Abstract
Construction of next-generation sequencing (NGS) libraries involves RNA manipulation, which often creates noisy, biased, and artifactual data that contribute to errors in transcriptome analysis. In this study, a total of 19 whole transcriptome termini site sequencing (WTTS-seq) and seven RNA sequencing (RNA-seq) libraries were prepared from Xenopus tropicalis adult and embryo samples to determine the most effective library preparation method to maximize transcriptomics investigation. We strongly suggest that appropriate primers/adaptors are designed to inhibit amplification detours and that PCR overamplification is minimized to maximize transcriptome coverage. Furthermore, genome annotation must be improved so that missing data can be recovered. In addition, a complete understanding of sequencing platforms is critical to limit the formation of false-positive results. Technically, the WTTS-seq method enriches both poly(A)+ RNA and complementary DNA, adds 5′- and 3′-adaptors in one step, pursues strand sequencing and mapping, and profiles both gene expression and alternative polyadenylation (APA). Although RNA-seq is cost prohibitive, tends to produce false-positive results, and fails to detect APA diversity and dynamics, its combination with WTTS-seq is necessary to validate transcriptome-wide APA.
Keywords: 3′-termini sequencing, amplification detours, transcriptome distribution, missing data, Bayesian model
NEXT-generation sequencing (NGS) technologies are used routinely for transcriptome investigation. Libraries for NGS can be prepared to sequence full transcripts or just their 5′ or 3′ ends depending on project goals (Jiang et al. 2015). RNA sequencing (RNA-seq) uses NGS to collect short reads that cover full transcripts (5′ to 3′ ends) (Morin et al. 2008). Given current capabilities in gene expression profiling, splicing form detection, and expressed polymorphism compilation, the method has gradually become the gold standard in transcriptome analysis (Wang et al. 2009; Wilhelm and Landry 2009; Costa et al. 2010; Nagalakshmi et al. 2010). However, the RNA-seq assay is not always cost-effective because random sequencing of full-length transcripts is not necessary to determine gene abundance. In addition, short reads generated by RNA-seq might make it difficult to reconstruct full-length isoforms of transcripts (Steijger et al. 2013). Furthermore, profiling alternative transcript ends is problematic because 5′- and 3′-end biases are introduced during RNA-seq library preparation (Wang et al. 2009; Jiang et al. 2015). However, profiling only the 5′ ends of transcripts is not feasible because the library preparation involves many steps, which increases the possibility of errors (Takahashi et al. 2012).
As such, effort has been focused largely on the development of methods to profile 3′ ends of transcripts. Functionally, the 3′-untranslated regions (UTRs) are important because they harbor regulatory elements that play essential roles in the stabilization, localization, translation, and degradation of messenger RNA (mRNA) (Matoulkova et al. 2012). Technically, the poly(A) tails are used frequently in reverse transcription to convert RNA to complementary DNA (cDNA) that can be sequenced. The 3′-termini of transcripts have been collected in two ways: by digestion of mRNA with restriction enzymes and by random fragmentation. The reverse serial analysis of gene expression (rSAGE) technique (Richards et al. 2006) and the poly(A) tags (PATs) (Wu et al. 2011) with restriction endonuclease cut are two examples of the former strategy. There are several challenges associated with these methods (Jiang et al. 2015). None of the currently available restriction endonucleases can effectively fragment an entire transcriptome because some transcripts may lack recognition sites. To overcome this limitation, the PATs with restriction endonuclease cut method incorporates a specific enzyme recognition site into cDNA and ensures that every transcript can be cut by a distinct restriction enzyme. Unfortunately, this strategy also may increase the length of some products, which can subsequently decrease PCR amplification efficiency and introduce artificial biases in whole transcriptome profiling (Jiang et al. 2015).
As for profiling 3′-termini using random fragmentation, the 3′ poly(A) site mapping using cDNA circularization (3PC) (Mata 2013), 3′-region extraction and deep sequencing (3′READS) (Hoque et al. 2013), and PATs with RNA fragmentation methods (Ma et al. 2014) all enrich fragmented poly(A)+ RNA, while the 3′T-fill (Pelechano et al. 2012; Wilkening et al. 2013) and expression profiling through random sheared cDNA tag sequencing (EXPRSS) techniques (Rallapalli et al. 2014) enrich fragmented cDNA. In comparison, the poly(A) site sequencing (PAS-seq) (Shepard et al. 2011; Yao and Shi 2014) and polyadenylation sequencing (PolyA-seq) approaches (Derti et al. 2012) use custom oligo(dT) primers to collect and sequence 3′-termini regions. These poly(A) site sequencing methods are not without drawbacks. When Ma et al. (2014) compared three different methods, for example, they found that 47.2–98.2% of reads could not be mapped to the 3′-UTRs.
The aforementioned difficulties in producing clean, usable data from NGS platforms clearly provide evidence that library construction for 3′-termini sequencing methods can and should be improved. In this study, we developed a procedure that we call whole transcriptome termini site sequencing (WTTS-seq). Our WTTS-seq approach starts with total RNA, followed by chemical fragmentation and enrichment of both poly(A)+ RNA and poly(A)+ cDNA. During assay development, we tested three types of primers used in PCR for synthesis of second-strand cDNA to complete construction of the NGS libraries. We found that primer design is a very important factor for accurate coverage of the entire transcriptome. By using poly(A)-anchored primers, we reduced noisy data to <0.1%. We also discovered that reduced PCR cycle numbers and lower primer concentrations improved transcriptome coverage. Moreover, we analyzed the same samples using traditional RNA-seq and examined WTTS-seq data of biological and technical replicates to reveal their strengths and weaknesses. Overall, our WTTS-seq method successfully collected poly(A) sites as signatures for global profiling of gene expression and examination of APA with one pipeline.
Materials and Methods
Experimental design
Animals and RNA extraction:
Three adult male and three adult female frogs (Xenopus tropicalis) (>6 months of age) were purchased from Nasco (Fort Atkinson, WI). Immediately on arrival, frogs were humanely killed, rinsed briefly with deionized water, wrapped in aluminum foil, immersed in liquid nitrogen until all tissues were completely frozen, and stored at −80°. Later the frogs were removed from storage and placed in a bath of liquid nitrogen, and tissues were broken into smaller pieces with a hammer. All tissue pieces were kept in liquid nitrogen and subsequently ground into a powder with a mortar and pestle. Ground tissues were thoroughly mixed, and a subsample was removed for total RNA extraction with Trizol reagent according to the manufacturer’s instructions. Contaminating DNA was removed by treating total RNA with DNase (AM1906, Ambion). RNA quantity and quality were assessed by NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE) and nondenaturing agarose gel electrophoresis, respectively. Equal amounts of total RNA from each frog were subsequently pooled and used for RNA-seq and WTTS-seq. In addition, total RNA from one of the female frogs was used as a technical replicate to test the variability of our WTTS-seq method.
WTTS-seq assay development:
We conducted seven trials to develop and improve our WTTS-seq method. As shown in Supplemental Material, File S1A, these trials mainly differed in (1) primers [OP, outer primer; IP, ion primer; and PAAP, poly(A)-anchored primer], (2) number of cDNA synthesis runs (two vs. one run) and PCR cycles (variable), (3) size selection (variable), and (4) amount of total RNA starting material (10, 5, or 2 µg). Oligo(dT20) was used only in trial 1, while the remaining trials used oligo(dT10) for reverse transcription. Oligo sequences are listed in File S1B.
RNA-seq:
Poly(A)+ RNA was selected from the pooled total RNA sample with a Poly(A) Purist Kit (Ambion) according to directions supplied by the manufacturer. Briefly, residual salts were removed by adding 0.1 vol of 5 M ammonium acetate and 2.5 vol of 100% ethanol. Total RNA was recovered by incubating the solution overnight at −80° in a freezer, centrifuging at ≥12,000 × g, and washing with 70% ethanol. The RNA pellet was resuspended in nuclease-free water and combined with binding buffer and oligo(dT) cellulose. The poly(A)+ sequences were hybridized to the oligo(dT) cellulose by incubating at room temperature for 30–60 min. The mixture was subsequently transferred to a spin column and washed to remove nonspecifically bound material and ribosomal RNA. The poly(A)+ RNA was eluted from the oligo(dT) cellulose with an aliquot of the warm solution provided with the kit. A second round of oligo(dT) selection was subsequently performed, and poly(A)+ RNA was recovered by precipitation, as described previously. The final poly(A)+ RNA pellet was resuspended in the solution provided with the kit. An RNA-seq library was constructed using the Ion Total RNA-Seq Kit v2 (Thermo Fisher Scientific) and sequenced on the Ion PGM Sequencer at Washington State University.
