Abstract
The phosphoinositide 3-kinase (PI3K) signaling pathway is a complex and tightly regulated network that is critical for many physiological processes such as cell growth, proliferation, metabolism and survival. Aberrant activation of this pathway can occur through mutation of almost any of its major nodes and has been implicated in a number of human diseases including cancer. The high frequency of mutations in this pathway in multiple types of cancer has led to the development of small molecule inhibitors of PI3K, several of which are currently in clinical trials. However, several feedback mechanisms either within the PI3K pathway or in compensatory pathways can render tumor cells resistant to therapy. Recently, targeting proteins of the bromodomain and extra-terminal (BET) family of epigenetic readers of histone acetylation has been shown to effectively block adaptive signaling response of cancer cells to inhibitors of the PI3K pathway, which at least in some cases can restore sensitivity. BET inhibitors also enforce blockade of the MAPK, JAK/STAT and ER pathways suggesting they may be a rational combinatorial partner for divergent oncogenic signals that are subject to homeostatic regulation. Here, we review the PI3K pathway as a target for cancer therapy and discuss the potential use of BET inhibition to enhance clinical efficacy of PI3K inhibitors.
Background
PI3Ks are a family of lipid kinases that phosphorylate the 3-hydroxyl group on phosphoinositides, generating second messengers that regulate several downstream pathways that are central in both normal physiology and disease (1, 2). In mammals, there are three classes of PI3Ks that differ in structure and substrate specificity but to date, mainly class IA PI3Ks has been implicated in the etiology of various diseases including cancer (3). Class IA PI3Ks are heterodimers composed of a p110 catalytic subunit (α, β and δ) and a p85/55 regulatory subunit (encoded by three different genes that are subject to alternative splicing) that can be activated downstream of Receptor tyrosine kinases (RTKs), G protein-coupled receptors (GPCRs) and small GTPases (4). Although PI3K was first linked to cancer almost thirty years ago when it was associated with the transforming activity of viral oncogenes (5), it wasn’t until the early 2000’s that PI3Ks were brought to the forefront of cancer research when PIK3R1 (6) and PIK3CA (7), the genes encoding p85α and p110α respectively, were found to be frequently mutated in several types of solid tumors. Since then, multiple studies have established that PIK3CA is one of the most, if not the most, frequently mutated oncogenes in human cancer. Mutations are mainly clustered in two hot-spots of the enzyme and can increase p110α activity through a variety of mechanisms (8–10).
In quiescent cells, p85 binds to p110, stabilizing it and inactivating its kinase activity (Fig. 1). Following growth factor stimulation, the PI3K complex is activated after binding to phosphotyrosines on receptors and adaptor proteins. The primary consequence of PI3K activation is the conversion of phosphatidylinositol-4,5-bisphosphate (PIP2) into the short-lived second messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3) on the inner leaflet of the plasma membrane. PIP3 recruits proteins to the membrane that contain a pleckstrin-homology (PH) domain, including AKT and its upstream activators PDK1 and mTORC2. At the membrane, PDK1 phosphorylates AKT at T308 (11) and mTORC2 phosphorylates it at S473 (12), which results in full activation of the enzyme. AKT acts as a major mediator of PI3K signaling by phosphorylating a wide range of substrates that regulate cell cycle entry, survival, protein synthesis, RNA translation, glucose metabolism and migration. PI3K activity is tightly controlled in cells and can be attenuated by lipid phosphatases, such as PTEN (13), INPP4B (14), and SHIP2 (15) that dephosphorylate phospholipids in positions 3, 4 and 5, respectively, on the inositol ring. PIP3 is also an important signal upstream of several pro-oncogenic signals including SGK3 (16, 17) and PREX1/ PREX2 (18, 19).
