Abstract
Genes encoding nuclear receptors (NRs) are attractive as candidates for investigating the evolution of gene regulation because they (1) have a direct effect on gene expression and (2) modulate many cellular processes that underlie development. We employed a three-phase investigation linking NR molecular evolution among primates with direct experimental assessment of NR function. Phase 1 was an analysis of NR domain evolution and the results were used to guide the design of phase 2, a codon-model-based survey for alterations of natural selection within the hominids. By using a series of reliability and robustness analyses we selected a single gene, NR2C1, as the best candidate for experimental assessment. We carried out assays to determine whether changes between the ancestral and extant NR2C1s could have impacted stem cell pluripotency (phase 3). We evaluated human, chimpanzee, and ancestral NR2C1 for transcriptional modulation of Oct4 and Nanog (key regulators of pluripotency and cell lineage commitment), promoter activity for Pepck (a proxy for differentiation in numerous cell types), and average size of embryological stem cell colonies (a proxy for the self-renewal capacity of pluripotent cells). Results supported the signal for alteration of natural selection identified in phase 2. We suggest that adaptive evolution of gene regulation has impacted several aspects of pluripotentiality within primates. Our study illustrates that the combination of targeted evolutionary surveys and experimental analysis is an effective strategy for investigating the evolution of gene regulation with respect to developmental phenotypes.
Keywords: ancestral gene reconstruction (AGR), codon models, hominid evolutionary survey, nuclear receptors, NR2C1, testicular receptor 2 (TR2), pluripotentiality
HUMAN evolutionary biology seeks to understand the origins of the defining characteristics of modern humans, such as our large brains, upright posture, obligatory bipedal gait, longevity, and extended juvenile period. While fossil morphology and artifacts recovered from archaeological sites are essential to inferring anatomical structure, function, and behavior in the past (Mcbrearty and Brooks 2000; Alemseged et al. 2006; Tryon et al. 2008; Jungers et al. 2009a,b; Braun et al. 2010; Ward et al. 2011), only through molecular genetic analyses can we make the ultimate connection between phenotype and genotype (Wood 1996; Allman et al. 2010; Boddy et al. 2012; Sherwood and Duka 2012). The eventual goal is to understand to what extent modern structures and functions are determined by different genetic systems and the extent to which the evolution of those systems has played a role in the evolution of the human lineage.
A superfamily of transcription factors called the nuclear receptors (NRs) are attractive candidates for a combined evolutionary and functional investigation of hominids (e.g., the clade that includes modern great apes and their last common ancestors). As transcription factors, NRs control many aspects of development, metabolism, reproduction, and endocrine signaling (Kohn et al. 2012). Their direct involvement in numerous physiological functions has motivated considerable research into their role in the evolution of hormone-mediated traits (Ketterson et al. 2009). Modern humans possess 48 NRs (Robinson-Rechavi et al. 2001) divided into six subfamilies known as NR 1–6 (Laudet 1997; Germain et al. 2006). Regulation of gene expression by NRs is typically induced by either endogenous or exongenous ligands, but there are some NRs, termed orphans, for which the ligand has yet to be identified or may not exist (Enmark and Gustafsson 1996; Benoit et al. 2006). Many functions of the NRs are sufficiently well characterized to permit the design of assays to investigate how amino acid changes could impact gene expression during key phases of primate development.
Because the NR family has been highly conserved for millions of years, an increase in the number of substitutions along a given lineage is often viewed as consistent with an underlying adaptive event. Such interpretations are premature, as amino acid changes also accumulate by neutral processes. In fact, there are only a handful of studies indicating that the dynamics of nonsynonymous evolution among primate NRs differ significantly from neutral expectations (Krasowski et al. 2005; Williamson et al. 2007; Chen et al. 2008). This is in part because sequence divergence among primates is characteristically low and it is challenging to differentiate adaptive substitutions from the background of neutral substitutions in such data. A full understanding of the phenotypic significance of primate NR evolution requires explicitly linking formal evolutionary analyses with experimental approaches focused on the functional impact of amino acid substitutions (Ugalde et al. 2004; Brayer et al. 2011; Kratzer et al. 2014).
Ancestral gene reconstruction (AGR), when combined with synthesis and assessment of the inferred ancestral protein in the laboratory, provides a powerful framework for investigating the contribution of extinct genetic systems to phenotypic change (Thornton 2001; Thornton et al. 2003; Bloch et al. 2015). Indeed, AGR has been used effectively to investigate the evolution of function in a wide variety of different proteins (Chang et al. 2002; Gaucher et al. 2003; Ugalde et al. 2004; Gaucher et al. 2008; Bridgham et al. 2009; Harms and Thornton 2010, 2013; Eick and Thornton 2011; Brayer et al. 2011). It is not feasible, however, to experimentally assess the functional consequences of even a subset of amino acids in each of the 48 primate NRs. Such a comprehensive survey would be prohibitively costly and time consuming. Molecular evolutionary modeling and analysis, on the other hand, is feasible on this scale and can provide information that can be used to identify candidate genes for further investigation (Chen et al. 2008). Here we describe an evolutionary analysis of NR sequence evolution, which identified NR2C1 as a candidate for AGR-based experimental assays. Our functional assays revealed NR2C1 as a potential modulator of pluripotentiality during hominid evolution.
Our investigation was divided into three phases. In the first phase, we employed fixed-effect (FE) codon models to characterize average rates and patterns of primate NR evolution with respect to its primary structural domains. Those results were used to guide the design of the second phase of analysis, which surveyed all 48 NRs for cases of hominid-specific alterations in the intensity of natural selection. We used a combination of branch-site codon models (developed to detect episodes of positive selection) and clade-site models (developed to detect any change in the distribution of selection pressures) to improve our capacity to detect functional divergence within NRs. Combining these models can uncover functionally relevant patterns of evolution that may not be apparent when they are used in isolation (Schott et al. 2014). Prior to interpreting the modeling results, we carried out a suite of reliability and robustness analyses, which led us to exclude a number of genes from further consideration. In the third phase, we selected a single gene (NR2C1) for further experimental investigation. We then inferred an ancestral amino acid sequence for NR2C1, generated an expression vector containing a synthetic open reading frame (ORF) that encodes this protein sequence, and tested in vitro whether evolutionary changes in its amino acid sequence impacted its function. Existing research on NR2C1 suggests it is involved in neural differentiation and can act as an activator of the pluripotency factors Oct4 and Nanog (Shyr et al. 2009). This information was used to design several in vitro assays, the results of which suggest that the function of the ancestral form of NR2C1 differed from both the human and chimpanzee forms. We hypothesize that NR2C1 may regulate aspects of the stem cell pluripotentiality, which could impact anatomical and physiological characteristics that distinguish humans from the other great apes.
Materials and Methods
Origin and processing of DNA sequences
The University of California Santa Cruz Genome Browser (Kent et al. 2002) contains 146 NR sequences for each of the 12 mammalian lineages (Figure 1) included in this study. We downloaded all the NR sequences (date of download: March 25, 2014) and obtained a provisional alignment for each using Multiple Alignment using Fast Fourier Transform (MAFFT) with default settings (Katoh 2002). These data were then filtered in a two-step process. First, all alignments were inspected for in-frame stop codons. These occurred within some alignments corresponding to splice variants at a single locus. We selected the alignment for the largest splice variant that did not contain an in-frame stop. This yielded one alignment per each human NR encoding gene (48 in total). Second, those 48 alignments were visually inspected and, where necessary, manually adjusted to improve the alignment or to exclude poorly aligned regions. Details for the original 146 sequences are provided in Supplemental Material, Table S1 and the final 48 alignments were deposited in the DRYAD data repository (datadryad.org: doi : 10.5061/dryad.bg3g3).
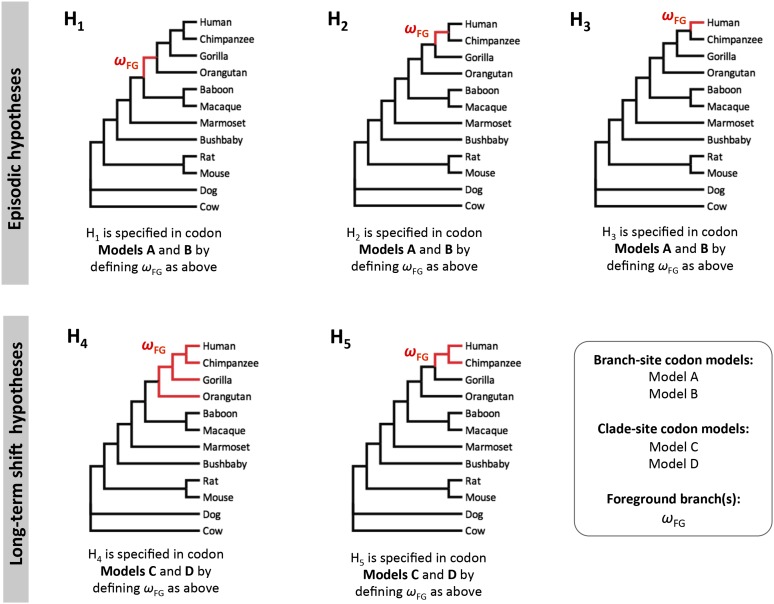
Figure 1.
Phylogenetic relationships of the 12 mammalian lineages included in this study, and the alternative hypotheses for branch-site (LRT-1 and LRT-2) and clade analyses (LRT-3 and LRT-4). The red branches in hypotheses 1–3 are specified as the foreground branch in branch-site models A and B, which are employed to carry out LRT-1 and LRT-2 for episodic evolution. The red clades in hypotheses 4 and 5 are specified as the foreground clades in clade-site models C and D, which are employed to carry out LRT-3 and LRT-4 for long-term shifts in selection pressure.
Assessing rates and patterns of evolution among NR structural domains
We investigated primate NR evolution with respect to four of the five major domains. We mapped each aligned site to (i) the N-terminal domain (NTD), (ii) the DNA binding domain (DBD), (iii) the flexible hinge domain (HD), and (iv) the ligand binding domain (LBD) using the Conserved Domain Database (Marchler-Bauer et al. 2011). The C-terminal domain (CTD) was excluded from this analysis, as it is not present in every NR (Bourguet et al., 2000). This structural information was added to a codon model as a FE partition of an alignment, and the different partitions were allowed to have heterogeneous evolutionary dynamics (Yang and Swanson 2002; Bao et al. 2007). The structural partitions are included in the files deposited in the DRYAD data repository (doi : 10.5061/dryad.bg3g3). We fit the FE models to each alignment and employed likelihood ratio tests (LRTs) to test for heterogeneity among domains in selection intensity (ω), overall rate of evolution via a branch-length scale parameter (c), transition–transversion ratio (κ), and equilibrium codon frequencies ((Bao et al. 2007). The parameter ω serves as a measure of the intensity of natural selection pressure, with purifying, neutral, or positive selection indicated by values of < 1, = 1, or > 1 (Bielawski et al., 2016). An expanded description of the approach is presented in File S1.
Surveying NRs for spatial and temporal variation in selective pressure
Detecting episodic evolution:
We employed branch-site codon models A and B to test for sites experiencing a short-term (episodic) shift in the intensity of natural selection pressure (Yang and Nielsen 2002; Zhang et al. 2005). Models A and B permit the intensity of selection pressure to vary both among sites and among branches, with a model for episodic change obtained by specifying unique selection along a single branch (ωFG) for a proportion of sites within a gene (pFG). We used these models to test three a priori hypotheses about episodic evolution (ωFG in Figure 1: H1–H3) at a fraction of sites, pFG, within primate NRs. Models A and B are mixture models, and they employ additional parameters for sites where the intensity of selection is not episodic. Further details about the other parameters of the ω distribution are provided in File S2. We employed two LRTs for episodic evolution at a fraction of sites. LRT-1 compares constrained model A (ωFG = 1) to unconstrained model A (ωFG > 1). This LRT is intended as a formal test for an episode of positive selection (i.e., a test for ωFG > 1). LRT-2 compares M3 (pFG = 0) to model B (pFG > 0). This LRT is employed to test for any episodic change in selection (i.e., the episode need not involve ωFG > 1). Both LRTs were applied to each of the three episodic hypotheses shown in Figure 1 (H1–H3). Further details about these LRTs, and the involved codon models, are provided in File S2.
Detecting a long-term shift:
We employed clade-site codon models C and D to test for a fraction of sites experiencing a long-term shift in the intensity of selection pressure (Yang and Nielsen 2002; Bielawski and Yang 2004). Models C and D permit the intensity of selection pressure to vary between entire clades, as well as among sites. A selective shift is permitted by including a parameter for the proportion of sites (pSHIFT) that have alternate ω’s (ωFG and ωBG) between subtrees. As mixture models, they have additional parameters for the intensity of selection at other sites (and selection is assumed to be homogenous over the tree at those other sites). Further details about the ω distributions for these models are provided in File S2. We used models C and D to test a priori hypotheses about clade-level selective shifts in primates (Figure 1, H4 and H5) within NR genes. We employed two different LRTs (LRT-3 and LRT-4) to test those hypotheses. LRT-3 compares model M2a-rel to clade-site model C (Weadick and Chang 2012). This LRT-3 is intended as a test for sites having positive selection across an entire clade (i.e., pSHIFT > 0 and ωFG > 1 or ωBG > 1). LRT-4 compares model M3 to clade-site model D (Bielawski and Yang 2004). LRT-4 is employed as a generalized test for any type of long-term shift in the intensity of selection (i.e., a test for pSHIFT > 0, regardless of the values of ωFG and ωBG). Further details about these LRTs, and the involved codon models, are provided in File S2. The codeml program from version 4.4 of the PAML package (Yang 2007) was used for likelihood calculation and parameter estimation under branch-site and clade-site codon models.