Five stages of X. tropicalis embryos as biological replicates:
X. tropicalis embryos were produced using two pairs of parents at The Chinese University of Hong Kong to test the repeatability of our WTTS-seq method. The embryos were cultured in 0.1× MMR at 25° and staged according to Khokha et al. (2002). Fifty embryos were collected and pooled from each parent family at stages 6 [before midblastula transition (MBT)], 8 (during MBT), 11 (gastrula), and 15 (neurula), while 30 embryos per family were pooled at stage 28 (tailbud). Once collected, these samples were immediately stored in 5 ml of Trizol reagent and then delivered directly to the Beijing Genome Institute (BGI), Hong Kong, for RNA extraction and quality control. RNA-seq libraries were prepared at BGI with in-house kits from 6 of the 10 pooled embryo samples and sequenced on an Illumina HiSeq 2000 with single 50-bp reads. All 10 pooled embryo samples also were used to construct WTTS-seq libraries by the Jiang Laboratory, which were sequenced on the Ion PGM Sequencer at Washington State University.
WTTS-seq library preparation
Fragmentation of total RNA and enrichment of poly(A)+ RNA:
The required amount of DNase I–treated total RNA (File S1A) was removed from storage at −80° and diluted to 9 μl with DNase/RNase-free water. Then 1 μl of 10× RNA fragmentation buffer (AM8740, Ambion) was added, and the sample was mixed and incubated for 15 min at 70°. The fragmentation reaction was terminated by adding 1 μl of stop solution (AM8740, Ambion), and the mixture was placed on ice until use. Next, Dynabeads Oligo(dT)25 (75 µg of beads; 61002, Ambion) were washed and prepared according to the manufacturer’s instructions. The fragmented total RNA was heated to 65° for 2 min to disrupt secondary structures, immediately placed on ice, added to the washed Dynabeads, and mixed thoroughly. The mixture then was rotated continuously for 5 min at room temperature to allow binding of the poly(A)+ RNA to the beads. Bead-bound poly(A)+ RNA was eluted with 10 µl of elution buffer (10 mM Tris-HCl, pH 7.5) as directed. The sample was incubated with Dynabeads an additional 5 min and eluted as described earlier to further enrich poly(A)+ RNA. The concentration of fragmented poly(A)+ RNA was measured with a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE).
Incorporation of 5′- and 3′-adaptors into first-strand cDNA with reverse transcription:
Fragmented poly(A)+ RNA was mixed with 1 μl each of 5′-adaptor (switching primer, 100 µM) and 3′-adaptor [containing oligo(dT10), 100 µM] (File S1B). The mixture was heated at 65° for 5 min and chilled on ice for 2 min to disrupt RNA secondary structure and repeated. After that, 4 μl of 5× First-Strand Buffer, 2.5 μl dNTPs (10 mM), 1 μl DTT (0.1 M), 1 μl RNase OUT (100 units/μl), and 1 μl SuperScript III Reverse Transcriptase (200 units/μl) (18080, Invitrogen) were added and the mixture incubated at 40° for 90 min. The reverse transcription reaction was terminated by heating the mixture to 70° for 15 min.
Optimization of second-strand cDNA synthesis by PCR:
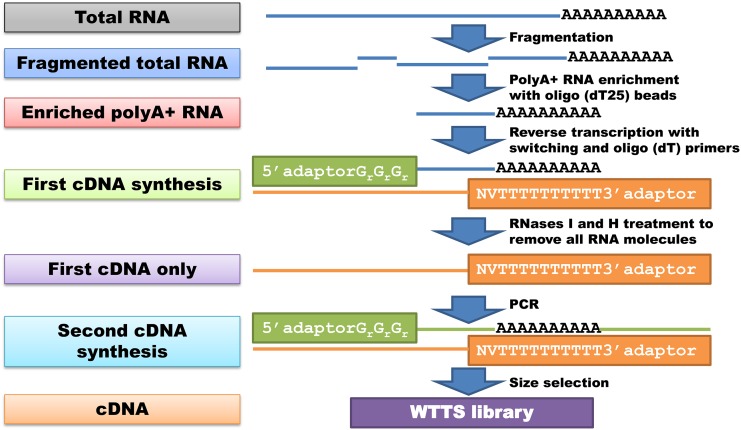
First-strand cDNA was used as a template to synthesize second-strand cDNA. Base PCR conditions were initial denaturation at 98° for 30 sec; PCR cycles of 98° for 10 sec, 50° for 30 sec, and 72° for 30 sec; and final extension at 72° for 10 min. The total PCR volume was 50 μl and contained size-selected cDNA-RNA, DNase/RNase-free water, 5× HF buffer, forward and reverse primers, dNTPs, and Phusion DNA Polymerase (M0530, New England Biolabs). Specific sizes of first- or second-strand cDNA fragments were selected by excision from agarose gels after electrophoresis or with solid-phase reversible immobilization beads (AMPure XP; A63880, Beckman Coulter). Final library quality determined the best preparation method and led to our conclusive procedures for WTTS-seq library construction, as shown in Figure 1.
Figure 1.
Illustration of our finalized WTTS-seq library preparation procedures. Total RNA serves as the starting material, followed by fragmentation and poly(A)+ RNA enrichment. Reverse transcription synthesizes the first-strand cDNA and adds both 5′- and 3′-adaptors into the library. Treatment with RNases I and H removes all RNA molecules and leaves the first-strand cDNA alone for second-strand synthesis by PCR. The library is then size selected and ready for NGS.
Data analysis
Read processing:
We trimmed all T nucleotides or T-rich stretches located at the 5′ end of each WTTS-seq raw read using in-house scripts (File S1C), as described by Shepard et al. (2011), but with modification. After T-trimming, sliding-window quality trimming was performed with Trimmomatic-0.33 (Bolger et al. 2014). Window size was set to 4 bp, and the minimum average quality score was set to 10. Next, only clean reads with sizes ≥16 bp were retained for further analysis (File S1A). While the T-trimming step was not conducted on RNA-seq reads, quality trimming was performed in the same manner.
X. tropicalis genome reference preparation:
X. tropicalis genome (v7.1) and the annotation file Xentr7_2_Stable.gff3 were downloaded from the Xenbase FTP site (ftp://ftp.xenbase.org/pub). In addition, 58,275 mRNA sequences (as of August 27, 2015) were downloaded from the National Center for Biotechnology Information (NCBI) Xenopus (Silurana) tropicalis (Western clawed frog) database. Gene quantification with our WTTS-seq method requires well-annotated 3′-UTR regions; therefore, we combined the current X. tropicalis genome annotation (Xenbase v7.2, Xentr7_2_Stable.gff3) with X. tropicalis mRNA sequences available from NCBI and generated a new annotation file combined.gtf (File S1D). First, the Genomic Mapping and Alignment Program for mRNA and EST Sequences (GMAP, v2014-10-22) was used to map NCBI mRNA sequences to the genome with the parameters –min-trimmed-coverage=0.8–min-identity=0.95 (Wu and Watanabe 2005). The mapping result was transformed into Gene Transfer Format (GTF) with script written in Perl, resulting in a file named mRNA.gtf. Second, the GTF file was combined with the Xentr7_2_Stable.gff3 annotation file, and a new genome annotation file (combined.gtf) was generated using Cuffmerge (Trapnell et al. 2012). Sequences in the combined.gtf file were then annotated with Cuffcompare (Trapnell et al. 2012) based on information from the Xentr7_2_Stable.gff3 and mRNA.gtf files. These data are shown in File S2.
Read mapping:
The CLC Genomics Workbench, v8.0.1 (CLC bio, a QIAGEN Company, Boston, MA), was used to process both WTTS-seq and RNA-seq data for read mapping and gene expression quantification. The workflow is illustrated in File S3A. Reads were first mapped to the X. tropicalis genome assembly (v7.1). The combined.gtf file described earlier then was used as reference for gene annotation with Joint Genome Institute (JGI) Gene_IDs as well as with NCBI gene symbols. After nuclear genome mapping, unmapped reads were aligned to the mRNA sequences described earlier. Finally, all remaining unassigned reads were used as inputs for a de novo assembly. Read mapping parameters were set to 95% similarity and 80% coverage for the first two mapping steps, while 92% similarity and 50% coverage were used as criteria for the de novo assembly step.
Gene annotation and quantification:
Genome and mRNA mapping results were combined to improve both the annotation rate of clean reads and quantification of gene expression. When reads were mapped to the nuclear genome, we calculated “unique gene reads” for each Gene_ID. In order to annotate NCBI mRNA sequences with Gene_ID, they were first mapped to the nuclear genome with GMAP (Wu and Watanabe 2005) and then annotated to sequences in the combined.gtf file using the tmap file generated by Cuffcompare (v2.2.1) (Trapnell et al. 2012) and Perl scripts. A final gene expression value was calculated by combining genome and mRNA gene expression data based on Gene_ID.
