detergent and protease inhibitors. After centrifugation at 16,100 g for 20 min, the pellet fraction containing aggregates and proteoliposomes was resuspended in buffer FB (5 mM Hepes-NaOH, pH 7.4, and 25 mM NaCl) containing 40% sucrose and transferred to an ultracentrifuge tube. 1.5 ml of 30% sucrose in FB, 500 µl of 10% sucrose in FB, and 175 µl of FB were overlaid, and the tube was spun at 200,000 g for 1.5 h. Proteoliposomes accumulated at 30/10% and 10/0% interfaces. The top fraction (1 ml) was collected, and mature Psd1 was quantified by SDS-PAGE analysis. To assess PS decarboxylase activity, proteoliposomes containing Psd1 (107 nM, lipid concentration set to 20 µM) were incubated with liposome K (180 µM; 40% DOPC, 20% tetraoleoyl-CL, 40% dioleoyl-PS, reconstituted in buffer FB) at 26°C in buffer M (20 mM Hepes-NaOH, pH 7.4, 92 mM NaCl, and 2 mM MgCl2). Reaction volume was set to 60 µl, and 10 µl of samples at each time point were subjected to lipid extraction and qMS. To assess spontaneous transfer of PS, liposome P was incubated with liposome K (harboring 2.5% 17:0 PC and filled with 20% sucrose) under identical conditions, and lipids in isolated liposome P were extracted and quantified by qMS. Contamination and recovery of liposomes were assessed by 17:0 PC and 1,2-dioleoyl-sn-glycero-3-phosphoglycerol and used to normalize values. Liposome fusion was assessed by monitoring dequenching of NBD fluorescence under identical conditions.
Construction of an artificial membrane tethering protein
The artificial tether was cloned into the vector YCplac22ADH by homologous recombination. Genes encoding mitochondrial targeting sequence of Neurospora crassa Atp9 (amino acid positions 1–69), Yme2 transmembrane sequence (amino acid positions 279–308), Tom70 transmembrane sequence (amino acid positions 10–30), and yeGFP protein were amplified with 20-bp homology between fragments. DNA encoding a linker region (e.g., AAAENLYFQGGG) between Yme2 and Tom70 sequences was introduced in primers. Yeast cells were transformed by the amplified fragments and linearized plasmids. Plasmids were isolated from cells growing on SC-TRP plates and verified by sequencing. To assess functional complementation of MICOS by the artificial tether, dpl1Δpsd2ΔMICOSΔ cells transformed by the YCplac22ADH-mTether construct were grown on SCD plates at 35°C for 1 d, serial dilutions of cells from the plate were spotted freshly on SCD plates, and growth of cells was monitored after incubation at 37°C for 2 d.
Statistical analysis
Error bars represent SD or SEM as indicated in the figure legends. Statistical analysis of differences between two groups was performed by Student’s t test.
Cell fractionation and isolation of mitochondria
Cell fractionation and isolation of mitochondria from yeast cells was performed according to standard procedures (Tatsuta and Langer, 2007). For further purification, crude mitochondria were washed, resuspended in buffer A (0.6 M sorbitol and 5 mM MES, pH 6.0), and loaded on a continuous sucrose gradient (20–50% [wt/vol] in buffer A). Sucrose gradient–purified mitochondria were collected from the lower third of the gradient after centrifugation at 100,000 g for 1 h and washed in cold SEM buffer (10 mM MOPS/KOH, pH 7.2, 1 mM EDTA, and 0.25 M sucrose). Fractionation of cell organelles and the purity of mitochondria were assessed by SDS-PAGE and Western blotting.