Assessing the quality of the signal for variation in selective pressure through a suite of reliability and robustness analyses
We inferred the subgroups of genes related by a given evolutionary event (H1–H5) by controlling the false discovery rate (FDR) within each subgroup according to the method of Storey (2002). For every gene within a subgroup, we carried out a series of additional analyses to assess the reliability of the signal and robustness to model assumptions. First, because incorrect positional homology can impact some inferences (Schneider et al. 2009; Fletcher and Yang 2010), two coauthors independently assessed each alignment. Second, because our analyses were carried out under the species tree, we also estimated gene trees with RAxML (Stamatakis 2014) and reanalyzed the data under that topology. Third, we reanalyzed the data assuming substitution probabilities are proportional to the equilibrium value of the target nucleotide (MG94-style codon model: Muse and Gaut 1994) rather than the equilibrium value of the target codon (GY94-style codon model: Goldman and Yang 1994). Fourth, we used the Genetic Algorithm for Recombination Detection-Multiple Breakpoint (GARD-MBP) method (Kosakovsky Pond et al. 2006) to test for evidence of within-gene recombination events, as these can negatively impact some LRTs (Anisimova et al. 2003). Fifth, we employed the multilayer codon model of Rubinstein et al. (2011) to investigate whether site variability in the baseline rate of DNA/RNA substitution (e.g., due to synonymous rate variability) impacted our initial estimates of ω.
Lastly, we employed nonparametric bootstrapping to quantify the uncertainty in the estimates obtained for the parameters of the ω distribution and to assess it for signs that statistical regularity conditions might not have been met (e.g., bimodal distributions). Bootstrapping is a procedure of “sampling with replacement” that is routinely employed to assess phylogenetic inference, but has only recently been applied to ω-based inference of selection pressure (Bielawski et al. 2016). In this procedure, site patterns within the original multisequence alignment are sampled at random (with replacement) to create many new alignments with different distributions of site patterns. To investigate the properties of the ω distribution, all model parameters (including branch lengths) are reestimated from each bootstrap dataset. We used 100 bootstrap datasets to approximate the maximum likelihood estimate (MLE) distribution for the pi and ωi parameters of a codon model. These data were used to obtain 95% C.I.s for those parameters. We also used these distributions to determine when standard regularity conditions might not have been met. Reliable interpretation of the LRT assumes that they have been met, but discretization of continuous ω distributions within the codon models can make this difficult for some datasets (Bielawski et al. 2016). Violation of regularity conditions can be diagnosed as nonstandard MLE behavior such as strongly bimodal distributions for the pi and ωi parameters.
Ancestral sequence reconstruction
Reconstruction of an ancestral sequence was carried out for both DNA and amino acid states using PAML 4.4 (Yang 2007). The DNA-based reconstructions were based on the general time reversible (GTR) model (Yang 1994), and the amino-acid-based reconstructions were based on the Whelan and Goldman (WAG) replacement matrix (Whelan and Goldman 2001). Additional details, including the reconstructed states and their posterior probabilities, are provided in File S3. These analyses generated almost identical ancestral sequences, with the exception of one amino acid. The ancestral sequence inferred using the amino acid model had higher posterior probabilities for each state, and thus was chosen for gene synthesis. The full ancestral sequences for the human, chimpanzee, and the inferred last common ancestor (LCA) are presented in File S3.
Sequencing of NR2C1 transcripts
We first used the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/) to search for polymorphisms. After confirming that the between-species differences were not associated with polymorphisms within either humans or chimpanzees (File S4), we partially sequenced complementary DNA (cDNA) samples prepared from cortex samples. The samples consisted of frozen postmortem brain tissue from the cortex of 11 nonhuman primate individuals. The fresh frozen brains were obtained from a variety of biomedical and zoological institutions. All animals were housed in accordance with National Institutes of Health, US Department of Agriculture, and the Animal Welfare Act regulations, and overseen by the Institutional Animal Care and Use Committees (IACUCs) of the respective institutions (and approved by George Washington University IACUC protocol A117). The time between death and tissue freezing was, for those cases in which this information was available, never longer than 24 hr. Frozen brain samples were stored at −80° until use. No neurological deficits were detected in any of the individuals included in this study, and all brains appeared normal on routine inspection at necropsy.
RNA was extracted and complementary DNA (cDNA) prepared as per Maynard et al. (2013); briefly, RNA was extracted by Trizol extraction (Invitrogen), and contaminating genomic DNA was removed by DNAse digestion (DNAfree Turbo, Ambion). cDNA was generated by random-hexamer-primed first-strand cDNA synthesis, using ImPromII Reverse Transcriptase (Promega). We PCR amplified four independent amplicons that overlapped the key polymorphisms identified in the AGR analysis. Amplification of each cDNA was performed using standard Taq polymerase (Qiagen), with PCR primers listed in Table S2. All primers were designed to regions where genomic sequencing shows complete conservation of the DNA sequence. Each PCR product was gel purified, quantified, and sent for sequencing using both the forward and reverse primers used for the initial PCR. Sequences were aligned using Sequencher software and each chromatogram was manually inspected to validate each point where sequences were divergent. To further validate that the amino acid substitutions were unique to extant humans and chimpanzees, we sequenced amplicons from additional primate samples from the following species: Pan troglodytes (3), Symphalangus syndactylus (1), Papio anubis (1), Macaca mulatta (3), Macaca nemestrina (2), and Pithecia pithecia (1). Additional details are provided in File S4. As a reference, and for comparison, we also sequenced a human NR2C1 coding sequence from a commercially available plasmid clone containing the NR2C1 ORF in the shuttle vector pFN21A (Promega). Accession nos. for the novel NR2C1 sequences generated in this study are: KT032104–KT032114.
Creation of expression vectors
We generated plasmid expression vectors containing the ORF of the human, chimpanzee, and inferred ancestral sequence reconstruction of NR2C1, by cloning these vectors into the pCINeo4 plasmid, which contains the composite cytomegalovirus (CMV)/chicken β-actin enhancer/promoter from pCAGG, the multicloning site and encephalomyocarditis virus–internal ribosomal entry site (EMCV–IRES) sequence from pIRES2–EGFP (Clontech), and a neomycin coding frame. The human ORF was cloned into the SalI–BamHI sites of this vector by PCR amplifying the ORF of the human NR2C1 clone described above, with a forward primer that adds a SalI site and canonical Kozak’s consensus site (GTCGACCACC) at the 5′ end of the ORF, immediately preceding the ATG start codon, and adding a BamHI site to the 3′ end, in frame with a human influenza hemagglutinin (HA) epitope tag in the vector. Chimpanzee and ancestral sequence expression vectors were generated by creating human codon-optimized sequences corresponding to their respective amino acid sequences, with similar SalI/Kozak’s consensus sites and BamHI sites added to the 5′ and 3′ ends, respectively. These DNA fragments were synthesized (GeneBlocks, Integrated DNA Technologies) and cloned in a similar fashion into the SalI and BamHI sites of pCINeo. A small interfering RNA (siRNA) knockdown vector was generated by cloning a synthetic oligonucleotide into a short-hairpin RNA expression vector derived from pSilencer (Invitrogen). This siRNA cassette was then subcloned into the backbone of an expression vector (pSCH) containing the composite CMV/chicken β-actin (pCAGG) enhancer/promoter fused to a mCherry–IRES–hygromycin cassette. Control plasmids containing the pCINeo plasmid without the NR2C1 insert, and containing the pSCH siRNA expression plasmid with a nonsense-sequence insert, were also generated. All plasmids were fully sequence verified before use. Endotoxin-free DNA preparations were made of each vector for use in embryonic stem (ES) cell experiments (EZNA Plasmid Maxi Kit, Omega Biotek).
ES cell cultures
Mouse ES cells [E14Tg2a, American Type Culture Collection (ATCC)] were cultured on a feeder layer in ES cell media containing Dulbecco’s minimum essential medium (Invitrogen), supplemented with 15% fetal calf serum (HyClone), 0.1 mM 2-mercaptoethanol, penicillin/streptomycin/amphotercin-B (Anti-Anti, Invitrogen), and 100 units/ml leukemia inhibitory factor (LIF) (Enzo). Feeder layers were generated from confluent cultures of STO fibroblast cells (ATCC) by mitotically inactivating the cells with a 2-hr treatment with 10 μg/ml mitomycin C and passaging the cells at a 1:2 ratio onto gelatin-coated tissue culture plastic dishes. Stock ES cells were routinely maintained by trypsinizing and passaging confluent plates of ES cells at a 1:4 subculture ratio every 3–4 days.
Transfection of ES cells and clonal cultures
To transfect ES cells with expression vector plasmids, confluent plates of ES cells were trypsinized, washed twice in PBS, and resuspended in OptiMem (Invitrogen). The 10 × 106 ES cells were resuspended in 800 µl aliquots, and placed in a 4-mm cuvette, along with 5 µg each of two separate plasmids (a pCINeo plasmid expressing a variant of NR2C1 or control, and a pSCH plasmid expressing the siRNA knockdown or the nonsense control). A total of five plasmid conditions were assayed and annotated as follows: nonsense (nonsense siRNA plasmid + control pCINeo plasmid), knockdown (knockdown siRNA plasmid + control pCINeo plasmid), human (knockdown siRNA plasmid + pCINeo containing hNR2C1 ORF), chimpanzee (knockdown siRNA plasmid + pCINeo containing cNR2C1 ORF), and ancestral (knockdown siRNA plasmid + pCINeo containing the inferred aNR2C1 ORF). The cells were electroporated with two pulses of 500 V for 1 ms using a square-pulse electroporator (BTX). Electroporated cells were plated onto feeder layers with LIF containing media in six-well plates. After 24 hr, media were changed to fresh media containing both 300 µg/ml hygromycin and 300 µg/ml G418, to select for ES cells cotransfected with both expression vectors. After 18–21 days, single selected clones were apparent on the ES cell plates; we selected single clones, dissociated and counted the total cell numbers, and plated 2000 cells/well into individual wells of six-well plates, in the same selection media (ES media with hygromycin and G418). These clonal ES cell cultures were repropagated every 18–21 days by selecting 10 clones (chosen from a set of contiguous clones in a single field to minimize any potential selection bias), and then this set of cells was again dissociated in trypsin, counted on a hemocytometer, and replated at 2000 cells/well to facilitate analysis over multiple generations.
Quantitative real-time PCR analysis of expression
Trizol-extracted RNA was obtained from pools of ES cells grown during the first passage from six independent clonal lines for each of the five plasmid conditions. cDNA was generated as described above and quantitative real-time PCR (qPCR) was performed to assess expression levels for mouse NR2C1, Oct4, and Nanog using the primers listed in Table S2. Expression was normalized using Gapdh primers as an internal control (Table S2). Reactions were assembled using an EpMotion 5070 liquid handling system (Eppendorf) that combines forward and reverse gene-specific primers, with 7.5 µl of SsoFast EvaGreen Supermix (Bio-Rad) in a 14-µl reaction. qPCR analysis was performed using a CFX-384 Real-Time PCR Detection System.
Promoter activation assay
To assess the transcriptional activity of the NR2C1 variants, we created a luciferase reporter vector using a synthetically generated 465-bp fragment of the mouse Pepck promoter (Roesler et al. 1989). HEK-293 cells (ATCC) were plated in 24-well plates in standard cell culture media (DMEM with 10% FCS and antibiotics) at 50% confluency and cotransfected with a reporter plasmid and an NR2C1 variant using Effectene reagent (Qiagen) according to the manufacturer’s protocol. After overnight incubation, luciferase activity was measured using a modified luciferase reporter assay system (Gold Biotechnology), and normalized to cells cotransfected with the reporter and an empty expression vector. Each cotransfection was repeated six times, with each assayed in duplicate.
