Abstract
Despite longstanding interest in the genetic mechanisms that underlie behavioral evolution, very few genes that underlie naturally occurring variation in behavior between individuals or species are known, particularly in vertebrates. Here, we build on our previous forward genetic mapping experiments and use transgenic approaches to identify Ectodysplasin as a gene that causes differences in schooling behavior between wild populations of threespine stickleback (Gasterosteus aculeatus) fish. This work provides rare insight into the proximate mechanisms that have shaped the evolution of vertebrate behavior.
Keywords: genetics of behavioral evolution, natural variation, schooling behavior, threespine stickleback, transgenic
BIOLOGISTS have long recognized that there must be genetic contributions to the evolution of behavioral differences among animals. Although genes that are necessary to perform specific behaviors have been identified through laboratory studies of inbred animals, these may not be the genes that underlie differences in behavior between individuals or species in nature (Boake et al. 2002; Hoekstra 2010). Because many behaviors arise from a complex interaction between multiple genes and the environment, it has been difficult to identify mutations that cause natural variation in behavior. Consequently, little is known about the genetic changes that enable behavior patterns to change in response to evolutionary forces, particularly in vertebrates (Bendesky and Bargmann 2011; Martin and Orgogozo 2013). To overcome these challenges, we used a combination of forward genetic mapping and transgenesis to identify Ectodysplasin (Eda) as a gene that contributes to the evolution of schooling behavior in threespine stickleback fish (Gasterosteus aculeatus).
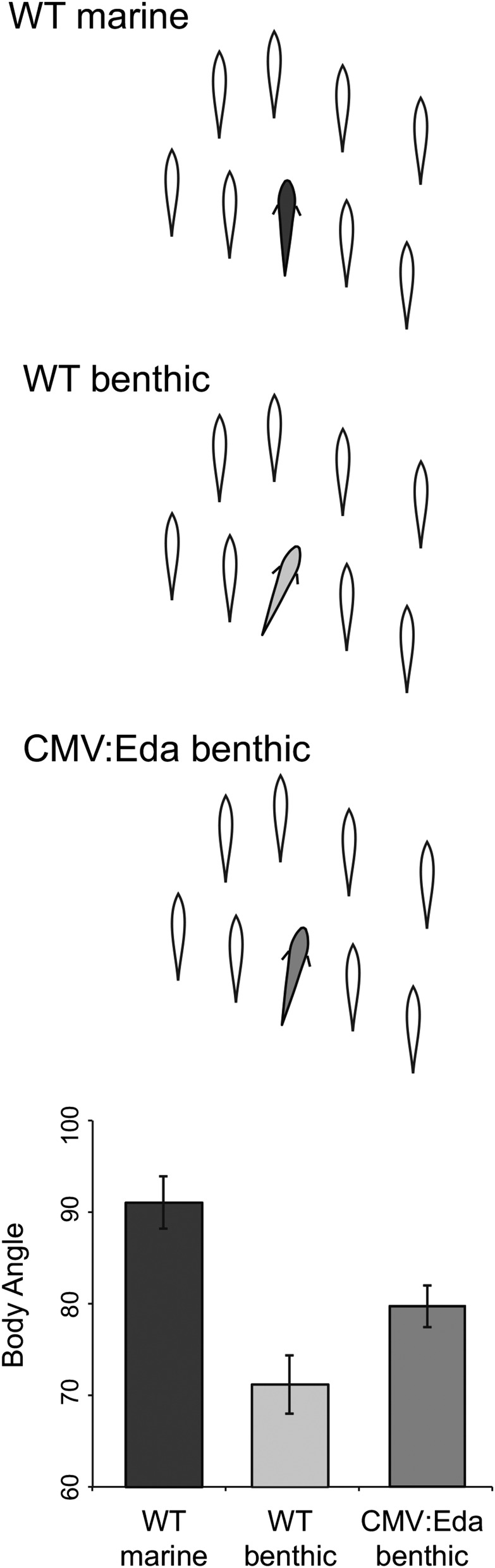
Schooling is a fascinating behavior known to vary widely among fish species, due to evolutionary trade-offs between the costs and benefits of group living (Krause and Ruxton 2002). To understand the proximate mechanisms that underlie the evolution of schooling behavior, we previously developed an assay to elicit naturalistic schooling in response to a controlled stimulus, a robotic school of model sticklebacks (Wark et al. 2011). This assay permits quantification of the two critical features of schooling behavior: motivation to school and ability to maintain an efficient body position within the school (Pitcher 1983). Marine sticklebacks from open-water, pelagic habitats school extensively both in the wild and in response to the model school (Wark et al. 2011; Di-Poi et al. 2014). They follow the school for extended durations and display parallel body position with the models when schooling (Figure 1). In contrast, sticklebacks from benthic lake habitats spend significantly less time with the model school (Wark et al. 2011). When benthic fish attempt to school, they are less capable of performing this complex behavior, exhibiting an inefficient body position characterized by a significantly less parallel angle with the model fish (Figure 1). Thus, this assay recapitulates the reduced schooling behavior observed in wild benthic sticklebacks, which is likely an adaptation to the abundant shelter in their highly vegetated lake environment (Larson 1976; Vamosi 2002; Wark et al. 2011).