qMS of phospholipids
Mass spectrometry was performed essentially as described (Connerth et al., 2012; Velázquez et al., 2016). Lipids were extracted from isolated pure mitochondria or whole yeast cells in the presence of internal standards of major phospholipids (PC 17:0-14:1, PE 17:0-14:1, PI 17:0-14:1, PS 17:0-14:1, PG 17:0-14:1, and PA 17:0-14:1; all from Avanti Polar Lipids) and CL (CL mix I, LM-6003; Avanti Polar Lipids). Extraction was performed according to Bligh and Dyer with modifications (Velázquez et al., 2016). Lipids were dissolved in 10 mM ammonium acetate in methanol and analyzed on a QTRAP 6500 triple-quadrupole mass spectrometer (Sciex) equipped with a nano-infusion splay device (TriVersa NanoMate; Advion) under the following settings: CUR, 20; CAD, medium; IHT, 90°C; EP, 10; mode: Highmass; step size, 0.1 D; setting time, 0 ms; scan rate, 200 D/s; pause 5 ms; CEM, 2300; sync, LC sync; and scan mode, Profile (for QT 6500); and sample infusion volume, 12 µl; volume of air to aspirate after sample, 1 µl; air gap before chip, enabled; aspiration delay, 0 s; prepiercing, with mandrel; spray sensing, enabled; temperature, 12°C; gas pressure, 0.4 psi; ionization voltage, 1.15 kV; polarity, positive; vent headspace, enabled; prewetting, 1×; volume after delivery, 0.5 µl; contact closure delay, 1 s; volume timing delay, 0 s; aspiration depth, 1 mm; prepiercing depth, 9 mm; and output contact closure, Rel 1/2.5 s duration (for NanoMate). The quadrupoles Q1 and Q3 were operated at unit resolution. PC analysis was performed in positive ion mode by scanning for precursors of m/z 184 at a collision energy (CE) of 50 eV. PE, PI, PS, PG, PA, and CDP-DAG measurements were performed in positive ion mode by scanning for neutral losses of 141, 277, 185, 189, 115, and 403 D at CEs of 25, 30, 20, 30, 25, and 40 eV, respectively. CL species were identified in positive ion mode by scanning for precursors of the masses (m/z 465.4, 467.4, 491.4, 493.4, 495.4, 505.5, 519.5, 521.5, 523.5, 535.5, 547.5, 549.5, 551.5, 573.5, 575.5, 577.5, 579.5, 601.5, 603.5, 605.5, 607.5, 631.5, 715.5, and 771.5 D) corresponding DAG-H2O fragments as singly charged ions at CEs of 40–50 eV. Mass spectra were processed by LipidView Software Version 1.2 (Sciex) for identification and quantification of lipids. Lipid amounts (pmol) were corrected for response differences between internal standards and endogenous lipids. Correction of isotopic overlap in CL species was performed according to Scherer et al. (2010).
Assays for lipid binding and transfer in vitro
Phospholipid binding by liposome flotation and lipid transfer assays were performed as described (Connerth et al., 2012; Miliara et al., 2015). To assess lipid binding, Ups2–Mdm35 complexes (5 µM) were incubated with liposomes (2 mM total lipids, extruded on 1-µm filter) in 50 µl buffer F (MES/NaOH, pH 5.5, 100 mM NaCl, and 2 mM EDTA) at 20°C for 10 min. After incubation, the sample was mixed with 100 µl of 60% sucrose in buffer FB, put into an ultracentrifuge tube, and overlaid with 1.35 ml buffer F30 (30% sucrose in F), 1 ml buffer F10 (10% sucrose in F), and 250 µl buffer F. Tubes were centrifuged at 200,000 g for 1.5 h. Four fractions of 700 µl were collected from the top, and proteins in the fractions were precipitated by TCA, analyzed by Tris-Tricine SDS-PAGE, and stained with colloidal Coomassie brilliant blue. In a standard assay, lipid transfer was monitored in the presence of Ups2–Mdm35 complexes (80 nM), donor liposomes (12.5 µM total lipids), and an acceptor liposome (50 µM total lipids) in 120 µl buffer TA (20 mM Tris/HCl, pH 7.4, 100 mM NaCl, and 1 mM EDTA) at 25°C.
Assessment of Psd1 enzymatic activity in isolated mitochondria
Mitochondria were resuspended in buffer B (0.1 M Tris-HCl, pH 7.4, 10 mM EDTA, and 2 µM 16:0-06:0 NBD PS [Avanti Polar Lipids, Inc.]) to a final concentration of 5 mg/ml and incubated at 25°C. A portion of sample (corresponding to 200 µg mitochondria) was taken at each time point and subjected to lipid extraction. Lipids were analyzed by TLC (developing solvent: chloroform/methanol/H2O/triethylamine 30:35:7:35 [vol/vol/vol/vol]). NBD signals were detected and quantified by fluorescence imaging (Typhoon Trio; GE Healthcare).
