Abstract
As personal genome sequencing becomes a reality, understanding the effects of genetic variants on phenotype—particularly the impact of germline variants on disease risk and the impact of somatic variants on cancer development and treatment—continues to increase in importance. Because of their clear potential for affecting phenotype, nonsynonymous genetic variants (variants that cause a change in the amino acid sequence of a protein encoded by a gene) have long been the target of efforts to predict the effects of genetic variation. Whole-genome sequencing is identifying large numbers of nonsynonymous variants in each genome, intensifying the need for computational methods that accurately predict which of these are likely to impact disease phenotypes. This review focuses on nonsynonymous variant prediction with two aims in mind: (1) to review the prioritization methods that have been developed to date and the principles on which they are based and (2) to discuss the challenges to further improving these methods.
Keywords: genetic variation, phenotypic effects, human disease, protein mutation
ADVANCES in sequencing technologies are rapidly making whole-genome sequencing of germline or somatic DNA routinely available for prognostic and diagnostic purposes. During the past decade and more, millions of single-nucleotide variants have been identified as the most common type of genetic difference, both among individuals (International HapMap Consortium 2005; Cotton et al. 2008; Abecasis et al. 2012) and between different somatic cells within an individual (Cancer Genome Atlas Research Network 2008; Campbell et al. 2015). Single-nucleotide variants are a substitution of one DNA base pair for another and may fall within genes (either protein-coding or functional RNA genes) in gene regulatory regions or in intergenic regions. Substitutions in the coding sequence of protein-encoding genes can be either synonymous (i.e., they encode the same amino acid due to redundancy/degeneracy in the genetic code and so have no effect on the protein product of a gene) or nonsynonymous (i.e., they change a single amino acid in the protein). Here, we focus specifically on nonsynonymous genetic variants (NSVs), of which there are an average number of ∼3000 per individual genome (Abecasis et al. 2012).
Proteins, either alone or in complex with other cellular molecules, comprise molecular “machines” that function at the biochemical level. An NSV by definition changes the sequence of a protein. However, only a subset of NSVs have a damaging functional effect (i.e., affecting the biochemical activity or regulatory control of a protein), as proteins are large molecules and their structures can be quite robust to single-site mutations. Note that the term “damaging” does not necessarily imply an impairment of a protein’s biochemical activity—in some cases a NSV that increases a protein’s biochemical activity can have a negative effect on the protein’s ability to properly serve one of its biological roles. In turn, some, but not all, damaging NSVs will be deleterious, meaning that they result in a phenotype at the organism level that is subject to natural selection (specifically, negative selection). Disease-causing, or pathogenic, NSVs obviously have a phenotypic effect, which may be subject to natural selection but is not necessarily so. Thus pathogenic NSVs are very often but not necessarily deleterious in the strict sense. Finally, most common (high frequency in a population) NSVs, and many if not most rare NSVs, have no appreciable deleterious or pathogenic effect and are called “neutral.”
Thus, the challenge of NSV impact prediction can be stated simply as a needle-in-the-haystack problem: most NSVs carried by an individual are neutral, so we need ways to predict the relatively few NSVs that will, upon closer investigation, turn out to be deleterious or pathogenic. Of course, genetic variation outside of protein-coding regions can also have phenotypic consequence, and with projects such as ENCODE now generating hypotheses about potential regulatory regions of the human genome (Encode Project Consortium 2012), methods for identification of disease-relevant regulatory variants is currently a major focus. Nevertheless, because of the clear mechanism by which NSVs can impact biological function and therefore phenotype, NSV prioritization remains an active area of research in which improvements are still required to meet the demands of precision genomic medicine (Fernald et al. 2011; Shendure and Akey 2015).
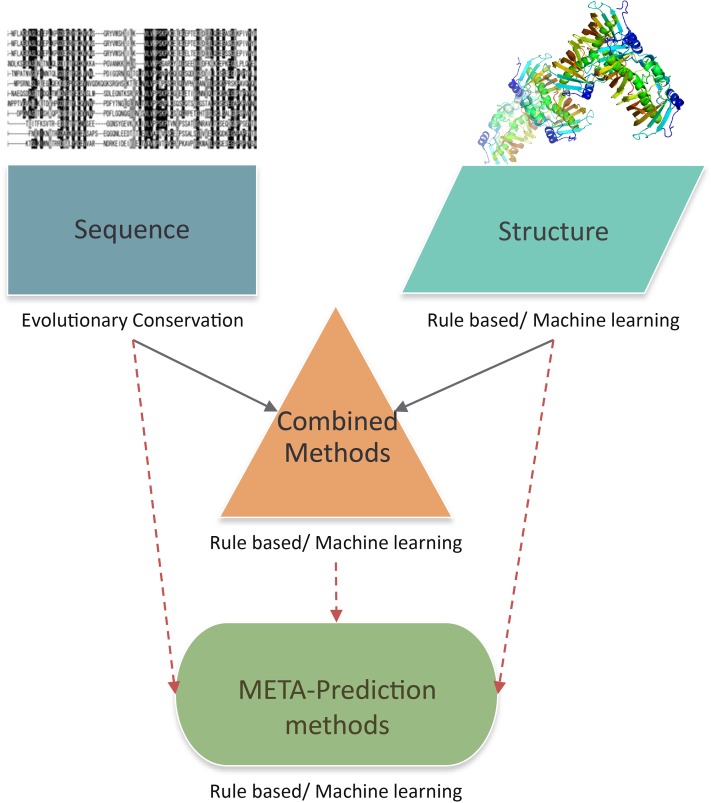
Computational methodologies for predicting the impact of NSVs fall into four main categories: sequence conservation-based, structure analysis-based, combined (including both sequence and structure information), and meta-prediction (predictors that integrate results from multiple predictors) approaches (Figure 1). We first review the foundations of SNV prediction methods in protein sequence and structure analysis. We then discuss each of the categories of computational prediction method in more detail, describing the basic principles underlying each approach and the differences between specific computational tools that have been developed in each area. We try to place particular emphasis on advances in methodology. Finally, after reviewing how NSV prediction method accuracy is assessed, we outline remaining challenges in the field, and prospects for further advancement.
Figure 1.
An overview of methods for predicting effects of NSVs. “Core” methods based on sequence (evolutionary conservation) and structure analysis can be combined into a multi-feature prediction method, or predictions from different individual methods can be used to make an overall meta-prediction.
Methods for Predicting Effects of NSVs: Overview and Background
The theoretical underpinnings of NSV prioritization methods were arguably developed at the dawn of molecular biology. Based on the limited structural studies available at the time on hemoglobin (primarily the peptide segments located near the heme iron), as well as partial amino acid sequences of hemoglobins from a number of other mammalian species, Zuckerkandl and Pauling first proposed the principle that disease can arise through a change in a protein’s amino acid sequence that affects its functioning as molecular machine and the corollary that different amino acids contribute to that functioning to different degrees (Zuckerkandl and Pauling 1962). They formulated the hypotheses that now underlie the two primary approaches to NSV prediction: that the amino acids of greatest functional importance can be identified either (1) directly by inspection of the protein structure or (2) indirectly by comparing the sequences of related proteins from different species and noting the positions that display evolutionary conservation. These hypotheses were borne out, and elaborated upon, by seminal large-scale protein mutagenesis-fitness studies on HIV-1 protease (Loeb et al. 1989), T4 lysozyme (Pazdrak et al. 1997), and Escherichia coli lacI protein (Markiewicz et al. 1994). In these studies, nearly every position in the protein was mutated to multiple different amino acids, and the relative fitness of organisms bearing the mutation was measured.
In protein structure analysis, the effect of a NSV is predicted from its likely effects on protein stability or function. As Zuckerkandl and Pauling (1962) observed, a small number of positions in hemoglobin are in direct physical contact with oxygen or heme, and, as NSVs at these sites could change the distribution of electrons critical for catalysis, they would likely have effects on hemoglobin’s ability to perform its oxygen delivery function. A much larger number of positions were predicted to play an important role in the stability of the overall structure of each monomeric subunit as well as the functional tetramer, which stably positions the few directly functional sites. Thus most of the effects of NSVs would be on protein stability. In either case, protein structure analysis requires consideration of physical mechanisms by which an NSV might affect its functioning, which would then presumably result in phenotypic effects.