Data availability
DRYAD data repository for alignments and structural partitions is as follows: doi : 10.5061/dryad.bg3g3. GenBank accession nos. for the novel NR2C1 sequences generated in this study are as follows: KT032104–KT032114. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results and Discussion
Phase 1: Characterizing NR domain evolution
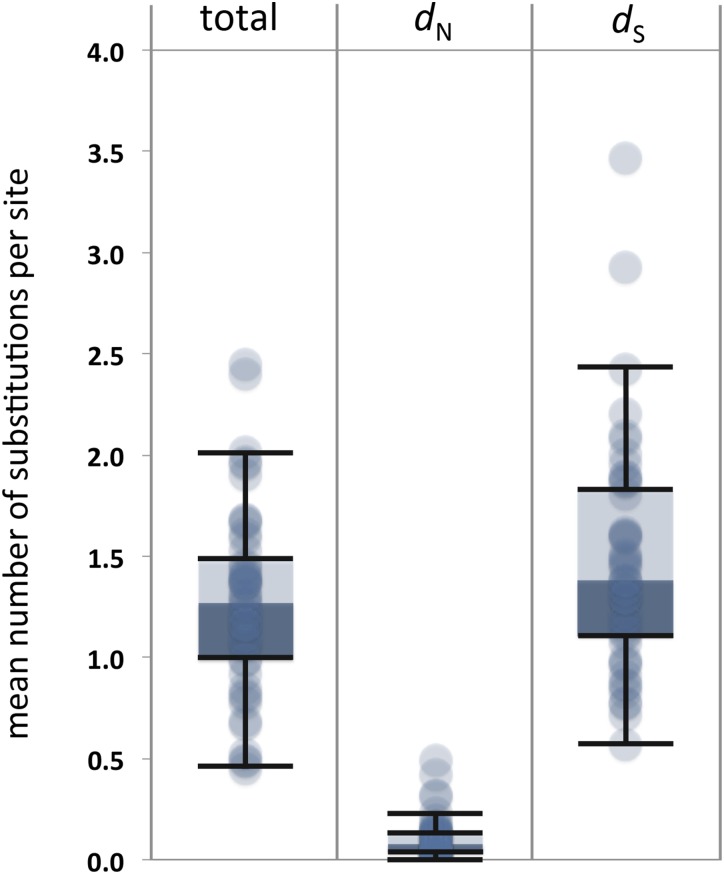
We assessed sequence evolution in the NR family as represented by a set of 12 mammalian lineages (Figure 1). The data were composed of 48 NR alignments. Only moderate levels of sequence divergence were observed. The median tree length (across the 48 NR genes) was just 1.27 substitutions per codon, and partitioning total sequence divergence into synonymous and nonsynonymous components revealed that the majority of change was synonymous (Figure 2). The low levels of sequence divergence, and the sparseness of nonsynonymous change in particular, raise the possibility that statistical regularity conditions might not be relied upon to justify inference under the complex models employed in phase 2. For this reason, we added a novel use of the bootstrap to assess inference under those models in phase 2 (Bielawski et al. 2016).
Figure 2.
Distribution of maximum likelihood estimates of total tree length (t), the nonsynonymous rate (dN), and the synonymous rate (dS) among the 48 NR gene sequences. Sequence divergence statistics are derived from maximum likelihood estimates of parameter values under codon model M0. The scale for the total tree length is the number of substitutions per codon site, which consists of three nucleotides. The scale for both the nonsynonymous and synonymous rates is the number of substitutions per single nucleotide site of the relevant type. The distributions are summarized as box and whisker plots. Data points outside the top and bottom fences are interpreted as outliers.
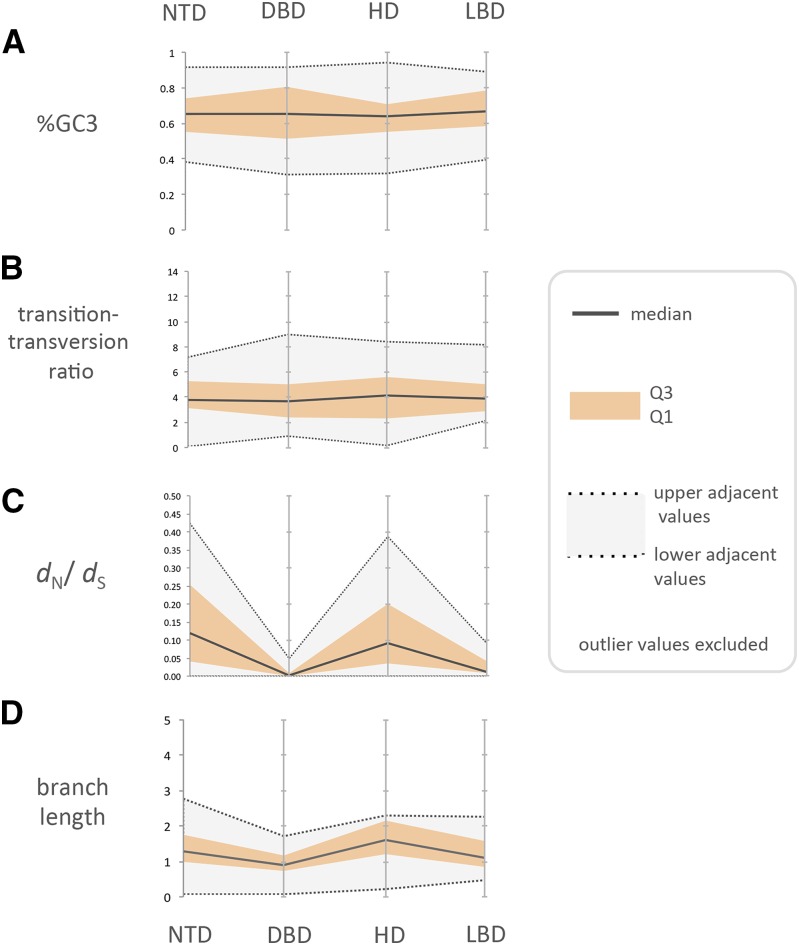
Most NRs share a modular structure (Figure 3), and we investigated the extent to which evolutionary constraints were associated with those domain structures. We did not include the CTD, due to its absence from some NRs. In two cases (NR0B1 and NR0B2) we were unable to assign sites to structural domains, so they were excluded from this analysis. We fit a fully heterogeneous codon model (denoted FE1 in Bao et al. 2007) to the remaining 46 genes in order to estimate the amount of among-domain variability in (i) the dN/dS ratio, ω; (ii) the transition-to-transversion rate ratio, κ; (iii) the branch length scale factor, c; and (iv) codon bias, πj’s. In this study we are primarily interested in ω as a measure of selection intensity. Aggregation of domain-specific parameter values indicated that NR domain evolution is relatively homogenous for both the transition-to-transversion rate ratio and the codon usage bias (Figure 4, A and B). In contrast, the intensity of selection (ω) differed substantially among domains; the DBD and the LBD were highly constrained by purifying selection and exhibited almost no nonsynonymous changes (Figure 4C). The DBD was the most conserved, with median ω of just 0.004 (Q1 = 0.001; Q3 = 0.009), and the LBD had median ω of 0.014 (Q1 = 0.007; Q3 = 0.041). Although still dominated by purifying selection, the NTD (median ω = 0.11) and the HD (median ω = 0.07) tended to exhibit more nonsynonymous changes. Moreover, for some genes, these two domains exhibited considerable divergence among primates (NTD Q3: ω = 0.25; HD Q3: ω = 0.20). Interestingly, we also observed signs of among-domain variability in scale parameters (c’s), suggesting the possibility of different synonymous rates among domains, or a history of recombination (Figure 4D). Application of LRTs to directly assess model complexity for each gene (Bao et al. 2007) yielded similar results (File S1). In response to the observed variation in c’s, we added two more robustness analyses to phase 2. Specifically, we applied (i) a genetic algorithm to screen alignments for evidence of recombination (Kosakovsky Pond et al. 2006) and (ii) a multilayer (DNA/RNA and protein) codon model to assess among-site variability in the baseline rate of DNA/RNA substitution (Rubinstein et al. 2011).
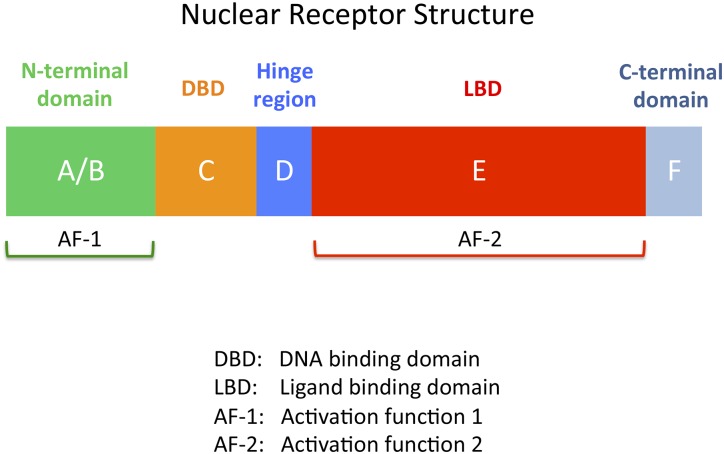
Figure 3.
The structural domains of nuclear receptors (NRs). Sites within NRs are typically classified into six regions (called regions A–F), which correspond to five structural domains. The A/B region contains activation function 1, and corresponds to the N-terminal domain (NTD) of the protein. Region C is the highly conserved DNA binding domain (DBD). Region D is a flexible hinge region that connects the DBD to region E. Region E corresponds to the ligand binding domain (LBD) and contains activation function 2. The C-terminal domain (CTD, region F) varies in length between the different NRs and is nonexistent in some NRs.
Figure 4.
Summary of the variability in the evolutionary process among domains of NR proteins as inferred from codon models where the domain structure was specified as a fixed effect in the model. Data are from 46 NR genes partitioned into four categories: N-terminal domain (NTD); DNA binding domain (DBD); flexible hinge domain (HD); and the ligand binding domain (LBD). Codon model FE1 was used to obtain partition-specific estimates (A) transition–transversion ratio (κ); (B) equilibrium codon frequencies, which are approximated by %GC3 in this figure; (C) selection intensity (ω = dN/dS); and (D) the overall rate of evolution (via a branch-length scale parameter, c). Each figure summarizes the distribution of the estimates over 46 NR genes. The dotted lines on the boundary of the gray band are equivalent to the top and bottom fences in a box and whisker plot [i.e., Q3 + (1.5 × interquartile range; IQR) and Q1 − (1.5 × IQR)], and are referred to as the upper and lower adjacent values. Values beyond the upper and lower adjacent values in the plot are considered outliers and for clarity are not displayed in these plots.
Phase 2: Survey for hominid-specific alterations in the intensity of natural selection
Survey design:
The objectives of this phase of the research were to (i) identify NRs that could have played a role in hominid evolution and (ii) choose the best single candidate for AGR-based laboratory investigation. We designed a survey to detect signal for sites having either an episodic change in selection pressure (Figure 1, H1−H3), or a long-term shift in selection pressure (Figure 1, H4 and H5), associated with the evolution of hominids. The criterion for an episodic change was a significant result under LRT-1 or LRT-2, and the criterion for a long-term shift was a significant result under LRT-3 or LRT-4. Because we tested each hypothesis in 48 different alignments, the single test significance level will not provide adequate control over the probability of making one or more type I errors. We used the FDR criterion of Storey (2002) to identify the subgroup of genes related by a given evolutionary scenario (Figure 1, H1–H5). The alignments for every gene within a subgroup were visually inspected and verified a second time by a different coauthor. The subgroups were then screened according to a suite of reliability and robustness analyses.
Assessing the signal for linage specific episodes of adaptive evolution:
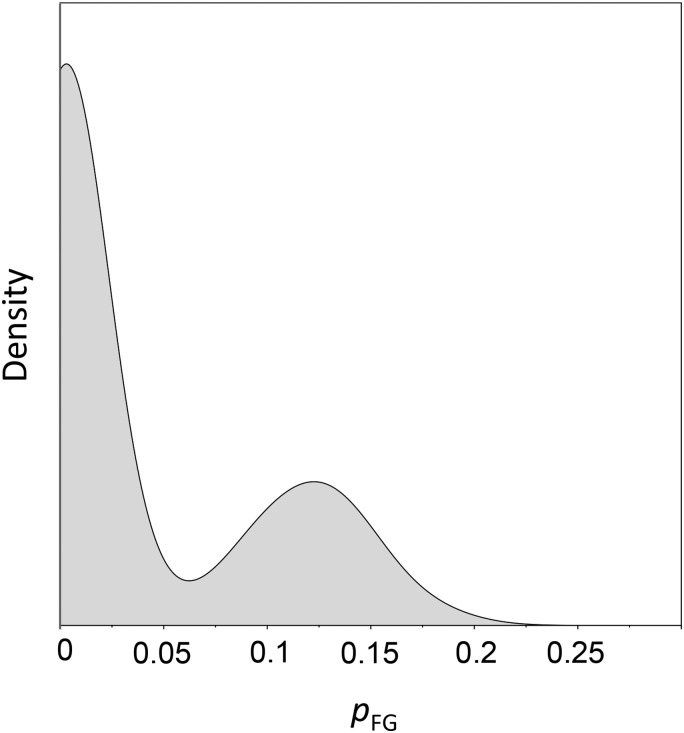
We formally tested each NR for sites having an episode of altered selection pressure at the origin of the great apes (H1), the origin of the human–chimpanzee clade (H2), or along the human lineage (H3) (Figure 1). LRT-1 and LRT-2 revealed little evidence of episodic evolution (Table 1). One gene (NR0B2) was significant for a change in selection pressure at the origin of the great apes; no genes were significant at the origin of human–chimpanzee clade; and four genes (NR1D1, PPARG, NR2C1, and PGR) were significant for a change along the human lineage. However, only one (NR1D1) was significant after controlling for false discoveries (using a q-value of <0.05 as the selection criterion). We used nonparametric bootstrapping to quantify uncertainty and check for instabilities in the MLEs of parameter values for NR1D1. The estimated distribution for the fraction of sites under episodic evolution (pFG) was strongly bimodal, with considerable density at zero (Figure 5), indicating a clear departure from the expected limiting properties of this MLE. This instability indicates that the conditions necessary for reliable MLEs and LRTs (see Self and Liang 1987) cannot be assumed for NR1D1. Thus, we found no reliable evidence for episodic adaptive evolution in primate NRs.