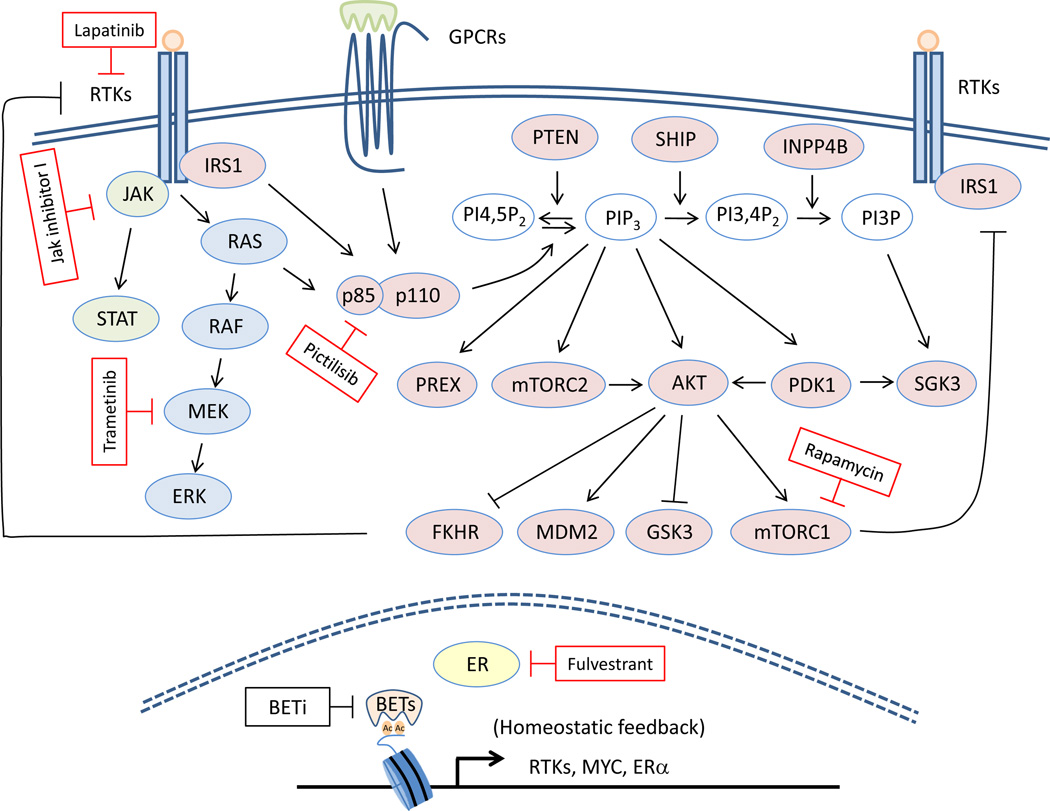
Figure 1.
Schematic representation of nodes of therapeutic blockade in the PI3K and other signaling pathways that synergize with BET inhibition. PI3Ks are a family of heterodimeric proteins that can be activated downstream of RTKs, GPCRs and small-GTPases. They catalyze the conversion of PIP2 to the second messenger PIP3 which helps recruit to the membrane proteins that contain a PH-domain such as AKT, PDK1, mTORC2 and PREX1/2. PI3K activity can be attenuated by several phosphatases including PTEN, SHIP1/2 and INPP4B. AKT acts as a major mediator of PI3K signaling by phosphorylating a wide range of substrates that regulate cell growth, proliferation, metabolism and survival. Given the high frequency of PI3K pathway activation in human cancers several inhibitors targeting kinases throughout the pathway are currently being evaluated in clinical trials. However their efficacy as monotherapies can be limited due to a variety of mechanisms including the unleashing of FOXO- and mTORC1-mediated feedback loops that reactivate the pathway. Inhibition of BET proteins has recently been shown to effectively block adaptive signaling response of cancer cells to inhibitors of the PI3K pathway and other signaling pathways (shown in red rectangles) suggesting they may be rational combinatorial partners for multiple oncogenic signals.
Overactivation of the PI3K signal is one of the most frequent events in human cancers and can be achieved through alterations in most of the major nodes of the pathway (4). Activating mutations and/or amplification of the genes encoding the PI3K catalytic subunits p110α (7) and less frequently p110β (20), mutations in the p85α regulatory subunit that abrogate its p110-inhibitory activity (6, 21) as well as loss of function of the lipid phosphatases PTEN (22, 23) and INPP4B (14) can all promote PI3K activity and cellular transformation. Mutations in genes acting both upstream of PI3Ks such as RTKs (for example EGFR and HER2) and the PI3K activator K-RAS as well as the downstream PI3K effectors AKT1–3, PDK1, TSC1/2 and mTOR are variably present in human tumors and should also result in aberrant activation of the PI3K pathway.
