Abstract
Generating a detailed description of human T cell repertoire diversity is an important goal in the study of human immunology. The circulation is the source of most T cells used for studies in humans. Here we use high throughput sequencing of TCR BV19 transcripts from CD8 T cells derived from unmanipulated PBMC from an older HLA-A2 individual to provide a quantitative and qualitative description of the clonotypic CDR3 nucleotide and amino acid composition of the TCR β-chain from this subset of circulating CD8 T cells. Aggregated samples from six time points spanning ~ 1.5 years were analyzed to smooth possible temporal fluctuation. BV19 encompasses the well studied RS-encoding clonotypes involved in recognition of the M158–66 epitope from influenza A in HLA-A2 individuals. The clonotype distribution was diverse, complex and self-similar. The amino acid composition was generally skewed in favor of glycines and there were specific amino acids observed at higher frequency at the NDN start position. The motif repertoire distribution was also diverse, complex and self-similar with respect to CDR3 length, NDN start and length.
INTRODUCTION
Infancy and childhood are characterized by the development of adaptive immune memory to environmental pathogens, eliminating the need for innate based responses and the inflammation that accompanies them. In the case of T cells, the generation of adaptive memory proceeds to such a point that an individual can become functionally athymic post-puberty and maintain general good health. The memory T cell repertoires that are developed during the early period are complex and finely tuned to surveillance for recurring exposures. They most likely represent a complexity and optimization that approaches that of the nervous system. However, memory T cell repertoires are still poorly understood and not extensively studied.
With the advent of next-generation sequencing it became important to analyze repertoires ex vivo and a number of researchers have done so (1–3). However, a detailed quantitative and qualitative analysis of a subset of the repertoire which contains a well studied pathogen epitope would be useful. While a true repertoire analysis at the clonotype level would include sequencing both TCR chains from the CD8 cells, current single cell approaches are not yet compatible with large scale analyses. Therefore, we analyzed the BV19 β-chain repertoire as a proxy for a full clonotype analysis. The choice of BV19 expressing CD8 T cells stems from our interest in CD8 T cell memory to influenza. In antigen experienced HLA-A2 individuals, the recall response to the influenza epitope, M158–66, predominantly involves CD8 T cells expressing the BV19 β-chain gene (4,5) and a restricted number of α-chain genes (6). We went on to show that the recall repertoire (7) and functional T cells therein are polyclonal (8). This includes the subset of CD8 BV19 cells that bind HLA-A2:M158–66 tetramer (8). Rank frequency analysis of the repertoire or various subsets thereof show power law-like distributions, and thus the repertoire can be considered a self-similar fractal (8, 9). The complexity of the repertoire in part reflects the complexity of a pathogen encounter, and different clonotypes may be invoked at different stages of pathogen density (10). The repertoire is also relatively cross-reactive with ~ 50% of clonotypes capable of recognizing a substituted epitope with the extent of cross-reactivity (number of substituted epitopes recognized) also showing a power law-like distribution (11).
We expected that the complexity and self-similarity of the flu-specific subset of the BV19 repertoire will extend to the BV19 entire repertoire, both at the clonotype (nucleotide sequence) and amino acid motif levels. Therefore, BV19-specific amplification emulsion PCR and pyrosequencing was used to generate the BV19 clonotype data. After cleaning the data we generated both a quantitative and qualitative analysis of circulating CD8 T cells. After a basic clonotypic description we analyzed the amino acid composition of the repertoire as well as the distribution and complexity of the amino acid sequence motifs contributed by the NDN region. The distribution of the motifs was analyzed over a series of motif lengths.
METHODS
The human research conducted here was authorized by Institutional Review Board of BloodCenter of Wisconsin under BC 05–11, “Generation and Decay of Memory T Cells in Older Populations,” which is still open for data analysis. Written consent was obtained.
PBMC corresponding to six different time points (8/21/06, 10/16/06, 2/5/07, 4/2/07, 9/24/07, and 3/17/08) spanning ~ 1.5 years, were thawed and CD8 positive cells were isolated using Dynal CD8 positive isolation kit, and mRNA isolation used Dynal Oligo (dT) Beads according to the manufacturer’s instruction (Invitrogen, Carlsbad, CA). cDNA was prepared using Poly T primer and M-MLV reverse transcriptase (Invitrogen) and used as template for amplification. The amplification was done using βV19 coded primers and Fam labeled βC coded primers (see below). The PCR products were purified using AMPure PCR purification kit according to manufacturer’s instructions. The concentration of purified PCR products was measured using NanoDrop-1000 spectrophotometer and 6 to 12 purified PCR products were mixed to obtain a total of 2500 ng. The samples were further amplified and prepared for high throughput sequencing at the Human and Molecular Genomic Center (HMGC) Sequencing Facility (www.hmgc.mcw.edu) of Medical College of Wisconsin. Sequence data was generated by emulsion-based PCR in which the initial PCR products have a linker sequence which allows attachment to beads at low concentrations so that each bead only binds on average one molecule. The beads are suspended in a PCR buffer and an emulsifier. Each bead is covered by thin buffer solution and separated from the other beads by the emulsifier. Additional cycles of PCR generate a clonally expanded population of molecules on the bead with one strand attached to the linker on the bead and the other available to continue to hybridize to available linkers. The beads are then washed and placed in their own well of a sequencing plate and subject to multiple rounds of additive sequencing. In this way the emulsion PCR replaces bacterial plasmid subcloning in previous approaches and provides a clonal sequence. The number of beads with the same sequence can be used to provide quantitative information about the repertoire. The sequencing was performed on the Roche GS-FLX Genome Sequencer using a two chamber gasket. Samples were coded by identifier sequences embedded in the primers. After decoding, sequences derived from each sample were downloaded in fasta format and analyzed using our proprietary “CDR3Reader” software, which assigns clonotype names according to the naming convention described by Yassai et al (12). These names can be used to reverse translate the amino acid sequence to the clonotypic nucleotide sequence.