Figure 1.
Eda transgene alters schooling behavior of benthic sticklebacks. The average body angle of marine (dark gray), benthic (light gray), and CMV:Eda transgenic benthic fish (medium gray) when schooling with a fixed school of stickleback models (open silhouettes). Graph depicts the mean ± SEM body angle of marine (n = 8), benthic (n = 18), and CMV:Eda benthic (n = 44) sticklebacks. Marines have a significantly more parallel body angle than benthics (Mann–Whitney–Wilcoxon test, P = 7.796e-04). CMV:Eda benthic fish have a significantly more marine-like body angle than their wild-type (WT) benthic siblings (Mann–Whitney–Wilcoxon test, P = 0.0296).
Our previous genome-wide linkage mapping of schooling behavior in a benthic × marine F2 intercross demonstrated that the ability to school and the motivation to school map to distinct genomic regions. Schooling ability, as measured by body position when schooling with the models, shows significant linkage to a region on chromosome 4 (Greenwood et al. 2013). Interestingly, this genomic region is also genetically linked to the presence of bony armor and the patterning of the sensory neuromasts of the lateral line (Wark et al. 2012). Both of these phenotypes are controlled by the Eda gene, which is contained within the schooling locus on chromosome 4 (Colosimo et al. 2005; Mills et al. 2014).
Here, we tested whether variation in Eda might also contribute to differences in schooling ability between marine and benthic sticklebacks by manipulating Eda expression in benthic sticklebacks. Benthics have significantly lower expression of Eda than marine sticklebacks in the developing flank (Mills et al. 2014; O’Brown et al. 2015) and in the brain (Supplemental Material, Figure S1). We had previously generated transgenic benthic sticklebacks that express the marine allele of the Eda complementary DNA (cDNA) under the control of a cytomegalovirus (CMV) promoter, which should drive constitutive expression of Eda throughout the fish (Mills et al. 2014). Here, we bred six CMV:Eda founder individuals to wild-type benthics to establish six independent stable benthic CMV:Eda transgenic lines. These crosses yielded a mix of wild-type and transgenic offspring. We then tested the offspring in the model school assay, comparing the body position of wild-type and CMV:Eda benthic siblings. As predicted, the CMV:Eda benthic fish exhibited a significantly more marine-like schooling position compared with their wild-type siblings (Figure 1; Mann–Whitney–Wilcoxon test, W = 256, P = 0.0296). The magnitude of this effect is consistent with the fact that schooling position is a complex trait and that the Eda locus had a relatively small effect (13% variance explained) in our previous mapping study (Greenwood et al. 2013). There were no founder effects on schooling behavior among the transgenic lines (Figure S2; Kruskal–Wallis test, χ2 = 2.25, P = 0.521). Consistent with the fact that the motivation to school is not genetically linked to chromosome 4 (Greenwood et al. 2013, 2015), there was no difference between wild-type and transgenic fish in the latency to join the model school (Mann–Whitney–Wilcoxon test, W = 317, P = 0.22). These results reveal that Eda contributes to variation in the ability to school between marine and benthic sticklebacks and provide one of the only examples in which the manipulation of a single gene can recapitulate behavioral differences seen in wild vertebrate species.
