Abstract
Mechanical forces play increasingly recognized roles in T cell receptor (TCR) signal transduction. Hu and Butte (2016. J. Cell Biol. http://dx.doi.org/10.1083/jcb.201511053) demonstrate that actin is required for T cells to generate forces at the TCR and that exogenous application of force can emulate these cytoskeletal forces and trigger T cell activation.
Physiological T cell stimulation requires a specific peptide–major histocompatability complex (pMHC) interaction and an intact F-actin cytoskeleton, and is further enhanced by myosin II–based contractility (Valitutti et al., 1995; Ilani et al., 2009). This concept of chemical specificity enhanced by force sensing has captured the imagination of investigators from fields of immunology, engineering, physics, and cell biology, but experimentally testing this concept is challenging. In this issue, Hu and Butte show that they can replace the role of F-actin in T cell triggering with forces delivered by an atomic force microscope (AFM). Several studies over the past decade converge on the idea that T cell recognition is based at least in part on mechanotransduction, the conversion of a force into a biochemical signal (Vogel and Sheetz, 2009), pointing to an important role for physical forces in T cell responses. Hu and Butte (2016) demonstrate force-based T cell receptor (TCR) triggering in a system with a crippled F-actin cytoskeleton. This system constrains potential mechanisms through which forces can be linked to signaling events and thus these findings offer an exciting advance.
TCR signaling has been traditionally studied using bivalent anti-CD3ε antibodies that bind the CD3ε signaling subunits of the TCR and activate TCR signaling pathways by artificially aggregating the receptors in the membrane. This artificial method of initiating TCR transduction made studying the TCR response to force a challenge and seemingly argued against the idea that T cells need to apply a force to the TCR to trigger a signal. However, the physiological ligand of the TCR is a monomeric pMHC on the surface of antigen presenting cell, which creates an opening for force in extending the sensitivity, and perhaps also the specificity of the TCR. It is clear that monovalent pMHC binding to CD4+ T cells does not trigger signaling despite the potential of CD4 to recruit the kinase Lck to the TCR. Thus, experimental efforts to study force-actuated signaling in T cells must establish initial conditions to engage the TCR without triggering signaling in the absence of an applied force. Such conditions have been achieved, demonstrating force-actuated TCR signaling in multiple configurations (Kim et al., 2009; Li et al., 2010; Liu et al., 2014). However, how an applied force converts nonproductive TCR engagement into a productive one and whether this is a fundamental mechanism of natural pMHC recognition by T cells in vivo remain unresolved. This is the context in which Hu and Butte (2016) explored TCR responses with an AFM to address the linked questions of how triggered T cells generate force and if force can actuate TCR signaling.
In their study, Hu and Butte (2016) coupled anti-CD3ε antibodies or pMHC complexes to an AFM tip and used a specially designed AFM to both measure forces on the T cell and to apply complex patterns of force onto the T cell. The AFM tip was a single silicon crystal 4–6 µm high and with a ∼6-nm radius of contact (Fig. 1 A). Although the coating density is not explicitly determined, the coupling chemistry used should be able to achieve high-density, close packing of the anti-CD3ε antibodies or pMHC (Fig. 1 B). Thus, the ligand presentation by this device is likely to be similar to earlier studies where MHC monomers were closely packed onto quantum dots, which were highly effective at T cell activation (Anikeeva et al., 2006). Hu and Butte (2016) showed that contact with the anti-CD3ε– or pMHC-coated AFM tip induced similar Ca2+ signaling in T cells, which in turn generated pushing and pulling forces on the AFM, consistent with previous results (Husson et al., 2011; Bashour et al., 2014). Treatment of the cells with latrunculin A, which sequesters G-actin and prevents its polymerization to F-actin, prevented Ca2+ flux and force generation, illustrating the importance of the cytoskeleton in TCR triggering. In this context, Hu and Butte (2016) discovered that applying an oscillating force to the anti-CD3ε–coated AFP tip in contact with the latrunculin A–treated T cell surface restored Ca2+ signaling. Control antibodies coated on the AFM tip in contact with T cells didn’t induce a Ca2+ flux, demonstrating that specificity was preserved. It remains to be determined if similar effects can be seen with pMHC, which appeared to be less able to sustain high pulling forces.