Clinical-Translational Advances
Given the high frequency of PI3K pathway activation in human cancers, a significant amount of effort has been put into the development of drugs targeting several kinases throughout the pathway, especially PI3Ks. Several inhibitors targeting all PI3K isoforms (Pan-PI3K) are currently in clinical trials but although they have turned out to be reasonably well tolerated, they have shown limited efficacy as single agents (24). Partial inhibition of the signal due to limitations in the dosing, compensatory feedback mechanisms as well as off-target effects could all account for the limited clinical responses that were observed in patients treated with Pan-PI3K inhibitors. Recent preclinical studies have highlighted the divergent roles of class I PI3K catalytic isoforms depending on the genetic context. For example, HER2/Neu- and KRAS-driven tumors have been shown to rely mainly on p110α (25, 26) whereas p110β has been shown to be important in certain PTEN-deficient tumors (27–29). In light of these findings PI3K isoform-selective inhibitors have been developed and are being tested in clinical trials with the hope to maximize target-inhibitory doses while sparing the adverse toxic effects of Pan-PI3K inhibitors. In 2014 Idelalisib (CAL-101) became the first p110 isoform-specific inhibitor (against p110δ) that has been approved by the FDA for the treatment of chronic lymphocytic leukemias (30) and B cell lymphomas (31). Alpelisib (BYL719) is a p110α-specific PI3K inhibitor that has shown some encouraging results in early phase clinical trials. As a single agent it resulted in some tumor regressions and prolonged disease control in heavily pretreated patients with various tumor types carrying a PIK3CA mutation (32). Preliminary results from the combination of BYL719 and Fulvestrant, an estrogen receptor antagonist, also indicate an encouraging activity in patients with PIK3CA-mutant breast cancer, with 3 patients achieving confirmed partial responses (33). GDC-0032, a β isoform-sparing PI3K inhibitor targeting PI3Kα/δ/γ, is another example of next generation PI3K inhibitors that has shown promising preliminary clinical activity in PIK3CA mutant cancers (34) and is currently being evaluated in solid tumors both as a single agent and in combination with endocrine therapies and other anticancer therapies. Given these early signals of clinical activity more PI3K inhibitors could be FDA approved for the use in human cancer, especially as part of combination therapies. As PI3K inhibitors are making their way into trials, it is critical to identify biomarkers that will help future selection of patients that are most likely to benefit from these targeted therapies.
While Herceptin (Trastuzumab), a monoclonal antibody against HER2, has shown remarkable activity in HER2-postitive breast cancer patients, the efficacy of kinase-based therapeutics that act downstream has been limited due to a variety of adaptive and compensatory responses to the drugs within the cancer cell. Early preclinical studies with inhibitors against mTOR identified that blockade of the mTOR-mediated activation of S6 kinase released inhibition of IRS1, resulting in aberrant PI3K/AKT activation (35). To deal with unleashing of this natural break in the pathway several dual PI3K/mTOR inhibitors have been developed and are being tested in early phase clinical trials. As these compounds exhibit a broad activity profile and significantly higher toxicity, they might be more suitable for the treatment of patients with more than one alteration in the PI3K pathway. Multiple studies have also shown that feedback activation of the PI3K pathway in response to PI3K inhibitors can also be achieved by suppression of the FOXO-dependent activation of expression of RTKs (36–38). Blocking PI3K, along with different upstream RTKs has been shown to block this adaptive response and resensitize cancer cells (36, 39–41).