Table 1. Significant LRTs for a subset of sites having experienced an episodic alteration of selection pressure.
| Gene | 2δl | P | q | pi | ωi |
|---|---|---|---|---|---|
| H1: Great Ape | |||||
| LRT-1: model A (ωFG = 1) vs. model A (ωFG > 1) | |||||
| None | N.A. | N.A. | N.A. | N.A. | N.A. |
| LRT-2: M3(k = 2) vs. model B | |||||
| NR0B2 | 7.07 | 0.0292 | 0.7012 | PFG(a+b) = (0.35 + 0.65) | ωFG = 0.0 |
| p1 = 0.00 | ω1 = 0.59 | ||||
| p0 = 0.00 | ω0 = 0.05 | ||||
| H2: Human–Chimpanzee | |||||
| LRT-1: model A (ωFG = 1) vs. model A (ωFG > 1) | |||||
| None | N.A. | N.A. | N.A. | N.A. | N.A. |
| LRT-2: M3(k = 2) vs. model B | |||||
| None | N.A. | N.A. | N.A. | N.A. | N.A. |
| H3: Human | |||||
| LRT-1: model A (ωFG = 1) vs. model A (ωFG > 1) | |||||
| NR1D1 | 10.27 | 0.0013 | 0.0650 | PFG(a+b) = 0.002 | ωFG = 99 |
| p1 = 0.05 | [ω1 = 1] | ||||
| p0 = 0.95 | ω0 = 0.04 | ||||
| LRT-2: M3 (k = 2) vs. model B | |||||
| NR1D1 | 15.25 | 0.0005 | 0.0234 | PFG(a+b) = 0.01 | ωFG = 99 |
| P1 = 0.05 | ω1 = 0.86 | ||||
| p0 = 0.94 | ω0 = 0.04 | ||||
| PPARG | 9.71 | 0.0077 | 0.1867 | PFG(a+b) = 0.01 | ωFG = 35 |
| p1 = 0.11 | ω1 = 0.46 | ||||
| p0 = 0.88 | ω0 = 0.0 | ||||
| NR2C1 | 8.70 | 0.0129 | 0.2063 | PFG(a+b) = (0.24 + 0.76) | ωFG = 99 |
| p1 = 0.00 | ω1 = 0.33 | ||||
| p0 = 0.00 | ω0 = 0.02 | ||||
| PGR | 7.67 | 0.0215 | 0.2586 | PFG(a+b) = (0.12+0.05) | ωFG = 6.23 |
| p1 = 0.24 | ω1 = 0.58 | ||||
| p0 = 0.59 | ω0 = 0.05 | ||||
Genes having a q-value of <0.05 are shown in boldface type. The foreground (FG) branches are fully specified for each hypothesis in Figure 2. The null model for all LRTs assumes homogenous selection pressure for all branches (ωBG = ωFG). LRT-1 has d.f. = 1. LRT-2 has d.f. = 2. The q-value is the expected proportion of false discoveries expected if the single-test P-value is used as the boundary to control the FDR. The parameter PFG(a+b) represents the proportion of sites subject to a change in selection intensity.
Figure 5.
The sampling distribution for the proportion of sites evolving under episodic evolution (pFG) for NR1D1. The sampling distribution was estimated from 100 replications of the nonparametric bootstrap under branch-site model B. Because the estimated distribution for pFG is strongly bimodal, with considerable density at zero, the conditions necessary for reliable MLEs and LRTs cannot be assumed for NR1D1.
Assessing the signal for long-term shifts in the intensity of selection pressure in the great ape clade (H4):
In contrast to our analyses of episodic evolution, clade-site models (Bielawski and Yang 2004) uncovered evidence for a nontrivial subgroup of genes having a shift in selection pressure in some lineages. We employed both LRT-3 and LRT-4 to test for a shift at the origin of the great apes (Figure 1, H4). Nine genes were significant for LRT-3, with five remaining significant after controlling the FDR (Table 2: q-value <0.05). Ten genes were significant for LRT-4, but only four had a q-value of <0.05 (Table 2). The same four genes (RORA, RARG, ESRRB, and NR2C1) also had a q-value of <0.05 for LRT-3, suggesting strong signal for a long-term shift within this subgroup. The fraction of sites subject to a shift in selection pressure (pSHIFT in Table 2) ranged from ∼11% (RORA) to 90% (RARG) under model D. The estimate of pSHIFT for RARG is extreme, suggesting the possibility of a large estimation error, or even a failure to meet statistical regularity conditions.
Table 2. Significant LRTs for a subset of sites having experienced a shift in the intensity of selection pressure at the origin of the great apes (H4).
| Gene | 2δl | P | q | pSHIFT | ωBG | ωFG |
|---|---|---|---|---|---|---|
| LRT-3: M2a-rel vs. model C | ||||||
| RORA | 47.02 | 7.04e-12 | 1.55E-10 | 0.11 | 0.26 | 3.40 |
| RARG | 15.88 | 6.76e-05 | 0.0007 | 0.90 | 0.00 | 0.08 |
| ESRRB | 11.89 | 0.0006 | 0.0041 | 0.27 | 0.11 | 0.44 |
| NR2C1 | 9.52 | 0.0020 | 0.0112 | 0.38 | 0.23 | 0.91 |
| ESRRA | 6.66 | 0.0098 | 0.0433 | 0.75 | 0.00 | 0.08 |
| PGR | 5.67 | 0.0173 | 0.0633 | 0.71 | 0.05 | 0.18 |
| NR5A2 | 4.59 | 0.0321 | 0.0946 | 0.30 | 0.21 | 0.00 |
| NR4A3 | 4.47 | 0.0344 | 0.0946 | 0.19 | 0.39 | 0.10 |
| THRA | 4.02 | 0.0449 | 0.1098 | 0.025 | 1.91 | 0.00 |
| LRT-4: M3(k = 2) vs. model D(k = 2) | ||||||
| RORA | 47.02 | 7.04e-12 | 1.69e-10 | 0.11 | 0.26 | 3.40 |
| RARG | 15.88 | 6.76e-05 | 0.0008 | 0.90 | 0.00 | 0.08 |
| ESRRB | 11.89 | 0.0006 | 0.0045 | 0.28 | 0.11 | 0.44 |
| NR2C1 | 9.59 | 0.0020 | 0.0117 | 0.81 | 0.03 | 0.28 |
| ESRRA | 5.86 | 0.0155 | 0.0690 | 0.75 | 0.00 | 0.08 |
| PGR | 5.67 | 0.0173 | 0.0690 | 0.71 | 0.05 | 0.18 |
| NR5A2 | 4.57 | 0.0325 | 0.0967 | 0.28 | 0.23 | 0.00 |
| NR4A3 | 4.47 | 0.0344 | 0.0967 | 0.19 | 0.39 | 0.10 |
| NR1H2 | 4.38 | 0.0363 | 0.0967 | 0.89 | 0.02 | 0.00 |
| NR3C2 | 4.19 | 0.0407 | 0.0976 | 0.88 | 0.04 | 0.12 |
Genes having a q-value of <0.05 are shown in boldface type. The foreground (FG) branches are fully specified for each hypothesis in Figure 1. The null model for all LRTs assumes homogenous selection pressure for all branches (ωBG = ωFG). LRT-4 and LRT-5 have d.f. = 1. The q-value is the expected proportion of false discoveries expected if the single-test P-value is used as the boundary to control the FDR. pSHIFT is the fraction of sites subject to a shift in selection pressure.
Further interpretation of the MLEs should always be treated with caution without additional assessment. Therefore, we carried out a series of robustness analyses for the five genes that had a q-value of <0.05 under H4 (RORA, RARG, ESRRB, NR2C1, and ESRRA). None of these genes exhibited evidence of recombination, as inferred under the GARD-MBP method (File S5). All five genes had highly consistent MLEs and LRTs under the MG94-style codon models, as well as under gene trees that differed from the organismal tree (Table 3). Note that gene trees differed from the assumed organismal tree for all genes except NR2C1.
Table 3. Robustness of inferences about long-term shifts in the intensity of selection pressure at the origin of the great apes (H4).
| Model C | Model D | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Analysis | pSHIFT | ωBG | ωFG | LRT | pSHIFT | ωBG | ωFG | LRT |
| RORA | Original | 0.11 | 0.26 | 3.40 | P = 7.0e-12 | 0.11 | 0.26 | 3.40 | P = 7.0e-12 |
| MG94 | 0.11 | 0.30 | 3.90 | P = 8.0e-12 | 0.11 | 0.30 | 3.90 | P = 8.0e-12 | |
| Gene tree | 0.12 | 0.24 | 3.03 | P = 1.9e-11 | 0.12 | 0.24 | 3.03 | P = 1.9e-11 | |
| Bootstrap | [0.04–0.17] | [0.04–0.42] | [0.83–9.53] | N.A. | [0.06–0.17] | [0.11–0.43] | [1.44–8.32] | N.A. | |
| RARG | Original | 0.90 | 0 | 0.08 | P = 6.8e-05 | 0.90 | 0 | 0.08 | P = 6.8e-05 |
| MG94 | 0.89 | 0 | 0.10 | P = 7.1e-05 | 0.89 | 0 | 0.10 | P = 7.1e-05 | |
| Gene tree | 0.90 | 0 | 0.08 | P = 6.8e-05 | 0.90 | 0 | 0.08 | P = 6.8e-05 | |
| Bootstrap | [0.02–0.96]a | [0–6e-3] | [0.02–3.06] | N.A. | [0.77–0.99] | [0–8e-3] | [0.03–0.15] | N.A. | |
| ESRRB | Original | 0.27 | 0.11 | 0.44 | P = 0.0006 | 0.27 | 0.11 | 0.44 | P = 0.0006 |
| MG94 | 0.27 | 0.14 | 0.59 | P = 0.0007 | 0.27 | 0.15 | 0.59 | P = 0.0007 | |
| Gene tree | 0.27 | 0.11 | 0.44 | P = 0.0006 | 0.27 | 0.11 | 0.44 | P = 0.0006 | |
| Bootstrap | [0.14–0.75] | [0–0.16] | [0.07–1.00] | N.A. | [0.14–0.45] | [0.04–0.17] | [0.19–1.07] | N.A. | |
| NR2C1 | Original | 0.38 | 0.23 | 0.91 | P = 0.002 | 0.81 | 0.03 | 0.28 | P = 0.002 |
| MG94 | 0.37 | 0.25 | 1.02 | P = 0.001 | 0.36 | 0.27 | 1.0 | P = 0.002 | |
| Gene tree | Match | Match | Match | N.A. | Match | Match | Match | N.A. | |
| Bootstrap | [0.05–0.71] | [5e-3–0.36] | [0.21–2.86] | N.A. | [0.65–0.94] | [0–0.05] | [0–0.70] | N.A. | |
| ESRRA | Original | 0.75 | 0 | 0.08 | P = 0.010 | 0.75 | 0 | 0.08 | P = 0.015 |
| MG94 | 0.69 | 0 | 0.09 | P = 0.010 | 0.69 | 0 | 0.09 | P = 0.014 | |
| Gene tree | 0.75 | 0 | 0.08 | P = 0.008 | 0.75 | 0 | 0.08 | P = 0.013 | |
| Bootstrap | [0.24–1] | [0–0.03] | [0–0.21] | N.A. | [0.162–1]a | [0–0.02] | [0–0.35] | N.A. | |
Robustness and bootstrapping analyses were carried out for genes having at least one q-value of <0.05. For analyses under the gene tree, a match designation indicates that the gene tree was identical to the organismal tree.
Bimodal distribution.
Next we used bootstrapping to assess the MLE distribution of the five genes. The MLE distributions for three genes (RORA, ESRRB, and NR2C1) were unimodal and bell-shaped (allowing for boundaries such as the prohibition of negative frequencies). Although there is uncertainty associated with the point estimates for the model parameters (Table 3), the bootstrap distributions upheld the signal for sites subject to divergent selection pressure in all three genes (pSHIFT > 0), as well as the signal for positive selection within RORA (Table 3). In contrast, bootstrapping revealed that the MLE distribution for pSHIFT was bimodal for RARG and ESRRA. This indicates a clear departure from the asymptotic distributional properties expected when regularity conditions are satisfied for this MLE. We used multiple analyses started from different initial parameter values to confirm that these bimodal distributions were not due to suboptimal peaks in likelihood. This indicates that conditions necessary for reliable MLEs and LRTs cannot be assumed for RARG and ESRRA, despite having q-values of <0.05. We consider the signal for a shift in selection pressure at the origin of the great apes to be reliable for RORA, ESRRB, and NR2C1 only.
Assessing the signal for long-term shifts in the intensity of selection pressure in the human–chimpanzee clade (H5):
We also employed LRT-3 and LRT-4 to test for a shift in selection pressure at the origin of the human–chimpanzee clade (Figure 1, H5). As displayed in Table 4, eight genes were significant for LRT-3 and LRT-4. After controlling for false discovery (q-value <0.05), five genes remained under LRT-3 (NR2C1, NR1D1, PGR, NR2E3, and ESRRB) and three genes remained under LRT-4 (NR2C1, NR2E3, and PGR), suggesting a strong signal for a long-term shift within the latter subgroup. Interestingly, estimates of the ω distribution under clade-site models indicate a substantial fraction of sites (22–36%) evolving under positive selection since the origin of the human–chimpanzee clade.