Sequence conservation analysis, on the other hand, begins with observation of the effects of natural selection, which operates at the phenotypic level. Evolutionary conservation among homologs reflects the effects of negative selection against mutations that reduced the fitness of the individuals bearing those mutations. In a typical protein family, some positions in the protein are absolutely conserved among homologs, while other positions display varying degrees of tolerance for different amino acids. In this approach, observations of substitution patterns over macroevolutionary timescales, in which sequences typically differ at many different positions, are used to estimate effects of a single unit of potential microevolutionary change, a NSV in an individual. The prediction of impact will be accurate only insofar as the effects are similar on these timescales and population scales (e.g., a small selective effect can be enough to retain evolutionary conservation in a large population although the effect in a single individual lifetime may be difficult to detect).
Early Computational Methods: Quantitating Sequence Conservation and Rules for Structural Features
With the development of more rapid DNA-sequencing technologies in the late 1990s, databases began to be compiled of DNA sequence variation within the human population (Collins et al. 1998; Buetow et al. 1999; Cargill et al. 1999; Halushka et al. 1999), as well as between individuals of different species. The rate of discovery of such variants soon outstripped our abilities to analyze each one manually, and work began on computational methods. Miller and Kumar were the first to demonstrate the ability of sequence conservation to statistically distinguish between disease-causing variants and presumably mostly neutral common variants in a population (Miller and Kumar 2001), while Wang and Moult showed that structure information can also succeed at that task (Wang and Moult 2001). Chasman and Adams combined both sequence and structure information into a prediction method (Chasman and Adams 2001). The earliest computational methods available for broad use considered evolutionary conservation (Ng and Henikoff 2001; Thomas et al. 2003), structural effects (Wang and Moult 2001), or a combination of the two (Ramensky et al. 2002). Evolutionary conservation was treated as quantitative, while structural effects were handled as qualitative features for rule-based prediction.
The early sequence conservation methods borrowed from earlier work in assigning a probability to a replacement of one amino acid by another over evolutionary time (Dayhoff et al. 1978; Henikoff and Henikoff 1992; Jones et al. 1992). In this work, amino acid replacement probabilities were derived empirically from a database of known protein sequences. Homologous proteins were aligned to each other, providing one-to-one correspondences between presumably homologous amino acids within a group of related proteins, which were then used to generate statistics for pairwise amino acid substitutions (often expressed as a symmetric 20 × 20 matrix). These statistical matrices quickly came to replace earlier empirical methods of estimating amino acid replacement from physicochemical properties of amino acids, such as the Grantham scale (Grantham 1974), and reached widespread use in sequence database searching in algorithms such as FASTA (Pearson and Lipman 1988) and BLAST (Altschul et al. 1990). However, the major drawback in these pairwise matrices is that they estimate an average replacement probability over all proteins. As observed by Zuckerkandl and Pauling (1962), and made abundantly clear by mutagenesis studies (Loeb et al. 1989; Markiewicz et al. 1994; Pazdrak et al. 1997), some sites in proteins are apparently tolerant to mutation, while others are not, and it matters very much not only what the amino acid change is, but also where in the sequence it occurs. As protein databases became larger, it became statistically feasible in many cases to compute probabilities specifically for each position in an alignment, termed “profiles” (Gribskov 1994). A profile is expressed as an amino acid probability vector for each position in the alignment, and pairwise amino acid replacements can simply be derived from the two relative profile probabilities. Not surprisingly, position-specific probabilities significantly outperform average substitution scores at identifying deleterious NSVs (Ng and Henikoff 2001). All the early conservation-based NSV prediction methods utilized position-specific profiles, although in somewhat different ways.
The differences among these early methods were in three main areas. The first was in the construction of alignments, both in identifying a set of homologs and in the algorithm used to align them. SIFT used PSI-BLAST (Altschul et al. 1997), while PANTHER-subPSEC (Thomas et al. 2003) used hidden Markov models (Barrett et al. 1997). The second difference was in how amino acid probabilities were determined from the alignment: SIFT and PANTHER weighted each sequence equally at all positions in a given alignment (Henikoff and Henikoff 1994), while PolyPhen used position-specific sequence weighting (Sunyaev et al. 1999). In addition, SIFT gave more weight to the prior probabilities for amino acids compared to the weighting scheme used in PANTHER-subPSEC (Sjolander et al. 1996), potentially leading to larger differences between these methods when the alignment contains either relatively few, or relatively closely related homologs. The third difference lies in how these amino acid probabilities were used to determine a quantitative substitution effect score. PolyPhen used the ratio between the probabilities of the wild-type and substituted alleles, PANTHER-subPSEC used the absolute value of that ratio to focus on the magnitude rather than the directionality of the change (i.e., an NSV could be judged deleterious if it dramatically decreased or increased the probability compared to the wild type), and SIFT used the ratio between the substituted amino acid probability and that of the most probable amino acid at that position, in effect treating the NSV not in terms of change from one amino acid to another, but rather in terms of the fit between the observed amino acid and the profile.
Unlike sequence conservation, effects on protein structure are diverse and cannot be treated in a single unified formalism. Early methods for variant-effect prediction utilized multiple general features describing the fit between an amino acid and its local environment within a three-dimensional structure (and thus affecting protein stability), as well as features describing specific functional roles for a particular amino acid, such as an enzyme active site or ligand-binding site (Table 1). An NSV at a specific functional site is likely to be deleterious, but as discussed above this applies to relatively few amino acid sites in a typical protein. These sites—such as enzyme active sites and individual residues that bind metal ions, other cofactors, or ligands—can be identified from direct analysis of protein structures, or, more commonly, from prior analysis results that are captured in resources such as the Swiss-Prot database (Boeckmann et al. 2003; UniProt Consortium 2011). Of much broader applicability are predictions of NSVs that decrease protein stability by a few kcals/mol, as this can substantially affect the amount of the correctly folded and functional protein in the cell. The free energy costs of many perturbations arising from a particular amino acid change (e.g., increasing hydrophobic surface area exposed to water or introducing a charged amino acid in the nonpolar protein core) have been well studied. These early methods used these costs to rationally guide the selection of features that are likely to have significant stability effects on most proteins. To combine the various features into a prediction of functional effect, empirical rules were developed. Thus, different structure-based methods differ in which features they consider, and how they are combined into rules for predicting functional impact.
Table 1. List of structure features used by methods.
| Structure features | PolyPhen (Ramensky et al. 2002) | SNPs3D (Yue et al. 2006) | Chasman and Adams (2001) |
|---|---|---|---|
| Secondary structure | √ | ||
| Region of (phi, psi) map | √ | ||
| Loss of hydrogen bond/stabilizing energy of water bridges | √ | ||
| van der Waals force | √ | ||
| Overpacking | √ | √ | |
| Hydrophobic burial | √ | √ | |
| Surface accessibility/change in accessible surface propensity | √ | √ | √ |
| Crystallographic B-factor | √ | √ | √ |
| Cavity | √ | ||
| Electrostatic repulsion | √ | √ | |
| Backbone strain | √ | √ | |
| Buried charge | √ | √ | |
| Buried polar | √ | ||
| Breakage of a disulfide bond | √ | ||
| Turn breaking | √ | ||
| Helix breaking | √ | ||
| Near hetero (nonprotein) atom | √ | ||
| Near subunit interface | √ | ||
| Sidechain conformational entropy |
Further Developments in Evolutionary Conservation-Based and Structure-Based Methods
Sequence conservation-based methods
Conservation-based methods perform as well as structure-based methods on benchmarking sets (described in more detail below), but can be applied to a much larger number of human NSVs because they can be used even when there is no known three-dimensional structure of a homologous protein. Consequently, extensive efforts have been made to improve conservation-based methods. The MAPP algorithm (Stone and Sidow 2005) converts an amino-acid-probability profile into a multidimensional profile of physicochemical properties and assesses an NSV according to its fit to the physicochemical profile rather than to an amino acid profile. The same article also demonstrated the value of creating a conservation profile based only on orthologous proteins (Fitch 1970) (i.e., descended from the same gene in their common ancestor species, like human hemoglobin and horse hemoglobin) rather than also including paralogous proteins (i.e., genes that descend from homologous but distinct genes in their common ancestor species, like human hemoglobin and human myoglobin). This approach is based on the recognition that duplicated genes are likely to have diverged somewhat in function to become fixed in the genome (Ohno 1970) and that the functional divergence may arise from adaptive amino acid substitutions at some positions in the protein sequence of one or both duplicates. The PANTHER-subPSEC algorithm was improved by allowing paralogous proteins to be included in the profile only if they match the profile of orthologous proteins arising from the same duplication event in a phylogenetic tree (Thomas and Kejariwal 2004). Thus the profiles are subfamily-specific if the subfamily displays a significantly different profile from the entire family.