PI3K isoforms are able to compensate for each other as well. For example, preclinical studies have shown that cancer cells can induce expression of p110β to counteract the blockade of p110α and vice versa (42, 43). In an ER+/PIK3CA-mutant breast cancer patient treated with the p110α-specific inhibitor BYL719, loss of PTEN expression (which could likely switch dependence on the p110β isoform) was present in multiple metastatic lesions (44). Markedly, treatment of xenografts derived from the BYL719-resistant lesions with the p110β-selective inhibitor AZD6482 was able to restore sensitivity. Concurrent inhibition of multiple PI3K isoforms or the use of dual-isoform PI3K inhibitors might be necessary for the treatment of tumors that have become refractory to isoform-specific inhibitors. Compounds that would be active against only the mutant PIK3CA should minimize increased systemic insulin production that is seen in the clinic upon treatment with p110α inhibitors and are highly anticipated in the field. Finally, activation of several compensatory pathways has been documented to drive resistance to PI3K inhibitors and could therefore inform combinatorial treatments. Clinical trials are currently evaluating preclinical findings of synergism between PI3Ki and antiestrogen therapy (45) as well as inhibitors against poly-ADP-ribose polymerase (PARP) (46), Cyclin-dependent kinases 4 and 6 (CDK4/6) (47) and MEK (37, 48).
Within the past year inhibition of BET proteins (BETi) has emerged as a potential therapeutic approach to restore sensitivity to kinase inhibitors of the PI3K pathway. The BET family of proteins consists of 4 members, BRD2–4 which are ubiquitously expressed and BRDT that is only expressed in germ cells. They contain two tandem bromodomains (BRDs) located at the N-terminus that recognize and bind acetylated-lysine residues in nucleosomal histones to facilitate the recruitment of transcription factors and chromatin organizers required in transcription initiation and elongation. Initially, the concept of targeting a bromodomain was thought to be challenging because it would involve inhibiting a protein-peptide interaction. Moreover there was a lot of skepticism around the concept of epigenetic therapy as inhibition of BET proteins which are global regulators of gene transcription was expected to impact on the transcriptional activity across all active genes and be highly toxic. In 2010 two selective and potent BET protein inhibitors, JQ1 (49) and I-BET762 (50) were reported to have activity in a NUT midline carcinoma and an inflammatory disease model respectively. Mechanistically, BET inhibition was shown to result in significant reduction of the transcript levels of only a small number of key genes in a cell- and context-specific manner (51, 52). This recent discovery of small molecules capable of blocking their lysine-binding pocket is the first successful example of pharmaceutical inhibition of epigenetic readers and has sparked intense efforts to develop novel BRD antagonists. Several compounds are currently tested in early phase clinical trials (Table 1) for solid tumors and hematological malignancies where for example deregulated c-MYC, a major target of BET proteins in this context, is an important driver of tumorigenesis. Most of them share a similar thieno-diazepine scaffold whereas for some of them the exact chemical structure has not yet been disclosed (53). Notably, multiple studies have recently documented that BET inhibition can effectively block adaptive responses to inhibitors of the PI3K pathway that help cancer cells to evade the effects of the drug and develop resistance. For example, our group has shown that BET inhibitors can block BRD4 from binding to regulatory regions of genes encoding several RTKs that are frequently induced by treatment with Pictilisib (GDC-0941), a Pan-PI3K inhibitor (Fig. 1). As a result, combined PI3K and BET inhibition can prolong blockade of the PI3K signal and induce cell death in multiple tumor cell lines (54). BET proteins have also been shown to regulate sensitivity of breast cancer cells to treatment with Lapatinib, an EGFR/HER2 inhibitor, by blocking the expression/phosphorylation of kinases (55) and/or activation of the FOXO/c-MYC axis (56). Finally, inhibitors of BET proteins have shown synergism with mTOR inhibitors in multiple tumor models (57–59) suggesting that blockade of the epigenetic regulation of the PI3K pathway may present an opportunity to overcome resistance to kinase inhibitor therapy.
Table 1.
BET inhibitors currently in clinical trials for human cancers
| Drug | Sponsor | Solid tumors or hematological malignancies |
Most advanced clinical phase |
|---|---|---|---|
| I-BET762 | GlaxoSmithKline | Both | I/II |
| GSK2820151 | GlaxoSmithKline | Solid tumors | I |
| TEN-010 | Tensha | Both | I |
| OTX015 | Oncoethix/Merck | Both | II |
| BAY 1238097 | Bayer | Both | I |
| CPI-0610 | Constellation Pharmaceuticals | Hematological malignancies | I |
| GS-5829 | Gilead | Both | I |
| INCB054329 | Incyte | Both | I |
| ABBV-075 | Abbvie | Both | I |
| BMS-986158 | Bristol-Myers Squibb | Solid tumors | I/IIa |
| FT-1101 | Forma Therapeutics, Inc | Hematological malignancies | I |
Data taken from http://clinicaltrials.gov/.