Table 4. Significant LRTs for a subset of sites having experienced a shift in the intensity of selection pressure at the origin of the human–chimpanzee clade (H5).
| Gene | 2δl | p | q | pSHIFT | ωBG | ωFG |
|---|---|---|---|---|---|---|
| LRT-3: M2a-rel vs. model C | ||||||
| NR2C1 | 12.57 | 0.0004 | 0.0126 | 0.36 | 0.25 | 2.06 |
| NR1D1 | 8.70 | 0.0032 | 0.0384 | 0.002 | 0.00 | 126.89 |
| PGR | 8.47 | 0.0036 | 0.0384 | 0.30 | 0.54 | 2.57 |
| NR2E3 | 7.90 | 0.0049 | 0.0395 | 0.22 | 0.41 | 3.39 |
| ESRRB | 7.23 | 0.0071 | 0.0458 | 0.27 | 0.12 | 0.61 |
| PPARG | 5.01 | 0.0251 | 0.1341 | 0.12 | 0.43 | 1.86 |
| ESR2 | 4.07 | 0.0437 | 0.1820 | 0.74 | 0.04 | 0.40 |
| NR3C2 | 4.00 | 0.0455 | 0.1820 | 0.12 | 0.52 | 2.71 |
| LRT-4: M3(k = 2) vs. model D(k = 2) | ||||||
| NR2C1 | 12.55 | 0.0004 | 0.0111 | 0.35 | 0.26 | 2.13 |
| NR2E3 | 9.38 | 0.0022 | 0.0306 | 0.22 | 0.41 | 3.39 |
| PGR | 8.47 | 0.0036 | 0.0336 | 0.30 | 0.54 | 2.57 |
| ESRRB | 7.23 | 0.0071 | 0.0500 | 0.27 | 0.12 | 0.61 |
| PPARG | 5.01 | 0.0251 | 0.1408 | 0.12 | 0.43 | 1.86 |
| ESR2 | 4.12 | 0.0424 | 0.1629 | 0.79 | 0.05 | 0.40 |
| NR3C2 | 4.03 | 0.0448 | 0.1629 | 0.11 | 0.54 | 2.72 |
| NR1D1 | 3.96 | 0.0465 | 0.1629 | 0.06 | 0.81 | 4.88 |
Genes having a q-value of <0.05 are shown in boldface type. The foreground (FG) branches are fully specified for each hypothesis in Figure 1. The null model for all LRTs assumes homogenous selection pressure for all branches (ωBG = ωFG). LRT-4 and LRT-5 have d.f. = 1. The q-value is the expected proportion of false discoveries expected if the single-test P-value is used as the boundary to control the FDR. pSHIFT is the fraction of sites subject to a shift in selection pressure.
We performed additional assessments of robustness and bootstrapped the MLEs for the five candidate genes. Two genes (NR1DI and ESRRB) were excluded because bootstrapping revealed bimodal MLE distributions suggestive of parameter estimate instabilities (Table 5). NR2E3 was excluded due to significant signal for recombination obtained by using the GARD method (File S5). Inference of selection was generally robust to assumptions about among-site variation in the baseline DNA/RNA substitution rate (File S6). Although PPARG did not have a q-value of <0.05, it is noteworthy that ω’s were very sensitive to assumptions about the baseline DNA/RNA substitution rate in this gene (File S6). Such sensitivity highlights the importance of assessing this in all candidate genes.
Table 5. Robustness of inferences about long-term shifts in the intensity of selection pressure at the origin of the human–chimpanzee clade (H5).
| Model C | Model D | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Analysis | pSHIFT | ωBG | ωFG | LRT | pSHIFT | ωBG | ωFG | LRT |
| NR2C1 | Original | 0.36 | 0.25 | 2.06 | P = 0.0004 | 0.35 | 0.26 | 2.13 | P = 0.0004 |
| MG94 | 0.37 | 0.25 | 1.02 | P = 0.0015 | 0.36 | 0.27 | 1.04 | P = 0.0017 | |
| Gene tree | Match | Match | Match | N.A. | Match | Match | Match | N.A. | |
| Bootstrap | [0.17–0.57] | [0.11–0.42] | [0.31–8.32] | N.A. | [0.18–0.47] | [0.18–0.41] | [0.4–9.41] | N.A. | |
| NR1D1 | Original | 0.002 | 0 | 126.89 | P = 0.0032 | 0.06 | 0.81 | 4.88 | P = 0.0465 |
| MG94 | 0.002 | 0 | 162.68 | P = 0.0057 | 0.07 | 0.74 | 4.19 | P = 0.0511 | |
| Gene tree | Match | Match | Match | N.A. | Match | Match | Match | N.A. | |
| Bootstrap | [0–8e-3] | [0–2.12]a | [8.84–99] | N.A. | [0.02–0.13] | [0.38–1.43] | [0–22.31] | N.A. | |
| PGR | Original | 0.30 | 0.54 | 2.57 | P = 0.0036 | 0.30 | 0.54 | 2.57 | P = 0.0036 |
| MG94 | 0.30 | 0.59 | 2.70 | P = 0.0049 | 0.30 | 0.60 | 2.68 | P = 0.0049 | |
| Gene tree | 0.31 | 0.54 | 2.54 | P = 0.0011 | 0.31 | 0.54 | 2.54 | P = 0.0038 | |
| Bootstrap | [0.18–0.5] | [0.22–0.7] | [0.65–5.11] | N.A. | [0.22–0.42] | [0.41–0.68] | [0.77–8.37] | N.A. | |
| NR2E3 | Original | 0.22 | 0.41 | 3.39 | P = 0.0049 | 0.22 | 0.41 | 3.39 | P = 0.0022 |
| MG94 | 0.21 | 0.45 | 3.97 | P = 0.0051 | 0.21 | 0.45 | 3.94 | P = 0.0015 | |
| Gene tree | *** | *** | *** | N.A. | *** | *** | *** | N.A. | |
| Bootstrap | [0.13–0.36] | [0.26–0.71] | [0.36–99] | N.A. | [0.11–0.34] | [0.27–0.69] | [0.6–13.98] | N.A. | |
| ESRRB | Original | 0.27 | 0.12 | 0.61 | P = 0.0071 | 0.27 | 0.12 | 0.61 | P = 0.0071 |
| MG94 | 0.26 | 0.17 | 0.80 | P = 0.0081 | 0.26 | 0.17 | 0.80 | P = 0.0081 | |
| Gene tree | 0.27 | 0.12 | 0.61 | P = 0.0076 | 0.27 | 0.13 | 0.61 | P = 0.0076 | |
| Bootstrap | [0.15–0.84]a | [0–0.21]a | [0.07–1.92] | N.A. | [0.1–0.40] | [0.06–0.22] | [0.07–1.84] | N.A. | |
Robustness and bootstrapping analyses were carried out for genes having at least one q-value of <0.05. For analyses under the gene tree, a match designation indicates that the gene tree was identical to the organismal tree. Analyses that were impossible because the gene tree topology prevented specification of H5 are indicated by ***.
Bimodal distribution.
The remaining two candidates (NR2C1 and PGR) had unimodal and approximately bell-shaped MLE distributions. Parameter estimates suggesting positive selection in the human–chimpanzee clade were consistent across alternative codon models (models C and D), modeling frameworks (GY94 vs. MG94), and tree topologies (gene vs. organism) (Table 5). Bootstrapping corroborated the signal for a fraction of sites subject to divergent selection pressure among the human–chimpanzee clade, as the estimates of pSHIFT were always > 0. The bootstrap also indicated substantial density for ωFG > 1; however, it was not 100% of the distribution (models C/D: 90/94% ωFG > 1 for NR2C1; 95/98% ωFG > 1 for PGR). Because any change in the distribution of ω, even when ω < 1, is an indicator of functional divergence at the molecular level (Forsberg and Christiansen 2003; Bielawski and Yang 2004) we conclude that the signal for functional divergence is strong in NR2C1 and PGR under H5.
Identifying the top candidates for AGR-based experimental assays and selection of NR2C1 for further investigation:
When the H4 and H5 results are combined, we are left with a set of four genes (ESRRB, NR2C1, PGR, and RORA) whose evolution is statistically associated with the origin of the great ape and human–chimpanzee clades. AGR-based characterization of each gene is not within the scope of this study, nor is each one an equally promising candidate. Hence, we carried out a subjective ranking and chose NR2C1 as our top candidate for further AGR-based experimental assays. Our ranking was based on our assessment of the signal for a change in selection pressure, the extent to which the biological role of each gene has been characterized, and the capacity to carry out in vitro assays for functional effects of amino acid substitutions. The three genes that we did not select for further investigation (ESRRB, PGR, and RORA) remain, nonetheless, very strong candidates for future work. Further details about our ranking of those genes are provided in File S7.
Our top candidate, NR2C1, belongs to a subtype of NRs known as orphan receptors for which the endogenous ligand (if any) has yet to be identified (Lee and Chang 1995). Originally named testicular receptor 2 (TR2) because it was first isolated from human testis and prostate (Chang and Kokontis 1988; Anderson et al. 2012), its expression in ES cells and in pluripotent cell culture lines indicates it plays a role in early embryonic development (Hu et al. 2002). It is one of a handful of genes implicated in the regulation of the pluripotentiality of stem cell populations in the embryo and in neural stem cells in particular (Lee and Chang 1995; Hu et al. 2002; Lee et al. 2002; Shyr et al. 2009). In addition, NR2C1 has been shown to regulate the expression of Oct4 and Nanog, two transcription factors essential for maintaining the pluripotentiality of embryonic stem cells (Pikarsky et al. 1994; Niwa et al. 2000; Boiani 2002). Examination of the level of NR2C1 expression in induced pluripotent stem cells (iPSCs) generated from human melanocytes, fibroblasts, and hepatocytes from previously published experiments (Gene Expression Omnibus (GEO) accession no. GDS3867; Ohi et al. 2011) similarly shows that NR2C1 expression is increased in each iPSC line relative to its nonpluripotent parental cell type. Furthermore, expression of NR2C1 in chimpanzee iPSCs is increased relative to their undifferentiated parental cell lines (Gallego Romero et al. 2015). Given its role in maintaining the pluripotentiality of stem cells and in neural differentiation, and a signal for positive selection within the human–chimpanzee clade (H5), we hypothesized that NR2C1 has played a role in the evolution of a developmental system(s) relevant to the anatomical and physiological characteristics that distinguish humans and chimpanzees from the other great apes. In the next phase of this study, we take the first steps to evaluate this hypothesis by investigating if amino acid substitutions within NR2C1 since the LCA of humans and chimpanzees have affected its capacity to regulate pluripotentiality.
Phase 3: Inference of an ancestral NR2C1 sequence, synthesis of an ancestral protein, and in vitro assessment of its gene regulatory effects
Experimental design:
In this phase of the investigation, we set out to determine whether the NR2C1 amino acid substitutions unique to either modern humans (hNR2C1), chimpanzees (cNR2C1), or the inferred LCA of humans and chimpanzees–bonobos (aNR2C1) alter the ability of the gene to maintain pluripotentiality. More specifically, by heterologously expressing these proteins in embryonic stem cells, we tested whether these sequences differ (i) in their ability to maintain pluripotentiality, (ii) in their relative ability to regulate transcripts associated with pluripotentiality (i.e., Oct4 and Nanog), and (iii) whether they differentially regulate the transcription of a promoter element associated with NR-mediated signaling (Lucas et al. 1991) and differentiation (Zimmer and Magnuson 1990). Any change in the efficiency of NR2C1 as a transcriptional activator of pluripotentiality or of differentiation-related genes, or any change in its ability to maintain pluripotentiality in a stem cell pool, could be evolutionarily significant. Such change, even if relatively small, could underlie substantial changes in overall anatomical and physiological characteristics.
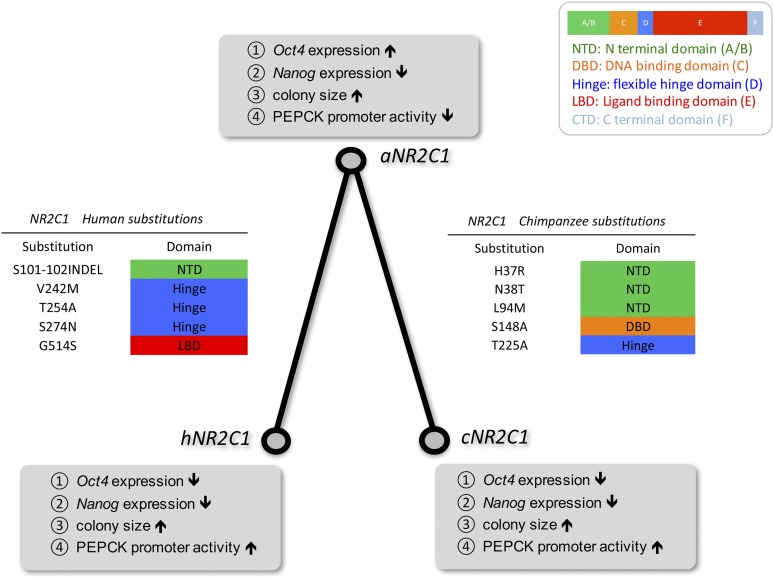
We used maximum likelihood estimates of model parameters to infer the LCA of humans and chimpanzees–bonobos (aNR2C1) according to empirical Bayes posterior probabilities. The ancestral amino acid sequence, along with additional details about the methods of ancestral state reconstruction, are provided in File S3. We then sequenced NR2C1 transcripts from multiple primates and used these in combination with available sequence data to exclude the possibility that some of the inferred amino acid substitutions might represent polymorphisms or errors in the prediction of the messenger RNA (mRNA) sequences (File S4). Figure 6 gives the amino acid substitutions along the human and chimpanzee lineages implied by the inferred ancestral sequence. The cDNA for the ancestral (aNR2C1) and extant (hNR2C1 and cNR2C1) genes was synthesized, and all subsequent assays reflect the amino acid states corresponding to those sequences.