Freezing and freeze substitution for ultrastructural studies
Electron microscopic analysis of yeast cells was done as previously described (Lefebvre-Legendre et al., 2005). In brief, pellets of yeast cells were placed on the surface of a copper EM grid (400 mesh) coated with formvar. Each loop was very quickly submersed in precooled liquid propane and held at –180°C by liquid nitrogen. The loops were then transferred to a precooled solution of 4% osmium tetroxide in dry acetone in a 1.8-ml polypropylene vial at –82°C for 48 h (substitution fixation), warmed gradually to RT, and washed three times in dry acetone. Specimens were stained for 1 h with 1% uranyl acetate in acetone at 4°C in a black room. After another rinse in dry acetone, the loops were infiltrated progressively with araldite (epoxy resin, Fluka; Sigma-Aldrich). Ultrathin sections were contrasted with lead citrate.
Online supplemental material
Fig. S1 shows the functionality of Ups2 variants in vivo, their purification from E. coli, and in vitro lipid transfer assays using Ups2–Mdm35 and SLMO2–TRIAP1, demonstrating the dependence of PS transport on negatively charged lipids. Fig. S2 shows ethanolamine prototrophy of cells lacking Ups/Preli family lipid transfer proteins and nonmitochondrial PE synthesis, control experiments to exclude liposome fusion during Psd1 activity assays, the accumulation of PE and Psd1 activity in MICOS-deficient cells, the respiratory growth of these cells upon expression of an artificial membrane tether, and experiments to assess the cellular localization of this tether. Fig. S3 shows respiratory growth of cells lacking MICOS subunits and/or Ups2 in the absence of extramitochondrial PE synthesis, the phospholipid profile and acyl-chain distribution in PE in mitochondria lacking Ups2 and/or MICOS subunits, and the respiratory growth of cells lacking MICOS and/or Psd1. Table S1 lists yeast strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201602007/DC1.
Supplementary Material
Acknowledgments
We thank Gudrun Zimmer for excellent technical support and Philipp Lampe for support in cell-free synthesis.
This work was supported by a grant of the Deutsche Forschungsgemeinschaft to T. Tatsuta and T. Langer (LA 918/14-1 and TA 1132/2-1) and a grant of the European Research Council to T. Langer (AdG No. 233078).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CDP
- cytidine diphosphate
- CE
- collision energy
- CL
- cardiolipin
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- IM
- inner membrane
- IMS
- intermembrane space
- MICOS
- mitochondrial contact site and cristae organizing system
- OM
- outer membrane
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- qMS
- quantitative mass spectrometry
- SC
- synthetic complete
References
- Alkhaja A.K., Jans D.C., Nikolov M., Vukotic M., Lytovchenko O., Ludewig F., Schliebs W., Riedel D., Urlaub H., Jakobs S., and Deckers M.. 2012. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol. Biol. Cell. 23:247–257. 10.1091/mbc.E11-09-0774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birner R., Bürgermeister M., Schneiter R., and Daum G.. 2001. Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol. Biol. Cell. 12:997–1007. 10.1091/mbc.12.4.997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.Y., Wu W.I., and Voelker D.R.. 2005. Phosphatidylserine decarboxylases as genetic and biochemical tools for studying phospholipid traffic. Anal. Biochem. 347:165–175. 10.1016/j.ab.2005.03.017 [DOI] [PubMed] [Google Scholar]
- Connerth M., Tatsuta T., Haag M., Klecker T., Westermann B., and Langer T.. 2012. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 338:815–818. 10.1126/science.1225625 [DOI] [PubMed] [Google Scholar]
- Dee C.T., and Moffat K.G.. 2005. A novel family of mitochondrial proteins is represented by the Drosophila genes slmo, preli-like and real-time. Dev. Genes Evol. 215:248–254. 10.1007/s00427-005-0470-4 [DOI] [PubMed] [Google Scholar]