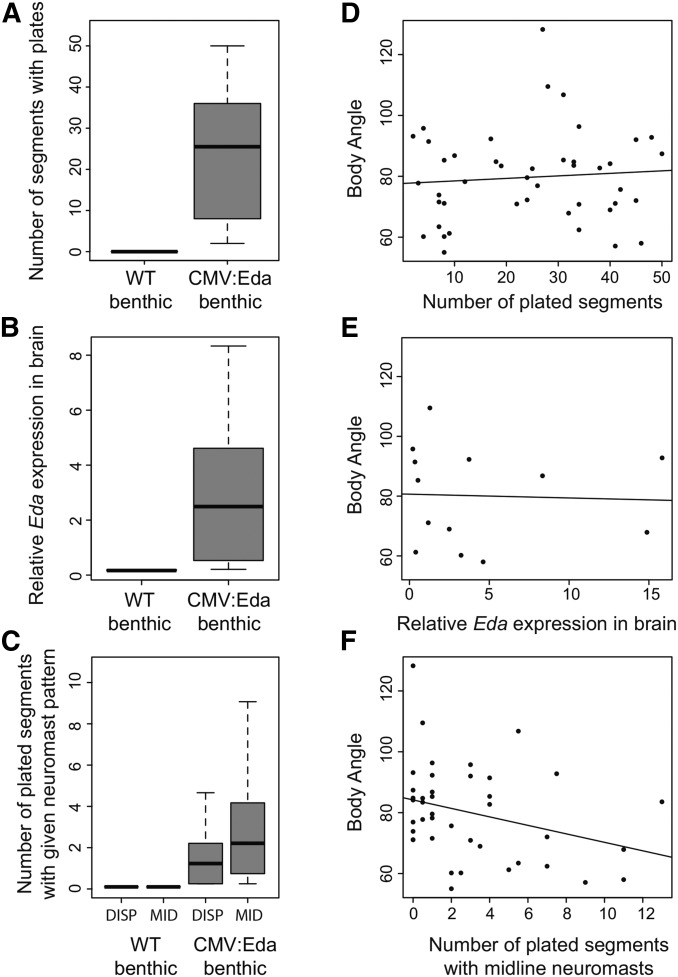
Consistent with the known effects of Eda on morphological traits (Colosimo et al. 2005; Mills et al. 2014), CMV:Eda transgenic benthics also have differences in bony plates and neuromast pattern, as well as Eda expression in the diencephalon (Figure 2). However, only variation in posterior lateral line patterning, but not bony plates or Eda expression in the diencephalon, predicted schooling body angle in the CMV:Eda benthics (Figure 2). Specifically, the number of plates with midline neuromasts showed an association with schooling ability in the CMV:Eda benthics (Figure 2; multiple regression, β = −0.311, P = 0.048). These data suggest that the effects of Eda on schooling behavior may be mediated by its effects on lateral line patterning. However, manipulating lateral line phenotypes by physical or chemical ablations had no effect on schooling behavior in marines, benthics, or CMV:Eda transgenics (data not shown). Furthermore, it is possible that Eda affects schooling behavior through expression at a specific time or in a specific tissue not examined here. Thus, extensive future work is required to reveal the precise mechanism by which Eda affects schooling behavior.
Figure 2.
Associations between individual variation in bony plates, Eda brain expression, lateral line pattern, and schooling behavior among CMV:Eda benthic transgenics. CMV:Eda fish have phenotypes significantly different from their wild-type (WT) siblings, including (A) more plates (Mann–Whitney–Wilcoxon test, W = 0, P = 5.2e-10; WT, n = 18; CMV:Eda, n = 44); (B) higher Eda mRNA expression in the diencephalon (Mann–Whitney–Wilcoxon test, W = 0, P = 7.4e-05; WT, n = 6; CMV:Eda, n = 13); and (C) more plates with dispersed neuromasts (DISP: Mann–Whitney–Wilcoxon test, W = 102, P = 6.5e-06; WT, n = 17; CMV:Eda, n = 41) or midline neuromasts (MID: Mann–Whitney–Wilcoxon test, W = 76.5, P = 1.13e-06; WT, n = 17; CMV:Eda, n = 41). Box plots in A–C show the median and 25% and 75% quartiles, and whiskers show the 1.5× interquartile range. In CMV:Eda benthics, there is no relationship between individual variation in median body angle and either (D) the number of plates (n = 41; multiple regression, β = 0.240, P = 0.146) or (E) Eda mRNA levels in the diencephalon (n = 13; Spearman’s correlation, Rho = −0.176, P = 0.565), but there is a significant correlation between individual variation in median body angle and (F) the number of plated segments that have only midline neuromasts (n = 41; multiple regression, β = −0.311, P = 0.048).