Figure 6.
Amino acid substitutions along the human and chimpanzee lineages and their impact on four different measures of molecular phenotype. Lineage-specific amino acid substitutions were inferred by comparing the ancestral state with the highest posterior probability to the extant state at a site in human and chimpanzee NR2C1. The location of each substitution is given with respect to alignment position and structural domain. The cDNA corresponding to the ancestral (aNR2C1) and extant (hNR2C1 and cNR2C1) gene sequences was synthesized, and experimental assays were employed to measure four different aspects of molecular phenotype (Oct4 expression, Nanog expression, colony size, and Pepck promoter activity). The results of those assays are summarized within the gray boxes at the three nodes of the two-taxon phylogeny. The up and down arrows indicate the relative effect of the amino acid substitutions on the molecular phenotype between the ancestral and extant variants.
Evidence for functional divergence in the transcriptional regulation of pluripotentiality genes by NR2C1 gene variants:
Pluripotency is the transient attribute of single embryonic cells to generate all cell lineages of the developing and adult organism. During embryonic development, pluripotent stem cell populations form organs and tissues by differentiating in a stepwise fashion. One way of assessing this process is by following the expression of molecular markers of differentiation, such as Oct4 and Nanog. Oct4 and Nanog are important regulators of pluripotency and cell lineage commitment and are essential for establishing and maintaining pluripotentiality (Nichols et al. 1998; Mitsui et al. 2003). As mouse NR2C1 (mNR2C1) has been shown to regulate the expression of Oct4 and Nanog in mouse ES cells (Pikarsky et al. 1994; Niwa et al. 2000; Boiani 2002), we investigated whether the amino acid divergence between the NR2C1s of human, chimpanzee, and the inferred LCA might alter its ability to regulate these key pluripotentiality genes.
We expressed hNR2C1, cNR2C1, and aNR2C1 in mouse ES cells under the control of a strong promoter along with a siRNA construct that knocks down endogenous expression of mNR2C1. Cotransfected cells were selected by addition of antibiotics (see Materials and Methods). The results of the knockdown/overexpression experiments were compared to both a negative knockdown control (which received the siRNA construct and an empty heterologous expression vector) and a nonsense positive control [which received a control (nonsense) siRNA and an empty heterologous expression vector]. Our goal was to knock down NR2C1 and then replace it with the same gene from different species. Mouse ES cell lines are very well-defined pluripotent lines, and the three tested variants are all effectively the same evolutionary distance from mouse. Using the mouse line also allowed us to knock down the endogenous NR2C1 expression using siRNAs, and then reexpress the test NR2C1 constructs without any interference from the siRNAs. The human codon optimization of the synthetically generated chimpanzee and ancestral NR2C1 constructs was performed to ensure that all constructs were expressed with equivalent efficiency and equally divergent from the endogenous mouse sequence that was being silenced from the mouse knockdown constructs.
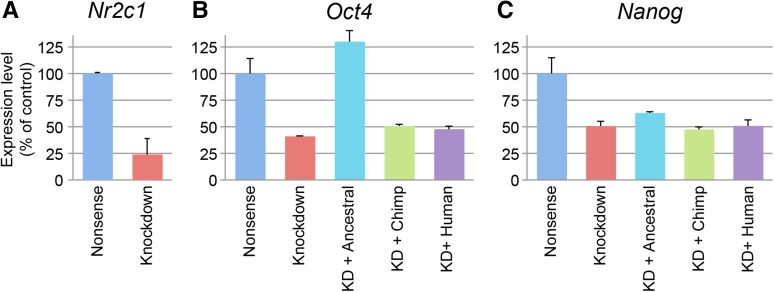
The results of our analysis demonstrate a 75% decrease in NR2C1 expression in the knockdown cohort, thus confirming NR2C1 was knocked down by our siRNA construct (Figure 7A). We also confirmed an earlier report (Shyr et al. 2009) that Oct4 and Nanog had significantly reduced expression in the knockdown cohort (41 and 51% reductions relative to the positive control). We then evaluated whether overexpression of the NR2C1 variants could rescue the reduced expression of Oct4 and Nanog. We found that the ancestral gene variant rescued Oct4, even reaching levels above that observed in control-transfected cells, with a 130% increase in expression compared to the negative control. We did not observe any rescue by the chimpanzee or human variants (Figure 7B), as the expression levels of Oct4 remained at the levels observed in the knockdown cohort. None of the NR2C1 variants rescued Nanog (Figure 7C).
Figure 7.
Effect of NR2C1 knockdown on pluripotentiality-related gene expression in ES cells. (A) ES cells transfected with a mNR2C1 knockdown vector show significantly reduced NR2C1 expression relative to ES cells receiving a nonsense control vector (24%, P < 0.01). (B) Cotransfection of aNR2C1 with knockdown vector rescues expression of Oct4 (130%), but hNR2C1 and cNR2C1 do not. (C) Normal expression of Nanog is not rescued by cotransfection with any of the NR2C1 plasmids.
These results suggest two key points. First, the lack of rescue by any primate variant suggests functional divergence in either the transcription factor or in the mouse Nanog promoter. Such divergence is plausible, given that primates and rodents have been evolving independently for ∼75 million years; however, it is by no means inevitable (Wasserman et al. 2000; Liu et al. 2004). Second, the fact that only the ancestral variant rescues Oct4 expression indicates that the ancestral and extant forms of NR2C1 differ in their ability to bind to, and/or transcriptionally activate the mouse Oct4 promoter. Our results raise the possibility that the inferred ancestral form of NR2C1 may have an activity level closer in function to the mouse variant than to its human and chimpanzee counterparts. Thus, our findings imply that considerable functional divergence in NR2C1 occurred sometime within the last 8–13 million years of hominid evolution and that amino acid substitutions at no more than five sites was sufficient to alter the transcriptional properties of this key pluripotentiality gene. Additional work is required to determine whether the divergence of NR2C1 was a unique evolutionary event or if the regulatory capacity of NR2C1 is relatively plastic and has been more widely modified within primates.
In vitro assays of pluripotentiality suggest differential activity of hNR2C1, cNR2C1, and aNR2C1:
Given that we demonstrated functional divergence between the ancestral and extant forms of primate NR2C1 to regulate Oct4, we predicted that the knockdown of endogenous mNR2C1 and its replacement with its primate variants could result in a detectable change in the pluripotentiality of transfected ES cells. As ES cells must remain pluripotent in order to undergo self-renewal under their standard culture conditions, the ability of ES cells to form new clonal colonies is a reliable proxy of their pluripotentiality (Chambers and Smith 2004). We exploited this property of ES cells to assess and compare the phenotype of pluripotentiality in the extant and ancestral forms of the three primate NR2C1 gene variants.
We selected 12 clonal ES cell cultures for each of the above plasmid combinations and plated these cells at a limiting dilution to assess their ability to generate new pluripotent clones. We assayed the number of clones formed for each replicate and then assessed the average size of the clonal colonies by harvesting, dissociating, and counting 10 random clones for each well. As it is likely that factors influencing the pluripotentiality of the ES cell population will act gradually over time by limiting the number of cell divisions over which the cells can successfully self-renew, we repeated this assay by replating the dissociated cells across four iterations. Of the initial 60 replicates, 58 survived through the duration of the experiment (two did not survive the initial passage from a single selected clone and are not included in the analysis). A representative photo of colonies is provided in Figure S1.
Although the knockdown of NR2C1 quantitatively and substantially reduces the expression of Oct4 and Nanog, by assessing the number of clones formed, we found that the lack of NR2C1 does not impair their self-renewal ability in our in vitro assays; indeed, even at the end of the experiment, numerous ES cell colonies were present in knockdown cultures, and both knockdown and negative control colonies were morphologically normal. Similarly, overexpression of the human, chimpanzee, and inferred ancestral variants of NR2C1 does not significantly alter their ability to self-renew. Thus, it appears that neither the loss of endogenous NR2C1 nor the ectopic expression of the primate variants leads to outright differentiation of ES cells as is observed following a direct knockdown of Oct4 or Nanog (Hay et al. 2004; Zaehres et al. 2005) and leads us to conclude that the role of rodent and primate NR2C1 in regulating pluripotentiality appears to be more complex than simply acting as an upstream regulator of Oct4/Nanog.
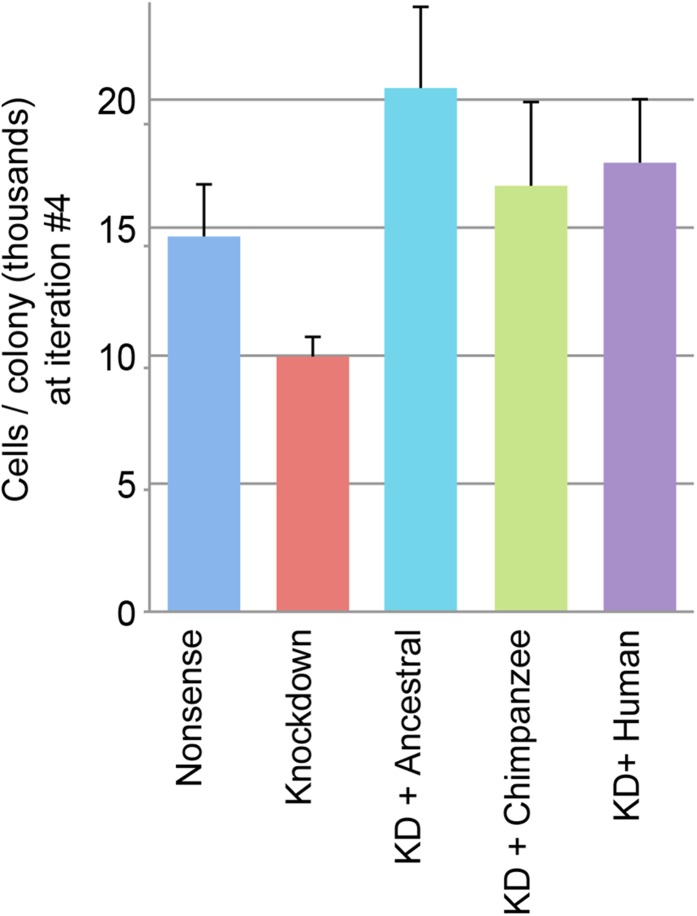
Interestingly, a difference was observed in the average size of the clonal colonies, which suggests that NR2C1 may regulate other aspects of differentiation and proliferation. Knockdown colonies were approximately one-third smaller than the positive control colonies (68%; P = 0.04 by Student’s t-test). Expression of any of the chimpanzee, human, or ancestral variants of NR2C1 reversed this effect (114, 120, and 140%, respectively, although differences among the variants are not significant by one-way ANOVA). As stem cells differentiate, their cell cycle times change (e.g., as neural stem cells progress through the stages of neural differentiation, the length of their cell-cycle increases). Therefore, for a given stem cell population, the rate of proliferation is effectively defined by their level of differentiation. Collectively, our results raise the possibility that evolutionary modulation of NR2C1 may be related to the fundamental properties of the stem cell population. It is interesting that the ancestral variant, which rescues Oct4 expression, exhibited the largest colony size in our assays (Figure 8).
Figure 8.
The loss of NR2C1 activity appears to impact the ES cells proliferative ability; knockdown colonies appear approximately one-third smaller than the positive control colonies (68%; P = 0.04 by Student’s t-test). Expression of any of the chimpanzee, human, or ancestral variants of NR2C1 reverses this effect (114, 120, and 140%, respectively).
In vitro assays suggest hNR2C1, cNR2C1, and aNR2C1 differentially regulate a differentiation-associated promoter:
The primary function of NR2C1 is to act as a transcription factor by binding to specific sites in the promoters of target genes and then either activating or repressing their expression. Previous studies identified one such candidate target of NR2C1 by showing the regulation of the phosphoenolpyruvate carboxykinase (Pepck/Pck1) promoter using a luciferase reporter assay (Shyr et al. 2009). Expression of the Pepck gene increases in numerous cell types as development proceeds, including in the developing neural tube (Zimmer and Magnuson 1990) and liver endothelial cells (Gruppuso et al. 1999). As Pepck is a key metabolic enzyme, changes in its expression likely reflect changes in metabolic activity during differentiation. Such changes in metabolic activity are evident even in the earliest stages of stem cell differentiation, and may in fact be intimately linked to pluripotentiality (Mandal et al. 2011).