Data were downloaded from CDR3Reader and analyzed using Microsoft Excel. Clonotype is used to refer to the unique CDR3 nucleotide sequence of the TCR β-chain gene. Clonotype data consists of the CDR3 sequence and its translation, the length of the CDR3 region (L), and the number of observations/sequences (M). Repertoire characteristics are the number of clonotypes (N), a simple estimate of abundance defined as observations per clonotype (M/N), the highest number of observations of a clonotype(s) which define the maximum rank (Rmax), the number of singleton clonotypes, i.e. observed once (S), and a simple clonotype diversity measure (Dc), that incorporates the abundance and distribution, Dc=(N/M * Rmax)-1. These have been defined previously (13, 14). Rank-frequency plots of the log transformed clonotype data were generated as described (9). Trend lines for rank-frequency summaries were generated by least squares fit of data subsets to maximize the R2 value of the first component of the plots. Shannon diversity of the first order was estimated for each studied composite of clonotype data (15). The Shannon diversity measure (Ds) was adapted to the rank frequency data and was estimated as the inverse product of the number of clonotypes NRi encoding the motifs at rank Ri corrected for the total number of clonotypes encoding the motifs Ni at rank Ri, so for Pi=(NRi/Ni), Ds= Πi(Pi)RiPi.
Rearrangement analysis (Spectratyping)
CDR3 length analysis was performed by amplification of the BV19 CDR3 region using cDNA as template and BV19 and Fam-labeled CB specific primers (16).
BV19 primer; 5’ CCAAAAGAACCCGACAGCTTTC
Fam-labeled BC primer; 5’ Fam-GCTTCTGATGGCTCAAACACAG
1–2 ul of amplified products were combined with 9 ul of Formamide/Liz 500 (900 ul Formamide + 50 ul Liz standard). Samples were heat denatured at 90°C for 3 minutes and then loaded on ABI 3130XL Gene Analyzer (Applied Biosystems). GeneScan software (Applied Biosystems) was used for the collection of the data. The files were analyzed using proprietary software which gives the relative frequency of each CDR3 length. The relative frequency of each CDR3 length was generated for each of the six PBMC samples and an average and the standard deviation calculated.
RESULTS
Description of sample source
UPN204 was 68 years old at time of enrollment. PBMC samples were obtained at multiple times. Unmanipulated samples of CD8 T cells were prepared for sequence analysis separately from six time points spanning ~ 1.5 years and the BV19-specific PCR products generated. The data from multiple sample times were combined to provide a global description irrespective of temporal variation. We chose an older subject because the age-based focusing of the CD8 repertoires could help facilitation of identifying highly selected CDR3 amino acid motifs corresponding to pMHC recognition elements.
Clonotype-based repertoire characteristics
The sequences generated were initially analyzed by estimation of the error rate. This was accomplished by using a number of high frequency clonotypes as benchmarks. The number of highly similar sequences observed that constituted obvious errors due to transitions, and less frequently transversions, were counted. These could result in a different encoding of the same amino acids, a change in amino acid sequence, or read-through into the J region. Insertion and deletion errors were less common and result in out-of-frame J region translation products. An example of the error analysis is shown in Supplementary Figure 1. In this manner we estimate that the average error rate for the dataset was ~2%. An alternative approach examined the error rate in the V region which should be identical in all sequences. This gave an estimated error rate of ~1%. The stricter 2% error rate implies that for any hundred sequences two would be false, independent of how these sequences are distributed among clonotypes. This interpretation led to disallowing one of any paired clonotypes where one of the pair differed by one bp from the other and the ratio was four observations of the former to one of the latter. If the count of the more frequent of the two was nine or less, its frequency would be increased by the number of exemplars of the clonotype with the presumed error. Sequences whose translation resulted in a chain-termination were eliminated from consideration.
The total number of sequences was 203185, and these identified 12269 clonotypes. The most frequent clonotype was observed 5782 times (2.85% of all observations) and 1835 clonotypes were only observed once (15.00% of clonotypes). The number of observations per clonotype, a simple measure of abundance, was 16. 56. The diversity, Dc, was 348. The complete cleaned sequence data used for the analyses, in the form of the clonotype name (12) and number of observations is available as Supplemental Table 1.
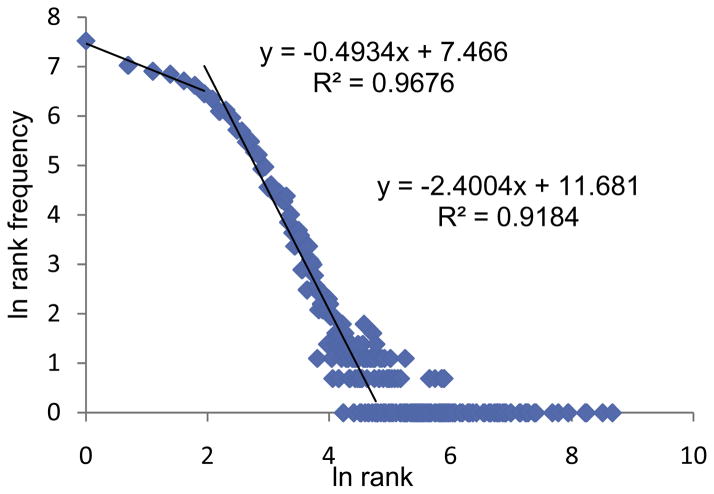
The overall shape of the repertoire can be best described by rank and rank frequency analysis of the clonotypes. Plotting the loge of the rank and of the rank frequency simplifies the repertoire description. In our previous analyses of BV19 recall repertoires such an approach yields single or multiple power law-like descriptions (8, 9,14). Most commonly the plot is hockey stick-like; with a power law-like component starting at with the lowest rank (clonotypes observed once) and decreasing until the data shows single clonotypes at high ranks. The second component represents a continuation of many single clonotypes at higher ranks. Previous large scale sequencing studies of PBMC have also shown power law-like components in the clonotype distribution (1). The rank – rank frequency plot of the BV19 data is shown in Figure 1. The ex vivo data is more complex than that observed for influenza-responsive in vitro recall repertoires. There is an initial component constituted of low ranks, for which a slope of −0.49 and an R2 of 0.97, a middle portion with a slope of −2.40 and an R2 of 0.92, and a large component of high ranking clonotypes, generally with one or occasionally two clonotype per rank.
Figure 1. Rank frequency distribution of BV19 clonotypes.
The data is plotted as the natural log of both components and treated as a three component distribution. The first two components low and mid ranking clonotypes show a good fit (R2 > 0.9) to a line, which would be expected of a power law like distribution. The third component represents high ranking (frequency) clonotypes, with generally only one clonotype per rank.