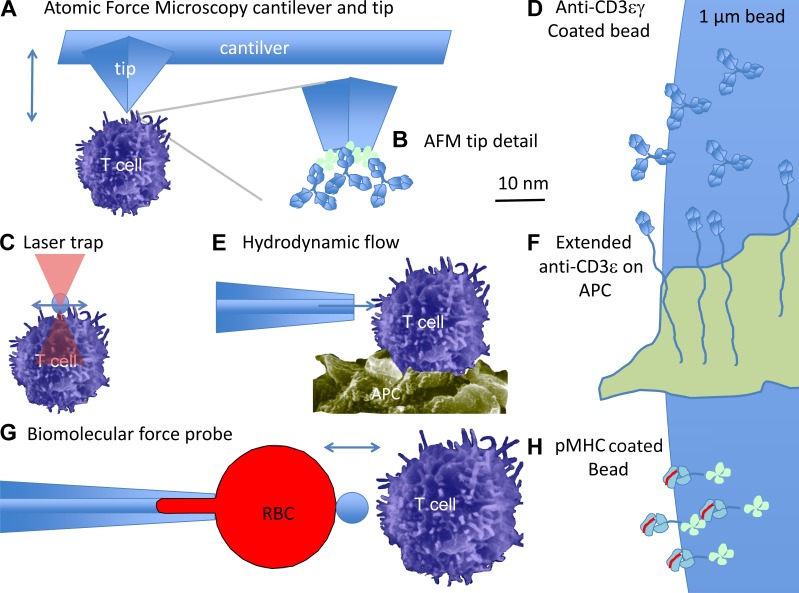
Figure 1.
Different experimental approaches that have been used to ligate the TCR and reveal force-actuated T cell signaling. (A) In AFM, a cantilever with a pyramidal tip comes into direct contact with a cell. The tip can be moved in three dimensions and reports the force applied to a cell. (B) The AFM tip is fully coated with anti-CD3ε antibodies. (C) The laser trap system uses infrared light to exert low nN forces on the bead. (D) The bead surface viewed on the same scale as the AFM tip is large and will likely contact a T cell with intact cytoskeleton on microvilli. (E) Hydrodynamic flow can apply tangential forces similar to the laser trap, with potentially greater magnitude, but less fine control. (F) Tethering of antibodies reduces activation, which can be overcome by application of force. (G) The BFP method uses a bead at the apex of an RBC captured by a suction pipette to test for interaction and apply calibrated forces. (H) The pMHC on the bead are titrated to provide only a single interaction at a time, at least statistically. These systems all generate results that suggest that the TCR combines immunological specificity and force sensing.