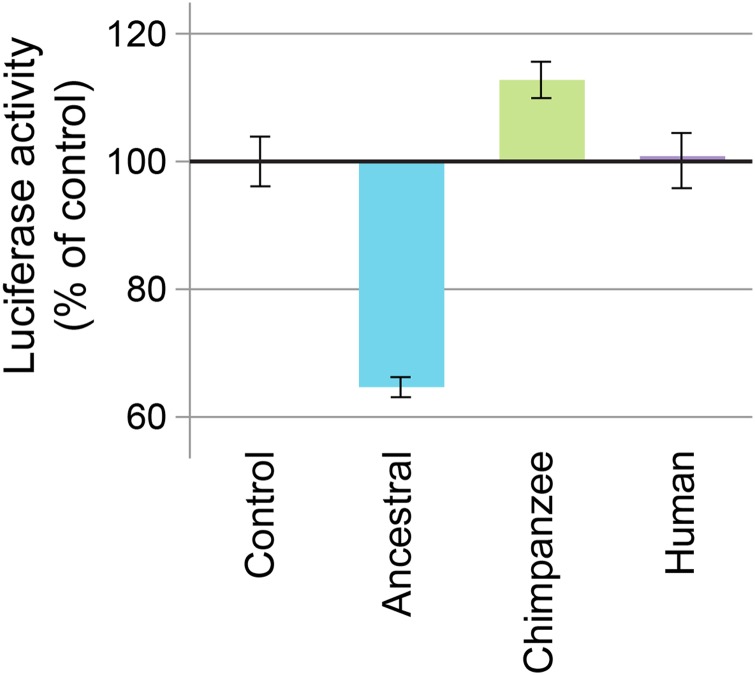
To examine whether this function differs among the three gene variants, we assayed the ability of the NR2C1 variants to modulate transcription using an in vitro luciferase reporter gene. We transfected HEK-293 cells with the plasmids encoding aNR2C1, cNR2C1, and hNR2C1 as well as the Pepck-LUC reporter plasmid. After an overnight culture we lysed the cells to measure luciferase activity. Higher light output in this assay is indicative of increased promoter activity. HEK-293 is a transformed human cell line that normally expresses modest levels of Pepck, as evidenced in this assay by significant basal activity of the Pepck promoter. We found that hNR2C1 shows virtually no ability to modulate this activity (Figure 9). In contrast, aNR2C1 showed a significant and substantial repression of the basal activity of the Pepck promoter (64.7% relative to control, P < 0.0001 by ANOVA with Dunnett’s post-test). This repression was unique to the ancestral variant, as the cNR2C1 transfection caused a significant increase (112% relative to control, P = 0.03 by ANOVA with Dunnett’s post-test). Thus, it appears that the three primate variants of NR2C1 have differential abilities to regulate transcription from this promoter element.
Figure 9.
Reporter assay used to assess regulation of a differentiation-associated promoter (Pepck-luciferase). While the human NR2C1 (hNR2C1) construct did not exhibit any ability to modulate the activity of the Pepck promoter, the ancestral NR2C1 (aNR2C1) displayed a significant repression of the basal activity of the promoter, and the chimpanzee NR2C1 (cNR2C1) transfection caused a significant increase in the activity of the Pepck promoter. Luciferase activity measurements are the average of eight assays and normalized to nonsense control assay values.
As there are relatively few amino acid changes between these variants, it is possible to speculate on which changes may underlie the differential transcriptional response. The human and the ancestral variants have identical DBDs, but different transcriptional activities. As the HD is thought to have minimal effect on NR function, substitutions at the three sites identified in that domain (Figure 6) are not likely to have a direct effect on transcriptional activity. In comparing the ancestral and chimpanzee variants, the candidate substitutions lie at sites in either the NTD or DBD. Because both ancestral and chimpanzee NR2C1 have transcriptional activity, it seems unlikely that the difference in activity is due to an inability of the DBD of one variant to bind to the Pepck promoter. Thus, it is likely that the three substitutions in the NTD are the strongest candidates for direct modulation of the transcriptional activity along the chimpanzee lineage.
Conclusions
We found that rigorous screening of candidate genes for (i) robustness to model assumptions and (ii) reliability of the MLEs are critical components of experimental design in an evolutionary survey. While it is common practice to employ alternative formulations of codon models to assess robustness of signal for positive selection (e.g., consistency between model A and model B), further assessment of the assumptions upon which such models are built is rarely pursued. We expect that robustness analyses will have a greater impact on studies of lineages more divergent than primates. More critical to future investigations of primate molecular evolution was the finding of instabilities in the MLE distributions for several genes [ESRRA, ESRRB (under H5), NR1D1, and RARG], which indicated that the conditions for reliable interpretation of the MLEs and LRTs could not be assumed. We suggest that consistency among alternative codon models, although desirable, should not be taken as a “talisman of reliability” because it provides no indication of whether the underlying requirements for inference have been met.
From an initial set of 48 NRs, we identified a set of four genes [ESRRB (under H4), PGR, RORA, and NR2C1] whose evolution was statistically associated with the origin of the great ape or the human–chimpanzee clades and that we consider good candidates for further investigation via ancestral state reconstruction, gene synthesis, and laboratory analyses. These results, which were derived from clade-site models C and D, contrast with those derived from branch-site models A and B, where we were unable to identify any candidates for further investigation. This difference is, in part, due to the different evolutionary assumptions made by the models. Models C and D assume that some sites experience a shift in selection pressure independent of whatever selection pressure happens to be acting on other sites. Models A and B, on the other hand, assume that some sites experience an episodic shift away from an ancestral level that is equivalent to the level at other sites. Thus, our evolutionary survey supports the notion that signal relevant to functional divergence can go undetected when only a single family of models is used to characterize their evolution (Schott et al. 2014).
Our experimental characterization of human, chimpanzee, and ancestral NR2C1’s transcriptional activities validated the evolutionary signal for functional divergence derived from the clade-site models. Comparison of Oct4 expression in the three gene variants that had NR2C1 knocked down demonstrate the ancestral variant has a 130% increase in expression of Oct4. This observation is consistent with divergence in the regulation of Oct4. As none of the primate variants rescued Nanog, our assays suggest additional divergence must have occurred between primates and rodents in either the promoter of the mouse Nanog gene or within the gene itself. Differences among the three NR2C1 variants also modulated the promoter activity for Pepck, an enzyme whose metabolic activities are closely linked to development and differentiation (Zimmer and Magnuson 1990; Gruppuso et al. 1999; Mandal et al. 2011). Our results indicate that differential abilities to bind and regulate transcription from this promoter element are a result of substitutions at a relatively small subset of sites, likely in the NTD. Lastly, our assays suggest that reduced NR2C1 levels in the knockdown construct are associated with reduced proliferation, suggesting that its activity positively regulates ES cell proliferation. The fact that the average colony size of the aNR2C1-transfected cells was the highest provides an intriguing hint that aNR2C1 may have a more potent ability to rescue this proliferation defect than hNR2C1 and cNR2C1.
Our results are consistent with the hypothesis that evolution of NR2C1 is associated with lineage-specific differences in several aspects of stem cell populations related to the developmental phenotype of pluripotentiality. Given that neurogenesis occurs over a very limited period of development, evolutionary modulation of pluripotentiality is likely to have more profound effects on the proliferation of central nervous system neurons than on other organs or tissues. Thus, we propose that evolution of NR2C1 could be associated with anatomical or physiological characteristics that distinguish humans from other great apes. Finally, analysis of NR2C1 expression in the mouse embryo illustrates that it is robustly expressed in placodally derived neuroectodermal stem cells and presumptive neural-crest-derived mesenchymal stem cells (Baker et al. 2016). Further investigation as to whether this gene can regulate self-renewal and/or pluripotentiality in populations of stem cells other than ES cells such as in cultures of neural stem cells, or in iPSCs generated from nonhuman primates (Marchetto et al. 2013; Wunderlich et al. 2014; Gallego Romero et al. 2015; Ramaswamy et al. 2015) is warranted. iPSCs are an incredibly useful tool and provide a great deal of insight into questions of pluripotentiality and differentiation (Takahashi and Yamanaka 2006); however, some questions remain regarding whether epigenetic reprogramming during the iPSC protocol results in full pluripotency potential (Bilic and Belmonte 2012; Robinton and Daley 2012; Halevy and Urbach 2014). There are also known differences between iPSCs and ES cells in epigenetic landscape, transcribed genes, mutational load, and differentiation potential (Bilic and Belmonte 2012). For these reasons, we decided to take the more conservative path and use traditional ES cells to reduce potential confounding factors while testing our original hypotheses.
By characterizing the translational activity of the ancestral form of NR2C1, we were able to polarize the evolution of regulation of Pepck promoter activity. Specifically, amino acid substitutions occurring at different sites in humans and chimpanzees since their LCA increased transcriptional activities in both lineages. The substitutions that occurred along the chimpanzee lineage appear to have a greater effect on transcriptional activation than those that occurred along the human lineage. In broad terms, this is an example of parallel evolution, as the phenotypic trajectory (increased transcriptional activation) was the same along both lineages. It is interesting that the MLE distribution for NR2C1 suggested positive selection, as this implies that parallel evolution of gene expression could have been driven by Darwinian positive selection.
Acknowledgments
The work described here was supported by a National Science Foundation (NSF) doctoral dissertation research improvement grant (BSC-1455625), a Wenner-Gren Foundation dissertation fieldwork grant (8735), an NSF Integrative Graduate Education and Research traineeship grant (DGE-0801634), an NSF-Human Origins Moving in New Directions (HOMINID) grant (BCS-0827546), a grant from the James S. McDonnell Foundation (220020293), George Washington (GW) Office of the Vice President for Research and the GW School of Medicine for the GW Institute of Neuroscience Biomarker Core Facility, National Institutes of Health grant (DC-001534), Natural Sciences and Engineering Research Council of Canada (DG298394), Centre for Comparative Genomics and Evolutionary Bioinformatics (funded by the Tula Foundation), and Canadian Institutes of Health Research (CMF-108026).
Footnotes
Communicating editor: J. M. Akey
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.183889/-/DC1.