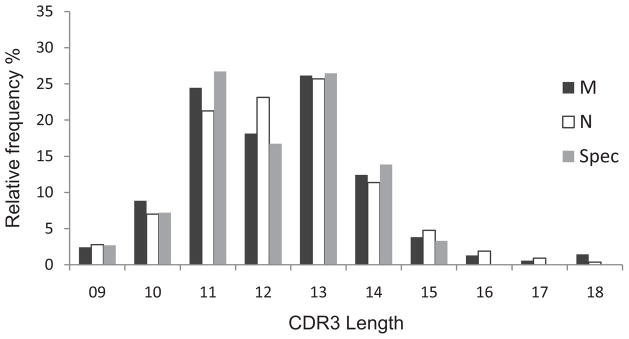
We use a definition of the CDR3 as the amino acids starting immediately after the conserved cysteine in the V region and extending to the amino acid immediately before the conserved phenylalanine-glycine in the J region. The observed CDR3 lengths spanned from 2 to 23, however CDR3 lengths representing cumulatively 99% of observations spanned from 9 to 18. The frequency of observations per CDR3 length was bimodal with lengths 11 and 13 most frequently observed (Fig 2, black bars). The frequency of clonotypes at each CDR3 length is more symmetric with the highest fraction of clonotypes at L13 (Figure 2, white bars). For comparison, the average BV19 CD8 spectratype data generated fromCD8 cells from the same samples is included. These data represent amplicons from the total cDNA and therefore are equivalent to the M data set.
Figure 2. CDR3 length distribution of the BV19 clonotypes.
The percent of clonotypes (open bars) or observations (closed bars) are shown for CDR3 lengths representing more than 1% of the data. Spectratype data from the same samples was averaged and shown as grey bars with standard deviations. The number of observation data is equivalent to a virtual spectratype .
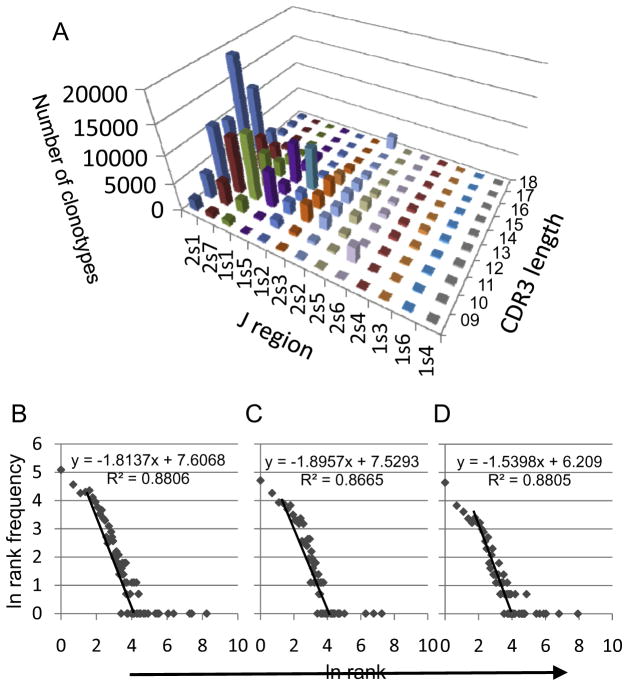
Deconvoluting the CDR3 length distribution on the basis of J region use, shows that the L13 skew is predominantly a function of J2s1, 1s5 and 1s2. The L11 skew is predominantly due to clonotypes expressing J2s7 and J1s1 (Fig 3A). The skews observed for J2s1, J2s7, J1s1, and J1s2 do not represent an underlying change in repertoire shape. This is shown by examining the log-transformed rank – rank frequency plots for these J and L based subsets. In the three cases where the number of clonotypes examined was greater than 500, the rank frequency plots were similar to that of the entire repertoire (Fig 3B). The outliers at L18 J2s2 and L10 J2s6 are due to one expanded clonotype, with one clonotype providing ~99% of the observations at L18 J2s2 and another providing ~96% of the observations at L10 J2s6. Skewing by such clonotypic expansion is expected in repertoires of older individuals (17,18).
Figure 3. J region analysis.
A. Frequency of observations of different J regions as a function of CDR3 length. The Y-axis (vertical) shows the number of clonotypes for each J region at each CDR3 length. J regions are colored differently and identified on the X-axis in order of decreasing frequency. CDR3 lengths are identified on the Z-axis. B-D. Loge rank vs loge rank frequency distribution of clonotypes expressing the three most frequent J regions. B. 2s1. C. 2s7. D. 1s1
CDR3 analysis
The CDR3 represents three genetic components; that derived from the V region, that from the D region, and that from the J region. In addition the CDR3 contains untemplated nucleotides flanking the D region. These are added by terminal transferase and perhaps polymerase mu (N-nucleotides), and less frequently by hairpin loop resolution (P-nucleotides); hence the name NDN region. For any V - J pair, the amino acids from these two components are fixed, with the diversity being a function of the NDN component.
The portion of the CDR3 derived from the NDN generates the most diversity in the TCR, but the length of the CDR3 is not fixed. We therefore examined the relation between CDR3 length and NDN length by plotting the number of clonotypes with each possible combination of values (Sup Figure 2). In general, the NDN length with the largest number of clonotypes corresponded to CDR3 lengths that were 7±1 amino acids longer. The clonotype distributions are distributed relatively equally in the CDR3 length dimension (row) and NDN length dimension (column).
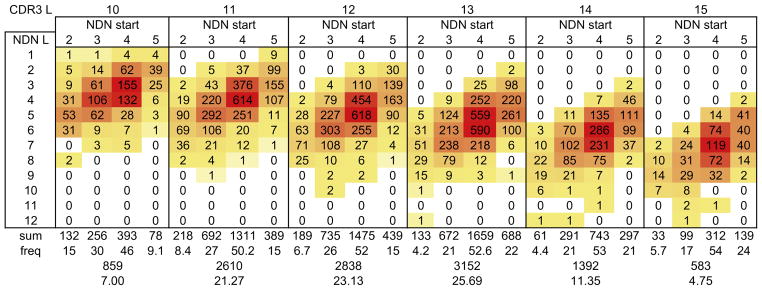
Another variable in CDR3 structure is the CDR3 position at which the NDN region starts. This corresponds to the rearrangement position. We examined the relation between the NDN length, CDR3 length, and the starting position of the NDN. BV19 can completely encode four amino acids after the cysteine (CASSI) and can encode the first two bases of either Asp or Glu. Thus, the NDN can start from CDR3 position 1 to 5. In the former case, the CDR3 is NDN encoded immediately after the Cys (C-NDN) and contains no V-region component. In the latter case all four V-region amino acids are included in the CDR3 and the NDN starts after the V-region component (CASSI-NDN). Interestingly, none of the clonotypes sequenced had an NDN start at CDR3 position 1. For each NDN start we examined the clonotype distribution as a function of CDR3 length and NDN length (Figure 4). The same relation between NDN length and CDR3 length (7±1 amino acids) is maintained but the NDN length decreases as the start position increase. Irrespective of CDR3 length, slightly more than half of NDN regions start at CDR3 position 4. Only 16.3% of NDN regions start at CDR3 Position 5.