The use of AFM presented by Hu and Butte (2016) builds on a series of studies demonstrating force actuation of TCR-mediated signals. These earlier studies used different strategies to achieve 0-force baselines without signaling and then activated signaling by applying a force (summarized in Fig. 1). Kim et al. (2009) generated an anti-CD3εγ–specific antibody that bound to only one site per TCR complex, such that it was unable to cross-link the TCR and trigger signals even when coated on 1-µm-diameter beads. When the cell-attached anti-CD3εγ beads were subjected to tangential forces on the order of 1 nN with an optical trap (Fig. 1 C), the TCR was triggered as indicated by a cytoplasmic Ca2+ increase. It was particularly important in this study that the force was applied tangentially and not vertically and this was explained in terms of the forces T cells would naturally generate during migration over the surface of an antigen-presenting cell. The coating of that anti-CD3εγ on the 1-µm bead likely generates a large surface area for the application of force (Fig. 1 D). Tangential movement might also “roll” the bead over the surface, generating a mixture of stretching and compressive forces, which would not be generated by vertical pulling. Subsequently, Li et al. (2010) used a set of recombinant anti-CD3ε–based single-chain Fv antibody domains that were presented on the surface of cells with long, flexible stalks composed of proteins such as the sialomucin CD43 (Fig. 1, E and F). T cells that came into contact with cells expressing the extended anti-CD3ε constructs were not activated. Similar constructs were reported to be unable to trigger T cell activation, but this was attributed to their failure to form close cell–cell contacts from which the large tyrosine phosphatase CD45 could be excluded (Chang et al., 2016). However, Li et al. (2010) could activate signaling by providing tangential shear forces with a micropipette. This result thus reinforced the work from Kim et al. (2009) that TCR signaling from suboptimal anti-CD3–based stimuli could be rescued by the application of large physical forces in a manner that would generate a mix of pulling, pushing, and sliding forces. The orientation of an AFM tip is comparatively fixed relative to the cell; therefore, Hu and Butte (2016) avoid the issue of rolling behavior and could better isolate the effect of vertical forces; their findings strengthen the earlier studies of Kim et al. (2009) and Li et al. (2010). The AFM approach also provides better control of applied forces than the cellular-level application of shear and avoids artifacts associated with optic trap illumination.
In a different approach, Liu et al. (2014) used a biomolecular force probe (BFP) both to interrogate the response of single TCR–pMHC interactions to force and to test the ability of serial, single interactions to be summed by the T cell to generate a robust Ca2+ response. In the BFP experiment, a low density of agonist pMHC is attached to a 1-µm bead, which was then docked on a red blood cell (RBC) held in a suction pipette (Fig. 1, G and H). RBCs were used in this study because their physical properties are uniform and well understood, which is an advantage over AFM technology. In the absence of cell contact, the bead attached at the apex of the RBC is imaged with nanometer resolution and high speed as it oscillates based on the thermally driven fluctuations in the RBC membrane. A T cell bearing the appropriate TCR is maneuvered into contact with the bead and when an interaction takes place, the linkage to the T cell dampens the oscillation of the bead to allow detection of the bond lifetime, its response to applied force, and eventual rupture. An exciting analytical finding from these studies was that the agonist pMHC forms “catch-bonds” with TCRs, such that the bond lifetime is actually increased as force is applied in the 10-pN range. Liu et al. (2014) then observed cytoplasmic Ca2+ elevation during repeated trials when T cells accumulated a series of single TCR–pMHC catch bonds totaling 10 s within a 60-s period. The identification of 10 pN as a critical threshold has been reinforced by recent studies with DNA-based force sensors and tension gauges (Liu et al., 2016). Hu and Butte (2016) explored this reinforcing behavior in more detail, applying periodic inputs to the T cell and in a configuration that is more accessible than BFP.
In all of these previous studies, the mechanosensing activity of the TCR is always taking place in a cell with an intact actin cytoskeleton to help interpret the external force. In this setting, there are many paradigms for mechanotransduction that might apply to the TCR and many possibilities to test. Hu and Butte (2016) were able to complement a pharmacologically induced cytoskeletal defect by applying an oscillating force to the TCR. Interestingly, they were not able to detect evidence for the accumulation of discontinuous signals, suggesting that this type of signal accumulation may require an intact F-actin cytoskeleton. Regardless, the success of applying oscillatory force to rescue TCR signaling in a system depleted of F-actin suggests that the necessary force sensing may very well be encoded in TCR proximal elements, if not being intrinsic to the TCR complex itself. AFM-based assays for F-actin–independent force sensing by the TCR should now enable the systematic testing of such hypotheses about the TCR by also taking advantage of classical models of early TCR signaling. Beyond forces generated by the T cell, this study further supports the idea that forces generated by the antigen presenting cell may influence T cell activation. A fuller story of the role of forces in immune cell function is thus developing, with complementary studies showing that dendritic cells modulate the mobility of ICAM-1, which in turn affects T cell activation and function (Comrie et al., 2015). Finally, the approach of Hu and Butte (2016) is part of a new generation of AFM-based methods, including the stiffness clamp method developed by Webster et al. (2011) that provide new levels of control over the presentation and response of biomolecular cues.