Several groups have investigated BETi as a potential therapeutic approach to potentiate the effect of targeted therapy against oncogenic drivers in other signaling pathways. For example, combinations of JQ1 and tyrosine kinase inhibitors such as Imatinib and JAK inhibitor I were shown to synergize and induce apoptosis in leukemias and lymphomas driven by constitutive STAT5 activation (60). Using an anti-cancer drug library containing 180 small molecule inhibitors, Jing et al identified that combined BETi/MEKi suppressed both cell proliferation and survival in an ovarian cancer model (61). In an ER+/tamoxifen-resistant breast cancer model, JQ1 induced a strong growth inhibitory effect when it was combined with fulvestrant (62). These studies highlight the ability of BETi to synergize with inhibition of diverse oncogenic signals which could be due to their unique ability to interfere with the adaptive feedback expression of a few relevant genes to restore signaling homeostasis depending on the genetic context (Fig. 1). However, it remains to be seen whether there will be a therapeutic window for any of the above combinatorial therapies in the clinic.
A striking development in BRDi research is the recent finding that BET proteins are unanticipated targets of certain widely used kinase inhibitors. The first such report was by Martin et al. who discovered that the potent CDK inhibitor Dinaciclib interacts with the acetyl-lysine recognition site of the bromodomain testis-specific protein BRDT (63). Soon after, two new studies (64, 65) identified more than a dozen kinase inhibitors that possess cross-reactivity towards BRD4 with nanomolar potencies, including the JAK2 inhibitor TG101209 and the PLK1 inhibitor BI2536 which is in Phase I/II Clinical trials for acute myeloid leukemia (AML) and non-small cell lung cancer (NSCLC). This finding has triggered the development of more potent dual BET/PLK1 inhibitors using a structure–activity relationship (SAR) study (66). Notably, LY94002 which has been routinely used to block PI3K activity was also recently shown to interact with BRD2–4 (67). The above studies provide the framework for the rational design of next-generation dual BET-Kinase inhibitors. The simultaneous inhibition of two structurally and functionally unrelated proteins by a single drug may provide a new opportunity when the application of combination therapies poses significant clinical challenges.
Conclusions
The concept of oncogene addiction described by the dependence of certain tumors for their growth and survival on a limited number of mutational events has fueled the development of targeted therapies. The PI3K signaling pathway has attracted much interest due to its involvement in a large fraction of human tumors and several inhibitors targeting oncogenic kinases throughout the pathway are currently being tested in clinical trials. However, preclinical studies and past experience with other kinase-based therapies suggest that escape mechanisms and drug resistance may eventually limit the efficacy of these compounds as monotherapies. Concurrent inhibition of PI3K and BET proteins may provide an alternative for durable inhibition of the oncogenic signal and clinical trials could test the safety and efficacy of such combinations.