Figure 4. Clonotype distribution as a function of CDR3 length, NDN start position and NDN length.
The CDR3 lengths are identified across the top of each set of four columns. The NDN starts are identified for each length. The NDN lengths are identified in the first column. The data are shaded to provide a relative heat map, with white being a minimum and red a maximum for each CDR3 L dataset. The sum and relative frequency for each NDN start are shown below each column, and the sum and percent are shown for each CDR3 L below each CDR3 length-based group
NDN amino acid sequence distributions
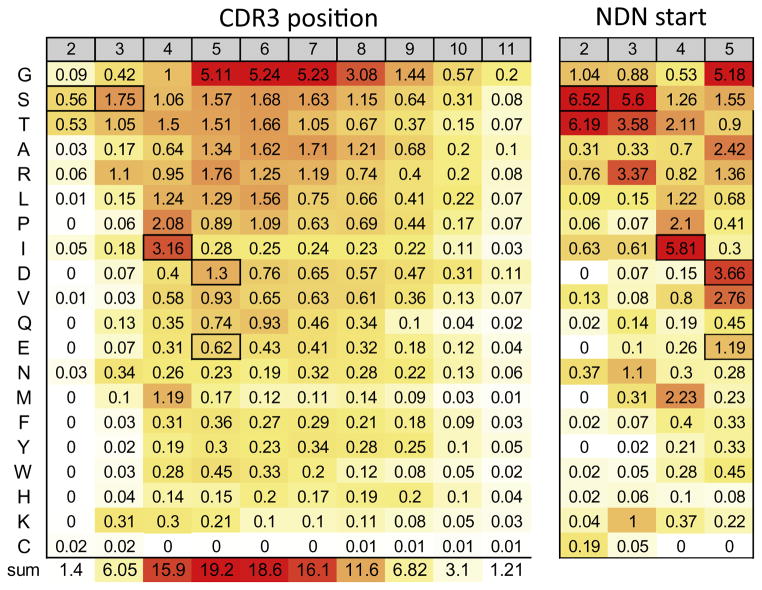
The amino acid composition of the NDN region of the TCR is of some importance as the specificity of the various BV19 clonotypes is determined by these residues, and a long term goal in immunology is to understand the details of TCR recognition of peptide-MHC. The measurements at this point are related to the number of clonotypes that encode the amino acid, and not simply the number of sequences in which the amino acid was observed. The percentage of clonotypes that encode the various amino acids at each CDR3 position is shown in Figure 5A, with the values highlighted to reflect relative frequency, with the highest as red and the lowest as white. Gly was the most frequent amino acid observed in the NDN (21.1%). It could be observed at all positions examined, but was especially frequent at positions 5 to 8. In general the frequency distribution of the amino acids across the CDR3 positions was highest at the central CDR3 positions (5 – 7) and dropped off at either edge. This is shown on the lowest row which sums the values at each position. There are some evident outliers; with S, T, or R at position 2, and P, I, or M at position 3.
Figure 5. Amino acid usage in the NDN region.
A. Amino acid usage is presented for each CDR3 position independent of NDN start. The amino acids identified by the one letter code are shown in the first column. The data are shaded to show relative frequency (white = 0, yellow = mid-frequency, red = maximum). The amino acids that would be encoded by the V gene are boxed. The sum of the values at each CDR3 position is shown in the bottom row with shading to indicate relative frequency. B. Amino acid usage at each NDN start position. The entire amino acid use data for each NDN start is given in Supplementary Figure 3. The data are shaded relative to the data in Supplementary Figure 3.
The amino acids expected from the V region at each position are boxed and they generally are among the most frequent at their corresponding position. This can be seen more readily if the amino acid frequency is examined as a function of NDN start position (Figure 5B). The NDN amino acids are ranked in the same order as in the total dataset for ease of comparison, but the relative frequency is calculated with respect to only those clonotypes where the NDN starts at the position identified. The data are only for the amino acid usage at the starting position itself. The entire amino acid utilization dataset for each NDN start position is provided in Supplementary Figure 3.
For NDN starts at position 2, there was a strong preference for Ser, Thr or Gly at the start. While this was seen to some extent in the overall data (Fig 5A), the effect was washed out by the relatively small number of clonotypes with NDN regions that start at position 2. Clonotypes encoding these three a.a. accounted for just over 80% of amino acids when the NDN starts at this position. A number of amino acids (D, E, Y or M) were never observed at this start position, and others were infrequent (L, P, Q, F, W, H or K). The high frequency of Ser at is due in part to rearrangements at the third codon position. Ser encoded as AGC, in lieu of the AGT encoded in the genome, constitutes 5.4% of the a.a. at position 2. However, selection on the amino acid itself also plays a role as Ser that are encoded as TCx (0.7%) are only surpassed at this position by Thr (6.19%), Gly (1.04%) and Arg (076%). The encoding of Arg, which is the other amino acid encoded by a third base change, is split relatively evenly between the AGA and AGG codons (20 vs 21 clonotypes). This equal encoding is expected if rearrangement was random and the selection was on the amino acid.
The same logic can be extended to the rearrangements at second base of the Ser codon; in this case G, changing to the other three possibilities. Rearrangement at the position equivalent to the second base of the codon would encode Asn or Lys, if the new base was A, Ile or Met if the base was T, and Thr if a G (Fig 5A). Ile and Asn are both represented at ~ 0.5% (Fig 5B). If we take this as the normal rate for rearrangement beginning at this nucleotide position, then the low values for Lys and Met indicate a negative selection and the high value for Thr could indicate a positive selection.
The data for starts at position 3 (Fig 5B, second column) resemble those for starts at position 2, which is not surprising since the V gene encodes a Ser with the same AGT codon at this position. Arg is now more frequent (3.24%) although not yet approaching the level of Ser (5.56%). Asn and Lys are present at similar levels (~1%). Ile is present in the same proportion as in position 2 starts but Met is increased to 0.3%. There is still a high level of Thr at this start (3.45%). Gly, Ala and Val are the other more frequent i.e. positively selected amino acids.
Ile is the most frequent amino acid at start position 4 (Fig 5B, third column) as would be expected since two of the three possible third base codon changes would encode Ile and the third change would encode Met, which is about half as prevalent (2.22%). If rearrangement took place at the second base of the codon, Ser, Arg, Asn, Lys and Thr are possible amino acids that would be generated, of which only Ser and Thr are observed at elevated levels. Interestingly Pro is third most frequent amino acid at this start.