MutationAssessor (Reva et al. 2011) introduced two advances: a new method of assessing the impact of a NSV based on the change in relative entropy of an alignment position (essentially the weighted diversity of amino acids observed at that position) upon adding the observed substitution, and a new way to identify divergent subfamilies within an alignment without first distinguishing orthologs from paralogs. The MutationAssessor impact score combines the subfamily-based entropy score with the global family-based entropy score so that both family and subfamily profiles are considered in the final prediction. The Ancestral Site Preservation algorithm (Marini et al. 2010; Tang and Thomas 2016) uses a multiple alignment in a way that is completely different from other methods. Rather than considering columns of a multiple alignment, a phylogenetic tree is reconstructed from the alignment and probabilistic ancestral protein sequences are inferred (Yang 1997) at each node in the tree, representing all common ancestors of extant proteins. The “preservation” of an amino acid is then traced back through its ancestors; the longer the trace-back, the greater the probability that the preservation reflects the effects of negative selection. To varying degrees, these latest developments address the main drawback of using homologous proteins to infer the effects of NSVs: the assumption that the constraints on amino acid replacement at a given position remain constant (or “equivalent”) over evolutionary time. Even close orthologs typically differ in sequence at multiple positions, and a change at one position can dramatically alter the probability that a change elsewhere will be tolerated (Bridgham et al. 2009). This phenomenon is often referred to as “compensatory mutation” and may be quite widespread (Kondrashov et al. 2002; Kulathinal et al. 2004; Liao and Zhang 2007). By considering evolutionary change within a family of related proteins, the methods discussed in this section begin to address the problem of correctly identifying the constraints on a particular protein of interest even if they differ from other related proteins.
Because of the reliance of sequence conservation methods on a multiple sequence alignment, it seems obvious that alignment quality would affect the prediction accuracy of these methods on benchmark tests (Ng and Henikoff 2006; Thusberg et al. 2011). Karchin suggested that alignment differences might partially explain differences in predictions from different algorithms (Karchin 2009). Hicks et al. tested these hypotheses by comparing SIFT and PolyPhen-2 on alignments constructed by four different methods (Hicks et al. 2011). While SIFT accuracy was slightly decreased by using the alignment generated by PolyPhen-2, this decrease was not statistically significant, and only use of an alignment that is composed of only distantly related homologs (<50% pairwise identity among all homologs) had a significant effect on SIFT performance. PolyPhen-2 accuracy, on the other hand, was unaffected by alignment methods. It is possible that this increased robustness to alignment differences may be due to PolyPhen’s use of several additional features other than conservation. Agreement between the predictions from SIFT and PolyPhen-2 was not increased regardless of which alignments were used, suggesting that differences in alignments do not contribute appreciably to prediction discrepancies between algorithms. These results also suggest that the alignments generated by both SIFT and PolyPhen-2 are generally of high quality and further improvement in this area may yield limited gains in predictive value. However, this study was limited to NSVs in four human proteins, and it is not clear how these conclusions will hold for more comprehensive test sets.
Structure-based methods
Further advances in structure-based methods have focused on predictions of impact on protein stability. Recent reviews have covered the development of methods in this field (Masso and Vaisman 2010; Compiani and Capriotti 2013), as well as assessment of relative performance (Potapov et al. 2009), so we focus here on the major recent methodological developments. Stability predictions are based on an explicit or implicit model of the change in stability (Δ-Δ-G or change in free-energy difference between folded and unfolded states) upon substitution with a different amino acid. For the purposes of NSV impact prediction, the main interest is in mutations that have a relatively large effect on protein stability and can thus be expected to have an appreciable effect on the amount of functional protein (i.e., in the conformation required for its function and stable enough to avoid degradation) present in vivo. Proteins vary in stability, but a Δ-Δ-G in the range of 2 kcal/mol is generally considered to result in a mutational “hot spot” of sufficient effect. Using this criterion, Potapov et al. found that the accuracy of predicting such hot spots was between 72 and 80% across six different commonly used methods (Potapov et al. 2009). While their initial assessment of one method, Rosetta (Rohl et al. 2004), suggested a somewhat lower accuracy, a later study has shown that this resulted from inappropriate parameter settings (Kellogg et al. 2011).
Most mutant stability change prediction programs use an explicit model of the energetics of the folded (requiring a 3D structure) and unfolded (generally assumed to depend only on the amino acid substitution) states of the protein. Protein backbone conformation may be assumed to remain unperturbed or to allow small changes upon mutation; sidechains may be allowed to rotate and repack within varying distances of the mutated amino acid. Energy functions, also called “potentials,” consist of linear combinations of terms to capture different interactions or entropic factors (e.g., solvation or conformational entropy) and can be physics-based or statistical (inferred from observed frequencies). The relative weights of the terms can derive from experimental measurements or theoretical calculations or can be optimized to solve a particular task. Fold-X (Guerois et al. 2002) is a mostly physics-based energy function (or “potential”) that uses a full atomic description of the structure of the proteins. Terms of the function were weighted to maximize the fit to experimentally measured Δ-Δ-G values for hundreds of point mutants. Rosetta (Rohl et al. 2004) computes energies using a potential that includes numerous terms, both statistical and physics-based, and can sample both protein backbone and sidechain rotamers to adjustable degrees. CC/PBSA (Benedix et al. 2009) performs conformational sampling, computes energies using an all-atom physics-based potential, and reports an average Δ-Δ-G over the sampled conformations. EGAD (Pokala and Handel 2005) uses an all-atom physics-based potential with a fixed native state conformation; however, the unfolded state is modeled explicitly.
Machine learning has also been applied to develop mutant stability prediction methods. Unlike the approaches based on explicit modeling of the energetics of folding, these methods consider only the folded state of the protein and result in an energy-like scoring that is a nonlinear function of a wide variety of features. I-Mutant (Capriotti et al. 2005) trained a support-vector machine (SVM, discussed in more detail below) on a database of experimentally assayed single substitutions, where for each substitution the SVM is given the quantitative known change in stability as well as a feature vector that encodes (1) the two variant amino acids and (2) the number of amino acids of each type in a 9Å radius within the three-dimensional structure, in essence allowing the SVM to determine an energy-like function that depends on the amino acids in a 9Å sphere. AUTO-MUTE (Masso and Vaisman 2010) uses an SVM and Random Forests (another machine-learning technique that we discuss more below) using known experimental Δ-Δ-G values with features that include the amino acid substitution and a “statistical potential” (Sippl 1990) calculated from combinations of four residues in mutual physical contact in a database of known protein structures.