Our combined approach of forward genetic mapping and transgenic analysis demonstrates for the first time that Eda is a gene that shapes evolutionary differences in behavior, thereby adding to a small list of genes known to influence behavioral variation in the wild (Bendesky and Bargmann 2011; Martin and Orgogozo 2013). Identification of additional genes and mechanisms that underlie behavioral variation in natural populations will ultimately provide key insights into the genetic changes that mediate the evolution of behavioral differences among individuals or species.
Materials and Methods
Transgenic fish
All experiments were conducted in accordance with the guidelines of the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee (protocol 1575). Mosaic CMV:Eda benthic fish were generated as previously described (Mills et al. 2014). Briefly, a transgene consisting of a CMV promoter driving expression of a marine allele of the Eda cDNA was incorporated into single-cell benthic embryos using Tol2 transgenesis (Kawakami 2007). Previous work has shown that regulatory but not coding differences in Eda are responsible for the development of bony plates (O’Brown et al. 2015). Individuals mosaic for transgene incorporation were identified by screening for presence of ectopic plates. For the current study, mosaic founders were crossed with wild-type benthic fish (derived from fish caught in Paxton Lake, British Columbia) to yield stable lines, comprising a mix of transgenic and wild-type progeny. Wild-type and transgenic identity was confirmed by phenotyping for presence of plates and PCR genotyping to identify the transgene [forward primer (in CMV): 5′-AGG CCT CTT CGC TAT TAC G-3′; reverse primer (in Eda): 5′-ATT GTA TCC CGC TTC TGG TG-3′]. Some phenotypically wild-type fish (i.e., no ectopic plates) were positive for the transgene and were excluded from the study. The final data set consisted of n = 44 CMV:Eda benthics and n = 18 genotypically wild-type benthic siblings. The transgenics were from six independent stable lines from six different founders (family 1, n = 1; family 2, n = 20; family 3, n = 6; family 4, n = 3; family 5, n = 13; family 6, n = 1). The single transgenic fish from family 1 and family 6 were included in statistical comparisons of wild-type vs. transgenic fish but were excluded from analyses of the effect of “founder” on phenotypes due to a sample size of one. Wild-type marine sticklebacks (n = 8; derived from fish caught in the Bekanbeushi River, Japan) were also tested in the model school assay for comparison in Figure 1.
Behavioral testing
Fish were tested in the model school assay as previously described (Wark et al. 2011). Briefly, the assay consists of a 61-cm diameter circular white tank in which a school of eight plastic sticklebacks (cast from a benthic-marine F2 hybrid individual) is moved in a counterclockwise circle. Transgenic and wild-type individuals were tagged with fluorescent elastomer (Northwest Marine Technologies, Shaw Island, WA), and individuals from the same family were housed together and tested on the same day. Fish were captured from their home tank and placed into 950-ml isolation chambers for 1.5 hr. Fish were then placed into the assay chamber and given 5 min to acclimate. The motor controlling the model school was then switched on remotely and fish were videotaped interacting with the model school for 5 min. Videos were then digitized, and when the model school was in a predefined position in the tank, frames were extracted for analysis of body angle. We then calculated the median body angle for each fish. Note that the circular configuration of the tank and the counterclockwise movement of the model school act to bias the direction of body-angle deviation. Specifically, the body angle tends to deviate from parallel with the models (90 degrees; marine-like) to an acute angle directed toward the center of the tank (benthic-like). Videos were also scored for the latency to join the school, which measures the motivation to school, a behavior that is unlinked to schooling body position (Greenwood et al. 2013).