Estimation of gene expression means:
Gene/locus expression was either quantified as a raw count of reads expressed or adjusted as reads per million (RPM). The former measurement was used to count the number of genes with evidence of at least one read, whereas the latter served as expression levels for determination of minimum cutoff points. However, counts are intrinsically linked to the library (status) size, which are not exactly comparable, and on the laboratory-observable scale, detecting no sequence (i.e., frequency rate = 0) for a specific gene does not necessarily indicate that its expression level is truly zero. Hence, to bypass this “frequentist dilemma,” the underlying expression mean of each gene was estimated with a Bayesian model setting. Let be a count of reads expressed, say, in the ith sample (or statuses), for , where K is the total number of samples (or statuses). This gene expression can be modeled as a Poisson event (variable)
| (1) |
where the parameter is a positive integer that corresponds to the expectation and variance of the variable . Under the heterogeneity assumption (), each gene expression has its own intrinsic mean. Let , where is the unobservable gene expression mean. Then Equation 1 can be rearranged as
In the Bayesian inference, a gamma prior distribution for is assumed: , where α and β are two hyperparameters with their values given arbitrarily. It can be shown that the posterior distribution of is also gamma:
and the posterior mean and variance are given as
Under the homogeneity assumption (), a common overall mean μ can be inferred instead by pooling the counts of all the samples:
and
The edgeR program (Robinson et al. 2010) was used to determine the number of differentially expressed genes (DEGs) in embryos at different developmental stages. Genes with greater than twofold changes were classified as upregulated genes, while genes with fold changes that were less than −2 were designated as downregulated genes. An online Venn diagram tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to draw Venn diagrams.
Data availability
The raw WTTS-seq and RNA-seq data for this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE74919. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
A brief workflow of WTTS-seq
Our WTTS-seq library preparation involved four major steps: fragmentation, poly(A)+ RNA enrichment, first-strand cDNA synthesis by reverse transcription, and second-strand cDNA synthesis by PCR (Figure 1). We conducted seven trials, but they were not initially designed based on any prior knowledge. In fact, techniques gradually evolved to solve problems with library quality and quantity. Library preparation conditions, read mapping outputs, and transcriptome parameters and coverage for each trial are listed in File S1A. By merging both the X. tropicalis genome assembly (v7.2) and the NCBI mRNA entries (58,275 as of August 27, 2015), the Cuffmerge program (Trapnell et al. 2012) identified a total of 27,836 loci (File S2), which served as a reference of genes/transcripts for all data analyses in this study.
Primer types and troubleshooting
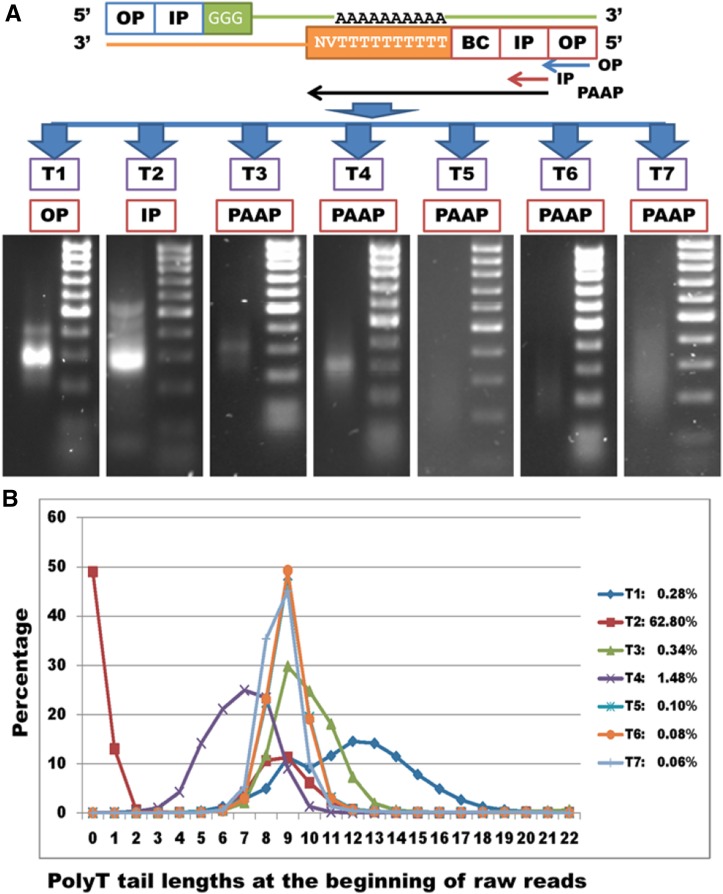
In trial 1, a library with a final concentration of ∼660 ng was constructed by PCR using OPs (Figure 2A and File S1B), but a regular full Ion PGM Sequencer run in which 50–100 pg was loaded generated only 13,309 reads. As such, two more runs were conducted, yielding 31,186,220 raw reads from the entire library (File S1A). A good library preparation should generate an average of 60–80 million reads per regular run. The low yield of reads indicated that most constructs generated with OP lacked an Ion adaptor sequencing region.
Figure 2.
Effect of adaptor design used for synthesis of second-strand cDNA in seven trials (T) on library quality and number of T nucleotides at the beginning of raw reads. (A) Adaptor design included OP (outer primer), IP (ion primer), BC (barcode), and PAAP [poly(A)-anchored primer] regions. T1 used OPs, T2 used IPs, and T3–T7 tested PAAPs in PCR reactions. Gel images are shown for library outputs (from concentrated bands to smooth distributions). Ladder was the ACTGene DNA marker 100 bp, including 100, 200, 300, 400, 500, 600, 700, 800, 900, and 1000 bp, respectively. (B) Poly(T) length distributions at the beginning of raw reads are plotted for T1–T7. Only T1 used an adaptor containing oligo(dT20) rather than oligo(dT10) for synthesis of the first-strand cDNA by reverse transcription. The percentage on the right is the proportion of reads with zero to three T’s in each trial.
The library in trial 2 used IPs (File S1B) with two rounds of PCR including 20 and 35 cycles, respectively, to increase read yield. This library had an adequate number of constructs (Figure 2A), and thus two regular, full Ion PGM Sequencer runs yielded 141,543,418 raw reads (File S1A), which implied that the IPs significantly enhanced sequencing efficiency. However, at least 49% (69,361,559/141,543,418) (Figure 2B) of the raw reads had no T’s at the 5′ ends, indicating that they were spurious products.
The library in trial 3 was constructed with PAAPs (File S1B) and reduced PCR cycles (2 and 20 in rounds 1 and 2, respectively) to minimize amplification of specious products. An initial survey of the data showed that 99.66% of the raw reads started with four or more T’s (Figure 2B), indicating that PAAPs efficiently anchored the poly(A) sites. However, read mapping revealed only 14,905 loci with evidence, which was fewer than the 15,961 and 19,242 loci discovered in trials 1 and 2, respectively (File S1A).
PCR runs and troubleshooting
To address the low transcriptome coverage issue encountered in trial 3, the library in trial 4 was constructed with one round of PCR with 25 cycles and the forward and reverse primer concentrations reduced from 25 to 5 µM and from 25 to 2.5 µM, respectively (File S1A). These modifications led to discovery of 17,289 loci with evidence and 15,544 loci with RPM ≥ 0.2, which was a significant improvement in transcriptome coverage compared to trial 3 (14,905 loci with evidence and 11,118 loci with RPM ≥ 0.2) but notably lower than coverage in trial 2 (19,242 loci with evidence and 16,679 loci with RPM ≥ 0.2) (File S1A).
Therefore, the concentrations of forward and reverse primers were further reduced to 0.8 and 0.4 µM, respectively (based on various tests; data not shown), with 20 PCR cycles in trial 5 (File S1A). These modifications created a library with evenly distributed products (Figure 2A) with significantly improved transcriptome coverage (19,695 loci with evidence and 17,339 loci with RPM ≥ 0.2).
The same strategy then was applied to library construction in trials 6 and 7, which examined the effects of less total RNA and different product size-selection methods on transcriptome coverage (File S1A). In both trials, 2 μg of total RNA was used to prepare libraries. First-strand cDNA products between 200 and 500 bp were selected by excision after gel electrophoresis, and second-strand cDNA products between 200 and 500 bp were selected with SPRI beads in trial 6. First- and second-strand cDNA products between 200 and 500 bp were selected with SPRI beads in trial 7, which resulted in a library with the best transcriptome coverage: 20,690 loci with evidence and 17,740 loci with RPM ≥ 0.2 (File S1A). Therefore, procedures used in trial 7 were adopted as our finalized WTTS-seq library preparation method and used in technical and biological replicate tests.
IPs and noisy/biased reads
In this study, “noisy” reads were defined as reads that were not derived from the 3′-end regions, while “biased” reads were overamplified 3′-end reads. There were 15 and 8 genes in trial 2 that produced the majority of noisy and biased reads, respectively, which accounted for 89.4% of the total mapped reads (File S4). In contrast, reads for the same set of genes accounted for only 0.72% (159,428/21,772,746) of the total mapped reads in trial 7 (File S4). Inclusion and exclusion of these noisy/biased reads in trial 2 significantly influenced transcriptome coverage: 11,073 and 16,679 loci, respectively, with RPM ≥ 0.2 (File S1A).