Combining Sequence Conservation with Structural Features
PolyPhen (Ramensky et al. 2002) was the first widely available software to combine sequence conservation with structural features. As with early structure-based methods (Wang and Moult 2001), it used a series of empirical rules to combine these various features into an overall prediction. These rules are rational and attempt to capture knowledge about the forces driving protein stability and function, but arbitrary in that there are many rational ways to combine the various features. Machine-learning techniques have been employed by many new NSV impact prediction tools to better integrate available features. The general approach is to first collect predetermined positive (typically pathogenic NSVs) and negative (neutral NSVs) examples and “train” a “machine-learning prediction classifier” that effectively distinguishes between the positive and negative training examples. Training is typically accomplished by iteratively adjusting a computational representation of the input “features” (e.g., sequence conservation and structural features), so as to separate the positive from negative training examples to the greatest extent possible. Parameters of the prediction algorithms are then optimized using one or more cross-validation sets. Finally, performance of the prediction algorithm is analyzed on a test set to estimate its general applicability to new data and compare it with other methods. The first work of this kind employed a Bayesian network learning algorithm (Cai et al. 2004), and the subsequent decade has seen many variations on this basic theme. A growing number of machine-learning-based NSV impact prediction tools are now available. They differ from each other primarily in three ways: the type of machine-learning algorithm used, the set of input features that are considered, and the sets of NSVs of known (or inferred) effect that are used as training and test sets. Table 2 lists the main algorithms that have been employed and some representative NSV impact prediction methods that have utilized these algorithms. Table 3 lists representative NSV data sets that are used as training or test data sets of some machine-learning-based methods. The selection of training set and test set is very important for development of machine-learning tools and correct assessment of these tools.
Table 2. Machine-learning algorithms used in NSV prediction and the specific tools developed using each algorithm.
| Machine-learning algorithm | Description | Example of NSV prediction tools utilizing algorithm |
|---|---|---|
| Support vector machine | Maps positive (pathogenic) and negative (neutral) training examples to a high-dimensional space (a transformation of the input features) in which the positive and negative examples can be distinguished from each other; predictions for a new SNV are made on the basis of where it lies in this space. | PhD-SNP (Capriotti et al. 2006) |
| Artificial neural networks | Trains a multi-layer network of nodes (“artificial neurons”), including one layer of input feature nodes and one layer of two output nodes (pathogenic/neutral) and one or more middle layers, where weights of input and output edges connecting nodes in adjacent levels are adjusted to maximize prediction accuracy on training examples. | PMUT (Ferrer-Costa et al. 2005) and SNAP (Bromberg and Rost 2007) |
| Random forests | Trains a large number (“ensemble”) of decision trees to distinguish positive from negative training examples, each tree utilizing a random set of input features; predictions for a new SNV is derived statistically from the ensemble of predictions from individual trees. | MutPred (Li et al. 2009) |
| Naive Bayes classifiers | Probabilistic classifier (i.e., assigns a probability of being damaging or neutral) that treats each feature as independent of the others; parameters are adjusted so as to maximize the probability of impact for positive examples and minimize probability for negative examples. | PolyPhen2 (Adzhubei et al. 2010) and MutationTaster (Schwarz et al. 2010) |
Table 3. Representative data sets used to evaluate the performance of NSV impact predictions.
| Data set (reference) | NSVs with impact | NSVs with no impact |
|---|---|---|
| HumDiv data set (Adzhubei et al. 2010) | Annotated in Swiss-Prot as human disease causing and Mendelian disease causing and as affecting protein molecular function | Amino acid differences between human proteins and closely related mammalian homologs |
| SP_human data set (Calabrese et al. 2009) | Annotated in Swiss-Prot as human disease causing | Annotated in Swiss-Prot as neutral, excluding hypervariable proteins of class I and II of the major histocompatibility complex |
| HumVar data set (Capriotti et al. 2006) | Annotated in Swiss-Prot as human disease causing | Human variants in Swiss-Prot not annotated as disease causing |
| VariBench (Thusberg et al. 2011; Sasidharan Nair and Vihinen 2013) | Annotated in Swiss-Prot as human disease causing and found in an LSDB [from PhenCode database (Giardine et al. 2007) or registries in IDbases (Piirila et al. 2006)] or 1 of 18 other LSDBs | dbSNP variants with MAF (minor allele frequency) > 0.01 and observed in at least 49 chromosomes |
| Protein Mutant Database (Kawabata et al. 1999) | Damaging variants from experiments | Nondamaging variants from experiments |
| SwissVar database (Mottaz et al. 2010), also called HUMSAVAR | Up-to-date Swiss-Prot annotations, disease causing | Up-to-date Swiss-Prot annotations, polymorphism |
| MutPred data set (Li et al. 2009) | Somatic mutations in genes resequenced in 22 cancer cell lines from Sjoblom et al. (2006); somatic kinase genes resequenced in 210 individual tumors (Greenman et al. 2007); annotated in HGMD (Dehouck et al. 2013) as disease causing; annotated from Swiss-Prot as disease causing | Annotated in Swiss-Prot as common polymorphism |
| SNAP data set (Bromberg and Rost 2007) | Annotated in PMD as changed in function | Annotated in PMD as no change; substitutions between pairwise-aligned Swiss-Prot homologs with same E.C. number (enzyme function) |
| Cancer LSDB data set (Hicks et al. 2011) | Annotated as pathogenic in an LSDB for one of the following cancer genes: BRCA1, MLH1, MSH2, TP53 | Annotated as pathogenic in an LSDB for one of the following cancer genes: BRCA1, MLH1, MSH2, TP53 |
| Meta-SVM testing data sets (Dong et al. 2015) | Reported as causing Mendelian diseases in 57 Nature Genetics publications 2011–2014 | Common variants (MAF > 0.01) and rare variants (singletons) in 900 healthy participants of the ARIC study (Abecasis et al. 2010) |
| CADD data set (Kircher et al. 2014) | Annotated in ClinVar (Baker 2012) as pathogenic | Common variants (derived allele frequency > 0.05) from exome sequencing (Fu et al. 2013) |
| SwissVarSelected, VariBenchSelected, et al. (Grimm et al. 2015) | Selected variants from these data sets that do not overlap with common training sets such as HumVar |
Machine-learning methods continue to dominate the recent literature and are the best-performing methods on a variety of test data sets. In general, many different machine-learning models perform equally well; for example, the latest version of SNAP averages >10 different predictors with similar performance, for which evolutionary conservation is the only feature in common to all 10 (Hecht et al. 2015). The advantage of machine-learning approaches is that they can include features of very different types, and potentially a large number of such features that can be combined in highly complex and nonlinear ways. The main disadvantage is that it is generally difficult to infer important principles driving prediction accuracy, such as the relationships of the most informative features combined. In addition, there is a possibility of overfitting because a model is typically trained by maximizing its performance on training data. The model may therefore be less accurate in predicting new data than training data. Furthermore, as pointed out by earlier analyses (Thusberg et al. 2011; Grimm et al. 2015), performance of some machine-learning methods can be overestimated when comparing with non-machine-learning methods because performance assessment data sets contain NSVs on which some machine-learning methods have been trained. Even if these estimates are accurate, the performance improvement of machine-learning methods over non-machine-learning methods with few features such as SIFT, PANTHER-subPSEC, and MAPP, while significant, is not dramatic (accuracy improvement on the order of a few percentage points). Indeed, after controlling for two types of circularity (see discussion of prediction assessment below), Grimm et al. (2015) found that the improvement of combined and meta-prediction methods, compared to conservation-based methods, is substantially less dramatic than originally reported.
In addition to structural features that reflect effects on protein stability, other biological features have proved useful in identifying NSVs that impact protein function. For instance, including Gene Ontology functional class information in an SVM classifier SNPs&GO (Calabrese et al. 2009) improved prediction rates by nearly 5% on the HumVar benchmark. MutPred (Li et al. 2009) includes a number of “functional” features that have been mapped to specific amino acids in the protein sequence, such as sites of post-translational modifications (e.g., phosphorylation sites), DNA-binding sites, and catalytic active sites.