Asp and Glu would be expected as preferred amino acids at position 5 since the last two bases of the V gene would encode one or the other. Asp expression is indeed elevated and is the second most frequent amino acid at start 5, with only Gly expressed more frequently. However, Glu is observed less frequently than Asp.
After position 5 the NDN sequence is a function of D-region and random addition. The amino acid usage in the central portion of the CDR3, positions 5 to 8 is relatively consistent (Fig 5A).
These data indicate that the V-encoded SSI sequence is favored at these positions when they are part of the NDN. After the initial four positions there is a relatively even usage of nine amino acids, with Gly being most frequent. As described above, NDN start and CDR3 position and NDN length have an effect on each other, so we examined the aa utilization independently for L11 to L15. Most of the above observations are CDR3 length independent (data not shown).
Distributions of NDN-encoded amino acid motifs
The NDN amino acid analysis presented above describes the CDR3 at a simple level. However, defining the combination of amino acids that generate the secondary structure needed for peptide-MHC recognition is the key reason for detailed CDR3 analysis. As described above, the focus on the BV19 repertoire was driven by the role of BV19 TCR chains in the HLA-A2 restricted response to the M158–66 epitope from influenza A. The major conserved NDN motif is the Arg–Ser doublet at CDR3 positions 5 and 6, in the context of CDR3 L11. This is the core of a more extended motif comprising one more amino acid on either side.
There are only 400 possible doublet motifs, which allows for a comprehensive description. Our dataset included examples of all but 13 of these (96.8%). The distribution of the doublet motifs is not easily described. Figure 6A shows the motifs ordered by the number of clonotypes in which they were observed. There are 39 motifs (~10%) that are observed in 301 to 2212 clonotypes (28602 total) represented by the red portion of the curve. The remaining 348 motifs (shown in green) are observed in 300 or less clonotypes. We assume that flexibility in the CDR3 can allow different arrangement in terms of contact with the pMHC. Thus an NDN region 3aa long can define two different doublet motifs, one 4aa long can define 3 doublet motifs, etc. This maps more than one doublet motif to one clonotype.
Figure 6. NDN motif distributions.
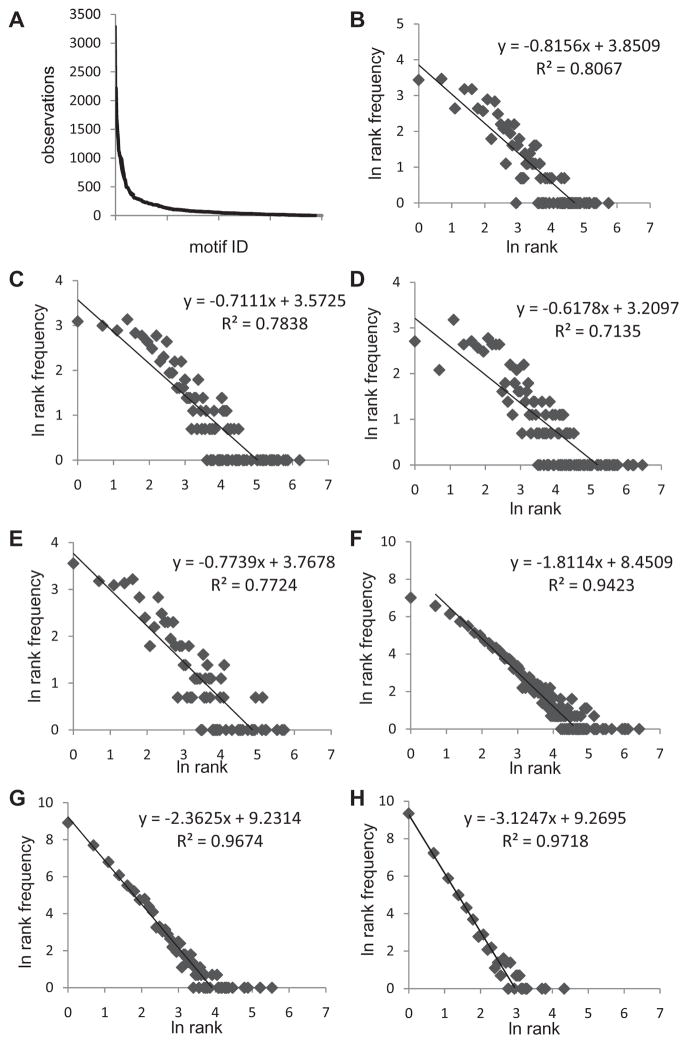
A. Plot of the various doublet motifs in descending frequency. The names of the motifs have been omitted for readability. B-E. Plot of the natural log of the rank vs that of the rank frequency data for the doublet motifs fractionated by CDR3 length. B = L11, C = L12. D = L13, and E = L14. F-H. Plot of the natural log of the rank vs rank frequency data for longer CDR3 amino acid motifs. F. Triplet. G. Quadruplet, H. Pentuplet. B-H. A line was fit to the data optimizing the R2 value. The R2, slope, and intercept are given for each graph.
The rank-frequency plot for the overall doublet motif data is not well behaved (Sup Fig 4A). Based on the relation between the number of possible motifs per clonotype and the CDR3/NDN length, we asked if the doublet data would generate a power law-like description when subsets based on CDR3 length are considered. The rank-frequency plots can be fit to a power law-like distribution for CDR3 lengths of 11 (Fig 6B), 12 (Fig 6C), and 14 (Fig 6E), all with R2 ≈ 0.8, but show a marginal fit for CDR3 L13 (Fig 6D, R2 = 0.7). The approach of analyzing subsets was extended to include the effect of NDN start position on the motif repertoire distribution. The distribution for the L11 to L14 doublet repertoires is shown in Supplemental Figure 4B-E. The problematic L13 subset (Supp Fig 4D) fractionated on this basis shows a better fit to a multi-component power law-like distribution (avg R2 ≈ 0.8).
If triplet motifs are considered the possible number of motifs is 8000, of which 4309 are represented in our data set (53.9%). These different triplet motifs were observed in 40700 clonotype exemplars. The rank-frequency of the entire dataset shows a good fit to a power law-like distribution, R2 = 0.94 (Fig 6F). As would be expected of a self-similar system, if the distributions are examined on the basis of CDR3 length the fit is also very good, average R2 = 0.94±0.02 (not shown). The 3aa motif distribution is beginning to approach the composite distribution (“hockey stick”) that we observe in recall responses to the M1 peptide.