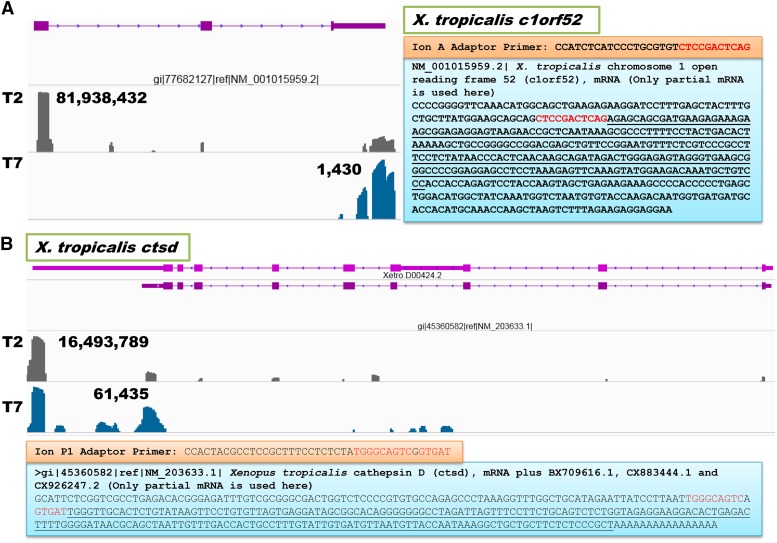
Of the 15 genes in trial 2 that produced noisy reads, 14 are well annotated in the current X. tropicalis genome assembly (v7.1) (File S4). Examination of sequence features revealed that half these genes had 8–12 internal nucleotide sequences identical to the Ion A Adaptor or the sequencing primer (5′-CCA TCT CAT CCC TGC GTG TCT CCG ACT CAG-3′), and the remaining genes contained mismatched sequences. The X. tropicalis c1orf52 gene is shown in Figure 3A to explain how noisy reads were generated. All eight genes with biased reads were created because they had internal nucleotide sequences that were highly similar to the Ion P1 Adaptor (5′-CCA CTA CGC CTC CGC TTT CCT CTC TAT GGG CAG TCG GTG AT-3′). The X. tropicalis ctsd (cathepsin D) gene is illustrated in Figure 3B as an example of a gene that produced biased reads in trial 2.
Figure 3.
Examples of genes that produced overwhelming numbers of noisy (A) and biased (B) reads in trial 2. (A) X. tropicalis c1orf52 gene had the highest number of noisy reads (81,938,432) produced because 11 internal nucleotides (red color) upstream of the amplified products (underlined; see NM_001015959.2) were identical to the 3′ end of the sequencing primer (Ion A Adaptor primer). (B) X. tropicalis ctsd gene had the highest number of biased reads (16,493,789) because it had 15 nucleotides highly similar to the 3′ end of the Ion P1 Adaptor with only one nucleotide mismatch (red color). The amplified product is underlined (see NM_203633.1). Reads from trial 2 (T2) and trial 7 (T7) are not proportionally visualized by the Integrative Genome Viewer (IGV) program.
Library size selection and read length distribution
Product size selection (200–300 bp in trial 1, 300–500 bp in trial 2, 250–450 bp in trials 3 and 4, and 200–500 bp in trials 5–7) was not uniform among the seven trials (File S1A). We observed that library product sizes were not necessarily associated with read sizes compiled by the Ion PGM Sequencer (File S3B). Based on clean reads (≥16 bp in length), we found that size distribution patterns were similar among trials, with the exception of trial 2 (File S3B). As described earlier, trial 2 had a few significant noisy and biased reads, which contributed to a high proportion of large fragment sizes.
RNA-seq and technical replicate tests
Gene abundances in the pooled total RNA sample also were profiled by RNA-seq. After data were normalized with the Bayesian model, we observed that the standard errors (SEs) for WTTS-seq trials 6 and 7 were similar but slightly higher (1.24- and 1.33-fold, respectively) than the SEs observed in the RNA-seq analysis (File S1A). In trials 1 and 5, SE estimates were 2.07- and 2.51-fold greater in WTTS-seq libraries than in RNA-seq libraries. In comparison, SEs in WTTS-seq libraries from trials 2–4 were 16.05-, 13.61-, and 11.48-fold higher, respectively, than SEs observed in RNA-seq, reflecting the noisy and biased data and PCR overamplification issues observed earlier. Despite the differences in transcriptome variations, Spearman’s rank correlations of estimated locus expression means between WTTS-seq trials and the RNA-seq library were well retained (File S3C). In particular, trial 7 had the highest Spearman’s rank correlation (ρ = 0.912) with the RNA-seq data set when all 27,836 loci were involved in the calculation.
Here we focus on a comparison between trial 7 and RNA-seq data sets. Both revealed that at least 2751 of 27,836 reference loci were not expressed in the pooled sample (File S1E). The trial 7 library had 7345 loci expressed at levels of 0 ≤ RPM < 0.2. Of these, 4694 (63.9%), 2454 (33.4%), and 197 (2.7%) were present at levels of 0 ≤ RPM < 0.2, 0.2 ≤ RPM < 10, and 10 ≤ RPM < 250 in the RNA-seq data set, respectively. These results indicated that an RNA-seq library with over 100 million reads improves the likelihood that transcripts with low expression levels are detected. Actually, the same principle also can be applied to WTTS-seq libraries. When we combined reads from all seven trials, the number of loci detected with evidence increased from 20,690 with evidence and 17,740 with RPM ≥ 0.2 (trial 7 alone) (File S1A) to 22,889 with evidence and 21,002 with RPM ≥ 0.2 (sum of all 7 trials) (File S5). However, 452 loci expressed at RPM ≥ 0.2 were detected in the trial 7 WTTS-seq library but were expressed at RPM < 0.2 in the RNA-seq analysis (File S1E).
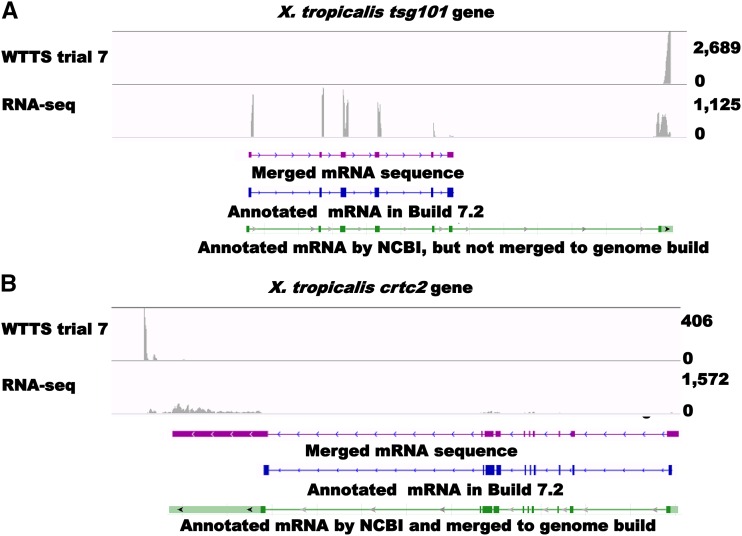
We selected 37 genes (File S6) to determine why we found significant discrepancies in gene expression between libraries created by WTTS-seq and RNA-seq methods. Of these genes, 33 were expressed at 50 ≤ RPM < 250 in RNA-seq but at 0 ≤ RPM < 0.2 in WTTS-seq (trial 7). These differences were caused by problems related to either incomplete genome sequencing/assembly (17 genes) or incomplete transcriptome annotations (16 genes) (File S6). The X. tropicalis tsg101 (Xetro.K02827 and NM_203935.1) and crtc2 (Xetro.K02136) (File S1F) genes are illustrated in Figure 4, A and B, to explain how genes can be detected in an RNA-seq library but missed in WTTS-seq libraries.
Figure 4.
Incomplete genome assembly (A), incomplete gene annotation (B), and missing data for WTTS-seq analysis. (A) Because of incomplete exon sequencing of the X. tropicalis tsg101 gene, the last exon region was not marked in the current genome assembly or in our merged data sets. A search of the NCBI database for the X. tropicalis tsg101 gene revealed a 1637-bp full-length mRNA sequence [NM_203935.1, including 60 bp of poly(A) tail] but only 1041 bp or 66% (94–706 and 1150–1577 bp) of this sequence aligned with the current genome assembly. Because the alignment cutoff criterion (80%) was not met for this gene, the Cuffmerge program did not replace XetroK02827 (681 bp in length) with the longer NCBI sequence. Therefore, the tsg101 gene was detected only by RNA-seq, even though WTTS reads were mapped to that region of the X. tropicalis genome. (B) The X. tropicalis crtc2 gene was not completely annotated and was missing the 3′-UTR sequence. Both RNA-seq and WTTS-seq reads provided clear evidence that this gene sequence can be extended another 920 to 6907 bp in length (File S7). In fact, an expressed sequence tag (EST) entry (CX401749.1) in the NCBI database with a poly(A) signal site (ATTAAA) and a poly(A) tail supports this unannotated 3′-UTR (File S7).
Among 37 genes, four genes were expressed at 50 ≤ RPM < 1100 in WTTS-seq but 0 ≤ RPM < 0.2 in RNA-seq owing to the artifacts produced during preparation of the WTTS-seq libraries (File S6). These artifacts were caused by overlapping loci oriented in opposite directions. The overlapped regions contained poly(T) stretches that were converted to poly(A) stretches after reverse transcription that were subsequently targeted by PAAPs. Different anchored PAAPs amplified those regions for sequencing because the sequencing primer was included at both 5′ and 3′ ends of the amplified products (Figure 5). After strand mapping, the reads were assigned to the overlapped genes without expression rather than to those with expression. These cases occurred only with the overlapped genes in the WTTS-seq library but not in the RNA-seq analysis.