Bony plate and lateral line phenotyping
Fish were treated with calcein to visualize bony armor plates and DASPEI (2-(4-(dimethylamino)styryl)-N-ethylpyridinium Iodide) to visualize lateral line neuromasts as previously described (Mills et al. 2014). Sticklebacks have plates in two developmentally separable regions on their flank: the anterior trunk comprises the first seven trunk segments and the posterior trunk is found in posterior body segments (Colosimo et al. 2004). We counted the number of segments containing plates in the posterior and anterior trunk. Following Mills et al. (2014), we quantified the number of plated and unplated segments in the posterior trunk that had dispersed (dorsal and ventral to the midline) or midline neuromasts. We also counted the number of neuromasts in the anterior and posterior trunk lateral line (Wark et al. 2012). All counts were performed on both the left and right sides of the fish, and counts from the two sides were combined for analyses. Some fish (n = 3 transgenics) died prior to DASPEI staining.
Quantitative PCR
We performed quantitative PCR (qPCR) to compare levels of Eda expression in the brain of larval and juvenile benthic and marine fish. We also compared Eda expression in a subset of adult transgenic CMV:Eda benthics. Benthic and marine larval brains were collected on the day after hatching. Brains were removed from 24 fish from each population, and three brains were pooled for each sample (n = 8 marine and n = 8 benthic samples). Larval brains were placed directly into Trizol (Life Technologies, Carlsbad, CA) and were homogenized immediately. For juveniles and transgenics, we removed whole brains of marines (n = 8), benthics (n = 8), CMV:benthic transgenics (n = 13), and matched wild-type benthic siblings (n = 6). Brains were placed into RNA later (Life Technologies) and stored at −20 C. Brains were then dissected into three portions: the telencephalon; the diencephalon and rostral midbrain; and the caudal midbrain, cerebellum, and hindbrain. For simplicity, we refer to these regions as the telencephalon, diencephalon, and hindbrain. Brain regions were homogenized in Trizol as above. For qPCR in the marine and benthic brain regions, total RNA was isolated using Trizol, and samples were DNAse-treated and subjected to cDNA synthesis as previously described (Mills et al. 2014). For qPCR in the CMV:Eda transgenics and wild-type siblings, we used only the diencephalon and performed an additional isolation of messenger RNA (mRNA) from total RNA using Dynabeads mRNA Purification kit (Life Technologies) to eliminate the possibility of amplifying the transgenic copy of the Eda cDNA present in genomic DNA. qPCR for Eda was performed in triplicate as previously described, using 17.5 ng of cDNA (RNA equivalent) in each reaction and Eef1b as a reference gene (Mills et al. 2014). Data are reported as relative Eda expression as a percentage of Eef1b levels (Figure 2 and Figure S1).
Statistical analyses
We first used histograms and normal quantile plots in R (http://www.r-project.org/) to examine whether the data fit a normal distribution (Whitlock and Schluter 2015). All traits except body angle showed departures from normality, even after log transformation. Thus, we conservatively used the nonparametric Mann–Whitney–Wilcoxon test in R for pairwise comparisons of all trait differences between transgenics and wild-type siblings and for analyses of qPCR data. However, all significant results reported remain significant when parametric mixed linear models that use population as a fixed effect and family as a random effect are used (data not shown). We used a nonparametric Kruskal–Wallis test in R to test for the effect of transgenic founder on body angle; similar results are observed with a parametric linear model (data not shown). We used multiple regression in SPSS 13.0 software (SPSS, Chicago) to test the effect of the following independent variables on body angle in CMV:Eda benthic transgenics: total plates in the anterior and posterior trunk region, numbers of plated and unplated segments with dispersed or midline neuromasts, and total numbers of neuromasts in anterior and posterior trunk region. Regression does not require that predictor variables are normally distributed as long as the residuals in the response variable (i.e., body angle) are normally distributed (Whitlock and Schluter 2015); visual inspection of the residuals with histograms and normal quantile plots confirms this assumption. Eda qPCR in the diencephalon was performed on a subset of transgenic individuals; thus the relationship between Eda mRNA levels and behavior was not included in the multiple regression but was analyzed in R using Spearman’s correlation.
Data availability
All behavioral, morphological, and qPCR data for CMV:Eda benthic transgenics and behavioral and morphological data for wild-type benthics and marines are provided in File S1. All qPCR data for wild-type benthics and marines are provided in File S2.
Acknowledgments
We thank Kim Hughes, David Kingsley, Harmit Malik, and Dolph Schluter for thoughtful comments on the manuscript and Shaugnessy McCann for fish care. This research was supported by National Science Foundation grant IOS 1145866 (to A.K.G. and C.L.P.).