Literature Cited
- Alemseged Z., Spoor F., Kimbel W. H., Bobe R., Geraads D., et al. , 2006. A juvenile early hominin skeleton from Dikika, Ethiopia. Nature 443: 296–301. [DOI] [PubMed] [Google Scholar]
- Allman J. M., Tetreault N. A., Hakeem A. Y., Manaye K. F., Semendeferi K., et al. , 2010. The von Economo neurons in frontoinsular and anterior cingulate cortex in great apes and humans. Brain Struct. Funct. 214: 495–517. [DOI] [PubMed] [Google Scholar]
- Anderson, A. M., K. W. Carter, D. Anderson, and M. J. Wise, 2012 Coexpression of nuclear receptors and histone methylation modifying genes in the testis: implications for endocrine disruptor modes of action. PLoS One 7: e34158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M., Nielsen R., Yang Z., 2003. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164: 1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J. L., Wood B., Karpinski B. A., LaMantia A.S., Maynard T. M., 2016. Testicular receptor 2, Nr2c1, is associated with stem cells in the developing olfactory epithelium and other cranial sensory and skeletal structures. Gene Expr. Patterns 20(1): 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Gu H., Dunn K. A., Bielawski J. P., 2007. Methods for selecting fixed-effect models for heterogeneous codon evolution, with comments on their application to gene and genome data. BMC Evol. Biol. 7: S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit G., Cooney A., Giguere V., Ingraham H., Lazar M., et al. , 2006. International Union of Pharmacology. LXVI. Orphan nuclear receptors. Pharmacol. Rev. 58: 798–836. [DOI] [PubMed] [Google Scholar]
- Bielawski J. P., Yang Z., 2004. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J. Mol. Evol. 59: 121–132. [DOI] [PubMed] [Google Scholar]
- Bielawski J. P., Baker J. L., Mingrone J., 2016. Inference of episodic changes in natural selection acting on protein coding sequences via CODEML. Curr. Prot. Bioinf. (in press). [DOI] [PubMed] [Google Scholar]
- Bilic J., Izpisua Belmonte J. C., 2012. Concise review: induced pluripotent stem cells vs. embryonic stem cells: Close enough or yet too far apart? Stem Cells 30: 33–41. [DOI] [PubMed] [Google Scholar]
- Bloch N. I., Morrow J. M., Chang B. S. W., Price T. D., 2015. SWS2 visual pigment evolution as a test of historically contingent patterns of plumage color evolution in warblers. Evolution 69: 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy A. M., McGowen M. R., Sherwood C. C., Grossman L. I., Goodman M., et al. , 2012. Comparative analysis of encephalization in mammals reveals relaxed constraints on anthropoid primate and cetacean brain scaling. J. Evol. Biol. 25: 981–994. [DOI] [PubMed] [Google Scholar]
- Boiani M., 2002. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev. 16: 1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguet W., Germain P., Gronemeyer H., 2000. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol. Sci. 21(10): 381–388. [DOI] [PubMed] [Google Scholar]
- Braun D. R., Harris J. W. K., Levin N. E., McCoy J. T., Herries A. I. R., et al. , 2010. Early hominin diet included diverse terrestrial and aquatic animals 1.95 Ma in East Turkana, Kenya. Proc. Natl. Acad. Sci. USA 107: 10002–10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayer K. J., Lynch V. J., Wagner G. P., 2011. Evolution of a derived protein-protein interaction between HoxA11 and Foxo1a in mammals caused by changes in intramolecular regulation. Proc. Natl. Acad. Sci. USA 108: E414–E420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham J. T., Ortlund E. A., Thornton J. W., 2009. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 461: 515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Smith A., 2004. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene 23: 7150–7160. [DOI] [PubMed] [Google Scholar]
- Chang B. S. W., Jonsson K., Kazmi M. A., Donoghue M. J., Sakmar T. P., 2002. Recreating a functional ancestral archosaur visual pigment. Mol. Biol. Evol. 19: 1483–1489. [DOI] [PubMed] [Google Scholar]
- Chang C., Kokontis J., 1988. Identification of a new member of the steroid receptor super-family by cloning and sequence analysis. Biochem. Biophys. Res. Commun. 155: 971–977. [DOI] [PubMed] [Google Scholar]
- Chen C., Opazo J. C., Erez O., Uddin M., Santolaya-Forgas J., et al. , 2008. The human progesterone receptor shows evidence of adaptive evolution associated with its ability to act as a transcription factor. Mol. Phylogenet. Evol. 47: 637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick G. N., Thornton J. W., 2011. Evolution of steroid receptors from an estrogen-sensitive ancestral receptor. Mol. Cell. Endocrinol. 334: 31–38. [DOI] [PubMed] [Google Scholar]
- Enmark E., Gustafsson J. A., 1996. Orphan nuclear receptors: the first eight years. Mol. Endocrinol. 10: 1293–1307. [DOI] [PubMed] [Google Scholar]
- Fletcher W., Yang Z., 2010. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol. Biol. Evol. 27: 2257–2267. [DOI] [PubMed] [Google Scholar]
- Forsberg R., Christiansen F. B., 2003. A codon-based model of host-specific selection in parasites, with an application to the influenza A virus. Mol. Biol. Evol. 20: 1252–1259. [DOI] [PubMed] [Google Scholar]
- Gallego Romero I., Pavlovic B. J., Hernando-Herraez I., Zhou X., Ward M. C., et al. , 2015. A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics. eLife 4: e07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher E. A., Thomson J. M., Burgan M. F., Benner S. A., 2003. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 425: 285–288. [DOI] [PubMed] [Google Scholar]
- Gaucher E. A., Govindarajan S., Ganesh O. K., 2008. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature 451: 704–707. [DOI] [PubMed] [Google Scholar]
- Germain P., Staels B., Dacquet C., Spedding M., Laudet V., 2006. Overview of nomenclature of nuclear receptors. Pharmacol. Rev. 58: 685–704. [DOI] [PubMed] [Google Scholar]
- Goldman N., Yang Z., 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11: 725–736. [DOI] [PubMed] [Google Scholar]
- Gruppuso P. A., Bienieki T. C., Faris R. A., 1999. The relationship between differentiation and proliferation in late gestation fetal rat hepatocytes. Pediatr. Res. 46: 14–19. [DOI] [PubMed] [Google Scholar]
- Halevy T., Urbach A., 2014. Comparing ESC and iPSC-based models for human genetic disorders. J. Clin. Med. 3(4): 1146–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M. J., Thornton J. W., 2010. Analyzing protein structure and function using ancestral gene reconstruction. Curr. Opin. Struct. Biol. 20: 360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M. J., Thornton J. W., 2013. Evolutionary biochemistry: revealing the historical and physical causes of protein properties. Nat. Rev. Genet. 14: 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay D. C., Sutherland L., Clark J., Burdon T., 2004. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells 22: 225–235. [DOI] [PubMed] [Google Scholar]
- Hu Y.-C., Shyr C.-R., Che W., Mu X.-M., Kim E., et al. , 2002. Suppression of estrogen receptor-mediated transcription and cell growth by interaction with TR2 orphan receptor. J. Biol. Chem. 277: 33571–33579. [DOI] [PubMed] [Google Scholar]
- Jungers W. L., Harcourt-Smith W. E. H., Wunderlich R. E., Tocheri M. W., Larson S. G., et al. , 2009a The foot of Homo floresiensis. Nature 459: 81–84. [DOI] [PubMed] [Google Scholar]
- Jungers W. L., Larson S. G., Harcourt-Smith W., Morwood M. J., Sutikna T., et al. , 2009b Descriptions of the lower limb skeleton of Homo floresiensis. J. Hum. Evol. 57: 538–554. [DOI] [PubMed] [Google Scholar]
- Katoh K., 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., et al. , 2002. The Human Genome Browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketterson E. D., Atwell J. W., McGlothlin J. W., 2009. Phenotypic integration and independence: hormones, performance, and response to environmental change. Integr. Comp. Biol. 49: 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn J. A., Deshpande K., Ortlund E. A., 2012. Deciphering modern glucocorticoid cross-pharmacology using ancestral corticosteroid receptors. J. Biol. Chem. 287: 16267–16275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond S. L., Posada D., Gravenor M. B., Woelk C. H., Frost S. D. W., 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22: 3096–3098. [DOI] [PubMed] [Google Scholar]
- Krasowski M. D., Yasuda K., Hagey L. R., Schuetz E. G., 2005. Evolutionary selection across the nuclear hormone receptor superfamily with a focus on the NR1I subfamily (vitamin D, pregnane X, and constitutive androstane receptors). Nucl. Recept. 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratzer J. T., Lanaspa M. A., Murphy M. N., Cicerchi C., Graves C. L., et al. , 2014. Evolutionary history and metabolic insights of ancient mammalian uricases. Proc. Natl. Acad. Sci. USA 111: 3763–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudet V., 1997. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J. Mol. Endocrinol. 19: 207–226. [DOI] [PubMed] [Google Scholar]
- Lee H. J., Chang C., 1995. Identification of human TR2 orphan receptor response element in the transcriptional initiation site of the simian virus 40 major late promoter. J. Biol. Chem. 270: 5434–5440. [DOI] [PubMed] [Google Scholar]
- Lee Y.-F., Lee H.-J., Chang C., 2002. Recent advances in the TR2 and TR4 orphan receptors of the nuclear receptor superfamily. J. Steroid Biochem. Mol. Biol. 81: 291–308. [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu X. S., Wei L., Altman R. B., Batzoglou S., 2004. Eukaryotic regulatory element conservation analysis and identification using comparative genomics. Genome Res. 14: 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas P. C., O’Brien R. M., Mitchell J. A., Davis C. M., Imai E., et al. , 1991. A retinoic acid response element is part of a pleiotropic domain in the phosphoenolpyruvate carboxykinase gene. Proc. Natl. Acad. Sci. USA 88: 2184–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S., Lindgren A. G., Srivastava A. S., Clark A. T., Banerjee U., 2011. Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cells 29: 486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto M. C., Narvaiza I., Denli A. M., Benner C., Lazzarini T. A., et al. , 2013. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 503: 525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., et al. , 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39: D225–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard T., Gopalakrishna D., Meechan D., Paronett E., Newbern J., et al. , 2013. 22q11 Gene dosage establishes an adaptive range for sonic hedgehog and retinoic acid signaling during early development. Hum. Mol. Genet. 22(2): 300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcbrearty S., Brooks A. S., 2000. The revolution that wasn’t: a new interpretation of the origin of modern human behavior. J. Hum. Evol. 39: 453–563. [DOI] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., et al. , 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642. [DOI] [PubMed] [Google Scholar]
- Muse S. V., Gaut B. S., 1994. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol. Biol. Evol. 11: 715–724. [DOI] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., et al. , 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95: 379–391. [DOI] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., Smith A. G., 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24: 372–376. [DOI] [PubMed] [Google Scholar]
- Ohi Y., Qin H., Hong C., Blouin L., Polo J. M., et al. , 2011. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 13(5): 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikarsky E., Sharir H., Ben-Shushan E., Bergman Y., 1994. Retinoic acid represses Oct-3/4 gene expression through several retinoic acid-responsive elements located in the promoter-enhancer region. Mol. Cell. Biol. 14: 1026–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy K., Yik W. Y., Wang X. M., Oliphant E. N., Lu W., et al. , 2015. Derivation of induced pluripotent stem cells from orangutan skin fibroblasts. BMC Res. Notes 16(8): 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Rechavi M., Carpentier A.-S., Duffraisse M., Laudet V., 2001. How many nuclear hormone receptors are there in the human genome? Trends Genet. 17: 554–556. [DOI] [PubMed] [Google Scholar]
- Robinton D. A., Daley G. Q., 2012. The promise of induced pluripotent stem cells in research and therapy. Nature 481: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein N. D., Doron-Faigenboim A., Mayrose I., Pupko T., 2011. Evolutionary models accounting for layers of selection in protein-coding genes and their impact on the inference of positive selection. Mol. Biol. Evol. 28(12): 3297–3308. [DOI] [PubMed] [Google Scholar]
- Roesler W. J., Vandenbark G. R., Hanson R. W., 1989. Identification of multiple protein binding domains in the promoter-regulatory region of the phosphoenolpyruvate carboxykinase (GTP) gene. J. Biol. Chem. 264(16): 9657–9664. [PubMed] [Google Scholar]
- Schneider A., Souvorov A., Sabath N., Landan G., Gonnet G. H., et al. , 2009. Estimates of positive Darwinian selection are inflated by errors in sequencing, annotation, and alignment. Genome Biol. Evol. 1: 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott R. K., Refvik S. P., Hauser F. E., López-Fernández H., Chang B. S. W., 2014. Divergent positive selection in rhodopsin from lake and riverine cichlid fishes. Mol. Biol. Evol. 31: 1149–1165. [DOI] [PubMed] [Google Scholar]
- Self S. G., Liang K. Y., 1987. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J. Amer. Stat. Assoc. 82(398): 605–610. [Google Scholar]
- Sherwood C. C., Duka T., 2012. Now that we’ve got the map, where are we going moving from gene candidate lists to function in studies of brain evolution. Brain Behav. Evol. 80: 167–169. [DOI] [PubMed] [Google Scholar]
- Shyr C.-R., Kang H.-Y., Tsai M.-Y., Liu N.-C., Ku P.-Y., et al. , 2009. Roles of testicular orphan nuclear receptors 2 and 4 in early embryonic development and embryonic stem cells. Endocrinology 150: 2454–2462. [DOI] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J. D., 2002. A direct approach to false discovery rates. J. R. Stat. Soc. B 64: 479–498. [Google Scholar]
- Takahashi K., Yamanaka S., 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. [DOI] [PubMed] [Google Scholar]
- Thornton J. W., 2001. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc. Natl. Acad. Sci. USA 98: 5671–5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton J. W., Need E., Crews D., 2003. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science 301: 1714–1717. [DOI] [PubMed] [Google Scholar]
- Tryon C. A., Roach N. T., Logan M. A. V., 2008. The Middle Stone Age of the northern Kenyan Rift: age and context of new archaeological sites from the Kapedo Tuffs. J. Hum. Evol. 55: 652–664. [DOI] [PubMed] [Google Scholar]
- Ugalde J. A., Chang B. S. W., Matz M. V., 2004. Evolution of coral pigments recreated. Science 305: 1433. [DOI] [PubMed] [Google Scholar]
- Ward C. V., Kimbel W. H., Johanson D. C., 2011. Complete fourth metatarsal and arches in the foot of Australopithecus afarensis. Science 331: 750–753. [DOI] [PubMed] [Google Scholar]
- Wasserman W. W., Palumbo M., Thompson W., Fickett J. W., Lawrence C. E., 2000. Human-mouse genome comparisons to locate regulatory sites. Nat. Genet. 26: 225–228. [DOI] [PubMed] [Google Scholar]
- Weadick C. J., Chang B. S. W., 2012. An improved likelihood ratio test for detecting site-specific functional divergence among clades of protein-coding genes. Mol. Biol. Evol. 29: 1297–1300. [DOI] [PubMed] [Google Scholar]
- Whelan S., Goldman N., 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18: 691–699. [DOI] [PubMed] [Google Scholar]
- Williamson S. H., Hubisz M. J., Clark A. G., Payseur B. A., Bustamante C. D., et al. , 2007. Localizing recent adaptive evolution in the human genome. PLoS Genet. 3: e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood B., 1996. Hominid palaeobiology: Have studies of comparative development come of age? Am. J. Phys. Anthropol. 99: 9–15. [DOI] [PubMed] [Google Scholar]
- Wunderlich S., Kircher M., Vieth B., Haase A., Merkert S., et al. , 2014. Primate iPS cells as tools for evolutionary analyses. Stem Cell Res. (Amst.) 12: 622–629. [DOI] [PubMed] [Google Scholar]
- Yang Z., 1994. Estimating the pattern of nucleotide substitution. J. Mol. Evol. 39: 105–111. [DOI] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19: 908–917. [DOI] [PubMed] [Google Scholar]
- Yang Z., Swanson W. J., 2002. Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol. Biol. Evol. 19: 49–57. [DOI] [PubMed] [Google Scholar]
- Zaehres H., Lensch M. W., Daheron L., Stewart S. A., Itskovitz-Eldor J., et al. , 2005. High-efficiency RNA interference in human embryonic stem cells. Stem Cells 23: 299–305. [DOI] [PubMed] [Google Scholar]
- Zhang J., Nielsen R., Yang Z., 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22: 2472–2479. [DOI] [PubMed] [Google Scholar]
- Zimmer D. B., Magnuson M. A., 1990. Immunohistochemical localization of phosphoenolpyruvate carboxykinase in adult and developing mouse tissues. J. Histochem. Cytochem. 38: 171–178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
DRYAD data repository for alignments and structural partitions is as follows: doi : 10.5061/dryad.bg3g3. GenBank accession nos. for the novel NR2C1 sequences generated in this study are as follows: KT032104–KT032114. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.