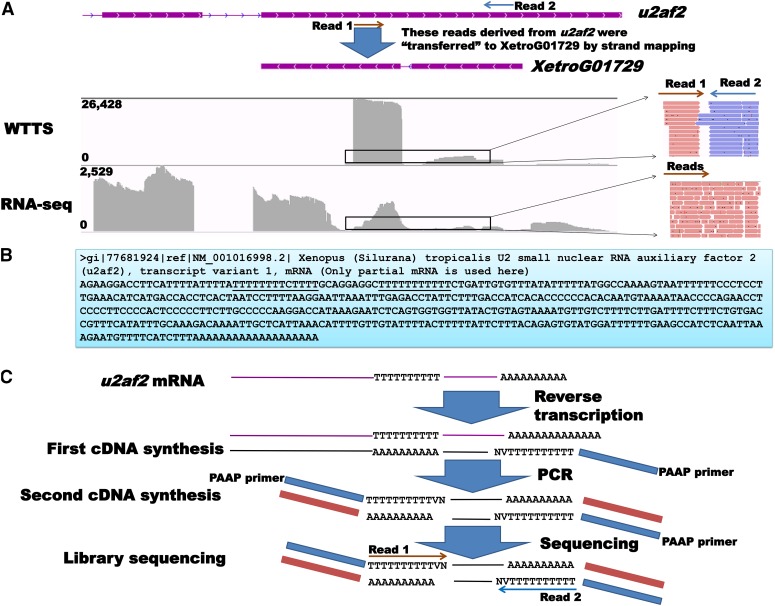
Figure 5.
An example of artifactual reads produced for XetroG01729 because of poly(T) stretches in the u2af2 gene. (A) XetroG01729 and u2af2 overlaps visualized by IGV. The WTTS-seq library produced two clusters of reads (read 1 and read 2) with opposite directions. The RNA-seq library had reads that covered the entire exon. (B) Based on u2af2 mRNA sequences (NM_001016998.2), we postulated that these two clusters of WTTS-seq reads were potentially derived from one gene (u2af2) rather than from each of these overlapped genes (XetroG01729 and u2af2). That is, the read 1 cluster originated from poly(T) stretches, while the read 2 cluster was derived from poly(A) junction sites. However, strand mapping assigned the read 1 cluster (artifacts) to XetroG01729 without evidence from the RNA-seq library. (C) Potential mechanism involved in production of artifactual reads with poly(T) stretches.
We also tested the repeatability of our finalized WTTS-seq protocol by preparing two technical replicates (rep 1 and rep 2) with a total RNA sample derived from a female frog. The replicates had different numbers of mapped reads: 11,403,853 for rep 1 and 22,287,985 for rep 2, representing 19,278 and 20,967 genes with evidence, respectively (File S7). Of these, 16,681 and 17,311 loci in rep 1 and rep 2 were retained, respectively, when RPM ≥ 0.2. Although the number of reads in rep 2 was almost twofold higher than the total reads collected in rep 1, the number of genes with evidence and RPM ≥ 0.2 increased by 1689 and 630, respectively. As such, the replicates had a Spearman’s rank correlation of 0.965, indicating that our method is very reproducible and stable (File S3C).
Biological replicate test
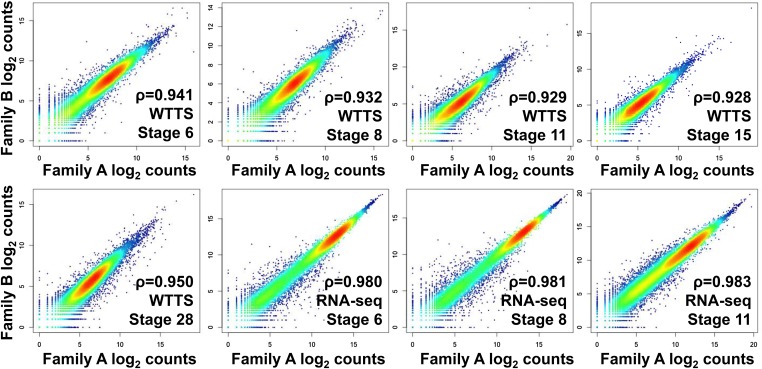
Ten pooled embryo samples of X. tropicalis from two families representing five developmental stages (6, 8, 11, 15, and 28) served as biological replicates in this study. For comparison, we also downloaded publicly available RNA-seq data for five similar developmental stages collected on an Illumina platform (NCBI Gene Expression Omnibus accession no. GSE37452) (Tan et al. 2013). Total raw reads, clean reads, combined mapped reads with annotation, numbers of genes with evidence and with RPM ≥ 0.2, and transcriptome means and SEs are summarized in File S1G. The number of genes detected with evidence ranged from 17,074 to 19,641 in our WTTS-seq libraries, from 20,599 to 23,400 in our RNA-seq libraries, and from 14,283 to 19,307 in Tan’s RNA-seq libraries. These results clearly indicated that the number of genes detected with evidence is highly correlated with the number of reads collected per library. However, when RPM ≥ 0.2 was employed, the number of genes collected decreased to 17,074–19,002, 14,570–17,979, and 12,980–17,708 for these three data sets, respectively. Further, Spearman’s rank correlations between replicates at all five stages ranged from 0.928 to 0.950 for WTTS-seq libraries (Figure 6). In comparison, the Spearman’s rank correlations between replicates of the first three stages were greater than 0.980 for RNA-seq libraries (Figure 6).
Figure 6.
Biological replicate test. Spearman’s rank correlations of WTTS-seq between estimated log2 counts in embryos collected from family A and family B at five developmental stages (6, 8, 11, 15, and 28) and RNA-seq between estimated log2 counts in embryos collected from family A and family B at three developmental stages (6, 8, and 11).
The number of DEGs in embryos at different developmental stages was determined in both WTTS-seq and RNA-seq data sets with the edgeR program (Robinson et al. 2010) (File S3D, A–I). No DEGs were detected between stages 6 and 8, but 1094 and 890 DEGs were found between stages 6 and 11 and between stages 8 and 11, respectively, in the WTTS-seq data sets (Bonferroni adjusted P < 0.05; File S3D, J). The numbers of DEGs between other pairs of stages also were compared and are presented in File S3D, J. Only three DEGs were detected between stages 6 and 8. However, between stages 6 and 11 and between stages 8 and 11, the numbers of DEGs increased dramatically to 4811 and 4662 (Bonferroni adjusted P < 0.05), respectively, in RNA-seq data sets. When pairwise data were combined, the WTTS-seq libraries contained 1158 DEGs, while the RNA-seq libraries had 5204 DEGs among the first three stages (File S3D, K). As such, 111 DEGs were exclusively identified by the former method, while 4157 DEGs were revealed by the latter method alone. Both methods shared a common set of 1047 DEGs, accounting for over 90% of total WTTS-seq DEGs but only approximately 20% of total RNA-seq DEGs (File S3D, K).
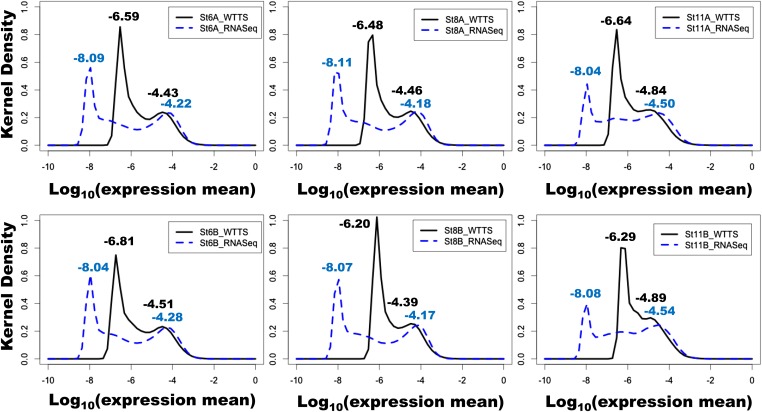
Why RNA-seq detected so many more DEGs than WTTS-seq prompted further investigation. As shown in File S8, the transcriptome means normalized by the Bayesian model were not dramatically different between WTTS-seq and RNA-seq data sets for embryos of two families at stages 6, 8, and 11. In contrast, the distributions of gene expression means were distinct (Figure 7). Kernel density plots clearly indicated that RNA-seq analysis resulted in much wider transcriptome distributions than WTTS-seq analysis. For RNA-seq data sets, the distances between abundantly and rarely expressed gene peaks spanned 3.54–3.93 log10 units (gene expression means). However, the same distances only varied from 1.40 to 2.30 units in WTTS-seq data sets (Figure 7). These results imply that the greater the distance between peaks, the greater is the chance that upregulated or downregulated genes reach statistical significance.
Figure 7.