Meta-prediction Methods
Because a large number of tools perform quite well on existing benchmark tests, it is perhaps not surprising that meta-prediction methods that integrate the scores from multiple prediction methods have recently been developed. This approach is largely justified by numerous studies showing that, despite similar overall prediction accuracy, individual methods can disagree substantially for the same NSV. CONDEL (Gonzalez-Perez and Lopez-Bigas 2011) was the first meta-prediction method, using a linear, weighted average of scores from five different prediction methods, to produce a final prediction. CAROL (Lopes et al. 2012) combines the predictions of SIFT and PolyPhen-2 using a linear, weighted Z-score. But because of their flexibility in handling multiple and diverse feature types, machine-learning approaches have become the most widely applied for constructing meta-predictors. An increasingly common approach has been to (1) evaluate multiple existing prediction methods, (2) select some number of top-performing methods relative to a given benchmark data set and a particular metric of performance, and (3) train a machine-learning classifier using as features the scores from each of the top-performing methods. PON-P was the first of these, integrating a conservation method (SIFT), a structural stability-based method (I-Mutant), and three combined machine-learning-based methods (SNAP, PolyPhen-2, PhD-SNP), using Random Forests to make a final prediction (Olatubosun et al. 2012). Numerous other studies have since followed this general approach, such as Meta-SNP (Capriotti et al. 2013a), CoVEC (Frousios et al. 2013), PredictSNP (Bendl et al. 2014), and Meta-SVM (Dong et al. 2015). Taking the idea of producing a meta-prediction from multiple individual prediction algorithms yet a step further, methods such as CADD (Kircher et al. 2014) treat the output of any NSV effect-prediction method as simply one type of “annotation” of a variant and then use an SVM-based approach to make a meta-prediction from a large list of diverse annotations.
Assessment of NSV Impact Predictions
Assessment of the accuracy of predictions is an increasingly important issue. Already in 2006, Ng and Henikoff observed that a wide variety of prediction methods were available, making it difficult for users such as medical geneticists to know which methods to use and how to interpret different predictions for the same NSV (Ng and Henikoff 2006). The problem has become far greater today. Publication standards in the field have generally demanded that a new method must outperform at least the most popular existing methods on one or more of the widely used benchmark data sets. It is worth discussing the aims of these benchmarks (Table 3) to explore how they might be better used to drive progress in the field. The benchmarks generally fall into two classes: those that distinguish presumed disease-causing variants from variants that have been observed but not associated with disease, and those that distinguish between mutations with and without effect in an experimental assay. In the first class, presumed disease-causing variants have been obtained from comprehensive databases, most commonly Swiss-Prot (Mottaz et al. 2010), but also OMIM (Hamosh et al. 2005) and HGMD (Stenson et al. 2003) or from locus-specific databases [LSDBs, typically focusing on only a single human gene, are reviewed in (Greenblatt et al. 2008)] such as the IARC TP53 database (Olivier et al. 2002) and the BIC database on BRCA1 and BRCA2 (Goldgar et al. 2004). Non-disease-associated variants have been obtained in diverse ways: from substitutions between closely related orthologs (Ramensky et al. 2002), from non-disease-associated human variants reported in Swiss-Prot (Boeckmann et al. 2003), or from either all or common (e.g., a minor allele frequency >1% in at least one population) human NSV alleles in a public resource like dbSNP (Sherry et al. 2001). These sets of non-disease-associated variants are relatively comprehensive, covering many genes and variants, but can be expected to have some degree of error simply because of our ignorance about the phenotypic effects of most alleles. Consequently, they are useful for statistical comparisons because they are expected to contain fewer NSVs with effects compared to the disease-associated sets, but any given NSV in the unassociated sets may actually have an impact on function.
Potential biases in these evaluation sets are important to identify, as they can lead to spurious assessments of the performance of NSV impact prediction methods. Grimm et al. (2015) recently characterized two such biases, which they call “type 1 and type 2 circularity.” Type 1 circularity is simply that the set of variants (both pathogenic and neutral) used to train a method can also appear in the evaluation set, leading to overinflation of prediction accuracy. Grimm et al. (2015) created filtered data sets for evaluation by removing variants that overlap with commonly used training sets, to create, for example, a SwissVarSelected and VariBenchSelected data set that minimizes the impact of type 1 circularity on prediction evaluation. Type 2 circularity is less obvious: evaluation sets tend to contain a large proportion of proteins for which the variants in the set are either all pathogenic or all neutral. Thus, a simple rule can outperform all existing prediction methods just by “gaming” the system, predicting all variants in a given protein as either pathogenic or neutral. This bias explains the large improvement in prediction accuracy achieved by FATHMM (Shihab et al. 2013) when it includes (in addition to conservation) a term based on a particular protein domain. Importantly, the bias leading to type 2 circularity is much more pronounced for some evaluation sets than for others: only 5% of variants in VariBenchSelected are found in proteins having both pathogenic and neutral variants in the set, while this is true of >25% of variants in SwissVarSelected. This analysis suggests that evaluations based on SwissVarSelected are likely to better reflect actual performance in real world applications, in which nearly all proteins can be expected to harbor both pathogenic and neutral variants. Taking this a step further, Grimm et al. (2015) also assess performance on subsets of SwissVarSelected, where proteins are grouped according to how well balanced the variants are in both pathogenic and neutral classes. Finally, this analysis suggests that LSDBs, the source of many VariBench variants, may suffer from systematic bias toward pathogenic variants.
The second class of evaluation sets, experimentally assayed effects of NSVs, is currently available for a very small number of proteins. These are mutagenesis studies, followed by an assay of function. The Protein Mutant Database contains a collection of >200,000 mutations, but unfortunately has not added data since 2003. Perhaps the most relevant assays are those based upon a direct fitness measurement, such as the growth of a microorganism containing either the wild-type or variant forms of a gene. Even when using such assays the number of NSVs impacting function may be underestimated: only a single fixed environment is typically assayed, so it is likely that some NSVs that are apparently neutral in the assay actually do have fitness effects under other conditions. Nevertheless such in-depth experimental studies afford the possibility of close inspection of predictions on individual variants to potentially identify systematic errors. In addition to the classic mutagenesis studies mentioned above, a few more recent studies have been carried out on specific proteins, such as cystathionine beta synthase (CBS) (Wei et al. 2010; Dimster-Denk et al. 2013) and methylenetetrahydofolate reductase (Marini et al. 2010). Wei et al. identified eight mutations—of the >200 NSVs that they tested in CBS—that were consistently incorrectly predicted by six different methods (Wei et al. 2010), suggesting a systematic overprediction of effects arising from NSVs that involve mutation of a cysteine residue. For a different set of CBS mutants, Dimster-Denk et al. (2013) found that predicted Δ-Δ-G changes of >4 kcal/mol from Rosetta (Kellogg et al. 2011) were predictive of loss of function, but for moderate changes no correlation was found between predicted values and functional assay results.
Several authors have previously suggested that the field could benefit greatly from the assessment of truly blinded predictions at regular intervals, similarly to the Critical Assessment of Structure Prediction experiments (Moult et al. 2014) that proved their utility in the field of protein structure prediction from primary amino acid sequence. The Critical Assessment of Genome Interpretation (CAGI) experiment, first held in 2012–2013, included a challenge for interpretation of genetic variants. Unfortunately, this first experiment has had limited impact on the field to date, with no general publication describing its results and few articles referencing it (a rare example is Chen et al. 2014), but a new CAGI experiment was just completed in February of 2016 (https://genomeinterpretation.org). While results were not available at the time of press, the experiment certainly holds promise in this area.
It is also important to consider which metrics are applied to assess accuracy on a given benchmark. As Cline and Karchin (2011) pointed out, the optimal comparisons use a method like the Receiver Operating Characteristic (ROC), which considers equally both true positive (sensitivity or recall) and false positive (specificity or precision) prediction rates and considers all possible thresholds in a quantitative manner. ROC analyses have dominated the field since its early days, with area under the curve (AUC) becoming the most common metric for the accuracy of predictions. It might be even more informative to follow the practice of other prediction fields that have focused on AUC for low false-positive rates [e.g., <20% as in (Gribskov and Robinson 1996)] rather than the entire ROC curve, as these are the predictions that are most likely to be used in real world applications. Other accuracy measures such as the Matthews Correlation Coefficient and Balanced Error Rate are also useful but can depend on the threshold employed (Wei et al. 2010). Finally, it is important to avoid conflating absence of a prediction with a prediction of absence of a functional effect. Nearly all prediction methods are unable to make predictions for some NSVs, and this number can vary widely between methods. While it is certainly useful to compare the prediction coverage for different methods, these comparisons should be kept distinct from the accuracy of predictions (e.g., Thusberg et al. 2011; Shihab et al. 2013) rather than included in a quasi-ROC analysis (e.g., Dong et al. 2015).