Author contributions: A.K.G., A.R.W., and C.L.P. designed the experiments; A.K.G., M.G.M., A.R.W., and S.L.A. performed experiments; A.K.G., S.L.A., and C.L.P. analyzed data; A.K.G. and C.L.P. wrote the manuscript. All authors gave final approval for publication.
Footnotes
Communicating editor: K. M. Nichols
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.188342/-/DC1.
Literature Cited
- Bendesky A., Bargmann C. I., 2011. Genetic contributions to behavioural diversity at the gene-environment interface. Nat. Rev. Genet. 12: 809–820. [DOI] [PubMed] [Google Scholar]
- Boake C. R. B., Arnold S. J., Breden F., Meffert L. M., Ritchie M. G., et al. , 2002. Genetic tools for studying adaptation and the evolution of behavior. Am. Nat. 160: S143–S159. [DOI] [PubMed] [Google Scholar]
- Colosimo P. F., Peichel C. L., Nereng K., Blackman B. K., Shapiro M. D., et al. , 2004. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol. 2: e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo P. F., Hosemann K. E., Balabhadra S., Villarreal G., Jr, Dickson M., et al. , 2005. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307: 1928–1933. [DOI] [PubMed] [Google Scholar]
- Di-Poi C., Lacasse J., Rogers S. M., Aubin-Horth N., 2014. Extensive behavioural divergence following colonisation of the freshwater environment in threespine sticklebacks. PLoS One 9: e98980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood A. K., Wark A. R., Yoshida K., Peichel C. L., 2013. Genetic and neural modularity underlie the evolution of schooling behavior in threespine sticklebacks. Curr. Biol. 23: 1884–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood A. K., Ardekani R., McCann S. R., Dubin M. E., Sullivan A., et al. , 2015. Genetic mapping of natural variation in schooling tendency in the threespine stickleback. G3 (Bethesda) 5: 761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra H. E., 2010. In search of the elusive behavior gene, pp. 192–210 in In Search of the Causes of Evolution: From Field Observations to Mechanisms, edited by Grant P., Grant R. Princeton University Press, Princeton, NJ. [Google Scholar]
- Kawakami K., 2007. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 8: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause J., Ruxton G. D., 2002. Living in Groups. Oxford University Press, New York. [Google Scholar]
- Larson G., 1976. Social behavior and feeding ability of two phenotypes of Gasterosteus aculeatus in relation to their spatial and trophic segregation in a temperate lake. Can. J. Zool. 54: 107–121. [Google Scholar]
- Martin A., Orgogozo V., 2013. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution 67: 1235–1250. [DOI] [PubMed] [Google Scholar]
- Mills M. G., Greenwood A. K., Peichel C. L., 2014. Pleiotropic effects of a single gene on skeletal development and sensory system patterning in sticklebacks. Evodevo 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brown N. M., Summers B. R., Jones F. C., Brady S. D., Kingsley D. M., 2015. A recurrent regulatory change underlying altered expression and Wnt response of the stickleback armor plates gene EDA. eLife 4: e05290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher T. J., 1983. Heuristic definitions of fish shoaling behavior. Anim. Behav. 31: 611–613. [Google Scholar]
- Vamosi S. M., 2002. Predation sharpens the adaptive peaks: survival trade-offs in sympatric sticklebacks. Ann. Zool. Fenn. 39: 237–248. [Google Scholar]
- Wark A. R., Greenwood A. K., Taylor E. M., Yoshida K., Peichel C. L., 2011. Heritable differences in schooling behavior among threespine sticklebacks revealed by a novel assay. PLoS One 6: e18316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wark A. R., Mills M. G., Dang L.-H., Chan Y. F., Jones F. C., et al. , 2012. Genetic architecture of variation in the lateral line sensory system of threespine sticklebacks. G3 (Bethesda) 2: 1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock M. C., Schluter D., 2015. The Analysis of Biological Data. Roberts and Company, Greenwood Village, CO. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All behavioral, morphological, and qPCR data for CMV:Eda benthic transgenics and behavioral and morphological data for wild-type benthics and marines are provided in File S1. All qPCR data for wild-type benthics and marines are provided in File S2.