Comparisons of embryo transcriptome distributions at stages 6, 8, and 11 between WTTS-seq and RNA-seq data sets in two families (A and B). The solid black curves represent gene expression detected by WTTS-seq, and the blue dotted curves represent gene expression detected by RNA-seq.
We also examined potential relationships between transcript length and number of detectable DEGs, particularly associated with RNA-seq analysis. We focused on four sets of genes: 27,836 loci representing the whole X. tropicalis transcriptome, 111 DEGs exclusively identified by WTTS-seq, 4157 DEGs exclusively collected by RNA-seq, and 1047 DEGs commonly discovered by WTTS-seq and RNA-seq (File S3D, B). Kernel density plots against transcript lengths clearly indicate that the length distributions of 111 DEGs detected by WTTS-seq and the whole transcriptome of 27,836 loci were similar, while DEGs detected by RNA-seq tended to be longer transcripts (File S3E). This provided evidence that expression levels of longer transcripts are somehow magnified by RNA-seq analysis. Such magnification even made it possible for RNA-seq to predict 1106 DEGs that were detected by WTTS-seq with data derived from the developmental stages 15 and 28 (File S3D, K).
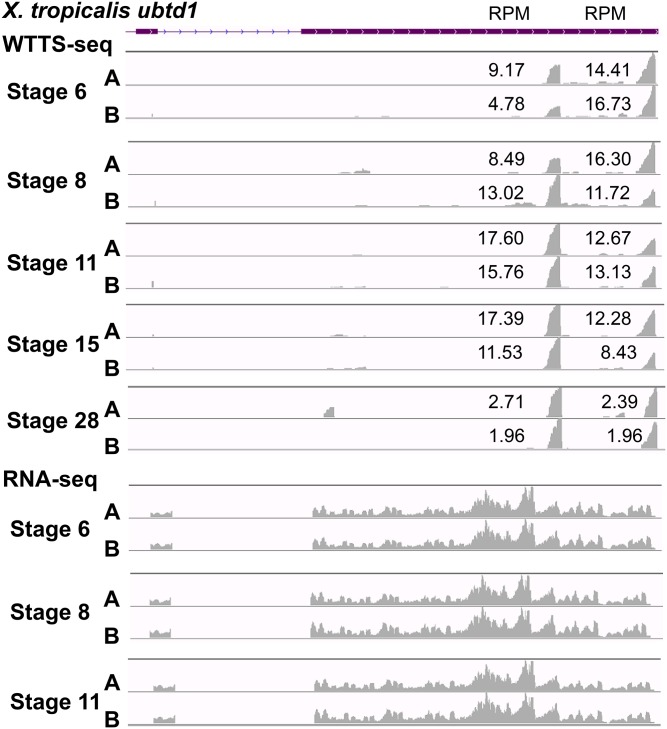
As shown in File S3D, B, WTTS-seq uniquely revealed 111 DEGs in X. tropicalis embryos from stages 6–11. Examination of the data set helped us to classify these DEGs into three major groups based on transcript properties. First, WTTS-seq can detect alternative poly(A) sites with different abundance levels that are specific to developmental stages (see an example in Figure 8). Second, unlike RNA-seq, WTTS-seq was able to detect short-transcript DEGs. Because RNA-seq is generally biased against both 5′ and 3′ ends (Wang et al. 2009), the numbers of reads for short transcripts are most likely underrepresented in an RNA-seq library (see an example in Figure 9). Third, overlapping genes complicate RNA-seq read mapping. Currently, the Illumina RNA-seq platform produces pair-end (PE) reads in one amplicon. Without strand restriction, PE reads derived from overlapped genes cannot be mapped correctly. However, this problem does not exist in WTTS-seq because all reads start from the 3′ end of the transcript (see an example in Figure 10).
Figure 8.
APA patterns during embryo development are revealed by WTTS-seq but not by RNA-seq. Partial genomic region of X. tropicalis ubtd1 gene including the last two exons is shown. WTTS-seq revealed that the distal APA site was dominant at stage 6, but usage switched to the proximal site at stage 11. At stage 28, however, both sites were used equally. Unfortunately, RNA-seq failed to reveal any differences in usage of proximal or distal APA sites among these five stages. The poly(A) site signals were presented proportionally for each family at each stage but disproportionally among different stages.
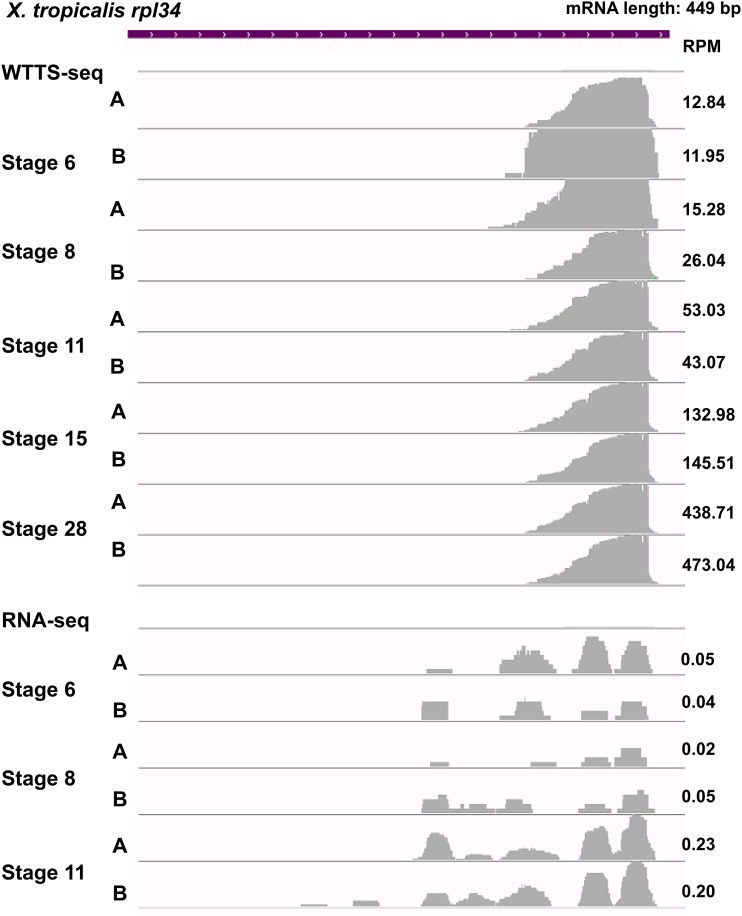
Figure 9.
Expression of short transcript is well detected by WTTS-seq but biased by RNA-seq. The X. tropicalis rpl34 gene has an mRNA sequence of 449 bp in the genome. WTTS-seq revealed that expression of rpl34 increased from stage 6 to stage 28 based on RPM values. However, rpl34 was not fully covered due to biases in RNA-seq libraries (see RPM values in the figure).
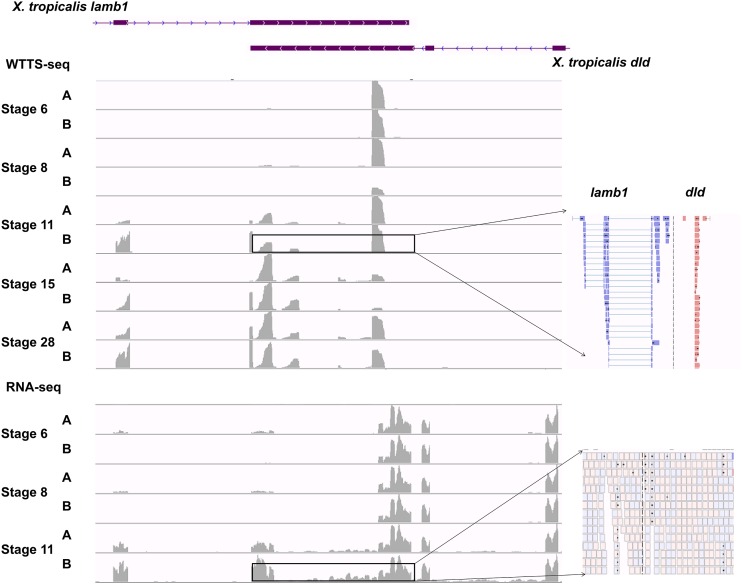
Figure 10.
Expression patterns of overlapping genes are well detected by WTTS-seq but not by RNA-seq. Partial genome regions of X. tropicalis lamb1 and dld genes overlap in opposite directions. The WTTS-seq libraries produced at least two major clusters of reads also with opposite directions in the overlapping region. The blue reads were derived from lamb1 and the red reads from dld. Reads in the RNA-seq libraries covered the overlapping region, but there was no way to allocate them to each gene. Furthermore, RNA-seq mapping quality in the overlapping region was quite low (see reads pointed out with arrows).