- Friedman J.R., Mourier A., Yamada J., McCaffery J.M., and Nunnari J.. 2015. MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. eLife. 4 10.7554/eLife.07739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohil V.M., Thompson M.N., and Greenberg M.L.. 2005. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. J. Biol. Chem. 280:35410–35416. 10.1074/jbc.M505478200 [DOI] [PubMed] [Google Scholar]
- Harner M., Körner C., Walther D., Mokranjac D., Kaesmacher J., Welsch U., Griffith J., Mann M., Reggiori F., and Neupert W.. 2011. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30:4356–4370. 10.1038/emboj.2011.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Collins S.R., Cassidy-Stone A., Hummel E., Devay R.M., Lackner L.L., Westermann B., Schuldiner M., Weissman J.S., and Nunnari J.. 2011. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195:323–340. 10.1083/jcb.201107053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S.E., Böttinger L., Vögtle F.N., Wiedemann N., Meisinger C., Becker T., and Daum G.. 2012. Processing and topology of the yeast mitochondrial phosphatidylserine decarboxylase 1. J. Biol. Chem. 287:36744–36755. 10.1074/jbc.M112.398107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S.E., Rampelt H., Oeljeklaus S., Warscheid B., van der Laan M., and Pfanner N.. 2015. Role of membrane contact sites in protein import into mitochondria. Protein Sci. 24:277–297. 10.1002/pro.2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre-Legendre L., Salin B., Schaëffer J., Brèthes D., Dautant A., Ackerman S.H., and di Rago J.P.. 2005. Failure to assemble the alpha 3 beta 3 subcomplex of the ATP synthase leads to accumulation of the alpha and beta subunits within inclusion bodies and the loss of mitochondrial cristae in Saccharomyces cerevisiae. J. Biol. Chem. 280:18386–18392. 10.1074/jbc.M410789200 [DOI] [PubMed] [Google Scholar]
- Lev S. 2010. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat. Rev. Mol. Cell Biol. 11:739–750. 10.1038/nrm2971 [DOI] [PubMed] [Google Scholar]
- Miliara X., Garnett J.A., Tatsuta T., Abid Ali F., Baldie H., Pérez-Dorado I., Simpson P., Yague E., Langer T., and Matthews S.. 2015. Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes. EMBO Rep. 16:824–835. 10.15252/embr.201540229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onguka O., Calzada E., Ogunbona O.B., and Claypool S.M.. 2015. Phosphatidylserine decarboxylase 1 autocatalysis and function does not require a mitochondrial-specific factor. J. Biol. Chem. 290:12744–12752. 10.1074/jbc.M115.641118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C., Haag M., Potting C., Rodenfels J., Dip P.V., Wieland F.T., Brügger B., Westermann B., and Langer T.. 2009. The genetic interactome of prohibitins: Coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 184:583–596. 10.1083/jcb.200810189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potting C., Tatsuta T., König T., Haag M., Wai T., Aaltonen M.J., and Langer T.. 2013. TRIAP1/PRELI complexes prevent apoptosis by mediating intramitochondrial transport of phosphatidic acid. Cell Metab. 18:287–295. 10.1016/j.cmet.2013.07.008 [DOI] [PubMed] [Google Scholar]
- Rabl R., Soubannier V., Scholz R., Vogel F., Mendl N., Vasiljev-Neumeyer A., Körner C., Jagasia R., Keil T., Baumeister W., et al. 2009. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J. Cell Biol. 185:1047–1063. 10.1083/jcb.200811099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch K.M., and De Camilli P.. 2015. SMP-domain proteins at membrane contact sites: Structure and function. Biochim. Biophys. Acta.:S1388-1981(15)00225-5 10.1016/j.bbalip.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer M., Schmitz G., and Liebisch G.. 2010. Simultaneous quantification of cardiolipin, bis(monoacylglycero)phosphate and their precursors by hydrophilic interaction LC-MS/MS including correction of isotopic overlap. Anal. Chem. 82:8794–8799. 10.1021/ac1021826 [DOI] [PubMed] [Google Scholar]