Extending the motif length to 4aa increases the possible motifs to 160000 of which only 12004 (7.5%) are represented in the dataset. The 4aa motif distribution in the circulating repertoire (Fig 6G) shows an excellent fit for the power law like component, R2 ≈ 0.97. The same is true for 5 aa motifs (Fig 6H). As the motif length increases, our data represent less of what is possible, with the 12474 identified pentamer motifs representing 0.4% of the possible 3.2 × 106 motifs. However, the number of clonotype exemplars of the motifs drops from 40700, to 28700 to 17600 for the three, four and five a.a. length motifs respectively. Interestingly the loss of motif exemplars does not greatly change the number of motifs represented by only one clonotype, (y-intercept in Figs 6, F, G, and H). The change is seen in the slope of the rank frequency data, representing the rapidity with which the higher ranking component is reached.
Summary of repertoire measures, characteristics and diversity
An experimental description of the repertoire is based on the measurements made and a number of calculated characteristics. Table 1 provides a summary of the measures and characteristics for the data set analyzed. In addition to the more straightforward characteristics we include three diversity parameters describing the power law-like component of the repertoires as well as a log-transformed Shannon diversity measure based on the rank frequency distribution of the clonotypes or motifs.
Table 1.
| Analysis | N | M | M/N | Rmax | Dc | ln(SDI) | slope | intercept | R2 | Figure |
|---|---|---|---|---|---|---|---|---|---|---|
| CLONOTYPE | ||||||||||
| total | 12269 | 203185 | 16.56 | 5782 | 348.1 | 155.64 | −2.4 | 7.47 | 0.92 | Fig 1 |
| J2S1 L13 | 590 | 8124 | 13.77 | 60 | 3.36 | 52.59 | −1.81 | 7.61 | 0.88 | Fig 3B |
| J2S7 L11 | 406 | 5407 | 13.32 | 49 | 2.68 | 36.68 | −1.82 | 7.53 | 0.87 | Fig 3C |
| J1S1L11 | 253 | 3762 | 14.87 | 48 | 2.23 | 28.77 | −1.54 | 6.21 | 0.88 | Fig 3D |
| AMINO ACID MOTIF | ||||||||||
| 2 aa length | 387 | 53598 | 138.5 | 2213 | 14.98 | 665.17 | SupFig4A | |||
| 3 aa length | 4309 | 40698 | 9.44 | 610 | 63.59 | 52.88 | −1.72 | 8.12 | 0.95 | Fig 6F |
| 4 aa length | 12938 | 28723 | 2.22 | 253 | 112.96 | 7.48 | −2.41 | 9.34 | 0.97 | Fig 6G |
| 5 aa length | 13474 | 17576 | 1.30 | 76 | 57.26 | 2.04 | −3.13 | 9.27 | 0.97 | Fig 6H |
| 6 aa length | 8592 | 9553 | 1.11 | 19 | 16.09 | 0.80 | −4.12 | 9.05 | 0.99 | NS |
| 7 aa length | 4179 | 4440 | 1.06 | 5 | 3.71 | 0.48 | −4.56 | 8.29 | 0.99 | NS |
| 2aa by CDR3 length | [400] | |||||||||
| CDR3 L10 | 287 | 2335 | 8.14 | 93 | 10.43 | 34.32 | −1.10 | 4.31 | 0.86 | NS |
| CDR3 L11 | 363 | 8647 | 23.82 | 320 | 12.43 | 115.19 | −0.82 | 3.85 | 0.81 | Fig 6B |
| CDR3 L12 | 363 | 11249 | 30.99 | 491 | 14.84 | 160.98 | −0.71 | 3.57 | 0.78 | Fig 6C |
| CDR3 L13 | 360 | 14312 | 39.76 | 640 | 15.10 | 210.16 | −0.62 | 3.23 | 0.71 | Fig 6D |
| CDR3 L14 | 353 | 7528 | 21.33 | 306 | 13.35 | 103.84 | −0.77 | 3.77 | 0.77 | Fig 6E |
| CDR3 L15 | 330 | 3794 | 11.50 | 154 | 12.39 | 52.14 | −0.99 | 4.27 | 0.88 | NS |
| 3aa by CDR3 length | [8000] | |||||||||
| CDR3 L10 | 879 | 1471 | 1.67 | 13 | 6.77 | 3.58 | −2.43 | 6.61 | 0.99 | NS |
| CDR3 L11 | 1994 | 5520 | 2.77 | 51 | 17.42 | 9.36 | −2.16 | 7.49 | 0.95 | NS |
| CDR3 L12 | 2249 | 8339 | 3.71 | 123 | 32.17 | 15.36 | −2.01 | 7.52 | 0.95 | NS |
| CDR3 L13 | 2621 | 11117 | 4.24 | 201 | 46.39 | 18.96 | −1.95 | 7.62 | 0.94 | NS |
| CDR3 L14 | 2046 | 6081 | 2.97 | 124 | 40.72 | 11.27 | −2.14 | 7.44 | 0.95 | NS |
| CDR3 L15 | 1360 | 2976 | 2.19 | 64 | 28.25 | 6.51 | −2.31 | 7.023 | 0.96 | NS |
| 4aa by CDR3 length [160000] | ||||||||||
| CDR3 L10 | 709 | 807 | 1.14 | 5 | 3.39 | 0.64 | −3.84 | 6.53 | 0.98 | NS |
| CDR3 L11 | 2603 | 3362 | 1.29 | 11 | 7.52 | 1.68 | −3.3 | 7.9 | 0.99 | NS |
| CDR3 L12 | 3781 | 5775 | 1.53 | 39 | 24.53 | 3.65 | −2.96 | 8.49 | 0.97 | NS |
| CDR3 L13 | 4997 | 8303 | 1.66 | 79 | 46.54 | 5.53 | −2.61 | 8.31 | 0.98 | NS |
| CDR3 L14 | 3313 | 4972 | 1.50 | 52 | 33.65 | 3.43 | −2.77 | 7.85 | 0.96 | NS |
| CDR3 L15 | 1921 | 2623 | 1.37 | 38 | 26.83 | 1.93 | −3.13 | 7.5 | 0.97 | NS |
| 5aa by CDR3 length [3200000] | ||||||||||
| CDR3 L10 | 295 | 308 | 1.04 | 2 | 0.92 | 0.42 | −4.44 | 5.64 | NS | |
| CDR3 L11 | 1311 | 1400 | 1.07 | 3 | 1.81 | 0.48 | −4.44 | 7.18 | 0.99 | NS |
| CDR3 L12 | 2818 | 3187 | 1.13 | 9 | 6.96 | 0.01 | −3.42 | 7.61 | 0.97 | NS |
| CDR3 L13 | 4426 | 5219 | 1.18 | 17 | 13.42 | −0.74 | −3.57 | 8.28 | 1 | NS |
| CDR3 L14 | 2989 | 3541 | 1.18 | 18 | 14.19 | 0.59 | −3.41 | 7.8 | 0.99 | NS |
| CDR3 L15 | 1689 | 1962 | 1.16 | 11 | 8.47 | 0.97 | −3.71 | 7.39 | 0.99 | NS |
| 2aa by NDN length | ||||||||||
| NDN L3 | 310 | 2693 | 8.69 | 111 | 11.80 | 36.81 | −1.08 | 4.35 | 0.84 | NS |
| NDN L4 | 355 | 7830 | 22.06 | 341 | 14.46 | 110.55 | −0.82 | 3.86 | 0.81 | NS |
| NDN L5 | 364 | 12680 | 34.84 | 525 | 14.07 | 181.48 | −0.67 | 3.38 | 0.71 | NS |
| NDN L6 | 362 | 12535 | 34.63 | 402 | 10.60 | 179.84 | −0.66 | 3.34 | 0.71 | NS |
| NDN L7 | 356 | 9024 | 25.35 | 393 | 14.55 | 129.26 | −0.76 | 3.66 | 0.79 | NS |
| NDN L8 | 446 | 4488 | 10.06 | 169 | 11.65 | 62.08 | −0.95 | 4.16 | 0.84 | NS |
Column headers: N-number of unique elements, M-total number of copies, M/N-number of copies per element; Rmax-maximum rank in a set; Dc-crude diversity measure; ln(SDI)-natural log of Shannon diversity index. The slope, intercept and R2 values are for the major power law-like component of the rank frequency data set. Figures associated with the rank frequency data are identified. Numbers in brackets show the maximum number of possible amino acid motifs. NS - not shown.