Discussion
Basic features of the finalized WTTS-seq method
WTTS-seq enriches both poly(A)+ RNA and poly(A)+ cDNA:
Currently available 3′-end sequencing methods enrich either poly(A)+ RNA or poly(A)+ cDNA during library preparation (Pelechano et al. 2012; Hoque et al. 2013; Wilkening et al. 2013; Mata 2013; Ma et al. 2014; Rallapalli et al. 2014). In our WTTS-seq assay, poly(A)+ fragments were enriched using oligo(dT25) beads (Figure 1). After first-strand cDNA synthesis, we removed single-stranded RNAs and RNA-DNA hybrids with RNases I and H. Second-strand cDNA was made during PCR using the PAAP. As such, our finalized WTTS-seq method involves enrichment of both poly(A)+ RNA and poly(A)+ cDNA (Figure 1). Unlike the poly(A) tail length profiling by sequencing (PAL-seq) method (Subtelny et al. 2014), our WTTS-seq technique was not designed to measure the length of poly(A) tails.
WTTS-seq simultaneously adds full-length 5′- and 3′-adaptors:
Generally speaking, reverse transcription and ligation are the two strategies employed to add 5′- and 3′-adaptors to library constructs. Internal priming issues may be responsible for up to 12% of the noisy data encountered in libraries prepared with the former strategy (Nam et al. 2002). To overcome this problem, a long oligo(dT20) primer was recommended (Shepard et al. 2011); however, others found the long-T stretch caused problems in the sequencing reaction (Wilkening et al. 2013). When libraries are sequenced with the Ion Torrent platform in particular, long homopolymers may increase the deletion error rate (Laehnemann et al. 2016). Ligation could successfully avoid internal priming (Jan et al. 2011; Hoque et al. 2013), but reaction efficiency and time required for the process can be challenging. As shown in Figure 1, we adapted the former strategy in our library preparation but used a short oligo(dT10) primer. We are currently reviewing the data generated in this study to further examine internal priming issues related to our WTTS-seq method.
WTTS-seq directs 3′-end sequencing:
In this study, the WTTS-seq libraries were sequenced with an Ion PGM Sequencer. As shown in File S1B, the reverse transcription adaptor included the Ion Torrent read sequence, a barcode sequence, and dT10VN. This design directed sequencing of the 3′ ends of transcripts because the adaptor anchored poly(A) junction sites. For instance, more than 99.9% of the reads derived from the trial 7 library began with four or more T’s (Figure 2B). Care must be taken to avoid creation of a “low-diversity library” when Illumina platforms are used for sequencing because they require libraries with equal proportions (25%) of A, C, G, and T at each base position (http://www.illumina.com/). Certainly, there are several strategies to ensure that a library will meet the Illumina requirements, such as using a custom primer for sequencing (Shepard et al. 2011; Derti et al. 2012; Yao and Shi 2014) or filling the T-stretch before sequencing (Pelechano et al. 2012; Wilkening et al. 2013). In comparison, the Ion PGM Sequencer has no requirements for library diversity.
Transcriptome analysis: challenges
There are two types of amplification detours:
In this study, we observed that inappropriate primer design resulted in both recessive and dominant “amplification detours” that produced noisy and biased reads. The recessive amplification detour occurred in the trial 1 library that was prepared with OPs, while the dominant detour occurred in the trial 2 library that was constructed with IPs. A noisy read issue also was reported by Ma et al. (2014) and Shepard et al. (2011). Therefore, our finalized method used adaptors that contain an IP region with a buffer zone added to the 5′-adaptor and a barcode (BC) + a PAAP region added to the 3′-adaptor to minimize both noisy and biased reads.
Overamplification of sequencing libraries may reduce transcriptome coverage:
Generally speaking, preparation of NGS libraries involves either exponential or linear amplification (Tang et al. 2009; Gertz et al. 2012; Hashimshony et al. 2012; Bhargava et al. 2013; Hou et al. 2015; Pan et al. 2013). Most 3′-end sequencing methods are based on the former strategy. However, results from trial 3 clearly showed that overamplification by PCR favors a subset of abundantly expressed transcripts in sequencing libraries, resulting in reduced representation of the remaining transcripts and failure to detect genes expressed at low levels (File S1A). To address the issue, we used only one round of PCR in combination with a low concentration of primers to synthesize and amplify second-strand cDNA. The modifications were very successful. Linear amplification of transcripts most likely would yield a library with an extremely low quantity of products, particularly when only 3′ ends are collected as profiling targets. As such, we favor exponential amplification by PCR at the moment.
Incomplete genome sequencing and incomplete gene annotation jeopardize transcriptome analysis:
No doubt the genome assembly of X. tropicalis has improved significantly from v4.1 (Hellsten et al. 2010) to v7.1 (http://www.xenbase.org/entry/). Unfortunately, Gilchrist (2012) estimated that 4610 transcripts did not contain UTR sequences and that 3396 transcripts did not have an annotated 3′-UTR in the latter assembly. Indeed, we had difficulties (Figure 4) assigning poly(A) sites to genes in this study when assembly v7.1 was used as a reference genome. Therefore, we plan to use v9.0 (http://www.xenbase.org/entry/) as the reference genome to improve transcriptome analysis of this and future research.
WTTS-seq vs. RNA-seq
Transcriptome profiles derived from WTTS-seq and RNA-seq are highly correlated:
In this study, the same pooled total RNA sample was used for both WTTS-seq and RNA-seq analyses. The Pearson correlation coefficient between two types of libraries was 0.80 (File S3F), while the Spearman ranking correlation coefficient was 0.91 (File S3C), indicating that the transcriptome profiles derived from WTTS-seq were more highly related to RNA-seq than other 3′-end sequencing methods. For instance, Pearson correlations were 0.7185 between 3′T-fill and RNA-seq and 0.7860 between PAT-seq and RNA-seq (Wilkening et al. 2013; Harrison et al. 2015). The high correlation between WTTS-seq and RNA-seq that we observed strongly indicates that WTTS-seq is a powerful and efficient tool that can be used for profiling transcriptomes and characterizing their diversities or dynamics among different biological samples or at physiologic time points.
RNA-seq detects more DEGs than WTTS-seq, but some of them are probably false positives:
When the 3′T-fill protocol was developed, Wilkening et al. (2013) extracted total RNA from Saccharomyces cerevisiae strain SLS045 cultured with either YPG (1% yeast extract, 2% peptone, and 1% glucose) or YPGal (1% yeast extract, 2% peptone, and 1% galactose). The authors observed that unlike RNA-seq, 3′T-fill captured a greater number of short transcripts, thus preventing size-biased counts of gene expression abundance. Interestingly, the number of DEGs detected between the two culture conditions were 2441 and 3401 for 3′T-fill and RNA-seq, respectively (adjusted P < 0.1). Our data also clearly showed that the WTTS-seq method is capable of capturing shorter transcripts (Figure 9). Moreover, we found that RNA-seq “exaggerates” DEG identification (File S3D, J and K) because it widens the distribution of transcriptomes and thus magnifies the fold changes for more DEGs (Figure 7). The correlation plots between biological replicates (Figure 6) provide further evidence that RNA-seq libraries had wider transcriptome distributions than WTTS-seq. That is, the centralized zones were usually below 10 (in log2 counts) for WTTS-seq libraries but ranged from 10 to 15 (log2 counts) in the RNA-seq libraries.
WTTS-seq, but not RNA-seq, can easily detect alternative polyadenylations:
In this study, we used the X. tropicalis ubtd1gene as an example to demonstrate how the WTTS-seq method can determine APA patterns across diverse developmental stages. Both proximal and distal polyadenylation signals of the gene were used from stages 6–28 during embryo development (Figure 8). While the distal APA site was dominant at stage 6, usage switched to the proximal site at stage 11. At stage 28, however, both sites were used equally. In addition, there was no switching harmony between the two families at stages 8 and 15. Unfortunately, RNA-seq failed to reveal any differences in usage of proximal or distal APA sites (Figure 8) during embryo development at these stages.
WTTS-seq is more cost-effective than RNA-seq:
The finalized procedures for construction of a WTTS-seq library (Figure 1) are not much different from those used for preparation of a library for RNA-seq, so library construction expenses should be similar. Therefore, the method that produces the greatest number of usable reads for accurate analysis can be classified as the most cost-effective. Our RNA-seq libraries produced an average of 143,163,201 reads per library, while Tan’s RNA-seq data averaged 11,685,268 per library (File S1G) (Tan et al. 2013). In comparison, our WTTS-seq libraries had only an average of 7,348,281 reads per library. As such, our RNA-seq runs identified the highest number (21,666 on average) of expressed genes with evidence, which was 3359 and 4236 more genes than those identified by WTTS-seq and Tan’s RNA-seq libraries, respectively. When RPM ≥ 0.2 was employed as a cutoff point, the average number of expressed genes in both RNA-seq libraries was similar (15,731 from our RNA-seq compared to 15,734 from Tan’s RNA-seq). In contrast, our WTTS-seq yielded an average of 17,930 genes expressed at RPM ≥ 0.2 (File S1G). These results imply that RNA-seq libraries with over 140 million reads are not required for gene detection but also demonstrate that 10 million reads per RNA-seq library may not be adequate. Wang et al. (2011) found that RNA-seq with 10 million (75-bp) reads per library detected up to 80% of annotated chicken genes but required at least 30 million (75-bp) reads to sufficiently cover all the genes in the chicken transcriptome. Results presented in Files S1, A and G, suggest that 5 to 10 million reads per WTTS-seq library should be sufficient for transcriptome analysis. Therefore, the cost of sequencing a library prepared by our WTTS-seq method is at least 67% cheaper than RNA-seq.