Acknowledgments
Grant Support: R.E. Parsons is supported by the NCI of the NIH under award number R01CA129432 and a Stand Up To Cancer Dream Team Translational Research Grant (SU2C-AACR-DT0209). Stand Up To Cancer is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 2.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nature Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 3.Vanhaesebroeck B, Waterfield MD. Signaling by distinct classes of phosphoinositide 3-kinases. Exp Cell Res. 1999;253:239–254. doi: 10.1006/excr.1999.4701. [DOI] [PubMed] [Google Scholar]
- 4.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitman M, Kaplan DR, Schaffhausen B, Cantley L, Roberts TM. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 6.Philp AJ, Campbell IG, Leet C, Vincan E, Rockman SP, Whitehead RH, et al. The phosphatidylinositol 3'-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res. 2001;61:7426–7429. [PubMed] [Google Scholar]
- 7.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 8.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 9.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 10.Hao Y, Wang C, Cao B, Hirsch BM, Song J, Markowitz SD, et al. Gain of interaction with IRS1 by p110alpha-helical domain mutants is crucial for their oncogenic functions. Cancer Cell. 2013;23:583–593. doi: 10.1016/j.ccr.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stokoe D, Stephens LR, Copeland T, Gaffney PR, Reese CB, Painter GF, et al. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 12.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 13.Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 14.Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clement S, Krause U, Desmedt F, Tanti JF, Behrends J, Pesesse X, et al. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature. 2001;409:92–97. doi: 10.1038/35051094. [DOI] [PubMed] [Google Scholar]
- 16.Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasser JA, Inuzuka H, Lau AW, Wei W, Beroukhim R, Toker A. SGK3 mediates INPP4B-dependent PI3K signaling in breast cancer. Mol Cell. 2014;56:595–607. doi: 10.1016/j.molcel.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donald S, Hill K, Lecureuil C, Barnouin R, Krugmann S, John Coadwell W, et al. P-Rex2, a new guanine-nucleotide exchange factor for Rac. FEBS Lett. 2004;572:172–176. doi: 10.1016/j.febslet.2004.06.096. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeldt H, Vazquez-Prado J, Gutkind JS. P-REX2, a novel PI-3-kinase sensitive Rac exchange factor. FEBS Lett. 2004;572:167–171. doi: 10.1016/j.febslet.2004.06.097. [DOI] [PubMed] [Google Scholar]
- 20.Pazarentzos E, Giannikopoulos P, Hrustanovic G, St John J, Olivas VR, Gubens MA, et al. Oncogenic activation of the PI3-kinase p110beta isoform via the tumor-derived PIK3Cbeta kinase domain mutation. Oncogene. 2016;35:1198–1205. doi: 10.1038/onc.2015.173. [DOI] [PubMed] [Google Scholar]
- 21.Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, Wang W, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16:463–474. doi: 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 23.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 24.Mayer IA, Arteaga CL. The PI3K/AKT Pathway as a Target for Cancer Treatment. Annu Rev Med. 2016;67:11–28. doi: 10.1146/annurev-med-062913-051343. [DOI] [PubMed] [Google Scholar]
- 25.Utermark T, Rao T, Cheng H, Wang Q, Lee SH, Wang ZC, et al. The p110alpha and p110beta isoforms of PI3K play divergent roles in mammary gland development and tumorigenesis. Genes Dev. 2012;26:1573–1586. doi: 10.1101/gad.191973.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castellano E, Sheridan C, Thin MZ, Nye E, Spencer-Dene B, Diefenbacher ME, et al. Requirement for interaction of PI3-kinase p110alpha with RAS in lung tumor maintenance. Cancer Cell. 2013;24:617–630. doi: 10.1016/j.ccr.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wee S, Wiederschain D, Maira SM, Loo A, Miller C, deBeaumont R, et al. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia S, Liu Z, Zhang S, Liu P, Zhang L, Lee SH, et al. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni J, Liu Q, Xie S, Carlson C, Von T, Vogel K, et al. Functional characterization of an isoform-selective inhibitor of PI3K-p110beta as a potential anticancer agent. Cancer Discov. 