DISCUSSION
The TCR BV19 CDR3 sequence data analyzed here describe a circulating repertoire that is the culmination of a long period of adaptive responses that relied on pre-existing T cells. We have smoothed out short term temporal fluctuations by pooling cells collected at six time points over a period of ~1.5 year. The repertoire was characterized both at the clonotype level and at the amino acid level in terms of individual amino acid usage at each CDR3 position and in terms of distributions of combinations of amino acids referred to as motifs.
The specific recognition of pMHC by the TCR is a function of the CDR3 length and sequence. Fine specificity is provided by the NDN-region contribution to the CDR3 in the case of the β-chain and the N region in the case of the α-chain. While the CDR3 lengths can vary from 2 to 23, lengths of 10 to 15 represented 5% or more of the clonotypes, with L11 to L14 accounting for ~ 80% of clonotypes. This short length is compatible with the CDR3 shortening reported during the positive selection step of thymic maturation (20) and the overall flat nature of the TCRαβ-pMHCI interface.
The variables in CDR3 structure also incorporate the point at which the NDN region starts in the context of the overall CDR3 length. Interestingly, the most frequent NDN start occurs at CDR3 position 4 (~51%), which formally could be a V-encoded position. Around 31% of the NDN starts occur at positions 2 or 3 and ~18% occur at position 5. The significance of the high number of clonotypes with NDN starts at CDR3 position 4 is under further study.
The analysis of amino acid utilization on the basis of NDN start and CDR3 length showed that for NDN starts internal to the end of BV19 V-gene, there is a propensity to encode the same or a related amino acid sequence as would be encoded for by the V gene. This argues that the 3’ V gene sequences have been selected to be useful in the pMHC recognition process. This constraint could prove helpful in modeling TCR-pMHC recognition. Glycine was the most common amino acid observed at most positions independent of NDN start and CDR3 length. There were certain increases in amino acid frequency that indicated possible selection.
Even though the repertoire analysis was of an older subject the repertoire was still diverse. This was observed for the overall repertoire as well as J-defined subsets and was evidenced using our simple diversity measure as well as by the highly correlated Shannon diversity measure (Table 1). The clonotype rank frequency analysis showed a three component plot with an extended range of single high-ranking clonotypes that are an expected outcome of a longer life (17, 18). The remaining clonotype components represent a large portion of mid-rank frequency clonotypes and a smaller low-frequency component. Both these components fit well to a power law-like distribution. The distribution represented by these two components bears a strong resemblance to our previous modeling of repertoire generation and selection in silico (Fig 10 in ref 14). The fit of the corrected clonotype data into a reasonable mathematical description that fits with expected scenarios of generation and selection is welcome as it indicates that the analysis platform is generally well-behaved, and that quantitative measurements can be made. We have made extended use of describing recall repertoires in the context of power law-like components in the analysis and modeling of repertoire changes between middle-aged and older individuals (20). Previous next-generation sequencing analyses of TCR repertoires had shown that the repertoires have distributional components that can be described as power law-like (1) and self-similar (21). Our more detailed analysis of the BV19 CD8 repertoire shows the same self-similarity in repertoire subset definitions as we first observed for the recall repertoires indicating the validity of this approach. Our argument for power law-like distributional behavior in recall was based on responder population assumptions incorporating expansion capabilities linked to a range of TCR avidities (9, 14). If the circulating repertoire in an older individual represents the history of similar in vivo expansions linked to a long and complex history of exposures, it should not be surprising that the overall circulating repertoire is similar to the recall repertoire for a particular peptide.
The diversity extends to the amino acid motifs encoded by the clonotypes. We define a motif as any serial combination of amino acids two or longer. Interestingly, the analysis revealed a relationship between the lengths of the NDN encoded motif and the ability to elegantly describe the diversity. The combined dimer motif dataset showed a poor fit to a power law-like distribution. Since dimers represent a very small set, 400 possibilities, we reasoned that the data were saturating the system. This was remedied in part by examining the dimer distributions as a function of CDR3 length, or CDR3 length and NDN start. Three and four amino acid-long motifs showed a very good fit to power law-like distributions. We propose that the ability to define the longer motifs in terms of a power law-like distribution is related to the optimum length of the NDN region for selection on pMHC and enrollment in the memory repertoire.