Acknowledgments
The authors declare no competing financial interests.
References
- Anikeeva N., Lebedeva T., Clapp A.R., Goldman E.R., Dustin M.L., Mattoussi H., and Sykulev Y.. 2006. Quantum dot/peptide-MHC biosensors reveal strong CD8-dependent cooperation between self and viral antigens that augment the T cell response. Proc. Natl. Acad. Sci. USA. 103:16846–16851. 10.1073/pnas.0607771103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashour K.T., Gondarenko A., Chen H., Shen K., Liu X., Huse M., Hone J.C., and Kam L.C.. 2014. CD28 and CD3 have complementary roles in T-cell traction forces. Proc. Natl. Acad. Sci. USA. 111:2241–2246. 10.1073/pnas.1315606111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang V.T., Fernandes R.A., Ganzinger K.A., Lee S.F., Siebold C., McColl J., Jönsson P., Palayret M., Harlos K., Coles C.H., et al. 2016. Initiation of T cell signaling by CD45 segregation at ‘close contacts’. Nat. Immunol. 17:574–582. 10.1038/ni.3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comrie W.A., Li S., Boyle S., and Burkhardt J.K.. 2015. The dendritic cell cytoskeleton promotes T cell adhesion and activation by constraining ICAM-1 mobility. J. Cell Biol. 208:457–473. 10.1083/jcb.201406120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K.H., and Butte M.J.. 2016. T-cell activation requires force generation. J. Cell Biol. 10.1083/jcb.201511053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson J., Chemin K., Bohineust A., Hivroz C., and Henry N.. 2011. Force generation upon T cell receptor engagement. PLoS One. 6:e19680 10.1371/journal.pone.0019680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilani T., Vasiliver-Shamis G., Vardhana S., Bretscher A., and Dustin M.L.. 2009. T cell antigen receptor signaling and immunological synapse stability require myosin IIA. Nat. Immunol. 10:531–539. 10.1038/ni.1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.T., Takeuchi K., Sun Z.Y., Touma M., Castro C.E., Fahmy A., Lang M.J., Wagner G., and Reinherz E.L.. 2009. The αβ T cell receptor is an anisotropic mechanosensor. J. Biol. Chem. 284:31028–31037. 10.1074/jbc.M109.052712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.C., Chen B.M., Wu P.C., Cheng T.L., Kao L.S., Tao M.H., Lieber A., and Roffler S.R.. 2010. Cutting edge: mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. J. Immunol. 184:5959–5963. 10.4049/jimmunol.0900775 [DOI] [PubMed] [Google Scholar]
- Liu B., Chen W., Evavold B.D., and Zhu C.. 2014. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell. 157:357–368. 10.1016/j.cell.2014.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Blanchfield L., Ma V.P., Andargachew R., Galior K., Liu Z., Evavold B., and Salaita K.. 2016. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc. Natl. Acad. Sci. USA. 113:5610–5615. 10.1073/pnas.1600163113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valitutti S., Dessing M., Aktories K., Gallati H., and Lanzavecchia A.. 1995. Sustained signaling leading to T cell activation results from prolonged T cell receptor occupancy. Role of T cell actin cytoskeleton. J. Exp. Med. 181:577–584. 10.1084/jem.181.2.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V., and Sheetz M.P.. 2009. Cell fate regulation by coupling mechanical cycles to biochemical signaling pathways. Curr. Opin. Cell Biol. 21:38–46. 10.1016/j.ceb.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster K.D., Crow A., and Fletcher D.A.. 2011. An AFM-based stiffness clamp for dynamic control of rigidity. PLoS One. 6:e17807 10.1371/journal.pone.0017807 [DOI] [PMC free article] [PubMed] [Google Scholar]