2012;2:425–433. doi: 10.1158/2159-8290.CD-12-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furman RR, Sharman JP, Coutre SE, Cheson BD, Pagel JM, Hillmen P, et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med. 2014;370:997–1007. doi: 10.1056/NEJMoa1315226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gopal AK, Kahl BS, de Vos S, Wagner-Johnston ND, Schuster SJ, Jurczak WJ, et al. PI3Kdelta inhibition by idelalisib in patients with relapsed indolent lymphoma. N Engl J Med. 2014;370:1008–1018. doi: 10.1056/NEJMoa1314583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez-Angulo AM, Juric D, Argiles G, Schellens JHM, Burris HA, Berlin J, et al. Safety, pharmacokinetics, and preliminary activity of the a-specific PI3K inhibitor BYL719: results from the first-in-human study. J Clin Oncol. 2013;31(suppl) (abstr 2531) [Google Scholar]
- 33.Juric D, Gonzalez-Angulo AM, Burris HA, Schuler M, Schellens J, Berlin J, et al. Preliminary safety, pharmacokinetics and anti-tumor activity of BYL719, an alpha-specific PI3K inhibitor in combination with fulvestrant: results from a phase I study. Cancer Res. 2013;73(24 Suppl) Abstract nr P2-16-14. [Google Scholar]
- 34.Juric D, Infante JR, Krop IE, Kurkjian C, Patel MR, Graham RA, et al. Evaluation of tolerability and anti-tumor activity of GDC-0032, a PI3K inhibitor with enhanced activity against PIK3CA mutant tumors, administered to patients with advanced solid tumors [abstract]. Proceedings of the European Cancer Congress 2013; 2013 Sep 27–Oct 1; Amsterdam, Netherlands. Brussels (Belgium): European CanCer Organisation; 2013. 2013. Abstract nr P017. [Google Scholar]
- 35.O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serra V, Scaltriti M, Prudkin L, Eichhorn PJ, Ibrahim YH, Chandarlapaty S, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene. 2011;30:2547–2557. doi: 10.1038/onc.2010.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garrett JT, Olivares MG, Rinehart C, Granja-Ingram ND, Sanchez V, Chakrabarty A, et al. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proc Natl Acad Sci U S A. 2011;108:5021–5026. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, et al. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov. 2011;1:248–259. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garrett JT, Sutton CR, Kurupi R, Bialucha CU, Ettenberg SA, Collins SD, et al. Combination of antibody that inhibits ligand-independent HER3 dimerization and a p110alpha inhibitor potently blocks PI3K signaling and growth of HER2+ breast cancers. Cancer Res. 2013;73:6013–6023. doi: 10.1158/0008-5472.CAN-13-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao JJ, Castel P, Radosevic-Robin N, Elkabets M, Auricchio N, Aceto N, et al. Antagonism of EGFR and HER3 enhances the response to inhibitors of the PI3K-Akt pathway in triple-negative breast cancer. Sci Signal. 2014;7:ra29. doi: 10.1126/scisignal.2005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz S, Wongvipat J, Trigwell CB, Hancox U, Carver BS, Rodrik-Outmezguine V, et al. Feedback suppression of PI3Kalpha signaling in PTEN-mutated tumors is relieved by selective inhibition of PI3Kbeta. Cancer Cell. 2015;27:109–122. doi: 10.1016/j.ccell.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa C, Ebi H, Martini M, Beausoleil SA, Faber AC, Jakubik CT, et al. Measurement of PIP3 levels reveals an unexpected role for p110beta in early adaptive responses to p110alpha-specific inhibitors in luminal breast cancer. Cancer Cell. 2015;27:97–108. doi: 10.1016/j.ccell.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature. 2015;518:240–244. doi: 10.1038/nature13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bosch A, Li Z, Bergamaschi A, Ellis H, Toska E, Prat A, et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci Transl Med. 2015;7:283ra51. doi: 10.1126/scitranslmed.aaa4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Juvekar A, Burga LN, Hu H, Lunsford EP, Ibrahim YH, Balmana J, et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2012;2:1048–1063. doi: 10.1158/2159-8290.CD-11-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vora SR, Juric D, Kim N, Mino-Kenudson M, Huynh T, Costa C, et al. CDK 4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer Cell. 2014;26:136–149. doi: 10.1016/j.ccr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Will M, Qin AC, Toy W, Yao Z, Rodrik-Outmezguine V, Schneider C, et al. Rapid induction of apoptosis by PI3K inhibitors is dependent upon their transient inhibition of RAS-ERK signaling. Cancer Discov. 2014;4:334–347. doi: 10.1158/2159-8290.CD-13-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung M, Gelato KA, Fernandez-Montalvan A, Siegel S, Haendler B. Targeting BET bromodomains for cancer treatment. Epigenomics. 2015;7:487–501. doi: 10.2217/epi.14.91. [DOI] [PubMed] [Google Scholar]