The repertoire analysis presented here presents an examination and enumeration of CDR3 amino acids and the distributions of CDR3 amino acid motifs. The data indicate a complex distribution of CDR3 motifs, that can be described in part as power-law like. The high-frequency component of the motif repertoire was populated by CDR3 that maintain amino acids that would be V-region encoded, and that are glycine rich. Arguing on the basis of the self-similar characteristic of the repertoire, we hypothesize that the observations made here for BV19 will generalize to the other BV genes in the CD8 repertoire.
Supplementary Material
Acknowledgments
This work was supported by NIH Contract NO1 AI-50032. We thank Dr Liz Worthy and Mike Tschannen at the Human and Molecular Genetics Center of the Medical College of Wisconsin for 454 sequencing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Robins HS, Campreghe PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T cell receptor β chain diversity in αβ T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C, Sanders CM, Yang Q, Schroeder HW, Jr, Wang E, Babrzadeh F, Gharizadeh B, Myers RM, Hudson JR, Jr, Davis RW, Han J. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci USA. 2010;107:1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qi Q, Liu Y, Cheng Y, Glanville J, Zhang D, Lee JY, Olshen RA, Weyand CM, Boyd SD, Goronzy JJ. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci U S A. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moss PA, Moots RJ, Rosenberg WM, WM, Rowland-Jones SJ, Bodmer HC, McMichael AJ, Bell JI. Extensive conservation of α-β-chains of the human T cell antigen receptor recognizing HLA-A2 influenza A matrix peptide. Proc Natl Acad Sci USA. 1991;88:8987–8990. doi: 10.1073/pnas.88.20.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehner PJ, Wang EC, Moss PA, Williams S, Platt K, Friedman SM, Bell JI, Borysiewicz LK. Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the Vβ17 gene segment. J Exp Med. 1995;181:79–91. doi: 10.1084/jem.181.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naumov YN, Naumova EN, Yassai MB, Kota K, Welsh RM, Selin LK. Multiple glycines in TCR alphachains determine clonally diverse nature of human T cell memory to influenza A virus. J Immunol. 2008;181:7407–7419. doi: 10.4049/jimmunol.181.10.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naumov YN, Hogan KT, Naumova EN, Pagel JT, Gorski J. A class I MHC-restricted recall response to a viral peptide is highly polyclonal despite stringent CDR3 selection: implications for establishing memory T cell repertoires in “real-world” conditions. J Immunol. 1998;160:2842–2852. [PubMed] [Google Scholar]
- 8.Zhou V, Yassai MB, Regunathan J, Box J, Bosenko D, Vashishath Y, Demos W, Lee F, Gorski J. The functional CD8 T cell memory recall repertoire responding to the influenza A M1(58–66) epitope is polyclonal and shows a complex clonotype distribution. Hum Immunol. 2013;74:809–817. doi: 10.1016/j.humimm.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naumov YN, Naumova EN, Hogan KT, Selin LK, Gorski J. A fractal clonotype distribution in the CD8+ memory T cell repertoire could optimize potential for immune responses. J Immunol. 2003;170:3994–4001. doi: 10.4049/jimmunol.170.8.3994. [DOI] [PubMed] [Google Scholar]
- 10.Naumov YN, Naumova EN, Clute SC, Watkin LB, Kota K, Gorski J, Selin LK. Complex T cell memory repertoires participate in recall responses at extremes of antigenic load. J Immunol. 2006;177:2006–2014. doi: 10.4049/jimmunol.177.3.2006. [DOI] [PubMed] [Google Scholar]
- 11.Petrova G, Naumova EN, Gorski J. The polyclonal CD8 T cell response to influenza M158–66 generates a fully connected network of cross-reactive clonotypes to structurally related peptides: a paradigm for memory repertoire coverage of novel epitopes or escape mutants. J Immunol. 2011;186:6390–6397. doi: 10.4049/jimmunol.1004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yassai MB, Naumov YN, Naumova EN, Gorski J. A clonotype nomenclature for T cell receptors. Immunogenetics. 2009;61:493–502. doi: 10.1007/s00251-009-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naumova EN, Gorski J, Naumov YN. Two compensatory pathways maintain long-term stability and diversity in CD8 T cell memory repertoires. J Immunol. 2009;183:2851–2858. doi: 10.4049/jimmunol.0900162. [DOI] [PubMed] [Google Scholar]
- 14.Naumova EN, Gorski J, Naumov YN. Simulation studies for a multistage dynamic process of immune memory response to influenza: experiment in silico. Ann Zool Fennici. 2008;45:369–384. doi: 10.5735/086.045.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Litwin S, Jores R. Shannon Information as a Measure of Amino Acid Diversity. Theoretical and Experimental Insights into Immunology. NATO ASI Series. 1992;66:279–287. [Google Scholar]
- 16.Maślanka K, Piatek T, Gorski J, Yassai M, Gorski J. Molecular analysis of T cell repertoires. Spectratypes generated by multiplex polymerase chain reaction and evaluated by radioactivity or fluorescence. Hum Immunol. 1995;44:28–34. doi: 10.1016/0198-8859(95)00056-a. [DOI] [PubMed] [Google Scholar]
- 17.Hingorani R, Choi IH, Akolkar P, Gulwani-Akolkar B, Pergolizzi R, Silver J, Gregersen PK. Clonal predominance of T cell receptors within the CD8+ CD45RO+ subset in normal human subjects. J Immunol. 1993;151:5762–5769. [PubMed] [Google Scholar]
- 18.Posnett DN, Sinha R, Kabak S, Russo C. Clonal populations of T cells in normal elderly humans: the T cell equivalent to “benign monoclonal gammapathy”. J Exp Med. 1994;179:609–618. doi: 10.1084/jem.179.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart JJ, Lee CY, Ibrahim S, Watts P, Shlomchik M, Weigert M, Litwin S. A Shannon entropy analysis of immunoglobulin and T cell receptor. Mol Immunol. 1997;34:1067–1082. doi: 10.1016/s0161-5890(97)00130-2. [DOI] [PubMed] [Google Scholar]
- 20.Naumov YN, Naumova EN, Yassai MB, Gorski J. Selective T cell expansion during aging of CD8 memory repertories to influenza revealed by modeling. J Immunol. 2011;186:6617–6624. doi: 10.4049/jimmunol.1100091. [DOI] [PubMed] [Google Scholar]
- 21.Meier J, Roberts C, Avent K, Hazlett A, Berrie J, Payne K, Hamm D, Desmarais C, Sanders C, Hogan KT, Archer KJ, Manjili MH, Toor AA. Fractal organization of the human T cell repertoire in health and after stem cell transplantation. Biol Blood Marrow Transplant. 2013;19:366–377. doi: 10.1016/j.bbmt.2012.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.