Conclusions and Prospects
Despite a growing interest in other types of genetic variation, predicting the impact of NSVs remains an area of active research and continual improvement (Capriotti et al. 2012). We have focused here on evolutionary conservation methods, combined methods using both conservation and structural features, and meta-prediction methods that make a unified prediction from multiple conservation, structural, or combined methods. These three classes are the most important for biomedical applications because they can be applied to a much larger number of SNVs, given that many human proteins currently have neither an experimentally determined structure nor a close homolog from which to build a model. Furthermore, in methods using both conservation-based and structure-based features, conservation has been repeatedly found to be the single most informative feature (Ramensky et al. 2002; Bromberg and Rost 2007; Li et al. 2009). Recent efforts to overcome the limitations of previous conservation-based metrics, such as considering amino acid physico-chemical similarity (Stone and Sidow 2005), subfamily-specific conservation (Thomas and Kejariwal 2004; Reva et al. 2011), and evolutionary reconstruction (Marini et al. 2010) have shown that further improvement in this approach is still possible. Incorporation of other potential improvements, including but not limited to modeling lineage-specific selection, may hold further promise. Combined and meta-prediction methods have a large space of potential combinations of features, and even development of novel feature types, yet to explore. Incorporation of the more recent conservation-based methods as a feature in machine-learning-based predictors would also be a natural next step.
In addition to methodological improvements, the field would benefit from advances in at least three more areas. The first area is reliable access to accurate predictions from multiple methods, which becomes increasingly important as the demand for variant interpretation grows. One could envision an integrated variant resource to address this need. Databases such as dbNSFP (Liu et al. 2011), SNPdbe (Schaefer et al. 2012), and the PON-P server (Niroula et al. 2015) have begun to make progress in this area by including predictions for an increasing number of methods on an increasing number of variants. An integrated variant data resource would also help to prevent problems in properly running each software package in a local environment, as well as issues with using an out-of-date version of a given software package. For example, we ran the PANTHER-subPSEC package locally on the same data set as reported in Shihab et al. (2013) and found that, surprisingly, the predictions for many variants did not match, possibly due to a bug or local installation problem with the software version used for the publication. Stable, shared data resources with persistent identifiers and versioning of predictions could have a dramatic effect on the accessibility, reproducibility, and utility of variant-effect prediction methods in biomedical applications.
The second area is further work on benchmark data sets, which are essential for accurate evaluation of prediction methods. As in nearly all domains of science, positive examples are easier to establish than negative examples, but gold standard sets of both pathogenic and neutral variants are required. The recent work of Grimm et al. (2015) represents a significant step toward developing such sets, but filtering out biased variants necessarily reduces the size and statistical power of the evaluation set. A focused effort in producing additional benchmark data sets, particularly those based on experimental mutagenesis under multiple conditions, could potentially address this issue. As described above, some more recent data sets of this kind have been generated (Marini et al. 2010; Dimster-Denk et al. 2013) but a more comprehensive set of mutations in a larger number of proteins is needed.
The third area is supporting more systematic approaches to new method development: developers of new prediction methods attempt, whenever possible, to critically assess why performance is improved over previous methods. This could entail comparing the performance of a new method to a baseline for that method that keeps all other variables fixed, such as the set of training examples, multiple alignments, etc. For machine-learning methods, this could also entail an effort to report and, ideally, standardize training sets and test sets. Several recent articles have demonstrated this principle particularly well. For example, for SNPs&GO (Capriotti et al. 2013b) the same learning procedure was followed with and without the additional Gene Ontology information (Ashburner et al. 2000) to quantitate the improvement in prediction accuracy from this one additional data source. SNAP2 (Hecht et al. 2015) reported results for multiple different machine-learning methods using the same set of features. The FATHMM article (Shihab et al. 2013) reported the performance of their method before and after incorporating protein domain-specific pathogenicity weights. There are numerous other cases in the literature, and consistent adherence to such systematic analyses will help drive further improvements, and even suggest fruitful combinations of existing methods. For example, the unweighted sequence, conservation-based method used by FATHMM performed worse than other conservation-based methods, suggesting that the same weighting (notwithstanding circularity issues discussed above) could be added to these other conservation-based methods to produce even better overall prediction rates.
Finally, it should be noted that the work on NSV impact prediction has paved the way for development of similar methods that emerged later, and are continuing to emerge, for treating variation in noncoding regions of the genome. Evolutionary conservation at the nucleotide level can be computed using methods such as GERP (Cooper et al. 2005), phyloP (Pollard et al. 2010), and phastCons (Siepel et al. 2005). Analogously to (and in addition to) protein structure features for NSVs, biochemical features such as transcription-factor-binding sites [either predicted (Macintyre et al. 2010; Zhao et al. 2011) or from experiments (Encode Project Consortium 2012)], open chromatin or DNA methylation (Barenboim and Manke 2013), and microRNA genes (Barenboim et al. 2010) are being used to develop machine-learning-based combined methods (Kircher et al. 2014). And all of these predictions are being incorporated as prior knowledge into larger statistical frameworks for prioritizing potentially causal variants in human disease (Lewinger et al. 2007; O’Fallon et al. 2013; Deo et al. 2014). The medical genetics applications envisioned in the early days of NSV prediction are only just beginning, and further improvements in prediction accuracy and availability will be required to help realize their full potential.
Acknowledgments
We thank Emidio Capriotti and Jasper Rine for critical reading of this manuscript and helpful discussions.