- 54.Stratikopoulos EE, Dendy M, Szabolcs M, Khaykin AJ, Lefebvre C, Zhou MM, et al. Kinase and BET inhibitors together clamp inhibition of PI3K signaling and overcome resistance to therapy. Cancer Cell. 2015;27:837–851. doi: 10.1016/j.ccell.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stuhlmiller TJ, Miller SM, Zawistowski JS, Nakamura K, Beltran AS, Duncan JS, et al. Inhibition of lapatinib-induced kinome reprogramming in ERBB2-positive breast cancer by targeting BET family bromodomains. Cell Rep. 2015;11:390–404. doi: 10.1016/j.celrep.2015.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matkar S, Sharma P, Gao S, Gurung B, Katona BW, Liao J, et al. An epigenetic pathway regulates sensitivity of breast cancer cells to HER2 inhibition via FOXO/c-Myc axis. Cancer Cell. 2015;28:472–485. doi: 10.1016/j.ccell.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong C, Laddha SV, Tang L, Vosburgh E, Levine AJ, Normant E, et al. The bromodomain and extra-terminal inhibitor CPI203 enhances the antiproliferative effects of rapamycin on human neuroendocrine tumors. Cell Death Dis. 2014;5:e1450. doi: 10.1038/cddis.2014.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee DH, Qi J, Bradner JE, Said JW, Doan NB, Forscher C, et al. Synergistic effect of JQ1 and rapamycin for treatment of human osteosarcoma. Int J Cancer. 2015;136:2055–2064. doi: 10.1002/ijc.29269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boi M, Gaudio E, Bonetti P, Kwee I, Bernasconi E, Tarantelli C, et al. The BET bromodomain inhibitor OTX015 affects pathogenetic pathways in preclinical B-cell tumor models and synergizes with targeted drugs. Clin Cancer Res. 2015;21:1628–1638. doi: 10.1158/1078-0432.CCR-14-1561. [DOI] [PubMed] [Google Scholar]
- 60.Liu S, Walker SR, Nelson EA, Cerulli R, Xiang M, Toniolo PA, et al. Targeting STAT5 in hematologic malignancies through inhibition of the bromodomain and extra-terminal (BET) bromodomain protein BRD2. Mol Cancer Ther. 2014;13:1194–1205. doi: 10.1158/1535-7163.MCT-13-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jing Y, Zhang Z, Ma P, An S, Shen Y, Zhu L, et al. Concomitant BET and MAPK blockade for effective treatment of ovarian cancer. Oncotarget. 2016;7:2545–2554. doi: 10.18632/oncotarget.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng Q, Zhang Z, Shea MJ, Creighton CJ, Coarfa C, Hilsenbeck SG, et al. An epigenomic approach to therapy for tamoxifen-resistant breast cancer. Cell Res. 2014;24:809–819. doi: 10.1038/cr.2014.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin MP, Olesen SH, Georg GI, Schonbrunn E. Cyclin-dependent kinase inhibitor dinaciclib interacts with the acetyl-lysine recognition site of bromodomains. ACS Chem Biol. 2013;8:2360–2365. doi: 10.1021/cb4003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ember SW, Zhu JY, Olesen SH, Martin MP, Becker A, Berndt N, et al. Acetyl-lysine binding site of bromodomain-containing protein 4 (BRD4) interacts with diverse kinase inhibitors. ACS Chem Biol. 2014;9:1160–1171. doi: 10.1021/cb500072z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ciceri P, Muller S, O'Mahony A, Fedorov O, Filippakopoulos P, Hunt JP, et al. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat Chem Biol. 2014;10:305–312. doi: 10.1038/nchembio.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen L, Yap JL, Yoshioka M, Lanning ME, Fountain RN, Raje M, et al. BRD4 structure-activity relationships of dual PLK1 kinase/BRD4 bromodomain inhibitor BI-2536. ACS Med Chem Lett. 2015;6:764–769. doi: 10.1021/acsmedchemlett.5b00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dittmann A, Werner T, Chung CW, Savitski MM, Falth Savitski M, Grandi P, et al. The commonly used PI3-kinase probe LY294002 is an inhibitor of BET bromodomains. ACS Chem Biol. 2014;9:495–502. doi: 10.1021/cb400789e. [DOI] [PubMed] [Google Scholar]