Both WTTS-seq and RNA-seq should be used together in transcriptome analysis:
Transcriptome analysis often requires verification and validation of DEGs using an independent method such as real-time quantitative reverse transcription PCR (qRT-PCR). After reviewing challenges in assay development, statistical analyses, reagents, and operator variability, however, Bustin (2002) concluded: “In reality, it is very difficult to answer the question of how quantitative, reproducible or informative real-time RT-PCR is” (p. 36). In addition, while there is no question that qRT-PCR can validate gene expression, the time and expense needed to verify all DEGs revealed by a whole transcriptome analysis would be insurmountable. Furthermore, extra challenges exist when qRT-PCR is used to validate alternative transcripts of a given gene revealed by WTTS-seq. In this study, six samples derived from X. tropicalis embryos at stages 6, 8, and 11 were profiled using both WTTS-seq and RNA-seq methods. Although RNA-seq cannot effectively detect APA sites, it can provide solid evidence to show that they are expressed within introns or that they switch from proximal to distal or from distal to proximal sites (see an example in File S3G for the intron case). Furthermore, RNA-seq can show initiation of distal site usage when the proximal site is consistently expressed. In the near future, we will examine cases where the proximal site is newly initiated, while the distal site is consistently expressed. Therefore, our data strongly suggest that both WTTS-seq and RNA-seq be used together to avoid further validation using other methods. The expression dynamics of alternative polyadenylated transcripts within a gene across developmental stages also can serve as mutual validation in addition to RNA-seq confirmation.
Conclusion
After serial adjustment and refinement with primer types and amount, PCR runs and cycles, and RNase types and combinations, we have successfully developed a WTTS-seq method that can be used to profile both gene expression and APA by sequencing the 3′ ends of transcripts. NGS library preparation, in fact, involves many steps, which, in turn, can produce biases, noisy data, and artifacts. Our finalized WTTS-seq assay radically addresses these challenges and serves as a powerful tool for the research community to investigate transcriptomes and reveal poly(A) site usages specific to complex phenotypes, disease stages, or biological processes in humans, animals, and plants.
Acknowledgments
We thank James Coulombe, National Institutes of Health/National Institute of Child Health and Human Development; Jay Shendure, University of Washington; Oliver Hobert, Columbia University; and two anonymous reviewers for their insightful comments and suggestions for improving the manuscript. WTTS-seq and RNA-seq were carried out on the Ion PGM Sequencer at the Genomics Core Laboratory, Washington State University, and the Illumina platforms at the Beijing Genome Institute (BGI), China. BGI, Hong Kong, extracted total RNA from X. tropicalis embryos and measured RNA quality, quantity, purity, and integrity. This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under award no. R21-HD076845 and the National Institute of Food and Agriculture, United States Department of Agriculture, under award no. 2016-67015-24470 to ZJ. The authors declare no conflict of interest.
Footnotes
Communicating editor: J. Shendure
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.188508/-/DC1.
Literature Cited
- Bhargava V., Ko P., Willems E., Mercola M., Subramaniam S., 2013. Quantitative transcriptomics using designed primer-based amplification. Sci. Rep. 3: 1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A., 2002. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29: 23–39. [DOI] [PubMed] [Google Scholar]
- Costa V., Angelini C., De Feis I., Ciccodicola A., 2010. Uncovering the complexity of transcriptomes with RNA-Seq. J. Biomed. Biotechnol. 2010: 853916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A., Garrett-Engele P., Macisaac K. D., Stevens R. C., Sriram S., et al. , 2012. A quantitative atlas of polyadenylation in five mammals. Genome Res. 22: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J., Varley K. E., Davis N. S., Baas B. J., Goryshin I. Y., et al. , 2012. Transposase mediated construction of RNA-seq libraries. Genome Res. 22: 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist M. J., 2012. From expression cloning to gene modeling: the development of Xenopus gene sequence resources. Genesis 50: 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P. F., Powell D. R., Clancy J. L., Preiss T., Boag P. R., et al. , 2015. PAT-seq: a method to study the integration of 3′-UTR dynamics with gene expression in the eukaryotic transcriptome. RNA 21: 1502–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T., Wagner F., Sher N., Yanai I., 2012. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Reports 2: 666–673. [DOI] [PubMed] [Google Scholar]
- Hellsten U., Harland R. M., Gilchrist M. J., Hendrix D., Jurka J., et al. , 2010. The genome of the Western clawed frog Xenopus tropicalis. Science 328: 633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M., Ji Z., Zheng D., Luo W., Li W., et al. , 2013. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat. Methods 10: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Jiang P., Swanson S. A., Elwell A. L., Nguyen B. K., et al. , 2015. A cost-effective RNA sequencing protocol for large-scale gene expression studies. Sci. Rep. 5: 9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan C. H., Friedman R. C., Ruby J. G., Bartel D. P., 2011. Formation, regulation and evolution of Caenorhabditis elegans 3′ UTRs. Nature 469: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Zhou X., Li R., Michal J. J., Zhang S., et al. , 2015. Whole transcriptome analysis with sequencing: methods, challenges and potential solutions. Cell. Mol. Life Sci. 72: 3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha M. K., Chung C., Bustamante E. L., Gaw L. W., Trott K. A., et al. , 2002. Techniques and probes for the study of Xenopus tropicalis development. Dev. Dyn. 225: 499–510. [DOI] [PubMed] [Google Scholar]
- Laehnemann D., Borkhardt A., McHardy A. C., 2016. Denoising DNA deep sequencing data-high-throughput sequencing errors and their correction. Brief. Bioinform. 17: 154–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Pati P. K., Liu M., Li Q. Q., Hunt A. G., 2014. High throughput characterizations of poly(A) site choice in plants. Methods 67: 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J., 2013. Genome-wide mapping of polyadenylation sites in fission yeast reveals widespread alternative polyadenylation. RNA Biol. 10: 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoulkova E., Michalova E., Vojtesek B., Hrstka R., 2012. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 9: 563–576. [DOI] [PubMed] [Google Scholar]
- Morin R., Bainbridge M., Fejes A., Hirst M., Krzywinski M., et al. , 2008. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques 45: 81–94. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U., Waern K., Snyder M., 2010. RNA-Seq: a method for comprehensive transcriptome analysis. Curr. Protoc. Mol. Biol. 11: 11–13. [DOI] [PubMed] [Google Scholar]
- Nam D. K., Lee S., Zhou G., Cao X., Wang C., et al. , 2002. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc. Natl. Acad. Sci. USA 99: 6152–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Durrett R. E., Zhu H., Tanaka Y., Li Y., et al. , 2013. Two methods for full-length RNA sequencing for low quantities of cells and single cells. Proc. Natl. Acad. Sci. USA 110: 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Wilkening S., Jarvelin A. I., Tekkedil M. M., Steinmetz L. M., 2012. Genome-wide polyadenylation site mapping. Methods Enzymol. 513: 271–296. [DOI] [PubMed] [Google Scholar]
- Rallapalli G., Kemen E. M., Robert-Seilaniantz A., Segonzac C., Etherington G. J., et al. , 2014. EXPRSS: an Illumina based high-throughput expression-profiling method to reveal transcriptional dynamics. BMC Genomics 15: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards M., Tan S. P., Chan W. K., Bongso A., 2006. Reverse serial analysis of gene expression (SAGE) characterization of orphan SAGE tags from human embryonic stem cells identifies the presence of novel transcripts and antisense transcription of key pluripotency genes. Stem Cells 24: 1162–1173. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard P. J., Choi E. A., Lu J., Flanagan L. A., Hertel K. J., et al. , 2011. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steijger T., Abril J. F., Engstrom P. G., Kokocinski F., Hubbard T. J., et al. , 2013. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 10: 1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny A. O., Eichhorn S. W., Chen G. R., Sive H., Bartel D. P., 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Lassmann T., Murata M., Carninci P., 2012. 5′ end-centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc. 7: 542–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M. H., Au K. F., Yablonovitch A. L., Wills A. E., Chuang J., et al. , 2013. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 23: 201–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., et al. , 2009. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6: 377–382. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ghaffari N., Johnson C. D., Braga-Neto U. M., Wang H., et al. , 2011. Evaluation of the coverage and depth of transcriptome by RNA-Seq in chickens. BMC Bioinformatics 12(Suppl. 10): S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M., 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm B. T., Landry J. R., 2009. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48: 249–257. [DOI] [PubMed] [Google Scholar]
- Wilkening S., Pelechano V., Jarvelin A. I., Tekkedil M. M., Anders S., et al. , 2013. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 41: e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. D., Watanabe C. K., 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875. [DOI] [PubMed] [Google Scholar]
- Wu X., Liu M., Downie B., Liang C., Ji G., et al. , 2011. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. USA 108: 12533–12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C. G., Shi Y. S., 2014. Global and quantitative profiling of polyadenylated RNAs using PAS-seq. Methods Mol. Biol. 1125: 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw WTTS-seq and RNA-seq data for this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE74919. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.