Footnotes
Communicating editor: R. A. Isaacson
Literature Cited
- Abecasis G. R., Altshuler D., Auton A., Brooks L. D., Durbin R. M., et al. , 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis G. R., Auton A., Brooks L. D., DePristo M. A., Durbin R. M., et al. , 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., et al. , 2010. A method and server for predicting damaging missense mutations. Nat. Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, 1990. Basic local alignment search tool. J. Mol. Biol. 215(3): 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., et al. , 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M., 2012. One-stop shop for disease genes. Nature 491: 171. [DOI] [PubMed] [Google Scholar]
- Barenboim M., Manke T., 2013. ChroMoS: an integrated web tool for SNP classification, prioritization and functional interpretation. Bioinformatics 29: 2197–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenboim M., Zoltick B. J., Guo Y., Weinberger D. R., 2010. MicroSNiPer: a web tool for prediction of SNP effects on putative microRNA targets. Hum. Mutat. 31: 1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett C., Hughey R., Karplus K., 1997. Scoring hidden Markov models. Comput. Appl. Biosci. 13: 191–199. [DOI] [PubMed] [Google Scholar]
- Bendl J., Stourac J., Salanda O., Pavelka A., Wieben E. D., et al. , 2014. PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations. PLOS Comput. Biol. 10: e1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedix A., Becker C. M., de Groot B. L., Caflisch A., Bockmann R. A., 2009. Predicting free energy changes using structural ensembles. Nat. Methods 6: 3–4. [DOI] [PubMed] [Google Scholar]
- Boeckmann B., Bairoch A., Apweiler R., Blatter M. C., Estreicher A., et al. , 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31: 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham J. T., Ortlund E. A., Thornton J. W., 2009. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 461: 515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y., Rost B., 2007. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 35: 3823–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buetow K. H., Edmonson M. N., Cassidy A. B., 1999. Reliable identification of large numbers of candidate SNPs from public EST data. Nat. Genet. 21: 323–325. [DOI] [PubMed] [Google Scholar]
- Cai Z., Tsung E. F., Marinescu V. D., Ramoni M. F., Riva A., et al. , 2004. Bayesian approach to discovering pathogenic SNPs in conserved protein domains. Hum. Mutat. 24: 178–184. [DOI] [PubMed] [Google Scholar]
- Calabrese R., Capriotti E., Fariselli P., Martelli P. L., Casadio R., 2009. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum. Mutat. 30: 1237–1244. [DOI] [PubMed] [Google Scholar]
- Campbell I. M., Shaw C. A., Stankiewicz P., Lupski J. R., 2015. Somatic mosaicism: implications for disease and transmission genetics. Trends Genet. 31: 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network , 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Fariselli P., Casadio R., 2005. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 33: W306–W310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Calabrese R., Casadio R., 2006. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 22: 2729–2734. [DOI] [PubMed] [Google Scholar]
- Capriotti E., Nehrt N. L., Kann M. G., Bromberg Y., 2012. Bioinformatics for personal genome interpretation. Brief. Bioinform. 13: 495–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Altman R. B., Bromberg Y., 2013a Collective judgment predicts disease-associated single nucleotide variants. BMC Genomics 14(Suppl 3): S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Calabrese R., Fariselli P., Martelli P. L., Altman R. B., et al. , 2013b WS-SNPs&GO: a web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genomics 14(Suppl 3): S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M., Altshuler D., Ireland J., Sklar P., Ardlie K., et al. , 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22: 231–238. [DOI] [PubMed] [Google Scholar]
- Chasman D., Adams R. M., 2001. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J. Mol. Biol. 307: 683–706. [DOI] [PubMed] [Google Scholar]
- Chen Y. C., Douville C., Wang C., Niknafs N., Yeo G., et al. , 2014. A probabilistic model to predict clinical phenotypic traits from genome sequencing. PLOS Comput. Biol. 10: e1003825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M. S., Karchin R., 2011. Using bioinformatics to predict the functional impact of SNVs. Bioinformatics 27: 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F. S., Brooks L. D., Chakravarti A., 1998. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 8: 1229–1231. [DOI] [PubMed] [Google Scholar]
- Compiani M., Capriotti E., 2013. Computational and theoretical methods for protein folding. Biochemistry 52: 8601–8624. [DOI] [PubMed] [Google Scholar]
- Cooper G. M., Stone E. A., Asimenos G., Green E. D., Batzoglou S., et al. , 2005. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15: 901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton R. G., Auerbach A. D., Axton M., Barash C. I., Berkovic S. F., et al. , 2008. GENETICS. The Human Variome Project. Science 322: 861–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff, M. O., R. M. Schwartz, and B. C. Orcutt, 1978 A model of evolutionary change in proteins. Atlas Prot. Sequence Struct. 5: 345–351. [Google Scholar]
- Dehouck Y., Kwasigroch J., Rooman M., Gilis D., 2013. BeAtMuSiC: prediction of changes in protein-protein binding affinity on mutations. Nucleic Acids Res. 41: W333–W339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo R. C., Musso G., Tasan M., Tang P., Poon A., et al. , 2014. Prioritizing causal disease genes using unbiased genomic features. Genome Biol. 15: 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimster-Denk D., Tripp K. W., Marini N. J., Marqusee S., Rine J., 2013. Mono and dual cofactor dependence of human cystathionine beta-synthase enzyme variants in vivo and in vitro. G3 (Bethesda) 3: 1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., et al. , 2015. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 24: 2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Project Consortium , 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernald G. H., Capriotti E., Daneshjou R., Karczewski K. J., Altman R. B., 2011. Bioinformatics challenges for personalized medicine. Bioinformatics 27: 1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Costa C., Gelpi J. L., Zamakola L., Parraga I., de la Cruz X., et al. , 2005. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics 21: 3176–3178. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., 1970. Distinguishing homologous from analogous proteins. Syst. Zool. 19: 99–113. [PubMed] [Google Scholar]
- Frousios K., Iliopoulos C. S., Schlitt T., Simpson M. A., 2013. Predicting the functional consequences of non-synonymous DNA sequence variants: evaluation of bioinformatics tools and development of a consensus strategy. Genomics 102: 223–228. [DOI] [PubMed] [Google Scholar]
- Fu W., O’Connor T. D., Jun G., Kang H. M., Abecasis G., et al. , 2013. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493: 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B., Riemer C., Hefferon T., Thomas D., Hsu F., et al. , 2007. PhenCode: connecting ENCODE data with mutations and phenotype. Hum. Mutat. 28: 554–562. [DOI] [PubMed] [Google Scholar]
- Goldgar D. E., Easton D. F., Deffenbaugh A. M., Monteiro A. N. A., Tavtigian S. V., et al. , 2004. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am. J. Hum. Genet. 75: 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez A., Lopez-Bigas N., 2011. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 88: 440–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., 1974. Amino acid difference formula to help explain protein evolution. Science 185: 862–864. [DOI] [PubMed] [Google Scholar]
- Greenblatt M. S., Brody L. C., Foulkes W. D., Genuardi M., Hofstra R. M., et al. , 2008. Locus-specific databases and recommendations to strengthen their contribution to the classification of variants in cancer susceptibility genes. Hum. Mutat. 29: 1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C., Stephens P., Smith R., Dalgliesh G. L., Hunter C., et al. , 2007. Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., 1994. Profile analysis. Methods Mol. Biol. 25: 247–266. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Robinson N. L., 1996. Use of receiver operating characteristic (ROC) analysis to evaluate sequence matching. Comput. Chem. 20: 25–33. [DOI] [PubMed] [Google Scholar]
- Grimm D. G., Azencott C. A., Aicheler F., Gieraths U., MacArthur D. G., et al. , 2015. The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum. Mutat. 36: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerois R., Nielsen J. E., Serrano L., 2002. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 320: 369–387. [DOI] [PubMed] [Google Scholar]
- Halushka M. K., Fan J. B., Bentley K., Hsie L., Shen N., et al. , 1999. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat. Genet. 22: 239–247. [DOI] [PubMed] [Google Scholar]
- Hamosh A., Scott A. F., Amberger J. S., Bocchini C. A., McKusick V. A., 2005. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33: D514–D517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M., Bromberg Y., Rost B., 2015. Better prediction of functional effects for sequence variants. BMC Genomics 16: S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G., 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89: 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G., 1994. Position-based sequence weights. J. Mol. Biol. 243: 574–578. [DOI] [PubMed] [Google Scholar]
- Hicks S., Wheeler D. A., Plon S. E., Kimmel M., 2011. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum. Mutat. 32: 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium, 2005. A haplotype map of the human genome. Nature 437: 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M., 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Karchin R., 2009. Next generation tools for the annotation of human SNPs. Brief. Bioinform. 10: 35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata T., Ota M., Nishikawa K., 1999. The Protein Mutant Database. Nucleic Acids Res. 27: 355–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E. H., Leaver-Fay A., Baker D., 2011. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins 79: 830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, M., D. M. Witten, P. Jain, B. J. O’Roak and G. M. Cooper, 2014 A general framework for estimating the relative pathogenicity of human genetic variants. 46: 310–315. [DOI] [PMC free article] [PubMed]