- Schwarz D., Junge F., Durst F., Frölich N., Schneider B., Reckel S., Sobhanifar S., Dötsch V., and Bernhard F.. 2007. Preparative scale expression of membrane proteins in Escherichia coli-based continuous exchange cell-free systems. Nat. Protoc. 2:2945–2957. 10.1038/nprot.2007.426 [DOI] [PubMed] [Google Scholar]
- Signorell A., Gluenz E., Rettig J., Schneider A., Shaw M.K., Gull K., and Bütikofer P.. 2009. Perturbation of phosphatidylethanolamine synthesis affects mitochondrial morphology and cell-cycle progression in procyclic-form Trypanosoma brucei. Mol. Microbiol. 72:1068–1079. 10.1111/j.1365-2958.2009.06713.x [DOI] [PubMed] [Google Scholar]
- Simbeni R., Paltauf F., and Daum G.. 1990. Intramitochondrial transfer of phospholipids in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 265:281–285. [PubMed] [Google Scholar]
- Simbeni R., Pon L., Zinser E., Paltauf F., and Daum G.. 1991. Mitochondrial membrane contact sites of yeast. Characterization of lipid components and possible involvement in intramitochondrial translocation of phospholipids. J. Biol. Chem. 266:10047–10049. [PubMed] [Google Scholar]
- Tamura Y., Endo T., Iijima M., and Sesaki H.. 2009. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J. Cell Biol. 185:1029–1045. 10.1083/jcb.200812018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Onguka O., Hobbs A.E., Jensen R.E., Iijima M., Claypool S.M., and Sesaki H.. 2012a Role for two conserved intermembrane space proteins, Ups1p and Ups2p, [corrected] in intra-mitochondrial phospholipid trafficking. J. Biol. Chem. 287:15205–15218. 10.1074/jbc.M111.338665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Onguka O., Itoh K., Endo T., Iijima M., Claypool S.M., and Sesaki H.. 2012b Phosphatidylethanolamine biosynthesis in mitochondria: phosphatidylserine (PS) trafficking is independent of a PS decarboxylase and intermembrane space proteins UPS1P and UPS2P. J. Biol. Chem. 287:43961–43971. 10.1074/jbc.M112.390997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T., and Langer T.. 2007. Studying proteolysis within mitochondria. Methods Mol. Biol. 372:343–360. 10.1007/978-1-59745-365-3_25 [DOI] [PubMed] [Google Scholar]
- Tatsuta T., Scharwey M., and Langer T.. 2014. Mitochondrial lipid trafficking. Trends Cell Biol. 24:44–52. 10.1016/j.tcb.2013.07.011 [DOI] [PubMed] [Google Scholar]
- Tavassoli S., Chao J.T., Young B.P., Cox R.C., Prinz W.A., de Kroon A.I., and Loewen C.J.. 2013. Plasma membrane–endoplasmic reticulum contact sites regulate phosphatidylcholine synthesis. EMBO Rep. 14:434–440. 10.1038/embor.2013.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G., Voelker D.R., and Feigenson G.W.. 2008. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9:112–124. 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez A.P., Tatsuta T., Ghillebert R., Drescher I., and Graef M.. 2016. Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J. Cell Biol. 212:621–631. 10.1083/jcb.201508102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Malsburg K., Müller J.M., Bohnert M., Oeljeklaus S., Kwiatkowska P., Becker T., Loniewska-Lwowska A., Wiese S., Rao S., Milenkovic D., et al. 2011. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell. 21:694–707. 10.1016/j.devcel.2011.08.026 [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pöhlmann R., and Philippsen P.. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 10:1793–1808. 10.1002/yea.320101310 [DOI] [PubMed] [Google Scholar]
- Wang K., Jin M., Liu X., and Klionsky D.J.. 2013. Proteolytic processing of Atg32 by the mitochondrial i-AAA protease Yme1 regulates mitophagy. Autophagy. 9:1828–1836. 10.4161/auto.26281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Tamura Y., Kawano S., and Endo T.. 2015. Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria. Nat. Commun. 6:7922 10.1038/ncomms8922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., He F., Yao H., Wang C., Wang J., Li J., Qi X., Xue H., Ding J., and Zhang P.. 2015. Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex. EMBO Rep. 16:813–823. 10.15252/embr.201540137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.