- Kondrashov A. S., Sunyaev S., Kondrashov F. A., 2002. Dobzhansky-Muller incompatibilities in protein evolution. Proc. Natl. Acad. Sci. USA 99: 14878–14883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathinal R. J., Bettencourt B. R., Hartl D. L., 2004. Compensated deleterious mutations in insect genomes. Science 306: 1553–1554. [DOI] [PubMed] [Google Scholar]
- Lewinger J. P., Conti D. V., Baurley J. W., Triche T. J., Thomas D. C., 2007. Hierarchical Bayes prioritization of marker associations from a genome-wide association scan for further investigation. Genet. Epidemiol. 31: 871–882. [DOI] [PubMed] [Google Scholar]
- Li B., Krishnan V. G., Mort M. E., Xin F., Kamati K. K., et al. , 2009. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 25: 2744–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B. Y., Zhang J., 2007. Mouse duplicate genes are as essential as singletons. Trends Genet. 23: 378–381. [DOI] [PubMed] [Google Scholar]
- Liu X., Jian X., Boerwinkle E., 2011. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 32: 894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb D. D., Swanstrom R., Everitt L., Manchester M., Stamper S. E., et al. , 1989. Complete mutagenesis of the HIV-1 protease. Nature 340: 397–400. [DOI] [PubMed] [Google Scholar]
- Lopes M. C., Joyce C., Ritchie G. R., John S. L., Cunningham F., et al. , 2012. A combined functional annotation score for non-synonymous variants. Hum. Hered. 73: 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintyre G., Bailey J., Haviv I., Kowalczyk A., 2010. is-rSNP: a novel technique for in silico regulatory SNP detection. Bioinformatics 26: i524–i530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini N. J., Thomas P. D., Rine J., 2010. The use of orthologous sequences to predict the impact of amino acid substitutions on protein function. PLoS Genet. 6: e1000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz P., Kleina L. G., Cruz C., Ehret S., Miller J. H., 1994. Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as “spacers” which do not require a specific sequence. J. Mol. Biol. 240: 421–433. [DOI] [PubMed] [Google Scholar]
- Masso M., Vaisman I. I., 2010. AUTO-MUTE: web-based tools for predicting stability changes in proteins due to single amino acid replacements. Protein Eng. Des. Sel. 23: 683–687. [DOI] [PubMed] [Google Scholar]
- Miller M. P., Kumar S., 2001. Understanding human disease mutations through the use of interspecific genetic variation. Hum. Mol. Genet. 10: 2319–2328. [DOI] [PubMed] [Google Scholar]
- Mottaz A., David F. P., Veuthey A. L., Yip Y. L., 2010. Easy retrieval of single amino-acid polymorphisms and phenotype information using SwissVar. Bioinformatics 26: 851–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moult J., Fidelis K., Kryshtafovych A., Schwede T., Tramontano A., 2014. Critical assessment of methods of protein structure prediction (CASP): round x. Proteins 82(Suppl 2): 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P. C., Henikoff S., 2001. Predicting deleterious amino acid substitutions. Genome Res. 11: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P. C., Henikoff S., 2006. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 7: 61–80. [DOI] [PubMed] [Google Scholar]
- Niroula A., Urolagin S., Vihinen M., 2015. PON-P2: prediction method for fast and reliable identification of harmful variants. PLoS One 10: e0117380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Fallon B. D., Wooderchak-Donahue W., Bayrak-Toydemir P., Crockett D., 2013. VarRanker: rapid prioritization of sequence variations associated with human disease. BMC Bioinformatics 14(Suppl 13): S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., 1970. Evolution by Gene Duplication. Springer-Verlag, New York. [Google Scholar]
- Olatubosun A., Valiaho J., Harkonen J., Thusberg J., Vihinen M., 2012. PON-P: integrated predictor for pathogenicity of missense variants. Hum. Mutat. 33: 1166–1174. [DOI] [PubMed] [Google Scholar]
- Olivier M., Eeles R., Hollstein M., Khan M. A., Harris C. C., et al. , 2002. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 19: 607–614. [DOI] [PubMed] [Google Scholar]
- Pazdrak K., Adachi T., Alam R., 1997. Src homology 2 protein tyrosine phosphatase (SHPTP2)/Src homology 2 phosphatase 2 (SHP2) tyrosine phosphatase is a positive regulator of the interleukin 5 receptor signal transduction pathways leading to the prolongation of eosinophil survival. J. Exp. Med. 186: 561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ, 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85: 2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piirila H., Valiaho J., Vihinen M., 2006. Immunodeficiency mutation databases (IDbases). Hum. Mutat. 27: 1200–1208. [DOI] [PubMed] [Google Scholar]
- Pokala N., Handel T. M., 2005. Energy functions for protein design: adjustment with protein-protein complex affinities, models for the unfolded state, and negative design of solubility and specificity. J. Mol. Biol. 347: 203–227. [DOI] [PubMed] [Google Scholar]
- Pollard K. S., Hubisz M. J., Rosenbloom K. R., Siepel A., 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapov V., Cohen M., Schreiber G., 2009. Assessing computational methods for predicting protein stability upon mutation: good on average but not in the details. Protein Eng. Des. Sel. 22: 553–560. [DOI] [PubMed] [Google Scholar]
- Ramensky V., Bork P., Sunyaev S., 2002. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30: 3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva B., Antipin Y., Sander C., 2011. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohl C. A., Strauss C. E., Misura K. M., Baker D., 2004. Protein structure prediction using Rosetta. Methods Enzymol. 383: 66–93. [DOI] [PubMed] [Google Scholar]
- Sasidharan Nair P., Vihinen M., 2013. VariBench: a benchmark database for variations. Hum. Mutat. 34: 42–49. [DOI] [PubMed] [Google Scholar]
- Schaefer C., Meier A., Rost B., Bromberg Y., 2012. SNPdbe: constructing an nsSNP functional impacts database. Bioinformatics 28: 601–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz J. M., Rodelsperger C., Schuelke M., Seelow D., 2010. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7: 575–576. [DOI] [PubMed] [Google Scholar]
- Shendure J., Akey J. M., 2015. The origins, determinants, and consequences of human mutations. Science 349: 1478–1483. [DOI] [PubMed] [Google Scholar]
- Sherry S. T., Ward M. H., Kholodov M., Baker J., Phan L., et al. , 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab H. A., Gough J., Cooper D. N., Stenson P. D., Barker G. L., et al. , 2013. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 34: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M., et al. , 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippl M. J., 1990. Calculation of conformational ensembles from potentials of mean force. An approach to the knowledge-based prediction of local structures in globular proteins. J. Mol. Biol. 213: 859–883. [DOI] [PubMed] [Google Scholar]
- Sjoblom T., Jones S., Wood L. D., Parsons D. W., Lin J., et al. , 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274. [DOI] [PubMed] [Google Scholar]
- Sjolander K., Karplus K., Brown M., Hughey R., Krogh A., et al. , 1996. Dirichlet mixtures: a method for improved detection of weak but significant protein sequence homology. Comput. Appl. Biosci. 12: 327–345. [DOI] [PubMed] [Google Scholar]
- Stenson P. D., Ball E. V., Mort M., Phillips A. D., Shiel J. A., et al. , 2003. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 21: 577–581. [DOI] [PubMed] [Google Scholar]
- Stone E. A., Sidow A., 2005. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 15: 978–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev S. R., Eisenhaber F., Rodchenkov I. V., Eisenhaber B., Tumanyan V. G., et al. , 1999. PSIC: profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng. 12: 387–394. [DOI] [PubMed] [Google Scholar]
- Tang, H., and P. D. Thomas, 2016 PANTHER-PSEP: predicting disease-causing mutations using position-specific evolutionary preservation. Bioinformatics. [DOI] [PubMed]
- Thomas P. D., Kejariwal A., 2004. Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: evolutionary evidence for differences in molecular effects. Proc. Natl. Acad. Sci. USA 101: 15398–15403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. D., Campbell M. J., Kejariwal A., Mi H., Karlak B., et al. , 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13: 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thusberg J., Olatubosun A., Vihinen M., 2011. Performance of mutation pathogenicity prediction methods on missense variants. Hum. Mutat. 32: 358–368. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium , 2011. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39: D214–D219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Moult J., 2001. SNPs, protein structure, and disease. Hum. Mutat. 17: 263–270. [DOI] [PubMed] [Google Scholar]
- Wei Q., Wang L., Wang Q., Kruger W. D., Dunbrack R. L., Jr, 2010. Testing computational prediction of missense mutation phenotypes: functional characterization of 204 mutations of human cystathionine beta synthase. Proteins 78: 2058–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yue P., Melamud E., Moult J., 2006. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics 7: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., W. T. Clark, M. Mort, D. N. Cooper, P. Radivojac et al, 2011 Prediction of functional regulatory SNPs in monogenic and complex disease. Hum. Mutat. 32: 1183–1190. [DOI] [PMC free article] [PubMed]
- Zuckerkandl, E., and L. Pauling, 1962 Molecular disease, evolution and genic heterogenetity, pp. 189–225 in Horizons in Biochemistry, edited by M. Kasha and B. Pullman. Academic Press, New York. [Google Scholar]